Abstract
Background
Esophageal cancer (EC) is a malignant tumor with high mortality. Correlations have been found between the expression level of tropomyosin 3 (TPM3) and the depth of tumor invasion, lymph node metastasis, and the 5-year survival rate. However, the specific mechanisms underlying EC remain unclear.
Methods
Stably transfected TPM3-overexpresing and TPM3-knockdown esophageal squamous cell carcinoma (ESCC) cell lines (ECa109 and EC9706) were constructed, and the association between TPM3 and the proliferation, invasion, and migration of ESCC was investigated using molecular biology methods. The associations between TPM3 and matrix metalloproteinase (MMP)2/9 or epithelial-mesenchymal transition (EMT)-related proteins were verified, and the potential tumor-promoting mechanism was explored by Gelatin Zymography Experiment.
Results
TPM3 was found to promote the proliferation, migration, and metastatic potential of ESCC in vivo and in vitro, and stimulate the expression of MMP2/9 and certain EMT markers other than E-cadherin. The replenishment of MMP2/9 restored the malignant behavior of ESCC caused by TPM3. A gelatinase assay showed that the expression of TPM3 was related to the activity of MMP9.
Conclusions
TPM3 promoted proliferation, migration, and metastatic potential in EC cells. Additionally, TPM3 promoted the EMT process. This function may be achieved via the regulation the expression of MMP2/9.
Keywords: Tropomyosin 3 (TPM3), matrix metalloproteinase 2 (MMP2), matrix metalloproteinase 9 (MMP9), epithelial-mesenchymal transition (EMT), esophageal cancer (EC)
Introduction
Esophageal cancer (EC) is a serious threat to human health worldwide. The main pathological type of EC in China is esophageal squamous cell carcinoma (ESCC) . Causing over 200,000 deaths each year, the morbidity and mortality of EC is high in China (1). Despite continued developments in the treatment of EC, nearly 50% of patients develop EC recurrence after systemic treatment, and the 5-year survival rate (~25%) has not improved significantly (2). These outcomes are closely associated with the high degree of malignancy and early metastasis of ESCC. Identifying the molecules associated with the invasion and metastasis of ESCC and their molecular mechanisms could be of great significance to the diagnosis, treatment, and prognosis of ESCC.
Tropomyosin (TPM) is a binding thin filament protein that can bind to actin. TPM3 is an important member of the TPM family and has the function of stabilizing cytoskeletal microfilaments. It is located on the 1q21.3 region and contains 1 exon (3). In studies of glioma, colon cancer, and liver cancer, TPM3 has been shown to affect the occurrence and development of tumors through gene fusion and epithelial-mesenchymal transition (EMT), and have high expression levels (4). Our previous study showed that TPM3 was increased in ESCC, and more specifically, that the expression of TPM3 was higher in patients with stage III ESCC than in those with stage I ESCC (5). In addition, TPM3 may be an independent factor, as it has been found to be statistically correlated with tumor prognosis (6). Additionally, the 5-year survival rate of patients with ESCC has been shown to be negatively correlated with the expression level of TPM3 (6). MMPs constitute unique proteins of the mesenchymal cell phenotype and can be used as important factors for EMT. As important members of the MMP family, MMP2/9 have a great impact on the occurrence and development of a variety of malignancies, including ovarian and breast cancer (4,7).
In the present study, stably transfected TPM3-overexpressing and TPM3-knockdown ESCC cell lines were constructed, and associations between TPM3 and the proliferation, invasion, and migration of ESCC cells were studied by performing in vitro and in vivo molecular biology experiments. The associations between TPM3 and matrix metalloproteinase (MMP)2/9 and EMT-related proteins were also verified on stable cell lines, and the potential tumor-promoting mechanism was explored. This article explores the possibility of using TPM3 as a new target and prognostic indicator for gene therapy in EC. A protocol was prepared before the study without registration
We present the following article in accordance with the ARRIVE reporting checklist (available at https://dx.doi.org/10.21037/atm-21-4043).
Methods
Cell lines
Human ECa109 and EC9706 ESCC cell lines were supplied by Fenghuishengwu Co., Ltd. 293T cells were provided by the Key Laboratory of Gastrointestinal Cancer of the Ministry of Education of China. Cells were cultured in Dulbecco’s modified eagle medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum (FBS) (Invitrogen; Thermo Fisher Scientific, Inc.) and incubated at 37˚C in a humidified incubator with 5% carbon dioxide (CO2).
Plasmids and the generation of stable ESCC cell lines
The open reading frame of the human TPM3 and MMP2/9 genes was amplified by polymerase chain reaction (PCR) and cloned into the pLKO.1-puro-GFP lentivirus expression vector (Shengminghulian). A semi-quantitative PCR was performed using a PCR kit (cat. No. 96871; Takara Bio, Inc.). The sense strand of TPM3 was 5'-GGAGACTTGGAACGCACAGA-3', while the antisense strand was 5'-TTCAGCAGCACTCAGACACTTC-3'. Subsequently, recombinant plasmids, including the sequence of TPM3 or an empty vector, were co-transfected with packaging plasmids into HEK293T cells. ESCC ECa109 or EC9706 cells were cultured in 6-cm dishes and infected with supernatant and collected 48 h after transfection. Puromycin-resistant clones were expanded into cell lines as TPM3-overexpressing cells (p-TPM3) or empty control cells (p-CDH). The expression level of TPM3 in the cell lines was evaluated by a western blot analysis. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
TPM3-overexpressing cells (ECa109 p-TPM3 or EC9706 p-TPM3) and empty control cells (ECa109 p-CDH or EC9706 p-CDH) were obtained via the expansion of puromycin-resistant clones. The shTPM3 RNA interference sequence was synthesized and cloned into pLKO.1-puro-GFP (Shengminghulian) to generate TPM3-knockdown clones. The recombinant plasmid or empty vector were co-transfected with pIK packaging plasmids (Invitrogen; Thermo Fisher Scientific, Inc.) into HEK293T cells, and the ECa109 or EC9706 cells were infected with the collected supernatants. TPM3-knockdown cells (ECa109 p-sRNA#1, ECa109 p-sRNA#2, EC9706 p-sRNA#1, or EC9706 p-sRNA#2) or empty control cells (ECa109 p-shRNA or EC9706 p-shRNA) were obtained through the expansion of the puromycin-resistant clones. A western blot analysis and reverse transcription-quantitative PCR (RT-qPCR) were used to evaluate the expression levels of TPM3 in the above-mentioned cell lines.
The recombinant plasmids, including the sequence of MMP2/9, (termed OE-MMP2 and OE-MMP9, respectively), and the empty vector (termed OE-NC) were co-transfected with packaging plasmids into HEK293T cells. EC9706 shRNA, EC9706 sRNA#1, or EC9706 sRNA#2 cells were infected with the supernatant and collected 48 h after transfection. The corresponding puromycin-resistant clones were expanded into cell lines as MMP2/9-overexpressing cells (EC9706 sRNA#1 + OE-MMP2, EC9706 sRNA#1 + OE-MMP9, EC9706 sRNA#2 + OE-MMP2, and EC9706 sRNA#2 + OE-MMP9) or empty control cells [EC9706 shRNA (p-shRNA)].
Western blot analysis
Cells were lysed in western and IP cell lysis buffer (GenStar BioSolutions) that was supplemented with phenylmethylsulfonyl fluoride (Roche Diagnostics) for 10 min at 4 °C, and then centrifuged at 12,000 ×g for 10 min at 4 °C. The total protein of the supernatants was measured with the Bicinchoninic Acid Protein Assay Kit (GenStar BioSolutions). The protein extracts were electrophoresed by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride membranes (GE Healthcare Life Sciences). After blocking, the membranes were incubated at 4 °C overnight with the following primary antibodies: Mouse anti-TPM3 (1:1,000; Abcam), rabbit anti-MMP2 (1:1,000; Abcam), rabbit anti-MMP9 (1:1,000; Abcam), mouse anti-tubulin (1:5,000; Abcam), mouse anti-β-actin (1:1,000; Abcam), and an Epithelial-Mesenchymal Transition (EMT) Antibody Sampler Kit (Cell Signaling Technology, Inc.). After 3 washes of 10 min each in tris-buffered saline and polysorbate (TBST; cat. No. 54124; Thermo Fisher Scientific, Inc.), the membranes were incubated with secondary antibodies (cat. No. 78543; Abcam, Inc.) at 1:2,000 dilution for 1 h at 25 °C, and an enhanced chemiluminescence reagent (GenStar BioSolutions) was used to develop the blots.
RNA extraction and RT-qPCR
Total ribonucleic acids (RNAs) were extracted from cultured cells with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and 1 mg of RNA was reverse transcribed into complementary deoxyribonucleic acid (cDNA) with the Ex Script RT-PCR Kit (Takara Bio, Inc.). Next, RT-qPCR was performed using a Mx3000P QPCR system (Agilent Technologies, Inc.) with the Ex Script RT-PCR Kit (Takara Bio, Inc.). The relative expression levels of the target genes were calculated using the 2−ΔΔCq method (7) with primers for human TPM3, MMP2, MMP9, vimentin, N-cadherin, E-cadherin, claudin-1, β-catenin, zonula occludens-1, Snail, Slug, and zinc finger E-box binding homeobox 1. The expression level was obtained by normalization to the endogenous glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Yingweichuangjin Biotechnology Co., Ltd. designed all the primers (cat. No. P2412S).
Wound healing assay
Cells were seeded into 6-well plates and grown to ~100% confluence. The monolayer culture was artificially scrape-wounded using a 20-μL disposable pipette tip to create a scratch of the same size. Each well was washed using Hank’s Balanced Salt Solution (Merck KGaA), and the cells were incubated at 37 °C in 5% CO2. Wound closure was photographed at 0 and 24 h. The assay was performed in a serum-free medium.
Transwell assay
A total of 1×105 cells in serum-free media were seeded into the Matrigel-coated top chamber of a Transwell insert with an 8-mm pore size (BD Biosciences). Equal numbers of cells were seeded into a Transwell insert that had not been coated with Matrigel. Medium containing 15% FBS was added to the bottom chamber. The cells were incubated for 24 h, and the cells in the top chambers were then cleaned, while the cells on the bottom chambers were stained with crystal violet (Merck KGaA) and counted under a Qimaging Micropublisher 5.0 RTV microscope camera (Olympus Corporation).
Colony formation assay
A total of 5×102 cells per well were plated in 6-well plates and incubated at 37 °C in the presence of 5% CO2 in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were continuously cultured for 2–3 weeks, during which time, cell growth in the well plates was observed under a microscope. When the cells grown in the well plates were fused to create a clone observable by the naked eye, the culture was stopped, and an appropriate quantity of 1% crystal violet solution (covering the cell plate) was added to each well for staining. After 30 min, the staining was completed, and the number of clones formed was evaluated under a microscope by counting the number of cell clones with >40 cells. The experiment was repeated 3 times.
Cell proliferation assay
The cell proliferation assay was carried out with cell counting kit-8 (CCK-8) (Dojindo Molecular Technologies, Inc.). Cells at a density of 5×102 cells per well were plated in 96-well plates and incubated for 24, 48, 72, 96, or 120 h at 37 °C with 5% CO2. Next, the cells in each well were incubated at 37 °C for additional 1 h with 10 μL CCK-8 solution. The absorbance was detected at 450 nm with a microplate reader (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Gelatinase assay
Equal quantities of protein from different cell lines were separated by 10% SDS-PAGE containing 1 mg/mL gelatin. The SDS was then eluted from the gels, which were incubated in buffer for 42 h at 37 °C. After being dyed with Coomassie Brilliant Blue (Thermo Fisher Scientific, Inc.) and discolored, the gels were analyzed in a GelDoc XR analysis system (Agilent Technologies, Inc.).
Subcutaneous tumorigenesis in nude mice
In total, male BALB/c nude mice (4 weeks old, Shanghai, China) were randomly divided into 4 groups. A total of 2×107 TPM3 upregulated and knockout Eca109 cells, and cells in the respective control groups were resuspended in 0.2 mL of PBS. The resulting xenograft tumors were established within 6 weeks. A total of 20 mice were randomly allocated to 5 experimental groups (ECa109 p-TPM3, ECa109 p-CDH, ECa109-psh, ECa109-ps-RNA#1, and ECa109 ps-RNA#2). After euthanizing the mice with limited pain, the weight and volume of each growing neoplasm was measured.
Tail vein injection
A total of 2×107 cells were intravenously injected via the lateral tail vein in male BALB/c nude mice (4 weeks old, Shanghai, China). Mice (n=10) were randomly allocated to the following 2 experimental groups: p-sRNA#1 and p-shRNA. At 8–9 weeks post-injection, the mice were sacrificed by cervical dislocation, and the lungs and liver of the nude mice were surgically removed for the metastasis analysis. Experiments were performed under a project license [No.: 2020 Association Ethics Examination No. (040)] granted by the Fujian Medical University Institutional Animal Car and Use Committee and were carried out according to institutional guidelines.
Statistical analysis
The statistical analysis was carried out using SPSS 22.0 software (IBM Corp.). A survival analysis was performed using a Kaplan-Meier analysis, while the Student’s t-test was used for statistical comparisons. Each experiment was performed 3 times, and all data are expressed as the mean ± standard deviation (SD). A value of P<0.05 was considered to indicate a statistically significant difference.
Results
The effects of TPM3 on the expression of MMP2/9 and EMT markers in ESCC
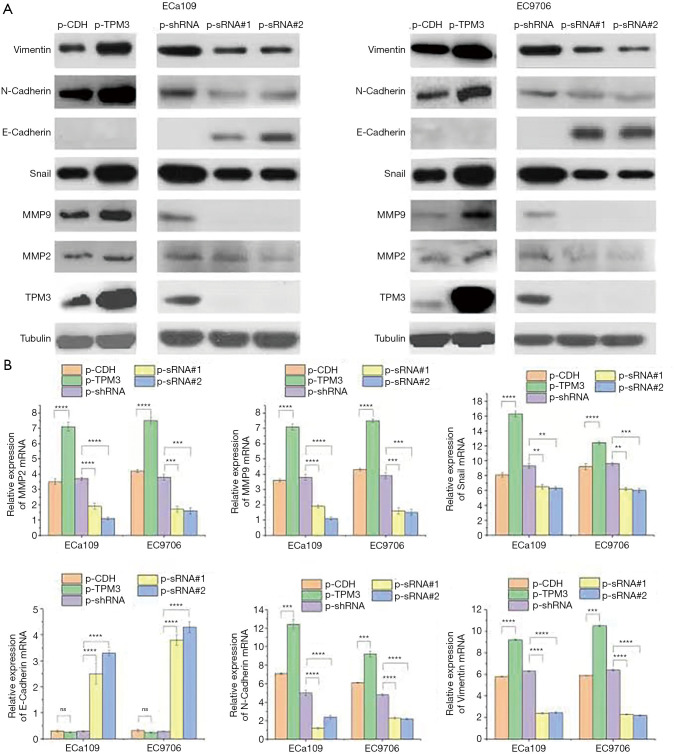
To determine whether the expression of TPM3 might be associated with the prognosis of EC, the present study examined the functional role of TPM3 in the malignant behavior of EC in vitro and in vivo. Gene silencing and overexpression strategies were used to specifically knockdown or overexpress TPM3 in ECa109 and EC9706 ESCC lines. Stable overexpression or knockdown of TPM3 in these cells was confirmed using western blotting (see Figure 1A) and RT-qPCR (see Figure 1B) analysis. The results of western blotting showed that overexpression of TPM3 promoted the expression of MMP2/9 (P<0.0001) and EMT markers; that is, N-cadherin (P<0.001), Snail (P<0.0001), and vimentin (P<0.0001) proteins. Conversely, TPM3 knockdown was shown to reduce the expression of MMP2/9 and EMT markers, including N-cadherin, Snail and vimentin proteins, but promote the expression of E-cadherin protein. Similarly, the RT-qPCR results indicated that the changes in mRNA expression of MMP2/9 and EMT markers, including N-cadherin, Snail, vimentin and E-cadherin, corresponded to those exhibited by TPM3.
Figure 1.
The effects of TPM3 on the expression of MMP2 and MMP2 and EMT markers in ESCC cells. (A) Western blot analysis. (B) RT-qPCR analysis. NS, P>0.05; **P<0.01; ***P<0.001; ****P<0.0001 vs. p-TPM3. TPM3, tropomyosin 3; MMP2, matrix metalloproteinase 2; MMP9, matrix metalloproteinase 9; p-CDH, empty control cells of TPM3 overexpression; p-TPM3, TPM3-overexpressing cells; p-shRNA, empty control cells of TPM3 knockdown; p-sRNA, TPM3-knockdown cells.
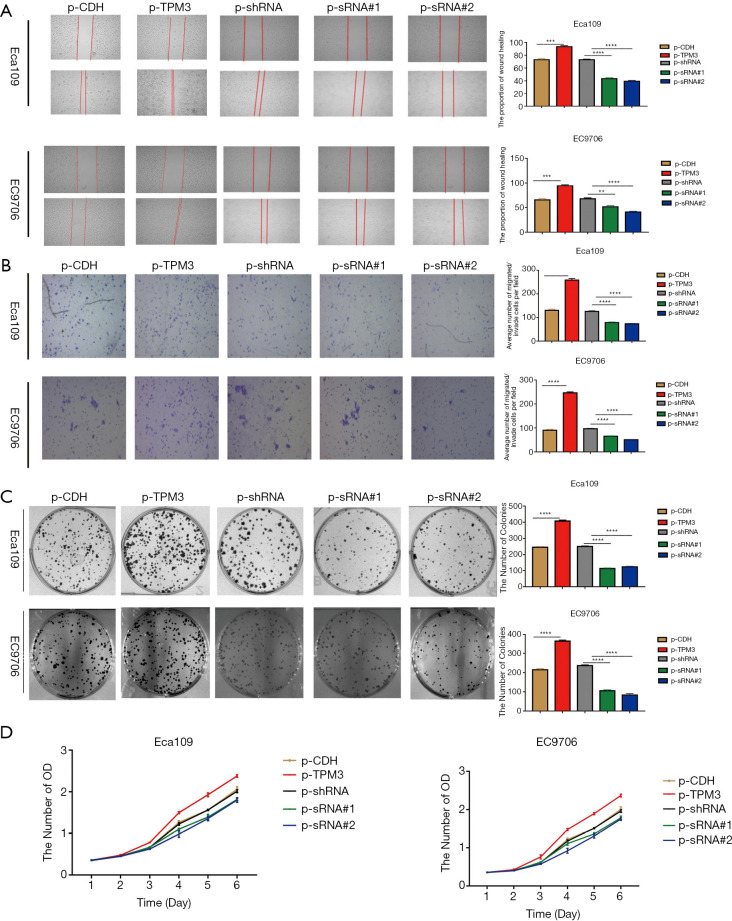
The effects of TPM3 on the migration, metastatic potential and proliferation of ESCC cells
Figure 2A shows the results of the wound healing assay in which TPM3-overexpressing cells (p-TPM3) were observed to close the wound more rapidly than the empty vector control cells (p-CDH). Additionally, the TPM3-knockdown cells (p-sRNA#1 and p-sRNA#2) closed the wound slower than the scrambled shRNA control cells (p-shRNA).
Figure 2.
The effects of TPM3 on the proliferation, migration, and metastatic potential of ESCC cells. (A) Wound healing assay (×50). (B) Transwell assay (×100; dyed by crystal violet). (C) Colony formation assay. (D) CCK-8 assay. **P<0.01, ***P<0.001, ****P<0.0001 vs. respective p-TPM3 or p-shRNA. TPM3, tropomyosin 3; p-CDH, empty control cells of TPM3 overexpression; p-TPM3, TPM3-overexpressing cells; p-shRNA, empty control cells of TPM3 knockdown; p-sRNA, TPM3-knockdown cells.
The effects of TPM3 on cell migration was analyzed by a Transwell assay (see Figure 2B). The overexpression of TPM3 enhanced the migration of the cells, while the knock down of TPM3 inhibited migration.
The overexpression of TPM3 promoted the proliferation of ECa109 and EC9706 cells in vitro, as evaluated by colony formation (see Figure 2C) and CCK-8 (see Figure 2D) assays. When TPM3 was knocked down, the proliferation of cells was suppressed.
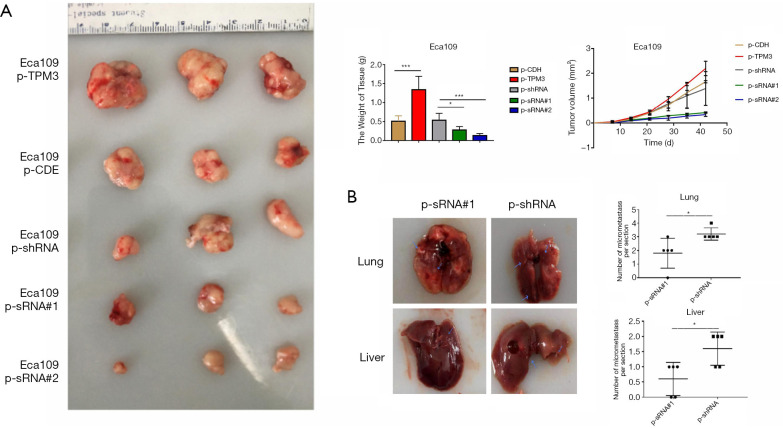
The effects of TPM3 on the proliferation of cells in vivo
The effects of TPM3 on the in vivo proliferation potential of cells was assessed by subcutaneously injecting ECa109 cells carrying p-TPM3, p-CDH, p-shRNA, p-sRNA#1, or p-sRNA#2 into BALB/c nude mice. The weight and volume of the generated neoplasms were measured after euthanizing the mice with limited pain after 6 weeks. As Figure 3A shows, the weight and volume of the xenograft tumor were enhanced by overexpression of TPM3 but suppressed when TPM3 was knocked down in ECa109 and EC9706 cells. As Figure 3B shows, the number of nodules in the liver and lungs of mice injected with TPM3-low expressing Eca109 p-sRNA#1 cells was significantly less than that of mice injected with control Eca109 p-shRNA cells (P<0.05).
Figure 3.
The effects of TPM3 on the in vivo proliferation of cells. (A) Subcutaneous tumor formation experiment (n=6). Three experiment mice are shown. (B) Tail vein injection assay (n=5). *P<0.05, ***P<0.001 vs. p-shRNA. TPM3, tropomyosin 3; p-CDH, empty control cells of TPM3 overexpression; p-TPM3, TPM3-overexpressing cells; p-shRNA, empty control cells of TPM3 knockdown; p-sRNA, TPM3-knockdown cells.
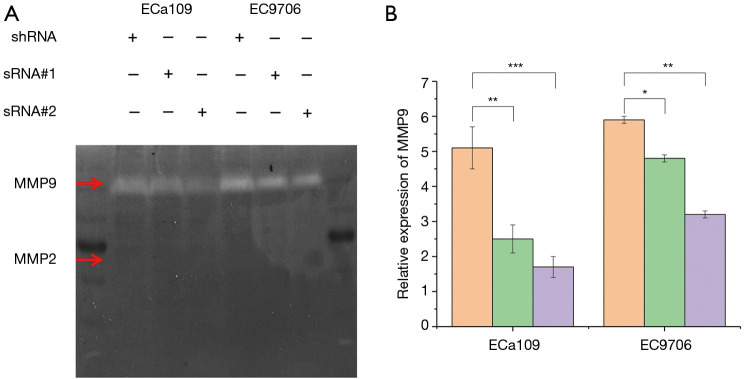
The effects of TPM3 on the activity and expression of MMP2/9 in ESCC cells
MMPs constitute unique proteins of the mesenchymal cell phenotype and can be used to affect the EMT process. As important members of the MMP family, MMP2 and MMP9 have a great effect on the occurrence and development of a variety of malignancies, including ovarian and breast cancer. The present study examined the expression of MMP2 and MMP9 following changes in the expression of TPM3 by western blotting and RT-qPCR (see Figure 1). The results clearly showed that the expression of MMP2 and MMP9 was positively correlated with TPM3 in both ECa109 and EC9706 cells. Further, the results of the gelatinase assay also showed that the knockdown of TPM3 could decrease the activity of MMP9 (see Figure 4). However, the expression of MMP2 was not detected in the gelatinase assay.
Figure 4.
The effects of TPM3 on the expression of MMP2/9 in EC cells. (A) Gelatinase assay. (B) RT-qPCR analysis. *P<0.05, **P<0.01, ***P<0.001 vs. p-shRNA. TPM3, tropomyosin 3; MMP2, matrix metalloproteinase 2; MMP9, matrix metalloproteinase 9; shRNA, empty control cells of TPM3 knockdown; sRNA, TPM3-knockdown cells. The red arrow points to the MMP2/MMP9 protein range
The effects of TPM3 on the regulation of the EMT process in ESCC cells
EMT provides motility and invasive characteristics to cancer cells, and is considered to be the initiating process of metastasis. To explore whether TPM3 is involved in the regulation of the EMT process in ESCC cells, the present study quantified the expression of markers associated with EMT in TPM3-overexpressing or TPM3-knockdown ECa109 and EC9706 cells. As Figure 1B shows, the knockdown of TPM3 improved the mRNA levels of E-cadherin, but reduced the expression of N-cadherin, Snail, and vimentin. The overexpression of TPM3 caused the opposite effect. Specifically, it significantly enhanced the expression of N-cadherin, Snail, and vimentin; however, no change in the level of E-cadherin was observed. A similar pattern of expression for the EMT markers was also revealed by the western blot analysis.
TPM3 regulates the EMT process via MMP2 and MMP9 in ESCC cells
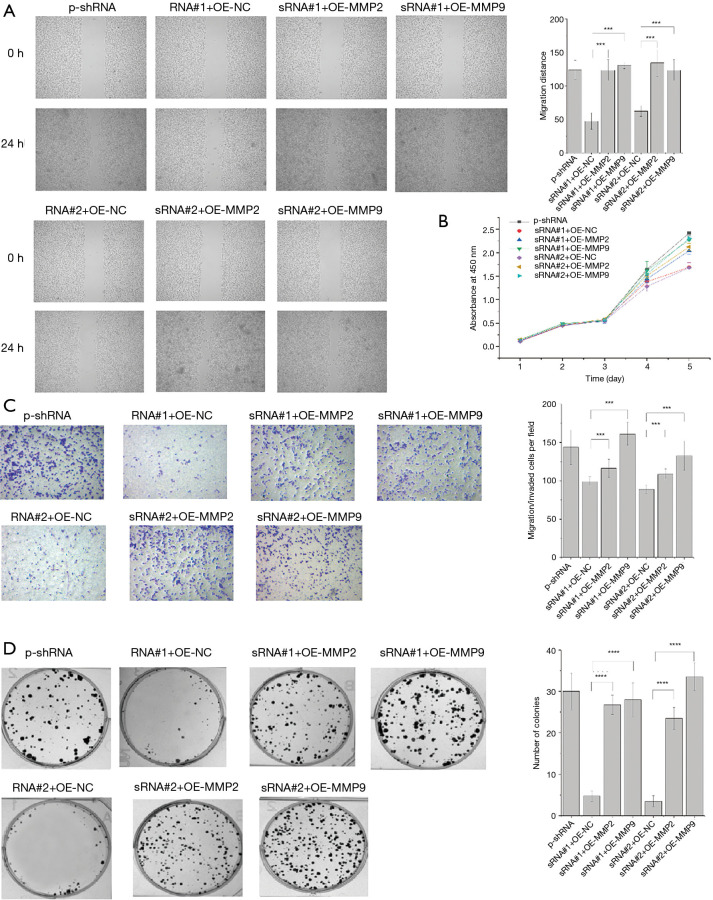
It is generally accepted that MMP2 and MMP9 play key roles in inducing EMT. Our study observed that when TPM3 was knocked down, the expression of MMP2 and MMP9 also decreased. Additionally, the cell functions of proliferation, migration, and metastasis were lowered due to this change. Next, MMP2 and MMP9 were replenished in EC9706 p-sRNA#1, EC9706 p-sRNA#2, and empty control EC9706 p-sh cells. The stable overexpression of MMP2 and MMP9 in these cells was confirmed using a western blot analysis (see Figure 5). As Figure 5 shows, the expression of E-cadherin returned to high levels, while that of N-cadherin, Snail, and vimentin returned to low levels. The effect of the replenishment of MMP2 and MMP9 on the migratory and invasive abilities of ESCC cells was measured. The recovery of the knocked down expression of TPM3 (EC9706 p-sRNA#1, EC9706 p-sRNA#2, and empty control EC9706 p-shRNA cells) with the over expression of MMP2 and MMP9 led to increased proliferation, migration, and metastatic capabilities, as determined by wound healing (see Figure 6A), CCK-8 (see Figure 6B), Transwell (see Figure 6C) and colony formation (see Figure 6D) assays.
Figure 5.
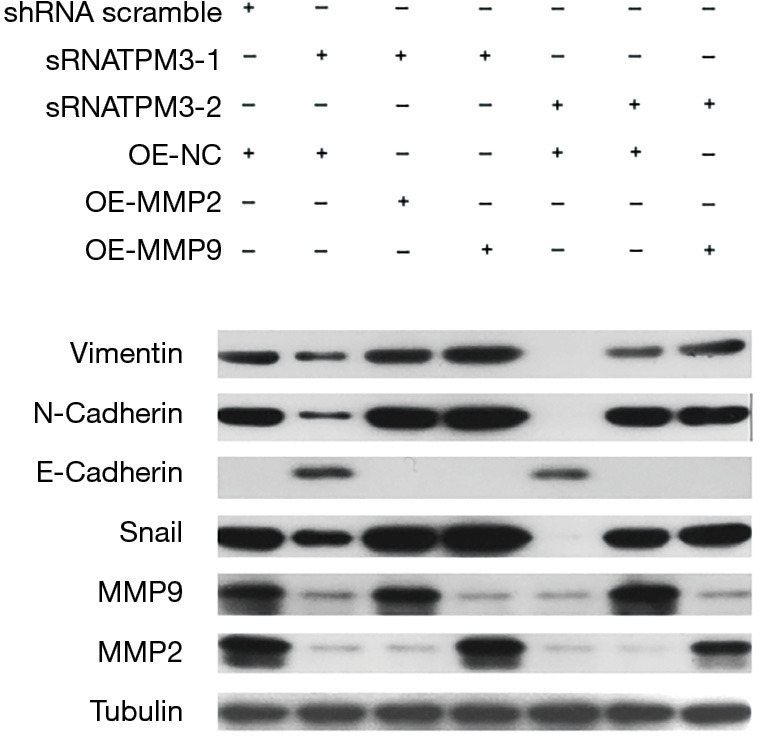
Replenishment of the expression of MMP2 and MMP9 in ESCC cells by western blot analysis. TPM3, tropomyosin 3; MMP2, matrix metalloproteinase 2; MMP9, matrix metalloproteinase 9; shRNA, empty control cells of TPM3 knockdown; sRNATPM3, TPM3-knockdown cells. OE-NC, empty control cells of MMP2/9 overexpression; OE-MMP2/9, MMP2/9-overexpressing cells.
Figure 6.
Replenishment of the expression of MMP2 and MMP9 on the proliferation, migration, invasion, and metastatic potential of Eca109 cells. (A) Wound healing assay (×50). (B) CCK-8 assay. (C) Transwell assay (×100, dyed by crystal violet). (D) Colony formation assay. ***P<0.001, ****P<0.0001 vs. sRNA#1 + OE-NC or sRNA#2 + OE-NC. TPM3, tropomyosin 3; MMP2, matrix metalloproteinase 2; MMP9, matrix metalloproteinase 9; p-shRNA, empty control cells of TPM3 knockdown; sRNATPM3, TPM3-knockdown cells; OE-NC, empty control cells of MMP2/9 overexpression; OE-MMP2/9, MMP2/9-overexpressing cells.
Discussion
This study showed that TPM3 increased the proliferation, migration, and metastasis of ESCC cells, indicating the oncogenic role of TPM3 in those cells. In the wound healing assay, TPM3 appeared to have a significant association with migration ability. In the study examining the effects of TPM3 on proliferation in vivo, the results showed that TPM3 appeared to function by promoting the migration and metastasis of cells. Both MMP2 and MMP9 were activated by TPM3. Further, as the gelatinase assay showed, a reduction in the expression of TPM3 inhibited the activity of MMP9. Thus, TPM3 appears to regulate the expression of MMP2 and MMP9, and affect the activity of MMP9. Additionally, the levels of markers of EMT, including N-cadherin, Snail, and vimentin, were observed to increase consistently with TPM3, while those of E-cadherin showed no significant changes. TPM3 appears to function by promoting the migration and invasion of EC cells via the activation of the EMT process. By replenishing MMP2 and MMP9 in the migratory and invasive abilities of ESCC cells, TPM3 was shown to promote the biological malignant behavior of EC9706 (ESCC) cells by regulating the expression and activity of MMP2 and MMP9.
The consistent changes observed in the expression levels of TPM3, MMP2, and MMP9 in the ESCC cells are consistent with those found by Tao et al. in gliomas (4). The MMP family plays a key role in the malignant behavior of tumors, including tumor growth, angiogenesis, proliferation, apoptosis, invasion, and metastasis. For example, MMP2 and MMP9 have been suggested to accelerate tumor growth and promote cellular invasion in endometrial cancer (8). MMP2 has been reported to hydrolyze intercellular matrix components and basement membrane type IV collagen, while MMP9 has been shown to degrade the extracellular matrix (ECM) or remodel its dynamic balance. Both MMP2 and MMP9 have been confirmed to be involved in the development and progression of numerous malignant tumors, including EC, breast cancer (9), non-small cell lung cancer (10) tongue squamous cell carcinoma (11), and renal cell carcinoma (12).
In previous studies on EC, Zeng et al. constructed an overexpressing and silencing model of erythrocyte membrane protein band 4.1-like 3 (Epb41I3), and found that Epb41I3 could reduce the expression of MMP2 and MMP9, and inhibit the malignant behavior of ESCC cells (13). Xuan et al. found that MMP2 was highly expressed in the tumor tissues of patients with ESCC, and promoted the invasion of ESCC cells as verified by in vitro experiments (14). The key pathway by which MMP2 and MMP9 are known to promote tumor progression is by mediating the EMT process. Notably, MMP2 and MMP9 have been shown to degrade EMT epithelial markers and ECM, leading to the inactivation of the cell adhesion system, which allows cells to acquire a mesenchymal phenotype that deviates from the basolateral membrane (15).
Our study showed that downregulating the expression of MMP2 and MMP9 led to a different expression of E-cadherin, N-cadherin, vimentin, and Snail, thereby inhibiting the progression of EMT. These results were further confirmed by MMP2 and MMP9 rescue experiments. Our findings are consistent with those of Wang et al. and Yang et al. in EC (16) and cholangiocarcinoma (17), respectively.
During EMT, cancer cells lose their epithelial phenotype and cell polarity, and gain a strong ability to degrade ECM, resist apoptosis, invasion, migration and induce an interstitial cell phenotype, including (18). This process has been reported to exhibit the following characteristics: The expression of cell adhesion molecules, represented by E-cadherin is decreased, while the expression of interstitial adhesion molecules, represented by N-cadherin, is increased, and the cytokeratin skeleton is transformed and is accompanied by increases in the expression of vimentin. Previous studies have shown that the occurrence of EMT might be associated with a variety of protein molecules, cell transcription factors (including Snail), ECM environment (e.g., MMP2 and MMP9) and mRNA. Further, multiple signaling pathways (e.g., transforming growth factor beta, receptor tyrosine kinases, Canonical Wn, and Notch Signaling pathway) and complex mechanisms might play key role in the malignant behavior of various cancer cells (19-22).
E-cadherin is the most well-studied member of the cadherin family and is important in the process of EMT. It has been shown to mediate adhesion, enable mutual recognition between cells, and exert adhesion functions. Wang et al. retrieved multiple databases, and collected and analyzed meta-analysis studies on E-cadherin, and concluded that the downregulated expression of E-cadherin promoted the progression of tumors and shortened the survival of patients (23). Snail is the first transcription factor found to induce the progression of EMT and affect signaling and transcriptional regulation (24). Previous studies have shown that the expression of Snail is upregulated in gastric, neuroendocrine lung, colon and pancreatic cancer, as well as other carcinomas (25,26). Snail may also influence tumor, nodes, and metastases staging, distant lymph node metastasis, and survival prognosis in malignant tumors. In a previous study, Liu et al. revealed that Snail was increased in ESCC cells, and further cell experiments showed that the downregulation of the expression of Snail inhibited their metastasis (27).
Research has shown that vimentin, functioning as a transcription factor, affects the progression of tumors and alters the tumor microenvironment in multiple tumor studies (28). Tanaka et al. showed that vimentin was upregulated in ESCC tumor tissues, and that vimentin is also an independent factor for predicting the mortality of patients with ESCC. Notably, the prognosis of patients was demonstrated to be worse than patients with vimentin-positive cells (29). Jin et al. also showed that vimentin in ESCC was an independent predictor of lymph node metastasis (30). The incidence of lymph node metastasis in vimentin-positive cells was significantly higher than that in negative cells, suggesting that vimentin increases the metastasis ability of tumors.
N-cadherin, the most prominent mesenchymal cell marker in the EMT process (31), has been shown to be highly expressed in some cancer types, including EC, breast cancer, ovarian cancer, glioma, and colon cancer. N-cadherin mainly enhances tumor invasion by mediating ETM in tumor cells and promoting tumor angiogenesis (32-35). Labernadie et al. found that reducing the expression of N-cadherin inhibited the malignant behavior of ESCC cells (36). Wang et al. suggested that the mechanism by which the low expression of N-cadherin disturbed the progression of ESCC might be related to the inhibition of vasculogenic mimicry (16).
In our study, TPM3 functioned as an oncogene in ESCC, and we further validated this finding. TPM3 also upregulated the expression of MMP2 and MMP9, induced the progression of EMT, and activated the proliferation, migration, and metastasis of ESCC cells. Thus, TPM3 could become a novel target to overcome ESCC, and may be a new indicator in the prognosis of ESCC patients.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was funded by the National Natural Science Foundation of China (grant No. 81773129 and 82070499), the Natural Science Foundation of Fujian Province (grant No. 2020J02055), the Fujian Young Teacher Fund (grant nos. JAT170230 and JAT180182), the Program for the Innovative Research Team in Science and Technology in Fujian Province University (grant No. 2018B053), the Family Planning Youth Health Project in Fujian Province (grant No. 2018-1-48) and the Startup Fund for Scientific Research at the Fujian Medical University (grant nos. 2019QH1031).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Experiments were performed under a project license [No.: 2020 Association Ethics Examination No. (040)] granted by the Fujian Medical University Institutional Animal Car and Use Committee and were carried out according to institutional guidelines.
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-4043
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-4043
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-4043). The authors have no conflicts of interest to declare.
(English Language Editor: L. Huleatt)
References
- 1.Smyth EC, Lagergren J, Fitzgerald RC, et al. Oesophageal cancer. Nat Rev Dis Primers 2017;3:17048. 10.1038/nrdp.2017.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. 10.3322/caac.21338 [DOI] [PubMed] [Google Scholar]
- 3.Lees JG, Bach CT, O'Neill GM. Interior decoration: tropomyosin in actin dynamics and cell migration. Cell Adh Migr 2011;5:181-6. 10.4161/cam.5.2.14438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao T, Shi Y, Han D, et al. TPM3, a strong prognosis predictor, is involved in malignant progression through MMP family members and EMT-like activators in gliomas. Tumour Biol 2014;35:9053-9. 10.1007/s13277-014-1974-1 [DOI] [PubMed] [Google Scholar]
- 5.Yu SB, Gao Q, Lin WW, et al. Proteomic analysis indicates the importance of TPM3 in esophageal squamous cell carcinoma invasion and metastasis. Mol Med Rep 2017;15:1236-42. 10.3892/mmr.2017.6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin W, Lin J, Chen B, et al. Tropomyosin3 is associated with invasion, migration, and prognosis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol 2016;9:11313-23. [Google Scholar]
- 7.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Qiu H, Hu W, et al. Over-expression of platelet-derived growth factor-D promotes tumor growth and invasion in endometrial cancer. Int J Mol Sci 2014;15:4780-94. 10.3390/ijms15034780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yousef EM, Tahir MR, St-Pierre Y, et al. MMP-9 expression varies according to molecular subtypes of breast cancer. BMC Cancer 2014;14:609. 10.1186/1471-2407-14-609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aparna M, Rao L, Kunhikatta V, et al. The role of MMP-2 and MMP-9 as prognostic markers in the early stages of tongue squamous cell carcinoma. J Oral Pathol Med 2015;44:345-52. 10.1111/jop.12245 [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Jiang T, Mao A, et al. Esophageal cancer stem cells express PLGF to increase cancer invasion through MMP9 activation. Tumour Biol 2014;35:12749-55. 10.1007/s13277-014-2601-x [DOI] [PubMed] [Google Scholar]
- 12.Jing SW, Wang YD, Kuroda M, et al. HIF-1α contributes to hypoxia-induced invasion and metastasis of esophageal carcinoma via inhibiting E-cadherin and promoting MMP-2 expression. Acta Med Okayama 2012;66:399-407. [DOI] [PubMed] [Google Scholar]
- 13.Zeng R, Huang JP, Li XF, et al. Epb41l3 suppresses esophageal squamous cell carcinoma invasion and inhibits MMP2 and MMP9 expression. Cell Biochem Funct 2016;34:133-41. 10.1002/cbf.3170 [DOI] [PubMed] [Google Scholar]
- 14.Xuan X, Li S, Lou X, et al. Stat3 promotes invasion of esophageal squamous cell carcinoma through up-regulation of MMP2. Mol Biol Rep 2015;42:907-15. 10.1007/s11033-014-3828-8 [DOI] [PubMed] [Google Scholar]
- 15.Jia LF, Wei SB, Mitchelson K, et al. miR-34a inhibits migration and invasion of tongue squamous cell carcinoma via targeting MMP9 and MMP14. PLoS One 2014;9:e108435. 10.1371/journal.pone.0108435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, He W, Fanghui P, et al. NF-κBP65 promotes invasion and metastasis of oesophageal squamous cell cancer by regulating matrix metalloproteinase-9 and epithelial-to-mesenchymal transition. Cell Biol Int 2013;37:780-8. 10.1002/cbin.10089 [DOI] [PubMed] [Google Scholar]
- 17.Yang H, Liang J, Zhou J, et al. Knockdown of RHOC by shRNA suppresses invasion and migration of cholangiocellular carcinoma cells via inhibition of MMP2, MMP3, MMP9 and epithelial-mesenchymal transition. Mol Med Rep 2016;13:5255-61. 10.3892/mmr.2016.5170 [DOI] [PubMed] [Google Scholar]
- 18.Lin F, Chengyao X, Qingchang L, et al. CRKL promotes lung cancer cell invasion through ERK-MMP9 pathway. Mol Carcinog 2015;54 Suppl 1:E35-44. 10.1002/mc.22148 [DOI] [PubMed] [Google Scholar]
- 19.McNiven MA. Breaking away: matrix remodeling from the leading edge. Trends Cell Biol 2013;23:16-21. 10.1016/j.tcb.2012.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu D, Pan H, Zhou Y, et al. microRNA-133b downregulation and inhibition of cell proliferation, migration and invasion by targeting matrix metallopeptidase-9 in renal cell carcinoma. Mol Med Rep 2014;9:2491-8. 10.3892/mmr.2014.2116 [DOI] [PubMed] [Google Scholar]
- 21.Zhang F, Wang Z, Fan Y, et al. Elevated STAT3 Signaling-Mediated Upregulation of MMP-2/9 Confers Enhanced Invasion Ability in Multidrug-Resistant Breast Cancer Cells. Int J Mol Sci 2015;16:24772-90. 10.3390/ijms161024772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Han H, Li Y, et al. Upregulation of long noncoding RNA HOTTIP promotes metastasis of esophageal squamous cell carcinoma via induction of EMT. Oncotarget 2016;7:84480-5. 10.18632/oncotarget.12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang C, Wang J, Chen Z, et al. Immunohistochemical prognostic markers of esophageal squamous cell carcinoma: a systematic review. Chin J Cancer 2017;36:65. 10.1186/s40880-017-0232-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hori S, Wadhwa K, Pisupati V, et al. Loss of hSef promotes metastasis through upregulation of EMT in prostate cancer. Int J Cancer 2017;140:1881-7. 10.1002/ijc.30604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurioka K, Wato M, Iseki T, et al. Differential expression of the epithelial mesenchymal transition factors Snail, Slug, Twist, TGF-β, and E-cadherin in ameloblastoma. Med Mol Morphol 2017;50:68-75. 10.1007/s00795-016-0149-0 [DOI] [PubMed] [Google Scholar]
- 26.Zhao L, Zhang P, Su XJ, et al. The ubiquitin ligase TRIM56 inhibits ovarian cancer progression by targeting vimentin. J Cell Physiol 2018;233:2420-5. 10.1002/jcp.26114 [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Zhou H, Zhu R, et al. SPSB3 targets SNAIL for degradation in GSK-3β phosphorylation-dependent manner and regulates metastasis. Oncogene 2018;37:768-76. 10.1038/onc.2017.370 [DOI] [PubMed] [Google Scholar]
- 28.Zheng M, Jiang YP, Chen W, et al. Snail and Slug collaborate on EMT and tumor metastasis through miR-101-mediated EZH2 axis in oral tongue squamous cell carcinoma. Oncotarget 2015;6:6797-810. 10.18632/oncotarget.3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka M, Kijima H, Shimada H, et al. Expression of podoplanin and vimentin is correlated with prognosis in esophageal squamous cell carcinoma. Mol Med Rep 2015;12:4029-36. 10.3892/mmr.2015.3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jin H, Morohashi S, Sato F, et al. Vimentin expression of esophageal squamous cell carcinoma and its aggressive potential for lymph node metastasis. Biomed Res 2010;31:105-12. 10.2220/biomedres.31.105 [DOI] [PubMed] [Google Scholar]
- 31.Abufaraj M, Shariat SF, Haitel A, et al. Prognostic role of N-cadherin expression in patients with non-muscle-invasive bladder cancer. Urol Oncol 2017;35:264-71. 10.1016/j.urolonc.2017.01.012 [DOI] [PubMed] [Google Scholar]
- 32.Yin T, Wang C, Liu T, et al. Expression of snail in pancreatic cancer promotes metastasis and chemoresistance. J Surg Res 2007;141:196-203. 10.1016/j.jss.2006.09.027 [DOI] [PubMed] [Google Scholar]
- 33.Galván JA, González MV, Crespo G, et al. Snail nuclear expression parallels higher malignancy potential in neuroendocrine lung tumors. Lung Cancer 2010;69:289-95. 10.1016/j.lungcan.2009.12.010 [DOI] [PubMed] [Google Scholar]
- 34.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci 2011;68:3033-46. 10.1007/s00018-011-0735-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal 2014;7:re8. 10.1126/scisignal.2005189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labernadie A, Kato T, Brugués A, et al. A mechanically active heterotypic E-cadherin/N-cadherin adhesion enables fibroblasts to drive cancer cell invasion. Nat Cell Biol 2017;19:224-37. 10.1038/ncb3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as