Abstract
Background
Long noncoding RNAs (lncRNAs) are important regulators in tumor progression. However, their biological functions and underlying mechanisms in hypoxia adaptation remain largely unclear.
Results
Here, we established a correlation between a Chr3q29-derived lncRNA gene and tongue squamous carcinoma (TSCC) by genome-wide analyses. Using RACE, we determined that two novel variants of this lncRNA gene are generated in TSCC, namely LINC00887_TSCC_short (887S) and LINC00887_TSCC_long (887L). RNA-sequencing in 887S or 887L loss-of-function cells identified their common downstream target as Carbonic Anhydrase IX (CA9), a gene known to be upregulated by hypoxia during tumor progression. Mechanistically, our results showed that the hypoxia-augmented 887S and constitutively expressed 887L functioned in opposite directions on tumor progression through the common target CA9. Upon normoxia, 887S and 887L interacted. Upon hypoxia, the two variants were separated. Each RNA recognized and bound to their responsive DNA cis-acting elements on CA9 promoter: 887L activated CA9’s transcription through recruiting HIF1α, while 887S suppressed CA9 through DNMT1-mediated DNA methylation.
Conclusions
We provided hypoxia-permitted functions of two antagonistic lncRNA variants to fine control the hypoxia adaptation through CA9.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12915-021-01112-2.
Keywords: Long noncoding RNA, Hypoxia, Carbonic anhydrase 9, Cancer, Hypoxia-induced factor, DNA methylation, Alternative promoter, Alternative splicing
Background
Oxygen is of fundamental importance for human cells. Reduced level of oxygen or hypoxia, generated by external and internal changes, may cause pathological consequences regionally and globally, or even life-threatening conditions. Tumor hypoxia, resulted from the rapid growth of solid tumor cells, is one of the most commonly observed features of tumor microenvironment [1–6]. To survive and gain growth advantage over normal cells, tumor cells developed a series of adaptive mechanisms under hypoxia. Hypoxia-induced factor alpha (HIFα) plays a key role in tumor hypoxia adaptation [2, 4, 6]. When the oxygen level falls, accumulated HIFα proteins will translocate to the nucleus, bind to the HIF response element (HRE) located in the 5’ regulatory regions of their downstream target genes and activate a broad range of gene transcription events [2, 4]. The HIFα downstream genes include carbonic anhydrase 9 (CA9), and many other oncoproteins [2, 4, 7–10]. Despite the positive influences on tumor progression, it has been proposed that prolonged activation of HIF may be potentially deleterious to the tumor cells per se and chronic reduction of HIFα proteins is to avoid “maladaptive” or “potentially deleterious effects” resulted from over-activation of the HIF pathway [11, 12]. Therefore, the fine control of HIFα downstream genes is critical for tumor cell progression.
Long noncoding RNAs (lncRNAs) represent a group of regulatory RNAs that are larger than 200 nucleotides. According to the GENCODE database (GRch38, version 32), more than 57,935 human lncRNAs have been identified and this number is still climbing due to the development of advanced sequencing techniques [13, 14]. Increasing evidence has shown that lncRNAs are important regulators in almost all the physical and pathological events, including tumor progression [15–18]. However, relative limited investigations have revealed the functions and molecular mechanisms of lncRNAs in tumor hypoxia [19, 20]. It remains to be a challenge to unveil the comprehensive picture of lncRNAs’ engagement in hypoxia adaptation.
In the present study, we identified that the activity of CA9, a well-known HIF1α target gene and oncoprotein, is regulated through a pair of functionally antagonistic lncRNA variants in tongue squamous cell carcinoma (TSCC). Our results shed light on an lncRNA-directed mechanism to fine controlled oncoprotein CA9’s expression during hypoxia adaptation.
Results
Identification of LINC00887 as a hypoxia-associated lncRNA in TSCC
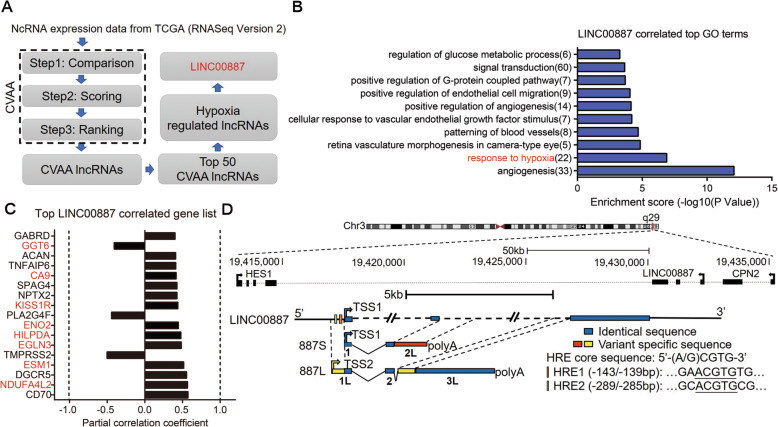
In order to identify the tumor-associated lncRNAs that might play roles in hypoxia adaptation, we first identified lncRNAs that were dysregulated in cancer patients by cross-value association analysis (CVAA), a normalization-free and nonparametric method that we recently developed [21], and searched those resulted lncRNAs whose expression levels were regulated by hypoxia (Fig. 1A). Briefly, we applied CVAA to the RNA-seq data-sets of 5540 patient samples retrieved from The Cancer Genome Atlas (TCGA), including 4907 tumor specimens from 13 types of primary solid tumor and 631 normal specimens from matching tissue type. The identified lncRNAs that were dysregulated in pan-cancer will be further referred as CVAA lncRNAs in this work. Next, to identify the hypoxia-regulated lncRNAs from the resulted top 50 CVAA lncRNAs, we searched the published reports on PubMed and found 6 CVAA lncRNAs whose expression levels were regulated by hypoxia treatment (Supplementary table 1, [22–31]). Among which, the involvement of LINC00887 in tumor hypoxia is currently unknown despite that it was initially reported to be induced by hypoxia in renal epithelial cell [24]. We then focused on LINC00887 and performed Partial Correlation Analysis (PCA) and Gene Ontology (GO) analysis using RNA-seq data-sets retrieved from TCGA. PCA and GO analysis showed that the function of LINC00887 was highly enriched in “response to hypoxia” (Fig. 1B). In addition, PCA analysis established significant correlation between LINC00887 and 17 genes (Fig. 1C). All of these genes have been shown to play important roles in tumor progression [32–47]. Interestingly, 8 of these 17 genes have been identified as hypoxia-associated genes, including CA9, gamma-glutamyltransferase 6 (GGT6), KISS1 receptor (KISS1R), enolase 2 (ENO2), hypoxia-inducible lipid droplet-associated protein (HILPDA), EGL-9 family hypoxia inducible factor 3 (EGLN3), endothelial cell specific molecule 1 (ESM1), and NDUFA4 mitochondrial complex-associated like 2 (NDUFA4L2) (Fig. 1C). Besides GGT6, that is negatively correlated with LINC00887, the other 7 hypoxia-associated genes showed to be positively correlated with LINC00887.
Fig. 1.
Identification of a hypoxia-regulated CVAA lncRNA gene, LINC00887, which generated two lncRNA variants 887S and 887L in TSCC. A A schematic view of experimental outline for screening hypoxia-regulated CVAA lncRNAs. B Top 10 biological processes that are related to LINC00887 and significantly enriched by Gene Ontology (GO) analysis. C LINC00887-correlated genes according to Partially Correlation Analysis (PCA) analysis (P<0.05). Hypoxia-associated genes are marked in red. D A schematic comparation of 887S, 887L, and LINC00887
We also performed a series of examinations and built a strong link between LINC00887 and TSCC: LINC00887 is derived from chromosome 3q29, a genomic region that is highly associated with squamous cancer by integrative large-scale analysis [48, 49]. Consistently, by using a set of cultured tumor cells for the expression levels of the top 10 CVAA lncRNAs, we found that LINC00887 was the only lncRNA to be upregulated in TSCC cells (including TSCC9, TSCC15, and TSCC25) (Fig. S1A). In addition, by analyzing the RNA-seq data from TCGA retrieved TSCC patients, we found that the expression level of LINC00887 was dramatically increased in TSCC and the upregulation of LINC00887 was positively correlated with poor patient survival rate (Fig. S1B, C). All together, our data suggested that LINC00887 might be a critical regulator to hypoxia response in TSCC.
LINC00887 generates two variants, 887S and 887L, in TSCC
Although LINC00887, also named as linc-ATP13A4-8 and HEIRCC [24, 50], has been reported as a long intergenic noncoding RNA derived from the antisense strand of chromosome (Chr.) 3q29 (Fig. 1D, upper panel), the genomic structure of this lncRNA has not yet been characterized. By performing 5′ and 3′ rapid-amplification of cDNA ends (RACE) assays, we found two previously unidentified variants of LINC00887 in TSCC (Fig. S2A-D). Due to their different length and the initial identification from TSCC, we designated these two variants as LINC00887_TSCC_Short (887S, 1593nt) and LINC00887_TSCC_Long (887L, 4202nt). For simplicity, we used “887S” and “887L” in the following manuscript and all datasets. The genome structure difference between 887S and 887L suggests that these two variants are generated by combined effects of alternative promoter (AP) selection and alternative splicing (AS). Compared with LINC00887 shown in NCBI database (Fig. 1D and S2E), 887S has an extra 1118bp at the 3′ of exon2 (named as exon2L for 887S) (Fig. 1D and S2E), and 887L has an extra 405bp at the 5′ of exon1 (named as exon1L for 887L) and an extra 776bp at the 5′ of exon3 (named as exon3L for 887L) (Fig. 1D and S2E).
Next, we conducted RT-PCR assay by using a set of primers that specifically recognized 887S, 887L, or LINC00887 based on NCBI database (Fig. S2C, D). The resulted PCR products were gel-purified and analyzed by Sanger sequencing. These results showed that the existence of 887S and 887L, but not the previously annotated LINC00887 in NCBI, in all three tested TSCC cell lines (TSCC9, TSCC15, and TSCC25; Fig. S2D). Using specific probes for 887S and 887L, our Northern blot results showed that the two investigated RNAs were relatively abundant in both TSCC15 and TSCC25 cell lines. In contrast, both RNAs were either undetectable or showed much weak signal in 293T cells (Fig. S2F-H). LINC00887 has additional 17 and 10 variants shown in LNCipedia and Ensemble databases, respectively. To test the existence of these variants, we designed specific primers for their common exons. In contrast to 887S and 887L in the same experiment, our RT-qPCR results showed that none of these variants, except LINC00887 variants 15 and 16, was expressed in TSCC (Fig. S2F, I). Although variants 15 and 16 were expressed in TSCC, the net expression levels of both variants were only about 1/3 of the expression level of 887S or 887L (Fig. S2F, I). These results indicated that 887S and 887L were two novel and predominate variants in TSCC cells. We further confirmed the existence of 887S and 887L in TSCC patient samples (Fig. S3C). Protein-coding potential analysis by CPC (coding potential calculator, http://cpc.cbi.pku.edu.cn) showed that both 887S and 887L had no typical protein-coding ORF (Fig. S4A). All together, our results suggested that the previously reported lncRNA LINC00887 can generate two lncRNA variants, 887S and 887L, in TSCC.
LncRNA-887S and LncRNA-887L exhibit differential response to hypoxia
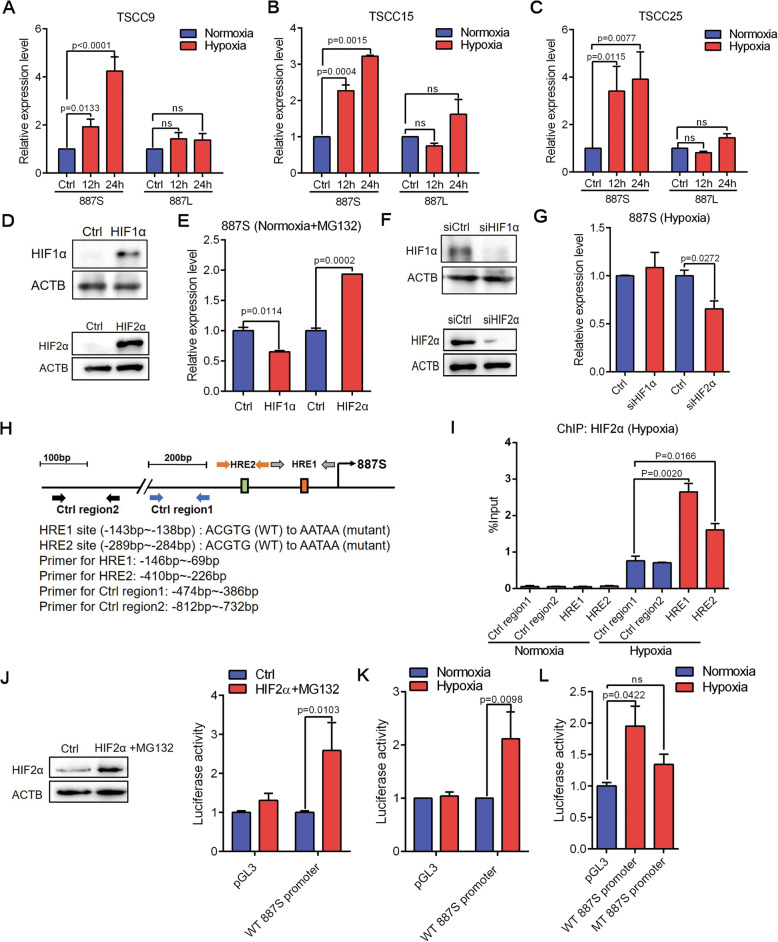
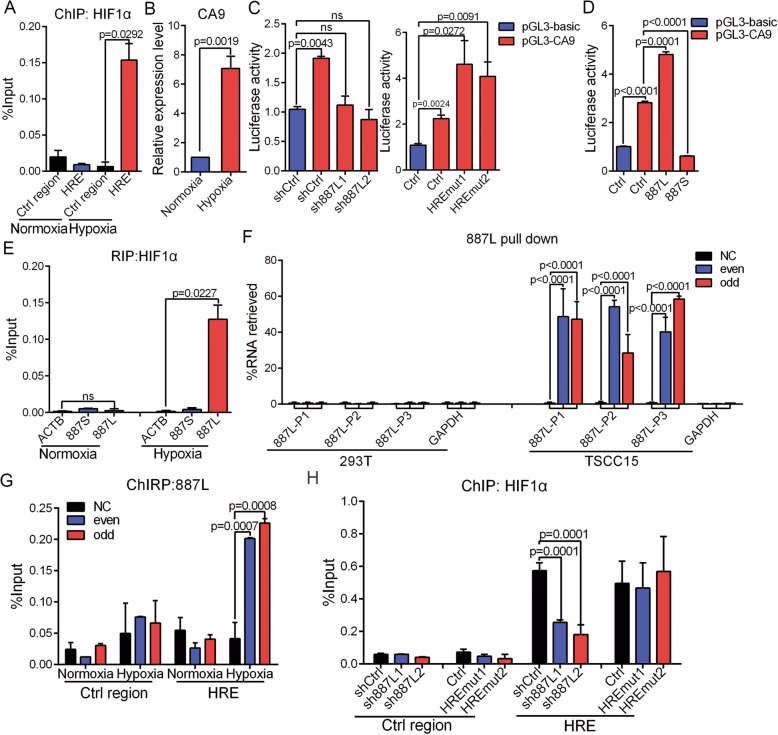
The differential transcriptional start sites (TSSs) of 887S and 887L resulted in an inclusion of two putative HRE sites exclusively located in the promoter of 887S (Fig. 1D, lower panel). In contrast, the promoter of 887L which has an extra 5′ sequences at the first exon did not contain the HRE sites (Fig. 1D, lower panel). HRE (5′-A/GCGTG-3′) is the cis-acting DNA element receiving the regulation of HIF family members [51]. Consistently, 887S, instead of 887L, could be significantly induced when the TSCC9, TSCC15, and TSCC25 cells were cultured under 1% O2 (Fig. 2A–C, S3A, B). HIF1α and HIF2α are accumulated under hypoxia or nomoxia but in the presence of MG132 to inhibit the proteasome degradation pathway [51, 52]. Our further experiments showed that the expression level of 887S was positively regulated by HIF2α, instead of HIF1α (Fig. 2D–G). Moreover, upon hypoxia, HIF2α was recruited to both HRE sites of 887S by ChIP assay (Fig. 2H, I). The luciferase reporter assay showed that either hypoxia or overexpression of HIF2α (in the presence of MG132) could enhance the activity of the wild type 887S promoter (Fig. 2J, K), which was abolished when we mutated the two HRE sites from 5′-CGTG-3′ to 5′-ATAA-3′ (Fig. 2L). These data suggested that the expression level of lncRNA-887L is hypoxia-independent, and lncRNA-887S is a HIF2α-inducible transcript upon hypoxia.
Fig. 2.
887S and 887L are differently modulated upon hypoxia. A–C Relative expression levels of 887S and 887L in TSCC9 (A), TSCC15 (B), and TSCC25 (C) in the presence of normoxia or hypoxia (n=3). D Overexpression efficiency of HIF1α (upper panel) or HIF2α (lower panel) with treatment of MG132 (10μM, 8h) in TSCC15 cells. (n=3). E Relative expression levels of 887S in response to HIF1α or HIF2α overexpression in the presence of MG132 (n=3). F Knockdown efficiency of HIF1α siRNA (upper panel) or HIF2α siRNA (lower panel) under hypoxia (n=3). G Relative expression levels of 887S in the presence of HIF1α or HIF2α siRNAs under hypoxia (n=3). H A schematic view of the 5’ regulatory regions of 887S with the relative location of HREs, wide-type, and mutant sequences of HREs, and the primers used in ChIP assays. I ChIP assay on the indicated regions of 887S promoter upon normoxia and hypoxia (n=3). J Overexpression efficiency of HIF2α with treatment of MG132 (10μM, 8h) in TSCC15 cells (left panel) and activity change of the pGL3 control (pGL3) or wild type (WT) 887S promoter after modulation of HIF2α (right panel). (n=4). K Activity change of the pGL3 control (pGL3) or wild type (WT) 887S promoter after modulation of oxygen levels (n=4). L Activity change of the WT or mutant (MT) 887S promoter in the presence of normoxia and hypoxia (n=4). Data are shown as means ± SEMs. P values are calculated using Student’s t test
LncRNA-887S and LncRNA-887L play antagonistic roles in TSCC through regulating CA9 in opposite directions
In order to investigate the subcellular location of lncRNA-887S and 887L, we performed RNA fluorescence in situ hybridization (FISH) and fractionationing assay in TSCC15 cells. Our results revealed a predominant nuclear expression pattern of both transcripts under both normoxia and hypoxia (Fig. S4B-D).
To further investigate the two lncRNAs’ biological functions, we took advantage of the identification that 887S was induced by hypoxia via HRE sites and established CRISPR-Cas9-mediated HRE mutant TSCC15 lines (HREmut1 and HREmut2) that could effectively abolish the induction of 887S upon hypoxia, without affecting the expression level of 887S under normoxia (Fig. S5A). Although the HRE mutation is also located within the first exon of 887L, the expression levels of 887L were not altered detected by multiple primer sets targeting the unique 887L exons (Fig. S5B, C). Therefore, the HRE mutant lines allowed us to investigate the role of 887S transcript exclusively for the hypoxic condition. For 887L, we designed two independent small hairpin RNAs (shRNAs) targeting the exon3L of 887L (sh887L1 and sh887L2) (Fig. S5D).
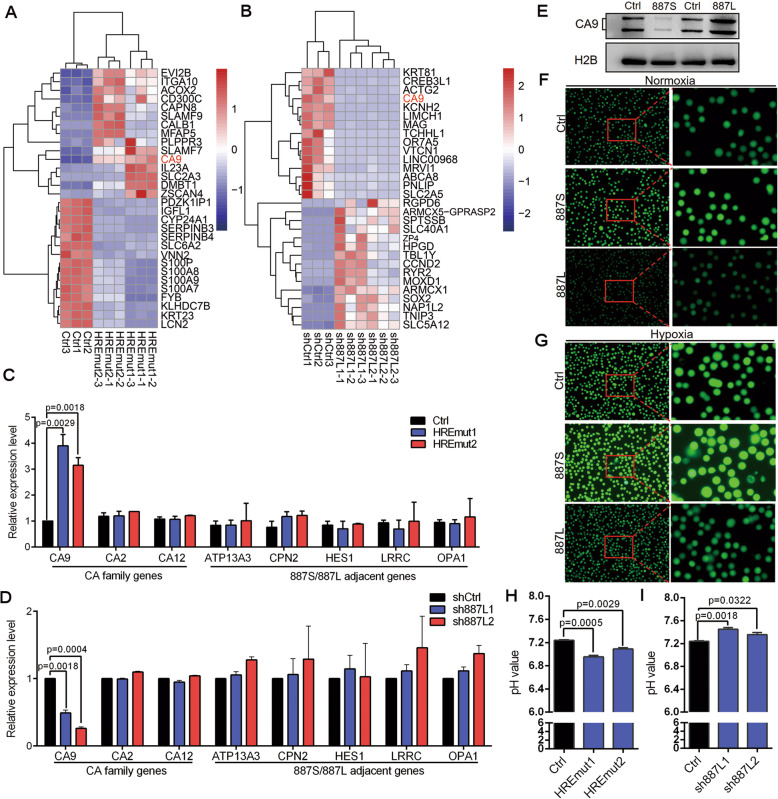
The nuclear expression suggested that 887S and 887L may function in transcriptional regulation. In order to identify the 887S and 887L regulated downstream genes, we performed RNA-sequencing (RNA-seq) in the 887S knockout cells (HREmut1 and HREmut2), 887L knockdown cells (sh887L1 and sh887L2), and their corresponding control cells. When compared with the corresponding controls, 318 or 855 genes were identified as common dysregulated genes for HREmut1 and HREmut2, or sh887L1 and sh887L2, respectively. Interestingly, among the top regulated genes, CA9, one of the 8 hypoxia-associated genes we identified by PCA (Fig. 1C), was shown to be a common downstream target of 887S and 887L in an opposite regulatory manner (Fig. 3A, B).
Fig. 3.
887S and 887L antagonistically regulate the expression and activity of CA9. A The top 15 upregulated or downregulated genes identified by RNA-seq in the two indicated 887S knockout cell lines (HREmut1 and HREmut2) and the corresponding control lines under hypoxic condition. B The top 15 upregulated or downregulated genes identified by RNA-seq in the two indicated 887L knockdown cell lines (sh887L1 and sh887L2) and the corresponding control lines under normoxic condition. C, D Relative expression levels of the indicated CA family genes and 887S/887L adjacent genes (ATP13A3, CPN2, HES1, LRRC, OPA1) in 887S knockout cells under hypoxic condition (C) or 887L knockdown cells under normoxic condition (D) (n=3). E The protein levels of CA9 in 887S or 887L overexpression cells (n=3). Related to Figure S5E, F. F, G Images of the fluorescence intensity detected by BCECF-AM assay in the indicated 887S or 887L overexpression cells in the presence of normoxia (F) and hypoxia (G). H Extracellular pH values of the indicated control (Ctrl) and 887S knockout cells under hypoxic condition. I Extracellular pH values of the indicated control (Ctrl) and 887L knockdown cells under hypoxic condition. Data are shown as means ± SEMs. P values are calculated using Student’s t test
CA9, a well-known hypoxia-induced gene, plays critical roles in promoting tumor progression and hypoxia adaptation [7]. Our analysis showed that the upregulation of CA9 was correlated with the poor survival rate in TCGA retrieved TSCC patients (Fig. S6A, B). The effect of CA9 on TSCC cells was also tested by colony formation and transwell assays. As shown in Fig. S6C-G, we found that knockdown of CA9 resulted in a remarkable decrease of tumor cell progression in TSCC15 cells. These results indicated that CA9 acted as a tumor promoter in TSCC. After validating the in-house RNA-seq data by RT-qPCR assay (Fig. 3C, D, and S7A), we performed western blot assay and showed that the protein level of CA9 was also reversely regulated by 887S and 887L (Fig. 3E, and S5E, F). CA9 is a zinc metalloenzyme that facilitates tumor acidification through hydration of carbon dioxide [8, 53]. To answer the question whether the function of CA9 as a pH modulator was altered as a result of the dysregulated mRNA and protein levels in either 887S or 887L modulated cells, we performed the BCECF-AM assay. As expected, our results showed that 887S decreased the value of intracellular pH, while 887L increased the value of intracellular pH (Figs. 3F, G, and S8). In consent with the BCECF-AM assay, the extracellular pH value was decreased when 887S was knocked out but increased when 887L was knocked down (Fig. 3H, I).
CA9 belongs to carbonic anhydrase family, which is composed of 16 members in human. Distinct from CA9, the other CA members are predominately expressed in normal tissues [54, 55]. Although TSCC tumor samples and TSCC15 cells also express CA2 and CA12, our in-house RNA-seq data showed that the expression levels of CA2 and CA12 were not altered when the expression of 887S or 887L was modulated. These results were further validated by the RT-qPCR tests (Fig. 3C, D). In addition, the direct (HES1, CPN2, as shown in Fig. 1A) and far neighboring genes of 887S/887L on Chr.3 (ATP13A, LRRC, and OPA1) were not regulated by the two lncRNAs (Fig. 3C, D). Together, our results indicated that 887S and 887L exhibited a strong specific regulation on CA9 among carbonic anhydrase family in TSCC.
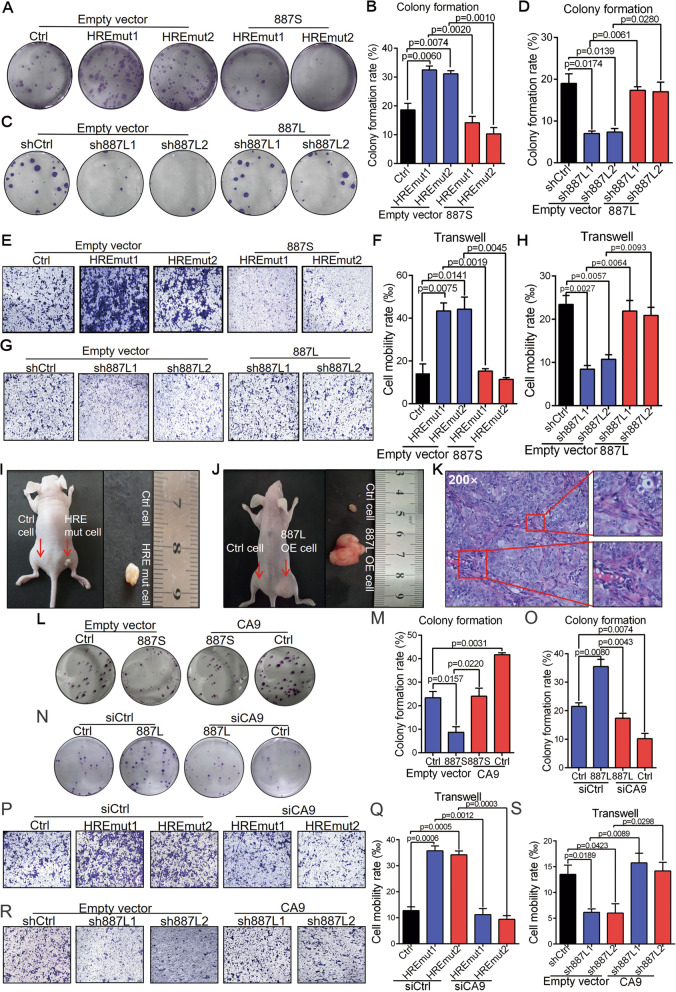
Next, we wanted to explore the biological roles of 887S and 887L on tumor progression under hypoxia. Knockout of 887S remarkably promoted tumor progression in TSCC15 cells observed by colony formation and transwell assays (Fig. 4A, B, E, F, and S5G). On the contrary, knockdown of 887L significantly inhibited tumor progression in TSCC15 cells (Fig. 4C, D, G, H, and S5H). To test whether the above identified roles were dependent on the two investigated lncRNAs, we overexpressed the 887S- or 887L-expressing plasmid in their corresponding knockout or knockdown cells and found that both overexpressions could significantly reverse the effects (Fig. 4A–H, and S5G, H). Consistently, modulating the expression levels of 887S and 887L dramatically affected TSCC growth in the opposite direction by xenograft experiments (Fig. 4I–K and S9).
Fig. 4.
887S and 887L drive TSCC progression in opposite directions through CA9 under hypoxia. A Representative images of colony formation assay in 887S knockout cells in the presence of control or 887S overexpressed plasmid. B Statistical analysis according to A (n=3). Related to Figure S5G. C Representative images of colony formation assay in 887L knockdown cells in the presence of control or 887L overexpressed plasmid. D Statistical analysis according to C (n=3). Related to Figure S5H. E Representative images of transwell assay in 887S knockout cells in the presence of control or 887S overexpressed plasmid. F Statistical analysis according to E (n=3). Related to Figure S5G. G Representative images of transwell assay in 887L knockdown cells in the presence of control or 887L overexpressed plasmid. H Statistical analysis according to G (n=3). Related to Figure S5H. I, J Representative images of xenografts from Balb/c (nu/nu) mice injected with 887S knockout (I) or 887L overexpressed (J) TSCC15 cells. The arrows denote tumors in situ. K HE staining image of tumor specimen dissected from the xenograft mice. The enlarged regions indicate the keratinized feature of TSCC15. L Representative images of colony formation assay in 887S overexpressed cells in the presence of control or CA9-overexpressed plasmid. M Statistical analysis according to L (n=3). Related to Figure S7B. N Representative images of colony formation assay in 887L overexpressed cells in the presence of control or siCA9. O Statistical analysis according to N (n=3). Related to Figure S7C. P Representative images of transwell assay in 887S knockout cells in the presence of control or siCA9. Q Statistical analysis according to P (n=3). R Representative images of transwell assay in 887L knockdown cells in the presence of control or CA9-overexpressed plasmid. S Statistical analysis according to R (n=3). Data are shown as means ± SEMs. P values are calculated using Student’s t test
Given that CA9 is a common downstream target of 887S and 887L, we sought to evaluate whether 887S and 887L regulated tumor cell progression via CA9. We introduced CA9-expression plasmid in 887S stable overexpression cells and found that the overexpression of CA9 completely blocked the effect caused by 887S (Fig. 4L, M, and S7B). Vice versa, knockdown of CA9 in 887S stable knockout cells also completely reversed the effects caused by 887S (Fig. 4P, Q). In addition, similar rescue effect was observed when we modulated the expression level of CA9 in 887L stably overexpression cells (Fig. 4N, O, and S7C) or 887L knockdown cells (Fig. 4R, S). To further validate the observed effects in 887S knockout cell lines, we also conducted ASO-mediated 887S knockdown assays (Fig. S10A). Our results showed that 887S ASO enhanced the promoter activity of CA9, increased the expression levels of CA9 mRNA and protein, and promoted TSCC15 cell proliferation and migration ability (Fig. S7E, F, and S10B-H). Together, our data suggested that, although 887S and 887L are both upregulated in TSCC, these two transcripts play antagonistic roles in TSCC through CA9.
LncRNA-887L is required for the HIF1α-induced activation of CA9 in TSCC
Previous work has reported that CA9 is transcriptionally activated by HIF1α in a broad spectrum of tumor cells [56–58]. In TSCC15 cells, we found that CA9 was also upregulated by HIF1α upon hypoxia when HIF1α was recruited to the HRE site of CA9 promoter (Fig. 5A, B). Furthermore, we constructed a luciferase reporter driven by CA9 promoter and performed luciferase assay. Our data showed that 887S and 887L could either inhibit or enhance the hypoxia-induced CA9 promoter activity, respectively (Fig. 5C, D), suggesting that both 887S and 887L regulated CA9 at the transcriptional level. Interestingly, our RNA immunoprecipitation (RIP) assay showed that only 887L interacted with HIF1α (Fig. 5E).
Fig. 5.
887L is required for the HIF1α-induced activation of CA9 under hypoxia. A HIF1α ChIP assay on the indicated HRE site and control region on CA9 promoter upon normoxia and hypoxia (n=3). B Relative expression levels of CA9 under normoxia and hypoxia (n=3). C Activity changes of CA9 promoter in the 887L knockdown cells or 887S knockout cells (n=3). D Activity change of CA9 promoter in the 887L overexpressed cells or 887S overexpressed cells (n=3). E HIF1α RIP assay showed the immunoprecipitation of the indicated β-actin (ACTB), 887S, and 887L RNAs upon normoxia and hypoxia (n=3). F Capture of 887L RNA by RNA pull-down assay in 293T cells, that do not express 887L, and TSCC15 cells. NC: non-probe control; even and odd: two separated pools of 887L probes containing either even or odd numbered probes based on their positions along the 887L sequence. 887L-P1, 887L-P2, and 887L-P3: primers for 887L detection; GAPDH: primers for GAPDH detection. (n=2). G ChIRP assay showed the association of the 887L RNA with the CA9’s HRE site under normoxia and hypoxia (n=2). H Recruitment of HIF1α to the control region and HRE site of CA9 promoter in 887S knockout and 887L knockdown cells (n=3). Data are shown as means ± SEMs. P values are calculated using Student’s t test
To test whether the 887L could be recruited to the HRE site of CA9, we synthesized antisense probes specifically targeting 887L and performed chromatin isolation by RNA pull-down (ChIRP) assays. The RT-qPCR following RNA pull-down was first performed to prove the specificity and efficiency of the 887L probes. Both the odd and even sets of 887L probes were shown to be highly effective in TSCC15 (Fig. 5F). In contrast, no PCR product was amplified using the control probes in TSCC15 cells or using 887L probes in the 293T cells that did not express 887L (Fig. 5F). These results indicated that the 887L probes were highly specific and effective. Then, we examined the 887L-associated chromatin and found that hypoxia induced a specific association of 887L RNA with the HRE site of CA9 (Fig. 5G). Moreover, the hypoxia-induced recruitment of HIF1α was found to be dramatically decreased when 887L was knocked down (Fig. 5H). In contrast, 887S knockouts showed no impact on the HIF1α recruitment (Fig. 5H). All together, our results indicated that 887L is required for the HIF1α-induced activation of CA9 in TSCC15 cells.
LncRNA-887S negatively regulates CA9 through DNA methyltransferase 1 (DNMT1)-mediated DNA methylation
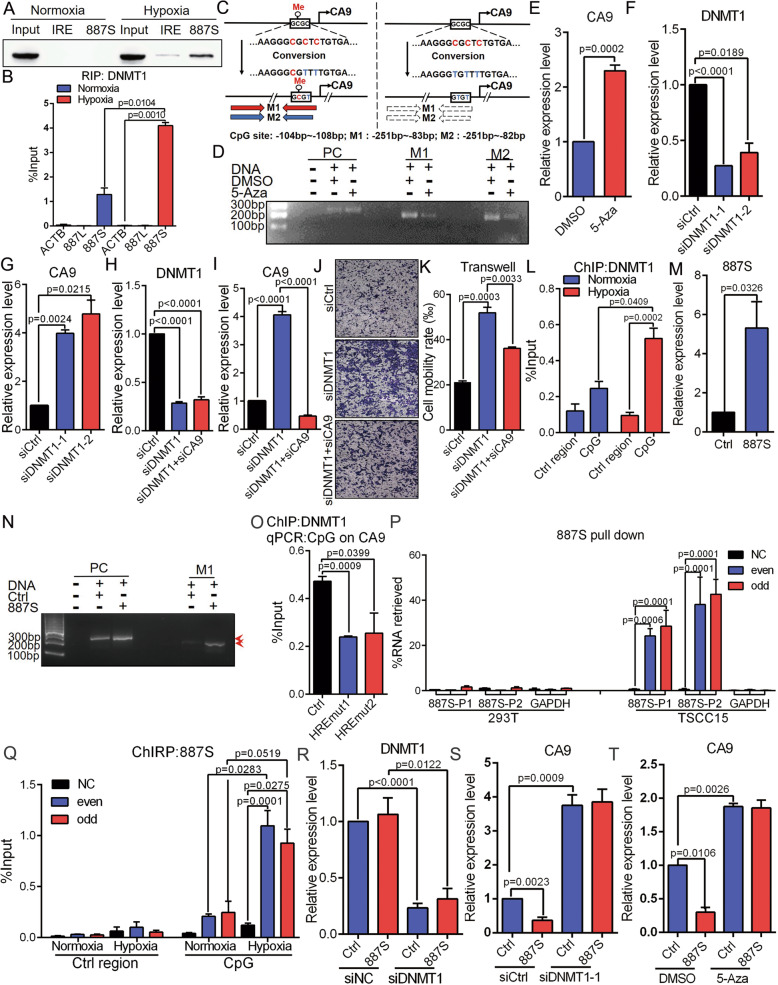
In Fig. 5E, H, we have showed that 887S negatively regulated CA9 in a HIF1α-independent manner. To further explore the molecular mechanism of 887S, we performed an RNA pull-down assay followed by mass spectrometry to identify the 887S-associated proteins. Among all the candidates, we focus on DNMT1 because a DNA methylation-involved regulation on CA9 has been recently suggested in a subset of tumor cells [59]. The interaction of 887S:DNMT1 was further confirmed by RNA immunoprecipitation (RIP) assay and RNA pull-down followed by western blot assay (Fig. 6A, B and S11). DNA methylation site was mapped to a CpG dinucleotide-containing region located at upstream of the TSS of CA9 with unknown mechanisms [59]. It has been known that DNMT1 regulates DNA methylation presumably in mammalian somatic cells, and its activity is sufficient to methylate CpG-poor regions [60–62]. These features make DNMT1 as a promising candidate of CA9 methylation.
Fig. 6.
887S inhibits CA9 expression through regulating DNMT1-mediated DNA methylation upon hypoxia. A DNMT1 was identified as 887S-associated protein in a hypoxia-dependent manner, by RNA pull down and Western blot. IRE: a control RNA provided by Pierce RNA 3′ End Desthiobiotinylation kit. Related to Figure S11. B DNMT1 RIP assay showed 887S, instead of 887L RNA was immunoprecipitated under hypoxia (n=3). C A schematic view of the process to detect methylation status of the CpG dinucleotides. M1 and M2: two primer sets to amplify the converted DNA sequence of CpG site on CA9 promoter. D Representative image of methylation status of the CpG site on CA9 promoter with or without 5-Aza treatment by the indicated PCR primer sets. PC: a primer set to detect the C to T conversion which is provided by the MSP kit. E Relative expression levels of CA9 in TSCC15 cells in the presence of carrier (DMSO) or 5-Aza (n=3). F, G Effects of the indicated DNMT1 siRNAs on DNMT1 (F) and CA9 (G) mRNA level (n=3). H, I Relative expression levels of DNMT1 (H) or CA9 (I) under the indicated treatments (n=3). J, K Transwell assay showed that knockdown of CA9 significantly inhibited the effect of DNMT1 knockdown in TSCC15 cells (n=3). L DNMT1 ChIP assay on the indicated CpG site and control region of CA9 upon normoxia and hypoxia (n=3). M Overexpression efficiency of 887S (n=3). N Representative image of methylation status of the CpG site on CA9 promoter detected by M1 in the control or 887S-overexpressed cells. O DNMT1 ChIP assay on the CpG site of CA9 promoter in 887S knockout cells under hypoxia (n=3). P Capture of 887S RNA by RNA pull-down assays in 293T cells, that do not express 887S, and TSCC15 cells. NC: non-probe control; even and odd: two separated pools of 887S probes containing either even or odd numbered probes based on their positions along the 887S sequence. 887S-P1 and 887S-P2: primers for 887S detection; GAPDH, primers for GAPDH detection. (n=2). Q ChIRP assay showed the association of the 887S RNA with the CA9’s CpG site under normoxia and hypoxia (n=2). R, S Relative expression level of DNMT1 (R) or CA9 (S) in control and 887S overexpressed TSCC15 in the presence of siDNMT1 (n=3). T Relative expression levels of CA9 in Ctrl and 887S overexpressed TSCC15 cells in the presence of carrier (DMSO) or 5-Aza (n=3). Data are shown as means ± SEMs. P values are calculated using Student’s t test
In order to investigate whether DNA methylation affects CA9’s expression in TSCC, we performed methylation specific PCR (MSP) assay and found a detectable DNA methylation status at the CpG site of CA9 promoter (Fig. 6C, D). Treatment of 5-Azacytidine (5-Aza), a specific inhibitor of DNA methylation, resulted in decreased DNA methylation of CA9 and a significant induction of CA9 mRNA level in TSCC15 cells (Fig. 6D, E).
Next, to further investigate whether DNMT1 is the yet-to-be found factor for CA9 methylation, we tested the effects of DNMT1 on CA9. By using two independent siRNAs specifically targeting DNMT1, we found that DNMT1 knockdown significantly increased the expression level of CA9 in TSCC15 cells (Fig. 6F, G). Moreover, knockdown of DNMT1 greatly enhanced the TSCC15 cells’ migration rate in a CA9-dependent manner (Fig. 6H–K). Consistently with these results, the ChIP assay showed a hypoxia-induced recruitment of DNMT1 specifically to the CpG dinucleotides containing region located on the CA9 promoter (Fig. 6L). The above results indicated that a DNMT1-mediated DNA methylation event occurs in the presence of hypoxia to negatively regulate the expression level of CA9.
To detect whether 887S regulates CA9 through DNMT1-meidiated DNA methylation, we first tested whether 887S would affect the DNA methylation and the recruitment of DNMT1 on CA9. As expected, we found DNA methylation of CA9 were dramatically increased upon 887S overexpression (Fig. 6M, N). The DNMT1 ChIP assay also indicated a positive answer because that the recruitment of DNMT1 to the CpG dinucleotides of CA9 promoter was abolished in 887S knockout cells (Fig. 6O). By using the similar ChIRP assay strategy as that of 887L, we found that 887S exhibited a hypoxia-enhanced interaction with the CpG site (Fig. 6P, Q). These results strongly suggested that the DNA methylation of CA9 and recruitment of DNMT1 to the CA9 promoter are dependent on the action of 887S.
Next, we wanted to investigate whether the 887S-mediated CA9 inhibition is through DNMT1-mediated DNA methylation. We treated the 887S-overexpressed TSCC15 cells with siDNMT1 or 5-Aza and found that 887S lost its inhibitory ability on the expression of CA9 when DNMT1 was knockdown or DNA methylation process was blocked (Fig. 6R–T). Collectively, our results suggested that the hypoxia-induced 887S mediates a DNMT1-dependent inhibitory effect on CA9 expression through DNA methylation in the presence of hypoxia.
The interaction between lncRNA-887S and 887L is regulated by the concentration of oxygen
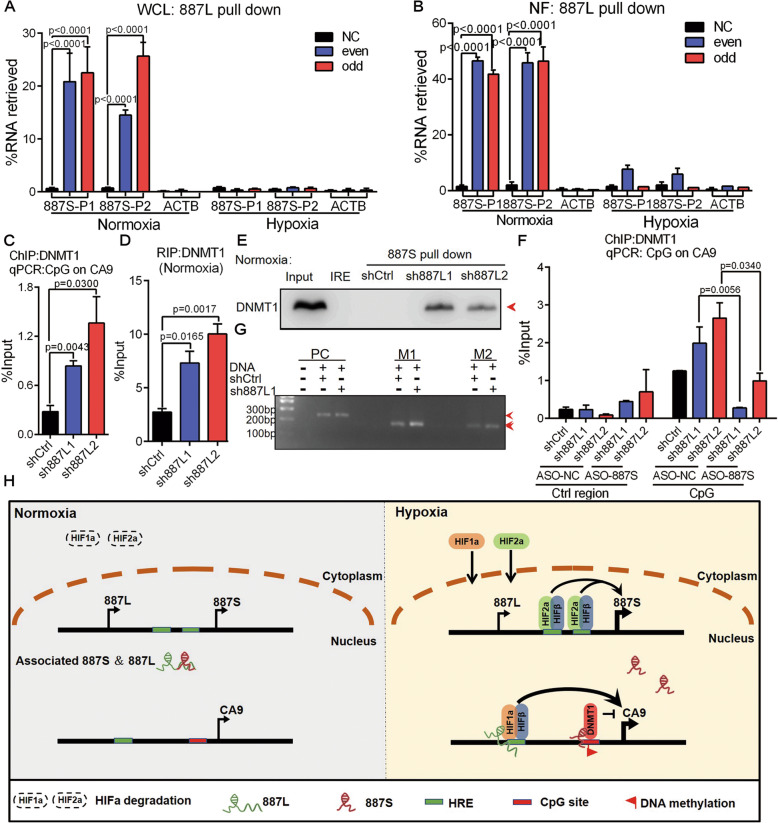
The observation that 887L-mediated HIF1α and 887S-mediated DNMT1 recruitments were both hypoxia-induced strongly suggested that dynamic change of oxygen level acts as a critical regulatory signal for 887S and 887L. It was interesting to further investigate the underlying mechanism of the hypoxia-induced regulation of 887S and 887L. The fractionationing assay showed that the majority of 887S and 887L were DNA-bound under hypoxia (Fig. S4B). Although some DNA-bound 887S and 887L could be detected under normoxia, the DNA-bound fractions for both RNAs had been dramatically decreased (Fig. S4B). By using the 887L antisense probes shown in Fig. 5F, G, we performed RNA antisense purification assay and RT-qPCR to investigate the potential interaction between 887S and 887L. These experiments were conducted using either whole cell lysates (Fig. 7A) or nuclear fractions (Fig. 7B). Our results detected an interaction between 887S and 887L exclusively under normoxia in both whole cell lysates and nuclear fractions (Fig. 7A, B). In addition, our results showed that 887S and 887L did not associate with their cis-acting elements under normoxia (Fig. 5G, and 6Q). Together, our results indicated that the interaction status between 887S and 887L is dynamically regulated by the concentration of oxygen. The interaction between 887S and 887L is likely to occur in the nucleoplasm.
Fig. 7.
887L interacts with 887S under normoxia and exhibits an inhibitory effect on 887S. A 887S was captured in the 887L-precipitated complex under normoxia, instead of hypoxia. 887S-P1 and 887S-P2: primer sets for 887S detection (n=2). WCL: Whole cell lysate. B 887S was captured in the 887L-precipitated complex under normoxia, instead of hypoxia. 887S-P1 and 887S-P2: primer sets for 887S detection (n=2). NF: Nuclear fraction. C DNMT1 ChIP assay showed the changes of DNMT1 recruitment to CA9’s CpG site in 887L knockdown cells under normoxia (n=3). D DNMT1 RIP assay showed the interaction of the DNMT1 and 887S in 887L knockdown cells under normoxia (n=3). E Representative image of RNA pull-down assay showed the association of DNMT1 and 887S in 887L knockdown cells under normoxia (n=3). IRE: a control RNA provided by Pierce RNA 3′ End Desthiobiotinylation kit. F Silencing of 887S RNA by antisense oligonucleotide (ASO) (ASO-887S) abolished the recruitment of DNMT1 to the CpG site of CA9 in 887L knockdown cells under normoxia, as determined by ChIP-qPCR assay (n=3). G Representative image of methylation status of the CpG sites on CA9 promoter in sh887L1 cells and control cells. PC: a primer set to detect the C to T conversion which is provided by the MSP kit. H A schematic illustration of the CA9 transcriptional regulation by interplay between 887S- and 887L-mediated pathways. Data are shown as means ± SEMs. P values are calculated using Student’s t test
In Fig. 5H, we have provided evidence that modulation of 887S expression level had no effect on HIF1α recruitment to CA9 promoter, suggesting that 887S is not an inhibitory factor to 887L-HIF1α axis. However, it did not exclude the possibility that 887L acted as a regulatory factor for 887S. To test our hypothesis, we knocked down 887L and performed a series of investigations. Our data showed that the recruitment of DNMT1 to the CA9 promoter was increased in both sh887L1 and sh887L2 cell lines (Fig. 7C). In addition, 887L knockdown dramatically increased the association of DNMT1:887S, observed by both DNMT1 RIP assay (Fig. 7D) and 887S RNA pull-down assay (Fig. 7E).
In order to investigate whether the observed DNMT1 recruitment in 887L knockdown lines shown in Fig. 7C was mediated through 887S, we knocked down 887S in the two sh887L lines and performed DNMT1 ChIP assay. Our data showed that DNMT1 recruitment was completely abolished after 887S knockdown (Fig. 7F). Moreover, we also observed a concomitant increase of DNA methylation in tested CpG region at CA9 promoter after 887L knockdown (Fig. 7G). All together, our data strongly indicated that the interaction between 887S and 887L is dependent on the oxygen concentration. In addition, 887L exhibits a one-directional inhibitory effect on 887S, which provides an indirect regulatory mechanism that explains the role of 887L on CA9 under normoxia (Fig. S12).
Discussion
In this work, we report the roles of two newly identified lncRNA variants in TSCC and the molecular mechanisms of how the activity of the oncoprotein CA9 is precisely maintained by these two antagonistic but interplaying lncRNA variants (Fig. 7H).
CA9 plays an essential role in hypoxia adaptation and oncogenic progression [8, 53, 63, 64]. However, the upstream regulatory mechanism of CA9 is largely unknown. In this work, the two investigated lncRNA variants, 887S and 887L, are both upregulated in TSCCs but function in opposite directions on CA9 transcription. Through recruiting HIF1α, 887L promotes CA9’s expression while 887S inhibits CA9 through DNMT1-DNA methylation under hypoxia.
CA9 is well known for being upregulated by hypoxia [56–58]. Surprisingly but intriguingly, both activation signals (887L-HIF1α) and repressive signals (887S-DNMT1-DNA methylation) to CA9 are observed under the same condition-hypoxia. Eukaryotic cells have developed sophisticated mechanisms to establish fine-controlled balance between activation and repression signals in a broad range of cellular events [65–68]. Chronic reduction of HIFα proteins has been observed in tumor cells after prolonged hypoxia treatment [11, 12]. It has been proposed that such chronic reduction of HIFα can protect tumor cells from unwanted deleterious consequences [11, 12]. We speculate that 887S-dependent inhibitory action is part of the fine control of CA9 expression to keep CA9 from being over-activated under hypoxia. As a result of this, the expression level of the inhibitory 887S should be proportional to that of 887L, the positive regulator of CA9. Supporting this, we consistently observed a synchronous expression pattern of 887S and 887L in the tumor cells (Fig. S3A, B) and the in-house collected TSCC patient specimens (Fig. S3C). Another interesting observation is that 887S is enhanced by HIF2α, instead of HIF1α upon hypoxia. It has been recently reported that HIF1α and HIF2α have unique and sometimes opposing target genes [52, 69]. Together, our results indicate that the expression level of CA9 is under fine and sophisticated control at the transcriptional level through an lncRNA-dependent manner.
Both HRE and DNA methylation sites are broadly distributed across human genome. It would be reasonable to speculate that the two investigated lncRNA variants may affect other targets through HIF1α and DNMT1 to reconcile the RNAs’ functions in tumor progression.
Enormous quantity in the human genome and high sensitivity to the environment change endow lncRNA transcripts as perspective regulators for human diseases [15–18, 70]. We [71] and other independent groups [72–74] have found that lncRNAs use AP, AS, and alternative polyadenylation (APA) as general strategies to further expand the genome diversity. Although several studies have been reported [75, 76], the biological significance of lncRNA variants generated by these upstream biogenesis signals are largely unknown. In this work, we found that the interaction between 887S and 887L, a pair of lncRNA variants, is dynamic and regulated by the oxygen concentration. In the presence of higher oxygen concentration, 887S and 887L interact. When the oxygen level falls, the two lncRNAs are separated from each other. Each individual transcript “talks” with their responsive DNA cis-acting elements on CA9. It would be of great interest to investigate how the dynamic interaction between these two RNAs is regulated in the future. LncRNAs are highly sensitive to their cellular environment [77]. It is very likely that certain hypoxia-triggered microenvironment alterations (e.g., pH) induce the signal-specific higher structural changes of either RNA variant or both to modulate their RNA-RNA interaction status, each RNA’s binding ability to the corresponding cis-acting element and thus their regulation on CA9 expression.
Growing clinical evidence has suggested that CA9 can serve as a prognostic biomarker and a therapeutic target [8, 53, 63, 64]. To date, about 20 CA inhibitors (CAIs) have been underwent clinical trials for anti-cancer therapies [78]. However, due to the similarity among CA family members in sequence, structure, and particularly within their active sites, it is difficult to design the isoform-specific CA inhibitors [54, 58, 79]. The specific effects of 887S and 887L on CA9 among CA family members indicate that targeting upstream molecules may offer a suitable alternative option for CA9-based anti-cancer drug design.
Together, our work provides a hypoxia-permitted “talk” between the two investigated lncRNA variants to their cis-acting elements and thus fulfill their direct regulation on CA9 transcription. Our findings bring up a novel lncRNA-mediated mechanism that lncRNAs act as environmental sensors and guide the appropriate cellular adaptation through precisely controlled transcription network.
Conclusion
In summary, we report the squamous cell carcinoma highly associated genomic locus Chr3q29 can generate a pair of alternative promoter-regulated lncRNA variants 887S and 887L. The two lncRNAs are differentially responded to hypoxia and oppositely control tumorigenesis through distinct but interplaying transcriptional regulatory axis on oncogene CA9. This integrated and coordinated collaboration between 887S and 887L results in a fine controlled expression level of CA9. Our work expands the current understanding on tumor hypoxia adaptation and provides a promising new therapeutic strategy in anti-cancer treatment.
Methods
Cell culture and treatment
All cell lines were purchased from the American Type Culture Collection (ATCC). The HEK 293T, Hela cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, USA). MCF-7, MDA-MB-231, SK-BR3, and HCT116 cells were cultured in RPMI1640 medium (Gibco, USA). TSCC9, TSCC15, TSCC25, and SH-SY5Y cells were cultured in Dulbecco’s modified Eagle’s medium/Nutrient Mixture F-12 (DMEM/F-12) medium (Sigma, USA), containing 10% fetal bovine serum (FBS, Gibco, USA) and 1% penicillin-streptomycin (WISENT Inc., CA). MCF-10A cells were cultured in DMEM/F-12 medium supplied with 5% horse serum (Gibco, USA), 10μg/ml insulin (Roche, USA), 20ng/ml EGF2 (Sigma-Aldrich, USA), 100ng/ml cholera toxin (Sigma, USA), 500ng/ml hydrocortisone (Melonepharma, CHINA), and 1% penicillin-streptomycin. All cells were cultured under humidified atmosphere of 5% CO2 at 37°C. For the 5-Azacytidine treatment, 1μM of 5-Azacytidine (5-Aza, MedChem Express) was added into culture medium when cell density was around 50%. After 48h treatment with 5-Aza, TSCC15 cells were collected and extracted for further analysis. For the hypoxia treatment, cells were cultured under 1% O2 at hypoxia station (Don Whitley Scientific:H35 hypoxystation, UK).
Xenograft mouse model
All animal studies were conducted with the approval from the Animal Research Ethics Committee of the University of Science and Technology of China (Approval number USTCACUC-1801020). Female BALB/C nude mice at the age of 5 to 6 weeks (Nanjing Biomedical Research Institute of Nanjing University) were used for model construction. Briefly, differently treated tumor cells were trypsinized and harvested, then 1x107 TSCC15 or 3x106 TSCC25 cells in serum-free medium containing 20% matrigel with a total volume of 0.2ml were injected subcutaneously to the inguina of the mice. Four weeks later, the mice were sacrificed by cervical dislocation and noticeable tumors were immediately excised. The tumor volumes were measured with a caliper and calculated using the equation V=0.5×L×W2, where V is the volume, L is the length (longest dimension), and W is the width (shortest dimension).
Clinical samples
TSCC specimens including paired paracancerous tissues and tumor samples were collected from the Department of Oral and Maxillofacial Surgery, Peking University School and Hospital of Stomatology. The research protocol was approved by the Ethics Committees for Human Experiments of Peking University School and Hospital of Stomatology. All participants signed and informed consent prior to sample collection (Approval number PKUSSIRB-2013009).
RNA-seq data resources and analysis
RNA expression data (RNA-seq) of The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov, RNA-seq Version 2) was downloaded. After excluding the cancer type with less than 10 normal tissue samples, we got 5540 samples in total, comprising 4907 primary solid tumor samples and 631 normal samples from 13 cancer types. The differentially expressed lncRNAs between tumor and normal samples were screened by performing cross-value association analysis (CVAA [21]). Briefly, the downloaded RNA-seq data sets were denoted as the matrix E with m rows (samples) and n columns (lncRNAs). Looping comparison was conducted as described before [21]. According to the log-linear model theory, Likelihood ratio test (LRT) value can statistically represent the significance score of genes which are differently expressed between samples [21]. And Logarithm 2 of odds ratio (LOD) values can indicate the overall change direction of a certain gene (upregulation: LOD > 0, downregulation: LOD < 0) [21]. Therefore, the lncRNAs running for looping comparison were scored and ranked according to LRT in descending order. The downregulated or upregulated lncRNAs were sorted by LOD. Next, to identify the hypoxia-regulated lncRNAs from the resulted top 50 CVAA lncRNAs, we searched the published reports on PubMed and found 6 CVAA lncRNAs whose expression levels were regulated by hypoxia treatment [22–31]. These 6 CVAA lncRNAs included CDKN2B-AS1, PVT1, HOTAIR, UCA1, MIR31HG, and LINC00887 and were listed as hypoxia-regulated CVAA lncRNAs in Supplementary Table 1.
Partial correlation analyses (PCA)
PCA was conducted as previously described [80]. Briefly, RNA-seq data-sets retrieved from TCGA was downloaded and then was mathematically calculated by the R (v 3.0.2) package “ggm” (v2.3) with the function “pcor” for given sets of TCGA expression data. The “pcor” function is to analyzed pair-wise gene expression correlation. With the cutoff of |cors| > 0.3, 634 genes showed to be LINC00887 co-expressed and the top correlated genes were listed in Fig. 1C.
Gene ontology (GO) analyses
The PCA generated LINC00887-correlated genes were applied to Database Annotation Visualization and Integrated Discovery (DAVID) (v6.8) (http://david.abcc.ncifcrf.gov/) for gene ontology (GO) enrichment analyses. The top 10 terms from the resulted significant GO terms (p < 0.01) were shown in Fig. 1B.
RNA isolation and reverse transcription (RT)—quantitative PCR (qPCR)
Total RNA was isolated using Trizol Reagent (Ambion, USA) according to the manufacturer’s instructions and treated with RNase-free DNase I (Thermo Scientific, USA). One Drop® OD-1000 Spectrophotometer (Nanjing Wuyi Corporation, CHINA) was used to measure RNA concentration and purity. Reverse-transcription was performed using HiScript II One Step RT-PCR Kit (Vazyme, CHINA) according to the manufacturer’s instructions. Reaction without transcriptase was performed as a no-RT control. Real-time PCR was performed using SYBR® Green Master Mix (Vazyme, CHINA) according to the manufacturer’s instructions on Light Cycle® 96 (Roche, USA). GAPDH RNA was used as internal control for Figure S1A, and 18S RNA was used as internal control for all the other RT-qPCRs. The primers used for the real-time PCR are listed in Supplementary Table S2. Fold changes were determined using the relative quantification 2-△△CT method.
Western blot
Cells were lysed in RIPA buffer (Vazyme, CHINA) for 45min on ice and centrifuged at 2500g for 5min at 4°C. After quantification using One Drop® OD-1000 Spectrophotometer (Nanjing Wuyi Corporation, CHINA), whole cell lysates were separated by SDS-PAGE under denaturing conditions and transferred to PVDF membranes (Millipore, USA). The membranes were blocked in 5% BSA (Sangon, CHINA) and then incubated with primary antibodies for β-ACTIN (Proteintech, USA), β-TUBULIN (Proteintech, USA), CA9 (Absolute Antibody, UK), FBL (Proteintech, USA), HIF1α (Novus, USA), HIF2α (BD, USA), and Histone H2B (Abcam, UK). The PVDF membranes were then incubated with secondary antibodies conjugated with horseradish peroxidase (Proteintech, USA). Immunoreactive proteins were visualized using the SuperSignal® West Femto Maximum Sensitivity Substrate (Thermo Scientific, USA) on ChemiDoc-It (UVP, UK).
Rapid amplification of cDNA ends (RACE)
RACE was performed with the 5′RACE System and 3′RACE System Kit (Invitrogen, USA) according to the manufacturer’s instructions. Briefly, the 5′RACE system is a set of prequalified reagents intended for synthesis of the first-strand cDNA, purification of the first-strand cDNA, homopolymeric tailing, and preparation of target cDNA for subsequent amplification by PCR. The 3′RACE procedure is also summarized as cDNA synthesis, RNA template degraded with RNase H, and amplificated by PCR. The primers used are as follows (GSP: Gene specific Primer):
5’RACE: cDNA GSP: TAAACAGGTGAAACACT;
PCR GSP: ACATTCGCAAGAGGGTGACAGT;
Nest PCR GSP: CGTCCCCAGGGCACCAGAATA;
3’RACE: GSP1: CAGGACCAGCCAGCCCTTTC;
GSP2: ATGGGAGCCTGGCCTTTGAG;
Primer-887 (1):
F: CACAGCAGCCTCCTCTTAAAC; R: CTTTTCTCTCCCATGCTGAGC
Primer-887 (2):
F: GGCCTTTGAGATTCCTGCGA; R: ATGCCTCAGTCGAAGGGAGA
Primer-887S: F: CTGCTCAGACACCGTTGC; R: GATGTGGTCTCACTCTGTTGC;
Primer-887L: F: CTGCTCAGACACCGTTGC; R: CTTGATGCTTTTACAGGCTCTC
siRNA and ASO oligonucleotides
siRNA and ASO oligos were designed and synthesized from Ribobio (Guangzhou, China). In each case, cells were plated at about 40% confluence before transfection. 20μM oligos were used for transient transfections into cells with Lipofectamine 3000 (Invitrogen) for 48h. The knockdown efficiency was validated with RT-qPCR. The sequences of siRNA and ASO are listed in Supplementary Table S3.
Plasmids construction for luciferase assay
The promoter of 887S (924bp) containing two hypoxic response elements (HREs) was amplified by PCR with the template of TSCC15 cell genomic DNA. Primers with the following restriction sites were used for 887S promoter: forward, 5′-CCCTCGAGGGCAAGTTTCCCTACTGCCTCC-3′ (XhoI); reverse, 5′-CCAAGCTTGGATTCTGTTTCTCATCAGGCG-3′ (HindIII). The clones were ligated into PGL3-basic vector’s upstream of the luciferase gene to construct the wild type 887S promoter reporter. The HRE sites in wild type 887S promoter reporter were mutated from 5′-CGTG-3′ to 5′-ATAA-3′ respectively by overlap extension PCR to generate mutant 887S promoter reporter. Similar, the promoter of CA9 (2024bp) was amplified using PCR with the template of TSCC15 cell genomic DNA. Primers with the following restriction sites were used for CA9 promoter: forward, 5′-CCGCTCGAGCGGAGTTCTGCATCAACCTGGTT-3′ (XhoI); reverse, 5′-CCCAAGCTTGGGTGTACGTGCATTGGAAACG-3′ (HindIII). The clones were ligated into PGL3-basic vector upstream of luciferase gene to construct the CA9 promoter reporter. The sequences of all plasmids were validated before use.
Dual Luciferase Reporter Assay
Cells were pre-seeded in a 24-well plate at a density of 1×105 cells/dish. On the following day, the cells were co-transfected with 1μg of constructed promoter reporter plasmids or control plasmids and 200ng of pRL-TK plasmid by Lipofectamine3000 (Invitrogen, USA). Twenty-four hours after transfection, cells were collected. Firefly and Renilla luciferase activities were sequentially measured by a Dual-LuciferaseTM Reporter Assay system (Promega). Luciferase activity was normalized by Renilla activity for each well. All assays and analyses were carried out in triplicate.
shRNA lentiviral transfection and stable cell lines establishment
Two independent shRNAs specifically targeting two different regions of 887L were designed and separately cloned into pLKO.1 vector to generate 887L shRNAs. 887L shRNAs and packaging vectors (pREV, pGag/pol, pVSVG) were co-transfected into HEK 293T cells using the lipofectamine 2000 (Invitrogen, USA). The medium was changed 6h after transfection with 20% FBS DMEM medium, and the supernatant containing lentivirus was collected 48h after transfection. Meanwhile, TSCC15 cells growing in the presence of 8μg/ml polybrene (Sigma, USA) were prepared to reach a confluency of 70–80% for viral infection. Stable monoclonal infectants were generated under the treatment of 1μg/ml puromycin (Gibco, USA), and monoclonal infectants with 887L knocking down efficiency more than 50% were selected for further tests. The shRNA sequences are as follows:
shRNA1:
F: ccggCTCACGTTGTCACCTTAGTggatccACTAAGGTGACAACGTGAGtttttg;
R: attcaaaaaCTCACGTTGTCACCTTAGTggatccACTAAGGTGACAACGTGAG;
shRNA2:
F: ccggCACTTCTCGTCACCACTA TggatccATAGTGGTGACGAGAAGTGtttttg;
R:aattcaaaaaCACTTCTCGTCACCACTATggatccATAGTGGTGACGAGAAGTG.
Plasmid overexpression and stable cell line establishment
887S or 887L was amplified using PCR with PhantaTM Super-Fidelity DNA Polymerase (Vazyme, CHINA) separately and subsequently cloned into the pcDNA3.1(+) (Invitrogen, USA) overexpression plasmid. Stable cell lines were constructed after transfection with the pcDNA3.1-887S or pcDNA3.1-887L plasmid and grew in the presence of 200 μg/ml G418 (WISENT Inc., CA). Samples with more than 2 times overexpression rate were selected for further tests. The PCR primers were used as follows:
887S:F: AGGAATTCCTGCTCAGACACCGTTGC;
887S:R: TAGGTACCGATGTGGTCTCACTCTGTTGC;
887L:F: AGGAATTCCTGCTCAGACACCGTTGC;
887L:R: TAGGTACCCTTGATGCTTTTACAGGCTCTC.
CRISPR-Cas9 and gRNA lentiviral transfection
All lentiviral vectors, including the CRISPR-Cas9 vector and guide RNA vector, were supplied by Cyagen Bioscience Corporation, USA (Service Agreement Number: MBS161230JX1). The experiment was performed according to the manufacturers’ instructions. Two independent guide RNAs sequences are sgRNA1: 5′-GTGAGCTGCAGAGGTAGCCG-3′; sgRNA2: 5′-AGCACGTGCGCTTGCTCTGC-3′. The resulted 887S knockout cell lines (HREmut1 and HREmut2) were further used in desired experiments under hypoxia.
Colony formation assay
200 or 400 cells collected during the logarithmic growth phase were seeded into 6-well cell culture plates (Corning, USA). The medium was replaced every 5 days. Cells were fixed with methanol and stained with 0.1% or 2.5% crystal violet after 7 or 14 days, then washed with 1×PBS. Colonies containing over 50 cells were counted manually (200×) under a microscope (Olympus, JAPAN). The colony formation rate was calculated as the percentage of colonies per numbers of inoculated cells (The colony formation rate (%) = (colonies per numbers / 200) ×100%).
Transwell assay
Transwell chambers inserting with an 8-μm pore size in 24-well cell culture plates (Corning, USA) were used. 1×105 or 5×104 of cells were suspended in 100ul serum-free media and added to the upper chamber. Complete medium containing 20% fetal bovine serum (0.7ml) was added to the bottom chamber as a chemo-attractant. The chambers were incubated in cell culture incubator for 24h. After incubation, the non-migrated cells in the upper chamber were removed with cotton swabs. The membranes were fixed with 4% paraformaldehyde (Sangon, USA) and stained with 0.1% crystal violet, and then cells from seven random fields (10×) were counted using light microscopy (Olympus, JAPAN). The relative mobility (‰) = (total cells of seven fields × 22.763)/(7 × initial incubated cells).
Intracellular and extracellular pH detection
BCECF-AM kit (Invitrogen, USA) was used to measure intracellular pH. BCECF-AM is a fluorescence probe sensitive to intracellular pH, which penetrates the cell membrane and is hydrolyzed into BCECF by esterase after entering the cell. BCECF can be excited to produce green fluorescence under the appropriate pH value [81]. Cell suspensions were prepared with HEPES at a concentration of 3×106 cells/ml. A final concentration of BCECF-AM at 3μM was obtained by adding 1 mM BCECF-AM/DMSO solution to the cell suspension (1/300 volume of the cell suspension). After 30min of incubation at 37°C, the cells were washed with HEPES buffer 3 times and adjust to 3×106 cells/ml. Finally, the fluorescence intensity of the cells was measured by fluorescence microscopy (Olympus, JAPAN). Detection of the extracellular pH value has been described previously [82, 83]. Briefly, the media from cultured cell were collected and detected by benchtop pH meter (Mettler Toledo, Switzerland).
Hematoxylin and eosin stain
The xenograft tumors were fixed with 4% paraformaldehyde, embedded in paraffin, and cut into 5μm sections. Then, the sections were transferred to adhesive-coated slides. All the sections were routinely deparaffinized and rehydrated, followed by incubation of hematoxylin (Sangon Biotech) for 8min at room temperature. After being rinsed with running water, the sections were treated by 1% ethanol hydrochloride to remove the excessive binding of hematoxylin dye and the adsorbed hematoxylin dye in cytoplasm. Next, the sections were respectively rinsed in running water and distilled water, then immersed in eosin dyes (Sangon Biotech) for 1min at room temperature. After the process of dehydration and fresh xylene treatment, the slides were examined and photographed by light microscopy (Olympus, JAPAN).
Cytosolic/nuclear fractionation
Cytosolic/nuclear fractionation assay was performed as previously described [84]. Briefly, TSCC15 cells were lysed in RSB-100 buffer (100mM Tris-HCl, 100mM NaCl, 2.5mM MaCl2, 40μg/ml digitonin). The supernatant fraction was collected as cytosolic fraction after centrifugation. The nuclear part was collected with supernatant after resuspended cell pellet in RSB-100T (100mM Tris-HCl, 100mM NaCl, 2.5mM MaCl2, 40μg/ml digitonin, 0.5% TritonX-100). After sonication, the soluble DNA-bound RNA fraction was collected.
Fluorescence in situ hybridization analysis (FISH)
Cy3-labeled FISH probes for 887L, β-ACTIN (ACTB), and Cy5 labeled FISH probes for 887S, β-ACTIN (ACTB) RNAs were designed and synthesized by GenePharma (Shanghai, China). The FISH assay was conducted according to the instruction of Fluorescent In Situ Hybridization Kit (GenePharma, Shanghai, China). Briefly, cells growing on glass coverslips were fixed with ice-cold 4% paraformaldehyde for 15 min and blocked with pre-hybridization buffer for 30min. The desired RNA probes (4μM) were incubated with hybridization solution for 5min at 73°C before hybridization. The cells were then incubated with the RNA probes (4μM) in the dark at 37°C for 12~16h, followed by three times of wash with 4×SSC containing 0.1% Tween-20. Finally, the cell nuclei were stained with Hochst33342 (Thermo, America). Images were taken by confocal fluorescence microscope (FV1200MPE-share, Olympus, Japan).
In vitro transcription and RNA pull-down
PCR was used to evaluate 887S, with the following primers that bound to the T7 promoter (887S: F: TAATACGACTCACTATAGGGCTCAGACACCGTTGC, R: GATGTGGTCTCACTCTGTTGC). The RNA products were transcribed by a T7 RNA polymerase kit (Invitrogen, USA) in vitro, treated with RNase-free TURBO DNase I (Invitrogen, USA), and labeled with the Pierce RNA 3′ End Desthiobiotinylation Kit (Thermo Scientific, USA). Cells were lysed in lysis buffer (50mM Tris-Cl, pH 7.0, 10 mM EDTA, 1% SDS, protease and phosphatase inhibitor cocktail, RNase inhibitor) on ice for 1–2h. At the same time, 3′ biotin-labeled RNA and magnetic beads were incubated for 30min at room temperature. Subsequently, the cell supernatants were collected after centrifugation at 13,000g for 10min at 4°C and added to the 3′ biotin-labelled RNA-magnetic beads mixture rotating for 2h at 4°C. After washing the beads for four times, the 887S-interacting proteins were subsequently identified by western blot.
RNA immunoprecipitation (RIP)
A total of 1×107 cells were harvested and suspended in 10 ml of PBS with 1% formaldehyde to fix for 10min at room temperature. Cross-linking was stopped by adding glycine to a final concentration of 0.25M, followed by incubation at room temperature for 5min. After pelleting cells at 1000rpm for 5min, cell pellet was isolated and lysed with 1ml of RIPA buffer (Vayzme, China) supplemented with RNase inhibitor (Vayzme, China) followed by sonication (6×, 6 s each time). After centrifuged at 14,000rpm for 10min at 4°C, the supernatant was pre-cleared with 25μl of Dynabeads Protein A/G at 4°C for 1h. Then, RNA-protein complexes were enriched by the beads conjugated with anti-HIF1α (Novus, USA; BD, USA), anti-DNMT1 (Abcam, UK), or IgG antibody at 4°C overnight. The RNA in beads complexes was isolated by TRIzol (Ambion, USA) and used to synthesize cDNA with SuperScript III Reverse Transcriptase (Vazyme, China), followed by RT-qPCR analysis.
Chromatin immunoprecipitation assay (ChIP)
ChIP assay was performed as previously described [84]. Briefly, TSCC15 cells were cross-linked with 1% formaldehyde for 10min. ChIP assays were performed using anti-HIF1α (Novus, USA; BD, USA) and anti-DNMT1 (Abcam, UK). Anti-IgG was used as a negative control. The DNA fragments were extracted and subjected to qPCR by primers detecting for corresponding cis-acting elements and negative control regions.
Chromatin isolation by RNA purification (ChIRP)
For each ChIRP experiment, 2×108 cells (293T or TSCC15) were used. The cells were cross-linked with formaldehyde and sonicated as previously described. Biotinylated RNA oligonucleotides of 887S or 887L were then hybridized with the cell lysates. The precipitated RNA or DNA was extracted and conducted for PCR by primers detecting for the desired regions and control regions. 293T is a cell line with no 887S or 887L expression and serves as a negative control. The sequences of probes are listed in Supplementary Table S3.
DNA methylation detection
The methylation status of the CpG site within CA9 promoter was determined by methylation-specific PCR (MSP) as reported before [85]. A total of 1μg of genomic DNA from TSCC15 cells was treated with sodium bisulfite using the bisulfite conversion kit (Active Motif, USA). A pair of positive control conversion-specific PCR primer was used to assess the success of the bisulfite conversion according to the manufacturer’s instructions. The design of specific primers targeting the bisulfite-modified methylated CpG site of CA9 promoter was done by Methprimer software (University of California, San Francisco, CA). The primer sequences used to detect methylated CA9 promoter are listed below. The presence of the respective PCR products was shown by Gel Red staining after electrophoresis in 2% agarose gels. In addition, a negative control reaction without template DNA was done together with each PCR experiment.
M primer1-F: TTTAAGTTGAGTTTATGGTTTCGA;
M primer1-R: AAAAAACAAACTAACTCACAAAACG;
M primer2-F: TTTAAGTTGAGTTTATGGTTTCGA;
M primer2-R: AAAAACAAACTAACTCACAAAACG.
Northern blot
RNAs from 293T, TSCC15, or TSCC25 cells were extracted with Trizol. Five microgram of RNA was separated on 1% formaldehyde agarose gel and blotted with biotin-labeled probes (General Biol, China). In vitro transcribed full length 887S and 887L RNA were used as positive controls and position markers. 293T is an 887S/887L non-expression cell line and serves as a negative control. The probes used for Northern blot were list in Supplementary table S2.
Quantification and statistical analysis
Each experiment was repeated independently at least three times, except for the ChIRP experiments (Fig. 5F, G, 6P, Q, and 7A, B) and the fractionationing assay (Fig. S4B) which were performed by two biological repeats. Statistical analysis was carried out using Microsoft Excel software and GraphPad Prism to assess the differences between experimental groups. Statistical significance was analyzed by two-tailed Student’s t test and expressed as a P value. P<0.05 were considered to be statistical significance.
Supplementary Information
Additional File 1: Supplementary Table1: Top 50 CVAA lncRNAs with hypoxia regulated information. Supplementary Table2: Primer sequence. Supplementary Table3: siRNA, ASO, ChIRP, FISH and Northern blot probe sequence
Additional File 2: Fig S1. Up-regulation of LINC00887 is correlated with poor survival rate of TSCC patients. Fig S2. The variety, existence and relative abundance of 887S, 887L and other LINC00887 variants. Fig S3. The expression pattern of 887S and 887L in the indicated cell lines. Fig S4. 887S and 887L subcellular localization and coding potential. Fig S5. Efficiency of gain-of-function or loss-of-function experiments in the indicated 887S and 887L knockdown or overexpression cells. Fig S6. CA9 promotes tumor progression in TSCC. Fig S7. Expression level of CA9 in the indicated 887S or 887L modulated cells. Fig S8. 887S and 887L regulates intracellular pH in TSCC. Fig S9. Effects of 887S and 887L on xenograft growth in Balb/c (nu/nu) mice. Fig S10. Effects of ASO-mediated 887S knockdown. Fig S11. In vitro transcribed 887S visualized by agarose gel with 1% formaldehyde. Fig S12. 887L promotes TSCC progression under normoxia.
Additional File 3. Raw blots and gel datasets
Additional File 4. The accession and information of TCGA-released RNA-seq datasets analysed in this paper
Acknowledgements
We thank Dr. Ge Shan from University of Science and Technology of China (USTC) and Dr. Xiaorong Zhang from Institute of Biophysics, Chinese Academy of Sciences (IBP) for their constructive suggestions. We also thank Dr. Xu Liu (USTC), Dr. Chunxiao Che (IBP), and Dr. Zhaoji Liu (USTC) for their technical help. We also thank Core Facility Center for life Sciences, USTC, for their technical support to this project. We are grateful for the data generated by the TCGA Research Network (https://www.cancer.gov/tcga), whose analysis formed the foundation of our study.
Authors’ contributions
X. W. and Q-P. K. initiated the project; X. W., T. S., and Q-P. K. designed the experiments. X. W. organized and supervised the whole project. T. S., W-X. X., F-H. S., and X-H. Z analyzed the NGS data. H-F. and Zh. supervised the hypoxia-related experiments. G-Y. Y., L-L. W., and Z-Y. W. supervised the clinical-related experiments. S-N. M., Y. Zh., and G-D. Sh. performed all the clinical related assays. T. S., Z-X. Y, and W. W. performed nude mice experiments. T. S., L-H., Ch., and Z-X. Y. performed cell-based tumor biological experiments. W. W. performed FISH assays. Z-X. Y, Q-X. Zh., and T. S. performed DNA methylation-related experiments. T.S. performed all the other experiments and analyzed the data. X. W. drafted the manuscript with input from Q-P. K., T. S., W-X. X., Z-X. Y., and all other authors. The authors read and approved the final manuscript.
Funding
This study was supported by grants from the Ministry of Science and Technology of China and National Natural Science Foundation of China (2016YFA0100502, 31970598, and 31671306 to X.W.), the Fundamental Research Funds for the Central Universities (YD2070002010 to X.W.), and the Major/Innovative Program of Development Foundation of Hefei Center for Physical Science and Technology (2018CXFX006 to X.W.). This study is also supported by grants from Key Research Program (KFZD-SW-221), Key Research Program of Frontiers Science (QYZDB-SSW-SMC020), Youth Innovation Promotion Association of the Chinese Academy of Sciences; Yunnan Applied Basic Research Project (2018FB137); Science and Technology Leading Talent Program of the Spring City (Kunming) and Yunling Scholar of the Yunnan Province; the National Natural Science Foundation of China (91749109 to Q-P.K. and 81671005 to G-Y.Y.).
Availability of data and materials
All data generated or analyzed during this study are included in this published article, its supplementary information files and publicly available repositories. The RNA-seq datasets that support the findings of this study are available in TCGA (https://confluence.broadinstitute.org/display/GDAC/Home) and listed in Additional File 4. The analytic codes and procedures that support the findings of this study have been deposited in GitHub with the identifier (https://github.com/liqg/CVAA) [21].
Declarations
Ethics approval and consent to participate
All animal studies were conducted with approval from the Animal Research Ethics Committee of the University of Science and Technology of China (Approval number USTCACUC-1801020). All participants signed and informed consent prior to sample collection (Approval number PKUSSIRB-2013009).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tao Shen, Wangxiao Xia, Sainan Min, Zixuan Yang and Lehua Cheng contributed equally to this work.
Contributor Information
Qing-Peng Kong, Email: kongqp@mail.kiz.ac.cn.
Xiangting Wang, Email: wangxt11@ustc.edu.cn.
References
- 1.Kaelin WG., Jr The VHL tumor suppressor gene: insights into oxygen sensing and cancer. Trans Am Clin Climatol Assoc. 2017;128:298–307. [PMC free article] [PubMed] [Google Scholar]
- 2.Schito L, Semenza GL. Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer. 2016;2(12):758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 3.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11(6):393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 4.Ratcliffe PJ. Oxygen sensing and hypoxia signalling pathways in animals: the implications of physiology for cancer. J Physiol. 2013;591(8):2027–2042. doi: 10.1113/jphysiol.2013.251470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jing XM, Yang FM, Shao CC, Wei K, Xie MY, Shen H, et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol Cancer. 2019;18(1):15. doi: 10.1186/s12943-019-1089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris AL. Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 7.Wykoff CC, Beasley NJ, Watson PH, Turner KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000;60(24):7075–7083. [PubMed] [Google Scholar]
- 8.Chiche J, Ilc K, Laferriere J, Trottier E, Dayan F, Mazure NM, et al. Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res. 2009;69(1):358–368. doi: 10.1158/0008-5472.CAN-08-2470. [DOI] [PubMed] [Google Scholar]
- 9.Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13(3):310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, Lin M, Yu H, Liu L, Levine AJ, Hu W, Feng Z. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013;4(1):2935. doi: 10.1038/ncomms3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavadas MA, Mesnieres M, Crifo B, Manresa MC, Selfridge AC, Scholz CC, et al. REST mediates resolution of HIF-dependent gene expression in prolonged hypoxia. Sci Rep. 2015;5(1):17851. doi: 10.1038/srep17851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruning U, Cerone L, Neufeld Z, Fitzpatrick SF, Cheong A, Scholz CC, Simpson DA, Leonard MO, Tambuwala MM, Cummins EP, Taylor CT. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol Cell Biol. 2011;31(19):4087–4096. doi: 10.1128/MCB.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22(9):1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, Bennett R, Bhai J, Billis K, Boddu S, Cummins C, Davidson C, Dodiya KJ, Gall A, Girón CG, Gil L, Grego T, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, Kay M, Laird MR, Lavidas I, Liu Z, Loveland JE, Marugán JC, Maurel T, McMahon AC, Moore B, Morales J, Mudge JM, Nuhn M, Ogeh D, Parker A, Parton A, Patricio M, Abdul Salam AI, Schmitt BM, Schuilenburg H, Sheppard D, Sparrow H, Stapleton E, Szuba M, Taylor K, Threadgold G, Thormann A, Vullo A, Walts B, Winterbottom A, Zadissa A, Chakiachvili M, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Yates AD, Zerbino DR, Flicek P. Ensembl 2019. Nucleic Acids Res. 2019;47(D1):D745–DD51. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet. 2014;48(1):433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Guo Q, Ding XP, Wang X. Mechanism of long noncoding RNAs as transcriptional regulators in cancer. RNA Biol. 2020;17(11):1680-92. 10.1080/15476286.2019.1710405. [DOI] [PMC free article] [PubMed]
- 19.Choudhry H, Harris AL, McIntyre A. The tumour hypoxia induced non-coding transcriptome. Mol Aspects Med. 2016;47-48:35–53. doi: 10.1016/j.mam.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 20.Shih JW, Kung HJ. Long non-coding RNA and tumor hypoxia: new players ushered toward an old arena. J Biomed Sci. 2017;24(1):53. doi: 10.1186/s12929-017-0358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li QG, He YH, Wu H, Yang CP, Pu SY, Fan SQ, Jiang LP, Shen QS, Wang XX, Chen XQ, Yu Q, Li Y, Sun C, Wang X, Zhou J, Li HP, Chen YB, Kong QP. A normalization-free and nonparametric method sharpens large-scale transcriptome analysis and reveals common gene alteration patterns in cancers. Theranostics. 2017;7(11):2888–2899. doi: 10.7150/thno.19425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu L, Zhang W, Huang C, Huang G, Su G, Xu J. lncRNA ANRIL protects H9c2 cells against hypoxia-induced injury through targeting the miR-7-5p/SIRT1 axis. J Cell Physiol. 2020;235(2):1175–1183. doi: 10.1002/jcp.29031. [DOI] [PubMed] [Google Scholar]
- 23.Yang J, Huang X, Hu F, Fu X, Jiang Z, Chen K. LncRNA ANRIL knockdown relieves myocardial cell apoptosis in acute myocardial infarction by regulating IL-33/ST2. Cell Cycle. 2019;18(23):3393–3403. doi: 10.1080/15384101.2019.1678965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin J, Zhang X, Xue C, Zhang H, Shashaty MG, Gosai SJ, et al. The long noncoding RNA landscape in hypoxic and inflammatory renal epithelial injury. Am J Physiol Renal Physiol. 2015;309(11):F901–F913. doi: 10.1152/ajprenal.00290.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grampp S, Platt JL, Lauer V, Salama R, Kranz F, Neumann VK, et al. Genetic variation at the 8q24.21 renal cancer susceptibility locus affects HIF binding to a MYC enhancer. Nat Commun. 2016;7:13183. doi: 10.1038/ncomms13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iden M, Fye S, Li K, Chowdhury T, Ramchandran R, Rader JS. The lncRNA PVT1 contributes to the cervical cancer phenotype and associates with poor patient prognosis. PLoS One. 2016;11(5):e0156274. doi: 10.1371/journal.pone.0156274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu F, Dong B, Dong P, He Y, Zheng J, Xu P. Hypoxia induces the activation of hepatic stellate cells through the PVT1-miR-152-ATG14 signaling pathway. Mol Cell Biochem. 2020;465(1-2):115–123. doi: 10.1007/s11010-019-03672-y. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y, Tian X, Wang T, Xia X, Cao F, Tian J, Xu P, Ma J, Xu H, Wang S. Long noncoding RNA Pvt1 regulates the immunosuppression activity of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. Mol Cancer. 2019;18(1):61. doi: 10.1186/s12943-019-0978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhan A, Deb P, Shihabeddin N, Ansari KI, Brotto M, Mandal SS. Histone methylase MLL1 coordinates with HIF and regulate lncRNA HOTAIR expression under hypoxia. Gene. 2017;629:16–28. doi: 10.1016/j.gene.2017.07.069. [DOI] [PubMed] [Google Scholar]
- 30.Xue M, Chen W, Xiang A, Wang R, Chen H, Pan J, Pang H, An H, Wang X, Hou H, Li X. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16(1):143. doi: 10.1186/s12943-017-0714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shih JW, Chiang WF, Wu ATH, Wu MH, Wang LY, Yu YL, Hung YW, Wang WC, Chu CY, Hung CL, Changou CA, Yen Y, Kung HJ. Long noncoding RNA LncHIFCAR/MIR31HG is a HIF-1alpha co-activator driving oral cancer progression. Nat Commun. 2017;8(1):15874. doi: 10.1038/ncomms15874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cvetkovic D, Babwah AV, Bhattacharya M. Kisspeptin/KISS1R system in breast cancer. J Cancer. 2013;4(8):653–661. doi: 10.7150/jca.7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Perdomo HA, Chaves MJ, Osorio JC, Sanchez A. Association between TMPRSS2:ERG fusion gene and the prostate cancer: systematic review and meta-analysis. Cent European J Urol. 2018;71(4):410–419. doi: 10.5173/ceju.2018.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo F, Yuan Y. Tumor necrosis factor alpha-induced proteins in malignant tumors: progress and prospects. Onco Targets Ther. 2020;13:3303–3318. doi: 10.2147/OTT.S241344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H, Zheng J, Shen N, Wang G, Zhou G, Fang Y, Lin J, Zhao J. Identification of pathways and genes associated with synovitis in osteoarthritis using bioinformatics analyses. Sci Rep. 2018;8(1):10050. doi: 10.1038/s41598-018-28280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobs J, Deschoolmeester V, Zwaenepoel K, Rolfo C, Silence K, Rottey S, Lardon F, Smits E, Pauwels P. CD70: An emerging target in cancer immunotherapy. Pharmacol Ther. 2015;155:1–10. doi: 10.1016/j.pharmthera.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74(1):115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- 38.Li C, Geng H, Ji L, Ma X, Yin Q, Xiong H. ESM-1: a novel tumor biomaker and its research advances. Anticancer Agents Med Chem. 2019;19(14):1687–1694. doi: 10.2174/1871520619666190705151542. [DOI] [PubMed] [Google Scholar]
- 39.Lucarelli G, Loizzo D, Franzin R, Battaglia S, Ferro M, Cantiello F, Castellano G, Bettocchi C, Ditonno P, Battaglia M. Metabolomic insights into pathophysiological mechanisms and biomarker discovery in clear cell renal cell carcinoma. Expert Rev Mol Diagn. 2019;19(5):397–407. doi: 10.1080/14737159.2019.1607729. [DOI] [PubMed] [Google Scholar]
- 40.Malemud CJ. Matrix metalloproteinases (MMPs) in health and disease: an overview. Front Biosci. 2006;11(1):1696–1701. doi: 10.2741/1915. [DOI] [PubMed] [Google Scholar]
- 41.Niu G, Deng L, Zhang X, Hu Z, Han S, Xu K, Hong R, Meng H, Ke C. GABRD promotes progression and predicts poor prognosis in colorectal cancer. Open Med (Wars). 2020;15(1):1172–1183. doi: 10.1515/med-2020-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddy RB, Khora SS, Suresh A. Molecular prognosticators in clinically and pathologically distinct cohorts of head and neck squamous cell carcinoma-a meta-analysis approach. PLoS One. 2019;14(7):e0218989. doi: 10.1371/journal.pone.0218989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shukla S, Pia Patric IR, Thinagararjan S, Srinivasan S, Mondal B, Hegde AS, Chandramouli BA, Santosh V, Arivazhagan A, Somasundaram K. A DNA methylation prognostic signature of glioblastoma: identification of NPTX2-PTEN-NF-kappaB nexus. Cancer Res. 2013;73(22):6563–6573. doi: 10.1158/0008-5472.CAN-13-0298. [DOI] [PubMed] [Google Scholar]
- 44.Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, Jiang Z, Zhang Y, Xu G, Zhang J, Zhou J, Sun B, Zou X, Wang S. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69(7):1193–1205. doi: 10.1136/gutjnl-2019-319639. [DOI] [PubMed] [Google Scholar]
- 45.Xue C, Chen C, Gu X, Li L. Progress and assessment of lncRNA DGCR5 in malignant phenotype and immune infiltration of human cancers. Am J Cancer Res. 2021;11(1):1–13. [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J, Liu B, Yang JA, Tang D, Wang X, Chen Q. Human sperm-associated antigen 4 as a potential biomarker of glioblastoma progression and prognosis. Neuroreport. 2019;30(6):446–451. doi: 10.1097/WNR.0000000000001226. [DOI] [PubMed] [Google Scholar]
- 47.Zou Y, Palte MJ, Deik AA, Li H, Eaton JK, Wang W, Tseng YY, Deasy R, Kost-Alimova M, Dančík V, Leshchiner ES, Viswanathan VS, Signoretti S, Choueiri TK, Boehm JS, Wagner BK, Doench JG, Clish CB, Clemons PA, Schreiber SL. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat Commun. 2019;10(1):1617. doi: 10.1038/s41467-019-09277-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, Akbani R, Bowlby R, Wong CK, Wiznerowicz M, Sanchez-Vega F, Robertson AG, Schneider BG, Lawrence MS, Noushmehr H, Malta TM, Stuart JM, Benz CC, Laird PW, Caesar-Johnson SJ, Demchok JA, Felau I, Kasapi M, Ferguson ML, Hutter CM, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Zhang J(J), Chudamani S, Liu J, Lolla L, Naresh R, Pihl T, Sun Q, Wan Y, Wu Y, Cho J, DeFreitas T, Frazer S, Gehlenborg N, Getz G, Heiman DI, Kim J, Lawrence MS, Lin P, Meier S, Noble MS, Saksena G, Voet D, Zhang H, Bernard B, Chambwe N, Dhankani V, Knijnenburg T, Kramer R, Leinonen K, Liu Y, Miller M, Reynolds S, Shmulevich I, Thorsson V, Zhang W, Akbani R, Broom BM, Hegde AM, Ju Z, Kanchi RS, Korkut A, Li J, Liang H, Ling S, Liu W, Lu Y, Mills GB, Ng KS, Rao A, Ryan M, Wang J, Weinstein JN, Zhang J, Abeshouse A, Armenia J, Chakravarty D, Chatila WK, de Bruijn I, Gao J, Gross BE, Heins ZJ, Kundra R, la K, Ladanyi M, Luna A, Nissan MG, Ochoa A, Phillips SM, Reznik E, Sanchez-Vega F, Sander C, Schultz N, Sheridan R, Sumer SO, Sun Y, Taylor BS, Wang J, Zhang H, Anur P, Peto M, Spellman P, Benz C, Stuart JM, Wong CK, Yau C, Hayes DN, Parker JS, Wilkerson MD, Ally A, Balasundaram M, Bowlby R, Brooks D, Carlsen R, Chuah E, Dhalla N, Holt R, Jones SJM, Kasaian K, Lee D, Ma Y, Marra MA, Mayo M, Moore RA, Mungall AJ, Mungall K, Robertson AG, Sadeghi S, Schein JE, Sipahimalani P, Tam A, Thiessen N, Tse K, Wong T, Berger AC, Beroukhim R, Cherniack AD, Cibulskis C, Gabriel SB, Gao GF, Ha G, Meyerson M, Schumacher SE, Shih J, Kucherlapati MH, Kucherlapati RS, Baylin S, Cope L, Danilova L, Bootwalla MS, Lai PH, Maglinte DT, van den Berg DJ, Weisenberger DJ, Auman JT, Balu S, Bodenheimer T, Fan C, Hoadley KA, Hoyle AP, Jefferys SR, Jones CD, Meng S, Mieczkowski PA, Mose LE, Perou AH, Perou CM, Roach J, Shi Y, Simons JV, Skelly T, Soloway MG, Tan D, Veluvolu U, Fan H, Hinoue T, Laird PW, Shen H, Zhou W, Bellair M, Chang K, Covington K, Creighton CJ, Dinh H, Doddapaneni HV, Donehower LA, Drummond J, Gibbs RA, Glenn R, Hale W, Han Y, Hu J, Korchina V, Lee S, Lewis L, Li W, Liu X, Morgan M, Morton D, Muzny D, Santibanez J, Sheth M, Shinbrot E, Wang L, Wang M, Wheeler DA, Xi L, Zhao F, Hess J, Appelbaum EL, Bailey M, Cordes MG, Ding L, Fronick CC, Fulton LA, Fulton RS, Kandoth C, Mardis ER, McLellan MD, Miller CA, Schmidt HK, Wilson RK, Crain D, Curley E, Gardner J, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton C, Shelton T, Sherman M, Thompson E, Yena P, Bowen J, Gastier-Foster JM, Gerken M, Leraas KM, Lichtenberg TM, Ramirez NC, Wise L, Zmuda E, Corcoran N, Costello T, Hovens C, Carvalho AL, de Carvalho AC, Fregnani JH, Longatto-Filho A, Reis RM, Scapulatempo-Neto C, Silveira HCS, Vidal DO, Burnette A, Eschbacher J, Hermes B, Noss A, Singh R, Anderson ML, Castro PD, Ittmann M, Huntsman D, Kohl B, le X, Thorp R, Andry C, Duffy ER, Lyadov V, Paklina O, Setdikova G, Shabunin A, Tavobilov M, McPherson C, Warnick R, Berkowitz R, Cramer D, Feltmate C, Horowitz N, Kibel A, Muto M, Raut CP, Malykh A, Barnholtz-Sloan JS, Barrett W, Devine K, Fulop J, Ostrom QT, Shimmel K, Wolinsky Y, Sloan AE, de Rose A, Giuliante F, Goodman M, Karlan BY, Hagedorn CH, Eckman J, Harr J, Myers J, Tucker K, Zach LA, Deyarmin B, Hu H, Kvecher L, Larson C, Mural RJ, Somiari S, Vicha A, Zelinka T, Bennett J, Iacocca M, Rabeno B, Swanson P, Latour M, Lacombe L, Têtu B, Bergeron A, McGraw M, Staugaitis SM, Chabot J, Hibshoosh H, Sepulveda A, Su T, Wang T, Potapova O, Voronina O, Desjardins L, Mariani O, Roman-Roman S, Sastre X, Stern MH, Cheng F, Signoretti S, Berchuck A, Bigner D, Lipp E, Marks J, McCall S, McLendon R, Secord A, Sharp A, Behera M, Brat DJ, Chen A, Delman K, Force S, Khuri F, Magliocca K, Maithel S, Olson JJ, Owonikoko T, Pickens A, Ramalingam S, Shin DM, Sica G, van Meir EG, Zhang H, Eijckenboom W, Gillis A, Korpershoek E, Looijenga L, Oosterhuis W, Stoop H, van Kessel KE, Zwarthoff EC, Calatozzolo C, Cuppini L, Cuzzubbo S, DiMeco F, Finocchiaro G, Mattei L, Perin A, Pollo B, Chen C, Houck J, Lohavanichbutr P, Hartmann A, Stoehr C, Stoehr R, Taubert H, Wach S, Wullich B, Kycler W, Murawa D, Wiznerowicz M, Chung K, Edenfield WJ, Martin J, Baudin E, Bubley G, Bueno R, de Rienzo A, Richards WG, Kalkanis S, Mikkelsen T, Noushmehr H, Scarpace L, Girard N, Aymerich M, Campo E, Giné E, Guillermo AL, van Bang N, Hanh PT, Phu BD, Tang Y, Colman H, Evason K, Dottino PR, Martignetti JA, Gabra H, Juhl H, Akeredolu T, Stepa S, Hoon D, Ahn K, Kang KJ, Beuschlein F, Breggia A, Birrer M, Bell D, Borad M, Bryce AH, Castle E, Chandan V, Cheville J, Copland JA, Farnell M, Flotte T, Giama N, Ho T, Kendrick M, Kocher JP, Kopp K, Moser C, Nagorney D, O’Brien D, O’Neill BP, Patel T, Petersen G, Que F, Rivera M, Roberts L, Smallridge R, Smyrk T, Stanton M, Thompson RH, Torbenson M, Yang JD, Zhang L, Brimo F, Ajani JA, Gonzalez AMA, Behrens C, Bondaruk, Broaddus R, Czerniak B, Esmaeli B, Fujimoto J, Gershenwald J, Guo C, Lazar AJ, Logothetis C, Meric-Bernstam F, Moran C, Ramondetta L, Rice D, Sood A, Tamboli P, Thompson T, Troncoso P, Tsao A, Wistuba I, Carter C, Haydu L, Hersey P, Jakrot V, Kakavand H, Kefford R, Lee K, Long G, Mann G, Quinn M, Saw R, Scolyer R, Shannon K, Spillane A, Stretch J, Synott M, Thompson J, Wilmott J, al-Ahmadie H, Chan TA, Ghossein R, Gopalan A, Levine DA, Reuter V, Singer S, Singh B, Tien NV, Broudy T, Mirsaidi C, Nair P, Drwiega P, Miller J, Smith J, Zaren H, Park JW, Hung NP, Kebebew E, Linehan WM, Metwalli AR, Pacak K, Pinto PA, Schiffman M, Schmidt LS, Vocke CD, Wentzensen N, Worrell R, Yang H, Moncrieff M, Goparaju C, Melamed J, Pass H, Botnariuc N, Caraman I, Cernat M, Chemencedji I, Clipca A, Doruc S, Gorincioi G, Mura S, Pirtac M, Stancul I, Tcaciuc D, Albert M, Alexopoulou I, Arnaout A, Bartlett J, Engel J, Gilbert S, Parfitt J, Sekhon H, Thomas G, Rassl DM, Rintoul RC, Bifulco C, Tamakawa R, Urba W, Hayward N, Timmers H, Antenucci A, Facciolo F, Grazi G, Marino M, Merola R, de Krijger R, Gimenez-Roqueplo AP, Piché A, Chevalier S, McKercher G, Birsoy K, Barnett G, Brewer C, Farver C, Naska T, Pennell NA, Raymond D, Schilero C, Smolenski K, Williams F, Morrison C, Borgia JA, Liptay MJ, Pool M, Seder CW, Junker K, Omberg L, Dinkin M, Manikhas G, Alvaro D, Bragazzi MC, Cardinale V, Carpino G, Gaudio E, Chesla D, Cottingham S, Dubina M, Moiseenko F, Dhanasekaran R, Becker KF, Janssen KP, Slotta-Huspenina J, Abdel-Rahman MH, Aziz D, Bell S, Cebulla CM, Davis A, Duell R, Elder JB, Hilty J, Kumar B, Lang J, Lehman NL, Mandt R, Nguyen P, Pilarski R, Rai K, Schoenfield L, Senecal K, Wakely P, Hansen P, Lechan R, Powers J, Tischler A, Grizzle WE, Sexton KC, Kastl A, Henderson J, Porten S, Waldmann J, Fassnacht M, Asa SL, Schadendorf D, Couce M, Graefen M, Huland H, Sauter G, Schlomm T, Simon R, Tennstedt P, Olabode O, Nelson M, Bathe O, Carroll PR, Chan JM, Disaia P, Glenn P, Kelley RK, Landen CN, Phillips J, Prados M, Simko J, Smith-McCune K, VandenBerg S, Roggin K, Fehrenbach A, Kendler A, Sifri S, Steele R, Jimeno A, Carey F, Forgie I, Mannelli M, Carney M, Hernandez B, Campos B, Herold-Mende C, Jungk C, Unterberg A, von Deimling A, Bossler A, Galbraith J, Jacobus L, Knudson M, Knutson T, Ma D, Milhem M, Sigmund R, Godwin AK, Madan R, Rosenthal HG, Adebamowo C, Adebamowo SN, Boussioutas A, Beer D, Giordano T, Mes-Masson AM, Saad F, Bocklage T, Landrum L, Mannel R, Moore K, Moxley K, Postier R, Walker J, Zuna R, Feldman M, Valdivieso F, Dhir R, Luketich J, Pinero EMM, Quintero-Aguilo M, Carlotti CG, Jr, Dos Santos JS, Kemp R, Sankarankuty A, Tirapelli D, Catto J, Agnew K, Swisher E, Creaney J, Robinson B, Shelley CS, Godwin EM, Kendall S, Shipman C, Bradford C, Carey T, Haddad A, Moyer J, Peterson L, Prince M, Rozek L, Wolf G, Bowman R, Fong KM, Yang I, Korst R, Rathmell WK, Fantacone-Campbell JL, Hooke JA, Kovatich AJ, Shriver CD, DiPersio J, Drake B, Govindan R, Heath S, Ley T, van Tine B, Westervelt P, Rubin MA, Lee JI, Aredes ND, Mariamidze A. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell. 2018;173(2):291–304. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MDM, Niu B, McLellan M, Uzunangelov V, Zhang J, Kandoth C, Akbani R, Shen H, Omberg L, Chu A, Margolin AA, van't Veer L, Lopez-Bigas N, Laird PW, Raphael BJ, Ding L, Robertson AG, Byers LA, Mills GB, Weinstein JN, van Waes C, Chen Z, Collisson EA, Cancer Genome Atlas Research Network. Benz CC, Perou CM, Stuart JM. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong J, Liu Y, Luo S, Jiang L, Zeng Y, Chen Z, Shi X, Lv B, Tang W. High expression of the long non-coding RNA HEIRCC promotes renal cell carcinoma metastasis by inducing epithelial-mesenchymal transition. Oncotarget. 2017;8(4):6555–6563. doi: 10.18632/oncotarget.14149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masoud GN, Li W. HIF-1alpha pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5(5):378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loboda A, Jozkowicz A, Dulak J. HIF-1 and HIF-2 transcription factors--similar but not identical. Mol Cells. 2010;29(5):435–442. doi: 10.1007/s10059-010-0067-2. [DOI] [PubMed] [Google Scholar]
- 53.Neri D, Supuran CT. Interfering with pH regulation in tumours as a therapeutic strategy. Nat Rev Drug Discov. 2011;10(10):767–777. doi: 10.1038/nrd3554. [DOI] [PubMed] [Google Scholar]
- 54.Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov. 2008;7(2):168–181. doi: 10.1038/nrd2467. [DOI] [PubMed] [Google Scholar]
- 55.McKenna R, Frost SC. Overview of the carbonic anhydrase family. Subcell Biochem. 2014;75:3–5. doi: 10.1007/978-94-007-7359-2_1. [DOI] [PubMed] [Google Scholar]
- 56.Ward C, Meehan J, Gray M, Kunkler IH, Langdon SP, Argyle DJ. Carbonic anhydrase IX (CAIX), cancer, and radiation responsiveness. Metabolites. 2018;8(1):13. 10.3390/metabo8010013. [DOI] [PMC free article] [PubMed]
- 57.Yang JS, Chen MK, Yang SF, Chang YC, Su SC, Chiou HL, Chien MH, Lin CW. Increased expression of carbonic anhydrase IX in oral submucous fibrosis and oral squamous cell carcinoma. Clin Chem Lab Med. 2014;52(9):1367–1377. doi: 10.1515/cclm-2014-0129. [DOI] [PubMed] [Google Scholar]
- 58.Lou Y, McDonald PC, Oloumi A, Chia S, Ostlund C, Ahmadi A, et al. Targeting tumor hypoxia: suppression of breast tumor growth and metastasis by novel carbonic anhydrase IX inhibitors. Cancer Res. 2011;71(9):3364–3376. doi: 10.1158/0008-5472.CAN-10-4261. [DOI] [PubMed] [Google Scholar]
- 59.Nakamura J, Kitajima Y, Kai K, Hashiguchi K, Hiraki M, Noshiro H, Miyazaki K. Expression of hypoxic marker CA IX is regulated by site-specific DNA methylation and is associated with the histology of gastric cancer. Am J Pathol. 2011;178(2):515–524. doi: 10.1016/j.ajpath.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gujar H, Weisenberger DJ, Liang G. The roles of human DNA methyltransferases and their isoforms in shaping the epigenome. Genes (Basel) 2019;10:2. doi: 10.3390/genes10020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19(2):81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 62.Zhang ZM, Liu S, Lin K, Luo Y, Perry JJ, Wang Y, Song J. Crystal structure of human DNA methyltransferase 1. J Mol Biol. 2015;427(15):2520–2531. doi: 10.1016/j.jmb.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lau J, Lin KS, Benard F. Past, present, and future: development of theranostic agents targeting carbonic anhydrase IX. Theranostics. 2017;7(17):4322–4339. doi: 10.7150/thno.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McDonald PC, Chafe SC, Brown WS, Saberi S, Swayampakula M, Venkateswaran G, et al. Regulation of pH by carbonic anhydrase 9 mediates survival of pancreatic cancer cells with activated KRAS in response to hypoxia. Gastroenterology. 2019;157(3):823–837. doi: 10.1053/j.gastro.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 65.Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, Zeng M, Song E. A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell. 2015;27(3):370–381. doi: 10.1016/j.ccell.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 66.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116(4):511–526. doi: 10.1016/S0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 67.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20(11):1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 68.Hangya B, Ranade SP, Lorenc M, Kepecs A. Central cholinergic neurons are rapidly recruited by reinforcement feedback. Cell. 2015;162(5):1155–1168. doi: 10.1016/j.cell.2015.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12(1):9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang X, Song X, Glass CK, Rosenfeld MG. The long arm of long noncoding RNAs: roles as sensors regulating gene transcriptional programs. Cold Spring Harb Perspect Biol. 2011;3(1):a003756. doi: 10.1101/cshperspect.a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu S, Wang Z, Chen D, Zhang B, Tian RR, Wu J, Zhang Y, Xu K, Yang LM, Cheng C, Ma J, Lv L, Zheng YT, Hu X, Zhang Y, Wang X, Li J. Annotation and cluster analysis of spatiotemporal- and sex-related lncRNA expression in rhesus macaque brain. Genome Res. 2017;27(9):1608–1620. doi: 10.1101/gr.217463.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ushijima T, Hanada K, Gotoh E, Yamori W, Kodama Y, Tanaka H, Kusano M, Fukushima A, Tokizawa M, Yamamoto YY, Tada Y, Suzuki Y, Matsushita T. Light controls protein localization through phytochrome-mediated alternative promoter selection. Cell. 2017;171(6):1316–1325. doi: 10.1016/j.cell.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 73.Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, Hartl C, Leppa V, Ubieta LT, Huang J, Lowe JK, Blencowe BJ, Horvath S, Geschwind DH. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540(7633):423–427. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, et al. An atlas of human long non-coding RNAs with accurate 5' ends. Nature. 2017;543(7644):199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14(7):496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 76.Yuan JH, Liu XN, Wang TT, Pan W, Tao QF, Zhou WP, Wang F, Sun SH. The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat Cell Biol. 2017;19(7):820–832. doi: 10.1038/ncb3538. [DOI] [PubMed] [Google Scholar]
- 77.Kuo TC, Kung HJ, Shih JW. Signaling in and out: long-noncoding RNAs in tumor hypoxia. J Biomed Sci. 2020;27(1):59. doi: 10.1186/s12929-020-00654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lomelino C, McKenna R. Carbonic anhydrase inhibitors: a review on the progress of patent literature (2011-2016) Expert Opin Ther Pat. 2016;26(8):947–956. doi: 10.1080/13543776.2016.1203904. [DOI] [PubMed] [Google Scholar]
- 79.Pinard MA, Mahon B, McKenna R. Probing the surface of human carbonic anhydrase for clues towards the design of isoform specific inhibitors. Biomed Res Int. 2015;2015:453543. doi: 10.1155/2015/453543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marchetti GM. Independencies induced from a graphical Markov model after marginalization and conditioning: the R package ggm. J Stat Softw. 2006;15(i06):498-526. 10.18637/jss.v015.i06.
- 81.Zheng G, Peng C, Jia X, Gu Y, Zhang Z, Deng Y, Wang C, Li N, Yin J, Liu X, Lu M, Tang H, He Z. ZEB1 transcriptionally regulated carbonic anhydrase 9 mediates the chemoresistance of tongue cancer via maintaining intracellular pH. Mol Cancer. 2015;14(1):84. doi: 10.1186/s12943-015-0357-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kondo A, Yamamoto S, Nakaki R, Shimamura T, Hamakubo T, Sakai J, Kodama T, Yoshida T, Aburatani H, Osawa T. Extracellular acidic pH activates the sterol regulatory element-binding protein 2 to promote tumor progression. Cell Rep. 2017;18(9):2228–2242. doi: 10.1016/j.celrep.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 83.Koyuncu I, Gonel A, Durgun M, Kocyigit A, Yuksekdag O, Supuran CT. Assessment of the antiproliferative and apoptotic roles of sulfonamide carbonic anhydrase IX inhibitors in HeLa cancer cell line. J Enzyme Inhib Med Chem. 2019;34(1):75–86. doi: 10.1080/14756366.2018.1524380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, et al. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454(7200):126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional File 1: Supplementary Table1: Top 50 CVAA lncRNAs with hypoxia regulated information. Supplementary Table2: Primer sequence. Supplementary Table3: siRNA, ASO, ChIRP, FISH and Northern blot probe sequence
Additional File 2: Fig S1. Up-regulation of LINC00887 is correlated with poor survival rate of TSCC patients. Fig S2. The variety, existence and relative abundance of 887S, 887L and other LINC00887 variants. Fig S3. The expression pattern of 887S and 887L in the indicated cell lines. Fig S4. 887S and 887L subcellular localization and coding potential. Fig S5. Efficiency of gain-of-function or loss-of-function experiments in the indicated 887S and 887L knockdown or overexpression cells. Fig S6. CA9 promotes tumor progression in TSCC. Fig S7. Expression level of CA9 in the indicated 887S or 887L modulated cells. Fig S8. 887S and 887L regulates intracellular pH in TSCC. Fig S9. Effects of 887S and 887L on xenograft growth in Balb/c (nu/nu) mice. Fig S10. Effects of ASO-mediated 887S knockdown. Fig S11. In vitro transcribed 887S visualized by agarose gel with 1% formaldehyde. Fig S12. 887L promotes TSCC progression under normoxia.
Additional File 3. Raw blots and gel datasets
Additional File 4. The accession and information of TCGA-released RNA-seq datasets analysed in this paper
Data Availability Statement
All data generated or analyzed during this study are included in this published article, its supplementary information files and publicly available repositories. The RNA-seq datasets that support the findings of this study are available in TCGA (https://confluence.broadinstitute.org/display/GDAC/Home) and listed in Additional File 4. The analytic codes and procedures that support the findings of this study have been deposited in GitHub with the identifier (https://github.com/liqg/CVAA) [21].