Abstract
Background and Aims
The C4Urochloa species (syn. Brachiaria) and Megathyrsus maximus (syn. Panicum maximum) are used as pasture for cattle across vast areas in tropical agriculture systems in Africa and South America. A key target for variety improvement is forage quality: enhanced digestibility could decrease the amount of land required per unit production, and enhanced lipid content could decrease methane emissions from cattle. For these traits, loss-of-function (LOF) alleles in known gene targets are predicted to improve them, making a reverse genetics approach of allele mining feasible. We therefore set out to look for such alleles in diverse accessions of Urochloa species and Megathyrsus maximus from the genebank collection held at the CIAT.
Methods
We studied allelic diversity of 20 target genes (11 for digestibility, nine for lipid content) in 104 accessions selected to represent genetic diversity and ploidy levels of U. brizantha, U. decumbens, U. humidicola, U. ruziziensis and M. maximum. We used RNA sequencing and then bait capture DNA sequencing to improve gene models in a U. ruziziensis reference genome to assign polymorphisms with high confidence.
Key Results
We found 953 non-synonymous polymorphisms across all genes and accessions; within these, we identified seven putative LOF alleles with high confidence, including those in the non-redundant SDP1 and BAHD01 genes present in diploid and tetraploid accessions. These LOF alleles could respectively confer increased lipid content and digestibility if incorporated into a breeding programme.
Conclusions
We demonstrated a novel, effective approach to allele discovery in diverse accessions using a draft reference genome from a single species. We used this to find gene variants in a collection of tropical grasses that could help reduce the environmental impact of cattle production.
Keywords: Urochloa brizantha, Urochloa decumbens, Urochloa humidicola, Urochloa ruziziensis, Megathyrsus maximum, cell wall digestibility, ecotilling, reverse genetics, forage energy content
INTRODUCTION
The environmental impact of cattle production could be decreased by reducing the amount of land required (e.g. land-sparing, sustainable intensification; IPCC, 2019) and the amount of methane (CH4) emitted per unit production (i.e. emission intensity). This could be achieved by genetic improvement of pasture grass on which they feed (O’Mara, 2012): an increase in digestibility and energy content would allow the same production to be achieved on a smaller land area, and an increase in lipid content in vegetative matter would decrease CH4 emitted per unit production (Hegarty et al., 2013), provided that these two traits could be improved without negative side effects, such as reduced growth or susceptibility to biotic or abiotic stresses.
Breeding of commercial tropical forage grass varieties in diploid and polyploid species and interspecific hybrids of Urochloa has been achieved by recurrent selection over many years, identifying superior-performing populations for key traits such as biomass production in different environments, resistance to pests and digestibility (Worthington and Miles, 2014). These targets increase the efficiency of forage grass, such that less land is required for production. Increasingly, environmental targets such as decreased nitrogen losses (Nuñez et al., 2018; Villegas et al., 2020) and reduced methane emissions from grazing cattle (Gaviria-Uribe et al., 2020) have become public breeding targets for improved pasture grasses. The diversity and species relationships of current breeding populations have been studied (Triviño et al., 2017), but continued improvement could be accelerated using genetic diversity that is available from accessions of the same genus in genebank collections such as that in the International Center for Tropical Agriculture (CIAT). An accession is simply a plant that was collected, which the CIAT genebank maintain by growing clonally where possible or as managed populations to try to preserve the original sexual composition as far as possible. For Urochloa accessions, it is often unknown whether they are completely or partially sexual or apomictic, and for polyploid accessions it is unknown whether they are allo- or autopolyploid or whether they are segmental polyploid. Recently, the ploidy and relatedness of 280 of Urochloa spp. accessions from the CIAT genebank have been defined for the first time (Tomaszewska et al., 2021).
Although they represent potentially useful diversity, introduction of such accessions into current breeding programmes is a major undertaking requiring evidence of likely benefit; breeding of Urochloa tropical forage grasses is particularly complicated by obligate outcrossing in sexual accessions and occurrence of apomixis in half of the progeny (Worthington and Miles, 2014). For traits where there are known key genes and an understanding of how variants of these might affect the traits, an allele mining approach may be feasible where sequencing of target genes rather than phenotyping can be used to find potentially useful alleles (Kumar et al., 2010; Vanholme et al., 2013; Greard et al., 2018). This reverse genetic approach can find useful loss-of-function (LOF) variation that would not be found by phenotyping as its effect is hidden due to gene/allele redundancy (Comai, 2005), particularly in polyploid or highly heterozygous material such as Urochloa, and provides the basis for perfect markers in crosses for following the alleles. Successful examples of allele mining for natural variation in known genes include studies on rice germplasm for starch synthesis genes (Butardo et al., 2017) and on Sorghum germplasm for a gene responsible for aluminium tolerance (Hufnagel et al., 2018).
Two traits where LOF alleles have been identified as beneficial are (1) digestibility where improvements have been gained by knockout or knockdown of genes involved in cell wall synthesis in grasses and (2) lipid content of vegetative tissue where improvements could be gained by knockout or knockdown of genes involved in lipid metabolism (literature summarized in Table 1). Increased lipid content of vegetative tissue of forage results in decreased CH4 emissions from cattle that feed on it, as well as benefiting meat and dairy fatty acid composition, as demonstrated by a transgenic approach in Lolium perenne (Winichayakul et al., 2020).
Table 1.
Evidence from the literature for selection of target genes to improve forage quality
| Target trait | Gene | Species | Suppression mode | Effect on trait | Pleiotropic effects | Reference |
|---|---|---|---|---|---|---|
| Digestibility | 4CL/Class I | Sorghum bicolor | Missense mutant bmr2 | 17 % increased saccharification | Saballos et al. (2008); Sattler et al. (2010) | |
| Saccharum officinarum | RNAi | 52 %, 76 % improved saccharification (field-grown) | 0 %, 30 % DM yield penalty (field-grown) | Jung et al. (2016) | ||
| BAHD01 | Setaria viridis, Saccharum officinarum | RNAi | 40–80 % increased saccharification | No growth penalty in GH | de Souza et al. (2018, 2019) | |
| BAHD05 | Setaria viridis | RNAi | 10–20 % increased saccharification | No growth penalty in GH | Mota et al. (2021) | |
| COMT | Zea mays | bm3 LOF mutant | Used in commercial hybrids with improved digestibility for cattle | Some reports yield penalty, sometimes no yield penalty | Sattler et al. (2010);Vignols et al. (1995) | |
| Sorghum bicolor | bmr12 LOF mutant | 30 % increased tract digestibility | 10 % DM yield penalty | Saballos et al. (2008); Sattler et al. (2010) | ||
| Panicum virgatum | RNAi | 30 % increased digestibility | No growth penalty in GH | Fu et al. (2011a) | ||
| Saccharum officinarum | TALEN induced LOF mutations in multiple paralogues | 40 % improved saccharification (field-grown) | No DM yield penalty (field-grown) | Kannan et al. (2018) | ||
| CAD/Group I | Brachypodium distachyon | Missense mutants | 40 % increased saccharification | No growth penalty in GH | Bouvier d’Yvoire et al. (2013) | |
| Sorghum bicolor | Missense mutant bmr6-3 | 20 % increased tract digestibility | 15 % DM yield penalty | (Saballos et al. (2009);Sattler et al. (2010) | ||
| Zea mays | bm1 | No yield penalty | Halpin et al. (1998); Sattler et al. (2010) | |||
| Panicum virgatum | RNAi | 20 % increased saccharification | Normal GH growth | Fu et al. (2011b) | ||
| CCR | Zea mays | RNAi | 20 % increased saccharification | Increased growth GH | Park et al. (2012) | |
| GT43A/IRX14 | Brachypodium distachyon | Missense mutant? | 10 % increased saccharification | No growth penalty in GH | Whitehead et al. (2018) | |
| CCoAOMT | Zea mays | Association with polymorphisms | Correlation with fibre digestibility | Brenner et al. (2010) | ||
| Lipid content | SDP1, SDP1-like | Arabidopsis | LOF mutant | Increase in leaf triacylglycerol | Poor seedling establishment in oilseeds | Kelly et al. (2013) |
| Medicago truncatula | VIGS | Increase in leaf lipid content | None | Wijekoon et al. (2020) | ||
| CGI-58 | arabidopsis | LOF mutant | Increase in leaf triacylglycerol | None | James et al. (2010) | |
| PXA1, PXA1-like | arabidopsis | LOF mutant | Increase in leaf triacylglycerol | poor seedling establishment in oilseeds, starvation sensitive, jasmonate deficient | Slocombe et al. (2009) | |
| Medicago truncatula | VIGS | Increase in leaf lipid content | none | Wijekoon et al. (2020) | ||
| TGD1 | arabidopsis | Leaky mutant | Increase in leaf triacylglycerol | embryo defect, growth penalty | Xu et al. (2005) | |
| TGD2 | arabidopsis | Leaky mutant | Increase in leaf triacylglycerol | embryo defect, growth penalty | Awai et al. (2006) | |
| TGD3 | arabidopsis | Leaky mutant | Increase in leaf triacylglycerol | Embryo defect, growth penalty | Lu et al. (2007) |
Here we compile a list of genes identified as targets from work in our labs or elsewhere with evidence of affecting these traits (Table 1). The cell wall genes identified as affecting digestibility are involved in monolignol synthesis (4CL, CCoAOMT, COMT, CCR, and CAD), glucuronoarabinoxylan (GAX) synthesis (GT43) or GAX feruloylation (BAHD01 and BAHD05). The lipid genes affecting oil content in vegetative tissue are involved in cytosolic triacylglycerol hydrolysis (SDP1, SDP1-like and CGI58), peroxisomal fatty acid β-oxidation (PXA1 and PXA1-like) or modulation of fatty acid synthesis mediated by lipid import into the plastid (TGD1, TGD2 and TGD3). We found the orthologues in U. ruziziensis diploid reference species. We then conducted a comprehensive screening of alleles for these genes in 104 diverse accessions of U. brizantha, U. decumbens, U. humidicola, U. ruziziensis and M. maximum using RNA sequencing (RNAseq) and bait capture genomic DNA sequencing (DNAseq).
MATERIALS AND METHODS
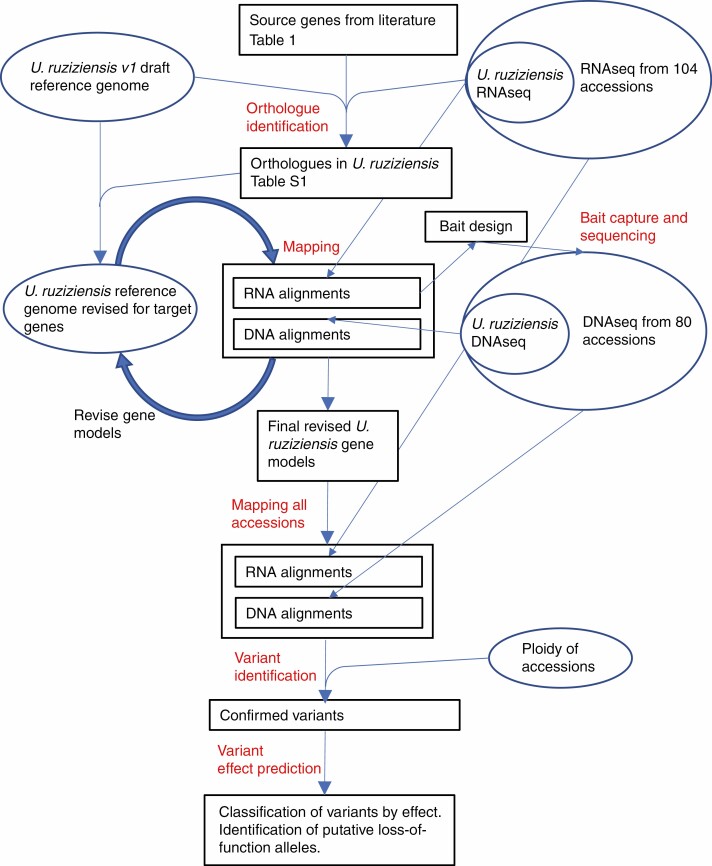
The sections below correspond to the steps shown in red in Fig. 1.
Fig. 1.
Workflow for analyses. Steps in red text are described in the Materials and Methods. The four boxes with thicker borders are the main outputs: improved gene models for target genes, alignments (BAM files of mapped reads) for each accession, variants identified and predicted effects of these on encoded proteins.
Plant materials and RNA sequencing
We collected leaf samples of 104 accessions (Supplementary data Table S1) from the field-grown genebank collection at the CIAT which were immediately frozen in liquid nitrogen. Samples were ground to a fine powder in liquid nitrogen and subsequently lyophilized. Total RNA was extracted as described in Pellny et al. (2012) with the difference that prior to DNase treatment the pellets were dried in a rotary evaporator (Eppendorf) and stored/transported at room temperature. Illumina sequencing using standard RNAseq library preparations with paired reads of length 150 bp was conducted by Novogene, Hong Kong. The raw reads were deposited in the Sequence Read Archive (SRA) under Bioproject PRJNA513453. We also collected leaf samples from an overlapping set of accessions for DNA extraction (Supplementary data Table S1), and 80 of these were used for DNA sequencing described below.
Orthologue identification
Here we use ‘orthologue’ as a convenient term to mean the most similar homologue in a different species. Some of these may not be true orthologues (as they could have arisen from a paralogue subsequently lost in one species) but they are likely to perform the same function. We searched for U. ruziziensis orthologues of the 16 target genes identified in other species selected from published evidence of affecting the traits and listed in Table 1. Firstly, we identified the putative orthologues of the target genes in the Setaria viridis (Setaria) v1.1 genome (Goodstein et al., 2012) as the closest reliably annotated genome using BLASTN with the coding sequences (CDS) of the target genes as original queries and source genomes (i.e. arabidopsis, maize, sorghum, Setaria or Brachypodium) of target genes for reciprocal BLASTN of hits. This identified one to one orthologues for all source genes except for arabidopsis SDP1 and PXA1 genes where there were two putative paralogues each in Setaria. We repeated this process for the draft U. ruziziensis v1.0 annotated genome (Worthington et al., 2021) and found the same orthologous relationships as for Setaria, except for one target gene CGI58 lipase, where an additional paralogue was found in U. ruziziensis v1.0. We compared the U. ruziziensis v1.0 gene models with the corresponding Setaria and source genes to judge whether they were correct; for ten of 22 they were incomplete. We compiled a set of 22 genes using the 12 complete U. ruziziensis v1.0 genes and ten Setaria genes for the others. We then mapped RNAseq reads from 11 U. ruziziensis accessions to this set. Two U. ruziziensis v1.0 genes with no equivalents in Setaria had almost zero mapped reads, and we removed these as likely pseudogenes, leaving a total of 20 U. ruziziensis target genes (Supplementary data Table S2). Baits were designed to these 20 U. ruziziensis genomic regions taking account of the mapped RNAseq to customize baits for each species–ploidy group. Bait capture was performed on genomic DNA isolated from 80 accessions. Resulting IonTorrent sequencing, RNAseq reads and, where required for finishing, targeted Sanger sequencing of amplicons for U. ruziziensis accessions were together used to check and refine gene models. We annotated the CDS by finding the longest open reading frame (ORF) and comparing it with that of orthologues. For 19 of these, we found thr complete CDS, but gene Ur.CGI58 lacks the first exon. We deposited the final annotated sequences for all 20 U. ruziziensis genes in GenBank/EMBL (accession numbers MW323383–MW323402).
Read mapping
We carried out the mapping and variant calling on the Galaxy platform (Giardine et al., 2005). We first mapped RNAseq of 11 U. ruziziensis accessions to the U. ruziziensis v1.0 reference genome using BWA-MEM (Li and Durbin, 2010). Taking these alignments into Geneious, for each target gene, we combined mapped reads with a set of all unmapped reads and conducted a de novo assembly. We compared the resulting contigs with U. ruziziensis and Setaria gene models and manually improved U. ruziziensis gene models. We substituted these gene models for the original versions as a first attempt at improving the reference, and designed baits based on these. After we completed sequencing of bait capture DNA, we mapped DNA and RNA reads of U. ruziziensis accessions to the modified reference to iteratively improve it until all reads mapped satisfactorily. We substituted the final version of the U. ruziziensis gene models (18 out of 21 were changed from the original version) into the U. ruziziensis genome annotation and mapped the RNA and DNA reads of all accessions using HiSAT2 (Kim et al., 2015) and the TMAP mapper within Torrent Suite 5.12.2 software (Thermofisher), respectively. For the latter, only reads >100 bp were used. The number of RNA reads mapped to each gene is shown in Supplementary data Table S3. We used the resulting BAM files (104 from RNAseq, 80 from DNAseq) to call variants and to manually inspect alignments on the Integrative Genomics Viewer (IGV; Robinson et al., 2011) for putative LOF alleles.
Bait capture
The CDS of the 20 genes of interest were targeted using myBaits Custom DNA-seq technology (Arbor Biosciences). A single bait set was designed to capture all genes in any of the species studied. To account for the likely diversity represented within and between species, consensus sequences were derived from the available RNAseq data for all genes in all individuals of each of the species. These were submitted to the design process performed by Arbor Biosciences which resulted in 20 346 baits of 70 nucleotide length with 3× tiling where each target base was covered by three different but overlapping baits where possible. Genomic DNA was extracted from frozen leaf tissue using a Plant DNeasy Kit (Qiagen) according to the supplied protocol. DNA quality was assessed by agarose gel electrophoresis and quantified using the Qubit dsDNA BR Assay Kit (Thermofisher). Whole-genome libraries for use in bait capture were prepared using Ion Plus Fragment Library kits according to the manufacturer’s instructions, with a target insert size of 400 bp and unique Ion Xpress barcodes for each sample. Libraries were then amplified using the library kit PCR reagents to generate sufficient DNA for bait hybridization. All libraries were quantified by qPCR using a Kapa Library Quantification Kit (Roche), and 16 equimolar pools were made, each comprising five libraries. For each pool, bait capture was performed according to the manufacturer’s myBaits® Manual v4. Libraries were then quantified by quantititave PCR (qPCR) as before, pooled and sequenced across two runs on an Ion Torrent PGM sequencer, using Ion Hi-Q View OT2 reagents for 400 bp templating and the Ion PGM Hi-Q View Sequencing Kit for 400 bp sequencing.
Variant calling
To call variants, we used FreeBayes (Galaxy Version 1.0.2.29-3), which is a haplotype-based variant calling program capable of dealing with polyploidy (Garrison and Marth, 2012). Both RNAseq and DNA capture BAM files (104 RNA, 80 DNA, overlap of accessions 74) were divided into 11 groups with the same species and ploidy (Supplementary data Table S1) using ploidy information from cytogenetics for the accessions (Tomaszewska et al., 2021). BAM files from each group were submitted together to FreeBayes with the appropriate setting for ploidy, and variant calling was limited to the target genes with default parameters for DNA reads; for RNA reads, the minimum fraction of observations supporting an allele (--min-alternate-fraction) was set to 0.05 to allow for low abundance due to nonsense-mediated decay of transcripts (Gutierrez et al., 1999) from LOF alleles. All other FreeBayes parameters were default. We retained variants with quality ≥20 using SNPsift v4.0 (Cingolani et al., 2012a). Using custom Perl scripts, we compared polymorphisms from DNA and RNA VCF files produced by FreeBayes for the same group. Polymorphisms observed from the RNAseq were filtered out unless they were also observed in the corresponding DNAseq bait capture sequences for the same accession, or, where this was not present, in another accession from the same species.
Variant effect prediction
We identified effects on function of the putative polymorphisms with SNPeff v4.0 (Cingolani et al., 2012b), which uses the CDS annotation to predict effects on encoded proteins. We compiled information on all unique variants using custom Perl scripts to process VCF files (summarized in Table 2 and Figs 2 and 3).
Table 2.
Total numbers of polymorphisms found in RNAseq and confirmed in bait capture DNAseq of 124 accessions for the 21 target U. ruziziensis genes
| Type | Effects | ||||||
|---|---|---|---|---|---|---|---|
| Gene | snp/mnp | Indel | Complex | Low | Moderate | High | |
| Cell wall genes | Ur.4CL | 200 | 0 | 44 | 186 | 58 (N: 15) | 0 |
| Ur.4CL_p1 | 195 | 3 | 39 | 169 | 67 (N: 17) | 1 | |
| Ur.BAHD01 | 146 | 1 | 33 | 144 | 35 (N: 5) | 1 | |
| Ur.BAHD05 | 183 | 0 | 29 | 159 | 53 (N: 19) | 0 | |
| Ur.CAD | 109 | 0 | 15 | 100 | 24 (N: 6) | 0 | |
| Ur.CAD_p1 | 117 | 1 | 17 | 88 | 46 (N: 11) | 1 | |
| Ur.CCoAOMT | 81 | 1 | 6 | 72 | 15 (N: 7) | 1 | |
| Ur.CCoAOMT_p1 | 49 | 0 | 22 | 64 | 7 (N: 3) | 0 | |
| Ur.CCR | 76 | 1 | 12 | 74 | 15 (N: 4) | 0 | |
| Ur.CGI58 | 54 | 0 | 14 | 39 | 29 (N: 7) | 0 | |
| Ur.CGI58_p1 | 68 | 0 | 15 | 47 | 35 (N: 11) | 1 | |
| Ur.COMT | 104 | 0 | 26 | 108 | 22 (N: 8) | 0 | |
| Lipid genes | Ur.GT43A | 169 | 0 | 30 | 159 | 40 (N: 13) | 0 |
| Ur.PXA1 | 284 | 0 | 42 | 228 | 97 (N: 26) | 1 | |
| Ur.PXA1-like | 337 | 0 | 21 | 228 | 130 (N: 50) | 0 | |
| Ur.SDP1 | 236 | 2 | 32 | 169 | 99 (N: 33) | 2 | |
| Ur.SDP1-like | 234 | 5 | 28 | 181 | 82 (N: 8) | 4 | |
| Ur.TGD1 | 81 | 0 | 12 | 78 | 15 (N: 3) | 0 | |
| Ur.TGD2 | 71 | 0 | 7 | 51 | 27 (N: 10) | 0 | |
| Ur.TGD3 | 118 | 0 | 17 | 90 | 45 (N: 0) | 0 | |
| total | 2912 | 14 | 461 | 2434 | 941 (N: 256) | 12 |
Genes are classified by type or predicted effect on protein. Type ‘snp/mnp’ includes SNPs and a small number of contiguous multiple nucleotide polymorphisms in the same haplotype; ‘complex’ denotes a mixture of SNPs and indels. ‘Low’ effects are synonymous variants, ‘Moderate’ are missense, in-frame indels, start or stop lost, and ‘High’ are frameshift or stop gained, predicted to cause loss of function (LOF). From moderate missense variants, counts of those predicted to be non-tolerated by the SIFT web server are shown ‘(N:)’.
Fig. 2.
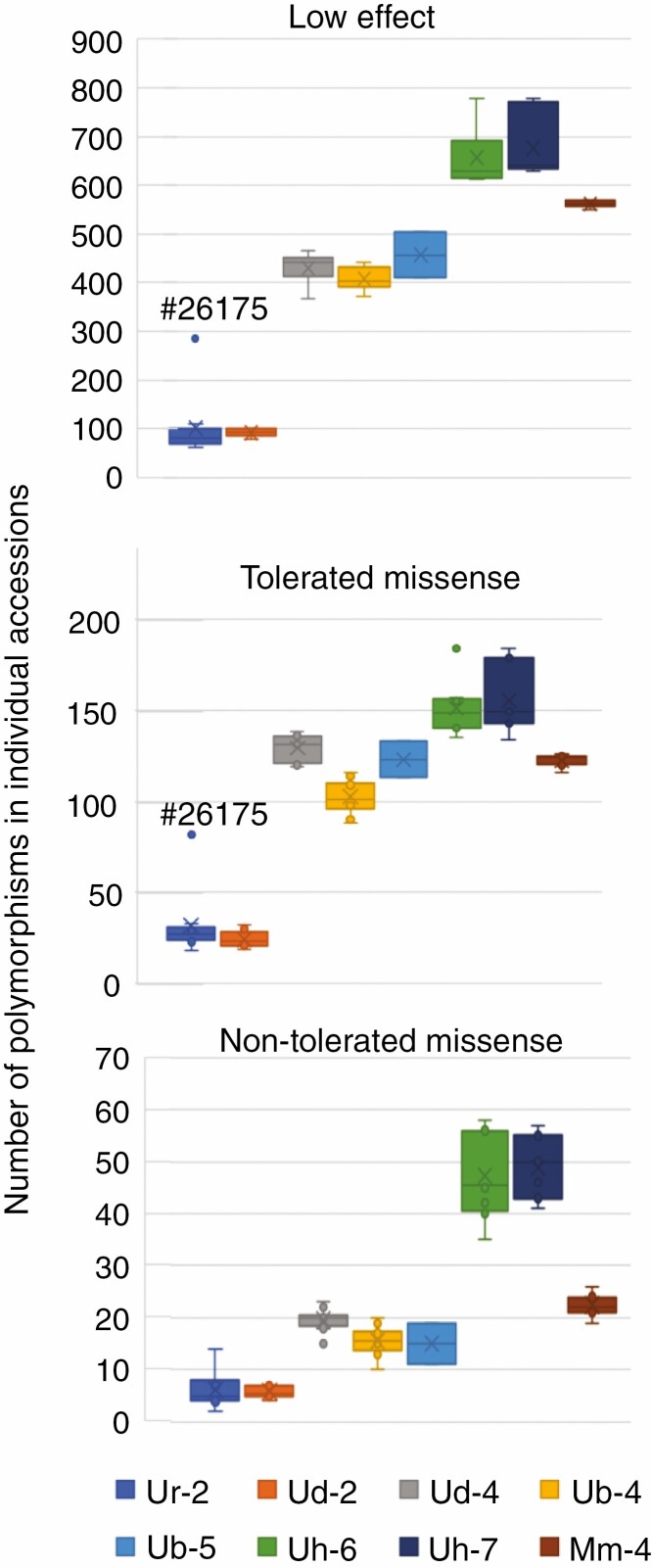
Number of variants in individual accessions grouped by species and ploidy. Only the 66 accessions with both RNA and DNA sequencing were used for this analysis. The key indicates the name of the group which is short code for the species followed by the ploidy level. Short codes are: Ur, U. ruziziensis; Ud, U. decumbens; Ub, U. brizantha; Uh, U. humidicola; Mm, M. maximus. Thus ‘Ur-2’ is the group with diploid U. ruziziensis accessions, ‘Ub-4’ is the group with tetraploid U. brizantha accessions, etc.
Fig. 3.
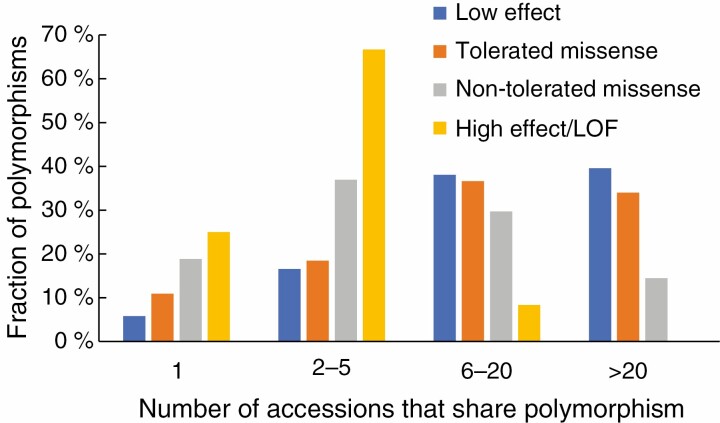
Numbers of accessions that share polymorphisms, grouped by predicted effect.
Classification of missense variants as tolerated or non-tolerated by SIFT
We downloaded protein sequences of orthologues for the 20 target genes in angiosperms with fully sequenced genomes from Phytozome v12 (Goodstein et al., 2012) and aligned them using Muscle (Edgar, 2004), with default parameters, together with our U. ruziziensis reference protein sequence. We assumed that at least one gene must be functional for each of the 50 species, with the exception of BAHD01 and BAHD04 genes, where we included only the 13 commelinid monocot species as their function is believed to be confined to these species (Mitchell et al., 2007). We removed any paralogues that did not align well. For each gene, we supplied these alignments and the discovered missense variants to the SIFT web server (Sim et al., 2012). We then used the SIFT prediction to classify the missense variants as tolerated (score > 0.05) or non-tolerated (score ≤ 0.05); non-tolerated predictions were all flagged as low confidence because of the small number of sequences available for alignment, while tolerated predicitons were regarded as reliable.
RESULTS
Identification of target genes
We identified genes from the literature, including published work from our own labs, where there was evidence that a loss of function in the gene would confer either increased digestibility (cell wall genes) or increased lipid content in vegetative tissue (lipid genes). This evidence is summarized in Table 1.
We identified orthologues of the genes in Table 1 in Setaria viridis, Setaria italica, Sorghum bicolor (as the most closely related fully sequenced genomes) and in the draft genome of U. ruziziensis (Supplementary data Table S2). We found additional putative paralogues in U. ruziziensis for 4CL, CAD, CCoAOMT and CGI58, and we included these in the analysis, naming them with the suffix ‘_p1’. We therefore had a final total of 11 cell wall genes and nine lipid genes as our target set identified in U. ruziziensis.
RNAseq was carried out on RNA collected from leaves of 104 accessions growing in fields at the CIAT. These data have been deposited at the NCBI under BioProject PRJNA513453. We took reads from U. ruziziensis accessions that mapped to the target U. ruziziensis genes (Supplementary data Table S2) and unmapped reads, and re-assembled these target genes. We compared the resulting sequences with target U. ruziziensis and S. viridis gene models to improve the U. ruziziensis gene models and design baits. We carried out bait capture of genomic DNA from a set of 80 accessions, 74 of which were in the RNAseq set. We used U. ruziziensis RNAseq, bait capture DNAseq and Sanger sequencing to iteratively improve the U. ruziziensis gene models. We submitted the final gene model versions to GenBank/EMBL and substituted them for the original versions into the U. ruziziensis reference genome. We then re-mapped RNAseq and bait capture DNAseq of all accessions to this updated reference. Nearly all genes in all accessions were expressed highly enough to give good coverage of RNAseq; an exception was CAD in some U. humidicola accessions which was noticeably expressed at a much lower level than CAD_p1 in this species, unlike in the other species (Supplementary data Table S3). We called variants on the resulting alignments and identified those that were found in RNAseq and confirmed as present in bait capture DNAseq in the same accession or, in cases where DNAseq was not available for the same accession, from any other accession of the same species. We revealed that 66 % of variants found in RNAseq that passed the quality score threshold (≥20) were confirmed in DNAseq (the remaining 33 % were not necessarily wrong but were often in regions of more complexity in bait capture due to several similar sequences). We also looked for special cases of loss of a splice donor or acceptor in DNAseq, which would be expected to change RNAseq read distribution, but did not find any instances of this. We present results below only for variants found in RNAseq and confirmed in bait capture DNAseq since these have good confidence as the two approaches have different sources of error.
The numbers of variants of different types in the target genes are summarized in Table 2 and the complete set is available in Supplementary data Table S4.
We were most interested in mutations that disrupt function, but found only 12 variants predicted to lose function (Table 1); however, among the 941 ‘moderate’ mutations (mostly missense non-synonymous mutations), the single nucleotide polymorphism (SNP) results in a different amino acid and it is expected that some of these changes will be disruptive. Using the SIFT web server (Sim et al., 2012), we supplied our protein alignments of orthologues from fully sequenced plant genomes and used resulting SIFT predictions to categorize missense mutations into tolerated and non-tolerated classes. From this analysis, 256 further variants that we discovered may disrupt gene function (Table 2).
We looked at the number of variants found in individual accessions, grouped by species and ploidy (presented as box and whisker plots in Fig. 2). As expected, we found more variants (identified with U. ruziziensis reference) in accessions from species that are more distantly related to U. ruziziensis, i.e. U. humidicola and M. maximus (Triviño et al., 2017). An outlier accession #26175 for group U. ruziziensis with high numbers of variants in these target genes is indicated in Fig. 2; this may be misclassified and is probably not U. ruziziensis according to a global analysis of the SNPs of all genes (J.D.V., unpubl. res.). We found very similar patterns for low effect and tolerated missense polymorphisms, suggesting that these both reflect relatedness to the reference. However, we found a different pattern for non-tolerated polymorphisms predicted to affect protein function by SIFT, which were much more common on groups with high ploidy (Fig. 2).
Many of the polymorphisms were shared between multiple accessions, and we present a summary of this in Fig. 3. We found that polymorphisms predicted to disrupt (non-tolerated missense) or eliminate (LOF) function were shared between fewer accessions than other polymorphisms. The more common polymorphisms may be characteristic of species or of sub-genomes in allopolyploid species. From our analysis, it appears that non-tolerated missense and LOF mutations are much less likely in these common polymorphisms (Fig. 3).
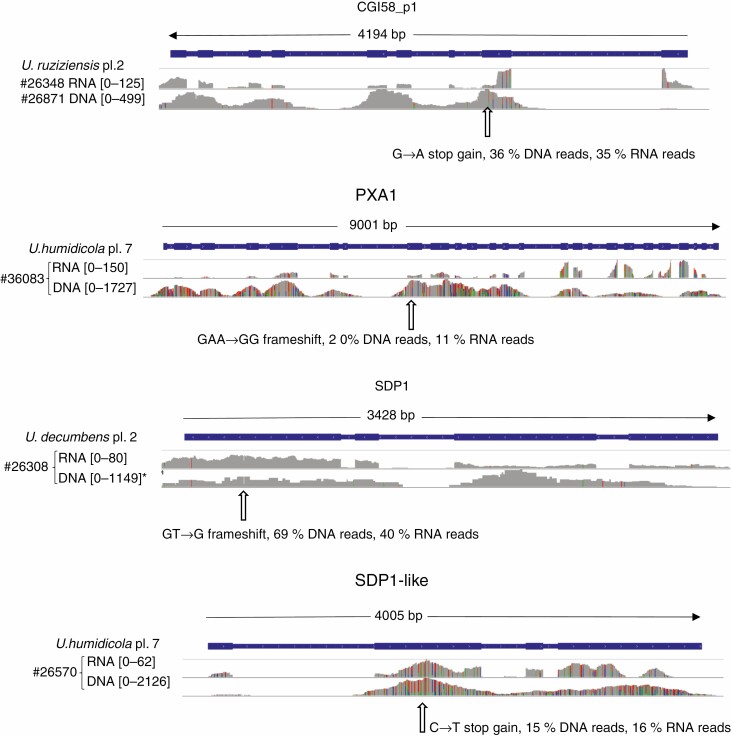
We manually examined the alignments for the 12 putative LOF alleles we discovered initially by our automated pipeline (Table 2) and found that three were frameshifts in stretches of homopolymers or of low complexity, with low coverage in some cases. It is likely that these are real since they were found in the same accessions in RNAseq and genomic DNA sequencing, but it is also possible they are artefacts due to systematic errors common to both DNAseq and RNAseq approaches. We found that two others were predicted to truncate the protein close to the C-terminus so were less certain to knock out function. We therefore designate these five as ‘low confidence’ and the remaining seven as ‘high confidence’. We show the alignments of these seven LOF alleles, three in cell wall genes in Fig. 4 and four in lipid genes in Fig. 5. From a breeding perspective, it is more difficult to transfer an allele from an accession with higher ploidy to a line with lower ploidy; since commercial varieties of these species are tetraploid, this may make the alleles of PXA1, SDP1-like and 4CL_p1 found in accessions with ploidy >4 of less immediate value. This leaves the putative LOF alleles in BAHD01 in tetraploid U. brizantha, in CAD_p1 in diploid U. ruziziensis and in SDP1 in diploid U. decumbens as of most potential interest. All these alleles appear to be present in heterozygous form, so further breeding would be required even in diploid accessions to achieve complete loss of function.
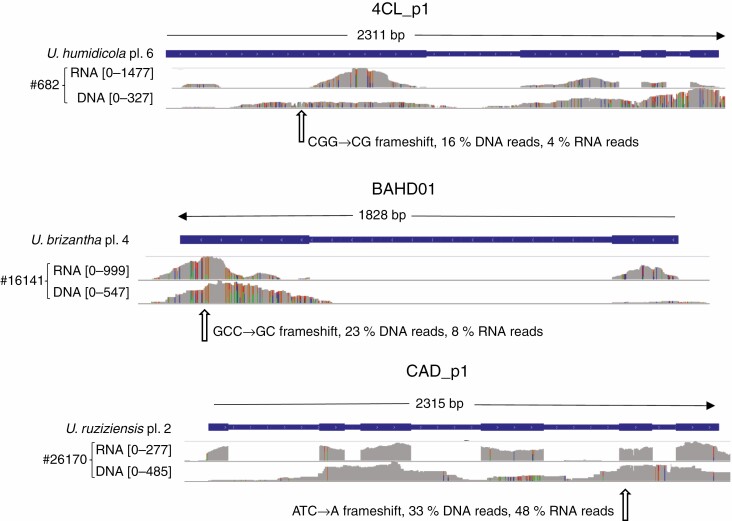
Fig. 4.
Read coverage for both RNAseq and DNAseq mapped to U. ruziziensis gene models of three cell wall genes for accessions which carry LOF alleles. Numbers within square brackets are the scale of coverage in number of reads. Gene model structure is indicated on the blue bar – wider parts are exons. Species name, ploidy level (pl.) and LOF polymorphisms are indicated. The numbers following ‘#’ are the CIAT accession numbers identifying the accessions carrying LOF alleles. Coverage graphs are grey when matching the reference; at variant positions, colours indicate bases (green A, blue C, orange G, red T).
Fig. 5.
Read coverage for both RNAseq and DNAseq mapped to U. ruziziensis gene models of four lipid genes for accessions which carry LOF alleles. Numbers within square brackets are the scale of coverage in the number of reads. Gene model structure is indicated on the blue bar – wider parts are exons. Species name, ploidy level (pl.) and LOF polymorphisms are indicated. The numbers following ‘#’ are the CIAT accession numbers identifying the accessions carrying LOF alleles. Coverage graphs are grey when matching a reference; at variant positions, colours indicate bases (green A, blue C, orange G, red T).
DISCUSSION
We developed a new methodology for allele discovery of candidate genes in a collection of diploid and polyploid accessions with only a draft genome sequence for one diploid species as reference. Our approach of combining RNAseq and bait capture (Fig. 1) provides a means of avoiding pseudogenes and resolving complexities. As part of the process, we improved gene models for 18 key genes and confirmed two more as accurate in the U. ruziziensis v1 genome. Our approach could be adapted for allele discovery in other plant collections or populations.
Our motivation in this work was the hope that breeding tropical forage grass with increased digestibility and lipid content could reduce the environmental impact of cattle production by, respectively, decreasing land requirement and CH4 emissions. We selected target genes from our work or the literature where reduction in function improves either digestibility or lipid content of vegetative tissue (Table 1). In the case of digestibility, the evidence comes directly from grass species, whereas the target genes for lipid content have so far only been tested in dicots. These genes differ substantially in the effects of knockdowns or knockouts, and in the evidence for any adverse pleiotropic effects. For some, it is thought that a complete knockout of function is required for the beneficial effect, and this causes little or no side effects (e.g. COMT in maize; Vignols et al., 1995). For the BAHD01 and BAHD05 genes putatively involved in addition of hydroxycinnamic acids to arabinoxylan, no complete knockouts have been reported, but knockdowns can have substantial effects (de Souza et al., 2018, 2019). In general, LOF alleles have less effect the greater the redundancy from other alleles and genes, so are recessive, and it can be necessary to stack LOF alleles in all of these to achieve a phenotype. In other cases, dosage effects can occur with increasing phenotype with an increasing proportion of redundant loci that have LOF alleles. (Comai, 2005).
We found 941 non-synonymous variants for our 20 target genes within 104 CIAT genebank accessions confirmed in RNAseq and DNAseq (Table 2). Most of these probably have little or no effect on function, but to gauge which ones are more likely to be detrimental we used the SIFT webserver (Sim et al., 2012) to identify a sub-set of 256 non-tolerated missense variants. Since these are predicted by SIFT based on alignments of all orthologues, they do not reflect relatedness to the U. ruziziensis reference and their frequency in accessions was principally dependent on ploidy (Fig. 2). This is most simply explained by the increasing copy number of the genes. An additional effect might be expected where detrimental mutations accumulate in lines with higher ploidy, as purifying selection will act less on highly redundant genes, but we could not judge this from our data. In fact, these non-tolerated missense variants were only predicted to be detrimental with low confidence by SIFT due to insufficient diversity of orthologues from fully sequenced plant genomes. These predictions could be improved in future as more genomes are sequenced and knowledge of the proteins improves.
The most secure predictions for disrupted function are the LOF variants with premature stop codons or frameshifts. We found that both non-tolerated missense and LOF variants tended to be shared between fewer accessions compared with other variants (Fig. 3); the rarity of these detrimental alleles means that complete functional knockout is very unlikely to occur naturally and requires artificial selection to bring them together in one plant.
On manual inspection of LOF variants, five were such that we were not completely confident they were real or were likely to knock out function. Of the other seven (Fig. 4), three were of particular interest since they occur in tetraploid or diploid accessions that are more easily incorporated into breeding programmes; these occurred in CAD_p1, BAHD01 and SDP1 genes. However, CAD_p1 is putatively redundant with CAD, and we have not found a report of effect of repressing CAD_p1 without also repressing CAD. No complete knockout of BAHD01 has been reported, but partial suppression had a substantial effect on digestibility in Setaria (de Souza et al., 2018), so it is possible that the dosage effects of this allele we found in CIAT accession #16141 might be observed even in tetraploid lines retaining some functional BAHD01 alleles. The SDP1 LOF allele occurs in a diploid U. decumbens accession (CIAT #26308) in heterozygous form. This accession is sexual so could be crossed to compatible diploid lines, the descendants of which could be crossed to produce a homozygous diploid line. Knockout of SDP1 alone increases storage lipid content by many fold in vegetative tissue of arabidopsis, (i.e. it is not redundant with SDP1-like) (Kelly et al., 2013), so such a line could be used to test for this effect in the Urochloa genus. If successful, the line could be crossed into tetraploid commercial breeding populations, e.g. using a chromosome doubling step.
In future, the allele mining approach we describe here could be applied to other genes and need not be confined to alleles detrimental to molecular function. For example, candidate genes underlying apomixis (Worthington et al., 2016) and spittlebug resistance (Ferreira et al., 2019) traits have recently been identified in Urochloa; with improving ability to predict consequences of variants in these, an allele mining approach could be of value. Also, for gene targets such as these where dominant alleles may affect phenotype, candidate gene association genetics could be a useful approach, as successfully applied for the FT gene and flowering time in Lolium perenne (Skøt et al., 2011). As knowledge of genes improves, allele mining of diverse germplasm will become an increasingly powerful tool to identify lines that could be beneficially brought into many crop breeding programmes.
CONCLUSION
Our successful use of bait capture, employed in parallel with RNAseq, is a highly cost-effective approach to allele discovery in diverse accessions of related species. Now that it is established, it could readily be applied to many more accessions of tropical grasses than we report here. However, even in this limited set, we discovered promising alleles that could be incorporated into breeding programmes of forage grasses to reduce the environmental impact of cattle production.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: CIAT genebank accessions of Urochloa spp. and Megathyrsus maximus used in this study. Table S2: target genes in U. ruziziensis. Table S3: number of mapped RNAseq reads and average coverage of 20 target genes for 104 accessions. Table S4: all variants discovered in the target genes from the 104 accessions, confirmed in RNAseq and bait capture.
FUNDING
This work was supported under the RCUK-CIAT Newton–Caldas Initiative ‘Exploiting biodiversity in Brachiaria and Panicum tropical forage grasses using genetics to improve livelihoods and sustainability’, with funding from the UK’s Official Development Assistance Newton Fund awarded by UK Biotechnology and Biological Sciences Research Council (BB/R022828/1). Additional funding for this study was received from the CGIAR Research Programs on Livestock; and Climate Change, Agriculture and Food Security (CCAFS).
LITERATURE CITED
- Awai K, Xu C, Tamot B, Benning C. 2006. A phosphatidic acid-binding protein of the chloroplast inner envelope membrane involved in lipid trafficking. Proceedings of the National Academy of Sciences, USA 103: 10817–10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier d’Yvoire M, Bouchabke-Coussa O, Voorend W, et al. 2013. Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon. The Plant Journal 73: 496–508. [DOI] [PubMed] [Google Scholar]
- Brenner EA, Zein I, Chen YS, et al. 2010. Polymorphisms in O-methyltransferase genes are associated with stover cell wall digestibility in European maize (Zea mays L.). BMC Plant Biology 10: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butardo VM, Anacleto R, Parween S, et al. 2017. Systems genetics identifies a novel regulatory domain of amylose synthesis. Plant Physiology 173: 887–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Patel VM, Coon M, et al. 2012a. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Frontiers in Genetics 3: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, et al. 2012b. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. 2005. The advantages and disadvantages of being polyploid. Nature Reviews. Genetics 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira RCU, Lara LAdC, Chiari L, et al. 2019. Genetic mapping with allele dosage information in tetraploid Urochloa decumbens (Stapf) R. D. Webster reveals insights into spittlebug (Notozulia entreriana Berg) resistance. Frontiers in Plant Science 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu CX, Mielenz JR, Xiao XR, et al. 2011a. Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proceedings of the National Academy of Sciences, USA 108: 3803–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu CX, Xiao XR, Xi YJ, et al. 2011b. Downregulation of cinnamyl alcohol dehydrogenase (CAD) leads to improved saccharification efficiency in switchgrass. Bioenergy Research 4: 153–164. [Google Scholar]
- Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing.https://ui.adsabs.harvard.edu/#abs/2012arXiv1207.3907G (1 July 2012).
- Gaviria-Uribe X, Bolivar DM, Rosenstock TS, et al. 2020. Nutritional quality, voluntary intake and enteric methane emissions of diets based on novel Cayman grass and its associations with two Leucaena shrub legumes. Frontiers in Veterinary Science 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B, Riemer C, Hardison RC, et al. 2005. Galaxy: a platform for interactive large-scale genome analysis. Genome Research 15: 1451–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu SQ, Howson R, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40: D1178– D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greard C, Barre P, Flajoulot S, Santoni S, Julier B. 2018. Sequence diversity of five Medicago sativa genes involved in agronomic traits to set up allele mining in breeding. Molecular Breeding 38: 141. [Google Scholar]
- Gutierrez RA, MacIntosh GC, Green PJ. 1999. Current perspectives on mRNA stability in plants: multiple levels and mechanisms of control. Trends in Plant Science 4: 429–438. [DOI] [PubMed] [Google Scholar]
- Halpin C, Holt K, Chojecki J, et al. 1998. Brown-midrib maize (bm1) – a mutation affecting the cinnamyl alcohol dehydrogenase gene. The Plant Journal 14: 545–553. [DOI] [PubMed] [Google Scholar]
- Hegarty M, Yadav R, Lee M, et al. 2013. Genotyping by RAD sequencing enables mapping of fatty acid composition traits in perennial ryegrass (Lolium perenne (L.)). Plant Biotechnology Journal 11: 572–581. [DOI] [PubMed] [Google Scholar]
- Hufnagel B, Guimaraes CT, Craft EJ, et al. 2018. Exploiting sorghum genetic diversity for enhanced aluminum tolerance: allele mining based on the AltSB locus. Scientific Reports 8: 10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC . 2019. Climate Change and Land: an IPCC special report on climate change, desertification, land degradation, sustainable land management, food security, and greenhouse gas fluxes in terrestrial ecosystems. https://www.ipcc.ch/site/assets/uploads/2019/11/SRCCL-Full-Report-Compiled-191128.pdf [Google Scholar]
- James CN, Horn PJ, Case CR, et al. 2010. Disruption of the Arabidopsis CGI-58 homologue produces Chanarin–Dorfman-like lipid droplet accumulation in plants. 107: 17833–17838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Kannan B, Dermawan H, Moxley GW, Altpeter F. 2016. Precision breeding for RNAi suppression of a major 4-coumarate:coenzyme A ligase gene improves cell wall saccharification from field grown sugarcane. Plant Molecular Biology 92: 505–517. [DOI] [PubMed] [Google Scholar]
- Kannan B, Jung JH, Moxley GW, Lee SM, Altpeter F. 2018. TALEN-mediated targeted mutagenesis of more than 100 COMT copies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnology Journal 16: 856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AA, van Erp H, Quettier AL, et al. 2013. The sugar-dependent1 lipase limits triacylglycerol accumulation in vegetative tissues of Arabidopsis. Plant Physiology 162: 1282–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nature Methods 12: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar GR, Sakthivel K, Sundaram RM, et al. 2010. Allele mining in crops: prospects and potentials. Biotechnology Advances 28: 451–461. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Xu C, Awai K, Jones AD, Benning C. 2007. A small ATPase protein of Arabidopsis, TGD3, involved in chloroplast lipid import. 282: 35945–35953. [DOI] [PubMed] [Google Scholar]
- Mitchell RAC, Dupree P, Shewry PR. 2007. A novel bioinformatics approach identifies candidate genes for the synthesis and feruloylation of arabinoxylan. Plant Physiology 144: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mota TR, Souza WRd, Oliveira DM, et al. 2021. Suppression of a BAHD acyltransferase decreases p-coumaroyl on arabinoxylan and improves biomass digestibility in the model grass Setaria viridis. The Plant Journal 105: 136–150. [DOI] [PubMed] [Google Scholar]
- Nuñez J, Arevalo A, Karwat H, et al. 2018. Biological nitrification inhibition activity in a soil-grown biparental population of the forage grass, Brachiaria humidicola. Plant and Soil 426: 401–411. [Google Scholar]
- O’Mara FP. 2012. The role of grasslands in food security and climate change. Annals of Botany 110: 1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Mei CS, Pauly M, et al. 2012. Downregulation of maize cinnamoyl-coenzyme A reductase via RNA interference technology causes brown midrib and improves ammonia fiber expansion-pretreated conversion into fermentable sugars for biofuels. Crop Science 52: 2687–2701. [Google Scholar]
- Pellny TK, Lovegrove A, Freeman J, et al. 2012. Cell walls of developing wheat starchy endosperm: comparison of composition and RNA-seq transcriptome. Plant Physiology 158: 612–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, et al. 2011. Integrative genomics viewer. Nature Biotechnology 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos A, Vermerris W, Rivera L, Ejeta G. 2008. Allelic association, chemical characterization and saccharification properties of brown midrib mutants of sorghum (Sorghum bicolor (L.) Moench). Bioenergy Research 1: 193–204. [Google Scholar]
- Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W. 2009. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181: 783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Funnell-Harris DL, Pedersen JF. 2010. Brown midrib mutations and their importance to the utilization of maize, sorghum, and pearl millet lignocellulosic tissues. Plant Science 178: 229–238. [Google Scholar]
- Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. 2012. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Research 40: W452–W457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skøt L, Sanderson R, Thomas A, et al. 2011. Allelic variation in the perennial ryegrass FLOWERING LOCUS T gene is associated with changes in flowering time across a range of populations. Plant Physiology 155: 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocombe SP, Cornah J, Pinfield-Wells H, et al. 2009. Oil accumulation in leaves directed by modification of fatty acid breakdown and lipid synthesis pathways. Plant Biotechnology Journal 7: 694–703. [DOI] [PubMed] [Google Scholar]
- de Souza WR, Martins PK, Freeman J, et al. 2018. Suppression of a single BAHD gene in Setaria viridis causes large, stable decreases in cell wall feruloylation and increases biomass digestibility. New Phytologist 218: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza WR, Pacheco TF, Duarte KE, et al. 2019. Silencing of a BAHD acyltransferase in sugarcane increases biomass digestibility. Biotechnology for Biofuels 12: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaszewska P, Vorontsova MS, Renvoize SA, et al. 2021. Complex polyploid and hybrid species in an apomictic and sexual tropical forage grass group: genomic composition and evolution in Urochloa (Brachiaria) species. bioRxiv: doi: 10.1101/2021.02.19.431966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triviño NJ, Perez JG, Recio ME, et al. 2017. Genetic diversity and population structure of Brachiaria species and breeding populations. Crop Science 57: 2633–2644. [Google Scholar]
- Vanholme B, Cesarino I, Goeminne G, et al. 2013. Breeding with rare defective alleles (BRDA): a natural Populus nigra HCT mutant with modified lignin as a case study. New Phytologist 198: 765–776. [DOI] [PubMed] [Google Scholar]
- Vignols F, Rigau J, Torres MA, Capellades M, Puigdomenech P. 1995. The brown midrib3 (Bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. The Plant Cell 7: 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas D, Arevalo A, Nunez J, et al. 2020. Biological nitrification inhibition (BNI): phenotyping of a core germplasm collection of the tropical forage grass Megathyrsus maximus under greenhouse conditions. Frontiers in Plant Science 11: 820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead C, Garrido FJO, Reymond M, et al. 2018. A glycosyl transferase family 43 protein involved in xylan biosynthesis is associated with straw digestibility in Brachypodium distachyon. New Phytologist 218: 974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijekoon C, Singer SD, Weselake RJ, et al. 2020. Down-regulation of key genes involved in carbon metabolism in Medicago truncatula results in increased lipid accumulation in vegetative tissue. Crop Science 60: 1798–1808. [Google Scholar]
- Winichayakul S, Beechey-Gradwell Z, Muetzel S, et al. 2020. In vitro gas production and rumen fermentation profile of fresh and ensiled genetically modified high-metabolizable energy ryegrass. Journal of Dairy Science 103: 2405–2418. [DOI] [PubMed] [Google Scholar]
- Worthington ML, Miles JW. 2014. Reciprocal full-sib recurrent selection and tools for accelerating genetic gain in apomictic Brachiaria. In: Budak H, Spangenberg G, eds. Molecular breeding of forage and turf. Cham: Springer International Publishing. [Google Scholar]
- Worthington M, Heffelfinger C, Bernal D, et al. 2016. A parthenogenesis gene candidate and evidence for segmental allopolyploidy in apomictic Brachiaria decumbens. Genetics 203: 1117–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington M, Perez JG, Mussurova S, et al. 2021. A new genome allows the identification of genes associated with natural variation in aluminium tolerance in Brachiaria grasses. Journal of Experimental Botany 72: 302–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan J, Froehlich JE, Awai K, Benning C. 2005. Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. 17: 3094–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.