Abstract
At a hospital in Halifax, Nova Scotia, Canada, three strains of Legionella pneumophila were detectable based on plasmid content, while the isolates collected at another hospital in Halifax had no plasmids. Genomic DNA was digested with BssHII, SalI, and SpeI and subjected to pulsed-field gel electrophoresis (PFGE). We found no relationship between plasmid profile and PFGE pattern.
Legionella pneumophila serogroup 1 has been present for up to 10 years in the potable water supply of five of six health care institutions in the city of Halifax, Nova Scotia, Canada (1). Nosocomial Legionnaires’ disease (LD) has occurred primarily at two of these hospitals (1, 4). At hospital B, all of the isolates from both patients and water samples have been L. pneumophila serogroup 1, which lacks plasmids (1). At hospital A, most of the isolates have contained one of the following plasmids: a 23-MDa plasmid (type II), 72- and 96-MDa plasmids (type III), and a 100-MDa plasmid (type VI). A few isolates had a 70-MDa plasmid (type IV), an 89-MDa plasmid (type V), and 58- and 103-MDa plasmids (type VII) (5). The potable water at both of these hospitals remained positive for Legionella from 1987 until the present for hospital A and until its demolition in 1996 for hospital B (where plasmidless isolates were obtained).
Our objectives in this study were to determine if the genomic DNA of the plasmidless isolates was identical to that of the plasmid-containing isolates, to determine the relatedness of the Legionella population at each institution, and to determine the genomic stability of isolates obtained by longitudinal sampling from the potable water supply.
L. pneumophila serogroup 1 strains which had been isolated and typed as previously described (5) were recovered from storage. These isolates were selected from our culture collection so as to be representative of the isolates collected from 1987 to 1992. Thirty of the isolates examined were from hospital A (plasmid-containing isolates), and 13 were from hospital B (plasmidless isolates). In addition, five isolates obtained from patients with community-acquired LD were examined to determine the degree of relatedness of these strains to the hospital strains.
Portions of the cultures grown after a 48-h incubation of isolates on buffered charcoal-yeast extract agar (BYCE) were suspended in 0.5 ml of TE buffer (0.5 M Tris-HCl [pH 8.0], 0.02 M EDTA). After pelleting and resuspension of the cells in 25 μl of TE buffer, plasmid DNA was extracted from the cells by a modified alkaline sodium dodecyl sulfate procedure (5). The contents of the extracts were determined by electrophoresis with vertical 0.75% agarose gels followed by ethidium bromide staining. Strains were classified as having no detectable plasmids (plasmid type 0), a 23-MDa plasmid (plasmid type II), 96- and 72-MDa plasmids (type III), and a 100-MDa plasmid.
Growth from a single plate was used to prepare high-molecular-weight DNA in agarose plugs for each strain (2). Agarose plugs were digested overnight with BssHII, SalI, or SpeI as recommended by the manufacturer (Boehringer Mannheim, Laval, Quebec, Canada). Electrophoresis was performed in 1% agarose gels for 12 h with a 5-s pulse and then for 12 h with a 10-s pulse with a contour-clamped homogeneous electronic field system (CHEF-DRIII; Bio-Rad Laboratories, Mississauga, Ontario, Canada). Pulsed-field gels were run with 5-kb-incremental molecular size standards ranging in size from 4.9 to 120 kb (Bio-Rad Laboratories) in every fifth lane in order to ensure accurate sizing of the bands. Gels were stained with ethidium bromide and digitized under UV illumination. Restriction fragment patterns were analyzed with Bio Image whole band analyzer software (Bio Image, Ann Arbor, Mich.) running on a SPARC 10 workstation (SUN Microsystems, Mountain View, Calif.). A dendrogram was generated for each group of samples in an enzyme category by the unweighted pair groups method with arithmetic mean. This method defines the intercluster distance as the average of all the pairwise distances for members of the two clusters (7).
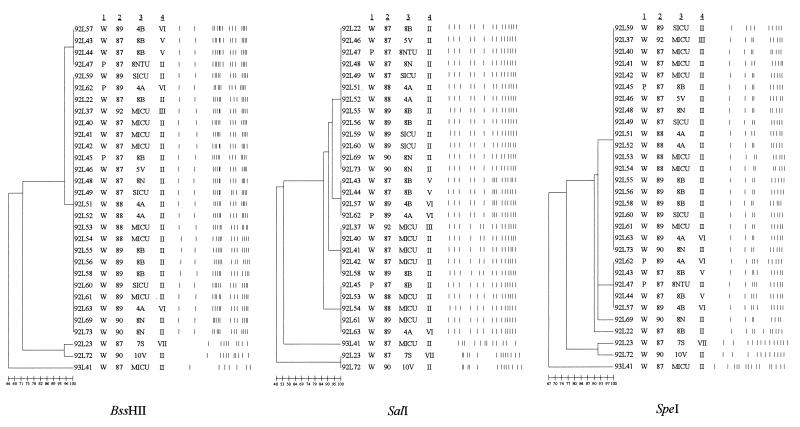
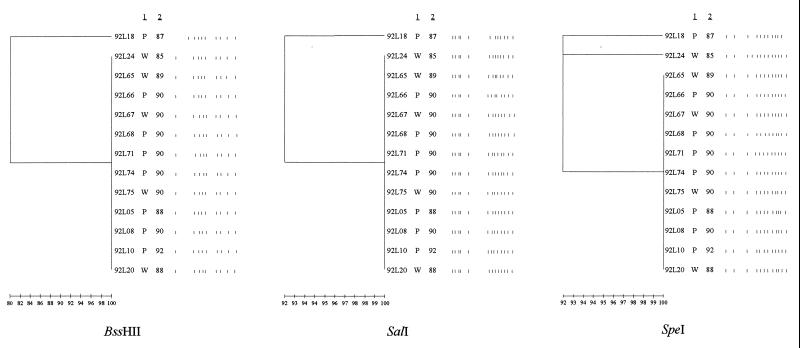
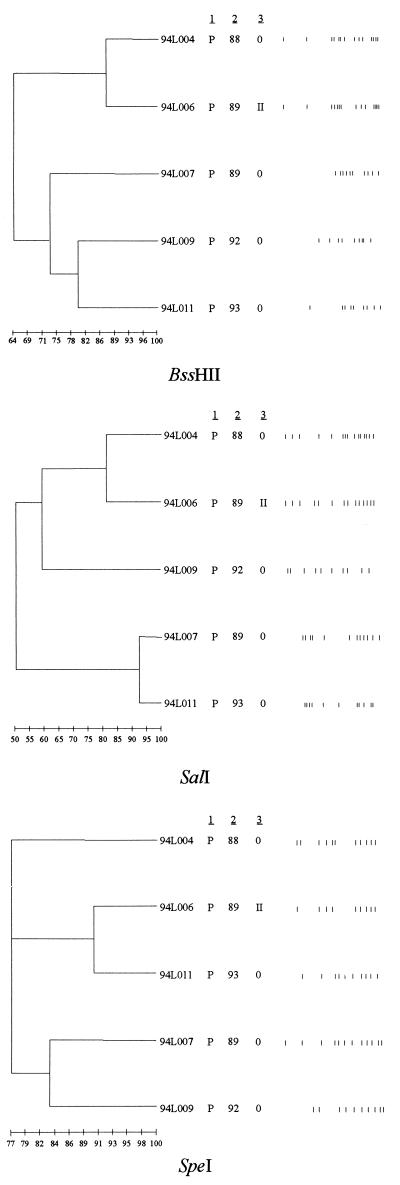
Thirty L. pneumophila serogroup 1 isolates recovered over 6 years from patients and potable water at hospital A fell into four clusters, with a few unrelated isolates remaining, when digested with SalI (Fig. 1). DNA from 23 plasmid type II-carrying isolates fell into three clusters consisting of 13, 4, and 4 isolates, with 2 single isolates remaining; each cluster had a different pulsed-field gel electrophoresis (PFGE) pattern (Fig. 1). Type II plasmid-containing strains had PFGE patterns identical to those of isolates with type III and type VI plasmids. An isolate with a type III plasmid profile recovered at hospital A was 100% related to the plasmidless isolates at hospital B (data not shown). Eleven of the 13 isolates from patients and potable water at hospital B were identical when digested with all three enzymes, and 12 of the 13 isolates produced identical profiles following digestion with BssHII and SalI (Fig. 2). These isolates had been collected over a period of 7 years. The results of digestion of five isolates from patients with community-acquired LD are shown in Fig. 3. In contrast to the isolates recovered at the two hospitals (Fig. 1 and 2), none of the isolates are identical.
FIG. 1.
PFGE patterns (after digestion with BssHII, SalI, or SpeI) of 30 isolates of L. pneumophila serogroup 1 recovered from the potable water supply or patients at hospital A during the period from 1987 to 1992. A dendrogram for patterns obtained with each enzyme is shown. The pedigree of each isolate is given. The designations to the left of column 1 refer to the laboratory identification of each isolate. Column 1 lists the source of the isolate (W, water; P, patient), column 2 gives the year the isolate was obtained, column 3 gives the location within the hospital (8NTU, 8 North Transplant Unit; SICU, surgical intensive care unit; MICU, medical intensive care unit) from which the isolate was recovered, and column 4 gives the plasmid type of each isolate. The vertical lines to the right of column 4 represent the digest pattern of each isolate, i.e., the PFGE pattern. The bar at the bottom of each dendrogram indicates the degree of genetic relatedness assessed by a distance matrix method.
FIG. 2.
PFGE patterns (after digestion with BssHII, SalI, or SpeI) of 13 isolates of L. pneumophila serogroup 1 recovered from the potable water supply or patients at hospital B during the period from 1987 to 1992. A dendrogram for patterns obtained with each enzyme is shown. The pedigree of each isolate is shown. Column 1 gives the source of the isolate (W, water; P, patient). Column 2 gives the year the isolate was obtained.
FIG. 3.
PFGE patterns (after digestion with BssHII, SalI, or Spe1) of five isolates of L. pneumophila serogroup 1 obtained from patients with community-acquired LD. A dendrogram for patterns obtained with each enzyme is shown. The pedigree of each isolate as shown on the figure. Column 1 lists the source of the isolate (P, patient). Column 2 gives the year each isolate was recovered. Column 3 is the plasmid type (0, no plasmid). The bar at the bottom of the figure indicates the degree of genetic relatedness.
This study shows that the strains of L. pneumophila recovered from the potable water of two Halifax hospitals have been remarkably stable in terms of genomic DNA over a period of several years. The plasmidless strains were more homogenous than the plasmid-containing strains and indeed, 12 of the 13 were identical despite isolation periods that spanned 7 years.
The plasmid-containing isolates from hospital A fell into two to four clusters that were 86 to 96% related when digested with all three enzymes. These clusters accounted for 27 of the 30 isolates examined. When the 21 plasmid type II-carrying isolates were examined they fell into three clusters of 10, 4, and 4 isolates and 3 single isolates with different PFGE patterns (Fig. 1). Furthermore, type II plasmid-carrying strains shared PFGE patterns with plasmid-containing isolates of type III, type II, and type VI. A type III plasmid-containing isolate from hospital A was 100% identical to the isolates with no plasmids from hospital B. This suggests that there is no correlation between PFGE pattern and plasmid profile.
We previously purified type II plasmid, cut it with BssHII, and found that two fragments (of 5 and 32.6 kb) were generated (3). The size limit for resolution under PFGE conditions is approximately 50 kb. Thus, for type II plasmid-containing isolates only chromosomal DNA was examined. Purified 100-MDa plasmid (type VI) was digested with SalI, and five fragments, of 43, 23, 15.5, 11.0, and 6.3 kb, were generated (6). This plasmid was not cut with BssHII.
REFERENCES
- 1.Bezanson G, Burbridge S, Haldane D, Yoell C, Marrie T J. Diverse populations of Legionella pneumophila present in the water of geographically clustered institutions served by the same water reservoir. J Clin Microbiol. 1992;30:570–576. doi: 10.1128/jcm.30.3.570-576.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson W M, Bernard K, Marrie T J, Tyler S D. Discriminatory genomic fingerprinting of Legionella pneumophila by pulsed-field electrophoresis. J Clin Microbiol. 1994;32:2620–2621. doi: 10.1128/jcm.32.10.2620-2621.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mansfield A D, Marrie T J, Bezanson G P. Characterization and cloning of a 37.6-kb plasmid carried by Legionella pneumophila recovered from patients and hospital water over a 12 year period. Can J Microbiol. 1997;43:193–197. doi: 10.1139/m97-025. [DOI] [PubMed] [Google Scholar]
- 4.Marrie T J, MacDonald S, Clark K, Haldane D. Nosocomial Legionnaires’ disease: lessons from a four year prospective study. Am J Infect Control. 1991;19:79–85. doi: 10.1016/0196-6553(91)90043-c. [DOI] [PubMed] [Google Scholar]
- 5.Marrie T J, Haldane D, Bezanson G, Peppard R. Each water outlet is a unique ecological niche for Legionella pneumophila. Epidemiol Infect. 1992;108:261–270. doi: 10.1017/s0950268800049736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marrie, T. J. Unpublished observations.
- 7.Weir B S. Genetic data analysis II. Sunderland, Mass: Sinauer Associates; 1996. p. 345. [Google Scholar]