Abstract
Background
Acute retroviral syndrome (ARS) is associated with human immunodeficiency virus type 1 (HIV-1) subtype and disease progression, but the underlying immunopathological pathways are poorly understood. We aimed to elucidate associations between innate immune responses during hyperacute HIV-1 infection (hAHI) and ARS.
Methods
Plasma samples obtained from volunteers (≥18.0 years) before and during hAHI, defined as HIV-1 antibody negative and RNA or p24 antigen positive, from Kenya, Rwanda, Uganda, Zambia, and Sweden were analyzed. Forty soluble innate immune markers were measured using multiplexed assays. Immune responses were differentiated into volunteers with stronger and comparatively weaker responses using principal component analysis. Presence or absence of ARS was defined based on 11 symptoms using latent class analysis. Logistic regression was used to determine associations between immune responses and ARS.
Results
Of 55 volunteers, 31 (56%) had ARS. Volunteers with stronger immune responses (n = 36 [65%]) had increased odds of ARS which was independent of HIV-1 subtype, age, and risk group (adjusted odds ratio, 7.1 [95% confidence interval {CI}: 1.7–28.8], P = .003). Interferon gamma-induced protein (IP)-10 was 14-fold higher during hAHI, elevated in 7 of the 11 symptoms and independently associated with ARS. IP-10 threshold >466.0 pg/mL differentiated stronger immune responses with a sensitivity of 84.2% (95% CI: 60.4–96.6) and specificity of 100.0% (95% CI]: 90.3–100.0).
Conclusions
A stronger innate immune response during hAHI was associated with ARS. Plasma IP-10 may be a candidate biomarker of stronger innate immunity. Our findings provide further insights on innate immune responses in regulating ARS and may inform the design of vaccine candidates harnessing innate immunity.
Keywords: HIV-1, acute infection, immune responses, acute retroviral syndrome, IP-10
A strong innate immune response was associated with symptoms of acute retroviral syndrome (ARS). Plasma induced protein (IP)-10 was profoundly activated, associated with most ARS symptoms, and may be a candidate biomarker to differentiate a stronger innate immunity during hyperacute human immunodeficiency virus type 1 (HIV-1) infection.
Virologic and immunologic events during acute human immunodeficiency virus type 1 (HIV-1) infection (AHI) are associated with disease pathogenesis. During the first 10 days of HIV-1 infection (eclipse phase), the virus replicates at the infection site and in local lymphoid tissues before undergoing systemic dissemination. Subsequent stages of AHI can be defined by detection of virus RNA only (Fiebig stage I), virus RNA and p24 antigen (Fiebig stage II), and development of HIV-1 specific antibodies (Fiebig stages III-V) [1]. In 40–90% of cases, acute retroviral syndrome (ARS) presents before or during peak plasma viremia 2–3 weeks after HIV-1 infection [2–5]. The number and severity of symptoms have been correlated with rapid disease progression [6–8]. AHI symptoms are also more common in HIV-1 subtype A1 compared to subtype C and D infections [4].
AHI is further characterized by an induction of innate immune responses, including activation of dendritic cells, macrophages, and natural killer (NK) cells [9–11]. The earliest systemic innate immune perturbations occur during the eclipse phase of AHI. As HIV-1 viremia increases, pro-inflammatory cytokines, and chemokines are induced with elevations in multiple analytes creating a cytokine storm [12–14]. This systemic activation has been shown to have antiviral activity, enhance other innate immune responses, and shape adaptive immunity to control virus replication [15–19]. However, the exaggerated response also has immunopathological consequences by promoting provirus transcription, enhancing HIV-1 replication and driving CD4+ T-cell depletion [20–22]. Little is known about involvement of innate immune responses in the regulation of AHI symptoms. We hypothesized that activation of strong innate immune responses are associated with AHI symptoms. We sought to test this hypothesis in a hyperacute HIV-1 infection (hAHI) study involving volunteers enrolled into cohorts from 4 African and 1 European countries.
METHODS
Study Design and Ethical Considerations
Data and samples were obtained from African and Swedish adult (≥18 years old) volunteers enrolled in acute and early HIV-1 infection studies. African volunteers were enrolled 2006–2011 at sites in Kenya, Rwanda, Uganda, and Zambia under IAVI’s protocol C [23]. Volunteers diagnosed with AHI before 2011 and identified from patient medical records at Skåne University Hospital in Lund/Malmö, Sweden, were included. Volunteers with hAHI diagnosis were eligible (defined as HIV-1 antibody negative and RNA [Fiebig stage I] or p24 antigen [Fiebig stage II] positive). Available matched preinfection samples from volunteers were also included in the analysis. All protocol C volunteers provided written informed consent for the use of their samples for biomedical research. Protocol C sites received approvals from respective country-specific ethics review boards [23]. Ethical approval for the Swedish cohort was obtained from the Lund University Ethical Review Board, Sweden (Diarienummer [Dnr] 2013/772).
Characteristics of Volunteers
Protocol C data were obtained from a centralized and curated repository. De-identified data from the Swedish volunteers were obtained from an electronic patient register (InfCareHIV) and medical records. Baseline variables included date of birth, sex, ethnicity, HIV-1 RNA, HIV-1 p24 antigen, and antibody test results, date of HIV-1 diagnosis, transmission risk group (serodiscordant, men who have sex with men [MSM], and heterosexual relationships), and AHI symptoms.
HIV-1 Subtyping
HIV-1 env sequences (V1-V3, approximately 940 base pairs) were generated, and HIV-1 subtype were phylogenetically determined as previously described [24]. In brief, HIV-1 RNA was extracted, reverse transcribed, polymerase chain reaction (PCR) amplified and sequenced using established protocols. Generated sequences were aligned with HIV-1 subtype reference sequences from the Los Alamos HIV-1 sequence database (https://www.hiv.lanl.gov/content/index). The general time reversible (GTR) model of nucleotide substitution with gamma distributed rate heterogeneity was used to infer maximum likelihood trees. Branch support was assessed using the approximate likelihood ratio test based on the Shimodaira-Hasegawa (aLRT-SH) method, with branch support of ≥ .90 considered significant [25].
Acute Retroviral Syndrome
For protocol C volunteers, AHI symptoms were ascertained 2–6 weeks following the estimated date of infection using a standardized questionnaire [4]. For Swedish volunteers, medical records were reviewed using the protocol C questionnaire as a template; documented AHI symptoms were extracted separately by 2 investigators. AHI symptoms reported at the incident visit or in follow-up visits (but within 6 weeks of incident visit) were extracted. Case files without a reported symptom were considered negative for that symptom. Inconsistencies were discussed and a consensus documented.
Acute retroviral syndrome (ARS) was defined based on 11 AHI symptoms. Previous studies have used various definitions for ARS including volunteers reporting any symptom, ≥2 symptoms, ≥3 symptoms, or a combination of fever with other symptom(s) [2, 26, 27]. A potential limitation of discrete classification methods is the inability to account for unobserved linkages between symptoms. Latent class analysis (LCA), a structural equation modelling approach, was therefore applied to group volunteers based on the number of AHI symptoms and other unobserved linkages, defined as those with or without ARS.
Innate Immune Responses
Cryopreserved plasma samples collected from whole blood treated with ethylenediaminetetraacetic acid (EDTA) were used. Multiplexed analysis was done in duplicates and measured by electrochemiluminescence using the MesoScaleDiscovery (MSD) multispot 40-plex assay (MesoScaleDiagnostics, LLC) according to manufacturer’s instructions [28]. All samples were processed from the IAVI Human Immunology Laboratory, London, United Kingdom. Analyte concentration read-outs, along with respective lower (LLOQ) and upper (ULOQ) limits of quantification, were inferred from standard curves. Analyte concentrations below the LLOQ were assigned half the LLOQ concentration, whereas those above the ULOQ were assigned the ULOQ value. The mean of the duplicate observations were analyzed.
Data Analysis
First, ARS was defined based on the number of AHI symptoms and other unobserved linkages using LCA, and volunteers disentangled into those with and without ARS. Second, associations between log10 analyte concentrations with AHI symptoms and with ARS were assessed using Wilcoxon rank sum tests. Analytes suggestive of an association with any of the 11 AHI symptoms and with ARS were further explored using principal component analysis (PCA). The PCA differentiated volunteers into those with stronger and comparatively weaker immune responses. Third, logistic regression was applied to determine associations between immune response and ARS. HIV-1 subtype, risk group, ethnicity, geographic region, and HIV-1 RNA were dichotomized to maximize precision. A threshold of P < .10 was used to select variables from univariable to multivariable models. Finally, individual analytes were assessed for prognostic performance in differentiating stronger immune responses using receiver operating characteristic area under the curve (ROC-AUC).
All analyses were done in Stata/IC version 15.1 (StataCorp LP, California), and outputs generated using GraphPad Prism version 8.4.2 (GraphPad Software, California). A 2-sided type 1 error of 5% was considered statistically significant. Due to sample size limitations and consistent with previous publications, no formal corrections for multiple comparisons were done [14, 26].
RESULTS
Characteristics of Study Volunteers
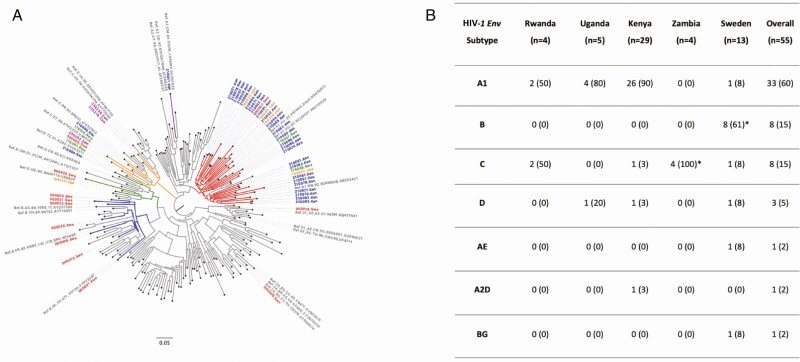
Fifty-five volunteers were eligible. The majority of volunteers were diagnosed with HIV-1 by p24 (Fiebig II, n = 42 [76%]) and had a matched preinfection sample (n = 31 [56%]), collected at median 47 (interquartile range [IQR], 22–120) days before HIV-1 infection. A complete set of baseline variables were available for all volunteers, except for HIV-1 RNA that was missing from 62% of volunteers (Table 1). No significant differences in baseline characteristics of volunteers with missing versus with HIV-1 RNA data were found (Supplementary Table 1), and HIV-1 RNA was therefore excluded from further analysis. Most volunteer HIV-1 genetic sequences clustered with subtype A1 (n = 33 [60%], Figure 1). The majority of HIV-1 subtype A1 sequences (n = 26) were from Kenya, whereas all HIV-1 subtype B sequences (n = 8) were from Sweden.
Table 1.
Characteristics of Volunteers Diagnosed With Hyperacute HIV-1 Infection From Africa (Rwanda, Uganda, Zambia, and Kenya) and Sweden by Fiebig Staging (N = 55)
| Baseline Variables | Fiebig I (N = 13) | Fiebig II (N = 42) | Overall (N = 55) | |
|---|---|---|---|---|
| Age (y) | Median (IQR) | 22.6 (20.9–27.1) | 29.3 (25.1–34.1) | 28.1 (23.0–34.1) |
| Age group (y) | 18.0–24.9 | 7 (54) | 10 (24) | 17 (31) |
| 25.0–34.9 | 4 (31) | 22 (52) | 26 (47) | |
| 35.0–44.9 | 1 (8) | 5 (12) | 6 (11) | |
| 45.0+ | 1 (8) | 5 (12) | 6 (11) | |
| Gender | Female | 0 (0) | 4 (10) | 4 (7) |
| Male | 13 (100) | 38 (90) | 51 (93) | |
| Year of infection | <2009 | 4 (31) | 15 (36) | 19 (35) |
| 2009–2010 | 7 (54) | 18 (43) | 25 (45) | |
| 2011+ | 2 (15) | 9 (21) | 11 (20) | |
| Ethnicity | African | 13 (100) | 29 (69) | 42 (76) |
| Arab/Asian | 0 (0) | 1 (2) | 1 (2) | |
| Caucasian | 0 (0) | 12 (29) | 12 (22) | |
| Risk groupa | DC | 2 (15) | 12 (29) | 14 (25) |
| HET | 1 (8) | 3 (7) | 4 (7) | |
| MSM | 10 (77) | 24 (57) | 34 (62) | |
| UNK | 0 (0) | 3 (7) | 3 (5) | |
| Country | Sweden | 0 (0) | 13 (31) | 13 (24) |
| Africanb | 13 (100) | 29 (69) | 42 (76) | |
| HIV-1 RNA (log10 cpm) | Median (IQR) | 4.0 (3.8–4.6) | 6.1 (5.9–7.0) | 5.0 (4.0–6.0) |
| HIV-1 RNA group (log10 cpm) | ≤5.0 | 11 (85) | 0 (0) | 11 (20) |
| >5.0 | 2 (15) | 8 (19) | 10 (18) | |
| Missing | 0 (0) | 34 (79) | 34 (62) | |
| Matched preinfection samplec | No | 2 (15) | 22 (52) | 24 (44) |
| Yes | 11 (85) | 20 (48) | 31 (56) | |
| Matched preinfection periodd | Median (IQR) | 85 (22–158) | 41 (22–81) | 47 (22–120) |
Abbreviations: cpm, copies per milliliter; HIV-1, human immunodeficiency virus type 1; IQR, interquartile range.
aRisk group data (DC [serodiscordant couples], HET [heterosexual], MSM (men who have sex with men] and UNK [unknown]).
bAfrican countries (protocol C sites from Rwanda [n = 4], Uganda [n = 5], Zambia [n = 4] and Kenya [n = 29]).
cMatched preinfection samples from Kenya (n = 29), Rwanda (n = 1) and Zambia (n = 1).
dAvailability of matched preinfection samples by days from sampling to the estimated date of infection (EDI).
Figure 1.
(A) Phylogenetic tree showing relatedness of HIV-1 partial envelope sequences (V1-V3 region) from individuals with hyperacute HIV-1 infection from Africa and Sweden (N = 55), with HIV-1 subtype reference sequences from the Los Alamos database (N = 158); and (B) summary table showing the distribution of HIV-1 partial env (V1-V3 region) subtypes of individuals with hyperacute infection from Africa and Sweden (N = 55). Branches are colored according to HIV-1 env subtype as follows: gray (references), red (subtype A1), dark blue (subtype B), orange (subtype C), green (subtype D), light blue (recombinant BG), purple (recombinant A2D), and brown (recombinant AE). Tip labels are colored according to the country of origin as follows: gray (references; only those clustering with volunteer sequences shown), green (Rwanda), orange (Uganda), blue (Kenya), purple (Zambia), and red (Sweden). *Unable to amplify HIV-1 env region from two volunteers, though previous gag/pol/nef sequence data suggests subtype C (Zambia, n = 1) and B (Sweden, n = 1) infections and included in the analysis as such. Abbreviation: HIV-1, human immunodeficiency virus type 1.
Acute Retroviral Syndrome
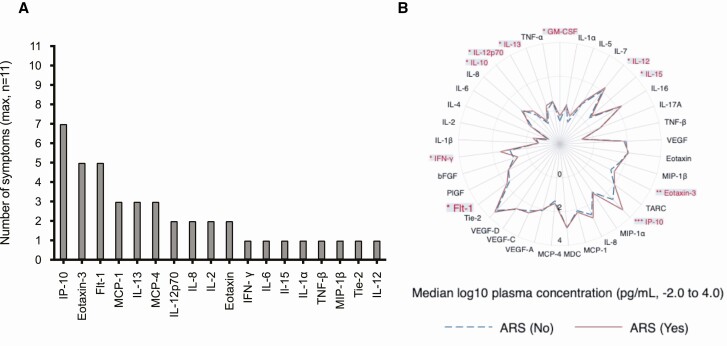
Overall, 43 (78%) volunteers had at least one AHI symptom. The median number of symptoms was 4 (IQR, 1–7). The most common symptom was fever (n = 37 [67%]; Figure 2A). Using LCA, volunteers were grouped into those with ARS (n = 31 [56%]; median symptoms per volunteer, 6 [IQR, 5–8]) and without ARS (n = 24 [44%]; 0.5 [IQR, 0–2.5], Figure 2B). Nine of the 11 AHI symptoms were significantly higher in volunteers with ARS. Myalgia demonstrated the strongest, whereas skin rash and pharyngitis demonstrated no discriminatory potential for ARS. Notably, fever was common in both groups (with and without ARS), suggesting a weak discriminatory ability for ARS (Figure 2C).
Figure 2.
(a) Distribution of volunteers diagnosed with hyperacute HIV-1 infection by AHI symptoms; (b) a schematic illustration of the distribution of AHI symptoms for all volunteers (black curve) and by grouping of volunteers resulting from latent class analysis (LCA, for those without acute retroviral syndrome, [ARS, blue] and for those with ARS [red]). The numbers denote median (interquartile ranges) symptoms per volunteer for all volunteers (in black), for those without ARS (in blue) and for those with ARS (in red); and (c) Graph comparing the distribution of AHI symptoms between volunteers that were defined to be with and without ARS (Fisher exact test P < .05 [*], P < .01 [**], and P < .001 [***], N = 55). ARS was defined based on the 11 AHI symptoms and other unobserved linkages between symptoms using LCA. Incremental latent group models were assessed to predict the goodness of fit. The model with 2 latent groups was the best fit as it had the lowest BIC value (660.5) compared to 3 (678.6), 4 (699.2), or 5 (714.7) groups. Volunteers were grouped based on their predicted posterior probabilities into those with ARS (n = 31 [56%]) and those without ARS (n = 24 [44%]). Abbreviations: AHI, acute human immunodeficiency virus type 1; ARS, acute retroviral syndrome; CI, confidence interval; HIV-1, human immunodeficiency virus type 1; LCA, latent class analysis.
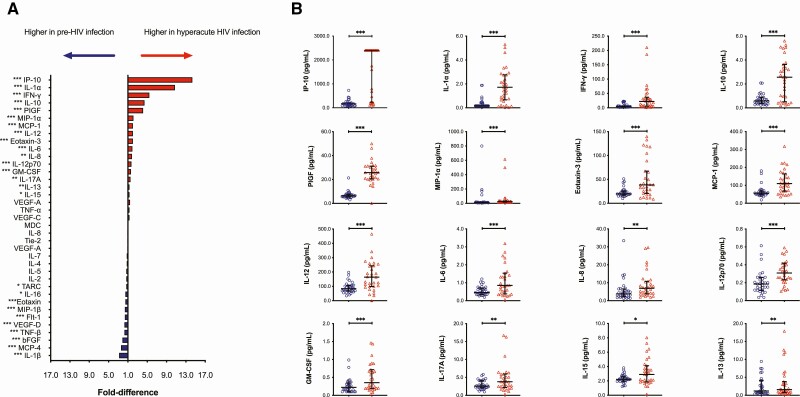
Innate Immune Responses During Hyperacute HIV-1 Infection
SAA, CRP, VCAM-1, and ICAM-1 concentrations were higher than the ULOQ in >90% of volunteers and therefore excluded from further analysis. Compared to matched preinfection responses, 16 analytes were significantly elevated during hAHI (Figure 3A; Supplementary Table 2). A 14-fold higher median level of Interferon gamma-induced protein (IP)-10 was observed during hAHI compared to HIV-1 preinfection (2400 [IQR, 251–2400] vs 168 [IQR, 86–204] pg/mL). Substantial between-individual variation in analyte responses was observed (Figure 3B). Macrophage inflammatory protein (MIP)-1β, Eotaxin-3, MIP-1α, interleukin (IL)-8, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-12p70, IL-13, and tumor necrosis factor (TNF)-α were positively correlated with each other (Supplementary Figure 1).
Figure 3.
(A) Plot illustrating fold-differences in median plasma biomarker concentration from matched pre-infection and hyperacute HIV-1 infection samples. Red bars demonstrate fold difference in analytes that had higher plasma concentrations during hyperacute HIV-1 infection compared to the pre-infection samples. Blue bars denote fold-difference in analytes that had higher concentrations in the pre-infection samples compared to hyperacute HIV-1 infection samples. (B) Graphs illustrating analytes that were significantly higher during hyperacute HIV-1 infection compared to preinfection (Wilcoxon paired signed-rank test P < .05 [*], P < .01 [**], and P < .001 [***], N = 31). Abbreviations: bFGF, basic fibroblast growth factor; Flt, FMS-like tyrosine kinase; GM-CSF, granulocyte-macrophage colony-stimulating factor; HIV-1, human immunodeficiency virus type 1; IP, interferon gamma-induced protein; IFN, interferon; IL, interleukin; MCP, monocyte chemoattractant protein; MDC, macrophage-derived chemokine; MIP, macrophage inflammatory protein; PlGF, placental growth factor; TARC, thymus and activation-related chemokine; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
Associations Between Innate Immune Responses and Acute Retroviral Syndrome
Overall, 18 analytes were higher in volunteers with any AHI symptoms compared to those without symptoms (Supplementary Figure 2). IP-10 activation stood out, with plasma concentrations being significantly higher across 7 of the 11 AHI symptoms (Figure 4A). Further, there were significantly higher IP-10 (P < .001), interferon (IFN)-γ (P = .020), granulocyte-macrophage colony-stimulating factor (GM-CSF) (P = .033), IL-12 (P = .043), IL-15 (P = .037), Eotaxin-3 (P = .002), Flt-1 (P = .025), IL-10 (P = .047), IL12p70 (P = .027) and IL-13 (P = .047) plasma concentrations amongst volunteers with versus without ARS (Figure 4B; Supplementary Table 3). After adjusting for HIV-1 subtype, age, and risk group, elevated IP-10 was independently associated with ARS (adjusted coefficient, 4.7 [95% confidence interval {CI}: 0.8–8.6], P = .002, Table 2).
Figure 4.
(A) Graph summarizing associations between log10 median plasma analyte concentration when comparing volunteers with and those without AHI symptoms (Supplementary Figure 2), and (B) plot illustrating differences in log10 median plasma biomarker concentration between hyperacute HIV-1 infected volunteers with ARS compared to mild ARS (Wilcoxon rank sum test P < .05 [*], P < .01 [**], and P < .001 [***], N = 55). Analytes suggestive of an association with any of the 11 AHI symptoms and with ARS (IL-12, IL-15, Eotaxin-3, IP-10, Flt-1, IFN-γ, IL-12p70 and IL-13) were further explored using principal component analysis (PCA). Abbreviations: AHI, acute human immunodeficiency virus type 1; ARS, acute retroviral syndrome; bFGF, basic fibroblast growth factor; Flt, FMS-like tyrosine kinase; GM-CSF, granulocyte-macrophage colony-stimulating factor; HIV-1, human immunodeficiency virus type 1; IFN, interferon; IP, interferon gamma-induced protein; IL, interleukin; MCP, monocyte chemoattractant protein; MDC, macrophage-derived chemokine; MIP, macrophage inflammatory protein; PlGF, placental growth factor; TARC, thymus and activation-related chemokine; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
Table 2.
Logistic Regression Analysis Demonstrating the Association Between Specific Analytes and Acute Retroviral Syndrome (ARS) Among Volunteers With Hyperacute HIV-1 Infection From Africa (Rwanda, Uganda, Zambia, and Kenya) and Sweden (N = 55)
| Analyte | Crude Coef. (95% CI) | P-value | Adjusted Coef. (95% CI)a | P-value |
|---|---|---|---|---|
| IL-12 | .24 (−.89 to 1.37) | .672 | … | … |
| IL-15 | .83 (−.98 to 2.64) | .355 | … | … |
| Eotaxin-3 | 3.20 (1.06–3.81) | <.001 | 3.57 (−.98 to 8.11) | .078 |
| IP-10 | 2.44 (1.07–3.81) | <.001 | 4.71 (.84 – 8.57) | .002 |
| Flt-1 | −.07 (−1.12 to .98) | .891 | … | |
| IFN-γ | 1.13 (.15 – 2.11) | .016 | 0.58 (-2.10 – 3.26) | .671 |
| IL-12p70 | 1.10 (−.44 to 2.64) | .161 | … | … |
| IL-13 | 1.39 (.14–2.63) | .020 | 1.37 (−1.77 to 4.51) | .379 |
Abbreviations: CI, confidence interval; Coef, coefficient; Flt, FMS-like tyrosine kinase; HIV-1, human immunodeficiency virus type 1; IFN, interferon; IL, interleukin; MSM, men who have sex with men.
aAdjusted for age group (18.0–24.9 vs >25.0 years), HIV-1 env subtypes (Non-A1 vs A1) and risk group (Non-MSM vs MSM).
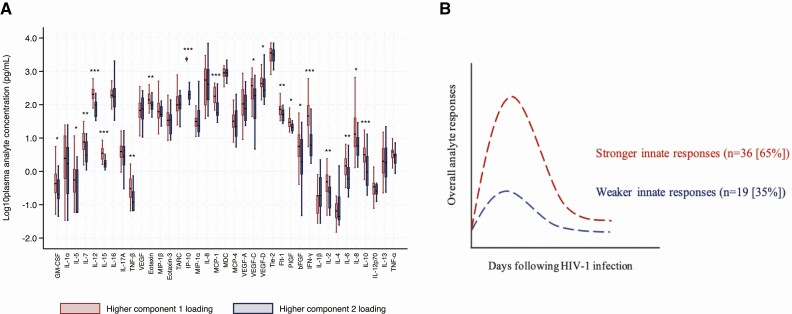
Significantly elevated analytes in volunteers with any of the 11 symptoms and in those with ARS were analyzed by PCA. Two principal components (PCs) were determined: PC1 (IP-10, IFN-γ, IL-12, IL-15, and Flt-1); and PC2 (Eotaxin-3, IL-13, and IL-12p70, Supplementary Figure 3). Both PCs were significantly associated with ARS, suggesting a synergistic effect of multiple analytes on ARS (more so for Eotaxin-3, IL-13 and IL-12p70, which were not significantly associated with ARS when assessed independently, Supplementary Table 4).
Furthermore, volunteers were grouped into those with higher PC1 loading and those with higher PC2 loading. Volunteers with higher PC1 loading had significantly higher median plasma concentration for the majority of the 40 analytes compared to those with higher PC2 loading (Figure 5A, Supplementary Table 5). Hence, PC1 analytes differentiated volunteers into those with stronger immune responses (n = 36 [65%]) and comparatively weaker immune responses (n = 19 [35%], Figure 5B). After controlling for HIV-1 subtype, age and risk group, volunteers with stronger innate immune responses had increased odds of ARS (adjusted odds ratio, 7.1 [95% CI: 1.7–28.8], P = .003, Table 3).
Figure 5.
(A) Graph demonstrating the differentiation of analyte responses by principal component analysis loadings. Volunteers with higher principal component (PC) 1 loadings had significantly higher plasma concentration in a majority of the analytes compared to those with higher PC2 loadings (Wilcoxon rank sum test P < .05 [*], P < .01 [**], and P < .001 [***], N = 55); (B) A schematic illustration of the differentiation of analyte responses into volunteers with and without a cytokine storm; and Wilcoxon signed rank test (P < .05 [*], P < .01 [**], P < .005 [***], and P < .0005 [****]; paired/matched signed rank test in green, N = 31). Abbreviations: hbFGF, basic fibroblast growth factor; Flt, FMS-like tyrosine kinase; GM-CSF, granulocyte-macrophage colony-stimulating factor; hAHI, hyperacute HIV-1 infection; HIV-1, human immunodeficiency virus type 1; IFN, interferon; IL, interleukin; IP, interferon gamma-induced protein; MCP, monocyte chemoattractant protein; MDC, macrophage-derived chemokine; MIP, macrophage inflammatory protein; PlGF, placental growth factor; TARC, thymus and activation-related chemokine; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
Table 3.
Logistic Regression Analysis Demonstrating the Association Between Innate Immune Responses and Acute Retroviral Syndrome (ARS) Among Volunteers With Hyperacute HIV-1 Infection From Africa and Sweden (N = 55)
| Particulars | ARS, n (%) | Crude OR (95% CI) | P-value | Adjusted OR (95% CI) | P-value | |
|---|---|---|---|---|---|---|
| Innate immune responses | Weaker responses | 6/19 (32) | Ref | .007 | Ref | .003 |
| Stronger responses | 25/36 (69) | 4.9 (1.5 – 16.3) | 7.1 (1.7–28.8) | |||
| Fiebig staging | Fiebig I | 5/13 (38) | Ref | .137 | … | … |
| Fiebig II | 26/42 (62) | 2.6 (.7 – 9.3) | ||||
| Gender | Male | 28/51 (55) | Ref | .422 | … | … |
| Female | 3/4 (75) | 2.5 (.2 – 25.3) | ||||
| Age group (y) | 18.0 – 24.9 | 13/17 (76) | 3.6 (1.0 – 13.1) | .040 | 2.4 (.5–12.1) | .264 |
| 25.0 + | 18/38 (47) | Ref | Ref | |||
| Year of infection | <2009 | 11/19 (58) | Ref | .712 | … | … |
| 2009 – 2010 | 15/25 (60) | 1.1 (.3 – 3.7) | ||||
| 2011+ | 5/11 (45) | .6 (.1 – 2.7) | ||||
| HIV-1 env subtype | Non-A1a | 9/22 (41) | Ref | .059 | Ref | .176 |
| A1 | 22/33 (67) | 2.9 (1.0 – 8.8) | 2.5 (.6 – 9.3) | |||
| Ethnicity | Non-Africanb | 7/13 (54) | Ref | .834 | … | … |
| African | 24/42 (57) | 1.1 (.3 – 4.0) | ||||
| Risk group | Non-MSMc | 8/21 (38) | Ref | .031 | Ref | .084 |
| MSM | 23/34 (68) | 3.4 (1.1 – 10.6) | 3.3 (.8–12.8) | |||
| Country | Sweden | 7/13 (54) | Ref | .834 | … | … |
| Africand | 24/42 (57) | 1.1 (.3–3.9) |
Abbreviations: CI, confidence interval; HIV-1, human immunodeficiency virus type 1; MSM, men who have sex with men; OR, odds ratio; Ref, reference.
aHIV-1 env subtypes B (n = 8), C (n = 8), D (n = 3), BG (n = 1), A2D (n = 1), and AE (n = 1).
bCaucasian (n = 12), Asian/Arabian (n = 1).
cDiscordant couples (n = 14), heterosexual (n = 4), and unknown (n = 3).
dRwanda (n = 4), Uganda (n = 5), Zambia (n = 4), and Kenya (n = 29).
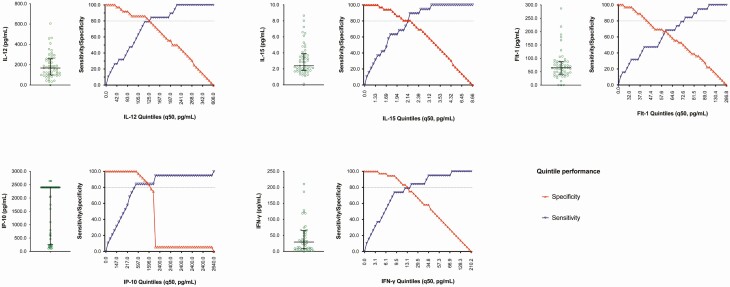
Next, analytes with higher PC1 loading (IP-10, IFN-γ, IL-12, IL-15, and Flt-1) were analyzed independently to assess their prognostic performance in differentiating a stronger innate immune response. All analytes demonstrated high prognostic value (ROC range, 0.73–0.90), with no significant differences in the AUCs (Supplementary Figure 4). The best performing analyte and threshold to differentiate a stronger innate immune response was IP-10 (466 pg/mL; AUC: 0.92 [95% CI: 0.84–1.00]; sensitivity: 84.2% [95% CI: 60.4–96.6] and specificity: 100.0% [95% CI: 90.3–100.0], Figure 6; Supplementary Table 6).
Figure 6.
Graphs demonstrating distribution, performance (sensitivity and specificity) and threshold of analytes (IL-12, IL-15, Flt-1, IP-10, and IFN-γ) indicative of a stronger innate immune response. Analytes and threshold with both sensitivity and specificity of more than 80% were further explored. Abbreviations: Flt, FMS-like tyrosine kinase; IFN, interferon; IL, interleukin; IP, interferon gamma-induced protein.
Fiebig II Subpopulation Analysis
Due to the dynamic nature of innate immune responses during hAHI, we performed a sensitivity analysis to address the potential confounding effect of combining Fiebig I and II samples. The analysis was restricted to Fiebig II samples (n = 42) and followed the analytical pipeline that was used for the full data set. Overall, the results were consistent with the analysis of the full data set. Briefly, the most common symptom was fever. Volunteers were grouped by LCA into those with (n = 26 [62%]) and without ARS (n = 16 [38%], Supplementary Figure 5). Fifteen analytes were significantly elevated during hAHI, with IP-10 having 9-fold higher levels compared to preinfection levels (Supplementary Figure 6). IP-10, Eotaxin-3, and Flt-1 were elevated in volunteers with ARS (Supplementary Figure 7). IP-10 was independently associated with ARS after controlling for HIV-1 subtype, age, and risk group (Supplementary Table 7). Analytes significantly altered during hAHI and with any symptom were analyzed by PCA. These clustered into 3 PCs indicative of stronger (PC1 and PC2, n = 28 [67%]) and comparatively weaker innate immune responses (PC3, n = 14 [33%], Supplementary Figures 8 and 9; Supplementary Table 8). Volunteers with stronger innate immune responses had increased odds of ARS (adjusted odds ratio [aOR] 11.9 [95% CI: 1.1–130.4], P = .019, Supplementary Table 9). Finally, IP-10 was the best prognostic performing analyte (threshold: 1752 pg/mL; AUC of 0.82 [95% CI: 0.69–0.95]; sensitivity: 71.4% [95% CI: 41.9–91.6] and specificity: 92.9% [95% CI: 76.5–99.1], Supplementary Figures 10 and 11; Supplementary Table 10).
DISCUSSION
Our findings are consistent with previous studies reporting striking elevation of innate immune markers resulting in the well-established cytokine storm during AHI [12–14]. In the absence of a gold standard, we applied a holistic systems approach to differentiate innate immune activation, and observed that a stronger immune response during hAHI was independently associated with ARS. To our knowledge, only one other study has reported associations between innate immune responses during AHI and ARS [26]. Crowell et al included 430 volunteers with AHI, a majority (62%) of whom were enrolled at Fiebig stage III or later. Five of the 10 analyzed analytes were included in our analysis (TNF-α, IL-6, IL-7, IP-10, and MCP-1). In contrast to our findings, TNF-α was elevated in ARS, whereas no association with IP-10 was reported. Several factors may explain these differences. First, because our volunteers were enrolled at Fiebig stages I/II, we were able to observe the earliest innate immune perturbations, including the cytokine storm, which may have been missed by a majority of the volunteers in the study by Crowell et al. Second, Crowell et al studied participants enrolled in Bangkok, Thailand (an HIV-1 CRF01_AE predominant region), whereas we studied participants from Africa and Europe infected by various strains of HIV-1 (subtypes A, B, C, and D, representing >70% of HIV-1 infections globally) [29]. Indeed, varying rates of disease progression have been suggested for different subtypes and circulating recombinant forms (CRFs) [30]. Third, given the different study settings, it is possible that differences in host genetic factors impacted innate immune responses differently [31]. Finally, Crowell et al defined ARS as ≥3 symptoms, whereas we considered both the number of symptoms and unobserved relatedness between symptoms. This may have resulted in a more sensitive differentiation of ARS.
We identified 16 analytes that had significantly higher plasma concentrations during hAHI compared to preinfection values. Systemic upregulation of innate immune analytes during AHI is consistent with literature, and the association with ARS may be explained by multiple but related events [12–14]. In brief, and during AHI, the virus targets innate immune cells including dendritic cells, macrophages, and NK cells [9–11]. The result is significant destruction of these cells, widespread systemic virus dissemination, and a subsequent immunological milieu resulting in activation of innate and adaptive immune responses. Further, debris released from infected cells and other microbial products are translocated across mucosal barriers causing additional systemic inflammation and cytokine production [9, 13]. Altogether, these processes result in profound damage to cells, blood capillaries and mucosal barriers which may explain subsequent systemic (eg, fever), lymphatic (eg, lymphadenopathy), gastrointestinal (eg, diarrhea), musculoskeletal (eg, myalgia), and neurologic (eg, headache) symptoms. Notably, cytokine responses are activated and peak within 2 weeks of HIV-1 infection [13], whereas symptoms start manifesting 2–3 weeks after infection [3, 32], suggesting that systemic innate immune activation is a precursor for symptomatic manifestations.
The observed IP-10 elevation among volunteers with ARS is consistent with literature [14, 33]. However, its role in the induction of AHI symptoms is less understood. IP-10 is induced during the eclipse phase of AHI by innate immune cells in response to IFN-γ stimulation [34]. It is therefore not surprising that IP-10 clustered closely with IFN-γ during PCA analysis. IP-10 induction increases exponentially and in parallel with HIV-1 RNA but prior to symptom manifestation [12]. In turn, IP-10 induces chemotaxis, apoptosis, cell growth inhibition, and angiostasis of different immune cells contributing to the damaging immunological milieu as described above. The role of IP-10 in disease pathogenesis is not unique to HIV-1; elevated IP-10 has been documented in a diverse range of virus infections, such as hepatitis C virus (HCV) infection, respiratory infections, and the more recent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection [35–37]. The utility of IP-10 as a diagnostic marker for infectious diseases has also been explored. For instance, and in a meta-analysis, IP-10 has been suggested as a diagnostic marker for differentiating active from latent tuberculosis [38]. IP-10 has also been suggested as a marker for detection of AHI and for virus load monitoring [33, 39]. Yet the role of IP-10 as a biomarker in the differentiation of stronger immune responses remains undocumented. We propose consideration of IP-10 as a candidate marker in the differentiation of stronger innate immune responses at a threshold of >466 pg/mL.
Our study is not without limitations. First, the majority of our volunteers were missing HIV-1 RNA data. HIV-1 RNA increases and peaks 2–3 weeks after infection [2–5]. As HIV-1 viremia increases, pro-inflammatory cytokines and chemokines are induced, suggesting a possible correlation, as has been demonstrated between HIV-1 RNA and IP-10 [12]. HIV-1 RNA is also associated with ARS [26]. Further studies to disentangle the relationship between HIV-1 RNA, innate immune perturbations, and ARS are therefore warranted. Second, although our strict Fiebig I/II inclusion criteria enabled us to elucidate very early immune responses, it is possible that peak activation of analytes with delayed response may have been missed [12, 13]. Third, it is possible that volunteers seeking hospital care are more likely to present with ARS compared to those from a routine longitudinal cohort. However, there was no significant difference in the number of symptoms reported by the hospital volunteers compared to routine cohort volunteers (median, 5 [IQR, 3–6] vs 4 [IQR, 0–7] symptoms per volunteer respectively, P = .610). Finally, our small sample size and potential bias toward male volunteers (93% of the study population) warrants validation of our findings in larger and more sex-balanced AHI studies.
In conclusion, we applied holistic systems approaches to elucidate ARS from AHI symptoms and to disentangle the myriad of complex but related pathways of soluble innate immune markers. We found compelling evidence of an association between stronger innate immune responses and ARS. Specifically, plasma IP-10 was profoundly activated during AHI and associated with ARS. Furthermore, and in the absence of a gold standard, we also identified plasma IP-10 as a candidate biomarker in the differentiation of a stronger innate immune response. Our findings provide further insights into early immune responses during hAHI, their role in the regulation of ARS, and may have implications for the design of vaccine candidates that harness innate immunity for virus neutralization or replication control.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. The authors thank IAVI for supporting HIV-1 research studies and capacity building initiatives in Kenya, Uganda, Rwanda, and Zambia. They are also grateful to staff and volunteers from IAVI’s protocol C sites in Africa and from the Department of Infectious Diseases at Skåne University Hospital in Sweden, without whom this work would not have been possible. They also acknowledge the following people for their generous guidance, contributions, and support: Esben Poulsen (MSD, Copenhagen, Denmark), Julia Makinde (IAVI-HIL, London, UK), Jan Albert (Karolinska Institute, Sweden), and Bengt Löfgren, Bertil Christensson, and Karin Behrens (all from SUS Skåne, Sweden).
Disclaimer. This report was published with permission from the Kenya Medical Research Institute (KEMRI).
Funding. This work was supported by the generous support of the American people through the United States Agency for International Development (USAID). The contents are the responsibility of the study authors and do not necessarily reflect the views of USAID, the National Institutes of Health (NIH), or the US government. This work was also supported by funding from the Swedish Research Council (grant number 2016–01417) and the Swedish Society for Medical Research (grant number SA-2016). The authors are also grateful for the support of the Sub-Saharan African Network for TB/HIV-1 Research Excellence (SANTHE), a DELTAS Africa Initiative (grant number DEL-15–006). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (grant number 107752/Z/15/Z) and the UK government.
Potentialconflicts of interest. A. S. H. was supported by a Training fellowship from the Wellcome Trust (209294/Z/17/Z). P. Bo. is a Jenner Institute Investigator and was supported by funding from the MRC (MR/K0120/37), NIH, NIAID, DAIDS, and the Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) (UM1 AI 00645). T. N. reports grants from South African National Research Foundation and Victor Daitz Foundation. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Fiebig EW, Wright DJ, Rawal BD, et al. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 2003; 17:1871–9. [DOI] [PubMed] [Google Scholar]
- 2.Braun DL, Kouyos RD, Balmer B, Grube C, Weber R, Günthard HF. Frequency and spectrum of unexpected clinical manifestations of primary HIV-1 infection. Clin Infect Dis 2015; 61:1013–21. [DOI] [PubMed] [Google Scholar]
- 3.Robb ML, Eller LA, Kibuuka H, et al. ; RV 217 Study Team . Prospective study of acute HIV-1 infection in adults in East Africa and Thailand. N Engl J Med 2016; 374:2120–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanders EJ, Price MA, Karita E, et al. Differences in acute retroviral syndrome by HIV-1 subtype in a multicentre cohort study in Africa. AIDS 2017; 31:2541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tindall B, Barker S, Donovan B, et al. Characterization of the acute clinical illness associated with human immunodeficiency virus infection. Arch Intern Med 1988; 148:945–9. [PubMed] [Google Scholar]
- 6.Ghosn J, Deveau C, Chaix ML, et al. ; ANRS PRIMO Cohort . Despite being highly diverse, immunovirological status strongly correlates with clinical symptoms during primary HIV-1 infection: a cross-sectional study based on 674 patients enrolled in the ANRS CO 06 PRIMO cohort. J Antimicrob Chemother 2010; 65:741–8. [DOI] [PubMed] [Google Scholar]
- 7.Lavreys L, Baeten JM, Overbaugh J, et al. Virus load during primary human immunodeficiency virus (HIV) type 1 infection is related to the severity of acute HIV illness in Kenyan women. Clin Infect Dis 2002; 35:77–81. [DOI] [PubMed] [Google Scholar]
- 8.Socías ME, Sued O, Laufer N, et al. ; Grupo Argentino de Seroconversión Study Group . Acute retroviral syndrome and high baseline viral load are predictors of rapid HIV progression among untreated Argentinean seroconverters. J Int AIDS Soc 2011; 14:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alter G, Malenfant JM, Delabre RM, et al. Increased natural killer cell activity in viremic HIV-1 infection. J Immun 2004; 173: 5305–11. [DOI] [PubMed] [Google Scholar]
- 10.Kazer SW, Aicher TP, Muema DM, et al. Integrated single-cell analysis of multicellular immune dynamics during hyperacute HIV-1 infection. Nat Med 2020; 26:511–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabado RL, O’Brien M, Subedi A, et al. Evidence of dysregulation of dendritic cells in primary HIV infection. Blood 2010; 116:3839–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stacey AR, Norris PJ, Qin L, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol 2009; 83:3719–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nat Rev Immunol 2010; 10:11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muema DM, Akilimali NA, Ndumnego OC, et al. Association between the cytokine storm, immune cell dynamics, and viral replicative capacity in hyperacute HIV infection. BMC Med 2020; 18:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ansari AA, Mayne AE, Sundstrom JB, et al. Administration of recombinant rhesus interleukin-12 during acute simian immunodeficiency virus (SIV) infection leads to decreased viral loads associated with prolonged survival in SIVmac251-infected rhesus macaques. J Virol 2002; 76:1731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenton-May AE, Dibben O, Emmerich T, et al. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology 2013; 10:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyer SS, Bibollet-Ruche F, Sherrill-Mix S, et al. Resistance to type 1 interferons is a major determinant of HIV-1 transmission fitness. Proc Natl Acad Sci U S A 2017; 114:E590–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mueller YM, Do DH, Altork SR, et al. IL-15 treatment during acute simian immunodeficiency virus (SIV) infection increases viral set point and accelerates disease progression despite the induction of stronger SIV-specific CD8+ T cell responses. J Immun. 2008; 180: 350–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirazi Y, Pitha PM. Interferon alpha-mediated inhibition of human immunodeficiency virus type 1 provirus synthesis in T-cells. Virology 1993; 193:303–12. [DOI] [PubMed] [Google Scholar]
- 20.Osborn L, Kunkel S, Nabel GJ. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc Natl Acad Sci U S A 1989; 86:2336–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin RH, Hwang YW, Yang BC, Lin CS. TNF receptor-2-triggered apoptosis is associated with the down-regulation of Bcl-xL on activated T cells and can be prevented by CD28 costimulation. J Immun 1997; 158: 598–603. [PubMed] [Google Scholar]
- 22.Swingler S, Mann A, Jacqué J, et al. HIV-1 Nef mediates lymphocyte chemotaxis and activation by infected macrophages. Nat Med 1999; 5:997–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamali A, Price MA, Lakhi S, et al. ; IAVI Africa HIV Prevention Partnership . Creating an African HIV clinical research and prevention trials network: HIV prevalence, incidence and transmission. PLoS One 2015; 10:e0116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esbjörnsson J, Mild M, Månsson F, Norrgren H, Medstrand P. HIV-1 molecular epidemiology in Guinea-Bissau, West Africa: origin, demography and migrations. PLoS One 2011; 6:e17025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010; 59:307–21. [DOI] [PubMed] [Google Scholar]
- 26.Crowell TA, Colby DJ, Pinyakorn S, et al. ; RV254/SEARCH010 Study Group . Acute retroviral syndrome is associated with high viral burden, CD4 depletion, and immune activation in systemic and tissue compartments. Clin Infect Dis 2018; 66:1540–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorrucci M, Rezza G, Vlahov D, et al. Clinical characteristics and prognostic value of acute retroviral syndrome among injecting drug users. Italian Seroconversion study. AIDS 1995; 9:597–604. [DOI] [PubMed] [Google Scholar]
- 28.{. V-PLEX Human Biomarker 40-Ples Kit. Available at: https://www.mesoscale.com/en/products/v-plex-human-biomarker-40-plex-kit-k15209d/. Accessed 8 February 2020.
- 29.Hemelaar J, Elangovan R, Yun J, et al. Global and regional molecular epidemiology of HIV-1, 1990–2015: a systematic review, global survey, and trend analysis. Lancet Infect Dis 2019; 19: 143–55. [DOI] [PubMed] [Google Scholar]
- 30.Palm AA, Esbjörnsson J, Månsson F, et al. Faster progression to AIDS and AIDS-related death among seroincident individuals infected with recombinant HIV-1 A3/CRF02_AG compared with sub-subtype A3. J Infect Dis 2014; 209:721–8. [DOI] [PubMed] [Google Scholar]
- 31.Sobieszczyk ME, Lingappa JR, McElrath MJ. Host genetic polymorphisms associated with innate immune factors and HIV-1. Curr Opin HIV AIDS 2011; 6:427–34. [DOI] [PubMed] [Google Scholar]
- 32.Lindbäck S, Karlsson AC, Mittler J, et al. Viral dynamics in primary HIV-1 infection. Karolinska Institutet primary HIV infection study group. AIDS 2000; 14:2283–91. [DOI] [PubMed] [Google Scholar]
- 33.Pastor L, Casellas A, Carrillo J, et al. IP-10 levels as an accurate screening tool to detect acute HIV infection in resource-limited settings. Sci Rep 2017; 7:8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol 2005; 23:307–36. [DOI] [PubMed] [Google Scholar]
- 35.Grebely J, Feld JJ, Applegate T, et al. Plasma interferon-gamma-inducible protein-10 (IP-10) levels during acute hepatitis C virus infection. Hepatology 2013; 57:2124–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayney MS, Henriquez KM, Barnet JH, et al. Serum IFN-γ-induced protein 10 (IP-10) as a biomarker for severity of acute respiratory infection in healthy adults. J Clin Virol 2017; 90:32–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang Y, Shen C, Li J, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J Allergy Clin Immunol 2020; 146:119–127.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qiu X, Tang Y, Zou R, et al. Diagnostic accuracy of interferon-gamma-induced protein 10 for differentiating active tuberculosis from latent tuberculosis: a meta-analysis. Sci Rep 2019; 9:11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pastor L, Casellas A, Rupérez M, et al. Interferon-γ-inducible protein 10 (IP-10) as a screening tool to optimize human immunodeficiency virus RNA monitoring in resource-limited settings. Clin Infect Dis 2017; 65:1670–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.