Abstract
The AML1/core binding factor β (CBFβ) transcription factor is essential for definitive hematopoiesis; however, the downstream pathways through which it functions remain incompletely defined. Using a differential cloning approach to define components of this pathway, we have identified a novel gene designated HERF1 (for hematopoietic RING finger 1), whose expression during development is dependent on the presence of functional AML1/CBFβ. HERF1 contains a tripartite RING finger–B box–α-helical coiled-coil domain and a C-terminal region homologous to the ret proto-oncogene-encoded finger protein. Expression of HERF1 during embryogenesis coincides with the appearance of definitive erythropoiesis and in adult mice is restricted to erythroid cells, increasing 30-fold during terminal differentiation. Importantly, inhibition of HERF1 expression blocked terminal erythroid differentiation of the murine erythroleukemia cell line MEL, whereas its overexpression induced erythroid maturation. These results suggest an important role for this protein in erythropoiesis.
The development of the hematopoietic system is regulated by a series of lineage-restricted transcription factors that control critical cell fate decisions, including the formation of primitive and definitive hematopoietic stem cells from embryonic mesoderm and the survival, expansion, lineage commitment, and differentiation of more committed progenitors. Our laboratory (29) as well as those of others (42, 43) has demonstrated that the AML1/core binding factor β (CBFβ) transcription factor complex, the most common target of chromosomal translocations in human leukemia (reviewed in reference 23), is essential for the formation of the definitive hematopoietic system. Null mutations in either AML1 or CBFβ, or expression of the dominant inhibitory t(8;21)-encoded leukemia protein, AML1-ETO (27, 47), result in an embryonic lethal phenotype, with embryos dying during the midpoint of development from a complete absence of fetal liver-derived hematopoiesis and lethal central nervous system hemorrhages. Although primitive yolk sac-derived erythropoiesis appears normal in these mutants, no definitive hematopoietic progenitors of any lineage are present. Thus, AML1/CBFβ appears to function as a critical element that controls the development of hematopoietic cells of the definitive lineages. This function is achieved by binding of AML1/CBFβ to the core enhancer DNA sequence and regulating the transcription of essential target genes (reviewed in reference 39).
A large number of transcriptional targets of AML1/CBFβ have been identified, including granulocyte-macrophage colony-stimulating factor (GM-CSF), the receptor for CSF-1, myeloperoxidase, neutrophil elastase, interleukin 3, and the α, β, γ, and δ subunits of the T-cell antigen receptor (reviewed in reference 39). Although each of these gene targets provide critical functions in hematopoietic cells, experimental data suggest that none are essential for the establishment of definitive hematopoiesis (20–22, 35). Therefore, it is clear that additional AML1-regulated target genes that play critical roles in the signaling pathways required for the establishment of definitive hematopoiesis must exist. In addition, the downstream mechanistic pathways through which the AML1/CBFβ-mediated transcription cascade functions remain incompletely defined.
To identify components of the downstream pathways initiated by AML1/CBFβ, we attempted to clone genes whose expression both occurs during the initial development of definitive hematopoietic progenitors and is dependent on the presence of a functional AML1/CBFβ transcription factor complex. To accomplish this, we performed representational difference analysis (RDA) using mRNA isolated from wild-type and AML1-deficient embryonic stem (ES) cells differentiated in vitro to a point in development at which the earliest commitment to definitive hematopoietic progenitors occurs. We and others have previously demonstrated that this in vitro ES cell differentiation assay accurately replicates the in vivo phenotype that results from the loss of AML1, that is, a complete lack of definitive hematopoiesis (29, 42). Using this approach, we cloned a novel erythroid cell-specific gene designated HERF1 (for hematopoietic RING finger 1), whose expression depends on the presence of functional AML1/CBFβ. Functional analysis indicates that HERF1 plays an important role in the maturation of definitive erythroid cells.
MATERIALS AND METHODS
EB cultures.
The procedure for in vitro differentiation of embryoid bodies (EBs) was as previously described (29). In brief, 3 × 102 wild-type or AML1-deficient ES cells were plated in 1 ml of Iscove modified Eagle medium containing 1.2% methylcellulose, 15% fetal calf serum, 2 mM glutamine, 450 μM monothioglycerol, and a mixture of hematopoietic growth factors including human erythropoietin (2 U/ml; Amgen), murine stem cell factor (10 ng/ml; Genzyme), murine interleukin 3 (1.8 ng/ml; R&D Systems), and murine GM-CSF (10 ng/ml; Genzyme). Cultures were grown for 7 days at 37°C under humidified conditions with 5% CO2. EBs from 30 individual cultures were then pooled, washed in phosphate-buffered saline (PBS), and used for RNA isolation. The morphology of the developing EBs was monitored by microscopic examination of cytocentrifuge preparations of single dispersed EBs.
RDA.
Total RNA was extracted from pooled EBs by using a modified acid-guanidinium-thiocyanate-phenol-chloroform extraction method (5), and poly(A)+ RNA was isolated from the extracted nucleic acid by using a FastTrack 2.0 kit (Invitrogen) according to the manufacturer’s directions. Oligo(dT)-primed double-stranded cDNA was synthesized from 5 μg of poly(A)+ RNA by using a cDNA synthesis system (GIBCO BRL) according to the manufacturer’s instructions.
RDA was performed essentially as described by Hubank and Schatz (14). Briefly, double-stranded cDNAs prepared from wild-type or AML1-deficient EBs were digested with DpnII and ligated to the oligonucleotide adapters 5′ R-Bgl-24 (5′-AGCACTCTCCAGCCTCTCACCGCA-3′) and 3′ R-Bgl-12 (5′-GATCTGCGGTGA-3′). Representative amplicons were then generated by PCR amplification for 20 cycles using the R-Bgl-24 oligonucleotide as a primer. Following amplification, the oligonucleotide adapters were removed from the amplified DNA by digestion with DpnII. Amplicons derived from AML1-deficient EBs (driver) were used directly following digestion. By contrast, digested amplicons prepared from wild-type EBs (tester) were size fractionated between 200 and 1,500 bp by gel purification and then ligated to a second pair of oligonucleotides adapters, 5′ J-Bgl-24 (5′-ACCGACGTCGACTATCCATGAACA-3′) and 3′ J-Bgl-12 (5′-GATCTGTTCATG-3′). Tester and driver amplicons were mixed at a ratio of 1:100 and incubated for 20 h at 67°C. Following this incubation, an aliquot of the hybridization mixture was subjected to 10 cycles of PCR using the J-Bgl-24 oligonucleotide as an amplification primer. The PCR products were then digested with mung bean nuclease (New England Biolabs) at 30°C for 35 min and then further amplified for an additional 18 cycles. The amplified PCR products were digested with DpnII and then used in additional rounds of subtraction. In the second round, the PCR products were ligated to a third pair of oligonucleotide adapters, 5′ N-Bgl-24 (5′-AGGCAACTGTGCTATCCGAGGGAA-3′) and 3′ N-Bgl-12 (5′-GATCTTCCCTCG-3′), and hybridized with driver amplicons at a ratio of 1:800. The adapters were removed from the PCR amplicons obtained following this hybridization, and the products were religated to the J-Bgl-12/24 adapters and rehybridized to the driver amplicons at a ratio of 1:8,000. After PCR amplification with the J-Bgl-24 primer, several bands were visible following electrophoresis in 2% agarose gels containing ethidium bromide. These individual bands were isolated, digested with DpnII, cloned into the BamHI site of pBluescriptII SK(+), and sequenced.
Library screening, cloning, Northern blot analysis, and in situ hybridization.
A lambda ZAP II mouse spleen cDNA library (Stratagene) was screened with the [α-32P]CTP-labeled 0.49-kb HERF1 fragment isolated by RDA. Positive phage were isolated by plaque purification, and HERF1-containing pBluescript phagemid were purified from positive phage by excision in vivo from the lambda ZAP II vector, using ExAssist helper phage (Stratagene). The DNA sequence of a full-length HERF1 cDNA was obtained by the dideoxy-chain termination method. Throughout the complete coding region, at least two independent clones of the same region were sequenced on both strands. Northern blot analysis and in situ hybridization were performed essentially as described elsewhere (28, 36).
Tetracycline-regulated expression of HERF1 and α-sense HERF1.
The mouse erythroleukemia cell line MEL (41) was maintained in complete medium (Dulbecco modified Eagle medium supplemented with 10% fetal calf serum and 2 mM glutamine). MEL cells were stably transfected by electroporation with 5 μg of BstXI-linearized pTET.TAK.HYG vector (generous gift of Brian Van Ness, University of Minnesota, Minneapolis) containing a tetracycline-inducible fusion of the tet repressor DNA binding domain and the VP16 activation domain, (Tet-VP16 fusion protein) as well as a constitutively expressed hygromycin-selectable marker. Cells were electroporated at 975 μF and 276 V with a Gene Pulser (Bio-Rad, Richmond, Calif.) in a 0.4 cm cuvette at a density of 2 × 107 cells/ml and a volume of 0.4 ml of medium. Cells were allowed to recover for 24 h before selection in hygromycin (1 mg/ml; Calbiochem) at a density of 5 × 104 cells/ml in the presence of tetracycline (0.5 μg/ml; Sigma) in a 1 ml volume in 24-well plates. Inducible expression of the Tet-VP16 fusion protein was assayed by Western blot analysis of protein extracts from hygromycin-resistant clones cultured in the presence and absence of tetracycline. The polyclonal antibody used for Western blot analysis was raised against the yeast GAL4-VP16 activation domain (Upstate Biotechnology Inc.). Clones expressing low levels of uninduced protein and high levels of induced activity were identified, and one was selected for subsequent stable transfection with AhdI-linearized pTET.HERF1.NEO or pTETα.HERF1.NEO, both of which were derived from the pTET.TAK.NEO vector. Conditions for selection of stable integration of these vectors were identical to those described above except that selection was carried out in the presence of G418 (0.8 mg/ml; Life Technologies), hygromycin (1 mg/ml), and tetracycline (0.5 μg/ml). Clones expressing inducible HERF1 and antisense (α-sense) HERF1 were identified by Northern and Western blot analyses.
Preparation of HERF1 antisera and Western blot analysis.
A unique SalI site was created immediately 5′ to the ATG codon in the HERF1 cDNA by PCR, and a 698-bp SalI-BglII restriction fragment corresponding to the N-terminal 230 amino acids was cloned in frame into the pGEX-4T-2 vector (Gibco). The resultant plasmid encodes a glutathione S-transferase (GST)–HERF1 fusion protein that contains a novel alanine residue at the point of fusion. Escherichia coli DH5α was transformed with this plasmid, and the GST-HERF1 fusion protein was isolated by a modification of established methods (38). Briefly, a 2-liter culture of bacteria in log growth phase was induced with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and grown for an additional 20 h at 25°C. The bacterial cell pellets were then collected and resuspended in 80 ml of bacterial protein extraction reagent (Pierce) containing 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml, 10 mM β-glycerophosphate, 1 mM NaF, and 5 μg of leupeptin per ml. The bacterial suspension was sonicated and then added to 2 ml of a 50% slurry of glutathione-Sepharose 4B (Pharmacia Biotech), and the mixture was incubated at 4°C for 30 min with constant rocking. The Sepharose beads were then washed three times with bacterial protein extract reagent and three times with cold PBS and finally resuspended in a final volume of 2 ml of PBS. Cleavage of the HERF1 protein from GST was achieved by incubating the beads with 100 U of thrombin (Pharmacia Biotech) at 22°C for 16 h with constant rocking. The purified polypeptide was injected into New Zealand White rabbits to produce anti-HERF1 polyclonal antibodies (Rockland). Western blot analysis was performed as previously described (27).
Nucleotide sequence accession number.
The HERF1 DNA sequence has been submitted to the GenBank database (accession no. BankH258518 AF134811).
RESULTS
Isolation of HERF1.
To clone genes whose expression requires functional AML1/CBFβ, we performed RDA using an in vitro ES cell differentiation assay that accurately mimics the in vivo definitive hematopoietic defect resulting from the loss of AML1 (17, 29, 42). In this assay, differentiation of either wild-type or AML1-deficient ES cells as EBs for 6 days results in the development of primitive hematopoietic progenitors. By contrast, when EBs are grown for 10 days, wild-type but not AML1-deficient ES cells generate definitive hematopoietic cells. We therefore took advantage of this system to clone genes that were expressed in wild-type but not AML1-deficient cells. To accomplish this, RDA was performed on RNA isolated from EBs grown for 7 days in methylcellulose-containing cultures. At this point in development, primitive erythropoiesis is well established, whereas only the earliest commitment to definitive hematopoiesis has occurred (17).
Three rounds of hybridization and PCR amplification were performed, and the differentially expressed products were cloned. Sequence analysis revealed a number of known hematopoiesis-specific genes that were differentially expressed between the tester (wild-type) and driver (AML1-deficient) amplicons, as well as a single PCR fragment that corresponded to a novel gene. A full-length 2.2-kb cDNA (see below) for this novel gene was isolated from a murine spleen cDNA library and sequenced. We identified an open reading frame that encoded a 489-amino-acid protein that contains an N-terminal cysteine-rich C3HC4 zinc finger, termed a RING finger domain after the first protein identified with this motif, RING1 (really interesting new gene 1) (Fig. 1) (24). Based on the presence of this motif, we have termed this gene HERF1, for hematopoietic RING finger 1. The human HERF1 gene (previously referred to as RFB30) was independently cloned and sequenced as part of an effort to map the human major histocompatibility complex class I region on chromosome 6p21.3 (12). The deduced sequence of the coding region of the human mRNA encoded by this locus showed 81% identity throughout its sequence to murine HERF1.
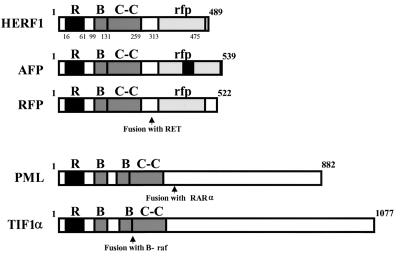
FIG. 1.
Structural organization of selected members of the RING (R), B box (B), coiled-coil (C-C) family of proteins. RAR, retinoic acid receptor alpha.
Immediately adjacent to the RING finger domain of HERF1 is a second distinct zinc-binding motif known as a B box, followed by a leucine α-helical coiled-coil domain and a C-terminal region referred to as the ret proto-oncogene-encoded finger protein (RFP)-like (rfp) or B30.2 domain (13). The tripartite RING–B box–coiled-coil (RBCC) domain defines a unique subfamily of proteins. The members of this subfamily that are most closely related to HERF1 are illustrated in Fig. 1. These include (i) the acid finger protein AFP, a nuclear protein of unknown function (6); (ii) RFP, which was initially identified as part of the RFP-RET chimeric oncoprotein (15, 40); (iii) promyelocytic leukemia protein (PML), which is involved in the PML-retinoic acid receptor alpha fusion protein produced as a result of the t(15;17) translocation (7, 16); and (iv) TIF1α, initially identified as a fusion with B-raf in a chemically induced hepatoma (25) and subsequently shown to function as a ligand-dependent coactivator of the retinoic acid family of transcriptional factors (19). The individual RING, B-box, coiled-coil, and rfp domains of HERF1 have between 30 and 56% identity to the homologous regions of AFP and RFP.
Pattern of HERF1 gene expression.
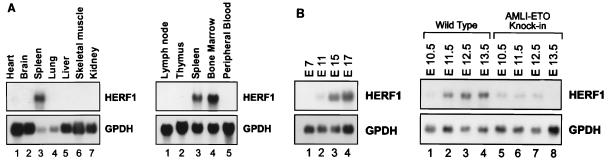
As an initial approach to define the pattern of HERF1 expression, we performed Northern blot analysis using RNA isolated from adult mouse tissues. As shown in Fig. 2A, HERF1 was expressed as a single 2.3-kb transcript in the spleen but was not detected in any of the nonhematopoietic tissues examined (heart, brain, lung, liver, skeletal muscle, and kidney). A closer examination of hematopoietic tissues revealed HERF1 expression in the bone marrow and spleen but not in hematopoietic organs that were composed primarily of lymphoid cells, including lymph node, thymus, and peripheral blood (Fig. 2A). Thus, these data suggest that HERF1 is expressed exclusively in hematopoietic tissues that contain developing myeloid, erythroid, and/or megakaryocytic progenitors.
FIG. 2.
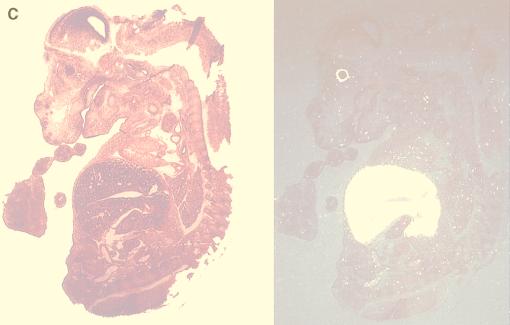
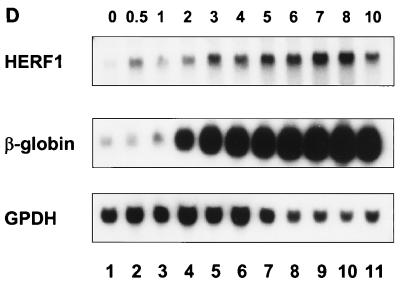
Expression pattern of HERF1. Northern blots of adult tissues (A), murine embryos (B), or MEL cells following treatment with the differentiation-inducing agent DMSO (D) were hybridized with a full-length HERF1 cDNA. Filters were stripped and rehybridized with a probe for glycerol-3-phosphate dehydrogenase (GPDH) to assess the integrity and amount of RNA and with a probe for β-globin (D; numbers above the lanes represent days in DMSO). (C) In situ hybridization was performed with a 271-bp restriction fragment from the unique 3′ region of HERF1 labeled with [33P]UTP. The left side shows the bright-field view of a 12-μm sagittal section from an E15 embryo stained with hematoxylin and eosin; the right side is a dark-field view of the same section hybridized with the HERF1 probe. Sense control shows no specific hybridization (data not shown).
To examine the pattern of HERF1 expression during murine development, RNA was isolated from developing embryos and examined by Northern blot analysis. As shown in Fig. 2B, HERF1 expression was first detected around embryonic day 11.5 (E11.5) at the start of definitive hematopoiesis and increased with further development. More importantly, the level of HERF1 expression was markedly reduced in embryos heterozygous for an AML1-ETO knock-in allele. These embryos are phenotypically identical to AML1-deficient embryos with a complete absence of normal definitive fetal liver-derived hematopoiesis (27, 47). Similarly, the level of HERF1 expression was also markedly reduced in AML1-deficient embryos (data not shown). To further characterize the pattern of HERF1 expression during embryogenesis, we performed in situ hybridization on sections of embryos between E10.5 and E15.5 (Fig. 2C). As shown in a representative section from an E15.5 embryo, HERF1 expression was confined to the fetal liver (the signal detected in the eye is an artifact during dark-field illumination of the retinal pigment layer). Collectively, these data suggest that HERF1 is a hematopoiesis-specific gene whose expression coincides with development of the definitive hematopoietic system and whose normal expression requires functional AML1/CBFβ.
During embryogenesis, the majority of the hematopoietic activity within the fetal liver is committed to cells of the erythroid lineage. To more precisely define the hematopoietic lineages in which HERF1 was expressed, we examined a panel of 27 murine leukemic cell lines by Northern blot analysis. This panel encompassed a variety of hematopoietic lineages including lymphoid (B- and T-cell), myeloid, erythroid, monocytic, and mast cells. HERF1 expression was detected only in the murine erythroleukemia cell line MEL (Fig. 2D and data not shown). No expression was detected in any of the lymphoid, myeloid, or monocytic cell lines examined. To extend these observations, we also examined the human erythroid leukemia cell line TF1 and again detected HERF1 expression (data not shown).
The MEL cell line is derived from Friend virus-transformed cells and is capable of indefinite proliferation. It is normally blocked in differentiation at the proerythroblast stage of development; however, these cells can be induced to undergo terminal erythroid differentiation by treatment with a number of inducing agents including dimethyl sulfoxide (DMSO) (41). Following treatment with DMSO, MEL cells undergo a coordinate program of differentiation that includes a limitation of their proliferative potential, transcription of β-major globin, hemoglobin accumulation, and morphologic differentiation into polychromatic normoblasts. Interestingly, only a low level of HERF1 expression was detected in MEL cells grown in the absence of DMSO; however, within 12 h of DMSO treatment, the level of HERF1 mRNA increased over 30-fold, and it continued to increase with further differentiation (Fig. 2D). We have also observed a similar increase in HERF1 expression following erythroid differentiation of the human leukemia cell line TF1 (data not shown). Thus, HERF1 expression is up-regulated in concert with the commitment to terminal erythroid differentiation in these two cell systems.
Critical role of HERF1 in erythroid differentiation.
To investigate the role of HERF1 in erythroid differentiation, we determined what effect loss of HERF1 would have on DMSO-induced MEL cell differentiation. To perform this experiment, MEL cells were first transfected by electroporation with a plasmid containing a tetracycline-regulated promoter that drives expression of a Tet-VP16 fusion protein (11, 37). In the presence of tetracycline, the Tet-VP16 fusion protein fails to bind to DNA and is thus unable to induce the transcription of either itself or any other cotransfected genes that are under the control of a tetracycline-regulated promoter. By contrast, in the absence of tetracycline, the Tet-VP16 protein binds DNA and strongly transactivates its own expression, resulting in the rapid accumulation of transcriptionally active Tet-VP16. This, in turn, results in the transcription of any other tetracycline-regulated gene (11, 37). Individual MEL cell clones that express the Tet-VP16 chimeric protein after removal of tetracycline were identified by Western blot analysis using antibodies against VP16 and were then assessed for the ability to undergo DMSO-induced differentiation, either in the presence or in the absence of tetracycline (data not shown). Several clones that maintained the ability to differentiate into terminal erythroid cells after treatment with DMSO were isolated and used in the following experiments.
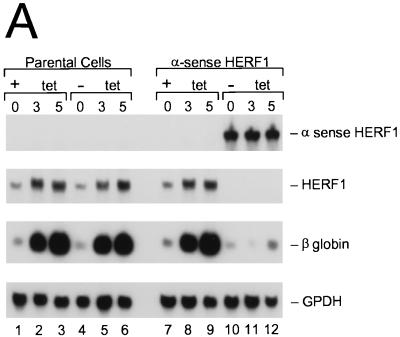
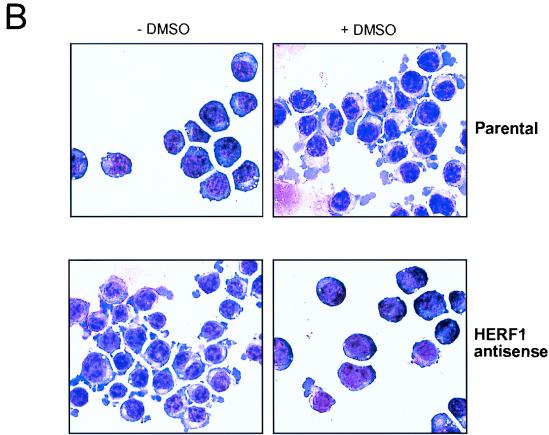
To determine the consequences of loss of HERF1 for DMSO-induced MEL cell differentiation, we expressed a HERF1 α-sense construct under the control of the tetracycline-regulated promoter. Eight independent clones expressing α-sense HERF1 were isolated, and representative results from a single clone are illustrated in Fig. 3. DMSO treatment of parental Tet-VP16-expressing (MEL-Tet-VP16) cells (which lack the α-sense construct) for 3 to 5 days results in the induction of endogenous HERF1 mRNA and terminal differentiation, as assessed by β-globin expression (Fig. 3A, lanes 1 to 6). This effect was not influenced by the presence or absence of tetracycline. By contrast, removal of tetracycline from MEL cells containing the α-sense HERF1 construct resulted in a high level of expression of the α-sense HERF1 mRNA (compare lanes 7 to 9 with lanes 10 to 12). This, in turn, resulted in both the elimination of endogenous HERF1 expression and a complete block in the ability of DMSO to induce terminal differentiation as assessed by β-globin expression (lanes 10 to 12). In addition to β-globin expression, terminal differentiation was assessed morphologically. As shown in Fig. 3B, DMSO treatment of the parental MEL-Tet-VP16 cells resulted in morphologic differentiation to the polychromatic normoblast stage and positive staining for benzidine (data not shown). By contrast, MEL cells expressing the α-sense HERF1 construct failed to show evidence of morphologic differentiation following treatment with DMSO.
FIG. 3.
Inhibition of DMSO-induced MEL cell differentiation by expression of α-sense HERF1. MEL cells expressing the tetracycline-regulated Tet-VP16 fusion protein, stably transfected with either an empty vector (parental) or a tetracycline-regulated α-sense HERF1-containing plasmid (α sense HERF1), were treated for 3 to 5 days with DMSO in the presence (+) or absence (−) of tetracycline (tet). Following the indicated treatments, cells were isolated and assessed by Northern blot analysis (A) and morphology (B). Northern blots of total RNA were sequentially hybridized with probes specific for the genes listed on the right side.
To ensure that expression of the α-sense HERF1 construct did not result in global changes in the ability of these cells to progress through the cell cycle, we used flow cytometry to assess the percentage of cells in each phase of the cell cycle as a function of time following DMSO treatment. No significant differences were observed between the parental MEL-Tet-VP16 cells and those expressing α-sense HERF1 (data not shown). In addition, no changes were observed in the level of mRNA expression for the related RBCC-containing genes, PML and RFP, or the RING-containing gene BMI1 following the induction of α-sense HERF1, suggesting that this α-sense construct specifically modulated the expression of its cognate transcript (data not shown). Taken together, these data suggest that induction of HERF1 expression is necessary for terminal erythroid differentiation.
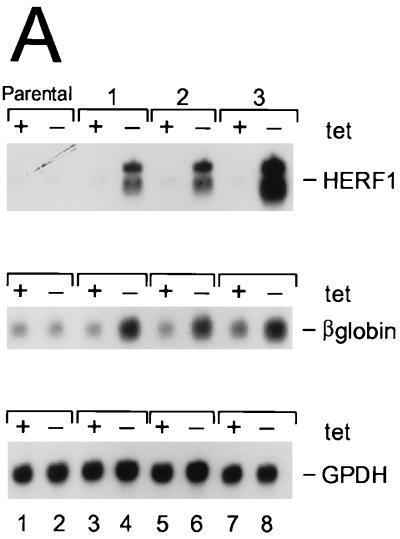
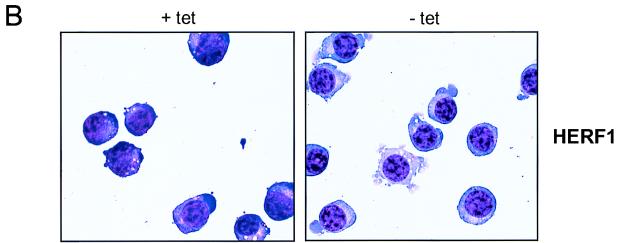
To determine whether HERF1 was sufficient to promote erythroid differentiation, we examined the consequences of enforced expression of wild-type HERF1 for undifferentiated MEL cells. The parental MEL-Tet-VP16 cells were transfected with a plasmid containing a murine HERF1 cDNA under the control of the tetracycline-regulated promoter. Eight independent cell lines were isolated, and representative results from three clones are illustrated in Fig. 4A. In the presence of tetracycline, the promoter was silent and only basal levels of HERF1 mRNA were detected. By contrast, removal of tetracycline resulted in the induction of a high level of HERF1 mRNA. As shown, HERF1 expression in the absence of DMSO treatment resulted in the induction of erythroid differentiation as measured by β-globin expression (Fig. 4A), morphologic differentiation (Fig. 4B), and benzidine positivity (data not shown). The degree of differentiation as assessed by both morphology and level of β-globin mRNA induction was consistently less than that observed after treatment of MEL cells with DMSO.
FIG. 4.
Induction of MEL cell differentiation by enforced HERF1 expression. MEL cells expressing the tetracycline-regulated Tet-VP16 fusion protein, either with an empty plasmid vector (parental) or with a tetracycline-regulated HERF1-containing plasmid (clones 1 to 3), were grown in the presence (+) or absence (−) of tetracycline (tet). Following the indicated treatments, cells were isolated and analyzed by Northern blot analysis (A) and morphology (B). Northern blots were sequentially hybridized with probes specific for the genes listed on the right side.
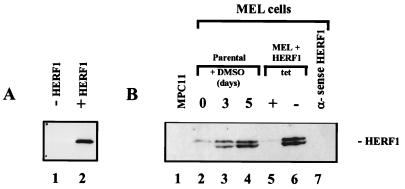
To examine the level of the HERF1 protein in MEL cells, we developed a rabbit antiserum against a recombinant GST-HERF1 fusion protein. Western blot analysis of cell lysates prepared from Cos cells transfected with a HERF1 expression plasmid demonstrated that this antiserum efficiently recognized the 56-kDa HERF1 protein (Fig. 5A, lane 2). Similarly, this antiserum was effective at immunoprecipitating HERF1 from transfected Cos cells (data not shown). To examine the level of HERF1 in MEL cells, total cell lysates were electrophoretically separated on a sodium dodecyl sulfate-polyacrylamide gel and Western blotted with the anti-HERF1 serum. As shown in Fig. 5B, HERF1 was not detected in the nonerythroid cell line MPC11 and was present at only a low level in undifferentiated MEL cells (lanes 1 and 2). By contrast, high levels of the HERF1 protein were observed after treatment with DMSO for 5 days (lane 4). The size of the endogenous protein was the same as that of the protein expressed from our cloned cDNA, confirming that the cDNA encoded a full-length protein. Interestingly, both the endogenous and transfected HERF1 proteins were expressed as a tight doublet. The nature of these two forms remains to be determined. Analysis of MEL cells transfected with the tetracycline-regulated HERF1 plasmid revealed only basal levels of HERF1 in cells grown in the presence of tetracycline (lane 5) but readily detectable levels of HERF1 after the removal of tetracycline from the growth medium (lane 6). The level of HERF1 expressed under these conditions was similar to that of the endogenous protein following DMSO-induced differentiation. Importantly, DMSO-treated MEL cells containing the tetracycline-regulated α-sense HERF1 construct failed to express detectable levels of endogenous HERF1 after growth in the absence of tetracycline (lane 7).
FIG. 5.
Western blot analysis of HERF1, performed with an anti-HERF1 rabbit serum on total cell lysates prepared from Cos cells transfected with an empty vector (−HERF1) or a HERF1 expression plasmid (+HERF1) (A) or the B-lineage leukemic cell line MPC11, parental MEL cells treated with the differentiation-inducing agent DMSO, and MEL cells transfected with a tetracycline (tet)-regulated sense or α-sense HERF1 cDNA (B).
DISCUSSION
We have identified a novel gene encoding a hematopoietic specific RING finger-containing protein, termed HERF1, whose expression is markedly reduced in the absence of AML1. The expression of HERF1 during embryogenesis coincided with the initiation of definitive erythropoiesis and, in leukemic cell lines from adult mice, was restricted to the erythroid lineage, increasing over 30-fold during terminal differentiation. Importantly, we have demonstrated that inhibition of HERF1 expression blocked terminal erythroid differentiation, whereas its overexpression in erythroid cells induced β-major globin expression and morphologic maturation. Taken together, these results suggest that HERF1 plays an important role in the development of mature erythroid cells.
The HERF1 protein contains an N-terminal tripartite RBCC domain and a C-terminal rfp region. The RING finger domain, identified in well over 80 different proteins to date, defines a gene family that encodes proteins with widely varying functions (reviewed in references 2 and 34). These proteins include integral components of peroxisomes, transcriptional coregulators, modulators of the signaling capacity of the tumor necrosis factor receptor, members of the polycomb group of homeotic gene repressors, regulators of the cell cycle, proto-oncogenes, and tumor suppressor genes. The RING finger domain is characterized by a cysteine-rich motif of the general structure C3HC4 and binds two atoms of zinc, with each zinc atom ligated tetrahedrally by either four cysteines or three cysteines and a histidine. Although similar in structure to other zinc-binding motifs, the RING finger does not appear to mediate DNA binding but alternatively mediates protein-protein interactions that are critical to the function of these proteins.
Although our data do not address the mechanism through which HERF1 mediates its biologic activity, several possibilities are suggested from a comparison to the known activities of a number of closely related family members. The best-characterized members of this subfamily are proteins that contain the tripartite RBCC domain but lack the rfp motif; they include TIF1α (19), TIF1β (10, 18, 26), and PML (7, 16). Each member of this subfamily is a nuclear protein that functions as a critical component of intracellular regulatory pathways, often as an integral element of multisubunit protein complexes. Recent studies suggest that these RBCC domain-containing proteins either directly or indirectly regulate gene transcription. For example, TIF1α functions as a ligand-dependent coactivator for members of the retinoic acid family of transcription factors (19), whereas TIF1β functions as a transcriptional repressor in conjunction with members of the Krüppel family of transcription factors (10, 18, 26). PML functions as a nuclear protein in distinct subnuclear organelles referred to as PML nuclear bodies or PML oncogenic domains (4, 8, 9, 46). Expression of PML is essential for retinoic acid-mediated signaling during normal myeloid cell differentiation (44) and also has a direct growth suppressive function possibly due to its ability to induce apoptosis (13, 32, 45). Thus, the presence of the RBCC domain in HERF1 suggests that this gene product functions as part of a pathway that controls erythroid differentiation.
In addition to the RBCC domain, HERF1 contains a C-terminal 170-amino-acid rfp domain. The presence of this domain may provide further insight into the potential mechanism of action of HERF1. In addition to AFP and RFP, whose functions remain unknown, several other RBCC proteins contain an rfp domain: MID1, which is mutated in Opitz syndrome, an inherited human multiorgan disorder primarily affecting midline structures (31); three Xenopus nuclear proteins (xnf7, XL43, and XL75), which are involved in early development (3, 30); and the amphibian PwA33 protein, which binds to nascent transcripts on lampbrush chromosome loops in oocytes (1). The rfp domain is also found associated with an immunoglobulin domain in butyrophilin, a secreted protein found in milk (13). Although the biologic functions of each of the RBCC and rfp domain-containing proteins remain to be defined, studies on butyrophilin suggest that the rfp domain functions as a protein-binding domain, interacting with regulatory ligands. Thus, the rfp domain in HERF1 may function as a regulatory domain that controls the intrinsic activity of the protein. Taken together, the data from studies of other RBCC and rfp domain-containing proteins suggest that HERF1 functions as an integral component of a multisubunit protein complex that is required for the maturation of erythroid cells. Although we can only speculate on the function of this complex, likely possibilities include a direct role in the regulation of transcriptional signaling pathways and a mechanistic role in the morphologic changes that occur during terminal maturation of erythroid cells such as nuclear condensation and enucleation. Ultimately, determining the mechanistic function of HERF1 will require a direct assessment of the biologic consequences that result from its loss during murine development, as well as the identification of its subcellular location and the proteins with which it interacts.
Although HERF1 was cloned as a potential downstream target of AML1/CBFβ, our data do not address whether it is a direct transcriptional target or an element far downstream of this transcriptional cascade. Since the loss of AML1 results in a complete absence of definitive hematopoietic cells, HERF1 may have been differentially expressed in this system simply because of the absence of definitive erythroid cells. Supporting this notion are the observations that in differentiating MEL cells the level of AML1 decreases while HERF1 levels increase (11a). However, determining whether AML1 plays an essential role in the basal expression of HERF1 will require the characterization of the HERF1 promoter/enhancer regulatory sequences.
In summary, we have identified a novel RBCC and rfp domain-containing gene, HERF1, which appears to regulate critical steps required for the normal maturation of erythroid cells. Little is presently known about the regulatory pathways involved in the terminal maturation of cells of the erythroid lineage. Elucidation of the biochemical mechanism through which HERF1 functions should provide important insights into this process.
ACKNOWLEDGMENTS
We thank Shouli Yang, Noel Lenny, and Zhongling Cai for excellent technical assistance and A. Thomas Look, Gerard Grosveld, Gerard Zambetti, and John Cleveland for helpful discussions and critical reading of the manuscript.
This work was supported by National Institutes of Health (NIH) grant P01 CA71907-03, NIH Cancer Center CORE grant CA-21765, and the American Lebanese Syrian Associated Charities (ALSAC), St. Jude Children’s Research Hospital.
REFERENCES
- 1.Bellini M, Lacroix J-C, Gall J G. A putative zinc-binding protein on lampbrush chromosome loops. EMBO J. 1993;12:107–114. doi: 10.1002/j.1460-2075.1993.tb05636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borden K L B, Freemont P S. The RING finger domain: a recent example of a sequence-structure family. Curr Opin Struct Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 3.Borden K L B, Martin S R, O’Reilly N J, Lally J M, Reddy B A, Etkin L D, Freemont P S. Characterisation of a novel cysteine/histidine-rich metal binding domain from Xenopus nuclear factor XNF7. FEBS Lett. 1993;335:255–260. doi: 10.1016/0014-5793(93)80741-c. [DOI] [PubMed] [Google Scholar]
- 4.Chang K S, Fan Y H, Andreeff M, Liu J, Mu Z M. The PML gene encodes a phosphoprotein associated with the nuclear matrix. Blood. 1995;85:3646–3653. [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Chu T W, Capossela A, Coleman R, Goei V L, Nallur G, Gruen J R. Cloning of a new “finger” protein gene (ZNF173) within the class I region of the human MHC. Genomics. 1995;29:229–239. doi: 10.1006/geno.1995.1236. [DOI] [PubMed] [Google Scholar]
- 7.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 8.Downing J R, Head D R, Raimondi S C, Carroll A J, Curcio-Brint A M, Motroni T A, Hulshof M G, Pullen D J, Domer P H. The der(11)-encoded MLL/AF-4 fusion transcript is consistently detected in t(4;11)(q21;q23)-containing acute lymphoblastic leukemia. Blood. 1994;83:330–335. [PubMed] [Google Scholar]
- 9.Dyck J A, Maul G G, Miller W H, Chen J D, Kakizuka A, Evans R M. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 10.Friedman J R, Fredericks W J, Jensen D E, Speicher D W, Rauscher III F J, Neilson E G. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- 11.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a.Harada, H., Y. Harada, and J. Downing. Unpublished data.
- 12.Henry J, Ribouchon M-T, Depetris D, Mattei M-G, Offer C, Tazi-Ahnini R, Pontarotti P. Cloning, structural analysis, and mapping of the B30 and B7 multigenic families to the major histocompatibility complex (MHC) and other chromosomal regions. Immunogenetics. 1997;46:383–395. doi: 10.1007/s002510050292. [DOI] [PubMed] [Google Scholar]
- 13.Henry J, Ribouchon M-T, Offer C, Pontarotti P. B30.2-like domain proteins: a growing family. Biochem Biophys Res Commun. 1997;235:162–165. doi: 10.1006/bbrc.1997.6751. [DOI] [PubMed] [Google Scholar]
- 14.Hubank M, Schatz D G. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994;22:5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isomura T, Tamiya-Koizumi K, Suzuki M, Yoshida S, Taniguchi M, Matsuyama M, Ishigaki T, Sakuma S, Takahashi M. RFP is a DNA binding protein associated with the nuclear matrix. Nucleic Acids Res. 1992;20:5305–5310. doi: 10.1093/nar/20.20.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kakizuka A, Miller W H J, Umesono K, Warrell R P J, Frankel S R, Murty V V, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 17.Keller G, Kennedy M, Papayannoupoulou T, Wiles M V. Hematopoietic commitment during embryonic stem cell differentiation in culture. Mol Cell Biol. 1993;13:473–486. doi: 10.1128/mcb.13.1.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S S, Chen Y M, O’Leary E, Witzgall R, Vidal M, Bonvetre J V. A novel member of the RING finger family, KRIP-1, associates with the KRAB-A transcriptional repressor domain of zinc finger proteins. Proc Natl Acad Sci USA. 1996;93:15299–15304. doi: 10.1073/pnas.93.26.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Douarin B, Zechel C, Garnier J-M, Lutz Y, Tora L, Pierrat B, Heery D, Hinrich G, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieschke G J. CSF-deficient mice—what have they taught us? Ciba Found Symp. 1997;204:60–74. doi: 10.1002/9780470515280.ch5. [DOI] [PubMed] [Google Scholar]
- 21.Lieschke G J, Grail D, Hodgson G, Metcalf D, Stanley E, Cheers C, Fowler K J, Basu S, Zhan Y F, Dunn A R. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 1994;84:1737–1746. [PubMed] [Google Scholar]
- 22.Lieschke G J, Stanley E, Grail D, Hodgson G, Sinickas V, Gall J A, Sinclair R A, Dunn A R. Mice lacking both macrophage- and granulocyte-macrophage colony-stimulating factor have macrophages and coexistent osteoporosis and severe lung disease. Blood. 1994;84:27–35. [PubMed] [Google Scholar]
- 23.Lo C F, Pisegna S, Diverio D. The AML1 gene: a transcription factor involved in the pathogenesis of myeloid and lymphoid leukemias. Haematologica. 1997;82:364–370. [PubMed] [Google Scholar]
- 24.Lovering R, Hanson I M, Borden K L, Martin S, O’Reilly N J, Evan G I, Rahman D, Pappin D J, Trowsdale J, Freemont P S. Identification and preliminary characterization of a protein motif related to the zinc finger. Proc Natl Acad Sci USA. 1993;90:2112–2116. doi: 10.1073/pnas.90.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miki T, Fleming T P, Crescenzi M, Molloy C J, Blam S B, Reynolds S H, Aaronson S A. Development of a highly efficient expression cDNA cloning system: application to oncogene isolation. Proc Natl Acad Sci USA. 1991;88:5167–5171. doi: 10.1073/pnas.88.12.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moosmann P, Georgiev O, Le Douarin B, Bourquin J P, Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okuda T, Cai Z, Yang S, Lenny N, Lyu C, van Deursen J A, Harada H, Downing J R. Expression of a knocked-in AML1-ETO leukemia gene inhibits the establishment of normal definitive hematopoiesis and directly generates dysplastic hematopoietic progenitors. Blood. 1998;91:3134–3143. [PubMed] [Google Scholar]
- 28.Okuda T, Cleveland J L, Downing J R. PCTAIRE-1 and PCTAIRE-3, two members of a novel cdc2/CDC28-related protein kinase gene family. Oncogene. 1992;7:2249–2258. [PubMed] [Google Scholar]
- 29.Okuda T, van Deursen J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 30.Perrin K, Lacroix J-C. XL43 and XL75: two novel RING finger-containing genes expressed during oogenesis and embryogenesis in Xenopus laevis. Gene. 1998;210:127–134. doi: 10.1016/s0378-1119(98)00052-3. [DOI] [PubMed] [Google Scholar]
- 31.Quaderi N A, Schweiger S, Gaudenz K, Franco B, Rugarli E I, Berger W, Feldman G J, Volta M, Andolfi G, Gilgenkrantz S, Marion R W, Hennekam R C M, Opitz J M, Muenke M, Ropers H H, Ballabio A. Opitz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat Genet. 1997;17:285–291. doi: 10.1038/ng1197-285. [DOI] [PubMed] [Google Scholar]
- 32.Quignon F, De Bels F, Koken M, Feunteun J, Ameisen J C, de The H. PML induces a novel caspase-independent death process. Nat Genet. 1998;20:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- 33.Sasaki K, Yagi H, Bronson R T, Tominaga K, Matsunashi T, Deguchi K, Tani Y, Kishimoto T, Komori T. Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor beta. Proc Natl Acad Sci USA. 1996;93:12359–12363. doi: 10.1073/pnas.93.22.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saurin A J, Borden K L, Boddy M N, Freemont P S. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 35.Seymour J F, Lieschke G J, Grail D, Quilici C, Hodgson G, Dunn A R. Mice lacking both granulocyte colony-stimulating factor (CSF) and granulocyte-macrophage CSF have impaired reproductive capacity, perturbed neonatal granulopoiesis, lung disease, amyloidosis, and reduced long-term survival. Blood. 1997;90:3037–3049. [PubMed] [Google Scholar]
- 36.Sheldon M, Rice D S, D’Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell B W, Cooper J A, Goldowitz D, Curran T. Scrambler and yotari disrupt the disabled gene and produce a reeler-like phenotype in mice. Nature. 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- 37.Shockett P, Difilippantonio M, Hellman N, Schatz D G. A modified tetracycline-regulated system provides autoregulatory, inducible gene expression in cultured cells and transgenic mice. Proc Natl Acad Sci USA. 1995;92:6522–6526. doi: 10.1073/pnas.92.14.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 39.Speck N A, Terryl S. A new transcription factor family associated with human leukemias. Crit Rev Eukaryot Gene Expr. 1995;5:337–364. doi: 10.1615/critreveukargeneexpr.v5.i3-4.60. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi M, Cooper G M. ret transforming gene encodes a fusion protein homologous to tyrosine kinases. Mol Cell Biol. 1987;7:1378–1385. doi: 10.1128/mcb.7.4.1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka M, Levy J, Terada M, Breslow R, Rifkind R A, Marks P A. Induction of erythroid differentiation in murine virus infected erythroleukemia cells by highly polar compounds. Proc Natl Acad Sci USA. 1975;72:1003–1006. doi: 10.1073/pnas.72.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Q, Stacy T, Miller J D, Lewis A F, Gu T L, Huang X, Bushweller J H, Bories J C, Alt F W, Ryan G, Liu P P, Wynshaw-Boris A, Binder M, Marin-Padilla M, Sharpe A H, Speck N A. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell. 1996;87:697–708. doi: 10.1016/s0092-8674(00)81389-6. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z G, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi P P. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 45.Wang Z G, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi P P. PML is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- 46.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 47.Yergeau D A, Hetherington C J, Wang Q, Zhang P, Sharpe A H, Binder M, Marin-Padilla M, Tenen D G, Speck N A, Zhang D E. Embryonic lethality and impairment of haematopoiesis in mice heterozygous for an AML1-ETO fusion gene. Nat Genet. 1997;15:303–306. doi: 10.1038/ng0397-303. [DOI] [PubMed] [Google Scholar]