Abstract
Esophageal cancer is one of the most common malignancy in China with high mortality. Understanding pathogenesis and identifying early diagnosis biomarkers can significantly improve the prognosis of patients with esophageal cancer. Exosomes are small vesicular structures containing a variety of components (including DNA, RNA, and proteins) mediating cell-to-cell material exchange and signal communication. Growing evidences have shown that exosomes and its components are involved in growth, metastasis and angiogenesis in cancer, and could also be used as diagnostic and prognostic markers. In this review, we summarized recent progress to elucidate the significance of exosomes in the esophageal cancer progression, microenvironment remodeling, therapeutic resistance, and immunosuppression. We also discuss the utility of exosomes as diagnostic and prognostic biomarkers and therapeutic tool in esophageal cancer.
Keywords: exosome, esophageal cancer, biomarkers, liquid biopsy, prognosis
Introduction
Esophageal cancer (EC) is one of the most common diseases worldwide with high mortality.1 China accounts for more than 50% of the world’s newly diagnosed patients of EC.2 Surgery is the cornerstone treatment for EC, with a median survival time of 13.6–19.3 months.3 However, approximately 30% of newly diagnosed patients are suitable for curative resection. Although the multimodality treatment of EC has made significant progress in recent decades, the prognosis of patients still remains poor. There is no effective treatment method for the advanced patients. Early diagnosis can significantly reduce the mortality in patients with EC. It is urgently needed to identify the biomarkers to early diagnosis and accurately prognosis prediction of EC.
Liquid biopsy allows detection of tumor-derived molecular biomarkers through biofluid sample with advantage of non-invasive, sensitive, and real-time analysis for early diagnosis and prognosis prediction.4–6 Currently, circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), and extracellular vesicles (EVs) are the main targets of liquid biopsy. According to the size and pathway of generation, EVs are classified into exosomes, microvesicles (MVs), and apoptotic bodies. Exosomes are nano-sized (30–150nm) vesicles generated by the invagination of the plasma membrane. Exosomes have received tremendous attention in recent years because of convenient collection from most biofluids and the stability of cargoes.7,8
Exosomes were first discovered in 1983 in serum medium.9 Exosomes are 30–150 nm nanovesicles secreted from various cell types, containing lipids, proteins and nucleic acids.10,11 Previous studies have shown that exosomes played important roles in cell-to-cell material exchange and signal communication.7,12 Recently, exosomes are found to be participated in tumorigenesis and progression.13,14 In this review, we summarize the significance of exosomes in the EC progression, microenvironment remodeling, therapeutic resistance, and immunosuppression. The utility of exosomes as EC diagnostic, prognostic and predictive biomarkers is reviewed. Furthermore, application of exosomes in the treatment of EC is also discussed.
The Relationship Between Exosomes and EC
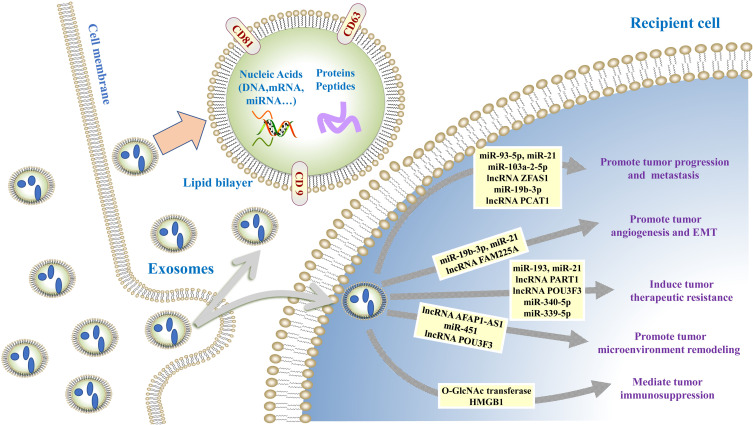
Exosomes play important roles in different stages of tumor development cascade (Figure 1) including tumor proliferation, angiogenesis, epithelial–mesenchymal transition (EMT), migration, microenvironment remodeling, and therapeutic resistance, which are described below (Table 1).
Figure 1.
The role of exosomes in esophageal cancer. Exosomes mediate cell-cell and cell-microenvironment communication through transferring the biologically components (DNA, RNA, and proteins) as well as contribute to esophageal cancer progression by regulating angiogenesis, epithelial–mesenchymal transition (EMT), immune escape, microenvironment remodeling, and therapeutic resistance.
Table 1.
The Role of Exosomes in EC
| Cargo Type | Exosomal Cargo | Originated Cells | Recipient Cells | Target | Function | Reference |
|---|---|---|---|---|---|---|
| miRNA | miR-93-5p | Overexpressed EC9706 | EC9706 | PTEN/PI3K/AKT | Promote proliferation | [15] |
| miR-103a-2-5p | Overexpressed TE-1 | TE-1 and KYSE-150 | CDH11/NR3C1 | Promote proliferation and migration | [16] | |
| miR-21 | Overexpressed EC9706 | EC9706 | PDCD4/JNK | Promote migration and invasion | [17] | |
| miR-19b-3p | Overexpressed EC9706 | EC9706 | PTEN | Promote EMT and migration, invasion | [19] | |
| miR-21 | KYSE-150 | HUVEC | SPRY1/VEGF | Promote angiogenesis | [27] | |
| miR-451 | HFSF-PI3 | KYSE-30 | Extracellular matrix | Promote migration and progression | [39] | |
| miR-21 | Cisplatin-resistant Eca109/DDP | TE-1 | PDCD4 | Confer cisplatin resistance | [55] | |
| miR-193 | Cisplatin-resistant TE-1/DDP | TE-1 | JAK-STAT | Confer cisplatin resistance | [56] | |
| miR-340-5p | Hypoxia TE-13, TE-1 and Eca109 | TE-13, TE-1 and Eca109 | KLF10/UVRAG | Confer radioresistance | [57] | |
| lncRNA | ZFAS1 | Overexpressed Eca109 | Eca109 | miR-124 | Promote proliferation and migration | [18] |
| PCAT1 | KYSE-150 | NE3 | miR-326 | Promote proliferation | [21] | |
| FAM225A | Eca109 and TE-1 | HUVEC | miR-206 | Promote angiogenesis and growth | [29] | |
| AFAP1-AS1 | M2 Macrophage | KYSE-410 | miR-26a/ATF2 | Promote migration and invasion | [37] | |
| POU3F3 | KYSE-450 and TE-12 | Het-1a | CAFs | Promote progression and confer cisplatin resistance | [40] | |
| PART1 | Gefitinib-resistant TE-1/GR and KYSE-450/GR | TE-1 and KYSE-450 | miR-129 | Confer gefitinib resistance | [54] | |
| FMR1-AS1 | ECSCs | Eca109 and KYSE-150 | TLR7/NFkappaB/c-Myc | Maintain cancer stem-like cell phenotype | [62] | |
| Protein | OGT | ECSCs | CD8+ T | PD-1 | Promote immunosuppression | [50] |
| HMGB1 | Eca109 and TE-13 | Monocyte | PD-1/IL-10 | Promote immunosuppression | [53] | |
| circRNA | circ-0048117 | Eca109 and TE-1 | THP-1 | miR-140 | Remodel microenvironment and promote progression | [52] |
Abbreviations: AKT, protein kinase B; ATF2, activating transcription factor 2; CAFs, cancer-associated fibroblasts; CDH11, Cadherin-11; EC, esophageal cancer; EMT, epithelial–mesenchymal transition; NR3C1, nuclear receptor subfamily 3 group C member 1; HUVECs, human umbilical venous endothelial cells; JAK, janus kinases; JNK, c-Jun N-terminal kinase; KLF10, kruppel-like factor 10; NF-κB, nuclear factor kappa-B; PDCD4, programmed cell death 4; OGT, O-GlcNAc transferase; PD‐1, programmed cell death protein 1; PI3K, phosphatidylinositol 3-kinase; PTEN, phosphatase and tensin homolog deleted on chromosome 10; SPRY1, sprouty homolog 1; STAT, signal transducer and activator of transcription; TLR 7, toll like receptor 7; UVRAG, UV radiation resistance-associated gene protein; VEGF, vascular endothelial growth factor.
Exosomes Regulate the Growth and Metastasis of EC
Growing evidences demonstrated that exosomes could regulate the growth and metastasis of EC through secreting biologically active components. Tumor-derived exosomal miR-93-5p could be transferred to EC9706 EC cells, which might inhibit the expression of p21 and Cyclin D1 through the phosphatase and tensin homolog deleted on chromosome 10 (PTEN)/phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) pathway to promote tumor cell proliferation.15 Gao et al identified the esophageal squamous cell carcinoma (ESCC)-derived exosomal miR-103a-2-5p promoting proliferation and migration of ESCC cells by targeting Cadherin-11 (CDH11) and nuclear receptor subfamily 3 group C member 1 (NR3C1).16 Tumor-derived exosome containing miR-21 could target programmed cell death 4 (PDCD4) protein and activate the downstream c-Jun N-terminal kinase (JNK) signaling pathway to promote the migration and invasion of recipient EC cells.17 Tumor-derived exosomal lncRNA ZFAS1 favored the tumorigenesis in EC cells through promoting proliferation, migration, and invasion but suppressing apoptosis by upregulating signal transducer and activator of transcription 3 (STAT3) and downregulating miR-124.18 Tumor-derived exosome containing miR-19b-3p could target PTEN to promote invasion and inhibit apoptosis of EC cells.19 Matsumoto et al demonstrated that tumor‐derived exosomes could promote tumor progression and malignant transformation by altering gene expression and phenotype of tumor cells.20 ESCC cell-derived exosomes containing lncRNA prostate cancer-associated transcript 1 (PCAT1) promoted ESCC cell proliferation by sponging tumor suppressor miR-326.21 Knockdown of PCAT1 could induce cell cycle arrest at G2/M phase, and increased paclitaxel sensitivity. Furthermore, studies have demonstrated that exosomes-derived from cancer-associated fibroblasts (CAFs) favored the development and progression of tumors.22,23 Zhao et al revealed CAFs-derived exosomes can improve ESCC cells growth and migration through Hedgehog signaling pathway.24 The facts mentioned above highlight that exosomes play a significant role in growth and metastasis of EC.
Exosomes Participate in EC Angiogenesis and EMT
Angiogenesis is a multistep process promoting the formation of new blood vessels and it is essential for tumor invasiveness and metastasis.25,26 Emerging evidences indicate exosomal involves in EC angiogenesis. Zhuang et al found that ESCC cell-derived exosome containing miR-21 could stimulate the angiogenic ability of human umbilical venous endothelial cells (HUVECs) by sprouty homolog 1 (SPRY1) downregulation and vascular endothelial growth factor (VEGF) upregulation.27 A recent study reported that exosomes derived from ESCC cells under hypoxia played a more important role in promoting tube formation of HUVECs than exosomes from ESCC cells under normoxia.28 Moreover, hypoxic exosomes significantly promoted tumor growth and lung metastasis compared with normoxic exosomes in nude mice models. Zhang et al also showed that ESCC derived exosome-mediated transfer of lncRNA FAM225A could participate in ESCC progression and angiogenesis by sponging miR-206 to upregulate neuropilin and tolloid like 2 (NETO2) and forkhead box prote1 (FOXP1) expression.29
During EMT, epithelial cells lose polarity and acquire aggressive mesenchymal phenotype favoring tumor cells progression.30–32 Min et al reported that exosomes derived from irradiated T cells could promote metastasis of EC TE13 cells by promoting EMT through regulating β-catenin and the nuclear factor kappa-B (NF-κB)/snail pathway.33 Tumor-derived exosomal miR-19b-3p could induce the expression of downstream EMT-related proteins through the PTEN pathway and promote the migration of EC9706 EC cells.19 Altogether, the studies mentioned above indicate that exosomes can induce the angiogenesis and EMT by regulating various gene expression and signal transduction in EC.
Exosomes Play Key Role in EC Microenvironment Remodeling
Fibroblasts, endothelial cells and infiltrating immune cells are the important component of the tumor microenvironment.34–36 Exosomes from cancer cells, immune cells, and CAFs mediated signaling pathway play central role in tumor progression by remodeling the microenvironment. M2 macrophage-derived exosomes transferred lncRNA AFAP1-AS1 to downregulate miR-26a and upregulate ATF2, thus promoting the invasion and metastasis of EC.37 Targeting M2 macrophages and the lncRNA AFAP1-AS1/miR-26a/activating transcription factor 2 (ATF2) signaling axis represents a promising therapeutic strategy for EC. Nouraee et al explored the role of exosomal miRNAs in the tumor microenvironment by a co-culture system of EC cells and normal fibroblasts.38 They found the 18 exo-miRNAs abnormal expression involved in cell adhesion, endocytosis and cell junctions pathways, suggesting CAFs through secretion of exo-miRNAs remodeling tumor microenvironment to favor tumor progression. Khazaei et al showed that CAFs cell-derived exosomes containing miR-451 played a stromal signaling role to facilitate extracellular matrix niche microenvironment for tumor cell migration.39 Recently, it has been revealed that ESCC derived exosomes containing lncRNA POU3F3 triggered normal fibroblasts (NFs) reprogramming into CAFs for creating a suitable tumor progression microenvironment.40
Additionally, exosome and its components participate in the cross-talks between cancer cells and mesenchymal cells under hypoxia microenvironment.41,42 Chen et al performed high-throughput analysis of ESCC cell-derived exosomal miRNAs during normoxia and hypoxia.43 They found 50 up-regulated miRNAs and 34 down-regulated miRNAs under hypoxic environment. These miRNAs mainly involved in regulation of phospholipase D signaling pathway, autophagy, focal adhesion and advanced glycation end products (AGE) signaling pathways. The hypoxic tumor cells might influence the surrounding microenvironment in a exosome-mediated paracrine manner. The above studies have shown that EC microenvironment remodeling is closely related to the regulation of various gene expression and signal transduction by exosome.
Exosomes Mediate EC Immunosuppression
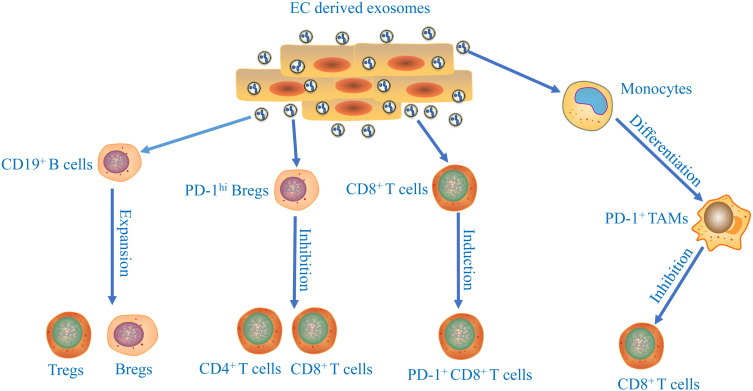
In tumor progression, tumor cells comprehensively induce an immunosuppressive microenvironment to fight against anticancer immunity.44,45 Recent studies have shown that tumor-derived exosomes participate in the development of immunosuppressive microenvironment (Figure 2). In tumor microenvironment, the proliferation and differentiation of B cells could be regulated by cancer cells-derived exosomes in glioma and Burkitt’s lymphoma.46,47 ESCC‐derived exosomes suppressed the proliferation of CD19+ B cells and mediated their differentiation into B10 cells.48 The induced B10 cells might further promote the suppressive status on other immune cells and increase the proportion of regulatory B cells (Bregs) and regulatory T cells (Tregs) by producing IL‐10. Furthermore, ESCC‐derived exosomes promoted the programmed cell death protein 1 (PD‐1) expression in B cells through toll like receptor 4 (TLR 4) and mitogen-activated protein kinases (MAPK) signaling pathways, which mediated the induction of PD‐1hi Bregs. PD‐1hi Bregs inhibited the proliferation and regulatory effects of CD4+ and CD8+ T cells through the PD‐1/ programmed death ligand 1 (PD‐L1)‐dependent pathway in thyroid tumors.49 Yuan et al showed that exosome derived from EC stem cells (ECSCs) could transfer nutrient sensor O-GlcNAc transferase (OGT) to neighboring CD8+ T cells promoting the immune escape of ECSCs via up-regulation of PD-1 in CD8+ T cells.50
Figure 2.
The immune suppressive effects of esophageal cancer cells-derived exosomes. The underlying mechanisms of exosomes released from tumor cells participate in the development of immunosuppressive microenvironment of esophageal cancer.
Macrophages are one of the most important components of immune cells.51 Tumor-derived exosomes played an important role in affecting macrophage polarization during development of ESCC.52 Moreover, ESCC-derived exosomal high mobility group box 1 (HMGB1) are involved in triggering differentiation of monocytes into PD-1+ tumor-associated macrophages (TAMs).53 The phenotype and function of PD-1+ TAMs displayed features of M2 macrophages, including enhancement in CD206 and IL-10 expression, reduction in human leukocyte antigen DR (HLA-DR), CD64, and IL-12 expression, and ability to repress CD8+ T-cell proliferation. Thus, combined immunotherapy with targeting PD-1+ TAMs and tumor-derived exosomal HMGB1 to restore immune function represents a potential therapeutic strategy for EC patients. The facts mentioned above show that exosomes play essential roles in the regulation of EC immunosuppression.
Exosomes Promote EC Therapy-Resistance
Therapy resistance remains a major obstacle for improving prognosis of EC. The exosomes derived from resistant tumor cells may confer resistance phenotype to the sensitive cells through cell-to-cell information transmission. Kang et al demonstrated that exosomes from gefitinib-resistant cells containing lncRNA PART1 promoted gefitinib resistance in ESCC via the miR-129/blc-2 axis.54 Exosomes containing miR-21 from cisplatin-resistant cells promoted the formation of cisplatin resistance in ESCC through targeting PDCD4.55 In addition, tumor derived exosome-mediated miR-193 delivery to ESCC cells increase cisplatin resistance via activation of janus kinases (JAK)-STAT signaling pathways.56 ESCC-derived exosomal lncRNA POU3F3 transformed NFs into CAFs, then IL-6 secreted from these CAFs boosted resistance to cisplatin in ESCC cells.40 Recently, it has been shown that hypoxic tumor cell-derived exosomal miR-340-5p conferred radioresistance in ESCC by targeting kruppel-like factor 10 (KLF10)/UV radiation resistance-associated gene protein (UVRAG).57 The level of miR-340-5p in the plasma exosomes was closely related to radiotherapy response and prognosis. miR-340-5p might be a therapeutic target for overcoming radioresistance in ESCC. Luo et al demonstrated that tumor-derived exosomal miR-339-5p enhanced radiosensitivity of ESCC cells by targeting Cdc25A.58 In addition, growing evidences have shown that cancer stem cells (CSCs) play pivotal role in tumor therapy-resistance.59–61 Exosomal FMR1-AS1 secreted from ECSCs transferred stemness phenotypes to recipient non-CSCs through activating TLR7/NF-κB/c-Myc signaling pathway.62 These studies suggest that exosomes play a pivotal role in regulating EC therapy-resistance. Further detailed investigation will offer new opportunities to develop more efficient exosome-based strategies reversing therapy resistance.
Application of Exosomes in the Diagnosis and Prognosis of EC
Growing evidences have suggested that early diagnosis and accurate therapy efficacy prediction could significantly improve the prognosis of ESCC patients. Exosome may be an attractive cancer biomarker due to easily obtained from most biofluids, high stability, and carrying dynamic information about tumor state. In the next part, exosomes used as diagnostic and prognostic biomarkers in EC is reviewed (Table 2).
Table 2.
Exosomes as Diagnostic and Prognostic Biomarkers for EC
| Cargo Type | Exosomal Cargo | Biofluids | Extraction Method | Identification Method | Detection Method | Clinical Value | Reference |
|---|---|---|---|---|---|---|---|
| miRNA | miR-652-5p | Serum | Differential centrifugation | Flow cytometry | qRT-PCR | Reduced expression in EC patients | [66] |
| miR-21 | Serum | Commercial kit | Not shown | qRT-PCR | Related to lymphatic metastasis and stage | [67] | |
| miR-766-3p | Serum | Commercial kit | TEM and Western blot | miRNA profiling, qRTPCR | Indicator for poor prognosis | [68] | |
| miR-182 | Serum | Commercial kit | Not shown | qRT-PCR | Indicator for poor prognosis | [69] | |
| miR-340-5p | Plasma | Differential centrifugation | TEM, NTA and Western blot | qRT-PCR | Indicator for radioresistance and poor prognosis | [57] | |
| miR-339-5p | Serum | Differential centrifugation | TEM and Western blot | qRT-PCR | Predicts response to preoperative radiotherapy | [58] | |
| lncRNA | PCAT1 | Serum | Ultracentrifugation | TEM and NTA | qRT-PCR | Higher expression in EC patients | [21] |
| UCA1 | Plasma | Ultracentrifugation | TEM, NTA and Western blot | qRT-PCR | Biomarker for diagnosis of EC | [73] | |
| UCA1, POU3F3, ESCCAL-1 and PEG10 | Serum | Commercial kit | TEM, NTA and Western blot | qRT-PCR | Biomarker for diagnosis of EC | [74] | |
| circRNA | circ-0026611 | Serum | Commercial kit | TEM and Western blot | circRNA microarray, qRT-PCR | Related to lymphatic metastasis and stage | [77] |
| Protein | Stathmin-1 | Serum | Ultracentrifugation | TEM, NTA and Western blot | ELISA | Indicator for poor prognosis | [79] |
| RNA | seG-chiRNA | Salivary | Commercial kit | TEM, NTA and Western blot | qRT-PCR | Predicts recurrence and therapeutic response of EC | [82] |
Abbreviations: EC, esophageal cancer; ELISA, enzyme-linked immunosorbent assay; NTA, nanoparticle tracking analysis; qRT-PCR, quantitative real time polymerase chain reaction; TEM, transmission electron microscope.
Exosomes Amount
Zhao et al found the level of circulating exosomes (CEs) from ESCC patients are significantly upregulated.63 The sensitivity and specificity of CEs to distinguish ESCC patients from healthy individuals were 75% and 85%, respectively. The level of CEs was an independent prognostic marker for ESCC patients. The increased level of CEs was associated with unfavourable overall survival and progression free survival in ESCC patients. However, the result is inconsistent with previous report. Matsumoto et al revealed that the 3-year overall survival rate in patients with high plasma exosome amount was significantly higher than those with low plasma exosome amount.64 Low levels of exosome predicted a poor prognosis. The prognostic significance of exosome amount in EC still needs further research.
Exosomal miRNAs
Warnecke-Eberz et al identified “Exosomal onco-miRs” was overexpressed in EC cells, such as miR-223-5p, miR-223-3p, miR-483-5p, miR-409-3p, miR-196b-5p, miR-192-5p, miR-146a-5p, and miR-126-5p.65 “Exosomal onco-miRs” may represent promising marker for diagnosis and monitoring efficacy of therapy for EC. The significant decrease in expression of exosomal miR-652-5p was detected in ESCC patients compared with healthy controls, suggesting that the circulating miR-652-5p is feasibility to be a novel biomarker for ESCC diagnosis.66 Tanaka et al revealed that circulatory serum exosomal miRNA-21 level was significantly increased in ESCC patients and closely related with advanced tumor stage and lymph node metastasis.67 Serum exosomal miRNA-21 might be a potential diagnostic biomarker for ESCC. Liu et al revealed that upregulation of serum exosomal miR‐766‐3p levels had close relationship with advanced TNM stage and poor prognosis (for overall survival, hazard ratio 2.21; for disease-free survival, hazard ratio 2.15) for patients with ESCC.68 Qiu et al found that serum exosomal miR-182 level was significantly higher in pre-operative ESCC patients compared with the level in post-operative ESCC patients.69 High serum exosomal miR-182 expression was also strongly associated with worse clinicopathologic profile including poor differentiation, lymph node metastasis, and advanced TNM stage, suggesting that the serum exosomal miR-182 accurately reflected the burden of ESCC. Furthermore, serum exosomal miR-182 might serve as a promising biomarker for predicting the unfavorable prognosis of ESCC. The patients with high serum exosomal miR-182 had poor overall survival and relapse free survival than those with low serum exosomal miR-182.
Lymph node metastasis is a significant risk factor for ESCC prognosis. Accurate monitoring of lymph node metastasis plays crucial roles in developing treatment strategies and predicting patient prognosis. Recently, Liu et al established integrated nomogram combining exosomal miRNA-based model and clinical characteristics could increase the accuracy prediction of lymph node status.70 In addition, circulatory exosomal miR-340-5p and miR-339-5p might be promising biomarkers for prediction of radiotherapy response and prognosis in patients with ESCC.57,58 Exosomal miRNAs will have great potential application in tumor individualized treatment.
Exosomal lncRNAs
Recently, Tian et al demonstrated that dysregulated exosomal lncRNAs were associated with different status of disease in esophagus.71 These exosomal lncRNAs potentially serve as a diagnostic or prognostic biomarker for EC patients. The ESCC cells-derived exosomes containing lncRNA PCAT1 were higher in the serum of ESCC patients, and PCAT1 might serve as a non-invasive biomarker for ESCC.21 Jiao et al found that expression levels of the lncRNAs NR_039819, NR_036133, NR_003353, ENST00000442416.1, and ENST00000416100.1 were significantly higher in exosomes from ESCC patients than controls.72 The levels of the five plasma exosome lncRNAs were significant decline after surgery in ESCC patients. These plasma exosome lncRNAs could be used as novel diagnostic markers for ESCC. Exosomal lncRNA UCA1 was a promising biomarker for the diagnosis of EC with 86.7% sensitivity and 70.2% specificity.73 Yan et al demonstrated that circulating exosomal 4-lncRNAs (UCA1, POU3F3, ESCCAL-1 and PEG10) had potential value for diagnosis of ESCC.74 Moreover, this lncRNA panel also had a higher sensitivity (73.40%) to detect stage I–II tumors than squamous cell carcinoma antigen 1 (SCCA1). The exosomal lncRNA panel provides a more accurate diagnosis of ESCC, especially for the early stage disease.
Exosomal circRNAs
Studies have confirmed that the level of circRNAs was significantly enriched in exosomes compared to exosome-derived cells.75 The circRNAs also play a critical role in the progression of ESCC. Fan et al validated circRNA has-circ-0001946 and has-circ-0043603 secreted by ESCC cells might be potential diagnostic biomarkers for ESCC.76 The sensitivity and specificity of has-circ-0001946 and has-circ-0043603 were 92% and 80%, 64% and 92% respectively, according to receiver operating characteristic (ROC) analysis. The combined detection might have a better diagnostic accuracy than that of single detection. Liu et al suggested an association between high expression of serum exosomal has-circ-0026611 with lymph node metastasis and advanced T stage in ESCC.77 Overexpression of serum exosomal has-circ-0026611 was closely associated with poor overall survival and disease-free survival. Serum exosomal has-circ-0026611 may be a novel, promising predictor of ESCC prognosis.
Others
Stathmin-1, a microtubule-destabilizing cytosolic phosphoprotein, plays an important role in tumor cell proliferation and migration.78 Study has shown that Stathmin-1 could be delivered by exosomes promoting the growth, migration, and invasion of ESCC.79 Knockdown stathmin-1 expression also enhanced the sensitivity of ESCC cells to docetaxel and radiation. Moreover, exosomal Stathmin-1 could be as a new predictor for unfavorable prognosis in ESCC patients.
Salivary exosomes might be considered a novel and convenient source of cancer biomarker discovery.80,81 Lin et al demonstrated that salivary exosomal GOLM1-NAA35 chimeric RNA (seG-NchiRNA) could predict recurrence or disease progression earlier than traditional radiological assessment in patients with ESCC.82 seG-NchiRNA was an independent predictor of progression-free-survival in patients with ESCC received chemoradiation. The sensitivity and specificity of seG-NchiRNA for ESCC diagnosis were 85.3–89.1% and 89.1%–91.4%, respectively. seG-NchiRNA assays might serve as a noninvasive biomarker for early diagnosis, reflecting of tumor burden, and prediction of therapy response to aid treatment decisions in ESCC.
Dicer plays pivotal roles in the pathway of miRNAs biogenesis, so monitoring exosomal Dicer may be more efficient and convenient than miRNA.83 The enrichment of Dicer can be monitored in exosomes derived from cancer cells and serum of cancer patients. Okuda et al reported that the noninvasive combined detection of Helicobacter pylori and serum exosomal Dicer provides a promising biomarker for identifying differentiated gastric cancer, especially for the early stage disease.84 The suitability of serum exosomal Dicer fit as a diagnostic biomarker for EC need further study.
Utility of Exosomes in the Treatment of EC
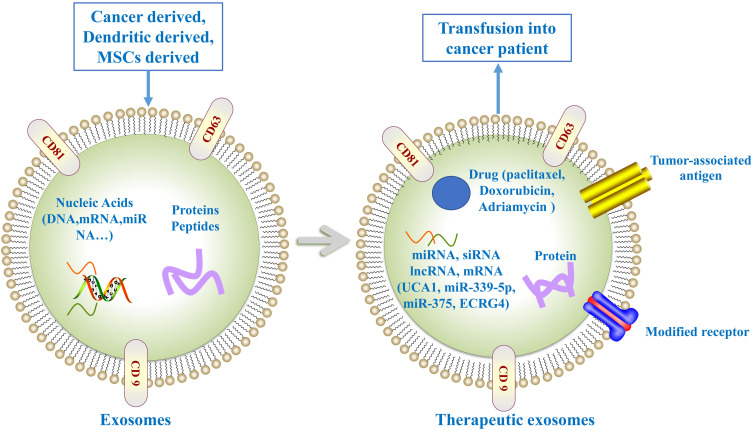
Exosomes have many advantages as delivery vehicles for drug, proteins, nucleic acids, and gene therapeutic agents (Figure 3), for example, low immunogenicity, high biocompatibility, long circulating half-life, less toxicity and ability to cross the biological barriers. Emerging evidence have shown the feasibility of exosomes used as carriers to deliver anti-tumor agents for EC therapy.
Figure 3.
Exosome as anti-tumor agents carriers for cancer therapy. Exosome can be loaded with drugs, miRNAs, or siRNAs exerting anti-tumor effect. Exosome-loaded tumor-associated antigen are able to promote anti-tumor immune response. Moreover, the expression of modified receptors in exosome can improve tumor-targeting ability.
Recently, exosomes-loaded chemotherapeutic drug represent a promising strategy for cancer therapy. Adriamycin and paclitaxel-loaded exosomes have been used for cancer therapy with low immunogenicity and toxicity.85,86 The utility of paclitaxel-loaded exosomes have been demonstrated to improve efficacy in the treatment of multidrug resistant cancer cells.87
Exosomes have been used to deliver RNAs such as siRNA and miRNAs for cancer therapy. Exosomal lncRNA UCA1 was taken up by EC cells and inhibited the progression of EC in vitro and tumor growth in vivo through targeting miRNA-613.73 miR-339-5p induced radiosensitivity in ESCC cells. Higher exosomal miR-339-5p levels were associated with radiosensitivity and favorable survival.58 Hence, miR-339-5p could be introduced into ESCC cells via exosomes to promote radiosensitivity and reverse radioresistance. Exosomal miRNAs from umbilical cord-mesenchymal stem cells (UCMSC) may represent novel therapeutic strategies for cancer treatment. UCMSCs derived exosomes could deliver miR-375 to ESCC cells, which promoted apoptosis and suppressed proliferation, invasion, migration, and tumorsphere formation via inhibiting enabled homolog (ENAH) expression.88 It has been shown EC related gene 4 (ECRG4) could inhibit cancer cells migration and invasion in EC.89 Mao et al revealed exosome is a suitable carrier for delivery of ECRG4 mRNA.90 Exosomes can transfer ECRG4 mRNA into recipient cells, which subsequently suppressed cell proliferation in vitro and inhibited tumor growth in vivo. Exosomal ECRG4 mRNA can be exploited as a target for EC gene therapy.
Furthermore, researchers have found that dendritic cell-derived exosomes-loaded tumor antigen could increase anti-tumor immune response by inducing antigen-specific CD8+ T cells and eliminating suppressive function of regulatory T cells. Dai et al found that autologous ascites-derived exosomes combined with granulocyte-macrophage colony stimulating factor (GM-CSF) could induce tumor antigen-specific cytotoxic lymphocyte (CTL) responses in patients with colorectal cancer, with no obvious toxicity.91 André et al reported that dendritic cell-derived exosomes transferred functional major histocompatibility complex (MHC) class I/peptide complexes could induce stable clinical responses in patients with melanoma.92 Morse et al revealed autologous dendritic cell derived-exosomes-loaded multiple melanoma-associated antigen (MAGE) peptides could induce a MAGE-specific T-cell response and increased NK lytic activity in patients with non-small cell lung carcinoma.93 Furthermore, Narita et al demonstrated that dendritic cells pulsed with squamous cell carcinoma antigen recognized by T-cells 1 (SART1) peptide possessed the ability to produce exosomes with an antigen presenting ability for inducing SART1 peptide-specific CTLs in patients with advanced EC.94
The induction of senescence may be a promising strategy for tumor therapy.95,96 Zheng et al identified sulforaphane (SFN) could inhibit ESCC cell proliferation via inducing senescence.97 Meanwhile, SFN caused autophagy dysfunction and exosome release in reactive oxygen species (ROS)-mammalian target of rapamycin (mTOR)-dependent manner participated in triggering paracrine senescence. The modulating autophagy and the exosome-mediated paracrine senescence represents a promising strategy for ESCC treatment. Altogether, the studies mentioned above indicate that exosomes are demonstrably an effective tool for carrying and delivering the anti-cancer therapeutics.
Conclusion and Future Prospects
EC is one of the most deadly malignant tumors, which seriously threatens human health and life. Due to the lack of effective early detection methods, EC patients often present at advanced stage with limited therapeutic interventions and high mortality. It is essential to discover novel molecular biomarkers for disease prediction or intervention. Exosomes are secreted by various cell types as various biological ingredients carriers and crucial tool for cell-to-cell communication. Growing evidences show that exosomes are involved in tumorigenesis and progression of many malignant tumors. Exosomes can be detected in many types of biofluids. Exosomes can be used as valuable biomarkers for diagnosis and prognosis and promising carriers for targeted delivery of drugs and biomolecules. In this review, we have attempted to summarize the role of exosomes in tumor growth, metastasis, microenvironment remodeling, therapeutic resistance, and immunosuppression in EC. Moreover, we discussed the application of exosomes as diagnostic, prognostic and predictive biomarkers and therapeutic tool in EC. Exosomes derived from EC are the most promising biomarkers for EC early diagnosis, prognosis, and response to treatment.
Although exosomes have great potential for application, there are some obstacles that hinder its utility. There is still no standardized method for the isolation and identification of exosomes from biological fluids. The methods used in reported studies lack repeatability and inconvenience, which limit their wide application. Furthermore, the ideal enrichment strategies of exosome with high purity and efficiency is currently unobtainable. The exosome-based engineering applications only carried out in cell or animal experiments due to lacking large scale exosomes for clinical research. Lastly, the mechanisms of exosomes involving in pathogenesis and development of tumor still lacks systematic and in-depth research.
In conclusion, there is still a long way to overcome the barriers for the implementation of exosome-based diagnostic and therapeutic strategies, but these strategies must be transformed into clinical applications for the benefit of EC patients in near future.
Acknowledgments
This work was supported by the Zhejiang Province Public Welfare Technology Application Research Project (grant numbers LGC21H160001).
Abbreviations
AGE, advanced glycation end products; AKT, protein kinase B; ATF2, activating transcription factor 2; Bregs, regulatory B cells; CAFs, cancer-associated fibroblasts; CDH11, Cadherin-11; CEs, circulating exosomes; CSCs, cancer stem cells; CTCs, circulating tumor cells; ctDNA, circulating tumor DNA; CTL, cytotoxic lymphocyte; EC, esophageal cancer; ECRG4, esophageal cancer-related gene 4; ECSCs, esophageal cancer stem cells; EMT, epithelial–mesenchymal transition; ENAH, enabled homolog; ESCC, esophageal squamous cell carcinoma; EVs, extracellular vesicles; FOXP1, forkhead box prote1; GM-CSF, granulocyte-macrophage colony stimulating factor; HMGB1, high mobility group box 1; HUVECs, human umbilical venous endothelial cells; MAPK, mitogen-activated protein kinases; mTOR, mammalian target of rapamycin; JAK, janus kinases; JNK, c-Jun N-terminal kinase; KLF10, kruppel-like factor 10; MAGE, melanoma-associated antigen; MHC, major histocompatibility complex; NETO2, neuropilin and tolloid like 2; NF-κB, nuclear factor kappa-B; NFs, normal fibroblasts; NR3C1, nuclear receptor subfamily 3 group C member 1; OGT, O-GlcNAc transferase; PCAT1, prostate cancer-associated transcript 1; PD‐1, programmed cell death protein 1; PDCD4, programmed cell death 4; PD‐L1, programmed death ligand 1; PI3K, phosphatidylinositol 3-kinase; PTEN, phosphatase and tensin homolog deleted on chromosome 10; ROC, receiver operating characteristic; ROS, reactive oxygen species; SART1, squamous cell carcinoma antigen recognized by T-cells 1; SCCA, squamous cell carcinoma antigen 1; SFN, sulforaphane; SPRY1, sprouty homolog 1; STAT3, signal transducer and activator of transcription 3; TAMs, tumor-associated macrophages; Tregs, regulatory T cells; TLR 4, toll-like receptor 4; UCMSC, umbilical cord-mesenchymal stem cells; UVRAG, UV radiation resistance-associated gene protein; VEGF, vascular endothelial growth factor.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Malhotra GK, Yanala U, Ravipati A, Follet M, Vijayakumar M, Are C. Global trends in esophageal cancer. J Surg Oncol. 2017;115(5):564–579. doi: 10.1002/jso.24592 [DOI] [PubMed] [Google Scholar]
- 2.Lin Y, Totsuka Y, Shan B, et al. Esophageal cancer in high-risk areas of China: research progress and challenges. Ann Epidemiol. 2017;27(3):215–221. doi: 10.1016/j.annepidem.2016.11.004 [DOI] [PubMed] [Google Scholar]
- 3.Huang FL, Yu SJ. Esophageal cancer: risk factors, genetic association, and treatment. Asian J Surg. 2018;41(3):210–215. doi: 10.1016/j.asjsur.2016.10.005 [DOI] [PubMed] [Google Scholar]
- 4.Pantel K, Alix-Panabières C. Liquid biopsy and minimal residual disease - latest advances and implications for cure. Nat Rev Clin Oncol. 2019;16(7):409–424. doi: 10.1038/s41571-019-0187-3 [DOI] [PubMed] [Google Scholar]
- 5.Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18(1):114. doi: 10.1186/s12943-019-1043-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Rubis G, Rajeev Krishnan S, Bebawy M. Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci. 2019;40(3):172–186. doi: 10.1016/j.tips.2019.01.006 [DOI] [PubMed] [Google Scholar]
- 7.Cui S, Cheng Z, Qin W, Jiang L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer. 2018;116:46–54. doi: 10.1016/j.lungcan.2017.12.012 [DOI] [PubMed] [Google Scholar]
- 8.Li G, Tang W, Yang F. Cancer liquid biopsy using integrated microfluidic exosome analysis platforms. Biotechnol J. 2020;15(5):e1900225. doi: 10.1002/biot.201900225 [DOI] [PubMed] [Google Scholar]
- 9.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33(3):967–978. doi: 10.1016/0092-8674(83)90040-5 [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9(1):19. doi: 10.1186/s13578-019-0282-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. doi: 10.1016/j.ceb.2014.05.004 [DOI] [PubMed] [Google Scholar]
- 12.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30(1):255–289. doi: 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- 13.Tai YL, Chen KC, Hsieh JT, Shen TL. Exosomes in cancer development and clinical applications. Cancer Sci. 2018;109(8):2364–2374. doi: 10.1111/cas.13697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu M, Gu J, Jiang P, Qian H, Xu W, Zhang X. Exosomes in gastric cancer: roles, mechanisms, and applications. Mol Cancer. 2019;18(1):41. doi: 10.1186/s12943-019-1001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu MX, Liao J, Xie M, et al. miR-93-5p transferred by exosomes promotes the proliferation of esophageal cancer cells via intercellular communication by targeting PTEN. Biomed Environ Sci. 2018;31(3):171–185. [DOI] [PubMed] [Google Scholar]
- 16.Gao DC, Hou B, Zhou D, et al. Tumor-derived exosomal miR-103a-2-5p facilitates esophageal squamous cell carcinoma cell proliferation and migration. Eur Rev Med Pharmacol Sci. 2020;24(11):6097–6110. [DOI] [PubMed] [Google Scholar]
- 17.Liao J, Liu R, Shi YJ, Yin LH, Pu YP. Exosome-shuttling microRNA-21 promotes cell migration and invasion-targeting PDCD4 in esophageal cancer. Int J Oncol. 2016;48(6):2567–2579. doi: 10.3892/ijo.2016.3453 [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Qin X, Bian W, et al. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J Exp Clin Cancer Res. 2019;38(1):477. doi: 10.1186/s13046-019-1473-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng Q, Zhu Z, Song L, He Z. Transferred by exosomes-derived MiR-19b-3p targets PTEN to regulate esophageal cancer cell apoptosis, migration and invasion. Biosci Rep. 2020;40(11):BSR20201858. doi: 10.1042/BSR20201858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumoto Y, Kano M, Murakami K, et al. Tumor-derived exosomes influence the cell cycle and cell migration of human esophageal cancer cell lines. Cancer Sci. 2020;111(12):4348–4358. doi: 10.1111/cas.14660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang L, Wang Y, Chen J, et al. Long noncoding RNA PCAT1, a novel serum-based biomarker, enhances cell growth by sponging miR-326 in oesophageal squamous cell carcinoma. Cell Death Dis. 2019;10(7):513. doi: 10.1038/s41419-019-1745-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jang I, Beningo KA. Integrins, CAFs and mechanical forces in the progression of cancer. Cancers (Basel). 2019;11(5):721. doi: 10.3390/cancers11050721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ham I-H, Lee D, Hur H. Role of cancer-associated fibroblast in gastric cancer progression and resistance to treatments. J Oncol. 2019;2019:6270784. doi: 10.1155/2019/6270784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao G, Li H, Guo Q, et al. Exosomal sonic hedgehog derived from cancer-associated fibroblasts promotes proliferation and migration of esophageal squamous cell carcinoma. Cancer Med. 2020;9(7):2500–2513. doi: 10.1002/cam4.2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sieczkiewicz GJ, Hussain M, Kohn EC. Angiogenesis and metastasis. Cancer Treat Res. 2002;107:353–381. [DOI] [PubMed] [Google Scholar]
- 26.Abe R, Fujita Y, Yamagishi S. Angiogenesis and metastasis inhibitors for the treatment of malignant melanoma. Mini Rev Med Chem. 2007;7(6):649–661. doi: 10.2174/138955707780859440 [DOI] [PubMed] [Google Scholar]
- 27.Zhuang H, Wang H, Yang H, Li H. Exosome-encapsulated MicroRNA-21 from esophageal squamous cell carcinoma cells enhances angiogenesis of human umbilical venous endothelial cells by targeting SPRY1. Cancer Manag Res. 2020;12:10651–10667. doi: 10.2147/CMAR.S259077 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Mao Y, Wang Y, Dong L, et al. Hypoxic exosomes facilitate angiogenesis and metastasis in esophageal squamous cell carcinoma through altering the phenotype and transcriptome of endothelial cells. J Exp Clin Cancer Res. 2019;38(1):389. doi: 10.1186/s13046-019-1384-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C, Luo Y, Cao J, Wang X, Miao Z, Shao G. Exosomal lncRNA FAM225A accelerates esophageal squamous cell carcinoma progression and angiogenesis via sponging miR-206 to upregulate NETO2 and FOXP1 expression. Cancer Med. 2020;9(22):8600–8611. doi: 10.1002/cam4.3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiori ME, Di Franco S, Villanova L, Bianca P, Stassi G, De Maria R. Cancer-associated fibroblasts as abettors of tumor progression at the crossroads of EMT and therapy resistance. Mol Cancer. 2019;18(1):70. doi: 10.1186/s12943-019-0994-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Weinberg RA. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front Med. 2018;12(4):361–373. doi: 10.1007/s11684-018-0656-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. doi: 10.1016/j.tcb.2018.12.001 [DOI] [PubMed] [Google Scholar]
- 33.Min H, Sun X, Yang X, et al. Exosomes derived from irradiated esophageal carcinoma-infiltrating T cells promote metastasis by inducing the epithelial-mesenchymal transition in esophageal cancer cells. Pathol Oncol Res. 2018;24(1):11–18. doi: 10.1007/s12253-016-0185-z [DOI] [PubMed] [Google Scholar]
- 34.Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79(18):4557–4566. doi: 10.1158/0008-5472.CAN-18-3962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deepak KGK, Vempati R, Nagaraju GP, et al. Tumor microenvironment: challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol Res. 2020;153:104683. [DOI] [PubMed] [Google Scholar]
- 36.Hui L, Chen Y. Tumor microenvironment: sanctuary of the devil. Cancer Lett. 2015;368(1):7–13. doi: 10.1016/j.canlet.2015.07.039 [DOI] [PubMed] [Google Scholar]
- 37.Mi X, Xu R, Hong S, Xu T, Zhang W, Liu M. M2 macrophage-derived exosomal lncRNA AFAP1-AS1 and MicroRNA-26a affect cell migration and metastasis in esophageal cancer. Mol Ther Nucleic Acids. 2020;22:779–790. doi: 10.1016/j.omtn.2020.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nouraee N, Khazaei S, Vasei M, Razavipour SF, Sadeghizadeh M, Mowla SJ. MicroRNAs contribution in tumor microenvironment of esophageal cancer. Cancer Biomark. 2016;16(3):367–376. doi: 10.3233/CBM-160575 [DOI] [PubMed] [Google Scholar]
- 39.Khazaei S, Nouraee N, Moradi A, Mowla SJ. A novel signaling role for miR-451 in esophageal tumor microenvironment and its contribution to tumor progression. Clin Transl Oncol. 2017;19(5):633–640. doi: 10.1007/s12094-016-1575-0 [DOI] [PubMed] [Google Scholar]
- 40.Tong Y, Yang L, Yu C, et al. Tumor-secreted exosomal lncRNA POU3F3 promotes cisplatin resistance in ESCC by inducing fibroblast differentiation into CAFs. Mol Ther Oncolytics. 2020;18:1–13. doi: 10.1016/j.omto.2020.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue M, Chen W, Xiang A, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer. 2017;16(1):143. doi: 10.1186/s12943-017-0714-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Z, Yang M, Li Y, Yang F, Feng Y. Exosomes derived from hypoxic colorectal cancer cells transfer Wnt4 to normoxic cells to elicit a prometastatic phenotype. Int J Biol Sci. 2018;14(14):2094–2102. doi: 10.7150/ijbs.28288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen F, Chu L, Li J, et al. Hypoxia induced changes in miRNAs and their target mRNAs in extracellular vesicles of esophageal squamous cancer cells. Thorac Cancer. 2020;11(3):570–580. doi: 10.1111/1759-7714.13295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molinier-Frenkel V, Castellano F. Immunosuppressive enzymes in the tumor microenvironment. FEBS Lett. 2017;591(19):3135–3157. doi: 10.1002/1873-3468.12784 [DOI] [PubMed] [Google Scholar]
- 45.Tsukamoto H, Fujieda K, Miyashita A, et al. Combined blockade of IL6 and PD-1/PD-L1 signaling abrogates mutual regulation of their immunosuppressive effects in the tumor microenvironment. Cancer Res. 2018;78(17):5011–5022. doi: 10.1158/0008-5472.CAN-18-0118 [DOI] [PubMed] [Google Scholar]
- 46.Han S, Feng S, Ren M, et al. Glioma cell-derived placental growth factor induces regulatory B cells. Int J Biochem Cell Biol. 2014;57:63–68. doi: 10.1016/j.biocel.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 47.Khare D, Or R, Resnick I, Barkatz C, Almogi-Hazan O, Avni B. Mesenchymal stromal cell-derived exosomes affect mRNA expression and function of B-lymphocytes. Front Immunol. 2018;9:3053. doi: 10.3389/fimmu.2018.03053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao Y, Wang Y, Dong L, et al. Circulating exosomes from esophageal squamous cell carcinoma mediate the generation of B10 and PD-1(high) breg cells. Cancer Sci. 2019;110(9):2700–2710. doi: 10.1111/cas.14122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Wang G, Wang Z, et al. PD-1-expressing B cells suppress CD4(+) and CD8(+) T cells via PD-1/PD-L1-dependent pathway. Mol Immunol. 2019;109:20–26. doi: 10.1016/j.molimm.2019.02.009 [DOI] [PubMed] [Google Scholar]
- 50.Yuan Y, Wang L, Ge D, et al. Exosomal O-GlcNAc transferase from esophageal carcinoma stem cell promotes cancer immunosuppression through up-regulation of PD-1 in CD8(+) T cells. Cancer Lett. 2021;500:98–106. doi: 10.1016/j.canlet.2020.12.012 [DOI] [PubMed] [Google Scholar]
- 51.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol. 2019;19(6):369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Q, Wang X, Zhu J, Fei X, Chen H, Li C. Hypoxic tumor-derived exosomal Circ0048117 facilitates M2 macrophage polarization acting as miR-140 sponge in esophageal squamous cell carcinoma. Onco Targets Ther. 2020;13:11883–11897. doi: 10.2147/OTT.S284192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B, Song TN, Wang FR, et al. Tumor-derived exosomal HMGB1 promotes esophageal squamous cell carcinoma progression through inducing PD1(+) TAM expansion. Oncogenesis. 2019;8(3):17. doi: 10.1038/s41389-019-0126-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kang M, Ren M, Li Y, Fu Y, Deng M, Li C. Exosome-mediated transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell carcinoma via functioning as a competing endogenous RNA. J Exp Clin Cancer Res. 2018;37(1):171. doi: 10.1186/s13046-018-0845-9 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Yang YC, Liu GJ, Yuan DF, Li CQ, Xue M, Chen LJ. Influence of exosome-derived miR-21 on chemotherapy resistance of esophageal cancer. Eur Rev Med Pharmacol Sci. 2019;23(4):1513–1519. [DOI] [PubMed] [Google Scholar]
- 56.Shi S, Huang X, Ma X, Zhu X, Zhang Q. Research of the mechanism on miRNA193 in exosomes promotes cisplatin resistance in esophageal cancer cells. PLoS One. 2020;15(5):e0225290. doi: 10.1371/journal.pone.0225290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen F, Xu B, Li J, et al. Hypoxic tumour cell-derived exosomal miR-340-5p promotes radioresistance of oesophageal squamous cell carcinoma via KLF10. J Exp Clin Cancer Res. 2021;40(1):38. doi: 10.1186/s13046-021-01834-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo A, Zhou X, Shi X, et al. Exosome-derived miR-339-5p mediates radiosensitivity by targeting Cdc25A in locally advanced esophageal squamous cell carcinoma. Oncogene. 2019;38(25):4990–5006. doi: 10.1038/s41388-019-0771-0 [DOI] [PubMed] [Google Scholar]
- 59.Talukdar S, Bhoopathi P, Emdad L, Das S, Sarkar D, Fisher PB. Dormancy and cancer stem cells: an enigma for cancer therapeutic targeting. Adv Cancer Res. 2019;141:43–84. [DOI] [PubMed] [Google Scholar]
- 60.Steinbichler TB, Dudás J, Skvortsov S, Ganswindt U, Riechelmann H, Skvortsova II. Therapy resistance mediated by cancer stem cells. Semin Cancer Biol. 2018;53:156–167. doi: 10.1016/j.semcancer.2018.11.006 [DOI] [PubMed] [Google Scholar]
- 61.Das PK, Islam F, Lam AK. The roles of cancer stem cells and therapy resistance in colorectal carcinoma. Cells. 2020;9(6):1392. doi: 10.3390/cells9061392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li W, Zhang L, Guo B, et al. Exosomal FMR1-AS1 facilitates maintaining cancer stem-like cell dynamic equilibrium via TLR7/NFkappaB/c-Myc signaling in female esophageal carcinoma. Mol Cancer. 2019;18(1):22. doi: 10.1186/s12943-019-0949-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao A, Guo L, Xu J, et al. Identification and validation of circulating exosomes-based liquid biopsy for esophageal cancer. Cancer Med. 2019;8(7):3566–3574. doi: 10.1002/cam4.2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsumoto Y, Kano M, Akutsu Y, et al. Quantification of plasma exosome is a potential prognostic marker for esophageal squamous cell carcinoma. Oncol Rep. 2016;36(5):2535–2543. doi: 10.3892/or.2016.5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warnecke-Eberz U, Chon SH, Hölscher AH, Drebber U, Bollschweiler E. Exosomal onco-miRs from serum of patients with adenocarcinoma of the esophagus: comparison of miRNA profiles of exosomes and matching tumor. Tumour Biol. 2015;36(6):4643–4653. doi: 10.1007/s13277-015-3112-0 [DOI] [PubMed] [Google Scholar]
- 66.Gao P, Wang D, Liu M, et al. DNA methylation-mediated repression of exosomal miR-652-5p expression promotes oesophageal squamous cell carcinoma aggressiveness by targeting PARG and VEGF pathways. PLoS Genet. 2020;16(4):e1008592. doi: 10.1371/journal.pgen.1008592 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Tanaka Y, Kamohara H, Kinoshita K, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119(6):1159–1167. doi: 10.1002/cncr.27895 [DOI] [PubMed] [Google Scholar]
- 68.Liu S, Lin Z, Zheng Z, et al. Serum exosomal microRNA-766-3p expression is associated with poor prognosis of esophageal squamous cell carcinoma. Cancer Sci. 2020;111(10):3881–3892. doi: 10.1111/cas.14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiu ML, Li X, Lin JB, Luo RG, Liu B, Feng Z. Serum exosomal miR-182 upregulation predicts unfavorable prognosis of esophageal squamous cell carcinoma. Eur Rev Med Pharmacol Sci. 2020;24(10):5412–5418. [DOI] [PubMed] [Google Scholar]
- 70.Liu T, Du LT, Wang YS, et al. Development of a novel serum exosomal MicroRNA nomogram for the preoperative prediction of lymph node metastasis in esophageal squamous cell carcinoma. Front Oncol. 2020;10:573501. doi: 10.3389/fonc.2020.573501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tian L, Yang L, Zheng W, et al. RNA sequencing of exosomes revealed differentially expressed long noncoding RNAs in early-stage esophageal squamous cell carcinoma and benign esophagitis. Epigenomics. 2020;12(6):525–541. doi: 10.2217/epi-2019-0371 [DOI] [PubMed] [Google Scholar]
- 72.Jiao Z, Yu A, Rong W, et al. Five-lncRNA signature in plasma exosomes serves as diagnostic biomarker for esophageal squamous cell carcinoma. Aging (Albany NY). 2020;12(14):15002–15010. doi: 10.18632/aging.103559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu Z, Wang H, Pang Y, Hu H, Zhang H, Wang W. Exosomal long non-coding RNA UCA1 functions as growth inhibitor in esophageal cancer. Aging (Albany NY). 2020;12(20):20523–20539. doi: 10.18632/aging.103911 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Yan S, Du L, Jiang X, et al. Evaluation of serum exosomal lncRNAs as diagnostic and prognostic biomarkers for esophageal squamous cell carcinoma. Cancer Manag Res. 2020;12:9753–9763. doi: 10.2147/CMAR.S250971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pan Y, Lin Y, Mi C. Cisplatin-resistant osteosarcoma cell-derived exosomes confer cisplatin resistance to recipient cells in an exosomal circ_103801-dependent manner. Cell Biol Int. 2020;45(4):858–68. doi: 10.1002/cbin.11532 [DOI] [PubMed] [Google Scholar]
- 76.Fan L, Cao Q, Liu J, Zhang J, Circular LB. RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol Cancer. 2019;18(1):16. doi: 10.1186/s12943-018-0936-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu S, Lin Z, Rao W, et al. Upregulated expression of serum exosomal hsa_circ_0026611 is associated with lymph node metastasis and poor prognosis of esophageal squamous cell carcinoma. J Cancer. 2021;12(3):918–926. doi: 10.7150/jca.50548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ni PZ, He JZ, Wu ZY, et al. Overexpression of stathmin 1 correlates with poor prognosis and promotes cell migration and proliferation in oesophageal squamous cell carcinoma. Oncol Rep. 2017;38(6):3608–3618. [DOI] [PubMed] [Google Scholar]
- 79.Yan L, Dong X, Gao J, et al. A novel rapid quantitative method reveals stathmin-1 as a promising marker for esophageal squamous cell carcinoma. Cancer Med. 2018;7(5):1802–1813. doi: 10.1002/cam4.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nonaka T, Wong DTW. Saliva-exosomics in cancer: molecular characterization of cancer-derived exosomes in saliva. Enzymes. 2017;42:125–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lau C, Kim Y, Chia D, et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem. 2013;288(37):26888–26897. doi: 10.1074/jbc.M113.452458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin Y, Dong H, Deng W, et al. Evaluation of salivary exosomal chimeric GOLM1-NAA35 RNA as a potential biomarker in esophageal carcinoma. Clin Cancer Res. 2019;25(10):3035–3045. doi: 10.1158/1078-0432.CCR-18-3169 [DOI] [PubMed] [Google Scholar]
- 83.Melo SA, Sugimoto H, O’Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Okuda Y, Shimura T, Iwasaki H, et al. Serum exosomal dicer is a useful biomarker for early detection of differentiated gastric adenocarcinoma. Digestion. 2020;102(4):640–649. doi: 10.1159/000510993 [DOI] [PubMed] [Google Scholar]
- 85.Schindler C, Collinson A, Matthews C, et al. Exosomal delivery of doxorubicin enables rapid cell entry and enhanced in vitro potency. PLoS One. 2019;14(3):e0214545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saari H, Lázaro-Ibáñez E, Viitala T, Vuorimaa-Laukkanen E, Siljander P, Yliperttula M. Microvesicle- and exosome-mediated drug delivery enhances the cytotoxicity of paclitaxel in autologous prostate cancer cells. J Control Release. 2015;220(Pt B):727–737. doi: 10.1016/j.jconrel.2015.09.031 [DOI] [PubMed] [Google Scholar]
- 87.Kim MS, Haney MJ, Zhao Y, et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2016;12(3):655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.He Z, Li W, Zheng T, Liu D, Zhao S. Human umbilical cord mesenchymal stem cells-derived exosomes deliver microRNA-375 to downregulate ENAH and thus retard esophageal squamous cell carcinoma progression. J Exp Clin Cancer Res. 2020;39(1):140. doi: 10.1186/s13046-020-01631-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li L, Zhang C, Li X, Lu S, Zhou Y. The candidate tumor suppressor gene ECRG4 inhibits cancer cells migration and invasion in esophageal carcinoma. J Exp Clin Cancer Res. 2010;29(1):133. doi: 10.1186/1756-9966-29-133 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 90.Mao L, Li X, Gong S, et al. Serum exosomes contain ECRG4 mRNA that suppresses tumor growth via inhibition of genes involved in inflammation, cell proliferation, and angiogenesis. Cancer Gene Ther. 2018;25(9–10):248–259. doi: 10.1038/s41417-018-0032-3 [DOI] [PubMed] [Google Scholar]
- 91.Dai S, Wei D, Wu Z, et al. Phase I clinical trial of autologous ascites-derived exosomes combined with GM-CSF for colorectal cancer. Mol Ther. 2008;16(4):782–790. doi: 10.1038/mt.2008.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.André F, Chaput N, Schartz NE, et al. Exosomes as potent cell-free peptide-based vaccine. I. Dendritic cell-derived exosomes transfer functional MHC class I/peptide complexes to dendritic cells. J Immunol. 2004;172(4):2126–2136. doi: 10.4049/jimmunol.172.4.2126 [DOI] [PubMed] [Google Scholar]
- 93.Morse MA, Garst J, Osada T, et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med. 2005;3(1):9. doi: 10.1186/1479-5876-3-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Narita M, Kanda T, Abe T, et al. Immune responses in patients with esophageal cancer treated with SART1 peptide-pulsed dendritic cell vaccine. Int J Oncol. 2015;46(4):1699–1709. doi: 10.3892/ijo.2015.2846 [DOI] [PubMed] [Google Scholar]
- 95.Cerella C, Grandjenette C, Dicato M, Diederich M. Roles of apoptosis and cellular senescence in cancer and aging. Curr Drug Targets. 2016;17(4):405–415. doi: 10.2174/1389450116666150202155915 [DOI] [PubMed] [Google Scholar]
- 96.Yuan L, Alexander PB, Wang XF. Cellular senescence: from anti-cancer weapon to anti-aging target. Sci China Life Sci. 2020;63(3):332–342. doi: 10.1007/s11427-019-1629-6 [DOI] [PubMed] [Google Scholar]
- 97.Zheng K, Ma J, Wang Y, He Z, Deng K. Sulforaphane inhibits autophagy and induces exosome-mediated paracrine senescence via regulating mTOR/TFE3. Mol Nutr Food Res. 2020;64(14):e1901231. doi: 10.1002/mnfr.201901231 [DOI] [PubMed] [Google Scholar]