This editorial refers to ‘Causal associations of short and long sleep durations with 12 cardiovascular diseases: linear and non-linear Mendelian randomization analyses in UK Biobank’, by S. Ai et al., doi:10.1093/eurheartj/ehab170.
In this issue of the European Heart Journal, Ai et al.1 address the important question of whether inadequate sleep duration increases risk of cardiovascular outcomes, or whether it is a biomarker of such an increased risk. They further studied the functional form of the association between sleep duration and risk of cardiovascular disease (CVD). They concluded that sleep duration is indeed causally associated with CVD risk, with an ‘L’-, rather than a ‘U’-, shaped association, i.e. that short sleep increases CVD risk while long sleep does not. Indeed, while many studies showed that long sleep duration is associated with increased CVD risk,2,3 there is very little experimental evidence supporting this association. In contrast, the experimental data and mechanisms relating short sleep to CVD more clearly implicate inflammatory and neurohumoral causal mechanisms. The findings of Ai et al. support a causal association of short but not long sleep with several CVDs (hypertension, atrial fibrillation, and ischaemic heart disease, as well as pulmonary embolism). However, an important limitation of the analysis is that fewer genetic variants are associated with long sleep, thus reducing the power of the instrumental variables (IVs) for assessing causal associations. Other important limitations are the use of self-reported sleep data, which may differentially under- and overestimate sleep duration,4 and the generalizability of the sample, which represents a generally healthier population than in the UK. In addition, in interpreting the short sleep duration findings, further information on the causes and subtypes of short sleep duration (e.g. secondary to job demands, associated with insomnia) would help target clinical sleep phenotypes for intervention. Here, we discuss several opportunities that may be pursued in expanding upon findings of causal association between sleep duration and CVD: increasing diversity and generalizability of estimates, employing additional analytical approaches to estimate causal effects, and progressing towards translation of findings to advance public health.
Other published papers used Mendelian randomization (MR) to study the causal effect of sleep duration on cardiovascular outcomes and cardiovascular risk factors.1 These studies often overlap in the CVD outcomes, and specific differences are in the genome-wide association study summary statistics used and/or specific analytical techniques. Critically, all of these studies used White British populations (mostly) or other European populations. It is important to study whether these results are generalized to other populations. Other populations may be different from White British UK Biobank (UKB) individuals both in genetic components and in environmental and lifestyle exposures, which may interact with genetic effects, resulting in different genetic effect sizes. A strength of using data from the UKB for genetic association studies is power derived from its large sample size and relative homogeneity. On the other hand, the UKB population suffers from selection bias, where UKB participants tend to be healthier and more educated than the general UK population, and genetic variants have been associated with participation in the UKB.5 Such selection may induce bias into exposure–outcome associations, including when using MR.6 To overcome these limitations and to strengthen our understanding of the causal association between sleep duration and CVD, future studies examining the causal effect of sleep duration on CVD should use data from additional cohorts with diverse populations. Such genetic datasets are increasing in size, but their optimal integration requires coordinated efforts from investigators across the globe. Some datasets also have deeper and objective sleep phenotypes (quantifying sleep fragmentation, quality, and sleep apnoea, and including validated insomnia scales) needed to better describe extreme sleep duration traits and infer causal mechanisms. By leveraging data across multiple studies, we can increase analytical power, expand the generalizability of results, and achieve stronger and more specific evidence, and thereby enhance our ability to translate findings to inform public health policies (Graphical Abstract).
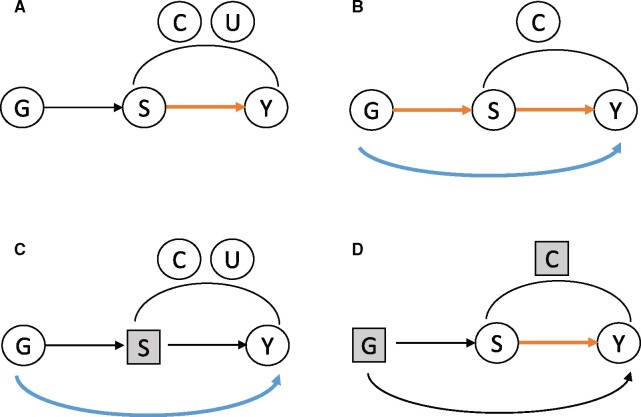
Graphical Abstract.
Causal diagrams demonstrating potential relationships between genetic causes (G) of sleep duration, sleep duration (S), and a cardiovascular outcome (Y). (A) G is a valid instrumental variable for the relationship of S and Y, and can be used to estimated the S–Y association (orange arrow). (B) G has a pleiotropic association with Y. Because there are no unmeasured confounders (U) of the S–Y association, causal mediation analysis can be used to estimate both the direct (pleiotropic) association of G and Y, and the indirect effect via S. (C) The pleiotropic effect of G on Y can be measured by blocking the pathway through S, which is done by conditioning on levels of S. (D) The effect of S on Y independently of G can be measured by blocking pathways through G and C (measured confounders) by conditioning or using matching methods.
MR analysis uses the IV framework, where genetic variants are IVs.7Graphical Abstract A visualizes the standard directed acyclic graph (DAG) representing the classic settings under which IVs are used to estimate causal associations: the exposure variable of interest is sleep duration S, and the outcome is Y, a CVD-related outcome. There are measured confounders of the S–Y associations, denoted by C, and unmeasured confounders U. The instrument, G, is assumed to be associated with S but not associated with Y, and it does not share unmeasured confounders of the S–Y association. The concern in MR analyses is that G is associated with Y via a pathway independent of Y, as in the settings demonstrated in Graphical Abstract B–D. Ai et al. appropriately carried out multiple sensitivity analyses using robust MR methods, such as MR-Egger8 and weighted median MR,9 to address potential pleiotropy. These methods remove the effects of genetic instruments that evidently have pleiotropic effects on the outcome, assuming that enough genetic variants are valid instruments. Genetic variables can be used with other causal inference approaches to estimate causal effects. For example, Graphical Abstract B demonstrates a DAG that is used when performing mediation analysis. There are no unmeasured confounders of the S–Y association, and two effects are estimated: the mediated effect of G on Y, through S, and the direct effect of G on Y. Thus, while the MR model based on Graphical Abstract A aims to estimate the association of sleep duration with Y only, the mediation analysis aims to estimate the effect of genetically predicted sleep duration, represented by G, on Y. In some settings, it is appropriate to assume no unmeasured confounding of the S–Y association and then estimate causal direct and indirect effects at the same time. For example, we previously used this approach when studying the effect of height on cardiometabolic and pulmonary outcomes,10 assuming that adult height is determined long before the outcomes of interest. However, this assumption may be unreliable for sleep duration, as it changes over time with other health and lifestyle exposures. An additional approach that is underutilized in the MR literature and could assess pleiotropy is that of negative outcome control, acknowledging that under valid instrument assumption, there could be no genetic instrument–outcome association.11 In the context of short sleep, one may restrict a dataset to individuals who consistently report sleeping the same sleep duration (e.g. 7 h a night). This approach is visualized in Graphical Abstract C, where conditioning on a specific level of S blocks the pathway in which the genetic instrument affects Y via S. If G is associated with Y in this restricted dataset, one may conclude that a pleiotropic association exists. Finally, Graphical Abstract D visualizes an approach to estimate the effect of S on Y that is independent of genetic predisposition, by blocking the effects of G and C (and assuming that there are no unmeasured confounders U), e.g. using matching methods.12 Matching methods attempt to create sets of ‘treated’ and ‘untreated’ individuals that have similar characteristics outside of ‘treatment’ (here, the treatment may be short sleep). It would be interesting to match individuals reporting short and adequate sleep duration using genetic variables relevant to sleep duration, e.g. by matching on alleles of top short sleep variants, or on values of a polygenic risk score for sleep duration, and compute causal effects. Such an approach treats genetics as a confounder rather than as an IV.
We now have an opportunity to further our understanding of the causal association between sleep duration and CVD. One challenge of studying a potentially heterogeneous measure such as sleep duration in its effect on CVD is the lack of specificity. How does short sleep increases risk of CVD, and do such effects vary according to the factors driving short sleep or by concomitant sleep quality? Research on this topic is abundant,13 with evidence for increased activity of proinflammatory cytokines, sympathetic activity, and oxidative stress, as well as other metabolic responses to short sleep. Can researchers now assess the causal effect of specific mechanistic pathways on CVD risk? In addition to expanding the use of ‘deeper’ sleep phenotypes, it will be useful to develop objective biomarkers of the effect of sleep duration that reflect causal effects of short sleep. Such biomarkers may be more strongly causally related to CVD, and may be useful when assessing the success of interventions designed to increase sleep duration.14 Finally, it is important to assess the public health implications of translating the causal findings via clinical trials.
In summary, MR has been used by Ai et al. and by others to show that short sleep duration is causally associated with CVD. No such association was shown for long sleep. While MR is uniquely appropriate for studying causal associations, relying on genetic variants determined at birth, it also has some limitations, e.g. because genetic variants may have pleiotropic associations with the outcome, and because it is not immune to selection bias. Also, the strength of the genetic IV affects the power of an MR analysis, weakening the finding about lack of causal effect of long sleep on CVD. The phenotypes examined in most MR studies of sleep are also limited in their specificity and sensitivity. There are opportunities to better characterize the causal relationship between sleep duration and CVD using additional causal inference methods, and, importantly, to expand those analyses to diverse populations. Finally, the mounting evidence of a causal association of sleep duration and CVD should motivate follow-up investigation of specific mechanisms behind the association, development of biomarkers, and studies of effective clinical and behavioural interventions.
Funding
All authors received NIH funding.
Conflict of interest: S.M.B. received grant funds from ApniMed and consulting fees from Merck and Eisai Inc. unrelated to this work. S.R. received grant funds from Jazz Pharma, and consulting fees from Eisai Inc. and Jazz Pharma unrelated to this work. The other authors have no conflicts to declare.
The opinions expressed in this article are not necessarily those of the Editors of the European Heart Journal or of the European Society of Cardiology.
References
- 1.Ai S, Zhang J, Zhao G, Wang N, Wang N, Li G, So HC, Liu Y, Chau SW, Chen J, Tan X, Jia F, Tang X, Shi J, Lu L, Wing YK.. Causal associations of short and long sleep durations with 12 cardiovascular diseases: linear and non-linear Mendelian randomization analyses in UK Biobank. Eur Heart J 2021;42:3349–3357. [DOI] [PubMed] [Google Scholar]
- 2.Yin J, Jin X, Shan Z, Li S, Huang H, Li P, Peng X, Peng Z, Yu K, Bao W, Yang W, Chen X, Liu L.. Relationship of sleep duration with all-cause mortality and cardiovascular events: a systematic review and dose–response meta-analysis of prospective cohort studies. J Am Heart Assoc 2017;6:e005947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kronholm E, Laatikainen T, Peltonen M, Sippola R, Partonen T.. Self-reported sleep duration, all-cause mortality, cardiovascular mortality and morbidity in Finland. Sleep Med 2011;12:215–221. [DOI] [PubMed] [Google Scholar]
- 4.Jackson CL, Ward JB, Johnson DA, Sims M, Wilson J, Redline S.. Concordance between self-reported and actigraphy-assessed sleep duration among African-American adults: findings from the Jackson Heart Sleep Study. Sleep 2020;43:zsz246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyrrell J, Zheng J, Beaumont R, Hinton K, Richardson TG, Wood AR, Davey Smith G, Frayling TM, Tilling K.. Genetic predictors of participation in optional components of UK Biobank. Nat Commun 2021;12:886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gkatzionis A, Burgess S.. Contextualizing selection bias in Mendelian randomization: how bad is it likely to be? Int J Epidemiol 2019;48:691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lawlor DA, Harbord RM, Sterne JAC, Timpson N, Davey Smith G.. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–1163. [DOI] [PubMed] [Google Scholar]
- 8.Bowden J, Davey Smith G, Burgess S.. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowden J, Davey Smith G, Haycock PC, Burgess S.. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sofer T, Moon J-Y, Isasi CR, Qi Q, Shah NA, Kaplan RC, Kuniholm MH.. Relationship of genetic determinants of height with cardiometabolic and pulmonary traits in the Hispanic Community Health Study/Study of Latinos. Int J Epidemiol 2018;47:2059–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemani G, Bowden J, Davey Smith G.. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet 2018;27:R195–R208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stuart EA.Matching methods for causal inference: a review and a look forward. Stat Sci 2010;25:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tobaldini E, Fiorelli EM, Solbiati M, Costantino G, Nobili L, Montano N.. Short sleep duration and cardiometabolic risk: from pathophysiology to clinical evidence. Nat Rev Cardiol 2019;16:213–224. [DOI] [PubMed] [Google Scholar]
- 14.Vasan RS.Biomarkers of cardiovascular disease: molecular basis and practical considerations. Circulation 2006;113:2335–2362. [DOI] [PubMed] [Google Scholar]