Abstract
Background
Ending the human immunodeficiency virus (HIV) epidemic requires knowledge of key drivers of spread of HIV infection.
Methods
Between 1996 and 2018, 1119 newly and previously diagnosed, therapy-naive persons with HIV (PWH) from San Diego were followed. A genetic distance–based network was inferred using pol sequences, and genetic clusters grew over time through linkage of sequences from newly observed infections. Cox proportional hazards models were used to identify factors associated with the rate of growth. These results were used to predict the impact of a hypothetical intervention targeting PWH with incident infection. Comparison was made to the Centers for Disease Control and Prevention (CDC) Ending the HIV Epidemic (EHE) molecular surveillance strategy, which prioritizes clusters recently linked to all new HIV diagnoses and does not incorporate data on incident infections.
Results
Overall, 219 genetic linkages to incident infections were identified over a median follow-up of 8.8 years. Incident cluster growth was strongly associated with proportion of PWH in the cluster who themselves had incident infection (hazard ratio, 44.09 [95% confidence interval, 17.09–113.78]). The CDC EHE molecular surveillance strategy identified 11 linkages to incident infections a genetic distance threshold of 0.5%, and 24 linkages at 1.5%.
Conclusions
Over the past 2 decades, incident infections drove incident HIV cluster growth in San Diego. The current CDC EHE molecular detection and response strategy would not have identified most transmission events arising from those with incident infection in San Diego. Molecular surveillance that includes detection of incident cases will provide a more effective strategy for EHE.
Keywords: phylogenetics, cluster growth, incident HIV, molecular surveillance, prevention
Incident infections drove growth of Human Immunodeficiency Virus (HIV) genetic clusters in San Diego over the past 20 years. Molecular surveillance that includes detection of incident cases will provide a more effective molecular detection and response strategy for ending the HIV epidemic.
The ambitious goal of the Ending the HIV Epidemic (EHE) initiative [1] includes reducing new infections by 75% within 5 years. Human immunodeficiency virus (HIV) molecular cluster detection and response is a core pillar of this plan, despite paucity of evidence to support its effectiveness in reducing HIV incidence. This strategy is intended to identify outbreaks that could be stopped by interventions, such as rapid antiretroviral therapy (ART) for newly diagnosed persons with HIV (PWH) and preexposure prophylaxis (PrEP) for persons at risk for HIV. Such efforts require analysis of HIV nucleotide sequences collected during routine drug resistance testing to define clusters to which interventions are directed by regional health departments. Successful interventions based on molecular surveillance require knowledge of which clusters are actively growing or are most likely to grow. One way to find out is to identify PWH with incident (newly acquired) infection who link to clusters; such events are denoted as “incident cluster growth.” In fact, the risk of transmission for persons with acute infection has been reported to be twice as high as for those with prevalent infection [2]. As an alternative to direct measures of incidence, the Centers for Disease Control and Prevention (CDC) strategy uses short genetic distance (0.5% genetic distance) and temporal information (cases diagnosed in the last 12 or 36 months) as a proxy for recent and rapid transmission [3–5].
We hypothesized that prioritizing PWH and incident vs prevalent HIV infection status would improve the performance of molecular detection and response. To test this hypothesis, we modeled the growth of clusters by addition of incidence cases and predicted the effect of theoretical prevention interventions that prioritized specific populations (ie, persons with incident infection) using fitted models. We analyzed data collected from persons newly diagnosed with incident and prevalent HIV infection in San Diego, California (1996–2018). We used statistical modeling to demonstrate that success of molecular cluster detection and response efforts depended on prioritization of persons with incident HIV infection.
METHODS
Study Population
Between 1 July 1996 and 31 March 2018, ART-naive adult and adolescent (≥16 years of age) persons with HIV were prospectively recruited to observational research studies at the University of California, San Diego (UCSD). Participants of the Primary Infection Resource Consortium (PIRC) were identified by a screening algorithm to detect incident HIV infection (≤1 year) and prevalent infection (>1 year) [6, 7], and an estimated date of infection (EDI) was calculated [8] (see Supplementary Materials). Participants of the Center for AIDS Research Network of Integrated Clinical Systems network were ≥18-year-old PWH newly entering care at UCSD and were assigned to “prevalent” infection status when no prospectively determined EDIs were available. All studies were approved by the UCSD Institutional Review Board, and all participants provided voluntary, written informed consent.
Baseline variables included HIV genotype, testing for bacterial sexually transmitted infections (STIs) (gonorrhea [GC], chlamydia [CT], and syphilis), and routine laboratory tests needed for clinical care. Positive STI screen was defined as a positive GC/CT nucleic acid test (pharyngeal, rectal, or urine) or evidence of active syphilis by a rapid plasma reagin titer >1:8 [9]. PIRC participants completed study visits at baseline (day 0); weeks 2, 4, 8, 12, 24, 36, and 48; and every 24 weeks thereafter. Some study variables were incompletely measured (see Table 1 footnotes).
Table 1.
Baseline Participant Characteristics
| Characteristic | PIRC Newly Diagnosed (n = 741) | PIRC Previously Diagnosed (n = 194) | CNICS Previously Diagnosed (n = 184) | Total (N = 1119) |
|---|---|---|---|---|
| Participant demographics | ||||
| Gendera | ||||
| Male | 719 (97.0) | 163 (92.1) | 168 (91.3) | 1050 (95.3) |
| Female | 19 (2.6) | 13 (7.3) | 16 (8.7) | 48 (4.4) |
| Transgender | 3 (0.4) | 1 (0.6) | … | 4 (0.4) |
| Race/ethnicity | ||||
| White non-Hispanic | 394 (53.2) | 80 (41.2) | 86 (46.7) | 560 (50.0) |
| Black non-Hispanic | 44 (5.9) | 25 (12.9) | 20 (10.9) | 89 (8.0) |
| Hispanic | 224 (30.2) | 54 (27.8) | 63 (34.2) | 341 (30.5) |
| Other/unknown | 79 (10.7) | 35 (18.0) | 15 (8.2) | 129 (11.5) |
| Transmission risk | ||||
| MSM | 666 (89.9) | 135 (69.6) | 123 (66.8) | 924 (82.6) |
| MSM + PWID | 28 (3.8) | 11 (5.7) | 14 (7.6) | 53 (4.7) |
| PWID | 1 (0.1) | 3 (1.5) | 9 (4.9) | 13 (1.2) |
| Heterosexual | 22 (3.0) | 6 (3.1) | 18 (9.8) | 46 (4.1) |
| Other/unknown | 24 (3.2) | 39 (20.1) | 20 (10.9) | 83 (7.4) |
| Age, y, median (IQR) | 32 (26–40) | 36 (29–42) | 38 (30–46) | 34 (27–42) |
| Months on study, median (IQR) | 18 (4–38) | 0 (0–1) | 37 (23–58) | 17 (3–39) |
| Risk characteristics reported for prior month of study entry | ||||
| Illicit drug useb,c | 125 (34.5) | 17 (30.9) | … | 142 (34.1) |
| Condomless anal sexd | 529 (76.9) | 43 (50.0) | … | 572 (73.9) |
| No. of sexual partnerse, median (IQR) | 2 (1–3) | 1 (1–2) | … | 2 (1–3) |
| HIV infection timeline at study entry | ||||
| Acutef | 172 (23.2) | … | … | 172 (15.4) |
| Recent | 483 (65.2) | … | … | 483 (43.2) |
| Prevalent | 86 (11.6) | 174 (89.7) | … | 260 (23.2) |
| Presumed to be prevalent | … | 20 (10.3) | 184 (100.0) | 204 (18.2) |
| STI diagnosis at study entryg | ||||
| Gonorrheah | 45 (8.4) | 3 (3.6) | 1 (0.8) | 49 (6.6) |
| Chlamydiah | 57 (10.7) | 3 (3.6) | 2 (1.6) | 62 (8.4) |
| Syphilish | 25 (4.5) | 3 (3.9) | … | 28 (3.8) |
| HIV laboratory resultsi at time of HIV sequencing | ||||
| Viral load, log10 copies/mL, median (IQR)i | 4.9 (4.1–5.6) | 4.3 (3.4–5.1) | 4.4 (3.9–4.9) | 4.7 (3.9–5.4) |
| CD4 count at study entry, cells/μL, median (IQR)i | 494 (360 642) | 410 (284 587) | 411 (238 617) | 477 (327 636) |
| Phylogenetic clustering, determined by HIV pol sequence | ||||
| Clustered | 377 (50.9) | 87 (44.8) | 68 (37.0) | 532 (47.5) |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: CDC, Centers for Disease Control and Prevention; CNICS, Center for AIDS Research Network of Integrated Clinical Systems; HIV, human immunodeficiency virus; IQR, interquartile range; MSM, men who have sex with men; PIRC, Primary Infection Resource Consortium; PWID, person who injects drugs; STI, sexually transmitted infection.
aGender was not assessed for 17 participants in the previously diagnosed PIRC cohort.
bIllicit drug use excluding cannabis.
cIllicit drug use was not assessed for the CNICS cohort and a subset of PIRC participants (379 newly diagnosed and 139 previously diagnosed participants).
dRecent condomless anal sex was not assessed for the CNICS cohort and a subset of PIRC participants (53 newly diagnosed and 108 previously diagnosed participants).
eNumber of recent sexual partners was not assessed for the CNICS cohort and a subset of PIRC participants (36 newly diagnosed and 66 previously diagnosed participants).
fAcute: HIV antibody negative, ≤10 days from estimated date of infection (EDI); recent: ≤1 year from EDI; prevalent: >1 year from EDI.
gAny STI classified as any positive gonorrhea; chlamydia confirmed by 3-site testing of urine, throat, and rectum; or presumed active syphilis (enzyme immunoassay + rapid plasma reagin titer >1:8).
hGonorrhea and chlamydia were not assessed for 61 CNICS participants (62 for chlamydia) and a subset of PIRC participants (206 newly diagnosed and 110 previously diagnosed participants). Syphilis was not assessed or incompletely assessed for 57 CNICS participants and a subset of PIRC participants (200 newly diagnosed and 117 previously diagnosed participants).
iViral RNA was unavailable for 25 previously diagnosed PIRC participants. CD4 count at sequence date was unavailable for a subset of PIRC participants (3 newly diagnosed and 45 previously diagnosed participants).
HIV Network Inference
HIV pol nucleotide sequences were derived for all participants (GenoSure MG, LabCorp Specialty Testing Group, South San Francisco, California; or Viroseq version 2.0, Celera Diagnostics, Alameda, California). If >1 HIV sequence was available for a participant, only the earliest was included in this analysis. We inferred the HIV network by computing all pairwise genetic distances between pol sequences from each participant (ie, network node) and connected nodes for which the corresponding genetic distance was <1.5% using HIV-TRACE [10] (hereafter, the UCSD method). For comparison, we reconstructed the network using the CDC criteria, which prioritizes only clusters that have at least 5 newly diagnosed cases identified within the previous year linked using a genetic distance threshold of 0.5% to cases diagnosed within the previous 3 years [5]. We then determined how many new incident cases linked to the clusters identified by the CDC vs UCSD methods during follow-up. We repeated the CDC method analysis using a genetic-distance threshold of 1.5% (vs 0.5%) to compare to outcomes derived using the UCSD method (with the identical temporal information used by the CDC method). There are 2 key differences in the way these methods prioritize clusters: The CDC method uses temporal thresholding (minimum number of nodes diagnosed within a specific timeframe), whereas the UCSD method uses incidence information (number of nodes in the cluster that are incident).
Incident Cluster Growth Analysis
We compared the number of linked incident cases (ie, incident cluster growth) and investigated the factors associated with incident cluster growth in the network using the UCSD method. To do so, we identified PWH (ie, nodes) that did not link to any earlier nodes in the network, which began in 1996. These were defined as “seeds” and followed over time. For each seed, or cluster that arose from a seed, we counted the number of incident nodes that subsequently linked to that seed or cluster (Supplementary Table 1A). The primary outcome for all cluster growth analyses was “incident cluster growth,” defined as linkage of an incident node to a node already in the network (Figure 1). While not counted as incident cluster growth events, newly linked nodes with prevalent infection (>1 year since infection, n = 148) did contribute to cluster size, which was analyzed as a covariate.
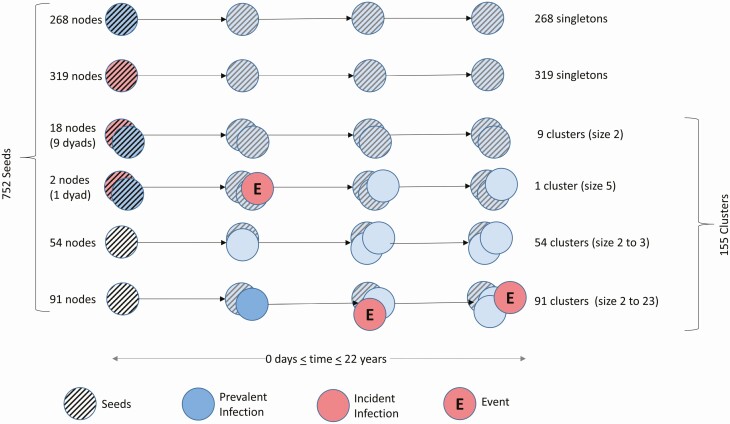
Figure 1.
Schematic of possible cluster outcomes. Prevalent (blue) and incident (red) infections are defined as nodes with estimated date of infection (EDI) >1 and ≤1 year, respectively. To investigate the factors associated with the linkage of a new linked incident node event (red node marked by “E”), we reconstructed the history of the network to identify the earliest node in each cluster “lineage,” including those nodes that never became part of a cluster during follow-up. We defined these nodes as seeds, indicated by the stripped pattern (n = 752). During the course of follow-up, 587 among the 752 seeds started as singletons and never acquired another node (268 prevalent seeds [illustrated in striped blue, row 1], 319 incident seeds [illustrated in striped red, row 2] that become prevalent during follow-up [illustrated in solid blue]). Eighteen seeds started as dyads and remained so (row 3). Fifty-four seeds acquired only prevalent nodes (row 5), whereas 2 seeds in a dyad (row 4) and 91 seeds (row 6) acquired at least 1 incident node during follow-up.
We used the Andersen-Gill extension to the Cox proportional hazards model [11] (“coxph” function in R with robust variance estimates) to analyze the time to recurrent, incident cluster growth arising from seeds (see Supplementary Materials for details). This model permitted estimation of mean number of links (averaged across clusters). To adjust for the time-varying rate of sampling, all analyses were weighted by the inverse of a smoothed density estimate of the number of sequences collected over time (see Supplementary Materials for details). This estimate was approximately proportional to the sampling rate, assuming that HIV prevalence stayed roughly constant over time (see Supplementary Materials). To model cluster growth, we identified predictors associated with incident events in separate univariable models and included them in a multivariable model if either they were significant at the α = .05 level or were considered factors that might potentially confound the relationship of other predictors to the outcome of cluster growth (cluster-level covariates 2 and 7 below).
Cluster-level covariates used to predict rate of incident cluster growth included (1) proportion of cluster members who had acquired HIV in the past year; (2) proportion of cluster members with Hispanic ethnicity; (3) proportion with unsuppressed viral load (>50 copies/mL); (4) median number of sex partners reported by cluster members in the 3 months preceding study enrollment; (5) cluster size; (6) median age of cluster members; and (7) indicator of a cluster start date (ie, enrollment of the first person in the cluster) before 2005. The year 2005 was chosen because the rate of identifying PWH was increasing before that date and remained nearly constant thereafter. Inclusion of item 7 in all models was intended to adjust for any potential residual confounding related to variation in characteristics of people included in samples that is not captured by the inverse weighting by the observed density [7].
All cluster-level covariates were time-varying, except the pre-2005 cluster start indicator. Since covariates were constructed based on features of cluster members, we used multiple imputation to address the issue of mismeasurement in time (ie, viral load could not be known for cluster members at the exact time of a new linkage) and missing covariate information. To investigate sensitivity of results to definition of incidence, we repeated all analyses using the threshold of 133 days since HIV infection, [8] which was chosen for easier identification of people with recent (ie, incident) infection using a limiting-antigen avidity assay [12] (see Supplementary Materials). Using the parameter estimates, we also predicted effects of hypothetical interventions targeting the proportions of PWH who were incident, unsuppressed, or both. We did so by multiplying the observed proportions of incident or suppressed cases within each cluster by 0.5 and 0.25 at each time point to emulate a hypothetical 50% and 75% reduction in this proportion (Supplementary Materials).
RESULTS
Study Population
We analyzed 1119 prospectively enrolled ART-naive PWH (741 were newly diagnosed and 378 were previously diagnosed and newly entering care). Of these, 655 (58.5%) had incident infection (acute [seronegative], n = 172 [15.4%]; or early, n = 483 [43.2%]) and 464 (41.5%) had prevalent infection at time of diagnosis. As with the overall HIV epidemic in San Diego [13], most participants were male (95.3%); 87.3% of male participants were men who had sex with men (MSM). Half of all participants were non-Hispanic white and 30.5% were Hispanic (Table 1). In the month before enrollment, 142 of 417 (34.1%) participants described using illicit substances, 572 of 774 (73.9%) reported engaging in condomless anal sex, and 833 reported a median of 2 sex partners (interquartile range [IQR], 1–3) (Table 1, see footnotes for incomplete entry variables). Screening for at least 1 bacterial STI was performed in 819 (73.2%) participants within 30 days of study entry; gonorrhea was documented in 6.6%, chlamydia in 8.4%, and syphilis in 3.8%. There was no difference in the rate of STIs between participants with incident and prevalent infection (P = .3). Participants with newly diagnosed HIV infection (88.4% had incident infection) had a higher (P < .01) median viral load (4.9 [IQR, 4.1–5.6] log10 copies/mL) than did those with previously diagnosed infection (4.3 [IQR, 3.8–5.0] log10 copies/mL).
Incident Cluster Growth Analysis by UCSD Criteria
A genetic network was inferred from all participant sequences with a range of follow-up from 0 days to 21.8 years (median, 8.8 years). The network comprised 752 nodes identified as seeds: 587 seeds never became part of a cluster (ie, remained as a singleton); the remaining 165 eventually linked to 1 or more nodes forming 155 clusters (20 nodes started as dyad-seeds). Among the 752 seeds, 147 (19.5%) linked to 1 or more nodes sequenced at a later date (ie, formed a cluster), and 93 seeds (12.4%) linked to at least 1 incident node, resulting in incident cluster growth. A total of 219 incident nodes linked to seeds or clusters. Since 2 incident nodes entered the network as a dyad, there were 218 linkage events observed during study. There were 117 incident-to-incident linkage events (54% of all linkage events) within 56 clusters; the median time between the EDI of putative transmission pairs was 2.6 (IQR, 1.4–4.9) months, demonstrating the short window of time available to intervene on incident cases.
Incident Cluster Growth Analysis by CDC Criteria
The CDC method prioritizes clusters that have a minimum of 3 or 5 (depending on jurisdiction) newly diagnosed cases identified within the previous year linked using a genetic distance threshold of 0.5% to cases diagnosed within the previous 12 months [5]. Applying these criteria to our data identified a total of 116 clusters (Supplementary Table 1B), including 3 priority clusters that would have triggered an investigation by CDC recommendations. During follow-up of these 3 clusters, 11 linked incident events would have been identified—all arising from 1 seed over a 1.2-year period (Supplementary Figure 1). Changing the genetic distance threshold to 1.5% resulted in identification of 4 clusters linked to 5 or more new cases within the previous year. During follow-up of these 4 clusters, 24 linked incident events would have been identified during a median follow-up of 4.4 (IQR, 1.2–8.2) years (Supplementary Figure 2). The CDC criteria uses close genetic distance (0.5%) and temporal information as a proxy for recent and rapid transmission; use of this distance threshold and temporal criteria when applied to our population identified only 11 of 218 (5.0%) linked incident events, which increased to 24 of 218 (11%) events when the genetic threshold was increased to 1.5% (Supplementary Table 1B and 1C).
Drivers of Incident Cluster Growth
In multivariable models, factors that increased incident cluster growth included proportion of persons with incident infection (duration of infection of ≤1 year) (hazard ratio [HR], 43.09 [95% confidence interval {CI}, 16.90–109.86]) (Table 2); cluster size (HR, 1.33 [95% CI, 1.29–1.37]); and cluster start date prior to 2005 (HR, 1.42 [95% CI, 1.02–1.97]). A weak association of proportion of cluster members with unsuppressed viral load with cluster growth was observed in the multivariate model (HR, 1.35 [95% CI, .98–1.86]), though the association was stronger in the univariate model (HR, 1.69 [95% CI, 1.33–2.14]). Both the proportion of Hispanic persons and median age in the cluster were negatively associated with cluster growth (HR, 0.63 [95% CI, .43–.92] and 0.96 [95% CI, .94–.97], respectively). Median number of sex partners reported during the month prior to study entry (HR, 0.99 [95% CI, .98–1.01], univariable model) was not associated with incident cluster growth. Results were qualitatively similar in multivariable analyses when duration of incident infection was defined to be within 133 days of infection (Table 3).
Table 2.
Univariable and Multivariable Drivers of Cluster Growth (Incident Infection Defined as Estimated Date of Infection ≤1 Year of Sequencing Date)
| Predictora | Univariable Model | Multivariable Model | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Coef | Coef SE | HR | (95% CI) | P Value | Coef | Coef SE | HR | (95% CI) | P Value | |
| Proportion of incidentb | 5.27 | 0.66 | 194.87 | (53.09–715.20) | <.001 | 3.76 | 0.48 | 43.09 | (16.90–109.86) | <.001 |
| Cluster size | 0.32 | 0.01 | 1.37 | (1.33–1.41) | <.001 | 0.29 | 0.01 | 1.33 | (1.29–1.37) | <.001 |
| Proportion of Hispanic ethnicityb | –0.35 | 0.14 | 0.70 | (.53–.92) | .011 | –0.46 | 0.19 | 0.63 | (.43–.92) | .017 |
| Median No. of sex partnersc | –0.01 | 0.01 | 0.99 | (.98–1.01) | .289 | … | … | … | … | |
| Cluster start pre-2005d | 0.90 | 0.15 | 2.45 | (1.81–3.31) | <.001 | 0.35 | 0.17 | 1.42 | (1.02–1.97) | .036 |
| Median age of clustere | –0.05 | 0.01 | 0.95 | (.93–.97) | <.001 | –0.04 | 0.01 | 0.96 | (.94–.97) | <.001 |
| Proportion of unsuppressedb | 0.52 | 0.12 | 1.69 | (1.33–2.14) | <.001 | 0.30 | 0.16 | 1.35 | (.98–1.86) | .069 |
Abbreviations: CI, confidence interval; Coef, coefficient; HR, hazard ratio; SE, standard error.
aAll predictors use clusters as the reference for evaluation.
bProportion as decimal, not percentage. For example, an increase in the proportion of incident infections from 0 to 50% would correspond to HR of exp(0.5 × 3.76) = 6.55, with the other predictors held constant.
cMedian number of sex partners in the past month at time of enrollment for each linked case.
dTime invariant.
eMedian age of cluster, which includes median ages of all individuals in cluster, up to but not including the individual linked at latest event time.
Table 3.
Model Fit for the Proportional Rates Recurrent Event Model (Incident Infection Defined as Estimated Date of Infection ≤133 Days of Sequencing Date)
| Predictora | Univariable Model | Multivariable Model | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Coef | Coef SE | HR | (95% CI) | P Value | Coef | Coef SE | HR | (95% CI) | P Value | |
| Proportion of incidentb | 1.84 | 0.58 | 6.27 | (2.02–19.40) | .001 | 1.66 | 0.54 | 5.24 | (1.80–15.23) | .002 |
| Cluster size | 0.31 | 0.01 | 1.37 | (1.33–1.41) | <.001 | 0.30 | 0.01 | 1.35 | (1.31–1.39) | <.001 |
| Proportion of Hispanic ethnicityb | –0.24 | 0.15 | 0.79 | (.58–1.06) | .113 | –0.35 | 0.20 | 0.71 | (.48–1.04) | .078 |
| Median No. of sex partnersc | –0.01 | 0.01 | 0.99 | (.97–1.01) | .306 | … | … | … | … | |
| Cluster start pre-2005d | 0.73 | 0.17 | 2.08 | (1.50–2.88) | <.001 | 0.24 | 0.18 | 1.27 | (.89–1.82) | .183 |
| Median age of clustere | –0.05 | 0.01 | 0.95 | (.93–.97) | <.001 | –0.05 | 0.01 | 0.96 | (.94–.97) | <.001 |
| Proportion of unsuppressedb | 0.59 | 0.13 | 1.81 | (1.40–2.35) | <.001 | 0.45 | 0.18 | 1.56 | (1.11–2.21) | .011 |
Abbreviations: CI, confidence interval; Coef, coefficient; HR, hazard ratio; SE, standard error.
aAll predictors use clusters as the reference for evaluation.
bProportion as decimal, not percentage. For example, a 50% increase corresponds to exp(0.5 β) times the hazard in the weighted model.
cMedian number of sex partners in past 3 months at time of enrollment for each linked case.
dTime invariant.
eMedian age of cluster, which includes median ages of all individuals in cluster, up to but not including the individual linked at latest event time.
Prevention Efforts
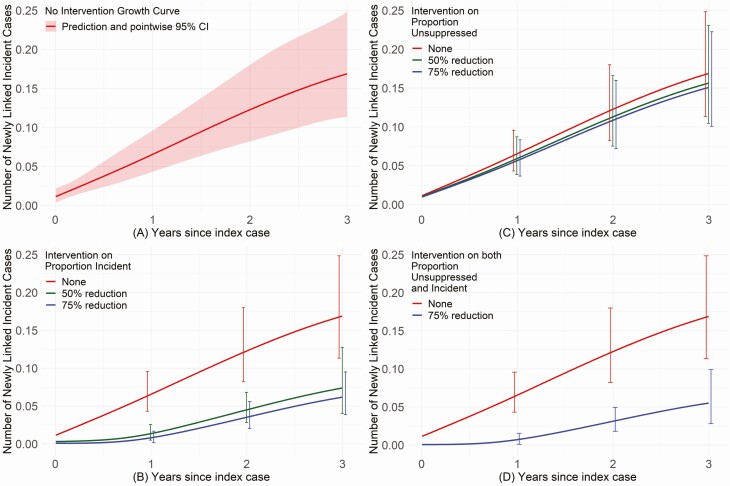
As incident infection was the characteristic most strongly associated with incident cluster growth (Table 2), we used model parameter estimates to evaluate how such growth would aid in prioritizing prevention efforts. The predicted mean number of newly linked incident cases per cluster was 0.17 at 3 years (Figure 2A) and 0.33 at 15 years (data not shown), implying that cluster growth slowed over time. If incident cases could be identified, and onward transmission from such cases reduced by a factor of 50% or 75% (eg, from rapid ART [or PrEP] prioritized to the index cases and their newly diagnosed infected contacts [PrEP to uninfected contacts]), we would predict a reduction in the mean number of newly linked cases of 56% and 63%, respectively, at 3 years (Figure 2B). By contrast, an intervention that reduced onward transmission from virologically unsuppressed persons by 50% or 75% would predict reductions by only 11% and 7% at year 3 (Figure 2C). Joint implementation of both interventions is predicted to be only slightly superior (67%) to a strategy that focused only on incident infections (Figure 2D).
Figure 2.
Predicted growth curves for the expected effect on cluster size with and without intervention. A, Without any intervention, the estimated mean number of newly linked incident cases per cluster would be 0.17 at 3 years after sequencing the original seed node, with 95% confidence intervals shown in the shaded red bands. B, Considers theoretical prevention and treatment interventions when toward persons with incident infection (estimated date of infection ≤1 year). An intervention that could reduce the number of transmissions that arise from incident infections by 50% (in green) or 75% (in blue) would decrease the cluster growth curve by 56% (estimated mean number of newly linked incident cases/cluster of 0.07) and 63% (estimated mean number of newly linked incident cases/cluster of 0.06), respectively, at 3 years. C, By comparison, interventions prioritized to reduce the transmission from virologically unsuppressed persons in a cluster (eg, through antiretroviral therapy) by 50% (in green) or 75% (in blue) would decrease the cluster growth curve by 7% (estimated mean number of newly linked cases/cluster of 0.16) and 11% (estimated number of linked cases/cluster of 0.15), respectively, at 3 years. D, No intervention (in red) compared to both interventions described in B and C above (to reduce onward transmissions by 75%; in blue) would be expected to reduce the human immunodeficiency virus growth rate by 67%, resulting in an estimated mean number of linked cases/cluster of 0.06 at the end of 3 years, mostly driven by changes in the number of incident infections in the population.
DISCUSSION
From data collected over 22 years, we found that people with incident HIV infection were the major driver of incident cluster growth in San Diego. As expected [14], we also found that larger cluster size was associated with incident cluster growth, albeit marginally given that the median cluster size was only 2 at the end of study follow-up. Interestingly, the proportion of cluster members with unsuppressed viral load showed only a weak association with incident cluster growth, and although the presence of bacterial STIs [15, 16], number of sex partners [16, 17], and use of illicit drugs [18] have all been shown to increase the risk of acquiring HIV infection, none of these factors were associated with new incident events.
The “Respond Pillar” of the EHE initiative seeks to rapidly detect and respond to growing HIV clusters to prevent new HIV infections [19]. Using the proposed CDC methods that prioritize clusters with a minimum of 3 or 5 cases diagnosed in the previous year connected through cases diagnosed in the previous 3 years [5] may fail to identify most new linked incident events in epidemics similar to that in San Diego (Supplementary Table 1B). When the CDC criteria were applied to our population, only 11 of 218 (5.0%) linked incident events were identified, which increased to 24 of 218 (11%) events even when the genetic threshold was relaxed to 1.5% (Supplementary Table 1C). The CDC strategy may well be effective for identifying larger unexpected outbreaks [20], identifying key populations for prevention resources [21] and monitoring the effect of prevention interventions [22], than for prospectively identifying and interrupting incident transmission events in epidemics similar to that in the San Diego region (ie, concentrated, established and not rapidly growing, predominantly MSM, relatively homogeneous); this strategy requires further evaluation.
This study also suggests that the strategy of using of cluster detection and response to reduce incidence may be suboptimal. Even if the “trigger” for prioritizing a cluster for evaluation and/or intervention were a single incident transmission event, its impact would likely be small, because the majority (58.9% [54/92]) of clusters with incident transmissions never grew to a size larger than 2 in our study population. Additionally, the window of opportunity to prevent an incident transmission from someone newly diagnosed with incident infection is limited (approximately 2.6 months). It seems unlikely that detect and respond methods could act rapidly enough to interrupt incident transmission events, even if sequence data were generated and analyzed within 3 weeks of diagnosis of PWH.
This is the first study to directly measure the number of linked incident events that arise within an HIV network and model the impact of potential prevention and treatment interventions. Consistent with the high degree of infectivity of persons with early HIV infection [23], intervention efforts to efficiently identify and rapidly treat persons with incident infection (Figure 2B) should provide an effective strategy to limit new transmissions. Implementation of the hypothetical intervention for persons with incident infection evaluated is feasible with current HIV screening technologies [24] and scale-up of rapid ART [25]. Our analyses found that total cluster growth was not a reliable proxy for incident cluster growth. However, identifying PWH with incident infection in conjunction with active molecular surveillance would allow direct measurement of the impact of an intervention designed to reduce HIV incidence. With adequate sampling depth (such as in our network [7]), randomized studies of prevention interventions that use incident cluster growth as outcomes would likely require fewer resources than do studies that measure incidence directly [26].
There are several limitations to this study. First, our sample was recruited from San Diego County, includes mostly MSM, and was enriched for persons with incident infection (58.5% of the sample); hence, results may not generalize to more diverse populations. Nonetheless, we believe that our concerns regarding CDC molecular surveillance analysis apply broadly, as the high proportion of incident infections does not drive these results but provides power to obtain them. Second, data on viral load and other covariates of interest were incomplete—a problem addressed through a multiple imputation procedure. Third, the lack of longitudinal assessment of sexual risk behavior limited investigation of its impact. Finally, sampling and data collection were not uniform across the 22 years of this observational study, hence our need to adjust for these factors.
Our findings raise several important issues. Current CDC molecular surveillance and response efforts will not likely identify the majority of incident transmission events within a concentrated epidemic, such as in San Diego. Efforts to limit incident cluster growth must include strategies to rapidly identify and treat persons with incident infection, which may provide a highly effective strategy to control HIV epidemic spread by limiting HIV transmission from people during the early stages of their infection.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by the US National Institutes of Health (grant numbers AI106039, AI136947, AI51164, AI036214, AI100665, MH100974, AI140970, and AI134384); and the James B. Pendleton Charitable Trust. Gilead Sciences provided antiretroviral therapy without cost to study participants.
Potential conflicts of interest. D. M. S. reports consulting fees from Bayer and Arena Pharmaceuticals, and scientific advisory board fees from FluxErgy and Linear Therapies, outside the submitted work. S. J. L. reports grants to her institution from Gilead Sciences during the conduct of the study, and speaker’s fees from Gilead Sciences, outside the submitted work. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Fauci AS, Redfield RR, Sigounas G, Weahkee MD, Giroir BP. Ending the HIV epidemic: a plan for the United States. JAMA 2019; 321:844–5. [DOI] [PubMed] [Google Scholar]
- 2.Li Z, Purcell DW, Sansom SL, Hayes D, Hall HI. Vital signs: HIV transmission along the continuum of care—United States, 2016. MMWR Morb Mortal Wkly Rep 2019; 68:267–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wertheim JO, Panneer N, France AM, Saduvala N, Oster AM. Incident infection in high-priority HIV molecular transmission clusters in the United States. AIDS 2020; 34:1187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oster AM, France AM, Panneer N, et al. . Identifying clusters of recent and rapid HIV transmission through analysis of molecular surveillance data. J Acquir Immune Defic Syndr 2018; 79:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. PS18-1802 detecting and responding to HIV transmission clusters: a guide for health departments. National Center for HIV/AIDS, Viral Hepatitis, STD, and TB Prevention. 2018. Available at: https://www.cdc.gov/hiv/pdf/funding/announcements/ps18-1802/CDC-HIV-PS18-1802-AttachmentE-Detecting-Investigating-and-Responding-to-HIV-Transmission-Clusters.pdf. Accessed 3 August 2020.
- 6.Morris SR, Little SJ, Cunningham T, Garfein RS, Richman DD, Smith DM. Evaluation of an HIV nucleic acid testing program with automated internet and voicemail systems to deliver results. Ann Intern Med 2010; 152:778–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Little SJ, Kosakovsky Pond SL, Anderson CM, et al. . Using HIV networks to inform real time prevention interventions. PLoS One 2014; 9:e98443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le T, Wright EJ, Smith DM, et al. . Enhanced CD4+ T-cell recovery with earlier HIV-1 antiretroviral therapy. N Engl J Med 2013; 368:218–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armour P, Dizikes G, Fakile Y, et al. . Suggested reporting language for syphilis serology testing. Silver Spring, MD: Association of Public Health Laboratories, 2015. [Google Scholar]
- 10.Kosakovsky Pond SL, Weaver S, Leigh Brown AJ, Wertheim JO. HIV-TRACE (TRAnsmission Cluster Engine): a tool for large scale molecular epidemiology of HIV-1 and other rapidly evolving pathogens. Mol Biol Evol 2018; 35:1812–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen PK, Gill RD. Cox’s regression model for counting processes: a large sample study. Ann Statist 1982; 10:1100–20. [Google Scholar]
- 12.Duong YT, Qiu M, De AK, et al. . Detection of recent HIV-1 infection using a new limiting-antigen avidity assay: potential for HIV-1 incidence estimates and avidity maturation studies. PLoS One 2012; 7:e33328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Macchione N, Wooten W, Waters-Montijo K, McDonald E.. HIV/AIDS epidemiology report—2016. San Diego, CA: County of San Diego Health and Human Services Agency Public Health Services, 2018. [Google Scholar]
- 14.Wertheim JO, Murrell B, Mehta SR, et al. . Growth of HIV-1 molecular transmission clusters in New York City. J Infect Dis 2018; 218:1943–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boily MC, Baggaley RF, Wang L, et al. . Heterosexual risk of HIV-1 infection per sexual act: systematic review and meta-analysis of observational studies. Lancet Infect Dis 2009; 9:118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menza TW, Hughes JP, Celum CL, Golden MR. Prediction of HIV acquisition among men who have sex with men. Sex Transm Dis 2009; 36:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoenigl M, Weibel N, Mehta SR, et al. . Development and validation of the San Diego early test score to predict acute and early HIV infection risk in men who have sex with men. Clin Infect Dis 2015; 61:468–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baggaley RF, Boily MC, White RG, Alary M. Risk of HIV-1 transmission for parenteral exposure and blood transfusion: a systematic review and meta-analysis. AIDS 2006; 20:805–12. [DOI] [PubMed] [Google Scholar]
- 19.Oster AM, France AM, Mermin J. Molecular epidemiology and the transformation of HIV prevention. JAMA 2018; 319:1657–8. [DOI] [PubMed] [Google Scholar]
- 20.Peters PJ, Pontones P, Hoover KW, et al. , Indiana HIV Outbreak Investigation Team . HIV infection linked to injection use of oxymorphone in Indiana, 2014–2015. N Engl J Med 2016; 375:229–39. [DOI] [PubMed] [Google Scholar]
- 21.German D, Grabowski MK, Beyrer C. Enhanced use of phylogenetic data to inform public health approaches to HIV among men who have sex with men. Sex Health 2017; 14:89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratmann O, Hodcroft EB, Pickles M, et al. , PANGEA-HIV Consortium . Phylogenetic tools for generalized HIV-1 epidemics: findings from the PANGEA-HIV methods comparison. Mol Biol Evol 2017; 34:185–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen MS, Gay CL, Busch MP, Hecht FM. The detection of acute HIV infection. J Infect Dis 2010; 202(Suppl 2):S270–7. [DOI] [PubMed] [Google Scholar]
- 24.Branson BM, Owen SM, Wesolowski LG, et al. . Laboratory testing for the diagnosis of HIV infection: updated recommendations. 2014. Available at: 10.15620/cdc.23447. Accessed 9 June 2020. [DOI]
- 25.Coffey S, Bacchetti P, Sachdev D, et al. . RAPID antiretroviral therapy: high virologic suppression rates with immediate antiretroviral therapy initiation in a vulnerable urban clinic population. AIDS 2019; 33:825–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdool Karim SS. HIV-1 epidemic control—insights from test-and-treat trials. N Engl J Med 2019; 381:286–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.