Abstract
We determined the resistance to quinolone of 309 Salmonella enterica subsp. enterica serotype Typhimurium strains isolated from humans and animals (cattle, pigs, or poultry) in 1995 or 1996. Nalidixic acid resistance increased from 8.5% in 1995 to 18.6% in 1996. The highest resistance levels correlated with a mutation at Ser-83 (or Asp-82). All strains remained ciprofloxacin susceptible. Human and animal isolates were compared by pulsed-field gel electrophoresis, and the banding patterns of the human isolates most closely matched those of the bovine isolates.
Salmonella infection is the most frequent food-borne gastrointestinal disease transmitted from animals to humans (5). Salmonella enterica subsp. enterica serotype Typhimurium (S. Typhimurium) is one of the most common serotypes both in animal and in human isolates (5). Fluoroquinolones are drugs of choice for treatment of human invasive salmonellosis, and some, namely, enrofloxacin, danofloxacin, and marbofloxacin, are also specifically approved for therapeutic veterinary use in France (4). The emergence of quinolone resistance in S. Typhimurium is a matter of concern (2), especially in animals whose products are often suspected sources of infection for human gastroenteritis. There have been several reports of human ciprofloxacin treatment failure (11, 12, 17, 20). Genetic analysis indicated that quinolone resistance often resulted from a Ser-83→Phe-83 mutation in the gyrA gene coding for the A subunit of the active DNA gyrase (9). This mutation leads to the loss of a HinfI restriction site (GANTC) present in the wild-type gyrA sequence (7). On HinfI digestion, the codon 82- or 83-mutated quinolone genes and the quinolone-sensitive genes yield 376- and 277-bp HinfI fragments, respectively.
The purpose of the present study was to investigate a potential link between human and animal S. Typhimurium isolates collected in the same rural area. The isolates were compared by pulsed-field gel electrophoresis (PFGE) (18), and quinolone-resistant strains were examined for the presence of a mutation at gyrase A codon 82 or 83 by restriction fragment length polymorphism (RFLP) analysis.
We studied a total of 309 S. Typhimurium strains isolated from humans (53 patients) and animals (181 cows, 34 pigs, and 41 chickens) in the department of Ille-et-Vilaine (France) in 1995 to 1996. The animal strains were provided by the Laboratoire Départemental Vétérinaire d’Ille-et-Vilaine. Antimicro-bial susceptibility to nalidixic acid, pefloxacin, and ciprofloxacin was assessed by the standardized disk diffusion method, and determination of the MICs was performed according to the recommendations of the Comité de l’Antibiogramme de la Société Française de Microbiologie (1). Statistical analysis of antimicrobial susceptibility was performed by chi-square analysis.
Preparation of whole cellular DNA for PFGE followed the protocol of Allardet-Servent et al. (3). Slices of DNA containing agarose plugs were incubated for 4 h in the presence of 20 U of XbaI (13). DNA fragments were separated by PFGE in a 1% agarose gel (Appligene) that was prepared and run in an 0.5× Tris-borate-EDTA buffer on a contour hexagonal electric field (Pharmaco-LKB; Gene Navigator). The pulse times were 5 to 15 s for 12 h and 15 to 40 s for 24 h. Electrophoresis was run at 150 V and at +8°C for 42 h. PFGE patterns were compared by calculation of the Dice correlation coefficient with the Gel Compar software (Applied Maths, BVAD, Kortrijk, Belgium) and were clustered into a dendrogram by the unweighted pair group method with the arithmetic average clustering technique.
HinfI RFLP analysis of the gyrase A gene (gyrA) in bacterial DNA was described by Fisher et al. (7). The following modification was made. The filter was hybridized to a digoxigenin-labelled 347-bp fragment of the gyrA gene which was prepared by PCR.
Of the 309 S. Typhimurium strains screened for resistance to quinolones, 41 isolates (13.3%) were resistant to nalidixic acid. They were found among cattle, pig, poultry, and human isolates. Twenty-four strains (7.8%) had a decreased pefloxacin susceptibility, but all were susceptible to ciprofloxacin. The distribution of these isolates according to their origin is shown in Table 1. Comparison of the total number of nalidixic acid-resistant strains between the two collection years revealed a significant increase in the percentage, from 8.5% in 1995 to 18.6% in 1996 (P < 0.01).
TABLE 1.
Susceptibilities to nalidixic acid of 309 S. Typhimurium isolates and number (percent) of strains with decreased susceptibility according to their origin
| Yr | Phenotypea | No. (%) of strains from:
|
Total | |||
|---|---|---|---|---|---|---|
| Humans | Cattle | Poultry | Pigs | |||
| 1995 | S | 21 | 107 | 10 | 12 | 150 |
| I or R | 1 (4.5) | 9 (8.6) | 3 (23) | 1 (7.7) | 14 (8.5) | |
| 1996 | S | 26 | 53 | 22 | 17 | 118 |
| I or R | 5 (16.7) | 12 (18.5) | 6 (21.4) | 4 (20) | 27 (18.6) | |
S, susceptible to nalidixic acid; I, intermediate; R, resistant.
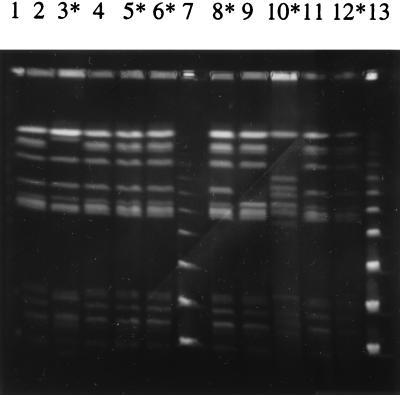
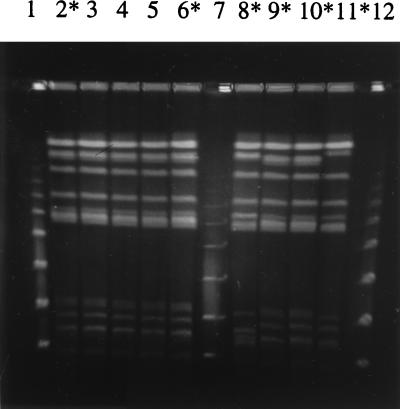
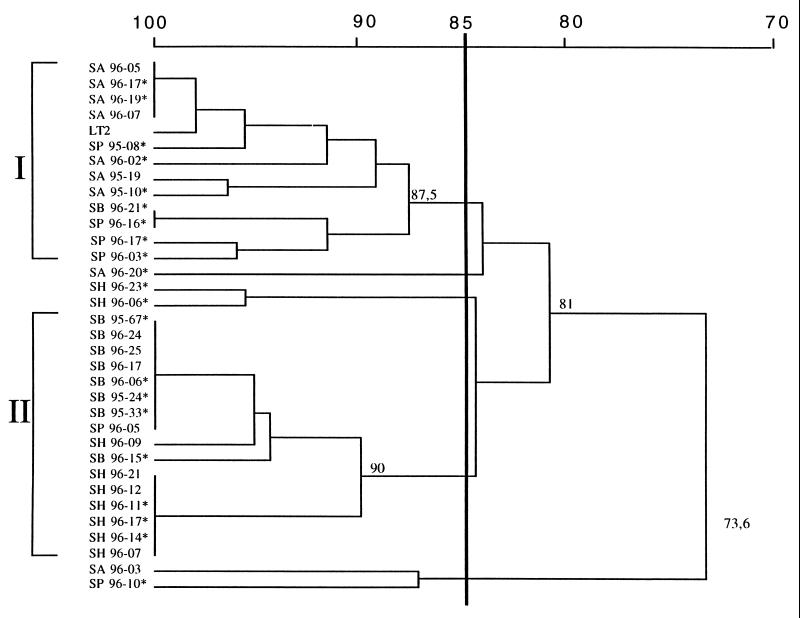
The 41 nalidixic acid-resistant and -susceptible strains of human and animal origin were tested by PFGE (Fig. 1 and 2). Comparison of the banding patterns of 30 S. Typhimurium strains (7 from humans, 8 from cattle, 8 from pigs, and 7 from poultry) showed a close relationship among human and bovine isolates (cluster I) with a Dice correlation coefficient of 90% (Fig. 3). The comparison of pig and poultry isolates showed a Dice coefficient of 87.5% (cluster II). These two clusters exhibited different patterns (Dice coefficient = 81%). The comparison of human and animal strains revealed clonal similarity between human and bovine strains and clonal diversity between human and poultry or pig strains. Resistant or susceptible strains cannot be distinguished by PFGE.
FIG. 1.
PFGE patterns of human S. Typhimurium isolates. Lanes: 2, SH96-21; 3, SH96-23; 4, SH96-12; 5, SH96-11; 6, SH96-17; 8, SH96-14; 9, SH96-9; 10, strain Haddar; 11, SH96-7; 12, SH95-18; 1, 7, and 13, molecular size markers which were successively larger concatamers of bacteriophage lambda DNA (Fluka-Biochemika). Lanes with asterisks indicate nalidixic acid-resistant strains.
FIG. 2.
PFGE patterns of bovine S. Typhimurium isolates. Lanes: 2, SB95-67; 3, SB96-24; 4, SB96-25; 5, SB96-17; 6, SB96-6; 8, SB96-21; 9, SB95-24; 10, SB95-33; 11, SB96-15; 1, 7, and 12, molecular size markers which were successively larger concatamers of bacteriophage lambda DNA (Fluka-Biochemika). Lanes with asterisks indicate nalidixic acid-resistant strains.
FIG. 3.
Dice similarity coefficients (percent). The dendrogram was obtained from cluster analysis of XbaI macrorestriction patterns of 30 S. Typhimurium isolates from humans (SH), cattle (SB), poultry (SA), and pigs (SP) isolated between 1995 and 1996. Asterisks indicate nalidixic acid-strains. SH96-6 is strain Haddar, SA96-3 is strain Bovis morbificans, and SP96-10 is nontypeable.
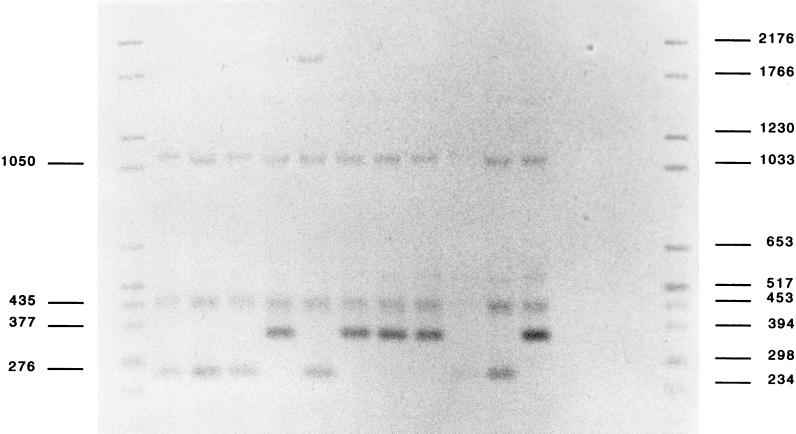
HinfI RFLP analysis of 36 nalidixic acid-resistant S. Typhimurium strains revealed two different patterns (Fig. 4): 19 strains had a 376-bp HinfI gyrA fragment whereas the other 17 had a 277-bp fragment like the LT2 reference strain. The 19 strains with a mutation at codon 82 or 83 were highly resistant to nalidixic acid with at least a 32-fold increase in MICs (MIC > 256 μg/ml). Many of these (17 of 19) had a decreased pefloxacin susceptibility (MIC = 2 μg/ml). MICs of ciprofloxacin were also increased at 0.25 μg/ml. Strains without a mutation at codon 82 or 83 were less resistant. Many of them (10 of 17) had only a twofold increase in the MIC of nalidixic acid (MIC = 16 μg/ml) and remained susceptible to fluoroquinolones (Table 2). The reason for their resistance maps elsewhere. Other mutations have been described in the gyrA gene of Escherichia coli (15, 16, 23) and Salmonella (9). The mutation at codon 83 of subunit A of DNA gyrase greatly reduces the binding of quinolone to the gyrase-DNA complex (22) and can explain the link between this mutation and the MIC (14, 23).
FIG. 4.
HinfI RFLP. Genomic DNA was digested with HinfI and probed on Southern blots with a digoxigenin-labelled 347-bp fragment of the gyrA gene. Quinolone-sensitive strains produced a 277-bp HinfI fragment, whereas nalidixic acid-resistant strains yielded a 376-bp fragment, indicating the loss of the HinfI site at position 244. Lanes: 2, SB96-65; 3, SA96-9; 4, SB96-52; 5, SH96-14; 6, LT2; 7, SB95-25; 8, SA95-10; 9, SB95-6; 10, SB96-11; 11, SB96-21; 12, SP95-111; 1 and 13, Marker VI (Boehringer) molecular size markers (in base pairs) labelled with digoxigenin. Lanes 5, 7, 8, 9, and 12 contain nalidixic acid-resistant strains.
TABLE 2.
Quinolone susceptibilities of 36 nalidixic acid-resistant Salmonella Typhimurium strains according to the presence of a mutation at Ser-83 (or possibly Asp-82)
| Quinolone | Mutation | No. of strains | MIC (μg/ml)
|
||
|---|---|---|---|---|---|
| Range | 50% | 90% | |||
| Nalidixic acid | At Ser-83 (or Asp-82) | 19 | 256–512 | 512 | 512 |
| No mutation at codon 82 or 83 | 17 | 16–256 | 16 | 256 | |
| Pefloxacin | At Ser-83 (or Asp-82) | 19 | 1–2 | 2 | 2 |
| No mutation at codon 82 or 83 | 17 | 0.112–1 | 0.5 | 1 | |
| Ciprofloxacin | At Ser-83 (or Asp-82) | 19 | 0.25–1 | 0.25 | 0.25 |
| No mutation at codon 82 or 83 | 17 | 0.03–0.25 | 0.06 | 0.12 | |
Quinolone-resistant strains have been reported in Asia and Europe. In the United States, multidrug-resistant S. Typhimurium definitive type 104 (DT 104) has become a widespread pathogen (8), but the prevalence of quinolone resistance in Salmonella remains low, although increasing from 0.1% in 1989 to 1991 to 0.5% in 1994 to 1995 (10). In Vietnam, nalidixic-acid-resistant Salmonella serotype Typhi, first described in 1993, is now associated with a treatment failure rate of 50% with short-course treatments which have proved remarkably effective for treatment of multidrug-resistant typhoid fever (21). In the United Kingdom, S. Typhimurium DT 104 was the second most prevalent strain of Salmonella (after Salmonella serotype Enteritidis PT4) isolated from humans in 1996. The incidence of fluoroquinolone resistance of this strain increased from 0% in 1994 to 7% in 1995 and 14% in 1996 (19). Emerging quinolone resistance in animal S. Typhimurium is worrying, since these resistant strains are responsible for human pathologies (6) and can lead to treatment failure. Continued monitoring of Salmonella for resistance to fluoroquinolone antimicrobial drugs is essential.
Acknowledgments
We thank A. Lacourt and M. Bonnier from the Laboratoire Vétérinaire Départemental d’Ille-et-Vilaine for giving us the animal strains of S. Typhimurium.
REFERENCES
- 1.Acar J F, Chardon H, Choutet P, Courvalin P, Dabernat H, Drugeon H, Dubreuil L, Goldstein F W, Morel F C, Philippon A, Rouveix B, Sirot J, Thabbaut A. Comité de l’Antibiogramme de la Société Française de Microbiologie. Communiqué 1997. Pathol Biol. 1997;44:I–VIII. [PubMed] [Google Scholar]
- 2.Acar, J. F., and F. W. Goldstein. 1997. Trends in bacterial resistance to fluoroquinolones. Clin. Infect. Dis. 24(Suppl. 1):67–73. [DOI] [PubMed]
- 3.Allardet-Servent A, Bouzigues N, Carles-Nurit M J, Bourg G, Gouby A, Ramuz M. Use of low-frequency cleavage restriction endonuclease for DNA analysis in epidemiological investigations of nosocomial bacterial infections. J Clin Microbiol. 1989;27:2057–2061. doi: 10.1128/jcm.27.9.2057-2061.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaslus-Dancla E, Martel J L. Résistance aux antibiotiques chez les animaux d’élevage. Bull Soc Fr Microbiol. 1997;12:152–159. [Google Scholar]
- 5.Desenclos J C, Bouvet P, Pierre V, Brisabois A, Fremy S, Lahellec C, Grimont F, Grimont P A D. Epidémiologie des infections àSalmonella: tendances récentes en France et en Europe. Bull Soc Fr Microbiol. 1996;11:209–215. [Google Scholar]
- 6.Feinman S E. Antibiotics in animal feed-drug resistance revisited. ASM News. 1998;64:24–30. [Google Scholar]
- 7.Fisher, L. M., J. M. Lawrence, I. C. Josty, R. Hopewell, and E. E. C. Maegerrison. 1989. Ciprofloxacin and the fluoroquinolones. New concepts on the mechanism of action and resistance. Am. J. Med. 87(Suppl. 5A):2–7. [DOI] [PubMed]
- 8.Glyn C, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo F J. Emergence of multidrug-resistant Salmonella enterica serotype typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:24–30. doi: 10.1056/NEJM199805073381901. [DOI] [PubMed] [Google Scholar]
- 9.Griggs D J, Gensberg K, Piddock L J. Mutations in gyrA gene of quinolone-resistant Salmonella serotypes isolated from humans and animals. Antimicrob Agents Chemother. 1996;40:1009–1013. doi: 10.1128/aac.40.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herikstad H, Hayes P, Mokhtar M, Fracaro M L, Threlfall E J, Angulo F J. Emerging quinolone-resistant Salmonella in the United States. Emerg Infect Dis. 1997;3:371–372. doi: 10.3201/eid0303.970316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howard A J, Joseph T D, Bloodworth L L, Frost J A, Chart H, Rowe B. The emergence of ciprofloxacin resistance in Salmonella typhimurium. J Antimicrob Chemother. 1990;26:296–298. doi: 10.1093/jac/26.2.296. [DOI] [PubMed] [Google Scholar]
- 12.McCarron B, Carron W L. A calculous nontyphoidal Salmonella cholecystitis requiring surgical intervention despite ciprofloxacin therapy: report of three cases. Clin Infect Dis. 1997;24:707–709. doi: 10.1093/clind/24.4.707. [DOI] [PubMed] [Google Scholar]
- 13.McClelland M, Jones R, Patel Y, Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genome. Nucleic Acids Res. 1987;15:5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oram M, Fisher L M. 4-Quinolone resistance mutations in the DNA gyrase of Escherichia coli clinical isolates identified by using the polymerase chain reaction. Antimicrob Agents Chemother. 1991;35:387–389. doi: 10.1128/aac.35.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouabdesselam S, Hooper D C, Tankovic J, Soussy C. Detection of gyrA and gyrB mutations in quinolone-resistant clinical isolates of Escherichia coli by single-strand conformational polymorphism analysis and determination of levels of resistance conferred by two different single gyrA mutations. Antimicrob Agents Chemother. 1995;39:1667–1670. doi: 10.1128/aac.39.8.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ozeki S, Deguchi T, Yasuda M, Nakano M, Kawamura T, Nishino Y, Kawada Y. Mutations in clinical strains isolated from patients with complicated urinary tract infections. J Clin Microbiol. 1997;35:2315–2319. doi: 10.1128/jcm.35.9.2315-2319.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piddock L J V, Griggs D J, Hall M C, Jin Y F. Ciprofloxacin resistance in clinical isolates of Salmonella typhimurium obtained from two patients. Antimicrob Agents Chemother. 1993;37:662–666. doi: 10.1128/aac.37.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz D C, Cantor C R. Separation of yeast chromosome-sized DNA by pulsed-field gel electrophoresis. Cell. 1984;37:67–76. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 19.Threlfall E J, Ward L R, Rowe B. Increasing incidence of resistance to trimethoprim and ciprofloxacin in epidemic Salmonella typhimurium DT 104 in England and Wales. Euro Surveillance. 1997;2:81–83. doi: 10.2807/esm.02.11.00187-en. [DOI] [PubMed] [Google Scholar]
- 20.Vasallo F J, Martin-Rabadan P, Alcala L, Garcia-Lechuz J M, Rodriguez-Creixems M, Bouza E. Failure of ciprofloxacin therapy for invasive nontyphoidal salmonellosis. Clin Infect Dis. 1998;26:534–535. doi: 10.1086/517087. [DOI] [PubMed] [Google Scholar]
- 21.Wain J, Hoa N T T, Chinh N T, Vinh H, Everett M J, Diep T S, Day N P J, Solomon T, White N J, Piddock L J V, Parry C H. Quinolone-resistant Salmonella typhi in Viet Nam: molecular basis of resistance and clinical response to treatment. Clin Infect Dis. 1997;25:1404–1410. doi: 10.1086/516128. [DOI] [PubMed] [Google Scholar]
- 22.Willmott C J R, Maxwell A. A single point mutation in the DNA gyrase A protein greatly reduces binding of fluoroquinolone to the gyrase-DNA complex. Antimicrob Agents Chemother. 1993;37:126–127. doi: 10.1128/aac.37.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]