Abstract
Despite massive government and private sector investments into prevention of cardiovascular disease, diabetes mellitus and obesity, efforts have largely failed, and the burden of cost remains in the treatment of downstream morbidity and mortality, with overall stagnating outcomes. A new paradigm shift in the approach to these patients may explain why existing treatment strategies fail, and offer new treatment targets. This review aims to provide a clinician-centred primer on metabolic memory, defined as the sum of irreversible genetic, epigenetic, cellular and tissue-level alterations that occur with long-time exposure to metabolic derangements.
Keywords: chronic kidney disease, diabetic kidney disease, epigenetics, metabolic memory, sirtuin 1
INTRODUCTION
Approximately 45% of the US population suffers from chronic disease, mainly pulmonary and cardiovascular diseases (CVD), diabetes mellitus (DM), obesity and cancer, while more than half of the annual deaths worldwide can be attributable to chronic conditions [1, 2]. We have moved past failed efforts at primary prevention to management of downstream morbidity and mortality, hopefully with detection of early biomarkers of progression [3].
Engerman and Kern [4], a relatively long time ago, described that diabetic retinopathy progressed despite good glycaemic control in dogs and coined the term metabolic memory. Despite almost 40 years of knowledge that tight glycaemic control does not completely prevent diabetic complications, we still follow glycated haemoglobin (HbA1c) to this day, with no better biomarkers in routine use.
Metabolic memory applies to chronic diseases [4] and describes microscopic to macroscopic irreversible changes that occur over time [5].
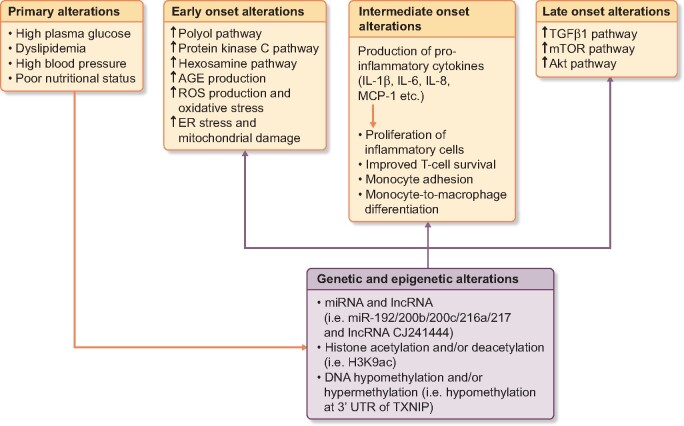
Various primary, early, intermediate, late-onset alterations with genetic and epigenetic interactions play a role in the metabolic memory (Figure 1).
FIGURE 1.
Triggering factors and mechanisms of development of metabolic memory. ER, endoplasmic reticulum.
Lead time bias in the natural history of metabolic memory likely accounts for treatment failures in some patients while others have better outcomes. As this affects patients seen in the clinic daily, this report aims to provide a clinician centred review of metabolic memory.
THE METABOLIC MEMORY OF DM
Diabetes causes more blindness (microvascular), myocardial infarction (macrovascular), stroke (macrovascular) and renal failure (microvascular) worldwide than any other disease [6, 7], and the outcomes of the microvascular complications are not completely addressed by HbA1c, which is still the main biomarker used to follow ‘successful’ diabetes management. Diabetes does not cause these effects overnight; rather, microvascular damage leads to macrovascular consequences [8].
In a study involving 1441 Type I DM patients undergoing either intensive (≥3 daily insulin injections adjusted via frequent glucose monitoring) or conventional therapy (1–2 daily insulin injections) for a mean duration of 6.5 years, the Diabetes Complications and Control Trial (DCCT) demonstrated that delayed onset of microvascular complications, as well as slower progression, could be achieved via intensive therapy [9, 10] but the microvascular complications did not reach zero, even with these unrealistically strict treatment guidelines.
Furthermore, after the study, all patients were placed on intensive therapy, and the group that had received conventional therapy previously had higher rates of microvascular complications at 8-year follow-up, 17-year follow-up for CVD and even 22-year follow-up for low glomerular filtration rate. The metabolic memory of the conventional therapy could not be corrected even with the strictest glycaemic control, suggesting that other treatment strategies must be developed for true microvascular prevention [11–15]. A separate study including 5102 newly diagnosed persons with Type II DM >10 years had similar findings [16, 17]. In patients with longstanding poor diabetic control, tight glycaemic control slightly improves cardiovascular outcomes compared with conventional management, a disappointing outcome for an intense intervention [18, 19]. Despite the best existing management, patients cannot escape the fate of their metabolic memory. In the below paragraphs, we give detailed description of mechanisms including immune mechanisms, oxidative stress, genetic and epigenetic changes in relation to tissue damage and pathological findings related to metabolic memory.
HISTOLOGICAL ALTERATIONS AND IMMUNE RESPONSE
The histopathological features of diabetic complications, most typically diabetic nephropathy, are mesangial expansion with increased extracellular matrix (ECM) production, formation of Periodic acid–Schiff (PAS) (+) diffuse thickening of glomerular basement membrane, effacement of podocyte food processes and hyaline arteriolosclerosis in both afferent and efferent arterioles of glomeruli and Kimmelstiel–Wilson nodules mostly as a post-mortem finding [20]. Similar patterns of histopathological changes are observed in both diabetic neuropathy and retinopathy with the addition of microaneurysm formation and punctate haemorrhages in retinopathy [21]. Expression of fibronectin and collagen mRNA, two predominant ECM proteins, has been shown to be significantly higher in human endothelial cells cultured in a hyperglycaemic environment [22, 23]. Elevated expression persisted after switch to normoglycaemic culture medium despite multiple cell divisions and replanting while cells with higher expression levels displayed proliferative advantage [22]. In addition to changes in ECM production, degradation of ECM is also impaired by hyperglycaemia leading to glycation of mesangial proteins that impair matrix metalloproteinase-2 (MMP-2), the primary MMP secreted in mesangium responsible for the degradation of Type IV collagen [24].
Hyperglycaemia also promotes inflammatory changes in tissues, predominantly via activation of nuclear factor kappa B (NF-κB) and Toll-like receptor pathways [25]. Changes include release of inflammatory cytokines [interleukin (IL)-1β, IL-6, IL-8 and monocyte chemoattractant protein (MCP-1)], enhancement of monocyte-endothelial or vascular smooth muscle cell binding, monocyte-to-macrophage transition and increased vascular permeability [26, 27]. Furthermore, hyperglycaemia may cause premature senescence of various cell types and accelerated cellular ageing processes that may lead to senescence-associated secretory phenotype that secretes pro-inflammatory cytokines, leading to a destructive loop. Microbiotic alterations occur in people with pro-inflammatory cytokine levels and inducing senescence-associated secretory phenotype [28]. Furthermore, hyperglycaemia reduces CXC chemokine receptor type 4 (CXCR4) expression in CD34 (+) haematopoietic stem cells via altered DNA methylation patterns, improving T-cell survival via the activity of advanced glycation end-product receptors (RAGE) [29, 30].
Endothelial dysfunction and inflammation-induced insulin resistance also cause diabetic progression [31–34]. High-carb, high-fat diets inflame feeding control neurons in the hypothalamus [35, 36]. In addition, HbA1c positively correlates with IL-6 and tumour necrosis factor-α [37].
Hyperglycaemia leads to increased production and impaired degradation of ECM proteins, persistent pro-inflammatory state and altered immune response, causing net fibrosis and thickening of basement membranes causing diabetic microvascular and macrovascular complications. These are also the treatment targets.
OXIDATIVE STRESS AND BIOCHEMICAL CHANGES
Diabetic rats have more oxidative stress than those with immediately controlled diabetes, measured via reduced levels of nitric oxide (NO) and reduced glutathione in the urine [38]. Human studies revealed similar outcomes, though, the negative effects of oxidative stress on endothelial cell function are reversible in patients with initial HbA1c <7% with glycaemic control and use of anti-oxidants—while damage is irreversible if initial HbA1c is >7% [32, 33]. This is due to cytoplasmic reactive oxygen damage upon mitochondrial function and structure, leading to instability of electron transport chain (ETC), thus, cyclically higher levels of oxidative stress.
Long-standing high plasma glucose levels are associated with excess production of reactive oxygen species (ROS), namely superoxide radicals that may be converted into other species, by mitochondrial ETC. Hyperglycaemic status leads to activation of glycolysis, formation of higher concentration of pyruvate, activation of tricarboxylic acid cycle and in turn higher levels of electron donors (nicotinamide adenine dinucleotide and flavine adenine dinucleotide). High rates of electron flux between ETC and electron donors lead to increase in the electrochemical gradient across inner mitochondrial membrane until a critical threshold, which leads to inhibition of complex III of ETC when reached. Electrons are accumulated in coenzyme Q, which are then transferred to molecular oxygen, leading to generation of superoxide radicals [39]. These changes are reversible with the inhibition of ETC or by the use of uncoupler molecules to eliminate electrochemical gradient across inner membrane [39, 40]. In addition, high cellular glucose levels lead to the inhibition of a glycolytic enzyme, glyceraldehyde 3-phosphate dehydrogenase, which leads to accumulation of upstream glycolytic intermediates. As a result, the protein kinase C (PKC) pathway, by formation of diacylglycerol, and the AGE products pathway, by the formation of main precursor called methylglyoxal, are activated.
In addition to increased ROS, many other pathways are used to describe biochemical changes occurring in hyperglycaemic states including the activation of the polyol pathway, AGEs, PKC pathway and hexosamine pathways. Activation of the polyol pathway, primarily the enzyme aldose reductase that converts glucose into sorbitol in the presence of nicotinamide adenine dinucleotide phosphate (NADPH), leads to depletion of NADPH stores of cells and may enhance the pre-existing status of oxidative stress observed in hyperglycaemia [41, 42]. However, in most cases of hyperglycaemia, the polyol pathway is not assumed to play major role since the requirement for cellular glucose concentration is too high according to the enzyme dynamics [43, 44].
Activation of the PKC pathway decreases NO production in vascular smooth muscle cells by inhibition of endothelial NO synthase, increase in the expression of transforming growth factor (TGF)-β1, NF-κB and plasminogen activator inhibitor-1, and increase in fatty acid oxidation in vascular endothelial cells, which may cause atherosclerosis [45–49]. Furthermore, increase in the expression of TGF-α, TGF-β1 and plasminogen activator inhibitor-1 is enhanced by the increase in O-GlcNAcylation of the transcription factor Sp1, as a result of activated hexosamine pathway [50, 51].
Increased levels of AGE in circulation leading to increased binding of AGE to its receptor referred to as RAGE result in increased expression of NF-κB, vascular cell adhesion molecule-1, trombomodulin, tissue factor, MCP-1 and vascular endothelial growth factor, and increased levels of ROS formation [52–58]. RAGE–NF-κB interaction has shown to be involved in the development of diabetic neuropathy and atherosclerosis as complications. Potential early biomarkers are summarized in Table 1 [59, 60].
Table 1.
Possible early biomarkers and alterations in patients with DM
| Possible early biomarker categories | Examples |
|---|---|
| miRNA | Upregulation of expression of certain miRNA:
|
Downregulation of expression of certain miRNA:
|
|
| lncRNA | Upregulation of expression of certain lncRNA:
|
Downregulation of expression of certain lncRNA:
|
|
| Signaling pathways | Over-activation of certain pathways:
|
| DNA methylation/demethylation | Hypomethylation at 3-UTR of TXNIP |
| Histone acetylation/deacetylation | Upregulation of certain regulators:
|
| Enzymatic control | Activation of certain enzymes:
|
Inhibition of certain enzymes:
|
|
| Upregulated molecules | Certain connective tissue regulators:
|
Biochemical outcomes:
|
|
Pro-inflammatory cytokines:
|
|
| Downregulated molecules |
|
NF-κB, nuclear factor-kappa light chain enhancer of activated B cells; CXCR4, C-X-C chemokine receptor Type 4; ncRNA, non-coding RNA; 3-UTR, three prime untranslated region.
GENETIC AND EPIGENETIC CHANGES
Genetics underpin maturity-onset diabetes of the young (MODY), and the development of Types I and II DM, with mutations at HNF4-alpha (MODY 1), glucokinase (MODY 2), HNF1-alpha (MODY 3) and HNF1-beta (MODY 4) [61]. On the other hand, epigenetic shifts (same DNA, different expression) likely play a more important role in metabolic memory. The main types of epigenetic modifications are DNA methylation, histone modifications, chromatin remodeling, non-coding ribonucleic acids (RNAs) as microRNA (miRNA), and lncRNA and RNA editing (Figure 1).
DNA methylation and histone acetylation, the two primary epigenetic mechanisms, are highly investigated in relation to their roles in metabolic memory in diabetic patients [62]. Monocyte and lymphocyte DNA analysis from patients in the DCCT and The Epidemiology of Diabetes Interventions and Complications (EDIC) trials showed elevated levels of H3k9ac, involved in the activation of affiliated genes, in many inflammatory genes in patients not receiving intensive therapy initially [63]. Another study demonstrated correlation between HbA1c levels and H3K9ac, thus, H3K9ac is one of the key contributors to metabolic memory. Another study utilizing both the initial and 17-year follow-up whole blood sample and monocytes of the participants of DCCT and EDIC trials demonstrated many hypo/hypermethylated DNA regions, for example, hypomethylation at the 3-untranslated region (UTR) of Thioredoxin-Interacting Protein (TXNIP) encoding thioredoxin-interacting protein, to be associated with diabetic complications [64]. Individualized analysis of the same data in accordance with the HbA1c levels during follow-ups revealed the persistent hypomethylation of 3′-UTR of the TXNIP gene, thus, epigenetic alterations of the TXNIP gene appear to be strongly related to metabolic memory [64]. In addition, a similar pattern has been observed in other studies in patients with high plasma glucose levels and hyperlipidaemia [65]. miRNAs, 20–25 nucleotides in length, are thought to regulate up to 60% of gene translation by binding to the 3′-UTR region of specific mRNAs. miRNAs are synthesized as a primary transcript by RNA polymerase II, the same enzyme involved in mRNA synthesis, and processed by Drosha-DGCR8 in the nucleus and Dicer in the cytoplasm into mature miRNA [66]. Despite being a highly investigated topic for many chronic diseases, the applicability to human diseases remains forthcoming with few actual human studies [67]. The main findings so far are in mice, where expression levels of five miRNAs (miR-192, miR-200b, miR-200c, miR-216a and miR-217) are enhanced in renal glomeruli in the early stages of induced-diabetic states compared with control [68]. Among those miRNA, miR-192 appeared to be the key regulator since it upregulated expression of the others and was involved in the down-regulation of mRNAs involved in collagen and ECM protein synthesis [69]. In addition, mice treated with miR-192 inhibitors and miR-192 knockout mice progressed to diabetic nephropathy more slowly, while miR-192 gene amplification resulted in glomerular basement membrane hypertrophy and increased accumulation of ECM proteins including collagen [70]. Similar expression status can be achieved through treatment with TGF-β1, thus, effects of hyperglycaemia appear to be mediated through TGF-β1 expression at the genomic level [71–75]. Other highly investigated miRNA changes include upregulation of miR-21 and miR-377 and downregulation of miR-29, all of which lead to ECM hypertrophy and accumulation of ECM proteins through TGF-β1, Akt and mammalian target of rapamycin (mTOR) signalling [76–83]. Another crucial miRNA involved in the pathogenesis of hyperglycaemia is downregulation of Let-7 family members that inhibit collagen production and TGF-β1 expression
miRNAs may be the treatment biomarkers of choice, with high specificity for downstream morbidities. A study of 700 miRNAs in 40 patients with Type I DM with chronic kidney disease (CKD) has been undertaken with this goal [84], and TGF-β1-regulated miRNAs and tissue-specific miRNA expression patterns have been proposed as biomarkers [68, 85–88].
Long non-coding RNA (lnc-RNA) are comprised of >100 nucleotides and regulate transcription and translation via histone modifications or regulating miRNAs [89]. Two miRNAs upregulated during hyperglycaemia and TGF-β1 signalling (miR-216a and miR-217) are regulated along with lncRNA RP23-298H6.1-001 [90, 91]. Similarly, coregulation of lncRNA CJ241444 and miR-192 has been reported. Diabetic nephropathy in mice is associated with downregulation of lncRNA Lin01619, and upregulation of both lncRNA Erbb4-IRand lncRNA Tug1 [92–97]. lncRNA NR_033515 upregulation is correlated with diabetic nephropathy and may be a treatment biomarker [98–100].
CLINICAL IMPLICATIONS
Obesity
Obesity is defined as a body mass index >30 kg/m2, and affects more than one-third of men and women in the USA. It is accompanied by various comorbidities including Type II DM, metabolic syndrome, hypertension, dyslipidaemia, obstructive sleep apnoea, hyperuricaemia CVD and strokes [101]. The concept of genetic background in obesity was first hypothesized by Von Noorden in 1907, while mutations in leptin, leptin receptor, pro-opiomelanocortin, melanocortin 4 receptor, proconvertase-1/2, Neurotrophic Receptor Tyrosine Kinase 2 (NTRK2)/Brain-derived neurotrophic factor (BDNF) and many others have been identified since then as a genetic aetiology [102, 103]. Studies performed on a Dutch cohort comprised participants exposed to Dutch famine demonstrated DNA hypomethylation at the IGF2 gene [104]. Similar outcomes have been observed in the Leningrad siege study, where participants were exposed to prenatal undernutrition and postnatal shortage of food supply [105]. Remarkable upregulation of genes involved in energy metabolism, inflammatory responses and cell growth/death via epigenetic mechanisms have been recorded on genome-wide association studies and epigenome studies. Human studies have demonstrated inverse correlation between the length of breast-feeding and DNA methylation levels of LEP promoter, a gene associated with low levels of plasma leptin and infant body mass index [106–108]. Histological, genetic, biochemical and epigenetic alterations have importance in the emergence and progression of obesity.
Parental nutritional status and hyperglycaemic exposure may be reflected in utero with an epigenetic signature, termed the Barker hypothesis. Association of high-fat maternal diet, low birthweight, short stature, hyperglycaemic state and undernutrition with epigenetic changes leading to obesity, insulin resistance and hypertension have been made. High-fat maternal diet leads to DNA hypermethylation at adipocytokine and leptin genes, leading to down regulation of those genes [109–117]. These changes persist over generations despite normal fat diet in the offspring and are not reversible with switches to normal fat diet prior to and during pregnancy [104, 118, 119]. Interestingly, paternal high fat diet also increases the risk of insulin resistance in the offspring. However, it is important to note that all studies are performed with animals in the absence of human studies, and there is uncertainty about their validity in humans. Similar effects are also indicated in cases of gestational DM and hyperglycaemia [120–123]. In addition to DNA hypermethylation of the adipocytokine and leptin genes, hypomethylation of mesoderm-specific transcript gene (MEST) and hypermethylation of ATP-binding cassette transporter A1 (ABCA1) gene may occur in response to those stimuli [121, 122, 124–126]. Even though those epigenetic changes have not been proven in human subjects yet, significant correlation between those epigenetic markers, including hypomethylation of MEST and obesity, has been reported [127].
Hypothalamic control of obesity through various genes involved in appetite and energy metabolism have been proposed as potential epigenetic alteration sites, although no human studies have been conducted yet [128].
CKD
CKD most commonly results from diabetes and hypertension, affects more than a tenth of the world population and causes significant morbidity and mortality in adults [129]. End-stage renal disease (ESRD) is defined by histopathological findings including fibrosis, chronic inflammatory infiltrates, sclerosis of the glomeruli and hyaline obliteration. Certain prenatal conditions can predispose to ESRD by decreasing the number of functional nephrons, including prematurity, low birthweight, placental insufficiency and maternal diabetes, smoking, alcohol abuse, drugs and hypoproteinaemia, may affect the nephron number [130–135].
Transforming growth factor beta-1 (TGFβ-1)/SMAD signalling is crucial in the histopathological changes including ECM accumulation in CKD and renal fibrosis, as with diabetic nephropathy. In addition to the number of nephrons, epigenetic alterations play a crucial role in CKD development and progression.
Renal fibrosis is related to the activation of histone acetyltransferase p300/CBP-associated factor, involved in the activation of pro-inflammatory molecules such as intercellular adhesion molecule-1 and vascular cell adhesion molecule-1, decreased histone acetylation at genes (HDAC1/2/5/6), involved in the downregulation of renal protective molecule BMP7, and upregulation of transcription factor ETS-1 (ETS-1 gene) via histone acetylation [136–139]. Furthermore, production of hypoxia-inducible factor 1 in cases with hypoxia results in the activation of histone demethylases mostly in genes involved in TGF-β1 pathway [140].
Increase in the production of Angiotensin Converting Enzyme-1 (ACE-1) in animal models in response to hypertension results in the enhancement of activatory epigenetic changes such as H3KAc and H3K4me and decline in repressive epigenetic modifications such as H3K9me2 at ACE promoter, thus inducing its over-production and a vicious cycle [141, 142]. Moreover, maternal low protein diet has been linked to ACE-1 overexpression.
As a model of RNA interference, regulation of miRNAs has been in the centre of epigenetics of CKD since animal studies involving downregulation of Dicer, the key enzyme in miRNA processing, were shown to cause to renal failure and death. miR-21, miR-29 and miR-200, downstream molecules in TGF-β signalling, have been shown to be important in renal fibrosis by pro-fibrotic effects of miR-21 via amplification of TGF-β signalling and anti-fibrotic effects of miR-29 and miR-200 via the inhibition of Epithelial to masenchymal transition (EMT). The latest studies identified other miRNAs with significant renal functions as miR-205, involved in apoptosis, and miR-146a, involved in ischaemia–reperfusion injury [80, 143–149].
CVD
CVD is the leading cause of morbidity and mortality in developed countries, developing as a consequence of either an embryological, anatomical, environmental and/or genetic conditions [150]. Despite the high prevalence and risk for mortality and morbidity in CVD, mostly secondary to ischaemic heart diseases, treatment modalities have limited influence on the progression of disease with significant efficiency and adverse reaction issues. Resistance to commonly prescribed coronary artery disease (CAD) medications such as nitrates, clopidogrel and aspirin have been reported [151–153]. The most common aetiologies of chronic heart diseases include atherosclerosis, dyslipidaemia and diabetes, all of which increases risk for vascular plaque formation and thrombosis. Similar histological and epigenetic alterations with diabetic patients have been reported in patients with chronic CVD.
DNA methylation levels vary significantly with gender, race and risk factors associated with CAD, while critical DNA methylation differences have been detected between atherosclerosis-prone and atherosclerosis-resistant arteries in the same CAD patients [154]. Alu and LINE-1, both retro-transposable elements involved in genomic rearrangements and control of genomic expression, are two commonly hypomethylated genes in patients with CAD. Also, certain CAD risk factors are proven to lead hypomethylation in those genes such as homocysteine, race, higher body mass index (for LINE-1) and height (for Alu), whereas no significant variation with gender or age has been determined [155–157]. Many other relevant genes have been identified, for example, ABCA1 methylation is highly correlated with atherosclerosis levels and is an important determinant of plasma high-density lipoprotein-cholesterol (HDL-C) levels [158]. The methylation status of ABCA1 has been considered a biomarker for early stages of atherosclerosis in patients with no other clinical or laboratory findings [159]. Additionally, methylation of forkhead box P3 (FOXP3) by mediating unstability in Treg cells and methylation at MMP-9, Collagen alpha-1(XIV) chain and INK4B (INK4B gene) have been proposed as candidates [160–163].
Nutritional intake as well as the medications used in the treatment of CAD influences the DNA methylation levels. Among these, homocysteine intake has crucial importance since it can be transformed into S-adenosyl methionine in the body, which is the primary methyl donor in biochemical pathways including DNA methylation [164]. Maternal nutritional status is an independent determinant of postnatal DNA methylation status of infants [165]. Age is another factor that leads to alterations in DNA methylation for which three age-related CpG islands have been identified, namely the genes ITGA2B, ASPA and PDE4C. Studies determining age by using the methylation status of those CpG islands have <5 years difference with the actual age [166]. To discriminate age and CAD-associated DNA methylation/hypomethylation another gene, ANGPTL2, has been proposed that has age-specific methylation status while elevated levels of methylation are observed in patients with CAD [167].
Although histone modifications have not been investigated in detail so far, few possible targets have been identified as downregulation of HDAC4, an enzyme that deacetylase histone molecules to alter gene expression levels. Over-expression of HDAC4 is known to induce mitochondrial dysfunction and apoptosis, while down-regulation has been shown to induce cardiac muscle and ECM hypertrophy and provides protection against oxidative stress and mitochondrial damage [168, 169].
On the other hand, miRNA in the context of CAD emerges as a possible site for earlier diagnosis and for intervention in order to postpone disease onset and progression. miR-1 and miR-133, which are involved in the control of cell cycle and death, are significantly elevated in plasma and urine in patients with acute coronary syndrome [170, 171]. Elevations occur in the first 2 h of acute coronary syndrome, especially ST elevated myocardial infarction, and positively correlated with other commonly used cardiac markers such as CK-MB and troponin [170–172]. In addition, miR-208 is a cardiac-specific miRNA only expressed in cardiac tissue and only elevated in serum in cases of acute heart diseases [173, 174]. Elevated levels of miR-208 in plasma have high sensitivity and specificity for acute coronary syndrome [175]. miR-126, miR-132, miR-146 and miR-499 have been proposed as candidates in CVD to have importance diagnostically or prognostically [176–182]. The role of lncRNA is unclear in the development and progression of CVD due a scarcity of studies, though lncRNA necrosis-related factor, carcinoma-associated 1 (UCA1) and LIPCAR may be involved [96, 183, 184].
FUTURE DIRECTIONS
Multimodal diabetic complication management ranges from lifestyle modifications, to drugs, to surgeries such as photocoagulation/vitrectomy for diabetic retinopathy. Complications are usually a late finding, and intervention at this stage has been met with limited success overall. However, cognizance of metabolic memory, and intervention on the metabolic memory, may open up new treatment pathways [185].
Epigenetic alterations have been utilized in the treatment of oncological, autoimmune and neurological diseases with promising outcomes, such as four histone deacetylase inhibitors (vorinostat, romidepsin, panobinostat and belinostat) and two DNA methylation inhibitors (azacytidine and decitabine) that are approved by the Food and Drug Administration (FDA) [186, 187]. After detection of downregulation of the targets of transcription factor EP300-related genes via computed analysis of microarray data from many studies, vascular endothelial cells of patients with DM have been treated with EP300 inhibitors and HDAC inhibitors experimentally [188, 189]. In addition to the results of that cell culture study, animal studies investigating the efficiency of EP300 or HDAC inhibitors in DM are promising. A class III HDAC [Sirtuin-1 (SIRT1)] inhibitor and HMT EZH2 inhibitors demonstrated promising outcomes in animal models [190–193]. Additionally, a bromodomain-containing protein, JQ1, which blocks the expression of Angiotensin-II related genes, is successful in mice studies to reverse AngII-induced hypertension in animal models [194]. miRNAs are another possible site for intervention. Mice with induced DM that are treated with miR-192 inhibitor illustrated downregulation of many miRNAs such as miR-192 itself and others (miR-200, miR-216a and miR-217, all of which are involved in DM) and expression of p53, thus reducing the risk for diabetic complications, mostly diabetic nephropathy. Other studies investigating the inhibition of miR-29c, miR-21 and miR-192 also demonstrate success [76, 77]. lncRNA, namely lnc-MGC, has also been investigated as a target in DM, while studies regarding that topic are limited (Table 2).
Table 2.
Possible novel therapeutic approaches proposed and/or investigating and targeting genetic and epigenetic modifications in DM
| Possible target for therapeutic approaches | Examples |
|---|---|
| miRNA |
|
| lncRNA | lnc-MGC |
| Histone acetylation/deacetylation |
|
| DNA methylation/demethylation |
|
| Genome editing | Not yet been investigated |
EP300, Adenovirus early region 1 A-associated protein p300.
As we develop a better understanding of genetic and epigenetic changes in chronic diseases including DM, CVD, CKD, metabolic syndrome and many others, genome editing may become a useful tool for treatment. Currently available techniques of genome editing include zinc-finger nucleases, first targeted nuclease developed by the fusion of FokI restriction endonuclease and custom-designed zinc finger domain, transcription activator-like effector nuclease, effector proteins derived from plant pathogenic bacteria genus Xanthomonas, homing endonuclease, referring to a number of endonucleases synthesized from the regions within introns and hydrolyzing genomic DNA within the same cell, and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9, the most commonly utilized method in gene editing [195–200]. In bacteria, the Type II CRISPR system is a protection mechanism against foreign DNA such as viruses and plasmids and provides some type of memory and adaptive immunity via RNA-guided DNA cleavage [201, 202].
CONCLUSION
Non-communicable cardio-metabolic chronic diseases are now a global health issue, spanning DM, CVD and CKD. Prevention strategies remain inadequate and the management of complications has not proven completely effective, so novel strategies are needed to approach these conditions. Metabolic memory, the sum of histological, biochemical, genetic, epigenetic and cellular alterations in response to specific stimuli including hyperglycaemia, dyslipidaemia or high blood pressure, have been identified and may provide new surrogate biomarkers for early detection and treatment response, as well as new targets for novel therapeutic actions.
ACKNOWLEDGEMENTS
M.K. gratefully acknowledges use of the services and facilities of the Koc University Research Center for Translational Medicine (KUTTAM), funded by the Presidency of Turkey, Presidency of Strategy and Budget. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Presidency of Strategy and Budget.
FUNDING
This study was not funded by any grant.
CONFLICT OF INTEREST STATEMENT
All authors declare that they have no conflict of interest.
ETHICAL APPROVAL
This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
- 1.Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017; 390: 1151–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamm NC, Pelletier L, Ellison J et al. Trends in chronic disease incidence rates from the Canadian chronic disease surveillance system. Health Promot Chronic Dis Prev Can 2019; 39: 216–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jing J, Gao Y. Urine biomarkers in the early stages of diseases: current status and perspective. Discov Med 2018; 25: 57–65 [PubMed] [Google Scholar]
- 4.Engerman RL, Kern TS. Progression of incipient diabetic retinopathy during good glycemic control. Diabetes 1987; 36: 808–812 [DOI] [PubMed] [Google Scholar]
- 5.Berezin A. Metabolic memory phenomenon in diabetes mellitus: achieving and perspectives. Diabetes Metab Syndr 2016; 10: S176–S183 [DOI] [PubMed] [Google Scholar]
- 6.Zimmet P, Alberti KG, Magliano DJ et al. Diabetes mellitus statistics on prevalence and mortality: facts and fallacies. Nat Rev Endocrinol 2016; 12: 616–622 [DOI] [PubMed] [Google Scholar]
- 7.American Diabetes Association. Economic costs of diabetes in the U.S. in 2017. Diabetes Care 2018; 41: 917–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bourne RR, Stevens GA, White RA et al. Causes of vision loss worldwide, 1990–2010: a systematic analysis. Lancet Glob Health 2013; 1: e339–e349 [DOI] [PubMed] [Google Scholar]
- 9.Nathan DM, Genuth S, Lachin J et al. ; Diabetes Control and Complications Trial Research Group. The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med 1993; 329: 977–986 [DOI] [PubMed] [Google Scholar]
- 10.Effect of intensive therapy on the microvascular complications of type 1 diabetes mellitus. JAMA 2002; 287: 2563–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nathan DM, Cleary PA, Backlund JY et al. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes. N Engl J Med 2005; 353: 2643–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Intensive diabetes treatment and cardiovascular outcomes in type 1 diabetes: the DCCT/EDIC study 30-year follow-up. Diabetes Care 2016; 39: 686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nathan DM, Lachin J, Cleary P et al. ; Epidemiology of Diabetes Interventions and Complications Research Group. Intensive diabetes therapy and carotid intima-media thickness in type 1 diabetes mellitus. N Engl J Med 2003; 348: 2294–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy: the Epidemiology of Diabetes Interventions and Complications (EDIC) study. JAMA 2003; 290: 2159–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Boer IH, Sun W, Cleary PA et al. Intensive diabetes therapy and glomerular filtration rate in type 1 diabetes. N Engl J Med 2011; 365: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holman RR, Paul SK, Bethel MA et al. 10-year follow-up of intensive glucose control in type 2 diabetes. N Engl J Med 2008; 359: 1577–1589 [DOI] [PubMed] [Google Scholar]
- 17.Holman RR, Paul SK, Bethel MA et al. Long-term follow-up after tight control of blood pressure in type 2 diabetes. N Engl J Med 2008; 359: 1565–1576 [DOI] [PubMed] [Google Scholar]
- 18.Reaven PD, Moritz TE, Schwenke DC et al. ; for the Veterans Affairs Diabetes Trial. Intensive glucose-lowering therapy reduces cardiovascular disease events in veterans affairs diabetes trial participants with lower calcified coronary atherosclerosis. Diabetes 2009; 58: 2642–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duckworth W, Abraira C, Moritz T et al. Glucose control and vascular complications in veterans with type 2 diabetes. N Engl J Med 2009; 360: 129–139 [DOI] [PubMed] [Google Scholar]
- 20.Najafian B, Alpers CE, Fogo AB. Pathology of human diabetic nephropathy. Contrib Nephrol 2011; 170: 36–47 [DOI] [PubMed] [Google Scholar]
- 21.Lechner J, O’Leary OE, Stitt AW. The pathology associated with diabetic retinopathy. Vision Res 2017; 139: 7–14 [DOI] [PubMed] [Google Scholar]
- 22.Roy S, Sala R, Cagliero E et al. Overexpression of fibronectin induced by diabetes or high glucose: phenomenon with a memory. Proc Natl Acad Sci USA 1990; 87: 404–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugliese G, Pricci F, Pugliese F et al. Mechanisms of glucose-enhanced extracellular matrix accumulation in rat glomerular mesangial cells. Diabetes 1994; 43: 478–490 [DOI] [PubMed] [Google Scholar]
- 24.McLennan SV, Martell SK, Yue DK. Effects of mesangium glycation on matrix metalloproteinase activities: possible role in diabetic nephropathy. Diabetes 2002; 51: 2612–2618 [DOI] [PubMed] [Google Scholar]
- 25.Thompson JA, Webb RC. Potential role of toll-like receptors in programming of vascular dysfunction. Clin Sci (Lond) 2013; 125: 19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka M, Masuda S, Matsuo Y et al. Hyperglycemia and inflammatory property of circulating monocytes are associated with inflammatory property of carotid plaques in patients undergoing carotid endarterectomy. J Atheroscler Thromb 2016; 23: 1212–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guarner V, Rubio-Ruiz ME. Low-grade systemic inflammation connects aging, metabolic syndrome and cardiovascular disease. Interdiscip Top Gerontol 2015; 40: 99–106 [DOI] [PubMed] [Google Scholar]
- 28.Johnson AR, Milner JJ, Makowski L. The inflammation highway: metabolism accelerates inflammatory traffic in obesity. Immunol Rev 2012; 249: 218–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vigorelli V, Resta J, Bianchessi V et al. Abnormal DNA methylation induced by hyperglycemia reduces CXCR 4 gene expression in CD 34(+) stem cells. J Am Heart Assoc 2019; 8: e010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durning SP, Preston-Hurlburt P, Clark PR et al. The receptor for advanced glycation endproducts drives T cell survival and inflammation in type 1 diabetes mellitus. J Immunol 2016; 197: 3076–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prattichizzo F, De Nigris V, Spiga R et al. Inflammageing and metaflammation: the yin and yang of type 2 diabetes. Ageing Res Rev 2018; 41: 1–17 [DOI] [PubMed] [Google Scholar]
- 32.Ceriello A, Esposito K, Ihnat M et al. Long-term glycemic control influences the long-lasting effect of hyperglycemia on endothelial function in type 1 diabetes. J Clin Endocrinol Metab 2009; 94: 2751–2756 [DOI] [PubMed] [Google Scholar]
- 33.Ceriello A, Esposito K, Ihnat M et al. Effect of acute hyperglycaemia, long-term glycaemic control and insulin on endothelial dysfunction and inflammation in Type 1 diabetic patients with different characteristics. Diabet Med 2009; 27: 911–917 [DOI] [PubMed] [Google Scholar]
- 34.Kanbay M, Yerlikaya A, Sag AA et al. A journey from microenvironment to macroenvironment: the role of metaflammation and epigenetic changes in cardiorenal disease. Clin Kidney J 2019; 12: 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y, Bielohuby M, Fleming T et al. Dietary sugars, not lipids, drive hypothalamic inflammation. Mol Metab 2017; 6: 897–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Souza CT, Araujo EP, Bordin S et al. Consumption of a fat-rich diet activates a proinflammatory response and induces insulin resistance in the hypothalamus. Endocrinology 2005; 146: 4192–4199 [DOI] [PubMed] [Google Scholar]
- 37.Saukkonen T, Mutt SJ, Jokelainen J et al. Adipokines and inflammatory markers in elderly subjects with high risk of type 2 diabetes and cardiovascular disease. Sci Rep 2018; 8: 12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du XL, Edelstein D, Dimmeler S et al. Hyperglycemia inhibits endothelial nitric oxide synthase activity by posttranslational modification at the Akt site. J Clin Invest 2001; 108: 1341–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trumpower BL. The protonmotive Q cycle. Energy transduction by coupling of proton translocation to electron transfer by the cytochrome bc1 complex. J Biol Chem 1990; 265: 11409–11412 [PubMed] [Google Scholar]
- 40.Nishikawa T, Edelstein D, Du XL et al. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature 2000; 404: 787–790 [DOI] [PubMed] [Google Scholar]
- 41.Chung SS, Ho EC, Lam KS et al. Contribution of polyol pathway to diabetes-induced oxidative stress. J Am Soc Nephrol 2003; 14 (8 Suppl 3): S233–S236 [DOI] [PubMed] [Google Scholar]
- 42.Lee AY, Chung SS. Contributions of polyol pathway to oxidative stress in diabetic cataract. FASEB J 1999; 13: 23–30 [DOI] [PubMed] [Google Scholar]
- 43.Ii S, Ohta M, Kudo E et al. Redox state-dependent and sorbitol accumulation-independent diabetic albuminuria in mice with transgene-derived human aldose reductase and sorbitol dehydrogenase deficiency. Diabetologia 2004; 47: 541–548 [DOI] [PubMed] [Google Scholar]
- 44.Bohren KM, Grimshaw CE, Gabbay KH. Catalytic effectiveness of human aldose reductase. Critical role of C-terminal domain. J Biol Chem 1992; 267: 20965–20970 [PubMed] [Google Scholar]
- 45.Craven PA, Studer RK, Felder J et al. Nitric oxide inhibition of transforming growth factor-beta and collagen synthesis in mesangial cells. Diabetes 1997; 46: 671–681 [DOI] [PubMed] [Google Scholar]
- 46.Kikkawa R, Haneda M, Uzu T et al. Translocation of protein kinase C alpha and zeta in rat glomerular mesangial cells cultured under high glucose conditions. Diabetologia 1994; 37: 838–841 [DOI] [PubMed] [Google Scholar]
- 47.Feener EP, Xia P, Inoguchi T et al. Role of protein kinase C in glucose- and angiotensin II-induced plasminogen activator inhibitor expression. Contrib Nephrol 1996; 118: 180–187 [DOI] [PubMed] [Google Scholar]
- 48.Williams B, Gallacher B, Patel H et al. Glucose-induced protein kinase C activation regulates vascular permeability factor mRNA expression and peptide production by human vascular smooth muscle cells in vitro. Diabetes 1997; 46: 1497–1503 [DOI] [PubMed] [Google Scholar]
- 49.Kuboki K, Jiang ZY, Takahara N et al. Regulation of endothelial constitutive nitric oxide synthase gene expression in endothelial cells and in vivo: a specific vascular action of insulin. Circulation 2000; 101: 676–681 [DOI] [PubMed] [Google Scholar]
- 50.Du XL, Edelstein D, Rossetti L et al. Hyperglycemia-induced mitochondrial superoxide overproduction activates the hexosamine pathway and induces plasminogen activator inhibitor-1 expression by increasing Sp1 glycosylation. Proc Natl Acad Sci USA 2000; 97: 12222–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen YQ, Su M, Walia RR et al. Sp1 sites mediate activation of the plasminogen activator inhibitor-1 promoter by glucose in vascular smooth muscle cells. J Biol Chem 1998; 273: 8225–8231 [DOI] [PubMed] [Google Scholar]
- 52.Pieper GM, Riaz Ul H. Activation of nuclear factor-kappaB in cultured endothelial cells by increased glucose concentration: prevention by calphostin C. J Cardiovasc Pharmacol 1997; 30: 528–532 [DOI] [PubMed] [Google Scholar]
- 53.Yerneni KK, Bai W, Khan BV et al. Hyperglycemia-induced activation of nuclear transcription factor kappaB in vascular smooth muscle cells. Diabetes 1999; 48: 855–864 [DOI] [PubMed] [Google Scholar]
- 54.Vlassara H, Fuh H, Donnelly T et al. Advanced glycation endproducts promote adhesion molecule (VCAM-1, ICAM-1) expression and atheroma formation in normal rabbits. Mol Med 1995; 1: 447–456 [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt AM, Hori O, Chen JX et al. Advanced glycation endproducts interacting with their endothelial receptor induce expression of vascular cell adhesion molecule-1 (VCAM-1) in cultured human endothelial cells and in mice. A potential mechanism for the accelerated vasculopathy of diabetes. J Clin Invest 1995; 96: 1395–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt AM, Crandall J, Hori O et al. Elevated plasma levels of vascular cell adhesion molecule-1 (VCAM-1) in diabetic patients with microalbuminuria: a marker of vascular dysfunction and progressive vascular disease. Br J Haematol 1996; 92: 747–750 [DOI] [PubMed] [Google Scholar]
- 57.Lu M, Kuroki M, Amano S et al. Advanced glycation end products increase retinal vascular endothelial growth factor expression. J Clin Invest 1998; 101: 1219–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirata C, Nakano K, Nakamura N et al. Advanced glycation end products induce expression of vascular endothelial growth factor by retinal Muller cells. Biochem Biophys Res Commun 1997; 236: 712–715 [DOI] [PubMed] [Google Scholar]
- 59.Soro-Paavonen A, Watson AM, Li J et al. Receptor for advanced glycation end products (RAGE) deficiency attenuates the development of atherosclerosis in diabetes. Diabetes 2008; 57: 2461–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bierhaus A, Haslbeck KM, Humpert PM et al. Loss of pain perception in diabetes is dependent on a receptor of the immunoglobulin superfamily. J Clin Invest 2004; 114: 1741–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Anik A, Catli G, Abaci A et al. Maturity-onset diabetes of the young (MODY): an update. J Pediatr Endocrinol Metab 2015; 28: 251–263 [DOI] [PubMed] [Google Scholar]
- 62.Miao F, Chen Z, Genuth S et al. Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes 2014; 63: 1748–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen Z, Miao F, Paterson AD et al. Epigenomic profiling reveals an association between persistence of DNA methylation and metabolic memory in the DCCT/EDIC type 1 diabetes cohort. Proc Natl Acad Sci USA 2016; 113: E3002–E3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shalev A. Minireview: thioredoxin-interacting protein: regulation and function in the pancreatic beta-cell. Mol Endocrinol 2014; 28: 1211–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walaszczyk E, Luijten M, Spijkerman AMW et al. DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case-control sample of the Lifelines study. Diabetologia 2018; 61: 354–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol 2018; 141: 1202–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kato M, Natarajan R. MicroRNAs in diabetic nephropathy: functions, biomarkers, and therapeutic targets. Ann NY Acad Sci 2015; 1353: 72–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Argyropoulos C, Wang K, McClarty S et al. Urinary microRNA profiling in the nephropathy of type 1 diabetes. PLoS One 2013; 8: e54662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kato M, Zhang J, Wang M et al. MicroRNA-192 in diabetic kidney glomeruli and its function in TGF-beta-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA 2007; 104: 3432–3437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Putta S, Lanting L, Sun G et al. Inhibiting microRNA-192 ameliorates renal fibrosis in diabetic nephropathy. J Am Soc Nephrol 2012; 23: 458–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deshpande SD, Putta S, Wang M et al. Transforming growth factor-beta-induced cross talk between p53 and a microRNA in the pathogenesis of diabetic nephropathy. Diabetes 2013; 62: 3151–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kato M, Putta S, Wang M et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat Cell Biol 2009; 11: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kato M, Dang V, Wang M et al. TGF-beta induces acetylation of chromatin and of Ets-1 to alleviate repression of miR-192 in diabetic nephropathy. Sci Signal 2013; 6: ra43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park JT, Kato M, Yuan H et al. FOG2 protein down-regulation by transforming growth factor-beta1-induced microRNA-200b/c leads to Akt kinase activation and glomerular mesangial hypertrophy related to diabetic nephropathy. J Biol Chem 2013; 288: 22469–22480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kato M, Wang L, Putta S et al. Post-transcriptional up-regulation of Tsc-22 by Ybx1, a target of miR-216a, mediates TGF-{beta}-induced collagen expression in kidney cells. J Biol Chem 2010; 285: 34004–34015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dey N, Das F, Mariappan MM et al. MicroRNA-21 orchestrates high glucose-induced signals to TOR complex 1, resulting in renal cell pathology in diabetes. J Biol Chem 2011; 286: 25586–25603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhong X, Chung AC, Chen HY et al. miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia 2013; 56: 663–674 [DOI] [PubMed] [Google Scholar]
- 78.Wang B, Komers R, Carew R et al. Suppression of microRNA-29 expression by TGF-beta1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol 2012; 23: 252–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen HY, Zhong X, Huang XR et al. MicroRNA-29b inhibits diabetic nephropathy in db/db mice. Mol Ther 2014; 22: 842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chau BN, Xin C, Hartner J et al. MicroRNA-21 promotes fibrosis of the kidney by silencing metabolic pathways. Sci Transl Med 2012; 4: 121ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qin W, Chung AC, Huang XR et al. TGF-beta/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J Am Soc Nephrol 2011; 22: 1462–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Q, Wang Y, Minto AW et al. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J 2008; 22: 4126–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Du B, Ma LM, Huang MB et al. High glucose down-regulates miR-29a to increase collagen IV production in HK-2 cells. FEBS Lett 2010; 584: 811–816 [DOI] [PubMed] [Google Scholar]
- 84.Satake E, Pezzolesi MG, Md Dom ZI et al. Circulating miRNA profiles associated with hyperglycemia in patients with type 1 diabetes. Diabetes 2018; 67: 1013–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pezzolesi MG, Satake E, McDonnell KP et al. Circulating TGF-beta1-regulated miRNAs and the risk of rapid progression to ESRD in type 1 diabetes. Diabetes 2015; 64: 3285–3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baker MA, Davis SJ, Liu P et al. Tissue-specific microRNA expression patterns in four types of kidney disease. J Am Soc Nephrol 2017; 28: 2985–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Higuchi C, Nakatsuka A, Eguchi J et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism 2015; 64: 489–497 [DOI] [PubMed] [Google Scholar]
- 88.Zampetaki A, Willeit P, Burr S et al. Angiogenic microRNAs linked to incidence and progression of diabetic retinopathy in type 1 diabetes. Diabetes 2016; 65: 216–227 [DOI] [PubMed] [Google Scholar]
- 89.Kumar MM, Goyal R. LncRNA as a therapeutic target for angiogenesis. Curr Top Med Chem 2017; 17: 1750–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kato M, Wang M, Chen Z et al. An endoplasmic reticulum stress-regulated lncRNA hosting a microRNA megacluster induces early features of diabetic nephropathy. Nat Commun 2016; 7: 12864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cabili MN, Trapnell C, Goff L et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011; 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leung A, Amaram V, Natarajan R. Linking diabetic vascular complications with LncRNAs. Vascul Pharmacol 2019; 114: 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Long J, Badal SS, Ye Z et al. Long noncoding RNA Tug1 regulates mitochondrial bioenergetics in diabetic nephropathy. J Clin Invest 2016; 126: 4205–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sun SF, Tang PMK, Feng M et al. Novel lncRNA Erbb4-IR promotes diabetic kidney injury in db/db mice by targeting miR-29b. Diabetes 2018; 67: 731–744 [DOI] [PubMed] [Google Scholar]
- 95.Li X, Zeng L, Cao C et al. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp Cell Res 2017; 350: 327–335 [DOI] [PubMed] [Google Scholar]
- 96.Liu Y, Zhou D, Li G et al. Long non coding RNA-UCA1 contributes to cardiomyocyte apoptosis by suppression of p27 expression. Cell Physiol Biochem 2015; 35: 1986–1998 [DOI] [PubMed] [Google Scholar]
- 97.Bai X, Geng J, Li X et al. Long noncoding RNA LINC01619 regulates microRNA-27a/Forkhead box protein O1 and endoplasmic reticulum stress-mediated podocyte injury in diabetic nephropathy. Antioxid Redox Signal 2018; 29: 355–376 [DOI] [PubMed] [Google Scholar]
- 98.Moran I, Akerman I, van de Bunt M et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab 2012; 16: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009; 23: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol 2011; 21: 354–361 [DOI] [PubMed] [Google Scholar]
- 101.Engin A. The definition and prevalence of obesity and metabolic syndrome. Adv Exp Med Biol 2017; 960: 1–17 [DOI] [PubMed] [Google Scholar]
- 102.Barker DJ. The fetal and infant origins of adult disease. BMJ 1990; 301: 1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kostovski M, Tasic V, Laban N et al. Obesity in childhood and adolescence, genetic fctors. Pril (Makedon Akad Nauk Umet Odd Med Nauki) 2017; 38: 121–133 [DOI] [PubMed] [Google Scholar]
- 104.Painter RC, Osmond C, Gluckman P et al. Transgenerational effects of prenatal exposure to the Dutch famine on neonatal adiposity and health in later life. BJOG 2008; 115: 1243–1249 [DOI] [PubMed] [Google Scholar]
- 105.Stanner SA, Bulmer K, Andres C et al. Does malnutrition in utero determine diabetes and coronary heart disease in adulthood? Results from the Leningrad siege study, a cross sectional study. BMJ 1997; 315: 1342–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Obermann-Borst SA, Eilers PH, Tobi EW et al. Duration of breastfeeding and gender are associated with methylation of the LEPTIN gene in very young children. Pediatr Res 2013; 74: 344–349 [DOI] [PubMed] [Google Scholar]
- 107.Sherwood WB, Bion V, Lockett GA et al. Duration of breastfeeding is associated with leptin (LEP) DNA methylation profiles and BMI in 10-year-old children. Clin Epigenet 2019; 11: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pauwels S, Symons L, Vanautgaerden EL et al. The influence of the duration of breastfeeding on the infant’s metabolic epigenome. Nutrients 2019; 11: 1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Masuyama H, Hiramatsu Y. Effects of a high-fat diet exposure in utero on the metabolic syndrome-like phenomenon in mouse offspring through epigenetic changes in adipocytokine gene expression. Endocrinology 2012; 153: 2823–2830 [DOI] [PubMed] [Google Scholar]
- 110.Whincup PH, Kaye SJ, Owen CG et al. Birth weight and risk of type 2 diabetes: a systematic review. JAMA 2008; 300: 2886–2897 [DOI] [PubMed] [Google Scholar]
- 111.Vanhees K, Vonhogen IG, van Schooten FJ et al. You are what you eat, and so are your children: the impact of micronutrients on the epigenetic programming of offspring. Cell Mol Life Sci 2014; 71: 271–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vickers MH, Breier BH, Cutfield WS et al. Fetal origins of hyperphagia, obesity, and hypertension and postnatal amplification by hypercaloric nutrition. Am J Physiol Endocrinol Metab 2000; 279: E83–E87 [DOI] [PubMed] [Google Scholar]
- 113.Bouchard L, Hivert MF, Guay SP et al. Placental adiponectin gene DNA methylation levels are associated with mothers' blood glucose concentration. Diabetes 2012; 61: 1272–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bouchard L, Thibault S, Guay SP et al. Leptin gene epigenetic adaptation to impaired glucose metabolism during pregnancy. Diabetes Care 2010; 33: 2436–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gould MM, Mohamed-Ali V, Goubet SA et al. Microalbuminuria: associations with height and sex in non-diabetic subjects. BMJ 1993; 306: 240–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rossing P, Tarnow L, Nielsen FS et al. Short stature and diabetic nephropathy. BMJ 1995; 310: 296–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rossing P, Tarnow L, Nielsen FS et al. Low birth weight: A risk factor for development of diabetic nephropathy? Diabetes 1995; 44: 1405–1407 [DOI] [PubMed] [Google Scholar]
- 118.Fu Q, Olson P, Rasmussen D et al. A short-term transition from a high-fat diet to a normal-fat diet before pregnancy exacerbates female mouse offspring obesity. Int J Obes 2016; 40: 564–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Heijmans BT, Tobi EW, Stein AD et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA 2008; 105: 17046–17049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Osmond DT, King RG, Brennecke SP et al. Placental glucose transport and utilisation is altered at term in insulin-treated, gestational-diabetic patients. Diabetologia 2001; 44: 1133–1139 [DOI] [PubMed] [Google Scholar]
- 121.Houde AA, Guay SP, Desgagne V et al. Adaptations of placental and cord blood ABCA1 DNA methylation profile to maternal metabolic status. Epigenetics 2013; 8: 1289–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.El Hajj N, Pliushch G, Schneider E et al. Metabolic programming of MEST DNA methylation by intrauterine exposure to gestational diabetes mellitus. Diabetes 2013; 62: 1320–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ruchat SM, Houde AA, Voisin G et al. Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics 2013; 8: 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Finer S, Mathews C, Lowe R et al. Maternal gestational diabetes is associated with genome-wide DNA methylation variation in placenta and cord blood of exposed offspring. Hum Mol Genet 2015; 24: 3021–3029 [DOI] [PubMed] [Google Scholar]
- 125.Binder AM, LaRocca J, Lesseur C et al. Epigenome-wide and transcriptome-wide analyses reveal gestational diabetes is associated with alterations in the human leukocyte antigen complex. Clin Epigenet 2015; 7: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rong C, Cui X, Chen J et al. DNA methylation profiles in placenta and its association with gestational diabetes mellitus. Exp Clin Endocrinol Diabetes 2015; 123: 282–288 [DOI] [PubMed] [Google Scholar]
- 127.Carless MA, Kulkarni H, Kos MZ et al. Genetic effects on DNA methylation and its potential relevance for obesity in Mexican Americans. PLoS One 2013; 8: e73950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yousheng J, Nguyen T, Desai M et al. Programmed alterations in hypothalamic neuronal orexigenic responses to ghrelin following gestational nutrient restriction. Reprod Sci 2008; 15: 702–709 [DOI] [PubMed] [Google Scholar]
- 129.Thomas MC, Cooper ME, Zimmet P. Changing epidemiology of type 2 diabetes mellitus and associated chronic kidney disease. Nat Rev Nephrol 2016; 12: 73–81 [DOI] [PubMed] [Google Scholar]
- 130.Brenner BM, Garcia DL, Anderson S. Glomeruli and blood pressure: Less of one, more the other. Am J Hypertens 1988; 1: 335–347 [DOI] [PubMed] [Google Scholar]
- 131.Zeman FJ. Effects of maternal protein restriction on the kidney of the newborn young of rats. J Nutr 1968; 94: 111–116 [DOI] [PubMed] [Google Scholar]
- 132.Toledo-Rodriguez M, Loyse N, Bourdon C et al. Effect of prenatal exposure to nicotine on kidney glomerular mass and AT1R expression in genetically diverse strains of rats. Toxicol Lett 2012; 213: 228–234 [DOI] [PubMed] [Google Scholar]
- 133.Smyth LJ, McKay GJ, Maxwell AP et al. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics 2014; 9: 366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.MacLennan NK, James SJ, Melnyk S et al. Uteroplacental insufficiency alters DNA methylation, one-carbon metabolism, and histone acetylation in IUGR rats. Physiol Genomics 2004; 18: 43–50 [DOI] [PubMed] [Google Scholar]
- 135.Gautier JF, Porcher R, Abi Khalil C et al. Kidney dysfunction in adult offspring exposed in utero to type 1 diabetes is associated with alterations in genome-wide DNA methylation. PLoS One 2015; 10: e0134654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang J, Wan D, Li J et al. Histone acetyltransferase PCAF regulates inflammatory molecules in the development of renal injury. Epigenetics 2015; 10: 62–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yuan H, Reddy MA, Sun G et al. Involvement of p300/CBP and epigenetic histone acetylation in TGF-beta1-mediated gene transcription in mesangial cells. Am J Physiol Renal Physiol 2013; 304: F601–F613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Marumo T, Hishikawa K, Yoshikawa M et al. Epigenetic regulation of BMP7 in the regenerative response to ischemia. J Am Soc Nephrol 2008; 19: 1311–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Marumo T, Hishikawa K, Yoshikawa M et al. Histone deacetylase modulates the proinflammatory and -fibrotic changes in tubulointerstitial injury. Am J Physiol Renal Physiol 2010; 298: F133–F141 [DOI] [PubMed] [Google Scholar]
- 140.Mimura I, Tanaka T, Nangaku M. Novel therapeutic strategy with hypoxia-inducible factors via reversible epigenetic regulation mechanisms in progressive tubulointerstitial fibrosis. Semin Nephrol 2013; 33: 375–382 [DOI] [PubMed] [Google Scholar]
- 141.Bogdarina I, Welham S, King PJ et al. Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circ Res 2007; 100: 520–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lee HA, Cho HM, Lee DY et al. Tissue-specific upregulation of angiotensin-converting enzyme 1 in spontaneously hypertensive rats through histone code modifications. Hypertension 2012; 59: 621–626 [DOI] [PubMed] [Google Scholar]
- 143.Nagalakshmi VK, Ren Q, Pugh MM et al. Dicer regulates the development of nephrogenic and ureteric compartments in the mammalian kidney. Kidney Int 2011; 79: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Matsuda S, Ichigotani Y, Okuda T et al. Molecular cloning and characterization of a novel human gene (HERNA) which encodes a putative RNA-helicase. Biochim Biophys Acta 2000; 1490: 163–169 [DOI] [PubMed] [Google Scholar]
- 145.Wilhide ME, Feller JD, Li B et al. Renal epithelial miR-205 expression correlates with disease severity in a mouse model of congenital obstructive nephropathy. Pediatr Res 2016; 80: 602–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang H, Zhang X, Yuan X et al. MicroRNA-205 inhibits renal cells apoptosis via targeting CMTM4. Iran J Basic Med Sci 2015; 18: 1020–1026 [PMC free article] [PubMed] [Google Scholar]
- 147.Muratsu-Ikeda S, Nangaku M, Ikeda Y et al. Downregulation of miR-205 modulates cell susceptibility to oxidative and endoplasmic reticulum stresses in renal tubular cells. PLoS One 2012; 7: e41462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Keller C, Kroening S, Zuehlke J et al. Distinct mesenchymal alterations in N-cadherin and E-cadherin positive primary renal epithelial cells. PLoS One 2012; 7: e43584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Amrouche L, Desbuissons G, Rabant M et al. MicroRNA-146a in human and experimental ischemic AKI: CXCL8-dependent mechanism of action. J Am Soc Nephrol 2017; 28: 479–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Di Lullo L, House A, Gorini A et al. Chronic kidney disease and cardiovascular complications. Heart Fail Rev 2015; 20: 259–272 [DOI] [PubMed] [Google Scholar]
- 151.Guirgis M, Thompson P, Jansen S. Review of aspirin and clopidogrel resistance in peripheral arterial disease. J Vasc Surg 2017; 66: 1576–1586 [DOI] [PubMed] [Google Scholar]
- 152.Helgason CM, Bolin KM, Hoff JA et al. Development of aspirin resistance in persons with previous ischemic stroke. Stroke 1994; 25: 2331–2336 [DOI] [PubMed] [Google Scholar]
- 153.Serebruany VL, Steinhubl SR, Berger PB et al. Variability in platelet responsiveness to clopidogrel among 544 individuals. J Am Coll Cardiol 2005; 45: 246–251 [DOI] [PubMed] [Google Scholar]
- 154.Weisenberger DJ, Campan M, Long TI et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005; 33: 6823–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wei L, Liu S, Su Z et al. LINE-1 hypomethylation is associated with the risk of coronary heart disease in Chinese population. Arq Bras Cardiol 2014; 102: 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yideng J, Jianzhong Z, Ying H et al. Homocysteine-mediated expression of SAHH, DNMTs, MBD2, and DNA hypomethylation potential pathogenic mechanism in VSMCs. DNA Cell Biol 2007; 26: 603–611 [DOI] [PubMed] [Google Scholar]
- 157.Perng W, Villamor E, Shroff MR et al. Dietary intake, plasma homocysteine, and repetitive element DNA methylation in the Multi-Ethnic Study of Atherosclerosis (MESA). Nutr Metab Cardiovasc Dis 2014; 24: 614–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Guay SP, Brisson D, Munger J et al. ABCA1 gene promoter DNA methylation is associated with HDL particle profile and coronary artery disease in familial hypercholesterolemia. Epigenetics 2012; 7: 464–472 [DOI] [PubMed] [Google Scholar]
- 159.Ma SC, Zhang HP, Kong FQ et al. Integration of gene expression and DNA methylation profiles provides a molecular subtype for risk assessment in atherosclerosis. Mol Med Rep 2016; 13: 4791–4799 [DOI] [PubMed] [Google Scholar]
- 160.Hansson GK. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med 2005; 352: 1685–1695 [DOI] [PubMed] [Google Scholar]
- 161.Huehn J, Polansky JK, Hamann A. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat Rev Immunol 2009; 9: 83–89 [DOI] [PubMed] [Google Scholar]
- 162.Jia L, Zhu L, Wang JZ et al. Methylation of FOXP3 in regulatory T cells is related to the severity of coronary artery disease. Atherosclerosis 2013; 228: 346–352 [DOI] [PubMed] [Google Scholar]
- 163.Zhuang J, Peng W, Li H et al. Methylation of p15INK4b and expression of ANRIL on chromosome 9p21 are associated with coronary artery disease. PLoS One 2012; 7: e47193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yu X, Ling W, Mi M. Relationship of impairment induced by intracellular S-adenosylhomocysteine accumulation with DNA methylation in human umbilical vein endothelial cells treated with 3-deazaadenosine. Int J Exp Pathol 2009; 90: 638–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dominguez-Salas P, Moore SE, Baker MS et al. Maternal nutrition at conception modulates DNA methylation of human metastable epialleles. Nat Commun 2014; 5: 3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Alisch RS, Barwick BG, Chopra P et al. Age-associated DNA methylation in pediatric populations. Genome Res 2012; 22: 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nguyen A, Mamarbachi M, Turcot V et al. Lower methylation of the ANGPTL2 gene in leukocytes from post-acute coronary syndrome patients. PLoS One 2016; 11: e0153920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kessler-Icekson G, Hochhauser E, Sinai T et al. A histone deacetylase inhibitory prodrug—butyroyloxymethyl diethyl phosphate—protects the heart and cardiomyocytes against ischemia injury. Eur J Pharm Sci 2012; 45: 592–599 [DOI] [PubMed] [Google Scholar]
- 169.Lee TM, Lin MS, Chang NC. Inhibition of histone deacetylase on ventricular remodeling in infarcted rats. Am J Physiol Heart Circ Physiol 2007; 293: H968– H977 [DOI] [PubMed] [Google Scholar]
- 170.Cheng Y, Tan N, Yang J et al. A translational study of circulating cell-free microRNA-1 in acute myocardial infarction. Clin Sci (Lond) 2010; 119: 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bauters C, Kumarswamy R, Holzmann A et al. Circulating miR-133a and miR-423-5p fail as biomarkers for left ventricular remodeling after myocardial infarction. Int J Cardiol 2013; 168: 1837–1840 [DOI] [PubMed] [Google Scholar]
- 172.Duan L, Xiong X, Liu Y et al. miRNA-1: functional roles and dysregulation in heart disease. Mol Biosyst 2014; 10: 2775–2782 [DOI] [PubMed] [Google Scholar]
- 173.Oerlemans MI, Mosterd A, Dekker MS et al. Early assessment of acute coronary syndromes in the emergency department: the potential diagnostic value of circulating microRNAs. EMBO Mol Med 2012; 4: 1176–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Cheng Y, Wang X, Yang J et al. A translational study of urine miRNAs in acute myocardial infarction. J Mol Cell Cardiol 2012; 53: 668–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xiao J, Shen B, Li J et al. Serum microRNA-499 and microRNA-208a as biomarkers of acute myocardial infarction. Int J Clin Exp Med 2014; 7: 136–141 [PMC free article] [PubMed] [Google Scholar]
- 176.Wang GK, Zhu JQ, Zhang JT et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J 2010; 31: 659–666 [DOI] [PubMed] [Google Scholar]
- 177.Ren J, Zhang J, Xu N et al. Signature of circulating microRNAs as potential biomarkers in vulnerable coronary artery disease. PLoS One 2013; 8: e80738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Olivieri F, Antonicelli R, Lorenzi M et al. Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int J Cardiol 2013; 167: 531–536 [DOI] [PubMed] [Google Scholar]
- 179.Long G, Wang F, Duan Q et al. Human circulating microRNA-1 and microRNA-126 as potential novel indicators for acute myocardial infarction. Int J Biol Sci 2012; 8: 811–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Guo M, Mao X, Ji Q et al. miR-146a in PBMCs modulates Th1 function in patients with acute coronary syndrome. Immunol Cell Biol 2010; 88: 555–564 [DOI] [PubMed] [Google Scholar]
- 181.Karakas M, Schulte C, Appelbaum S et al. Circulating microRNAs strongly predict cardiovascular death in patients with coronary artery disease-results from the large AtheroGene study. Eur Heart J 2017; 38: 516–523 [DOI] [PubMed] [Google Scholar]
- 182.Zhang L, Huang D, Wang Q et al. MiR-132 inhibits expression of SIRT1 and induces pro-inflammatory processes of vascular endothelial inflammation through blockade of the SREBP-1c metabolic pathway. Cardiovasc Drugs Ther 2014; 28: 303–311 [DOI] [PubMed] [Google Scholar]
- 183.Kumarswamy R, Bauters C, Volkmann I et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res 2014; 114: 1569–1575 [DOI] [PubMed] [Google Scholar]
- 184.Wang K, Liu F, Liu CY et al. The long noncoding RNA NRF regulates programmed necrosis and myocardial injury during ischemia and reperfusion by targeting miR-873. Cell Death Differ 2016; 23: 1394–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Reddy MA, Sumanth P, Lanting L et al. Losartan reverses permissive epigenetic changes in renal glomeruli of diabetic db/db mice. Kidney Int 2014; 85: 362–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Arrowsmith CH, Bountra C, Fish PV et al. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov 2012; 11: 384–400 [DOI] [PubMed] [Google Scholar]
- 187.Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet 2016; 17: 630–641 [DOI] [PubMed] [Google Scholar]
- 188.Rafehi H, Kaspi A, Ziemann M et al. Systems approach to the pharmacological actions of HDAC inhibitors reveals EP300 activities and convergent mechanisms of regulation in diabetes. Epigenetics 2017; 12: 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Advani A, Huang Q, Thai K et al. Long-term administration of the histone deacetylase inhibitor vorinostat attenuates renal injury in experimental diabetes through an endothelial nitric oxide synthase-dependent mechanism. Am J Pathol 2011; 178: 2205–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Huang K, Huang J, Xie X et al. Sirt1 resists advanced glycation end products-induced expressions of fibronectin and TGF-beta1 by activating the Nrf2/ARE pathway in glomerular mesangial cells. Free Radic Biol Med 2013; 65: 528–540 [DOI] [PubMed] [Google Scholar]
- 191.Hong Q, Zhang L, Das B et al. Increased podocyte Sirtuin-1 function attenuates diabetic kidney injury. Kidney Int 2018; 93: 1330–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zhou X, Xiong C, Tolbert E et al. Targeting histone methyltransferase enhancer of zeste homolog-2 inhibits renal epithelial-mesenchymal transition and attenuates renal fibrosis. FASEB J 2018; 32: fj756–767.R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Zhou X, Zang X, Ponnusamy M et al. Enhancer of zeste homolog 2 inhibition attenuates renal fibrosis by maintaining smad7 and phosphatase and tensin homolog expression. J Am Soc Nephrol 2016; 27: 2092–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Das S, Senapati P, Chen Z et al. Regulation of angiotensin II actions by enhancers and super-enhancers in vascular smooth muscle cells. Nat Commun 2017; 8: 1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kim YK, Wee G, Park J et al. TALEN-based knockout library for human microRNAs. Nat Struct Mol Biol 2013; 20: 1458–1464 [DOI] [PubMed] [Google Scholar]
- 196.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science 2003; 300: 763–763 [DOI] [PubMed] [Google Scholar]
- 197.Boch J, Scholze H, Schornack S et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009; 326: 1509–1512 [DOI] [PubMed] [Google Scholar]
- 198.Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure 2011; 19: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Stoddard BL. Homing endonucleases from mobile group I introns: discovery to genome engineering. Mob DNA 2014; 5: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010; 327: 167–170 [DOI] [PubMed] [Google Scholar]
- 201.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet 2010; 11: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012; 482: 331–338 [DOI] [PubMed] [Google Scholar]