Abstract
PURPOSE
This analysis evaluated the genomic landscape of premenopausal patients with hormone receptor–positive and human epidermal growth factor receptor 2–negative advanced breast cancer and the association of genetic alterations with response to ribociclib in the phase III MONALEESA-7 trial.
METHODS
Premenopausal patients were randomly assigned 1:1 to receive endocrine therapy plus ribociclib or placebo. Plasma collected at baseline was sequenced using targeted next-generation sequencing for approximately 600 relevant cancer genes. The association of circulating tumor DNA alterations with progression-free survival (PFS) was evaluated to identify biomarkers of response and resistance to ribociclib.
RESULTS
Baseline circulating tumor DNA was sequenced in 565 patients; 489 had evidence of ≥ 1 alteration. The most frequent alterations included PIK3CA (28%), TP53 (19%), CCND1 (10%), MYC (8%), GATA3 (8%), receptor tyrosine kinases (17%), and the Chr8p11.23 locus (12%). A treatment benefit of ribociclib was seen with wild-type (hazard ratio [HR] 0.45 [95% CI, 0.33 to 0.62]) and altered (HR 0.57 [95% CI, 0.36 to 0.9]) PIK3CA. Overall, patients with altered CCND1 had shorter PFS regardless of treatment, suggesting CCND1 as a potential prognostic biomarker. Benefit with ribociclib was seen in patients with altered (HR 0.21 [95% CI, 0.08 to 0.54]) or wild-type (HR 0.52 [95% CI, 0.39 to 0.68]) CCND1, but greater benefit was observed with altered, suggesting predictive potential of CCND1. Alterations in TP53, MYC, Chr8p11.23 locus, and receptor tyrosine kinases were associated with worse PFS, but ribociclib benefit was independent of alteration status.
CONCLUSION
In this study—to our knowledge, the first large study of premenopausal patients with hormone receptor–positive and human epidermal growth factor receptor 2–negative advanced breast cancer—multiple genomic alterations were associated with poor outcome. A PFS benefit of ribociclib was observed regardless of gene alteration status, although in this exploratory analysis, a magnitude of benefits varied by alteration.
INTRODUCTION
Breast cancer is the leading cause of cancer-related death in women worldwide.1 Approximately 20% of breast cancer cases in the United States are diagnosed in women under 50 years old.2 Globally, breast cancer comprises almost half of all cancer diagnoses in women under 50 years and the incidence of advanced breast cancer (ABC) in premenopausal women is increasing.3
CONTEXT
Key Objective
Premenopausal patients with hormone receptor–positive (HR+) and human epidermal growth factor receptor 2–negative (HER2–) advanced breast cancer (ABC) typically have a worse prognosis compared with postmenopausal women. Here, we sought to characterize genomic alterations detectable in plasma circulating tumor DNA and to determine their relationship with ribociclib treatment benefit in premenopausal patients with HR+ and HER2– ABC in the MONALEESA-7 trial.
Knowledge Generated
A progression-free survival benefit was observed with ribociclib treatment regardless of genomic alteration status, although a magnitude of benefits varied on the basis of specific alterations. Several were associated with a worse outcome in premenopausal patients with HR+ and HER2– ABC including alterations in TP53, MYC, Chr8p11.23 locus, and receptor tyrosine kinases.
Relevance
Understanding the impact of genomic alterations on prognosis or sensitivity to therapies could potentially inform treatment decisions in premenopausal patients with HR+ and HER2– ABC, and further confirmatory studies are warranted for clinical utility.
Approximately 65% of breast cancer cases in women under 50 years old are hormone receptor-positive (HR+) and human epidermal growth factor receptor 2–negative (HER2−).4 Premenopausal women with HR+ tumors, especially younger women, typically have a worse prognosis and are under-represented in clinical trials compared with postmenopausal women.3,5,6 Consequently, treatment approaches for premenopausal women with HR+ and HER2− ABC are usually extrapolated from data for postmenopausal patients.
To date, there is a lack of biomarker profiling, especially genomic profiling of premenopausal ABC, and the limited biomarker data available have been specific to the early disease setting.7 Previous gene expression, whole-exome, and transcriptome studies of early breast cancer from pre- and postmenopausal HR+ patients have shown that the molecular characteristics of premenopausal tumors are distinct from those of postmenopausal tumors.7 Understanding the genomic landscape of HR+ and HER2− ABC in premenopausal patients could inform treatment strategies in this group.
Ribociclib, a cyclin-dependent kinase (CDK) 4/6 inhibitor, is established as an effective treatment for many patients with HR+ and HER2− ABC; however, there are patients who exhibit de novo and/or acquired resistance to CDK4/6 inhibitors or endocrine therapy (ET), which may result from several mechanisms.8,9 ET resistance can be driven by dysregulation of the estrogen receptor (ER) pathway via modulation of receptor tyrosine kinase (RTK) signaling, which affects ER activity; alteration of GATA3, which can drive aberrant ER-mediated transcriptional activities; or dysregulation of FGFR1 signaling.10-17 Additionally, a study profiling the genomic landscape of endocrine-resistant ABCs indicated that genes involved in ER transcriptional machinery, including MYC, were enriched in endocrine-resistant tumors.9 Because various mechanisms can cause resistance to ET or CDK4/6 inhibition,8 identifying biomarkers predictive of sensitivity to these therapies can inform treatment decisions and future research. Thus, a key objective has been to identify biomarkers of resistance or response to ribociclib.
To our knowledge, the phase III MONALEESA-7 trial was the first trial of a targeted therapy performed exclusively in premenopausal patients with HR+ and HER2– ABC. The results of this trial demonstrated that treatment with ribociclib plus ET resulted in significantly longer median progression-free survival (PFS) and overall survival versus ET alone.18,19 Here, we report the results from the first (to our knowledge) analysis dedicated to characterizing the genomic landscape of premenopausal patients with HR+ and HER2– ABC. In this analysis, we evaluated molecular alterations detected in circulating tumor DNA (ctDNA) collected at baseline and their impact on PFS in MONALEESA-7. This is the largest data set of premenopausal patients with ABC receiving first-line endocrine-based therapy in the metastatic setting.
METHODS
Trial and Patients
The study population consisted of premenopausal patients enrolled in MONALEESA-7, the details of which have been previously published.18,19 In brief, MONALEESA-7 is a randomized, placebo-controlled, international, double-blind, phase III study evaluating ribociclib plus ET and goserelin versus placebo plus ET with goserelin in pre- or perimenopausal women with HR+ and HER2– ABC. The primary end point was investigator-assessed PFS, and the key secondary end point was overall survival; the results for these analyses were previously reported.18,19 In this analysis, PFS was defined as the time from random assignment to the first documented progression per local radiology assessment (RECIST version 1.1) or death from any cause. Genomic profiling by next-generation sequencing was an exploratory end point to characterize molecular alterations in ctDNA and correlate these alterations with outcomes in MONALEESA-7.
Biomarker Sample Collection and Assessment of Genetic Alterations
Baseline (before study treatment initiation) plasma samples were collected from 632 patients, and cell-free DNA was extracted. Total extracted cell-free DNA was used to generate next-generation sequencing libraries, which were enriched for a specific 2.9-Mb region of the human genome designed to contain approximately 600 genes relevant to cancer. Single-nucleotide variants were identified using MuTect.20 Copy number variants were called using PureCN, and indels were called using PINDEL.21,22 Germline mutations and artifacts were filtered out using publicly available databases dbSNP and ExAC and an internal database (Novartis Institutes for Biomedical Research) of normal circulating free DNA samples from healthy individuals without cancer.
Statistical Analysis
Correlative analyses with PFS were performed for genes altered in ≥ 8% of patients (leading to approximately 20 patients/arm), RTK genes, and the Chr8p11.23 amplicon (ZNF703, WHSC1L1, and FGFR1). Alterations in RTKs (FGFR1, EGFR, ERBB2, ERBB3, ERBB4, IGF1R, IGF1, KDR, KIT, PDGFRA, PDGFRB, and VEGFA) were grouped to evaluate infrequently altered genes with similar downstream signaling. The Chr8p11.23 amplicon was analyzed after a strong overlap of ZNF703, WHSC1L1, and FGFR1, which are known to be on the same amplicon, was observed.
For each gene or group in ctDNA, patients were classified as altered if ≥ 1 alteration, defined as the presence of a copy number alteration, short insertion or deletion, or mutation, was detected and as wild-type (WT) if no alterations were detected (excluding patients with zero alterations detected per assay limitation). Kaplan-Meier curves were generated, and median PFS (95% CI) was estimated by treatment and biomarker status. A Cox proportional hazards (PH) model was used to estimate the hazard ratios (HRs) of treatment benefit (ribociclib v placebo) for PFS by biomarker status. To test for a difference in treatment benefit by biomarker status, a gene-treatment interaction term was included in a Cox PH model. Cox PH models were stratified by the presence of liver or lung metastases, previous chemotherapy for advanced disease, and endocrine combination partner (tamoxifen or nonsteroidal aromatase inhibitor).18,19 No adjustments for multiple testing were made, as these analyses were hypothesis generating. Statistical analyses were performed using the R package.23
RESULTS
Patient Characteristics and Sample Collection and Analysis
Between December 17, 2014, and August 1, 2016, 672 premenopausal patients underwent 1:1 random assignment to receive ribociclib (n = 335) or placebo (n = 337).18,19 Baseline characteristics were balanced among treatment groups.18,19
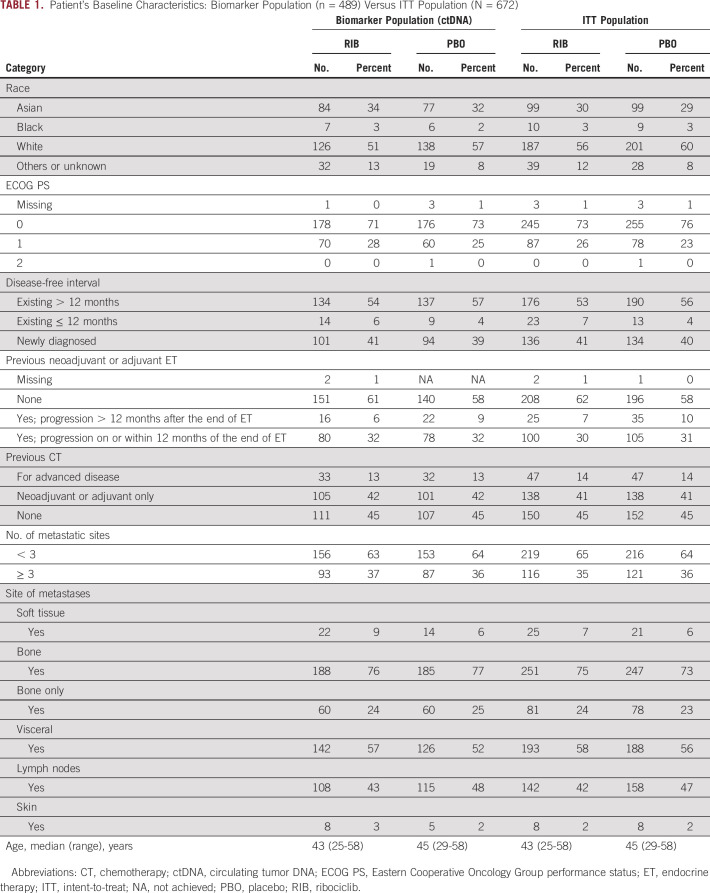
Baseline ctDNA was successfully sequenced in 565 patients (Fig 1); 489 patients with HR+ and HER2– ABC had ≥ 1 genetic alteration and were included in this analysis. The demographic and baseline characteristics of the biomarker population were balanced and generally representative of the intent-to-treat population (Table 1).
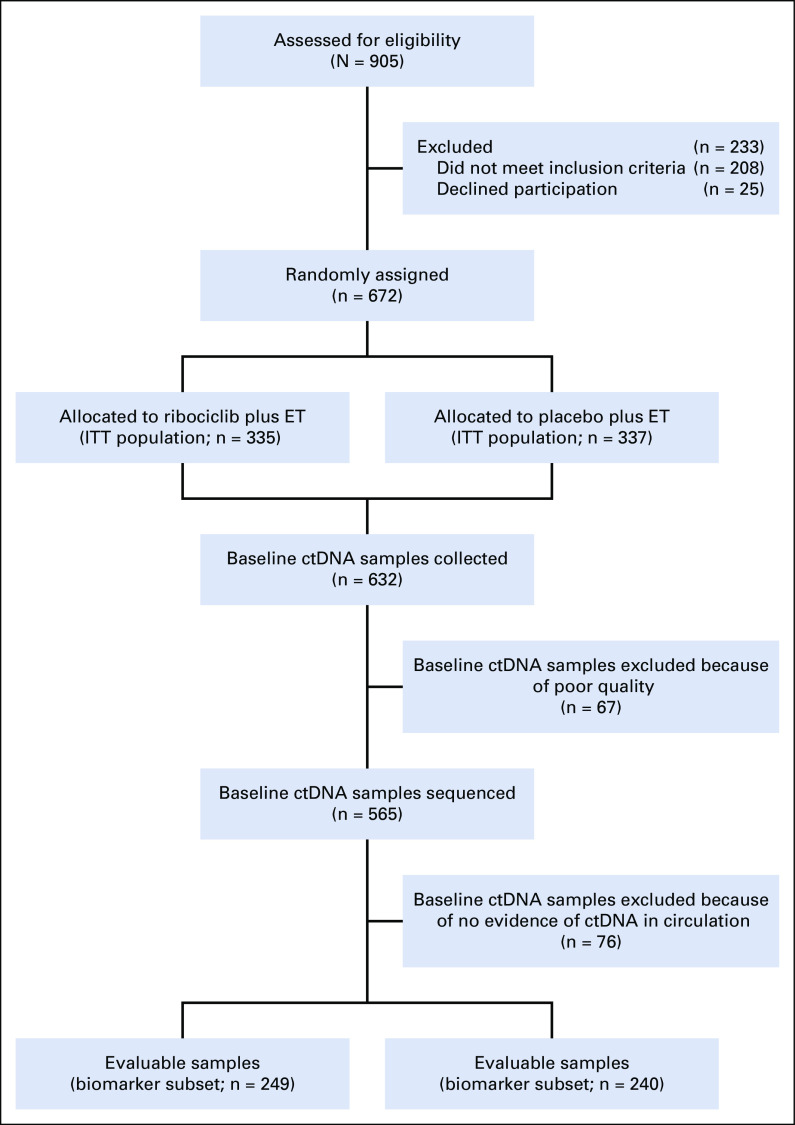
FIG 1.
CONSORT diagram of ctDNA sample collection and analysis. Baseline ctDNA was sequenced from 565 patients. All analyses focused on patients with evidence of tumor DNA in circulation with the presence of ≥ 1 genetic alteration. In all, 489 of 565 patients met all criteria and were subject to downstream analysis. ctDNA, circulating tumor DNA; ET, endocrine therapy; ITT, intention to treat.
TABLE 1.
Patient's Baseline Characteristics: Biomarker Population (n = 489) Versus ITT Population (N = 672)
Genomic Landscape of Premenopausal Metastatic HR+ and HER2− Breast Cancer in ctDNA
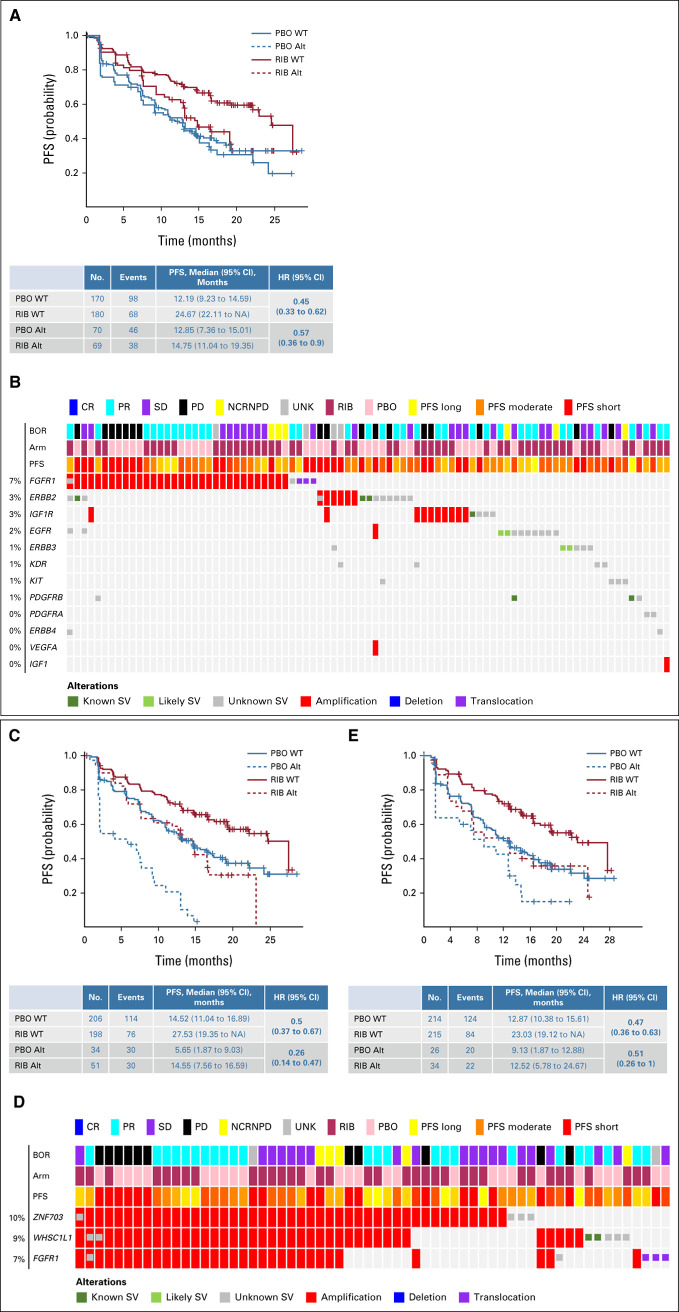
In these 489 patients, 32 genes were altered in ≥ 5% of patients (Fig 2). PIK3CA was the most frequently altered gene (28%), followed by TP53 (19%). Other frequently altered genes of interest, including CCND1, MYC, and GATA3, were altered in 10%, 8%, and 8% of patients, respectively (Fig 2). CCND1, FGF4, FGF3, and FGF19 were localized on Chr11q13.3 and were coamplified in 48 of 489 patients (10%; data not shown). FGFR1, ZNF703, and WHSC1L1 were localized on Chr8p11.23 and were altered in 12% of patients and coamplified in 28 of 489 patients (6%). Alterations in other genes of interest, including NF1 (6%), PTEN (4%), AKT1 (3%), ESR1 (3%), ERBB2 (3%), CDKN2A (3%), and RB1 (2%), were also identified (Fig 2).
FIG 2.
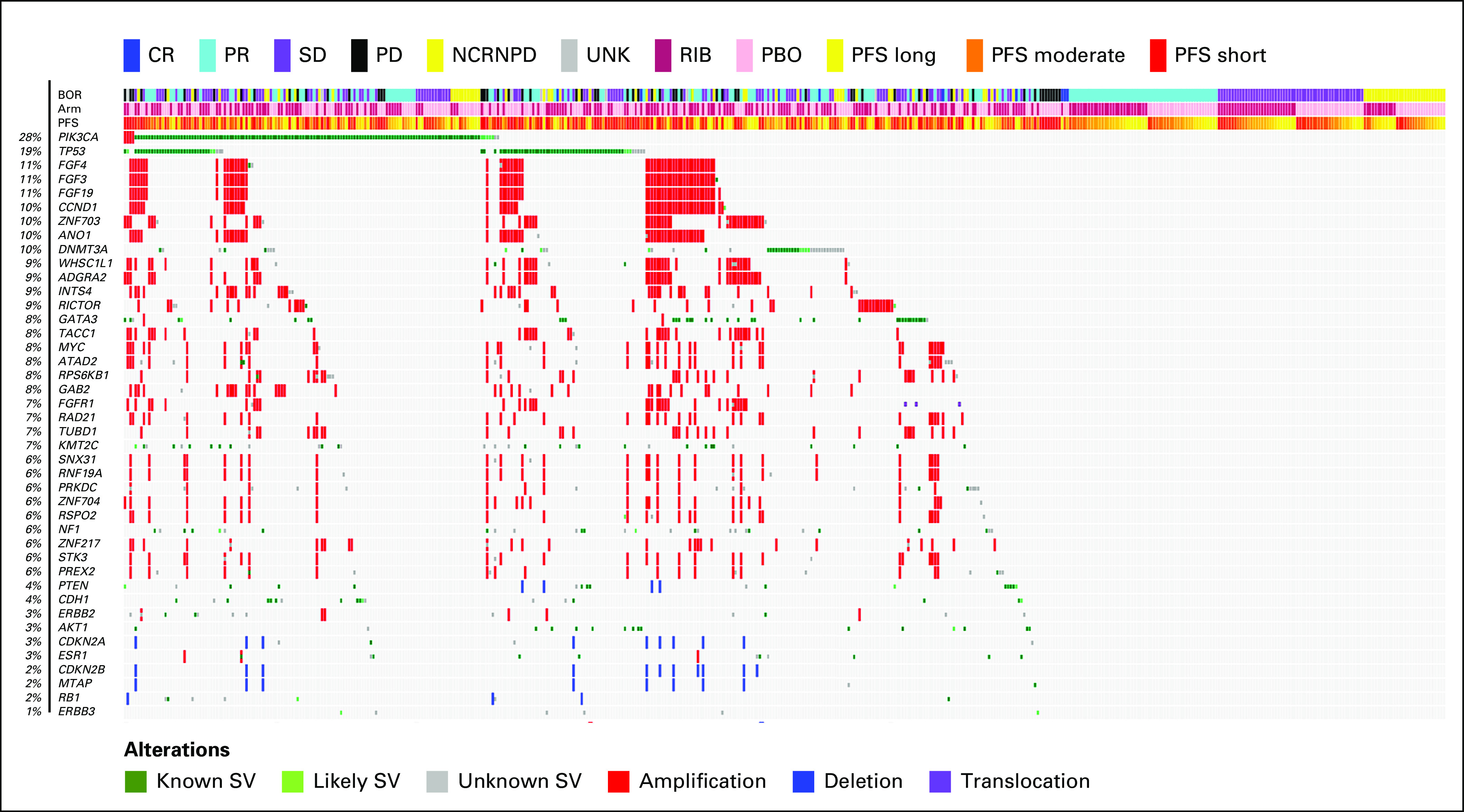
Genomic landscape of advanced breast cancer in premenopausal women. Oncoprint depicting the results of patient's ctDNA NGS data. MONALEESA-7 cfDNA at screening: Genes with frequency > 5% or genes of interest are included. BOR, best overall response; cfDNA, circulating free DNA; CR, complete response; ctDNA, circulating tumor DNA; NCRNPD, neither complete response nor progressive disease; NGS, next-generation sequencing; PBO, placebo; PD, progressive disease; PFS, progression-free survival; PR, partial response; RIB, ribociclib; SD, stable disease; SV, structural variations; UNK, unknown.
Association of PFS With Genes Involved in Cell Cycle Regulation
Median PFS was determined for each genetic subgroup, and a corresponding HR was calculated (Fig 3).
FIG 3.
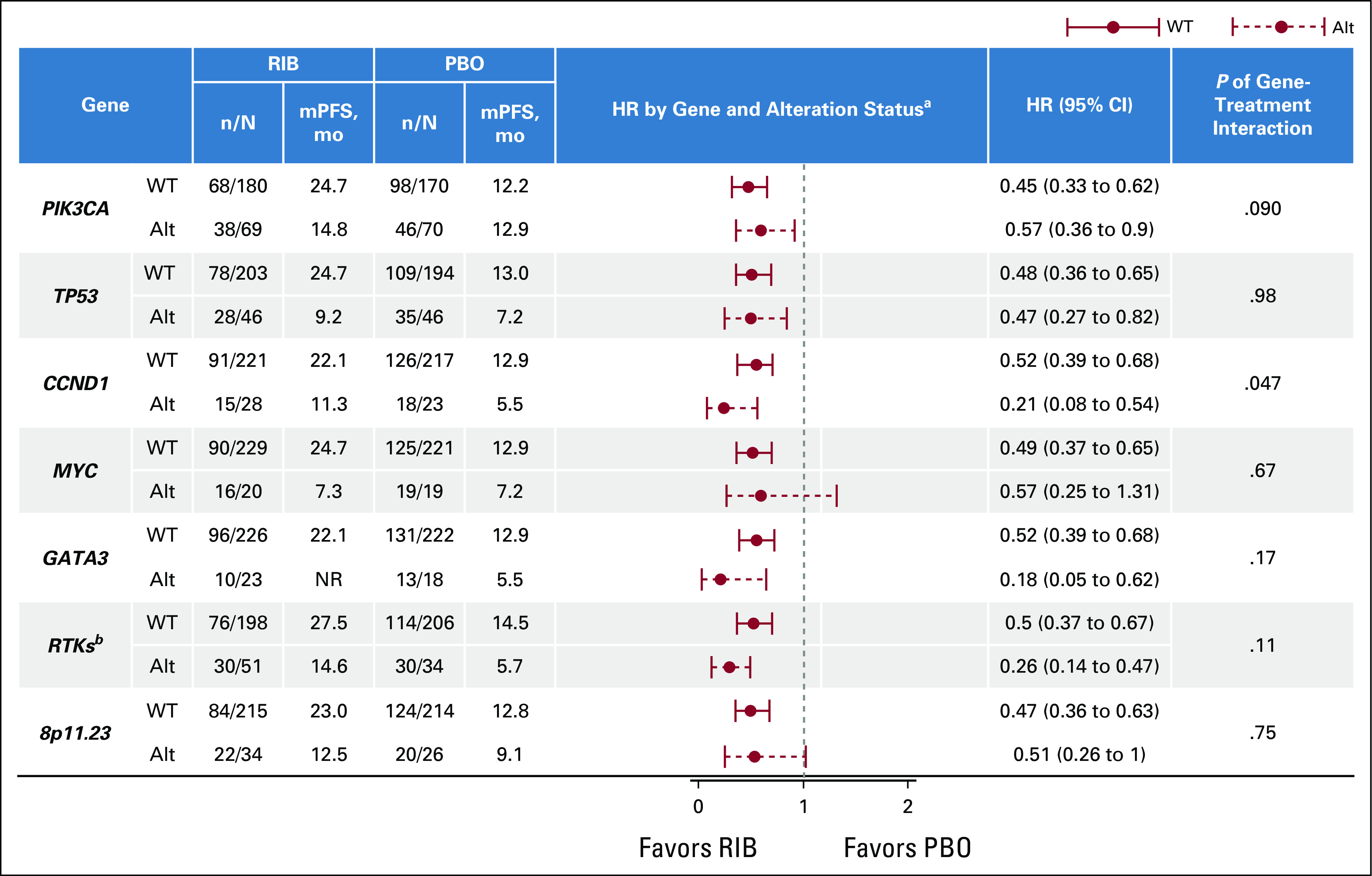
PFS by genetic subgroup. Forest plot analysis of PFS benefit from treatment with ribociclib. aStratified by the presence of lung or liver metastases, previous CT, and combination partner (NSAI/tamoxifen). bReceptor tyrosine kinase genes include EGFR, ERBB2, ERBB3, ERBB4, FGFR1, IGF1, IGF1R, KDR, KIT, PDGFRA, PDGFRB, and VEGFA. Alt, altered; CT, chemotherapy; HR, hazard ratio; mPFS, median PFS; n/N, events/total; NR, no response; NSAI, nonsteroidal aromatase inhibitor; PBO, placebo; PFS, progression-free survival; RIB, ribociclib; WT, wild-type.
CCND1.
Patients with altered CCND1 had a shorter PFS regardless of treatment (the median PFS in patients with WT and altered CCDN1 for ribociclib v placebo was 22.1 v 12.9 months and 11.3 v 5.5 months, respectively). Benefit with ribociclib was observed in both altered CCND1 and WT groups; although the magnitude varied, benefit with ribociclib appeared greater in patients with altered CCND1 (HR, 0.21 [95% CI, 0.08–0.54]) than in those with WT CCND1 (HR 0.52 [95% CI, 0.39 to 0.68]; P value of gene-treatment interaction = .047; Fig 4A).
FIG 4.
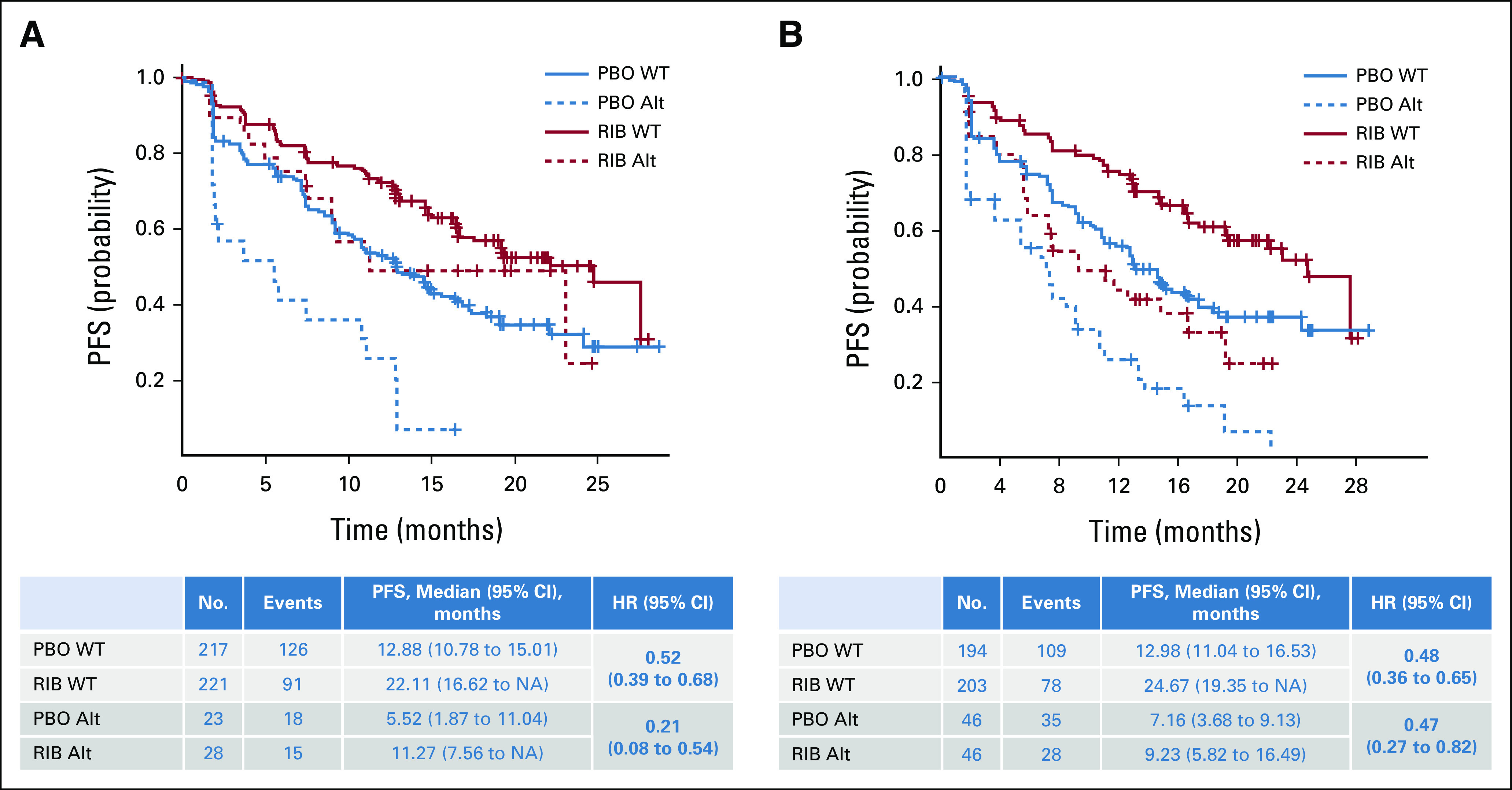
Association of PFS with genes involved in cell cycle regulation: (A) PFS by CCND1 alteration status and (B) PFS by TP53 alteration status. Kaplan-Meier curves of PFS in patients who exhibited alterations in the indicated genes in circulating tumor DNA. PFS in patients in the ribociclib treatment arm is shown in red; in patients in the placebo treatment arm, it is shown in blue. The WT subgroup is indicated by a solid line, and the altered subgroup is indicated by a dashed line. HR (95% CI) estimates and median PFS (95% CI) values are shown in the corresponding tables. Alt, altered; HR, hazard ratio; NA, not achieved; PBO, placebo; PFS, progression-free survival; RIB, ribociclib; WT, wild-type.
TP53.
A similar PFS benefit of ribociclib was observed in patients with WT TP53 (HR 0.48 [95% CI, 0.36 to 0.65]) and altered TP53 (HR 0.47 [95% CI, 0.27 to 0.82]; P = .98; Fig 4B). Although TP53 was not predictive of response to ribociclib, median PFS in patients with altered TP53 (ribociclib v placebo, 9.2 v 7.2 months) was numerically shorter than that in patients with WT TP53 (24.7 v 13.0 months, respectively) in both arms.
Association of PFS With Genes Relevant to the ER Pathway
GATA3.
Alterations in GATA3 were identified in 41 of 489 patients (8%) in this analysis.24 Alterations in GATA3 were associated with a shorter PFS in patients receiving placebo plus ET than in patients with WT GATA3 (the median PFS for WT v altered with placebo was 12.9 v 5.52 months). These findings suggest that patients with alteration of GATA3 might be resistant to ET and have a worse outcome than patients with WT GATA3. However, a PFS benefit of ribociclib was observed among patients with both WT (HR 0.52 [95% CI, 0.39 to 0.68]) and altered GATA3 (HR 0.18 [95% CI, 0.05 to 0.62]; P = .17; Fig 5A).
FIG 5.
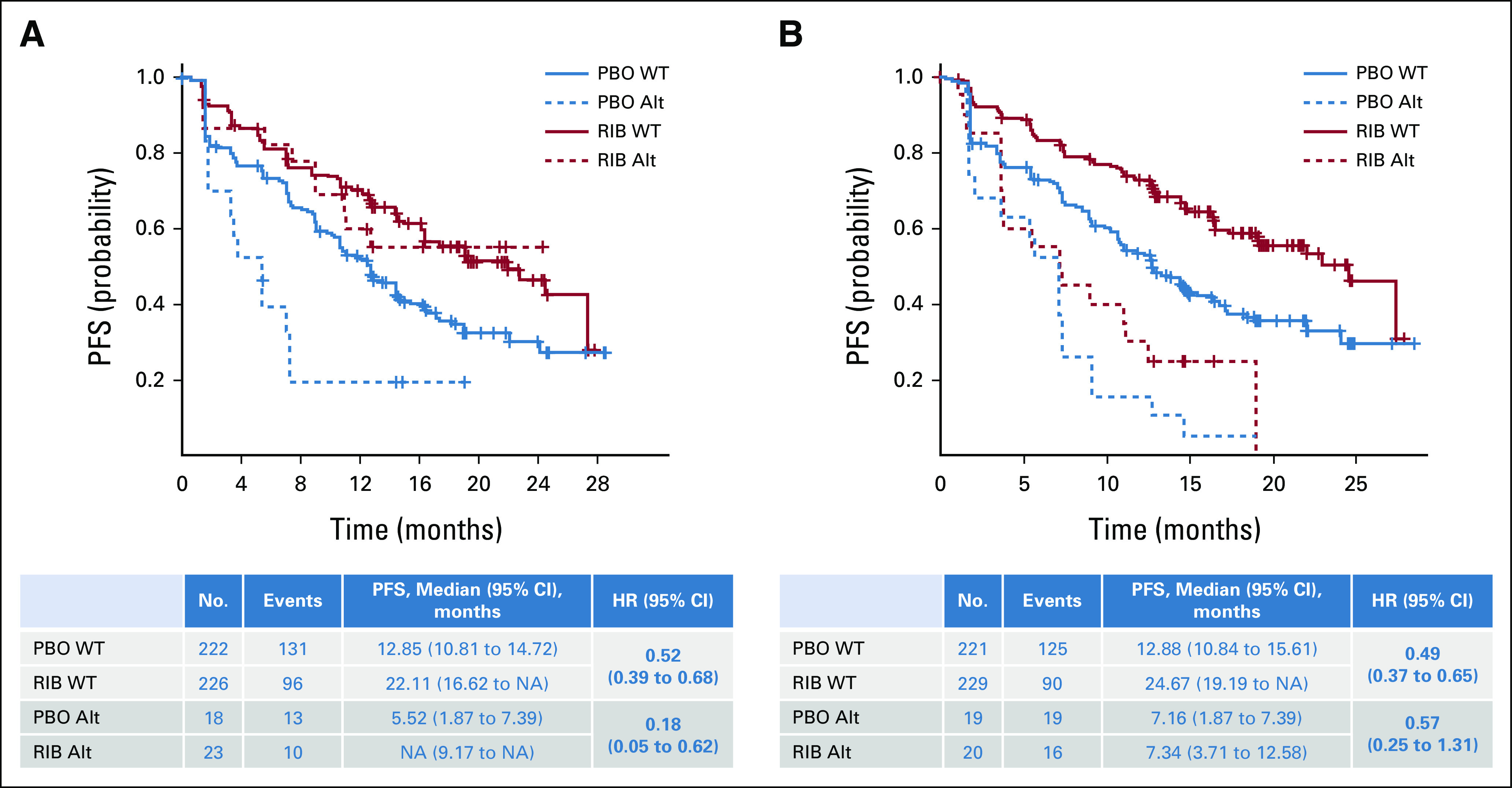
Association of PFS with genes relevant to the estrogen receptor pathway: (A) PFS by GATA3 alteration status and (B) PFS by MYC alteration status. Kaplan-Meier curves of PFS in patients who exhibited alterations in the indicated genes in circulating tumor DNA. PFS in patients in the ribociclib treatment arm is shown in red; in patients in the placebo treatment arm, it is shown in blue. The WT subgroup is indicated by a solid line, and the alteration subgroup is indicated by a dashed line. HR (95% CI) estimates and median PFS (95% CI) values are shown in the corresponding tables. Alt, altered; HR, hazard ratio; NA, not achieved; PBO, placebo; PFS, progression-free survival; RIB, ribociclib; WT, wild-type.
MYC.
Alterations in MYC were observed in 39 of 489 patients (8%).25,26 Patients with altered MYC had shorter PFS regardless of treatment, suggesting that these patients have a worse outcome (the median PFS in patients with WT and altered MYC for ribociclib v placebo was 24.7 v 12.9 months and 7.3 v 7.2 months, respectively; Fig 5B); these results were consistent with other studies in ABC demonstrating MYC as a prognostic marker.27 A similar PFS benefit of ribociclib was observed in patients with WT (HR 0.49 [95% CI, 0.37 to 0.65]) and altered MYC (HR 0.57 [95% CI, 0.25 to 1.31]; Fig 5B) on the basis of HRs (P = .67).
Association of PFS With Genes in the RTK Pathway
PIK3CA.
PIK3CA was altered in 139 of 489 patients (28%). The median PFS in patients with WT PIK3CA receiving ribociclib versus placebo was 24.7 (95% CI, 22.1 to not achieved) versus 12.2 months (95% CI, 9.2 to 14.6); in patients with altered PIK3CA, it was 14.8 (95% CI, 11.0 to 19.4) versus 12.9 months (95% CI, 7.4 to 15.0), respectively. A PFS benefit of ribociclib was observed in patients with WT (HR 0.45 [95% CI, 0.33 to 0.62]) and altered PIK3CA (HR 0.57 [95% CI, 0.36 to 0.90]; P = .09; Fig 6A).
FIG 6.
Association of PFS with genes or pathways in parallel to estrogen receptor and cyclin-dependent kinase 4/6 pathways: (A) PFS by PIK3CA alteration status, (B) oncoprint displaying altered RTKs (MONALEESA-7 cfDNA at screening: RTKs [87 of 489 samples]) identified in NGS analysis of patient's circulating tumor DNA, (C) PFS by RTK alteration status, (D) oncoprint displaying alterations (MONALEESA-7 cfDNA at screening: Chr8p11.23 [62 of 489 samples]) in the Chr8p11.23 region, and (E) PFS by Chr8p11.23 alteration status. (A, C, and E) Kaplan-Meier curves of PFS in patients who exhibited alterations in the indicated genes in ctDNA. PFS in patients in the ribociclib treatment arm is shown in red; in patients in the placebo treatment arm, it is shown in blue. The WT subgroup is indicated by a solid line; the Alt subgroup is indicated by a dashed line. HR (95% CI) estimates and median PFS (95% CI) values are shown in the corresponding tables. (B and D) Oncoprint depicting a subset of altered genes identified in NGS analysis of patient's ctDNA. Alt, altered; BOR, best overall response; cfDNA, circulating free DNA; CR, complete response; ctDNA, ctDNA, circulating tumor DNA; HR, hazard ratio; NA, not achieved; NCRNPD, neither complete response nor progressive disease; NGS, next-generation sequencing; PBO, placebo; PD, progressive disease; PFS, progression-free survival; PR, partial response; RIB, ribociclib; RTK, receptor tyrosine kinase; SD, stable disease; SV, structural variations; UNK, unknown; WT, wild-type.
Receptor tyrosine kinases.
RTK gene alterations (Fig 6B) were identified in 17% of patients. A PFS benefit was observed with ribociclib among patients with WT (HR 0.50 [95% CI, 0.37 to 0.67]) and altered RTKs (HR 0.26 [95% CI, 0.14 to 0.47]; P = .11; Fig 6C). Median PFS generally favored patients with WT versus altered RTKs (the median PFS in patients with WT and altered RTKs for ribociclib v placebo was 27.5 v 14.5 months and 14.6 v 5.7 months, respectively).
Chr8p11.23.
Previous studies demonstrated frequent amplification of the Chr8p11.23 locus driven by the FGFR1, ZNF703, and WHSC1L1 genes in patients with breast cancer.10 Alterations (mainly amplification) of FGFR1, ZNF703, and WHSC1L1 were identified in 60 of 489 patients (12%; Fig 6D). Alterations in Chr8p11.23 were prognostic of shorter PFS overall, irrespective of treatment (the median PFS for WT and altered Chr8p11.23 for ribociclib v placebo was 23.0 v 12.8 months and 12.5 v 9.1 months, respectively); these findings were in accordance with other studies demonstrating their prognostic relevance in HR+ ABC.28,29 However, on the basis of HRs, a similar PFS benefit of ribociclib was observed in patients with WT (HR 0.47 [95% CI, 0.36 to 0.63]) or altered Chr8p11.23 locus (HR 0.51 [95% CI, 0.26 to 1.0]; P = .75; Fig 6E).
Prognosis and Treatment Benefit in Patients Without Detectable Alterations
Compared with the biomarker population (patients with ≥ 1 alteration; n = 489), patients with no detected alterations (n = 76) had better Eastern Cooperative Oncology Group performance status and were less likely to progress on or within 12 months of completing ET (Data Supplement). Additionally, patients without detectable alterations exhibited a trend toward improved PFS with ribociclib and placebo compared with the biomarker population (Data Supplement). These findings suggest a better prognosis in patients without detectable alterations.30
DISCUSSION
Before this study, there were limited clinical trials with a focus on premenopausal patients with breast cancer and analyses of the association of genomic profiles with clinical outcomes in this patient population; thus, there was little information regarding biomarkers that were prognostic and/or predictive of sensitivity or resistance to therapies in premenopausal women with HR+ and HER2– ABC. To our knowledge, MONALEESA-7 was the first trial conducted exclusively in premenopausal patients with HR+ and HER2– ABC, and therefore, this is the first large-scale genomic biomarker profiling study to date. Frequent alterations were identified in genes that regulate the cell cycle, the endocrine pathway, and RTK pathways. These results indicate that an increase in PFS with ribociclib plus ET versus ET alone was observed regardless of biomarker alteration status. Particularly, the PFS benefit of ribociclib treatment is independent of alteration status of TP53, MYC, and genes on the Chr8p11.23 locus. Additionally, patients with WT and altered PIK3CA exhibited a PFS benefit with ribociclib. The ctDNA biomarker data presented here provide insight into the genomic landscape of premenopausal HR+ and HER2– ABC and impact on clinical outcomes.
CCND1 and TP53 are regulators of the cell cycle, and in this analysis, alterations in CCND1 and TP53 were frequent (10% and 19%, respectively) and associated with a worse outcome. CCND1 amplification and/or overexpression has been associated with ER+ breast cancer and was reported to render ER+ cancer cell lines more sensitive to CDK4/6 inhibition.31-33 Although data from PALOMA-1 and PALOMA-2 indicate that expression levels of cyclin D1 or CCND1 amplification were not associated with benefit from CDK4/6 inhibition by palbociclib in these studies, which comprised postmenopausal patients,34,35 our results suggest that premenopausal patients with an alteration of CCND1 experience a more pronounced PFS benefit with ribociclib, with a 79% difference in relative risk of progression with ribociclib versus placebo with altered CCND1 (HR 0.21 [95% CI, 0.08 to 0.54]) and only a 48% difference in relative risk of progression with ribociclib versus placebo with WT CCND1 (HR 0.52 [95% CI, 0.39 to 0.68]). Thus, it is possible that there may be a greater treatment interaction with CCND1 in premenopausal versus postmenopausal patients, but this requires confirmation.
TP53 has been established as a prognostic biomarker in breast cancer.36 In this analysis, there was a PFS benefit of ribociclib regardless of TP53 alteration status; however, patients with altered TP53 (19% of patients) versus WT TP53 had a shorter PFS regardless of treatment. In postmenopausal patients in MONARCH 3, there was a PFS benefit with abemaciclib plus nonsteroidal aromatase inhibitors in patients with WT and altered TP53 (26% of patients), but a greater benefit was observed in patients with WT TP53.37 In postmenopausal patients in MONALEESA-2, a PFS benefit with ribociclib was observed in patients with WT or altered TP53 (12% of patients), although a numerically shorter PFS was observed in patients with altered versus WT TP53 irrespective of treatment.38 Thus, TP53 represents a prognostic biomarker for ABC in both pre- and postmenopausal HR+ patient populations.
ESR1 mutations were only observed in 13 of 489 patients, and thus, a correlation between ESR1 alteration status and PFS could not be evaluated. GATA3 has been implicated in breast cancer development and ET resistance by driving ER-mediated transcriptional regulation of downstream gene expression.12 Our findings suggest that patients with altered GATA3 derived a more pronounced PFS benefit with ribociclib than patients with WT GATA3, although this difference was not statistically significant. Thus, GATA3 status may be predictive of response to ribociclib in premenopausal patients.
Previous studies have shown that patients with altered RTKs have poor outcomes with ET alone, as RTK pathway dysregulation can drive resistance to ET.11,14,15,39 The HRs from this analysis indicate that patients with altered RTKs derived a PFS benefit from ribociclib, suggesting that ribociclib was able to overcome ET resistance in patients harboring RTK gene alterations. Further investigation is needed to identify the specific RTK genes that are driving this effect. Additionally, because FGFR1, WHSC1L1, and ZNF703, located on Chr8p11.23 locus and mostly coamplified in this cohort, have been reported separately to play a role in mechanisms of resistance to ET,14,16,17 we investigated the association between treatment benefit and alterations of this Chr8p11.23 locus. Our results indicate that patients with WT and altered genes at the Chr8p11.23 locus had a similar PFS benefit with ribociclib. However, patients with gene alterations at the Chr8p11.23 locus appeared to have a poorer outcome overall, as these patients had a shorter PFS irrespective of treatment, highlighting the prognostic potential of this amplicon in HR+ ABC. Since the genes at the Chr8p11.23 locus are mostly coamplified, further research is needed to identify which individual genes are driving this effect.
In this study, PIK3CA was altered in 28% of patients, whereas a higher incidence of PIK3CA mutations (30%-40%) was reported in studies of CDK4/6 inhibitors in postmenopausal patients.37,40-42 The improvement in median PFS for ribociclib versus placebo was more pronounced in patients with WT versus altered PIK3CA (HR 0.45 [95% CI, 0.33 to 0.62] v 0.57 [95% CI, 0.36 to 0.9]; interaction P value = .09), although this difference was not statistically significant. However, studies in postmenopausal patients with ribociclib, palbociclib, and abemaciclib have shown increased benefit of study treatment versus ET alone regardless of PIK3CA mutation status, with the exception of MONARCH 3, which showed a more pronounced benefit in patients with WT PIK3CA.
This study has several limitations. First, this is a retrospective exploratory analysis. Second, some subgroup analyses included very small sample sizes, and the results have not been adjusted for multiple testing. Third, these analyses are hypothesis generating and should be confirmed in additional studies. Fourth, the sensitivity to detect DNA alterations is higher in samples with more tumor DNA in circulation, which has itself been shown to be prognostic.30 Finally, this study does not address acquired resistance and the role of other biomarkers, including epigenetic alterations and RNA and protein expression. Thus, further biomarker analyses are ongoing.
This analysis provides insight into the genomic landscape of premenopausal women with HR+ ABC and potential differences in the genetic landscape of HR+ and HER2– ABC in pre- and postmenopausal women. This study uncovered biomarkers associated with worse outcomes overall, but ribociclib treatment resulted in increased PFS benefit regardless of alteration status of these biomarker genes, although the magnitude of this benefit varied across subsets. Altogether, these findings highlight the potential role of genomic alterations in modulating clinical outcomes in premenopausal patients with ABC although these results are hypothesis generating and require confirmation in larger data sets.
ACKNOWLEDGMENT
We thank the patients who participated in this trial, their families and caregivers, data-monitoring committee members, study steering committee members, and the staff who assisted with the trial at each site. This study was sponsored by Novartis, which also provided financial support for medical editorial assistance. We thank Casey Nielsen, PhD, of MediTech Media, for medical editorial assistance with this manuscript. Ribociclib was discovered by Novartis Institutes for BioMedical Research in collaboration with Astex Pharmaceuticals.
Aditya Bardia
Consulting or Advisory Role: Novartis, Genentech, Pfizer, Spectrum Pharmaceuticals, BioTheranostics, Merck, Radius Pharma, Immunomedics, Sanofi, Puma Biotechnology, Daiichi Sankyo/Astra Zeneca, Foundation Medicine, Philips, Genentech/Roche, Radius Health, Innocrin Pharma
Research Funding: Genentech, Novartis, Pfizer, Merck, Sanofi, Radius, Immunomedics, AstraZeneca/Daiichi Sankyo
Open Payments Link: https://openpaymentsdata.cms.gov/physician/523675
Fei Su
Employment: Novartis
Stock and Other Ownership Interests: Novartis
Nadia Solovieff
Employment: Novartis
Stock and Other Ownership Interests: Novartis
Seock-Ah Im
Consulting or Advisory Role: AstraZeneca, Novartis, Roche/Genentech, Eisai, Pfizer, Amgen, Hanmi, Lilly, GlaxoSmithKline, MSD, Daiichi Sankyo
Research Funding: AstraZeneca, Pfizer, Roche/Genentech, Daewoong Pharmaceutical, Eisai
Other Relationship: Roche
Joohyuk Sohn
Research Funding: MSD, Roche, Novartis, Lilly, Pfizer, Bayer, Daiichi Sankyo, AstraZeneca, GlaxoSmithKline
Other Relationship: Roche
Keun Seok Lee
Consulting or Advisory Role: Lilly, MSD Oncology, Novartis, Roche, Pfizer
Research Funding: Dong-A ST
Saul Campos Gomez
Consulting or Advisory Role: Roche, Bristol Myers Squibb, MSD Oncology
Kyung Hae Jung
Consulting or Advisory Role: Roche, AstraZeneca, Celgene, Eisai, Takeda, Novartis
Marco Colleoni
Research Funding: Roche
Rafael Villanueva Vázquez
Consulting or Advisory Role: Novartis
Speakers' Bureau: Novartis, Lilly
Expert Testimony: Novartis
Travel, Accommodations, Expenses: Novartis, Lilly, Eisai, Daiichi Sankyo/AstraZeneca, Roche
Sara Hurvitz
Stock and Other Ownership Interests: Ideal Implant, ROM Tech
Research Funding: Genentech/Roche, Novartis, GlaxoSmithKline, Sanofi, Pfizer, Amgen, OBI Pharma, Puma Biotechnology, Dignitana, Bayer, Biomarin, Lilly, Merrimack, Cascadian Therapeutics, Seattle Genetics, Daiichi Sankyo, Macrogenics, Ambryx, Immunomedics, Pieris Pharmaceuticals, Radius Health, Arvinas, Zymeworks, Gilead Sciences, Phoenix Molecular Designs
Travel, Accommodations, Expenses: Lilly
Other Relationship: Roche, Pfizer
Nadia Harbeck
Stock and Other Ownership Interests: West German Study Group
Honoraria: Roche, Novartis, Amgen, Pfizer, AstraZeneca, Pierre Fabre, Daiichi Sankyo, Exact Sciences, MSD, Seagen
Consulting or Advisory Role: Roche/Genentech, Novartis, Pfizer, Lilly, Sandoz, Daiichi Sankyo, AstraZeneca, West German Study Group, Pierre Fabre, Seagen, MSD
Research Funding: Roche/Genentech, Lilly, MSD, AstraZeneca
Louis Chow
Research Funding: Novartis
Tetiana Taran
Employment: Novartis
Stock and Other Ownership Interests: Novartis
Karen Rodriguez Lorenc
Employment: Novartis, Regeneron
Stock and Other Ownership Interests: Novartis, Regeneron
Naveen Babbar
Employment: Novartis
Stock and Other Ownership Interests: Novartis
Debu Tripathy
Consulting or Advisory Role: Novartis, Pfizer, GlaxoSmithKline, Genomic Health, AstraZeneca, OncoPep
Research Funding: Novartis, Polyphor
Travel, Accommodations, Expenses: Novartis, AstraZeneca
Yen-Shen Lu
Honoraria: Pfizer, Roche, Merck Sharp & Dohme, Novartis, Lilly, Eisai, Eurofarma, Daiichi Sankyo/UCB Japan, AstraZeneca
Consulting or Advisory Role: Pfizer, Roche, Novartis, Lilly
Research Funding: Novartis, Roche, Merck Sharp & Dohme
Travel, Accommodations, Expenses: Roche, Pfizer, Eisai, Novartis
No other potential conflicts of interest were reported.
CLINICAL TRIAL INFORMATION
ClinicalTrials.gov identifier: NCT02278120.
AUTHOR CONTRIBUTIONS
Conception and design: Aditya Bardia, Fei Su, Seock-Ah Im, Saul Campos-Gomez, Fabio Franke, Louis Chow, Tetiana Taran, Karen Rodriguez Lorenc, Naveen Babbar, Yen-Shen Lu
Provision of study materials or patients: Aditya Bardia, Seock-Ah Im, Joohyuk Sohn, Kyung Hae Jung, Rafael Villanueva Vázquez, Sara Hurvitz, Nadia Harbeck, Louis Chow, Debu Tripathy
Collection and assembly of data: Aditya Bardia, Fei Su, Nadia Solovieff, Seock-Ah Im, Joohyuk Sohn, Keun Seok Lee, Saul Campos-Gomez, Marco Colleoni, Rafael Villanueva Vázquez, Sara Hurvitz, Nadia Harbeck, Louis Chow, Tetiana Taran, Naveen Babbar, Debu Tripathy
Data analysis and interpretation: Aditya Bardia, Fei Su, Nadia Solovieff, Seock-Ah Im, Saul Campos-Gomez, Kyung Hae Jung, Fabio Franke, Nadia Harbeck, Tetiana Taran, Karen Rodriguez Lorenc, Naveen Babbar, Debu Tripathy
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Aditya Bardia
Consulting or Advisory Role: Novartis, Genentech, Pfizer, Spectrum Pharmaceuticals, BioTheranostics, Merck, Radius Pharma, Immunomedics, Sanofi, Puma Biotechnology, Daiichi Sankyo/Astra Zeneca, Foundation Medicine, Philips, Genentech/Roche, Radius Health, Innocrin Pharma
Research Funding: Genentech, Novartis, Pfizer, Merck, Sanofi, Radius, Immunomedics, AstraZeneca/Daiichi Sankyo
Open Payments Link: https://openpaymentsdata.cms.gov/physician/523675
Fei Su
Employment: Novartis
Stock and Other Ownership Interests: Novartis
Nadia Solovieff
Employment: Novartis
Stock and Other Ownership Interests: Novartis
Seock-Ah Im
Consulting or Advisory Role: AstraZeneca, Novartis, Roche/Genentech, Eisai, Pfizer, Amgen, Hanmi, Lilly, GlaxoSmithKline, MSD, Daiichi Sankyo
Research Funding: AstraZeneca, Pfizer, Roche/Genentech, Daewoong Pharmaceutical, Eisai
Other Relationship: Roche
Joohyuk Sohn
Research Funding: MSD, Roche, Novartis, Lilly, Pfizer, Bayer, Daiichi Sankyo, AstraZeneca, GlaxoSmithKline
Other Relationship: Roche
Keun Seok Lee
Consulting or Advisory Role: Lilly, MSD Oncology, Novartis, Roche, Pfizer
Research Funding: Dong-A ST
Saul Campos Gomez
Consulting or Advisory Role: Roche, Bristol Myers Squibb, MSD Oncology
Kyung Hae Jung
Consulting or Advisory Role: Roche, AstraZeneca, Celgene, Eisai, Takeda, Novartis
Marco Colleoni
Research Funding: Roche
Rafael Villanueva Vázquez
Consulting or Advisory Role: Novartis
Speakers' Bureau: Novartis, Lilly
Expert Testimony: Novartis
Travel, Accommodations, Expenses: Novartis, Lilly, Eisai, Daiichi Sankyo/AstraZeneca, Roche
Sara Hurvitz
Stock and Other Ownership Interests: Ideal Implant, ROM Tech
Research Funding: Genentech/Roche, Novartis, GlaxoSmithKline, Sanofi, Pfizer, Amgen, OBI Pharma, Puma Biotechnology, Dignitana, Bayer, Biomarin, Lilly, Merrimack, Cascadian Therapeutics, Seattle Genetics, Daiichi Sankyo, Macrogenics, Ambryx, Immunomedics, Pieris Pharmaceuticals, Radius Health, Arvinas, Zymeworks, Gilead Sciences, Phoenix Molecular Designs
Travel, Accommodations, Expenses: Lilly
Other Relationship: Roche, Pfizer
Nadia Harbeck
Stock and Other Ownership Interests: West German Study Group
Honoraria: Roche, Novartis, Amgen, Pfizer, AstraZeneca, Pierre Fabre, Daiichi Sankyo, Exact Sciences, MSD, Seagen
Consulting or Advisory Role: Roche/Genentech, Novartis, Pfizer, Lilly, Sandoz, Daiichi Sankyo, AstraZeneca, West German Study Group, Pierre Fabre, Seagen, MSD
Research Funding: Roche/Genentech, Lilly, MSD, AstraZeneca
Louis Chow
Research Funding: Novartis
Tetiana Taran
Employment: Novartis
Stock and Other Ownership Interests: Novartis
Karen Rodriguez Lorenc
Employment: Novartis, Regeneron
Stock and Other Ownership Interests: Novartis, Regeneron
Naveen Babbar
Employment: Novartis
Stock and Other Ownership Interests: Novartis
Debu Tripathy
Consulting or Advisory Role: Novartis, Pfizer, GlaxoSmithKline, Genomic Health, AstraZeneca, OncoPep
Research Funding: Novartis, Polyphor
Travel, Accommodations, Expenses: Novartis, AstraZeneca
Yen-Shen Lu
Honoraria: Pfizer, Roche, Merck Sharp & Dohme, Novartis, Lilly, Eisai, Eurofarma, Daiichi Sankyo/UCB Japan, AstraZeneca
Consulting or Advisory Role: Pfizer, Roche, Novartis, Lilly
Research Funding: Novartis, Roche, Merck Sharp & Dohme
Travel, Accommodations, Expenses: Roche, Pfizer, Eisai, Novartis
No other potential conflicts of interest were reported.
REFERENCES
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries CA Cancer J Clin 68394–4242018 [DOI] [PubMed] [Google Scholar]
- 2.DeSantis CE, Ma J, Goding Sauer A, et al. Breast cancer statistics, 2017, racial disparity in mortality by state CA Cancer J Clin 67439–4482017 [DOI] [PubMed] [Google Scholar]
- 3.Bardia A, Hurvitz S.Targeted therapy for premenopausal women with HR(+), HER2(-) advanced breast cancer: Focus on special considerations and latest advances Clin Cancer Res 245206–52182018 [DOI] [PubMed] [Google Scholar]
- 4.Howlader N, Cronin KA, Kurian AW, et al. Differences in breast cancer survival by molecular subtypes in the United States Cancer Epidemiol Biomarkers Prev 27619–6262018 [DOI] [PubMed] [Google Scholar]
- 5.Azim HA, Jr, Partridge AH. Biology of breast cancer in young women. Breast Cancer Res. 2014;16:427. doi: 10.1186/s13058-014-0427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sheridan W, Scott T, Caroline S, et al. Breast cancer in young women: Have the prognostic implications of breast cancer subtypes changed over time? Breast Cancer Res Treat 147617–6292014 [DOI] [PubMed] [Google Scholar]
- 7.Liao S, Hartmaier RJ, McGuire KP, et al. The molecular landscape of premenopausal breast cancer. Breast Cancer Res. 2015;17:104. doi: 10.1186/s13058-015-0618-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoninger SF, Blain SW.The ongoing search for biomarkers of CDK4/6 inhibitor responsiveness in breast cancer Mol Cancer Ther 193–122020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Razavi P, Chang MT, Xu G, et al. The genomic landscape of endocrine-resistant advanced breast cancers Cancer Cell 34427–438.e62018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smid M, Rodriguez-Gonzalez FG, Sieuwerts AM, et al. Breast cancer genome and transcriptome integration implicates specific mutational signatures with immune cell infiltration. Nat Commun. 2016;7:12910. doi: 10.1038/ncomms12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giuliano M, Schifp R, Osborne CK, et al. Biological mechanisms and clinical implications of endocrine resistance in breast cancer Breast 20S42–S492011suppl 3 [DOI] [PubMed] [Google Scholar]
- 12.Ciocca V, Daskalakis C, Ciocca RM, et al. The significance of GATA3 expression in breast cancer: A 10-year follow-up study Hum Pathol 40489–4952009 [DOI] [PubMed] [Google Scholar]
- 13.Green AR, Aleskandarany MA, Agarwal D, et al. MYC functions are specific in biological subtypes of breast cancer and confers resistance to endocrine therapy in luminal tumours Br J Cancer 114917–9282016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner N, Pearson A, Sharpe R, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer Cancer Res 702085–20942010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Formisano L, Stauffer KM, Young CD, et al. Association of FGFR1 with ERalpha maintains ligand-independent ER transcription and mediates resistance to estrogen deprivation in ER(+) breast cancer Clin Cancer Res 236138–61502017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irish JC, Mills JN, Turner-Ivey B, et al. Amplification of WHSC1L1 regulates expression and estrogen-independent activation of ERalpha in SUM-44 breast cancer cells and is associated with ERalpha over-expression in breast cancer Mol Oncol 10850–8652016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang X, Mu X, Huang O, et al. Luminal breast cancer cell lines overexpressing ZNF703 are resistant to tamoxifen through activation of Akt/mTOR signaling. PLoS One. 2013;8:e72053. doi: 10.1371/journal.pone.0072053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tripathy D, Im SA, Colleoni M, et al. Ribociclib plus endocrine therapy for premenopausal women with hormone-receptor-positive, advanced breast cancer (MONALEESA-7): A randomised phase 3 trial Lancet Oncol 19904–9152018 [DOI] [PubMed] [Google Scholar]
- 19.Im SA, Lu YS, Bardia A, et al. Overall survival with ribociclib plus endocrine therapy in breast cancer N Engl J Med 381307–3162019 [DOI] [PubMed] [Google Scholar]
- 20.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples Nat Biotechnol 31213–2192013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riester M, Singh AP, Brannon AR, et al. PureCN: Copy number calling and SNV classification using targeted short read sequencing. Source Code Biol Med. 2016;11:13. doi: 10.1186/s13029-016-0060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye K, Schulz MH, Long Q, et al. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads Bioinformatics 252865–28712009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 24.Wilson BJ, Giguere V. Meta-analysis of human cancer microarrays reveals GATA3 is integral to the estrogen receptor alpha pathway. Mol Cancer. 2008;7:49. doi: 10.1186/1476-4598-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng AS, Jin VX, Fan M, et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters Mol Cell 21393–4042006 [DOI] [PubMed] [Google Scholar]
- 26.Wang C, Mayer JA, Mazumdar A, et al. Estrogen induces c-myc gene expression via an upstream enhancer activated by the estrogen receptor and the AP-1 transcription factor Mol Endocrinol 251527–15382011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deming SL, Nass SJ, Dickson RB, et al. C-myc amplification in breast cancer: A meta-analysis of its occurrence and prognostic relevance Br J Cancer 831688–16952000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwek SS, Roy R, Zhou H, et al. Co-amplified genes at 8p12 and 11q13 in breast tumors cooperate with two major pathways in oncogenesis Oncogene 281892–19032009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu XY, Ma D, Xu XE, et al. Genomic landscape and endocrine-resistant subgroup in estrogen receptor-positive, progesterone receptor-negative, and HER2-negative breast cancer Theranostics 86386–63992018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solovieff N, Su F, Leary R, et al. Association of tumor DNA in circulation with clinical characteristics and treatment response in HR+/HER2− advanced breast cancer. AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics, Boston, MA, 2019 (abstr A031)
- 31.Finn RS, Dering J, Conklin D, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spring L, Bardia A, Modi S.Targeting the cyclin D-cyclin-dependent kinase (CDK) 4/6-retinoblastoma pathway with selective CDK 4/6 inhibitors in hormone receptor-positive breast cancer: Rationale, current status, and future directions Discov Med 2165–742016 [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang TS, Han HS, Hong YC, et al. Prognostic value of combined analysis of cyclin D1 and estrogen receptor status in breast cancer patients Pathol Int 5374–802003 [DOI] [PubMed] [Google Scholar]
- 34.Finn RS, Liu Y, Zhu Z, et al. Biomarker analyses of response to cyclin-dependent kinase 4/6 inhibition and endocrine therapy in women with treatment-naive metastatic breast cancer Clin Cancer Res 26110–1212020 [DOI] [PubMed] [Google Scholar]
- 35.Finn RS, Crown JP, Lang I, et al. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): A randomised phase 2 study Lancet Oncol 1625–352015 [DOI] [PubMed] [Google Scholar]
- 36.Olivier M, Langerod A, Carrieri P, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer Clin Cancer Res 121157–11672006 [DOI] [PubMed] [Google Scholar]
- 37.Goetz MP. Efficacy of abemaciclib based on genomic alterations detected in baseline circulating tumor DNA from the MONARCH 3 study of abemaciclib plus nonsteroidal aromatase inhibitor. SABCS (2019) Poster #PD2-06, San Antonio, TX, 2019.
- 38.Hortobagyi G, Stemmer SM, Campone M, et al. First-line ribociclib + letrozole in hormone receptor-positive, HER2-negative advanced breast cancer: Efficacy by baseline circulating tumor DNA alterations in MONALEESA-2. SABCS (2017) Poster #PD4-06, San Antonio, TX, 2017.
- 39.Massarweh S, Osborne CK, Creighton CJ, et al. Tamoxifen resistance in breast tumors is driven by growth factor receptor signaling with repression of classic estrogen receptor genomic function Cancer Res 68826–8332008 [DOI] [PubMed] [Google Scholar]
- 40.Hortobagyi GN, Stemmer SM, Burris HA, et al. Updated results from MONALEESA-2, a phase III trial of first-line ribociclib plus letrozole versus placebo plus letrozole in hormone receptor-positive, HER2-negative advanced breast cancer Ann Oncol 291541–15472018 [DOI] [PubMed] [Google Scholar]
- 41.Cristofanilli M, Turner NC, Bondarenko I, et al. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): Final analysis of the multicentre, double-blind, phase 3 randomised controlled trial Lancet Oncol 17425–4392016 [DOI] [PubMed] [Google Scholar]
- 42.Tolaney SM. Clinical significance of PIK3CA and ESR1 mutations in ctDNA and FFPE samples from the MONARCH 2 study of abemaciclib plus fulvestrant. AACR Annual Meeting 2019, Atlanta, GA, 2019.