From July 22–23, 3 local COVID-19 cases were reported in Dalian City, Liaoning Province, China. All 3 patients reported that they did not leave Dalian 14 days before the onset of disease and had no COVID-19 case contact history and no foreign personnel contact history. Epidemiological investigation, medical isolation, and nucleic acid detection was immediately carried out in Dalian, and 12 asymptomatic infections were detected in close contacts of Patient 1. Because asymptomatic infections made up a large proportion of total infections, the outbreak was likely observed in the beginning stages. Most newly confirmed cases that had been detected were those that had been diagnosed as asymptomatic infections but had onset of symptoms during quarantine.
Throat swab samples were taken from COVID-19 patients and asymptomatic infections. Full length genomic sequences were acquired from the first four COVID-19 cases and found to have lower Ct values. The 4 SARS-CoV-2 genomes were completely identical and showed nucleotide similarity of 99.95% when compared to a reference strain (GenBank No. NC_045512). According to the phylogenetic tree based on the full-length genome of COVID-19 virus, all 4 genome sequences belonged to the L-Lineage European Branch 1. Using the latest classification principle (1), the Dalian strain could be defined as a new branch B.1.1.34 (Figure 1). This further confirmed that this outbreak in Dalian may have been caused by the introduction of an infectious source as it was found to be different from the virus that was prevalent in Wuhan in December 2019 that belonged to the S(A)-lineage and was likely not related to the continuous transmission of that virus.
Figure 1.
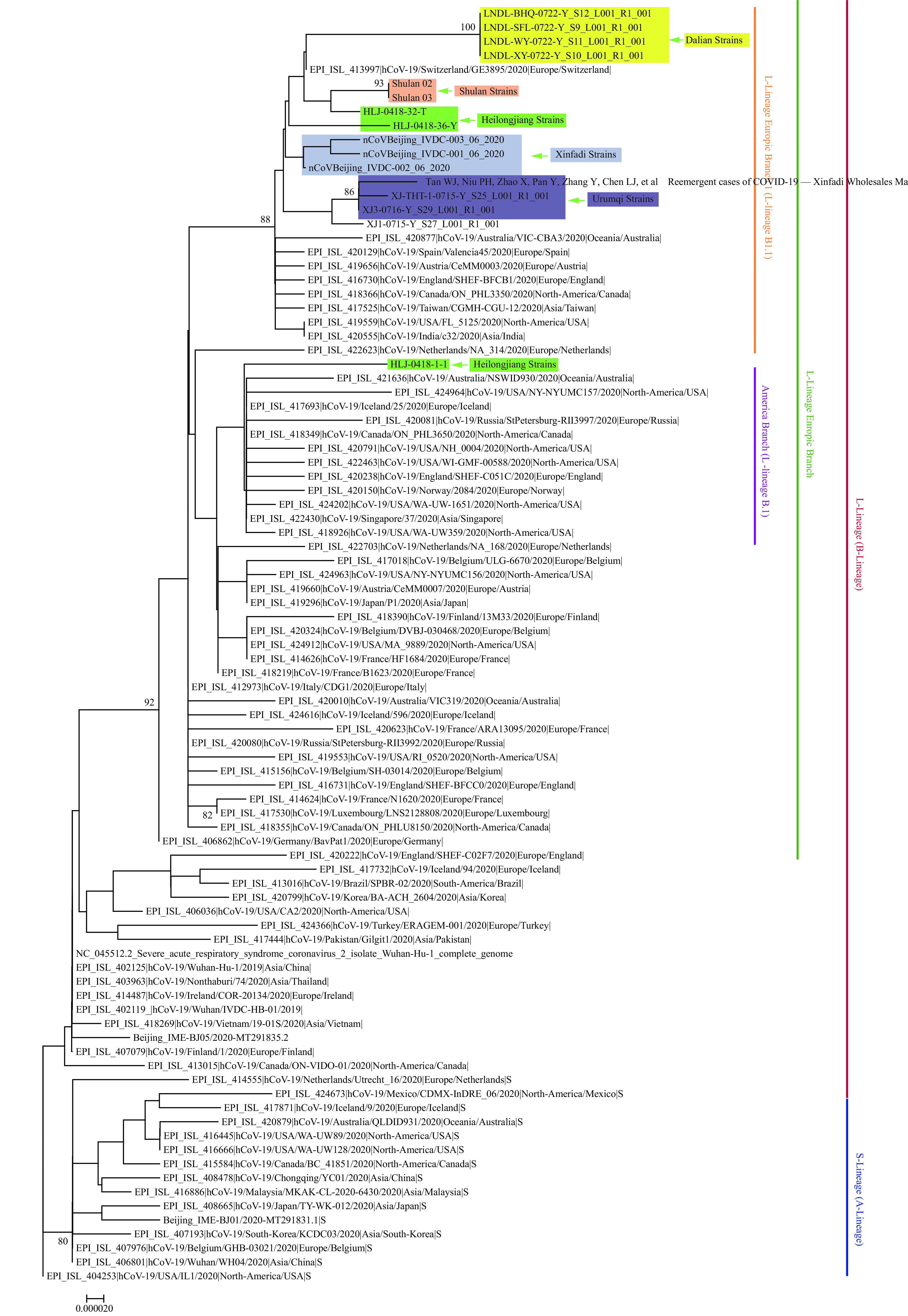
Phylogenetic tree based on the full-length genome sequences of the COVID-19 virus. The genomes of the COVID-19 virus from Dalian were highlighted in shades of yellow. The genomes of the reference COVID-19 virus from Wuhan (December 2019) were highlighted in shades of grey. The recent reemergence of COVID-19 virus in Beijing Xinfadi and Urumqi were highlighted in shades of green and blue, respectively, and the recent reemergence of COVID-19 virus in northeastern China (Shulan City and Heilongjiang Province) that was associated with imported cases was highlighted in shades of brown and ochre red, respectively. S(A)- or L(B)-lineage of the COVID-19 virus were marked and colored on the right.
Compared with the reference strain (GenBank No. NC_045512), all the four genome sequences shared the same substitutions at nt241 (C→T), nt3037 (C→T), nt14408 (C→T), nt23403 (A→G), nt28881 (G→A), nt28882 (G→A), and nt28883 (G→C), among which nt14408, nt23403, and nt28881-nt28883 were nonsynonymous substitutions in the ORF 1ab gene, S gene, and N gene, respectively. All 7 nucleotide substitutions were characteristic nucleotide substitutions of European Branch 1. All 4 genome sequences also shared another 7 unique nucleotide substitutions, nt2091 (C→T), nt5128 (A→G), nt8360 (A→G), nt13860 (C→T), nt19839 (T→C), nt19999 (G→T), and nt28905 (C→T), which were the characteristic nucleotide substitutions of COVID-19 virus in Dalian.
No other COVID-19 virus sequences with nucleotide substitutions at these 7 characteristic sites was found in the published databases at home and abroad, suggesting that the outbreak was caused by a new branch B.1.1.34 of L-Lineage European Branch 1, which was imported from abroad and likely from Europe. The complete genome sequence analysis of the COVID-19 virus in Dalian further confirmed that the source of the epidemic was not a new crossover event from a natural host or intermediate host. According to the dynamic variation rule of the COVID-19 viral genome and the characteristics of nucleotide substitutions in the genome, it was preliminarily judged that associations between this outbreak in Dalian and those in Beijing Xinfadi (2), Shulan (3), and Heilongjiang (4) were excluded
With the continuous emergence of next generation sequencing platforms and the rapid development of bioinformatics analytic technology, genomics has entered the field of epidemiological research and played an important role. Genomic epidemiology is a research method combining epidemiology with genomic information to evaluate the epidemiological significance of genomic information on infectious diseases. Cases of COVID-19 had not been reported in Dalian for more than 100 days before the first reemergent case occurred on June 22, 2020. Judging from the timing and phylogenetic analysis, the virus was likely imported from outside. According to the genomic epidemiological analysis, there was no clear relationship between the outbreak in Dalian and that in Beijing, and there was no evidence to suggest a definite link between the imported cases from other countries and Dalian.
The Dalian outbreak was likely related to the processing of cold chain seafood products, especially imported contaminated products. Therefore, the surveillance of imported COVID-19 should be strengthened, especially the detection and monitoring of nucleic acids of COVID-19 virus in imported foods, and a scientific and routine mechanism for entry detection should be implemented.
Acknowledgements
This study was supported by the National Key Technology R&D Programs of China (Project No. 2017ZX10104001, 2018ZX10102001, 2018ZX10711001, 2018ZX10713002).
Contributor Information
Dayan Wang, Email: dayanwang@cnic.org.cn.
Wenqing Yao, Email: wenqingyao@sina.com.
References
- 1.Rambaut A, Holmes EC, O'Toole Á, Hill V, McCrone JT, Ruis C, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol 2020. http://dx.doi.org/10.1038/s41564-020-0770-5.
- 2.Tan WJ, Niu PH, Zhao X, Pan Y, Zhang Y, Chen LJ, et al Reemergent cases of COVID-19 — Xinfadi Wholesales Market, Beijing Municipality, China, June 11, 2020. China CDC Wkeely. 2020;2(27):502–4. doi: 10.46234/ccdcw2020.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen C, Zhao X, Wang DY, Li J, Wang A, Wu DL, et al The initial case of COVID-19 — Shulan City, Jilin Province, China, May 8, 2020. China CDC Wkeely. 2020;2(25):458–9. doi: 10.46234/ccdcw2020.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J, Zhang Y, Zhao X, Wang DY, Dai WP, Jiao GY, et al A reemergent case of COVID-19 — Harbin City, Heilongjiang Province, China, April 9, 2020. China CDC Wkeely. 2020;2(25):460–2. doi: 10.46234/ccdcw2020.127. [DOI] [PMC free article] [PubMed] [Google Scholar]


