Abstract
Background
To expedite the development of new oral treatment regimens for visceral leishmaniasis (VL), there is a need for early markers to evaluate treatment response and predict long-term outcomes.
Methods
Data from 3 clinical trials were combined in this study, in which Eastern African VL patients received various antileishmanial therapies. Leishmania kinetoplast DNA was quantified in whole blood with real-time quantitative polymerase chain reaction (qPCR) before, during, and up to 6 months after treatment. The predictive performance of pharmacodynamic parameters for clinical relapse was evaluated using receiver-operating characteristic curves. Clinical trial simulations were performed to determine the power associated with the use of blood parasite load as a surrogate endpoint to predict clinical outcome at 6 months.
Results
The absolute parasite density on day 56 after start of treatment was found to be a highly sensitive predictor of relapse within 6 months of follow-up at a cutoff of 20 parasites/mL (area under the curve 0.92, specificity 0.91, sensitivity 0.89). Blood parasite loads correlated well with tissue parasite loads (ρ = 0.80) and with microscopy gradings of bone marrow and spleen aspirate smears. Clinical trial simulations indicated a > 80% power to detect a difference in cure rate between treatment regimens if this difference was high (> 50%) and when minimally 30 patients were included per regimen.
Conclusions
Blood Leishmania parasite load determined by qPCR is a promising early biomarker to predict relapse in VL patients. Once optimized, it might be useful in dose finding studies of new chemical entities.
Keywords: visceral leishmaniasis, parasitemia, biomarker, pharmacodynamics, qPCR
Blood Leishmania parasite load, determined by qPCR, is a promising early biomarker to predict relapse in visceral leishmaniasis patients and might particularly be useful in the context of dose finding studies of new chemical entities.
There is an urgent need to develop field-adapted oral efficacious treatments for the neglected tropical parasitic disease visceral leishmaniasis (VL), particularly in Eastern Africa. New candidates with different mechanisms of action have been identified and are progressing to clinical development [1]. To facilitate drug development, accurate tools are needed to evaluate treatment efficacy early after the treatment, which is a specific research priority for neglected tropical diseases according to the World Health Organization [2, 3]. This will allow early selection of promising drug regimens and will reduce the number of subjects exposed to regimens with poor efficacy.
Treatment evaluation is complicated because initially cured patients can relapse due to recrudescence of parasites, which is a long-term event that is particularly difficult to predict [4]. Therefore, definitive cure in Eastern African VL clinical trials is generally assessed at 6 months after completion of treatment, defined as a negative parasitological test of cure at the end of treatment (absence of Leishmania amastigotes in spleen or bone marrow aspirate smears by microscopy), lack of VL clinical symptoms, and no requirement for rescue treatment during 6 months’ follow-up. To speed up treatment evaluation, sensitive and specific biomarkers are needed to monitor treatment response and predict relapses. These biomarkers would be particularly useful in clinical trials with new chemical entities, where they could serve as a surrogate endpoint at an early time point after treatment.
Splenic aspiration is an invasive procedure, associated with risk of severe hemorrhage [5, 6], and cannot be performed in patients with unpalpable or reduced spleen size at the end of treatment. Quantification of blood parasite load by real-time quantitative polymerase chain reaction (qPCR) can be an alternative: previous results suggest that positive blood parasite load after treatment is associated with a higher risk of VL relapse [7–19]. In human immunodeficiency virus (HIV) co-infected patients, blood parasite load > 10 parasites/mL preceded clinical relapse [7]. However, risk of VL relapse in HIV co-infection is affected by other factors such as CD4 count [11]. In Eastern Africa, the region with the highest VL incidence globally, very limited Leishmania qPCR data have been published in the context of VL [20, 21]; only a small study in 11 patients focused on the relation with clinical outcome [19].
To evaluate the pharmacodynamic potential of blood parasite load as a predictor for clinical relapse, we longitudinally quantified the blood parasite load using qPCR in patients from 3 multicenter Eastern African clinical trials. The first objective was to identify the most optimal predictor for VL treatment outcome at 6 months in terms of absolute or relative blood parasite load and time of sampling. Second, blood parasite loads were compared with tissue aspirate parasite loads to assess whether the parasite biomass in whole blood is reflecting that in the primary infected organs. Third, the sensitivity of blood and tissue qPCR parasite loads were compared with microscopic readings of tissue samples. Last, the predictive power was quantified for different clinical trial scenarios with variable efficacy rates where this pharmacodynamic marker could hypothetically be used as early surrogate endpoint.
METHODS
Study Sites and Patients
Data originated from 3 phase 2 open-label randomized clinical trials to assess the safety and efficacy of different treatment regimens in the treatment of VL in Eastern Africa: LEAP0208 (NCT01067443 [21]); LEAP0714 (NCT02431143 [22]); and FEXI VL 001 (NCT01980199). Ethical approval was obtained from national and local ethics committees in Kenya, Sudan, and Uganda. Further patient and treatment details can be found in the Supplementary Material.
Clinical Assessment of Efficacy and Sample Collection
An initial cure was defined by improvement of clinical signs and symptoms of VL and a negative parasitological test of cure by microscopy at day 28. Patients who died or required rescue treatment before completion of study treatment were considered initial treatment failures. A definitive cure at day 210 (6 months) was defined as a patient who had initial cure and remained free of VL signs and symptoms (ie, no occurrence of relapse during the follow-up period and no requirement for rescue treatment).
Microscopic parasitological assessments on aspirate smears from lymph node, bone marrow, or spleen (LEAP0208 and FEXI VL 001), or spleen or bone marrow (LEAP0714) were performed at baseline and on day 28 in all studies; it was repeated at day 56, day 210, or in an unscheduled visit if clinically indicated because of the reappearance of VL signs and symptoms, which is suspicious for relapse. In LEAP0714 and FEXI VL 001, part of the tissue aspirate samples intended for microscopy were also collected to perform qPCR. Whole blood ethylenediaminetetraacetic acid samples with a volume of 200 µL were collected for pharmacodynamic assessment before treatment and nominally on day 3, 7, 14, 28, 56, and 210 (LEAP0208); day 3, 7, 14, 21, 28, and 56 (LEAP0714); and day 1, 3, 5, 8, 11, 14, 28, 56, and 210 (FEXI VL 001).
Microscopy and Molecular Methods
Parasitological assessments in the studies were adapted according to the practice of tissue aspiration (spleen, bone marrow, and lymph node) for VL diagnosis in the different countries. In LEAP0208, parasitological assessment by microscopy was done on lymph node aspirates (Dooka, Kassab), spleen aspirates (Kimalel), or bone marrow aspirates (all sites). In LEAP0714, spleen aspirates were collected or, under specific circumstances [22], bone marrow aspirates. In FEXI VL 001, lymph node or bone marrow samples were collected. Aspirates were smeared on 2 slides per sample, stained with Giemsa, and graded on a semiquantitative logarithmic scale from 0 (no parasites in 1000 microscopic fields) to 6+ (> 100 parasites per microscopic field). Measurements of the Leishmania parasite load in whole blood samples and tissue samples were performed using a qPCR method targeting Leishmania kinetoplastid DNA (kDNA). A detailed description of the DNA extraction, used primers, and qPCR protocol can be found in the Supplementary Material.
Data and Statistical Analysis
Data cleaning, statistical analysis, and clinical trial simulations were performed with R (version 3.5.1). qPCR data were excluded from the analysis for patients who were considered initial treatment failures, for samples collected after rescue treatment was given, or for samples considered unreliable. Absolute blood parasite concentrations and relative changes over time were evaluated for their ability to discriminate between cured and relapsed patients. Absolute and log-transformed data were checked for normality and equal variances using the Shapiro-Wilk test. Logistic regression was performed by an unpaired 1-sided Wilcoxon signed rank test to compare blood parasite loads at baseline, day 28, and day 56 after start of treatment. Subsequently, receiver-operating characteristic (ROC) curves were generated with the R packages “pROC” and “plotROC.” The area under the curve (AUC) was compared to find the most predictive parameter for clinical relapse in terms of follow-up day (day 14, day 28, or day 56 after start of treatment) and absolute parasite load or relative to baseline. Furthermore, the interplay between sensitivity and specificity of blood parasite load as a biomarker was evaluated and the optimal cutoff value was determined.
To evaluate the correlation between blood parasite load and tissue parasite load obtained by qPCR, Spearman’s rank correlation rho was determined. The relationship between the 2 sources was determined by linear regression on log-transformed data, excluding data below the limit of quantitation. The correlation between available matched qPCR blood and tissue loads and microscopy gradings of aspirate smears was evaluated visually. To evaluate the sensitivity of the qPCR and microscopy method, the percentage of samples having detectable parasites was compared.
Surrogate Endpoint Evaluation
Finally, clinical trial simulations were performed to evaluate the predictive performance and power associated with the use of qPCR blood parasite load on either day 28 or day 56 as a surrogate endpoint to predict final clinical outcome at 6 months. For this, we used noninferiority clinical trial scenarios where a control treatment arm (90% cure rate at 6 months), representing current standard of care [1], was compared with an alternative treatment arm with lower, varying, cure rates (20%, 40%, 60%, and 80%). Patient populations (n = 10, 20, 30, 40, 50) were sampled with replacement from the pool of cured (n = 143/147) and relapsed (n = 30/32) patients on day 28/56 in our original dataset. Although the actual cure rate was predefined, the predicted cure rate of both populations was derived based on blood parasite load at day 28 or 56, based on the optimal cutoff. To simulate the performance of the qPCR procedure realistically, previously excluded and missing samples were included in these simulations. Fisher’s exact test was used to test if these populations had significantly different predicted cure rates, based on blood parasite load. Per scenario, 1000 clinical trials were simulated. The power was defined as the number of times a significant difference was found between treatment arms and was considered adequate when > 80%.
RESULTS
In total, blood parasite loads were available from 177 patients (n = 134 for LEAP0208, n = 29 for LEAP0714, and n = 14 for FEXI-VL-001), treated with 5 different treatment regimens. Overall, 15.8% of blood and 16.3% of tissue qPCR data had to be excluded (Table 1). Main reasons for exclusion of data were an unreliable or incomplete DNA extraction of the sample (based on glyceraldehyde 3-phosphate dehydrogenase), bad sample quality, or insufficient sample material. None of the microscopic readings were excluded.
Table 1.
Overview of the Data Used for Logistic Regression (Days 0, 28, 56) and ROC Analysis (Days 14, 28, 56), Specifying Collected and Excluded qPCR Blood Samples, qPCR Tissue Samples, and Microscopy Scores Derived From Splenic or Bone Marrow Aspirates
| Blood qPCR | Tissue qPCR | Microscopy Score | ||||
|---|---|---|---|---|---|---|
| Day | Study | Collected Samples | Excluded Samples (%) | Collected Samples | Excluded Samples (%) | Available Readings |
| 0 | LEAP0208 | 131 | 14 (11) | N/A | N/A | 131 |
| LEAP0714 | 30 | 13 (43) | 30 | 0 (0) | 30 | |
| FEXI VL 001 | 14 | 5 (36) | 10 | 0 (0) | 14 | |
| Total | 175 | 32 (18) | 40 | 0 (0) | 174 | |
| 14 | LEAP0208 | 139 | 18 (13) | N/A | N/A | N/A |
| LEAP0714 | 30 | 12 (40) | N/A | N/A | N/A | |
| FEXI VL 001 | 14 | 5 (36) | N/A | N/A | N/A | |
| Total | 183 | 35 (19) | N/A | N/A | N/A | |
| 28 | LEAP0208 | 130 | 13 (10) | N/A | N/A | 126 |
| LEAP0714 | 29 | 5 (17) | 29 | 7 (24) | 28 | |
| FEXI VL 001 | 14 | 5 (36) | 13 | 5 (38) | 14 | |
| Total | 173 | 23 (13) | 42 | 12 (29) | 168 | |
| 56 | LEAP0208 | 136 | 12 (9) | N/A | N/A | 8 |
| LEAP0714 | 29 | 2 (7) | 1 | 0 (0) | 1 | |
| FEXI VL 001 | 13 | 4 (31) | 4 | 1 (25) | N/A | |
| Total | 178 | 18 (10) | 5 | 1 (20) | 9 | |
Abbreviations: N/A, not available; qPCR, quantitative polymerase chain reaction; ROC, receiver-operating characteristic.
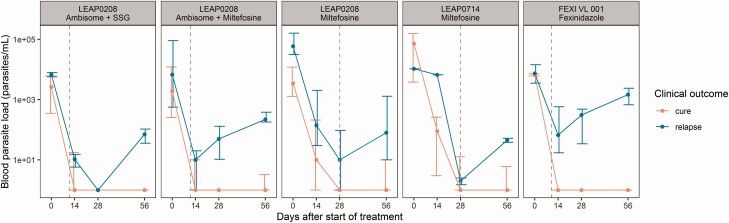
A difference in blood parasite load dynamics between cured and relapsed patients could be observed in all treatment groups (Figure 1). In total, cured patients had a significantly lower parasite load on day 28 (P = 3.91–06) and on day 56 (P = 2.58–14) (Table 2). Remarkably, cured patients also had a significantly lower baseline parasite load (P = .030). This correlation has been demonstrated earlier for tissue baseline parasite loads detected by microscopy in HIV co-infected patients [23]. Baseline parasite loads were not significantly different between treatment groups.
Figure 1.
Median absolute parasite load of cured patients (red line) and relapsed patients (blue line) at baseline and days 14, 28, and 56, stratified per treatment arm. Error bars represent the interquartile range. Gray dashed lines represent end of treatment.
Table 2.
Blood Parasite Loads Quantified by qPCR at Baseline, Day 28, and Day 56, Stratified by Clinical Outcome at 6 Months Follow-Up
| Total | Cure | Relapse | Difference | |||
|---|---|---|---|---|---|---|
| Day | N | N | Parasites/mLa | N | Parasites/mLa | P Valueb |
| 0 | 143 | 117 | 3070 (720–16 290) | 26 | 9760 (2574–63 195) | .030c |
| 28 | 150 | 123 | 0 (0–1.5) | 27 | 20 (0–230) | 3.91e-06c |
| 56 | 156 | 130 | 0 (0–2.75) | 26 | 270 (59.2–1242) | 2.58e-14c |
Abbreviation: qPCR, quantitative polymerase chain reaction.
aValues are given as median (interquartile range).
bWilcoxon test on absolute parasite concentrations.
cSignificant difference when P < .05.
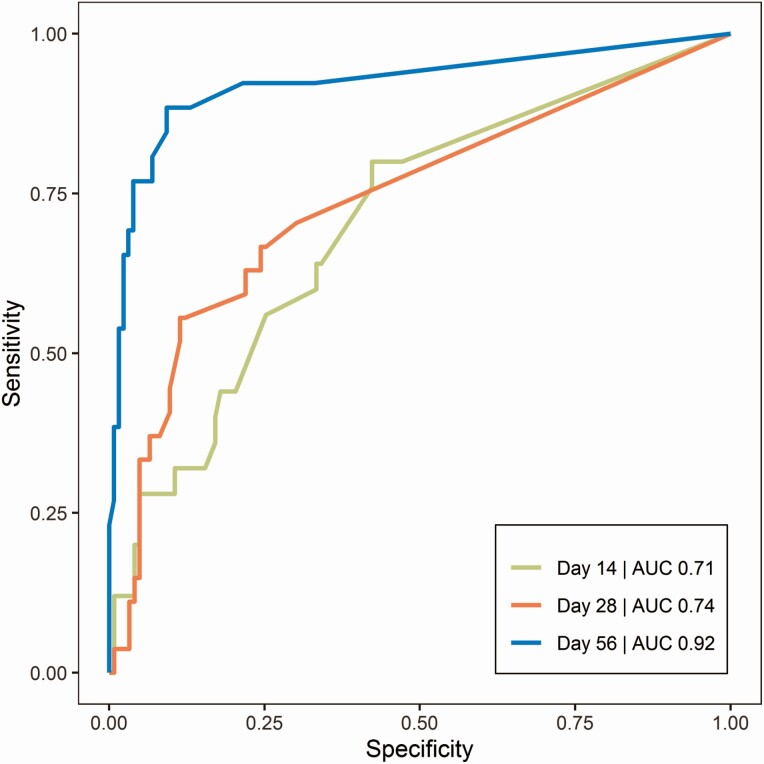
The ROC AUC for absolute blood parasite load classifying clinical relapse (Figure 2) was highest on day 56 (0.92) compared with day 14 (0.71) and day 28 (0.74). The optimal cutoff value on day 56 was 20 p/mL, corresponding to a sensitivity of 89% and a specificity of 91%. ROC curves of relative parasite load at day 14, 28, or 56 in relation to baseline were also evaluated, resulting in comparable AUCs (0.93 on day 56); thus, the absolute parasite load was preferred because only a single sample is needed. Based on a threshold of 20 p/mL on day 56, 67.6% of patients in this study were correctly categorized as relapsed for day 210 outcome, taking into account missing samples to evaluate the overall performance of the sampling procedure, extraction, and qPCR method. Without missing samples, 85.2% of patients were correctly categorized as relapsed, representing the performance of the qPCR method. Relapsed patients not predicted at day 56 relapsed at days 68, 86, 108, and 112, whereas correctly predicted relapsed patients relapsed at day 102 (median) (interquartile range 64.5–136.5).
Figure 2.
ROC curves of absolute parasite load as predictor of clinical relapse on days 14, 28, and 56 of follow-up. AUC represents the integrated area under the ROC curve. Green line: day 14 (AUC 0.71); red line: day 28 (AUC 0.74); blue line: day 56 (AUC 0.92). Abbreviations: AUC, area under the curve; ROC, receiver operating characteristic.
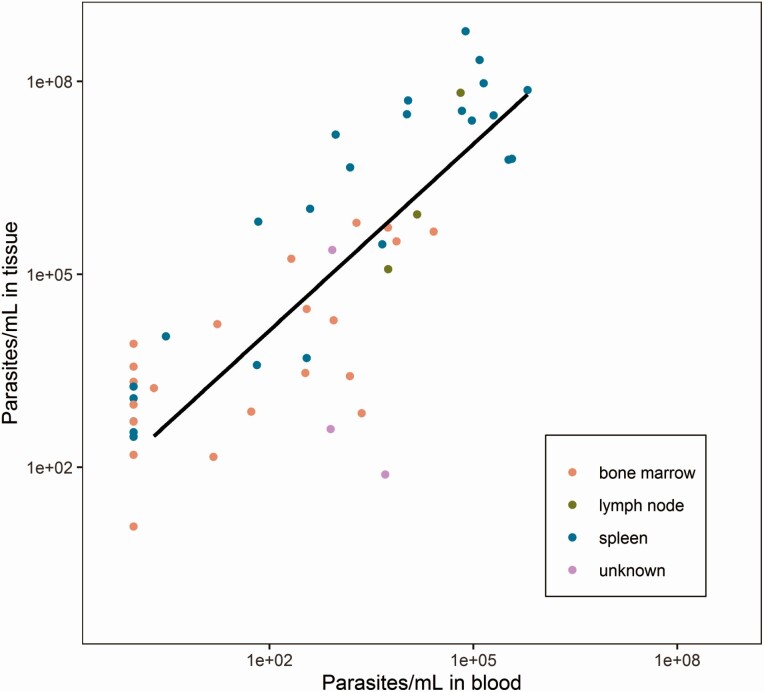
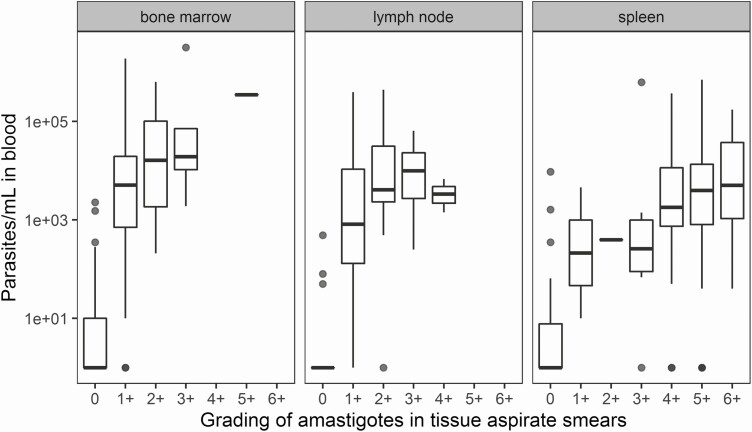
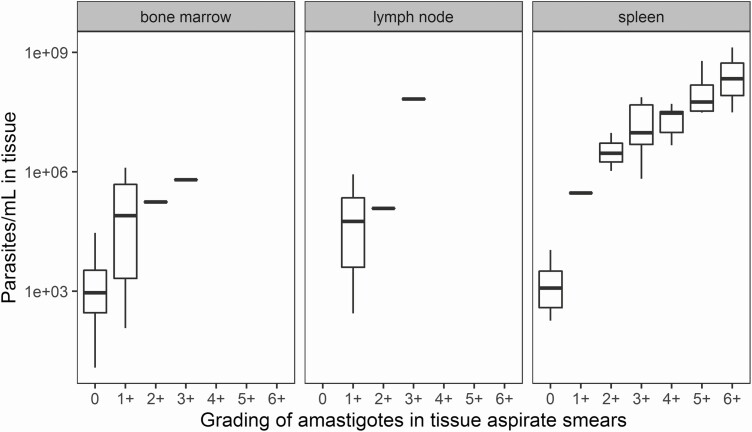
There was a significant correlation between matching log-transformed blood and tissue qPCR results (ρ = 0.80), indicating an approximately 2-log higher parasite load in spleen, bone marrow, or lymph node compared with whole blood (Figure 3). In total, 302 blood qPCR samples and 71 tissue qPCR samples were compared with matching microscopy gradings of tissue aspirate smears (Figures 4 and 5). When stratified by tissue source, there was a positive trend between the 2 scores, especially in samples from bone marrow and spleen. At start of treatment, parasites were detectable by microscopy in all tissue samples (microscopy grading > 0), whereas 6% of matching blood qPCR samples were negative (Table 3). When no parasites were detected by microscopy on day 28, parasites were still detected by qPCR in 36% of blood samples (Table 3). Parasites were detectable by qPCR in all of the available tissue samples (data not shown).
Figure 3.
Correlation between log-transformed qPCR blood and tissue parasite load (matching ID/timepoint) determined in bone marrow aspirates (red), lymph nodes (green), and spleen aspirates (blue). Tissue samples include 4 drops of bone marrow aspirate (~200 µL), or the remainder in the needle of the spleen or lymph node aspiration. Data below the limit of quantification are shown as 1 p/mL. Linear regression line (solid line) is based on the combined data, excluding data below the limit of quantification: y = 1.5 + 0.97x. Abbreviations: ID, identification; qPCR, quantitative polymerase chain reaction.
Figure 4.
Correlation between log-transformed qPCR blood parasite load and grading of amastigotes in aspirate smears by microscopy, stratified by parasite load according to tissue source. Abbreviation: qPCR, quantitative polymerase chain reaction.
Figure 5.
Correlation between log-transformed qPCR tissue parasite load and grading of amastigotes in aspirate smears by microscopy, stratified by parasite load according to tissue source. Tissue samples include 4 drops of bone marrow aspirate (~200 µL), or the remainder in the needle of the spleen or lymph node aspiration. Abbreviation: qPCR, quantitative polymerase chain reaction.
Table 3.
Number (%) of Positive and Negative Blood qPCR Loads Versus Microscopy Gradings for Matching Samples at Day 0 (N = 143) and Day 28 (N = 135)
| Day 0 | Day 28 | |||
|---|---|---|---|---|
| Microscopy Grading | Microscopy Grading | |||
| Positive | Negative | Positive | Negative | |
| Total (N) | 143 | 0 | 10 | 135 |
| Matching blood qPCR loads | ||||
| Positive, N (%) | 135 (94) | 0 (0) | 7 (70) | 48 (36) |
| Negative, N (%) | 8 (6) | 0 (0) | 3 (30) | 87 (64) |
Microscopy gradings > 0 were considered positive.
Abbreviation: qPCR, quantitative polymerase chain reaction.
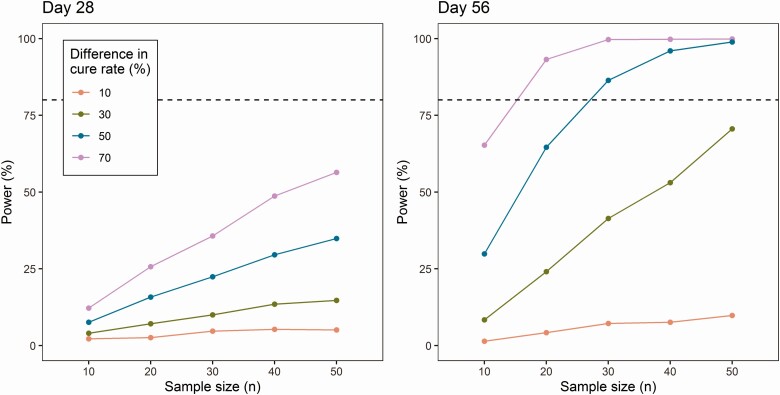
In the clinical trial simulations, absolute blood parasite load on either day 28 or day 56 was evaluated as a surrogate endpoint to predict clinical cure for various treatment regimens, with a threshold of ≤ 20 p/mL based on the ROC curves. The power of different simulation scenarios is shown in Figure 6. Clinical trial simulations demonstrated that the power to detect a difference in cure rate was higher when blood parasite load on day 56 was used, instead of day 28, in accordance with the ROC curves. When blood parasite load on day 56 was used, clinical trials only achieved a > 80% power when the difference in cure rate was high (> 50%) between the hypothetical investigational regimen and a standard of care regimen with an efficacy of 90% and when sufficient patients were included. For example, to identify an insufficient treatment regimen with 40% cure rate, at least 30 patients per treatment regimen need to be included. For alternative treatment regimens with higher cure rates, no adequate power was achieved with a sample size ≤ 50 subjects per treatment regimen.
Figure 6.
Predictive power of blood parasite load is shown for day 28 (left) and day 56 (right), with clinical cure defined as parasite load ≤ 20 p/mL. The difference in cure rate is the difference between the alternative treatment regimens (20%, 40%, 60%, or 80% cure rate) and the reference treatment regimen (90% cure rate). Sample size ranges from n = 10 to n = 50. Dotted horizontal line represents the 80% power cutoff.
DISCUSSION
In this study, various parasitemia parameters were evaluated for their sensitivity and specificity in classifying and predicting final treatment outcome in a large Eastern African VL patient population. Absolute parasite load on day 56 was a highly sensitive predictor of relapse at a cutoff of 20 p/mL. When compared with other approaches, the surrogate marker can be assessed early (day 56 instead of 6 months) compared with immunoglobulin G1 antigen detection [24] and more specific compared with antigen detection in urine [25]. The low cutoff value found in this study indicates that blood parasite loads as low as 20 p/mL are associated with a higher risk of disease relapse, even when patients do not yet present reoccurrence of clinical symptoms. Previously, this has only been demonstrated in HIV co-infected patients, in whom values ranging from 0.03 to 42 p/mL indicated relapse [7, 17, 18, 26].
A potential drawback of this biomarker is that blood represents only a proximal site for the total parasite biomass in the human host, of which the mainstay is resident in infected organs (eg, liver, spleen, bone marrow). This is in line with our findings because qPCR was approximately 2-log higher in tissue compared with whole blood. Another potential source of bias might be lingering kDNA of dead parasites in the circulation. However, a rapid clearance of circulating Leishmania kDNA immediately after treatment initiation has been shown previously, following clinical recovery [13]. Additionally, qPCR blood parasite load showed a good correlation with qPCR parasite load in tissue (ρ = 0.80), indicating that whole blood is a good proxy compartment to monitor the parasite biomass in the infected tissues.
qPCR has been shown to be a sensitive method to measure blood parasite load previously [16, 18, 19, 27, 28], as well as in this study. Both blood and tissue qPCR parasite loads showed a correlation with microscopy gradings from aspirate smears; the clearest trend was observed between spleen qPCR and microscopy gradings. The observed correlation is in line with previous data from India [29]. qPCR analysis seems to be a more sensitive method because parasites were detectable by qPCR in all tissue samples and in 76.7% of blood samples, compared with 60.5% of tissue samples by microscopy. The high sensitivity of qPCR on whole blood, as well as the convenience for the patient, suggest that qPCR is a suitable method for regular patient monitoring. Noteworthy is that detectable qPCR blood or tissue parasite loads at end of treatment or during follow-up were observed in patients considered clinically cured. This could indicate that patients can still harbor Leishmania parasites at low levels, but nevertheless remain asymptomatic. In the context of a clinical trial, this means negative blood qPCR loads cannot replace the gold standard of microscopic examination as a test of cure to define initial cure and the clinical value of a positive qPCR in a patient without clinical signs and symptoms of disease remains to be defined. It could indicate the need for closer follow-up but not directly rescue treatment, as for an immunocompetent patient the immune system is expected to control the infection, conferring long-lasting protection [30, 31].
To evaluate the usefulness of early blood parasite load as a surrogate endpoint for long-term clinical outcome in clinical trials evaluating novel drug regimens, clinical trial simulations were performed. The use of blood parasite load on day 56 might be suitable to identify insufficient treatment regimens or dose levels with a very poor cure rate of 40% or less and stop early for futility. However, the power will improve when the number of excluded samples can be reduced, for example, by improving the performance of DNA extraction.
With the introduction of new chemical entities as clinical candidates for VL treatment, there is a need for better and more accurate tools to evaluate their efficacy at early time points to allow for adaptive study design to select promising drug regimens and reduce the number of subjects exposed to regimens with poor activity. This is the first study that has evaluated the predictive value of qPCR for long-term clinical outcome and its use as a surrogate endpoint in clinical trials for VL, by using a large dataset from different studies in Eastern African VL patients, including treatment regimens with different cure rates. The absolute parasite load on day 56 was a highly sensitive predictor of relapse at a cutoff of 20 p/mL, and its potential application has been shown by clinical trial simulations. However, this cutoff value is based on the studied data only, and the exact threshold and time point may need to be optimized for future compounds, depending on their pharmacokinetic properties, treatment duration, and ultimately their effect on parasite dynamics. With the increase in molecular biology capacity in areas endemic for VL, we expect that it would be feasible to put this tool into practice in clinical trial settings. In the near future, validation of molecular biology tools in blood could be envisaged to replace the current invasive tissue aspiration procedures for parasitological diagnosis and treatment monitoring.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. Data originated from trials conducted by the Leishmaniasis East Africa Platform (LEAP), in collaboration with the trial sites, and coordinated and implemented by the Drugs for Neglected Diseases initiative (DNDi). The authors thank all members of the field teams, including nurses and laboratory technicians, in all study sites, and clinical monitors and Data Safety Monitoring Board for their contributions to the study. The authors also thank the Ministries of Health of Kenya, Uganda, and Gedaref State, Sudan, for their support. They sincerely thank the visceral leishmaniasis patients and the parents of the pediatric patients for their willingness to be enrolled in this study and their cooperation. The authors also acknowledge Erik van Werkhoven and Alwin Huitema for their input and discussion.
Financial support. This work was supported by the European Union Seventh Framework Programme Africoleish (grant number 305178); the World Health Organization—Special Programme for Research and Training in Tropical Diseases (WHO-TDR); the French Development Agency, France (grant number CZZ2062); UK aid, UK; the Federal Ministry of Education and Research through KfW, Germany; the Medicor Foundation, Liechtenstein; Médecins Sans Frontières, International; the Swiss Agency for Development and Cooperation (SDC), Switzerland (grant number 81017718); the Dutch Ministry of Foreign Affairs (DGIS), the Netherlands (grant number PDP15CH21); the French Ministry for Europe and Foreign Affairs (MEAE), France; The Rockefeller Foundation, USA; BBVA Foundation, Spain; the European Union—AfriKADIA project of the Second European and Developing Countries Clinical Trials Partnership Programme (EDCTP2) (grant number RIA2016S1635); and ZonMw/Dutch Research Council (NWO) Veni grant (project number 91617140 to T. P. C. D.).
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Alves F, Bilbe G, Blesson S, et al. Recent development of visceral leishmaniasis treatments: successes, pitfalls, and perspectives. Clin Microbiol Rev 2018; 31:1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Ending the neglect to attain the sustainable development goals—a road map for neglected tropical diseases 2021–2030. 2020:1–13. Available at: https://www.who.int/neglected_diseases/Revised-Draft-NTD-Roadmap-23Apr2020.pdf?ua=1. Accessed 25 August 2020.
- 3.World Health Organization. Control of the leishmaniases. Report of a meeting of the WHO expert committee on the control of leishmaniases, Geneva.2010: 49-51. Available at: https://apps.who.int/iris/bitstream/handle/10665/44412/WHO_TRS_949_eng.pdf?sequence=1. Accessed 10 October 2020. [PubMed]
- 4.Rijal S, Ostyn B, Uranw S, et al. Increasing failure of miltefosine in the treatment of Kala-azar in Nepal and the potential role of parasite drug resistance, reinfection, or noncompliance. Clin Infect Dis 2013; 56:1530–8. [DOI] [PubMed] [Google Scholar]
- 5.Chulay JD, Bryceson AD. Quantitation of amastigotes of Leishmania donovani in smears of splenic aspirates from patients with visceral leishmaniasis. Am J Trop Med Hyg 1983; 32:475–9. [DOI] [PubMed] [Google Scholar]
- 6.Chappuis F, Sundar S, Hailu A, et al. Visceral leishmaniasis: what are the needs for diagnosis, treatment and control? Nat Rev Microbiol 2007; 5:873–82. [DOI] [PubMed] [Google Scholar]
- 7.Bossolasco S, Gaiera G, Olchini D, et al. Erratum: real-time PCR assay for clinical management of human immunodeficiency virus-infected patients with visceral leishmaniasis. J Clin Microbiol 2004; 42:1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cascio A, Calattini S, Colomba C, et al. Polymerase chain reaction in the diagnosis and prognosis of Mediterranean visceral leishmaniasis in immunocompetent children. Pediatrics 2002; 109:e27. [DOI] [PubMed] [Google Scholar]
- 9.Pizzuto M, Piazza M, Senese D, et al. Role of PCR in diagnosis and prognosis of visceral leishmaniasis in patients coinfected with human immunodeficiency virus type 1. J Clin Microbiol 2001; 39:357–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudarshan M, Weirather JL, Wilson ME, Sundar S. Study of parasite kinetics with antileishmanial drugs using real-time quantitative PCR in Indian visceral leishmaniasis. J Antimicrob Chemother 2011; 66:1751–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cota GF, de Sousa MR, de Assis TSM, Pinto BF, Rabello A. Exploring prognosis in chronic relapsing visceral leishmaniasis among HIV-infected patients: circulating Leishmania DNA. Acta Trop 2017; 172:186–91. [DOI] [PubMed] [Google Scholar]
- 12.Cruz I, Cañavate C, Rubio JM, et al. ; Spanish HIV-Leishmania Study Group . A nested polymerase chain reaction (Ln-PCR) for diagnosing and monitoring Leishmania infantum infection in patients co-infected with human immunodeficiency virus. Trans R Soc Trop Med Hyg 2002; 96 Suppl 1:S185–9. [DOI] [PubMed] [Google Scholar]
- 13.Disch J, Oliveira MC, Orsini M, Rabello A. Rapid clearance of circulating Leishmania kinetoplast DNA after treatment of visceral leishmaniasis. Acta Trop 2004; 92:279–83. [DOI] [PubMed] [Google Scholar]
- 14.Fisa R, Riera C, Ribera E, Gállego M, Portús M. A nested polymerase chain reaction for diagnosis and follow-up of human visceral leishmaniasis patients using blood samples. Trans R Soc Trop Med Hyg 2002; 96 Suppl 1:S191–4. [DOI] [PubMed] [Google Scholar]
- 15.Kip AE, Balasegaram M, Beijnen JH, Schellens JH, de Vries PJ, Dorlo TP. Systematic review of biomarkers to monitor therapeutic response in leishmaniasis. Antimicrob Agents Chemother 2015; 59:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lachaud L, Dereure J, Chabbert E, et al. Optimized PCR using patient blood samples for diagnosis and follow-up of visceral leishmaniasis, with special reference to AIDS patients. J Clin Microbiol 2000; 38:236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mary C, Faraut F, Drogoul MP, et al. Reference values for Leishmania infantum parasitemia in different clinical presentations: quantitative polymerase chain reaction for therapeutic monitoring and patient follow-up. Am J Trop Med Hyg 2006; 75:858–63. [PubMed] [Google Scholar]
- 18.Mary C, Faraut F, Lascombe L, Dumon H. Quantification of Leishmania infantum DNA by a real-time PCR assay with high sensitivity. J Clin Microbiol 2004; 42:5249–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nuzum E, White F 3rd, Thakur C, et al. Diagnosis of symptomatic visceral leishmaniasis by use of the polymerase chain reaction on patient blood. J Infect Dis 1995; 171:751–4. [DOI] [PubMed] [Google Scholar]
- 20.Khalil EAG, Weldegebreal T, Younis BM, et al. Safety and efficacy of single dose versus multiple doses of AmBisome for treatment of visceral leishmaniasis in Eastern Africa: a randomised trial. PLoS Negl Trop Dis 2014; 8:e2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wasunna M, Njenga S, Balasegaram M, et al. Efficacy and safety of AmBisome in combination with sodium stibogluconate or miltefosine and miltefosine monotherapy for African visceral leishmaniasis: phase II randomized trial. PLoS Negl Trop Dis 2016;10:e0004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mbui J, Olobo J, Omollo R, et al. Pharmacokinetics, safety, and efficacy of an allometric miltefosine regimen for the treatment of visceral leishmaniasis in Eastern African children : an open-label, phase II clinical trial. Clin Infect Dis 2019; 68:1530–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abongomera C, Diro E, Vogt F, et al. The risk and predictors of visceral leishmaniasis relapse in human immunodeficiency virus-coinfected patients in ethiopia: a retrospective cohort study. Clin Infect Dis 2017; 65:1703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marlais T, Bhattacharyya T, Singh OP, et al. Visceral leishmaniasis IgG1 rapid monitoring of cure vs. relapse, and potential for diagnosis of post kala-azar dermal leishmaniasis. Front Cell Infect Microbiol 2018; 8:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogt F, Mengesha B, Asmamaw H, et al. Antigen detection in urine for noninvasive diagnosis and treatment monitoring of visceral leishmaniasis in human immunodeficiency virus coinfected patients: an exploratory analysis from ethiopia. Am J Trop Med Hyg 2018; 99:957–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molina I, Fisa R, Riera C, et al. Ultrasensitive real-time PCR for the clinical management of visceral leishmaniasis in HIV-infected patients. Am J Trop Med Hyg 2013; 89:105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams ER, Schoone G, Versteeg I, et al. Development and evaluation of a novel loop-mediated isothermal amplification assay for diagnosis of cutaneous and visceral leishmaniasis. J Clin Microbiol 2018; 56:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piarroux R, Gambarelli F, Dumon H, et al. Comparison of PCR with direct examination of bone marrow aspiration, myeloculture, and serology for diagnosis of visceral Leishmaniasis in immunocompromised patients. J Clin Microbiol 1994; 32:746–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudarshan M, Singh T, Chakravarty J, Sundar S. A correlative study of splenic parasite score and peripheral blood parasite load estimation by quantitative PCR in visceral leishmaniasis. J Clin Microbiol 2015; 53:3905–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khalil EA, Ayed NB, Musa AM, et al. Dichotomy of protective cellular immune responses to human visceral leishmaniasis. Clin Exp Immunol 2005; 140:349–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zijlstra EE, el-Hassan AM, Ismael A, Ghalib HW. Endemic kala-azar in eastern Sudan: a longitudinal study on the incidence of clinical and subclinical infection and post-kala-azar dermal leishmaniasis. Am J Trop Med Hyg 1994; 51:826–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.