Abstract
Extracellular vesicles (EVs), actively shed from a variety of neoplastic and host cells, are abundant in blood, and carry molecular markers from parental cells. For these reasons, EVs have gained much interest as biomarkers of disease. Among a number of different analytical methods that have been developed, surface plasmon resonance (SPR) stands out as one of the ideal techniques given its sensitivity, robustness, and ability to miniaturize. In this review, we compare different SPR platforms for EV analysis, including conventional SPR, nanoplasmonic sensors, surface-enhanced Raman spectroscopy, and plasmonic-enhanced fluorescence. We discuss different surface chemistries used to capture targeted EVs and molecularly profile their proteins and RNAs. We also highlight these plasmonic platforms’ clinical applications, including cancers, neurodegenerative diseases, and cardiovascular diseases. Finally, we discuss the future perspective of plasmonic sensing for EVs and their potentials for commercialization and clinical translation.
Keywords: surface plasmon resonance, nanoplasmonics, surface-enhanced Raman spectroscopy, plasmon-enhanced fluorescence, extracellular vesicles, biosensing, diagnostics, biomarkers
Graphical Abstract
“Extracellular vesicles” (EVs) is a catch-all term applied to various vesicles produced by host and neoplastic cells. EVs consist of molecularly and physically defined structures, including exosomes, microvesicles, apoptotic bodies, and ectosomes.1 While these vesicles are formed through biological processes, they have commonalities and can broadly serve as diagnostics biomarkers. For example, one recent study found that both exosomes and microvesicles carried similar diagnostic information.2 Today EV analysis has been used for the diagnosis of cancers,3-6 cardiovascular,7,8 neurodegenerative,9,10 and infectious diseases11,12 with hundreds of publications emerging.13 One attractive feature of EV analysis is that the vesicles are generally abundant, very stable, and can be obtained from any biofluid, particularly from blood. The current biggest challenge with EV analysis is the realization that EV subtypes (e.g., exosomes, microvesicles) are molecularly heterogeneous. For example, not all exosomes produced by normal cells contain the “ubiquitous” CD63, CD9, or CD81 markers. Conversely, only a very small fraction of exosomes shed from cancer cells will exhibit truly distinctive cancer markers that can be identified as such. For these reasons, future analytical techniques are desired to have a single EV or near-single EV resolution to detect rare subtypes. Bulk analysis (e.g., Western Blotting, enzyme-linked immunosorbent assay/ELISA) could have limited specificity since many cancer markers are also expressed by other tissues. Techniques that allow the analysis of individual vesicles or a combination of markers on individual EVs are likely going to add significant clinical value. Another challenge is what biomarker type to analyze: protein, miRNA, mRNA, and/or others. Analyzing bulk EVs from a certain cell type will carry most of the transcriptome from those cells (mRNA, miRNA, lncRNA, rRNA, tRNA, etc.); however, single EV analysis also adds some challenges. Single EVs are limited in what it can carry due to their small size. We know that a 100 nm retrovirus, in many ways, mimics an EV that can only package around 10 kb RNA. With this packaging constrain, each EV is only able to carry individual RNA transcripts14 and that mutant proteins are uncommon.2,15 Irrespective of the approach and type of amplification methods used, next generations of biosensors that leverage nanomaterials and microfluidic systems are needed.
Surface plasmon resonance (SPR) sensors have been widely used for detecting analytes and characterizing molecular interactions (e.g., antibody-antigens, proteins and small molecules.16,17 These sensors detect local refractive index changes induced by binding of target substances to a sensing surface, resulting in a shift of optical resonance. This mechanism allows for the label-free detection of target molecules captured by ligands immobilized on the sensor surface. Furthermore, SPR sensors have narrow sensing ranges from 10 - 300 nm from the surface.18-20 The size of the majority EVs, such as exosomes (50 - 200 nm), is well-covered by the evanescent field of surface plasmons (SPs), promoting a harmonious marriage of plasmonic sensing and EV analysis. SPR sensing has several advantages over radioactive and fluorescent labeling methods, namely (i) label-free and real-time investigation, (ii) kinetics and affinity measurement and (ii) low cost and less reagent use.21,22
In this article, we provide an overview of various SPR platforms and their latest technological development, including the surface chemistry used for EV capture and detection. Notably, we discuss how recent advances have bridged the gap from promising engineering platforms to practical tools positioned for translational investigation. To this end, we also present clinical applications of SPR platforms in EV analysis and discuss how SPR could favorably impact breakthrough pre-clinical and translational medical research and commercialization efforts.
OVERVIEW OF SPR PLATFORMS
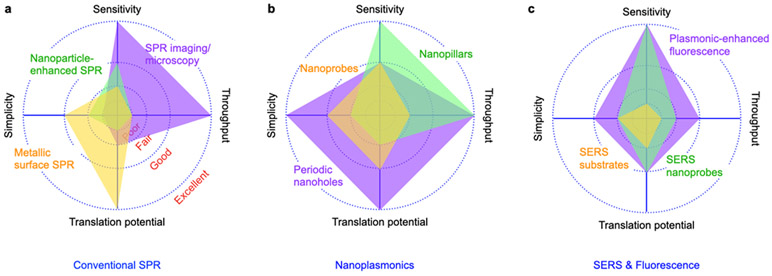
SPR platforms here we discuss include conventional SPR, nanoplasmonic sensors, surface-enhanced Raman scattering (SERS), and plasmonic-enhanced fluorescence (PEF). Comparative summaries of various SPR platforms developed for EV detection are shown in Figure 1 and Table 1.23-57 We compared the platforms in terms of sensitivity, throughput, simplicity, and translational potential exclusively for EV detection. Sensitivity is defined as the demonstrated limit of detection, ranging from >106 (Poor), 103 - 106 (Fair), 10 - 103 (Good), and single EV (Excellent). Throughput shows the demonstrated multiplexing capability, ranging from a single target (Poor), ~10 (Fair), ~100 (Good), and >1,000 targets (Excellent). Subsequently, simplicity is rated qualitatively based on the complexities of the detection system and chip fabrication. The translational potential is rated qualitatively based on demonstrated clinical validation and results with clinical samples.
Figure 1. Comparison of SPR platforms.
a Conventional SPR. b Nanoplasmonics. c SERS and PEF. System performances are based on demonstrated platforms for EV detection, as summarized in Table 1. Definition of sensitivity indicator: Poor (>106), Fair (~106), Good (~103), Excellent (single exosome). Definition of throughput (demonstrated multiplexing capability) indicator: Poor (single), Fair (~10), Good (~100), Excellent (>1000 arrays). Simplicity is rated qualitatively based on the complexities in the detection system and chip fabrication. Translational potential is rated qualitatively based on demonstrated clinical validation and results with clinical samples.
Table 1.
Summary of recent demonstrated plasmonic platforms with specifications.
| Platform | Source/detector | Target | Parameter | Sample volume |
LOD | Multiplex | Remarks | Ref | |
|---|---|---|---|---|---|---|---|---|---|
| Surface plasmon resonance | Conventional SPR | Commercial system | Exosome | SPR response unit | - | 10 - 100 pM | N | Au nanoparticle/exosome clustering | 23 |
| 20 μL | - | Y | 24 | ||||||
| Solid state laser/photodetector | Intensity | 50 μL | 2 × 1010 exosomes/mL | N | Compact SPR system | 25 | |||
| White light/spectrometer | Spectrum | - | 2.07 × 106 exosomes/mL | N | 26 | ||||
| Au nanoparticle amplification | Commercial system | Angle | - | 5 × 103 exosomes/mL | N | Low concentration EV detection Complex operation steps | 27 | ||
| Angle | - | 5.6 × 105 exosomes/mL | N | Complex operation steps | 28 | ||||
| Surface plasmon resonance imaging | SPR imaging | Commercial system | Exosome | SPR response unit | 500 μL | 1 μg/mL | Y | Sandwich assay | 29 |
| SPR imaging | 650-nm LED/ CCD camera | Exosome | SPR response unit | 130 μL | 2.4 × 107 exosomes/mL | Y | Amplification with antibody functionalized Au nanoparticles | 30 | |
| SPR microscopy | Solid state laser/CMOS camera | EV | Image | 10 μL | 1 exosome | N | Size distribution, concentration, affinity constant measurements | 31 | |
| Nanoplasmonics | Periodic nanoholes | Laser diode/imaging sensor | Exosome | Intensity | 0.3 μL | 3,000 exosomes | Y | Rapid (<30 min) High throughput (33 × 33 array) |
32 |
| White light/spectrometer | Exosome lysates | Spectrum | 0.5 μL | 104 exosomes | Y | High throughput (10 × 10 array) | 33 | ||
| Exosome | Spectrum | - | 200 exosomes | Y | 34 | ||||
| 3D photonics crystal | White light/spectrometer | Exosome | Spectrum | 10 μL | 100 exosomes | N | Complex fabrication processes | 35 | |
| Nanoislands | EV | Phase | 100 μL | 0.194 μg/mL | N | Complex microscopy system | 36 | ||
| Nanoellipsoids | Exosome | Spectrum | 50 μL | 1 ng/mL | N | 37 | |||
| Nanoshells | Pulsed laser/proteomics analyzer | Exosome | Mass spectrum | 0.5 μL | - | N | Laser desorption/ionization mass spectrometry | 38 | |
| Nanopillars | White light/ CCD spectrophotometer | Exosome | Intensity | - | 1 exosome | Y | High throughput (16 array, 400 nanopillars each) | 39 | |
| Interferometer | LED/ CCD | Exosome | Intensity | - | 3.86 × 108 exosomes/mL | N | 40 | ||
| Nanoprobes | Dark field/ digital camera | EV | Scattering intensity | 1 μL | 0.2 μg/mL | N | 41 | ||
| Nanoprisms | White light/spectrometer | Exosomal microRNA | Spectrum | 100 μL | 1 aM | N | 42 | ||
| Surface-enhanced Raman spectroscopy (Substrate) | Au nanoparticles | Raman laser/spectrometer | Exosome | Raman spectrum | 5 μL | - | N | Study on Raman peaks | 43 |
| Ag nanocubes on Au nanorods | 5 μL | - | N | 44 | |||||
| Ag coated CD/DVD | 5 μL | - | N | 45 | |||||
| Ag nanobowls | 20 μL | - | N | 46 | |||||
| Beehive macroporous | 50 μL | - | N | 47 | |||||
| Au nanopillars | Exosomal microRNA | 200 μL | 1 aM | N | 48 | ||||
| Surface-enhanced Raman spectroscopy (Probe with Raman reporter) | Au nanoparticles | Raman laser/spectrometer | Exosome | Raman intensity | 2 μL | 3.2 × 104 exosomes/mL | Y | Magnetic beads as capture beads | 49 |
| Ag coated Au nanoparticles | 4 μL | 1,000 exosomes/mL | N | 50 | |||||
| Au nanotrepangs | 2 μL | 2.6 × 104 exosomes/mL | Y | 51 | |||||
| Au nanoparticles | 2 μL | 500 exosomes/mL | N | Functionalized PDA substrate | 52 | ||||
| Au nanostars | 200 μL | 2.7 × 104 exosomes/mL | N | 53 | |||||
| Au nanoparticles | Raman laser/spectrometer and EMCCD | EV | 100 μL | 105 exosomes/mL | Y | Nanoscopic flow with alternating potential difference | 54 | ||
| Ag coated Au nanoparticles | Raman laser/spectrometer | Exosomal microRNA | - | 1 aM | N | Magnetic beads as capture beads | 55 | ||
| Plasmonic enhanced fluorescence detection | Diffraction grating | He-Ne laser/photodetector | EV | Fluorescence intensity | 50 μL | 3 × 10−4 RIU | N | 56 | |
| Periodic nanoholes | Fluorescence microscope | Exosome | 30 μL | Single exosome | Y | Molecular profiling based on single EV fluorescence image | 57 | ||
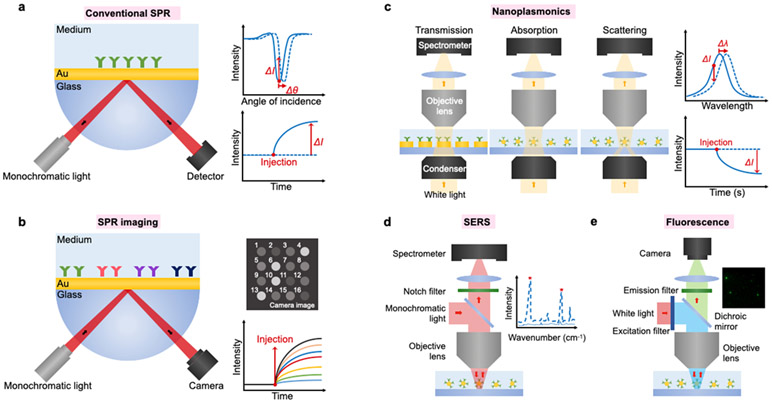
The experimental setups of these plasmonic platforms are presented in Figure 2. Conventional SPR sensors frequently employ attenuated total reflection coupler, also known as Kretschmann-Raether configuration, whereby SPs are induced by impinging obliquely incident light to a prism coated with a metallic film58 (Figure 2a). The reflectance SPR curve (reflectance vs. incident angle) has two distinctive characteristics: (i) The resonance condition is satisfied at an incidence angle with minimum reflectivity; (ii) the resonance angle shifts to a higher angle by the increase of refractive index (i.e., molecular binding). Conventional SPR measures changes in the SPR angle or intensity upon target molecular binding to the sensor surface. Conventional SPR technology is one of the most commonly used plasmonic biosensors and is employed in most commercially available SPR systems (e.g., Biacore by GE Healthcare, BioNavis). For EV detection, the conventional SPR systems measure real-time binding kinetics, i.e., equilibrium dissociation constant and association/dissociation rates, to search for promising antibodies24,26,59,60 and aptamers23,61-63 for EVs, quantify EV’s size and concentrations,64,65 and detect cancer-derived EVs.24,26,27
Figure 2. Representative equipment setups of different plasmonic platforms.
a Conventional SPR system using a photodetector to monitor a change in the intensity or resonance angle. b SPR imaging and microscopy using a camera to monitor intensity change in image. c Nanoplasmonic systems using optical transmission configuration to monitor intensity or wavelength change in absorption or scattering spectrum. d SERS system using monochromatic light for excitation and spectrometer to measure a Raman spectrum. e Plasmonic-enhanced fluorescence system using typical fluorescence microscopy for single EV fluorescence imaging.
However, the conventional SPR platforms perform bulk measurements with the limit of detection in the order of 106 - 107 EVs/mL.26,60 Metallic nanoparticles can be employed to enhance SPR signals,27,28 pushing the LOD down to 5 × 103 EVs/mL.27 They also have limited throughput often defined by the number of microfluidic channels in the system. To enable high-throughput, parallel sensing, SPR imaging platforms have been adapted in which spectral shifts from multiple sensing arrays can be detected simultaneously by monitoring intensity changes with a camera29-31,66 (Figure 2b). With SPR imaging, real-time binding kinetics of EVs in different sensing spots can be monitored, and high-throughput analysis in a microarray format was demonstrated. However, the total internal reflection system requires optical components such as a prism to couple an angled light into the metallic film plane, imposing the requirement of precise optical alignment and difficulties in miniaturization. Furthermore, the tuning of the sensing range is limited, motivating the development of nanoplasmonic platforms.
Nanoplasmonic platforms that use metallic nanostructures and/or nanoparticles to generate localized SPR have been introduced for EV sensing.32-42 In nanoplasmonic sensors, because their sensing range is tunable to match with EVs’ sizes, a higher EV detection sensitivity can be achieved by optimizing nanostructures and nanoparticles. Nanoplasmonic platforms based on periodic nanoholes32,34 or photonic crystals35 have demonstrated LODs of < 3,000 exosomes, which is much improved from the conventional SPR platforms, using a simpler optical configuration based on transmission, absorption, or scattering (Figure 2c). However, the fabrication of nanostructures often involves state-of-the-art nanofabrication technologies that increase the overall chip costs. Small variations in the nanostructure dimensions could also affect the optical resonance conditions and detection sensitivity, which is critical to achieving the excellent robustness and reproducibility for clinical translation.
Raman spectroscopy is a powerful technique used to determine molecular vibrational modes, providing information on chemical structure fingerprints. Generally, a laser beam illuminates a sample, and a monochromator detects inelastic photon scattering out of a sample (Figure 2d). Although Raman scattering signals from biological samples are very weak, it can be significantly amplified by metallic nanostructures with an enhancement factor >109. Such a technique, termed as surface-enhanced Raman spectroscopy (SERS), has demonstrated a detection sensitivity down to single molecules. By measuring different types of purified EVs, their distinctive SERS spectra can be used for cancer diagnosis and monitoring.43-48 However, SERS spectra are not as specific as ligand-based approaches in EV molecular profiling. For better specificity, Raman reporters are often conjugated metallic nanoparticles coated with affinity ligands (e.g., antibodies, aptamers) as detection probes.49,50,50-54 Raman reporters could be superior for high-dimensional multiplexing compared to fluorescence probes because of its narrower Raman signals and broad choices in Raman probes. These features make SERS suitable for multiplexed analysis without spectral overlapping, a common problem in fluorescence probes that limit its multiplexing capacity.
A fluorescence-based assay is one of the most popular detection methods in EV biosensing. Single EV fluorescence imaging is one of the promising techniques to understand the heterogeneity in EVs because the molecular properties of individual EVs can be acquired together with the number of EVs.2,15 Due to EVs’ small sizes below the diffraction limit, however, sensitive and robust fluorescence EV detection remains challenging, especially for multiplexed analysis and low-abundant marker detection. Weak fluorescence signals and poor signal-to-noise ratios from EVs often require sophisticated, multi-step amplification strategies to amplify fluorescence signals. It is well-known that plasmonic nanostructures can significantly amplify fluorescence signals over 100-folds especially when a resonance peak overlaps with fluorophore’s spectral profiles.67,68 Plasmon-enhanced fluorescence detection in the visible and near-infrared ranges have been demonstrated for various biosensing applications, targeting for DNAs, proteins, antibodies, cells, among others.69-74 Furthermore, plasmon-enhanced fluorescence platforms can be readily built on conventional fluorescence microscope systems (Figure 2e). A recent work demonstrated that the plasmon-enhanced fluorescence technique could be applied to EV detection with improved sensitivity compared to conventional fluorescence imaging.57 However, plasmonic resonances usually can only enhance fluorescence in the wavelengths that overlap with plasmonic resonances, limiting the level of multiplexing in EV molecular profiling.
CONVENTIONAL SPR SENSORS
In conventional SPR sensors, capturing EVs on a metal surface increases an effective refractive index, resulting in a shift of resonance angle. Early work focused on quantitative analysis of EV sizes and concentrations with appropriate calibration processes.64,65 While EV sizes and concentrations are important parameters, these tasks can be done by other approaches, such as dynamic light scattering and nanoparticle tracking analysis. Rather, the ability of the SPR sensors in measuring real-time binding kinetics could enable the quantification of the average amount of target proteins in EVs through multivalent binding analysis.75 To further facilitate the use of the conventional SPR systems for EV sensing, SPR approaches have been adapted to improve the detection sensitivity by using metallic nanoparticles and achieve higher throughput via imaging and microscopy.
Nanoparticle-enhanced SPR detection
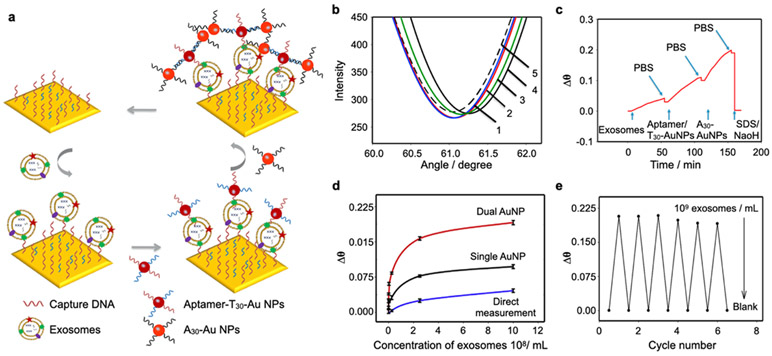
To improve the sensitivity of SPR sensors, signal amplification strategies using metallic nanoparticles as additional labeling have been suggested.76-78 Secondary labeling of captured EVs with metallic nanoparticles achieved larger shifts in SPR angle or changes in reflective intensity.79 Wang et al. amplified SPR signals by 4-orders of magnitudes through multiple amplification steps using two types of DNA-conjugated gold (Au) nanoparticle (Figures 3a-c).27 EVs were captured with DNA-based anti-CD63 aptamers, followed by consecutive injection of anti-CD63 aptamer/T30 (5′-SH-T30-3′) and A30 (5′-SH-A30-3′)-conjugated Au nanoparticles, which led to chains of AuNPs on the captured EVs. The SPR signal was significantly amplified through the dual Au nanoparticle conjugation approach (Figure 3d). Using this method, they achieved the LOD of 5 x 103 EVs/mL. Furthermore, the sensor chip could be reused after regeneration with 0.1% SDS/10 mM NaOH solution, maintaining the sensitivity for six cycles (Figure 3e). The same group recently introduced another nanoparticle-enhanced signal amplification strategy using polydopamine (PDA)-functionalized Au nanoparticle.28 PDA allows the reduction of HAuCl4, forming small Au nanoparticles on the surface of the PDA-functionalized Au nanoparticles. While the approach with PDA-functionalized Au nanoparticles is simpler than the previous approach based on DNA hybridization, the measured LOD of 5.6 × 105 EVs/mL was 100-times lower. In overall, the nanoparticle-enhanced signal amplification strategy could improve the detection sensitivity, but the additional steps increase the complexity of the assay, dimming the label-free detection capability of the SPR sensors.
Figure 3. Nanoparticle enhanced SPR platforms.
a Dual Au nanoparticle-assisted signal amplification for exosome detection. b SPR spectra of (1) aptamer modified Au film; (2) target exosomes; (3) reaction with T30 AuNPs; (4) reaction with A30 AuNPs; (5) regeneration. c SPR response of dual AuNP-assisted SPR sensor. d The relationship between Δθ and exosomes concentrations by using different sensing strategies: direct measurement, single AuNP amplified SPR aptasensor, and dual AuNP amplified SPR aptasensor. e Reproducibility investigation of the SPR biosensor. Reprinted with permission from ref. 27. Copyright 2019 Elsevier.
SPR imaging
SPR imaging was demonstrated by Rothenhäuslar and Knoll in 1988, where a photodetector was replaced with a camera to obtain a two-dimensional intensity map of refractive index distribution.80 SPR imaging has the capability of parallel measurement.81 Multiple capture ligands can be printed in an array for parallel detection by measuring intensity change at each spot through simultaneous acquisition of SPR sensorgrams.82,83 The number of sensing spots in a field-of-view of a camera defines the number of parallel measurements. Microfluidics channels or a microarray printer are typically used to generate an array of measurement spots with different ligands for high-throughput multiplexed detection with economical use of reagents.
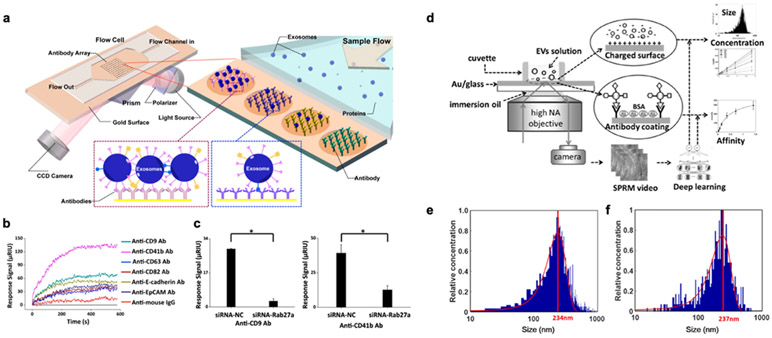
SPR imaging was applied to analyze exosomes from human hepatocellular carcinoma cell lines (MHCC97L, MHCC97H) and mouse melanoma cell lines (B16-F1, B16-F10).66 Antibodies against well-known exosome markers (CD9, CD63, CD81), cancer markers (CD41b, EpCAM, E-cadherin), and IgG as negative control were printed on the Au surface (Figure 4a). All channels were simultaneously measured after exosome incubation. In this experiment, anti-CD41b and anti-CD9 channels showed the high signals (Figure 4b), which means either there are more CD41b- or CD9-positive exosomes present or the levels of the markers are higher than others in a given sample. Subsequently, the signal levels of CD9 and CD41b were measured after transfecting the cells with siRNA-Rab27a to knock down the expression of Rab27a (a gene in the exosome secretion pathway). With the siRNA treatment, CD9 and CD41b signals were significantly reduced compared to a control group using negative control siRNA (Figure 4c). They showed there is a positive correlation between metastatic potential and EV secretion, implying that liquid biopsy-based on EV analysis can be used for clinical testing on the detection of cancer metastasis. Although SPR imaging has inherent multiplexing capability, the noise originated from the intensity fluctuation of a light source results in 1-2 orders worse sensitivity than conventional SPR based on angle interrogation.84
Figure 4. SPR imaging and microscopy.
a SPR imaging in combination with antibody microarray to capture and detect exosomes. b SPRi sensorgrams showing binding of exosomes to various antibodies. c Exosome secretion was suppressed by siRNA-Rab27a (siRNA-NC as a negative control). Reprinted with permission from ref. 66. Copyright 2014 American Chemical Society. d EVs detection with SPR microscopy with a high NA objective. A deep learning algorithm was developed for automatic SPRM image analysis. e-f Size measurement of EVs with SPRM (e) and nanoparticle tracking analysis (f). Mean values were located at 234 and 237 nm, respectively. Reprinted with permission from ref. 31. Copyright 2020 American Chemical Society.
SPR microscopy
SPR imaging fundamentally suffers from poor image resolution due to micrometer scale SP propagation length.85 Wang et al. proposed a single particle imaging apparatus for influenza virus detection where a high numerical objective was used for angled light illumination.86 Approaching a nanoparticle to an Au surface induces light scattering, and reflected light at the Au surface interferes with the scattering light. The interference between scattering and reflected light produces V-shaped patterns in an image. The intensity variation quantifies the approach of targeting nanoparticles.87
More recently, an EV analysis system based on SPR microscopy (SPRM) was introduced for the measurements of EV size, concentration, and binding affinity.31,88 EVs were captured on an Au-coated glass substrate by electrostatic interaction between negatively charged EVs and an amine-modified Au surface (Figure 4d). For size calibration, scattering patterns from silica nanoparticles with size ranging from 30 nm to 1 μm were measured. The EV scattering intensities were then transformed into EV sizes using a deep learning algorithm. They used 5,000 scattering images of silica nanoparticle as training sets in different imaging conditions. The EV detection accuracy was 85%, and the size measurements of A549 cell-derived EVs were well-matched with those measured by nanoparticle tracking analysis (Figure 4e-f). SPRM is a hybrid concept between single-particle imaging and SPR sensing. It was possible to measure specific antibody-EV binding at a concentration of 0.04 nM and in a quantized integer value. They showed that EV capture by anti-CD63 antibody reaches equilibrium within 10 minutes with a measured equilibrium dissociation constant of ~0.79 ± 0.12 nM. SPR microscopy provides single EV resolution sensing, but its clinical potential is relatively low due to the difficulty in parallel detection aroused from its small field-of-view and the complexity in the experimental setup (e.g., high NA objective lens). However, its real-time and multifunctional capabilities might be useful in research settings toward prospective biophysical applications.
NANOPLASMONICS
To further enhance the SPR signal and detection sensitivity, various metallic nanostructures and nanoprobes have been developed. Similar to conventional SPR sensors, EV binding to nanostructures or nanoprobes increases the effective refractive index on the metal surface, resulting in a resonance wavelength shift. Such shifts can be detected in light transmission, absorption, or scattering using a photodetector, imaging sensor, or spectrometer.
Periodic nanoholes
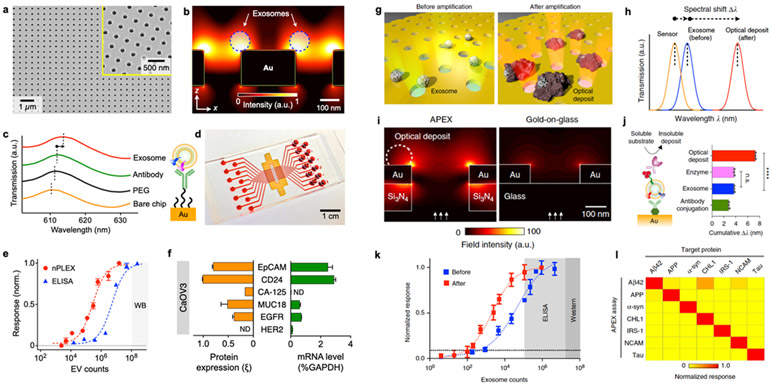
One of the demonstrated nanoplasmonic platforms for exosome detection is the nanoplasmonic exosome (nPLEX) technology (Figure 5a), which consisted of periodic nanohole arrays as sensing elements.32,33,89 The nanohole structure greatly confines the electromagnetic fields at the nanohole surface. It enhances the evanescence field within the exosome size range, leading to improved detection sensitivity (Figure 5b).32 For instance, the nanoholes had a 200 nm diameter with a 200-nm-thick Au film on a glass substrate and a probing depth <200 nm that matches the typical size of exosomes. Exosome binding to the nanohole surface led to a shift in the plasmonic resonance peak. The nPLEX platform operates in a transmission mode whereby the wavelength shift can be detected by a spectrometer or through intensity change using a complementary metal-oxide-semiconductor (CMOS) imaging sensor (Figure 5c). The nPLEX array had 36 sensing units for parallel and multiplexed detection of 12 potential exosomal markers in triplicate with a sample volume of 0.3 μL per marker (Figure 5d). nPLEX demonstrated a fast assay time of <30 min and a LOD of ~3,000 exosomes (670 aM). The sensitivity of the nPLEX platform was 100-fold higher than the chemiluminescence ELISA (Figure 5e), and the peak shift signal can be further enhanced by 20% and 300% through secondary labeling of spherical or star-shaped Au nanoparticles, respectively. In addition to exosome detection by targeting transmembrane proteins, the nPLEX platform can be used for direct exosomal protein detection (intravesicular and transmembrane proteins) from exosome lysates.33 Captured exosomes can also be eluted from the nPLEX chip for down-stream analysis, such as mRNA profiling (Figure 5f). Using interference lithography, the nPLEX chip can be scaled up for higher throughput in clinical applications. For instance, next-generation nPLEX chips with 100 sensing spots were developed for high-throughput clinical diagnosis of pancreatic malignancy through multi-marker EV analysis.89
Figure 5. Nanoplasmonics using periodic nanoholes.
a A scanning electron micrograph of the periodic nanoholes in the nPLEX sensor. b Finite-difference time-domain (FDTD) simulation shows the enhanced electromagnetic fields tightly confined near a periodic nanohole surface. c A representative schematic of changes in transmission spectra showing exosome detection with nPLEX, and scanning electron microscopy shows exosome captured by functionalized nPLEX. d Prototype with 12 × 3 detection spots. The nanohole arrays in the shaded area were integrated with microfluidics. e Comparison of the detection sensitivity of nPLEX and ELISA. f mRNA analysis of exosomes eluted from CaOV3 cells. Following nPLEX protein measurements, captured exosomes were released from the chip and subsequently analyzed for mRNA contents. Reprinted with permission from ref. 32. Copyright 2014 Springer Nature. g APEX assay for exosome profiling through an in situ enzymatic amplification. h Schematic of changes in the transmission spectra with APEX amplification. i FDTD simulations with back illumination. j Step-by-step APEX transmission spectral changes. The deposit formation led to ~400% signal enhancement. k Comparison of the detection sensitivities of APEX, ELISA, and western blotting. l specificity of APEX assays for measuring target proteins. Reprinted with permission from ref. 34. Copyright 2019 Springer Nature. Creative Common CC-BY.
Another nanohole-based nanoplasmonic platform, termed amplified plasmonic exosome (APEX) assay, was demonstrated using in situ enzymatic amplification to grow insoluble optical deposits over exosomes bound nanoholes34 (Figure 5g). The insoluble optical deposits increase the effective refractive index on the Au surface, leading to a larger wavelength shift of resonant peak (Figure 5h). To improve analytical stability for enzymatic amplification, direct illumination on the exosome-bound Au nanoholes should be avoided. Therefore, the APEX chip consists of periodic Au nanoholes that are suspended on a patterned silicon nitride (Si3N4) membrane. In contrast to the nPLEX chip’s Au-on-glass design that supports front illumination (from Au side), such a double-layered nanostructure facilitates back illumination (from Si3N4 side) to reduce temperature fluctuation (Figure 5i). With the enzymatic amplification, the detection sensitivity was enhanced by ~400% with a LOD of ~200 exosomes (Figure 5j-k) while maintaining good specificity (Figure 5l).
Zhu et al. have explored nanohole structures’ shape and depth to maximize the sensitivity.35 Asymmetrical quasi-3D plasmonic nanoholes showed improved sensitivity from 483 nm/RIU to 946 nm/RIU compared to the symmetrical one and narrowed the spectral line shape. This was because the asymmetrical structures further enhanced electromagnetic field intensity near the corners and generated additional quadrupole plasmon mode. To further increase the sensitivity, 3D plasmonic photonic crystal nanostructures were developed by introducing additional Au nanodots onto the quasi-3D Au nanoholes, which created a Fabry-Pérot cavity that couples with the plasmonic modes. The detection sensitivity was improved to 1,376 nm/RIU with a LOD of ~100 exosomes.
Other Nanostructures
Other explored nanoplasmonic structures include nanoellipsoids,37 nanoshells,38 ring-hole interferometer,40 among others. The simplest nanostructure was a self-assembled monolayer of Au nanoislands formed by depositing a thin layer of Au film on a glass slide and annealing at 550 °C.90 Au nanoislands have an average diameter of 40 nm and are partially embedded into the substrate for about 8 nm. EV capture on the nanoislands was detected by measuring differential phases through a common-path spectral interferometric system via a spectrometer. A LOD of 0.194 μg/ml was demonstrated.36 Larger Au nanoislands (~200 nm) can be made by shortening the annealing time to obtain a longer plasmonic penetration depth for higher sensitivity.91 A similar nanoplasmonic platform was also demonstrated by forming silver (Ag) nanoislands on a polydimethylsiloxane substrate.92 In this structure, resonance peak shifts were measured in absorption spectra.
To further improve the sensitivity down to single exosome detection, a periodic 90-nm-diameter quartz nanopillar array was fabricated. Au sensing tips were made on the elevated nanopillars (height of ~497 nm) to reduce background noises from non-specific binding.39 A CMOS camera was used to image 16 arrays of 20 × 20 nanopillars under a single field of view while spectra were collected simultaneously using optical fiber and spectrophotometer. Sharp temporal responses were observed by subsampling the image down to individual nanopillars. The digital responses occurred stochastically both in time and space, detecting single exosome binding on the nanopillars.
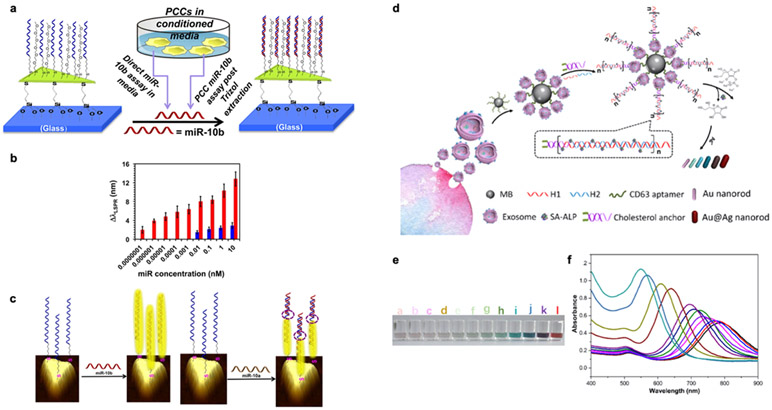
In addition to exosome detection, nanoplasmonic platforms can be easily modified to detect exosomal miRNAs using complementary oligonucleotides (Figure 6). Asymmetrical Au nanoprisms (Figure 6a) were chemically synthesized onto a silanized glass substrate and functionalized with complementary single-stranded DNAs (ssDNAs).42 Direct hybridization between the ssDNAs and target exosomal miRNAs red-shifts a resonance peak. Exosomal miRNA (miR-10b) over-expressed in pancreatic cancer cells was detected with a LOD of 83 aM (Figure 6b), which is more than 106-fold lower than a fluorescence detection. Using ssDNAs as a binding probe, specificity down to a single nucleotide can be achieved (miR-10b vs miR-10a) (Figure 6c). However, the plasmonic signal can be interfered with a high concentration of miR-10a, which forms partial hybridization with the ssDNA probe.
Figure 6. Nanoplasmonics using nanoprisms and nanoprobes.
a Schematic representation of Au nanoprisms for miR-10b detection. b Comparison of miR-10b (red bars) and miR-10a (blue bars) concentration-dependent plasmonic responses. c Schematic representation illustrates electron-transport through duplex DNA hybridization with miR-10b and miR-10a. Reprinted with permission from ref. 42. Copyright 2015 American Chemical Society. d Schematic illustration of multicolor visual detection of exosomes based on HCR and enzyme-catalyzed metallization of Au NRs. e-f Color variation (e) and UV–vis absorption spectra (f) of Au NRs in response to different concentrations of exosomes from 0 to 9 × 103 particles/mL. Reprinted with permission from ref. 94. Copyright 2019 American Chemical Society.
Nanoplasmonic platforms based on periodic nanoholes or nanostructures exploit localized, enhanced SPR to improve the sensitivity in EV detection down to hundreds of EVs. It is relatively difficult to achieve single EV sensitivity due to fabrication variations and uncontrollable EV binding position on the nanoholes. Along with introducing target-specific surface modification, single EV sensitivity can be achieved using more sophisticated nanostructures at the expense of more complex and non-standardized (i.e., slow and expensive) nanofabrication processes.
Nanoprobes
Designing nanoprobes by using metallic nanoparticles is an alternative approach in developing nanoplasmonic sensors. The nanoplasmon-enhanced scattering assay was demonstrated using antibody-bound Au nanospheres and nanorods as nanoprobes.41 Anti-CD81 antibody was conjugated on the silica surface as the capturing antibody of EVs. Subsequently, anti-CD63 Au nanospheres (AuNSs) and anti-CD9 Au nanorods (AuNRs) were added to form AuNS-EV-AuNR complexes, recognizing tumor-derived EVs. The AuNSs and AuNRs produce nanoplasmons that shift the scattering spectrum and can be detected through dark-field imaging. The signal-amplification plasmonic coupling effect of the dual-Au nanoparticles increases the platform’s sensitivity, leading to small volume plasma consumption (1 μL with dilution) and low LOD (0.2 μg/mL), which is ~385-fold lower than ELISA.
Colorimetric nanoplasmonic platforms use metallic nanoparticle aggregation or metallic deposition on nanoparticles to induce large plasmonic resonance shifts that can be distinguished by naked eyes or absorption spectroscopy for quantification.93,94 Non-covalent conjugation of aptamers on Au nanoparticles can prevent Au nanoparticles from aggregation. When exosomes bind to aptamers, the non-specific, weaker binding between aptamers and Au nanoparticles is broken. This causes aggregation of Au nanoparticles, resulting in a blue-shift of the absorption peak. Different aptamers can be used to target different exosomal surface proteins, and the intensity of Au nanoparticles aggregation correlates with the abundance of target proteins.93 To further amplify the signal and improve the detection sensitivity of the colorimetric approach, enzyme-induced Ag deposition on Au nanorods was suggested94 (Figure 6d). The multicolor detection of exosomes using hybridization chain reaction to catalyze Ag deposition on AuNRs is presented in Figure 6e. First, metallic beads coated with aptamers targeting CD63 were used to capture exosomes. Then, cholesterol-labeled DNA probes were integrated into the lipid bilayer of exosomes via hydrophobic interaction. Subsequently, two DNA hairpins labeled with alkaline phosphatase were introduced. In the presence of exosomes, hybridization chain reactions occur, leading to the enhanced alkaline phosphatase loading and boosting the ascorbic acid generation. Ag ions were then reduced by ascorbic acid with Ag deposition on Au nanorods, blue-shifting the longitudinal SPR peak (Figure 6f). Such reaction can be detected by naked eyes (LOD: 9 × 103 exosomes/μL) or more precisely using spectroscopy (LOD: 1.6 × 102 exosomes/μL). Although plasmonic-based nanoprobes offer a potential alternative to widely used fluorescence probes that are suffered from photobleaching, the limited multiplexing for EV molecular profiling hinders the use of such metallic nanoprobes in clinical applications.
SURFACE-ENHANCED RAMAN SPECTROSCOPY (SERS)
SERS platform measures specific chemical bonds that exist in EVs. Typical chemical bonds that can be detected from a Raman spectrum include proteins, lipids, phospholipid, C-C twist proteins, amino acids, and nucleic acids.
SERS substrates
Various SERS substrates have been exploited for EV detection, including super-hydrophobic surfaces decorated with silicon pillars,95 substrates with metallic nanoparticles,43,44,46,96,97 Ag coated compact disks,45 etc. In addition, a graphene layer overlaid onto periodic Au-nanopyramids was demonstrated as a hybrid SERS platform.98 The Au-nanopyramid surface creates enhanced surface plasmonic fields, amplifying Raman signals. On the other hand, the graphene layer provides a biocompatible surface that serves as a built-in gauge for quantitative Raman analysis. A SERS substrate can also be designed using cheap materials such as nata de coco (commercial coconut jelly).99 A bacterial cellulose membrane is created from drying and pressurizing nata de coco. Subsequently, in situ Ag nanoparticle synthesis was performed to form an active SERS layer on the membrane.
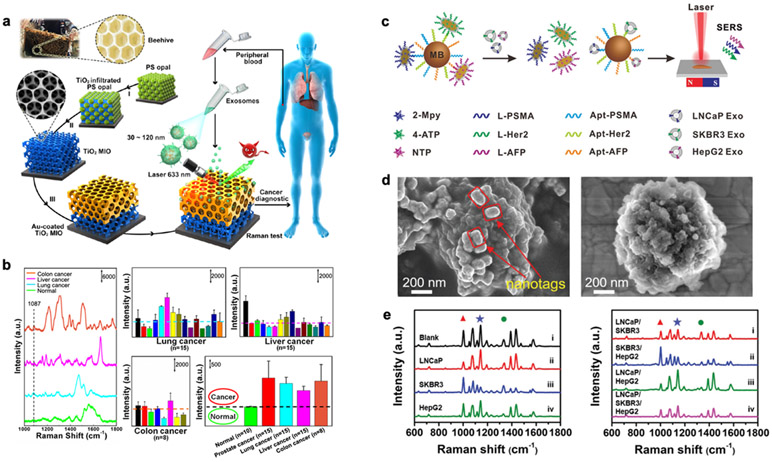
A more sophisticated 3D SERS substrate was demonstrated, inspired by natural beehive structures.47 A titanium dioxide (TiO2) microporous inverse opal (MIO) structure was first fabricated through a polystyrene opal template-based sol-gel method (Figure 7a). Subsequently, a thin layer of Au film was thermally evaporated and coated onto the top layer of the MIO structure. The pore size in MIO was designed to trap the 633-nm laser light (slow light effect) and enhance the Raman signals. The exosome Raman signals are further enhanced by the Au layer. SERS signals of exosomes derived from normal and cancer cells are distinctive. For example, the intensity of SERS peak at 1,087 cm−1 arises from the P-O bond within the phosphoproteins is higher in cancer-derived exosomes than those from healthy people (Figure 7b). Most studies measured the SERS spectra from different populations and applied principal component analysis to identify the major patterns for classification. However, most platforms relied on the non-specific binding of exosomes on SERS substrates. For specificity, surface functionalization to capture target-specific exosomes would be needed.
Figure 7. SERS platform.
a Detection process and design inspiration of the Au-coated TiO2 MIO SERS substrate. b SERS spectra of exosomes separated from plasma of different cancer patients and normal individuals. SERS intensity at 1,087 cm−1 is used for analysis. Reprinted with permission from ref. 47. Copyright 2020 American Chemical Society. c Multiplex exosome detection using SERS nanoprobes. Multiplexed capture probes were constructed by the co-modification of three types of aptamer DNAs. Followed by the recognition between aptamers and target exosomes, the SERS probes are released, and SERS signals are attenuated. d SEM images of the hybridized complexes of SERS probes–magnetic bead, and exosome-magnetic bead. e SERS spectra of the complexes obtained in the presence of single (left) and multiple (right) exosomes. Reprinted with permission from ref. 51. Copyright 2020 Royal Society of Chemistry.
Similar to nanoplasmonic sensors, a highly uniform plasmonic Au nanopillar SERS substrate was demonstrated to detect exosomal miRNAs.48 When a solution is applied and dried out, the plasmonic Au nanopillars lean towards each other due to capillary force. The head-flocked Au nanopillars create plasmonic hot spots and generate high Raman scattering signals, suitable for miRNA detection. A sandwich hybridization strategy is employed to use locked nucleic acid (LNA) capture and detection probes. Hybridization occurred with the presence of the target exosomal miRNAs, and Raman dye-labeled LNA probes (e.g., Cy3) were then added for further hybridization. Strong Cy3 SERS signals can be detected with demonstrated LOD of 1 aM and specificity down to single-base mismatch (miR-21 vs miR-21B). Various Raman dyes can be introduced on different LNA pairs to facilitate parallel detection of multiple exosomal miRNAs, which is useful in clinical applications.
SERS probes
Localized SERS sensing can be achieved using SERS probes made of different metallic nanoparticles,100,101 metallic nanoparticles with Raman reporters,49,50,53,54,102,103 multilayer metallic composite nanoparticles with Raman reporters,52,104 etc. For multiplexed exosome detection, bimetallic SERS nanotrepangs were designed with different Raman reporters (2-Mpy, 4-ATP, NTP) and decorated with linker DNAs that were complementary to the aptamers targeting exosomal membrane proteins (PSMA, HER2, and AFP).51 Figure 7c illustrates the detection scheme of bimetallic SERS nanotrepangs. Magnetic beads modified with aptamers were first coupled with the bimetallic SERS nanotrepangs via DNA hybridization. When target exosomes were added, the respective aptamers bound with the exosomes and released the bimetallic SERS nanotrepangs (Figure 7d). As a result, the attenuated SERS signal could be detected and correlated with exosome concentrations. The observed LOD for respective exosomes (LNCaP, SKBR3, and HepG2) were 26, 72, and 35 exosomes per μL, respectively (Figure 7e).
For exosomal miRNAs detection, a duplex-specific nuclease (DSN)-assisted dual SERS biosensor was developed.55 Ag-encapsulated magnetic beads were conjugated with complementary DNA probes, whereby the end of the DNA probes were bound with SERS tags. With the presence of the target miRNA-10b, hybridization with the complementary DNA probes occurred. DSN enzyme subsequently catalyzed the cleavage of the DNA probes, releasing the miRNAs and SERS tags from the magnetic bead complexes (attenuated SERS signal). Released microRNAs can be recycled to hybridize with other DNA probes and then declutter the DNA probes and SERS tags. With the usage of dual SERS tags and DSN-assisted recycling signal application, a LOD of 1 aM with single-base specificity could be achieved in a single processing step.
SERS provides a potential method to study exosomes’ interaction with cells as SERS presents superior spatial resolution and multiplexing capability. Chen et al. presented SERS-active (Raman reporter modified Au nanoparticles) exosomes to investigate the pathway involved in the internalization of exosomes in HeLa cells.102 Hydrogen tetrachloroaurate (III) trihydrate (HAuCl4·3H2O), 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) that shows a peak at 1,333 cm−1 band was used as a Raman reporter. SERS images were captured and created through the Raman intensity of the band using confocal Raman spectroscopy to track exosomes’ internalization into HeLa cells. The demonstrated SERS platform could potentially be applied in the development of exosome-based drug nanocarriers. With high flexibility in designing hybrid structures and compositions, various applications ranging from theranostics to photo-thermal therapy can also be explored.
In overall, SERS probes using Raman reporters enable multiplexing in EV molecular profiling due to its narrower Raman signals and broad choices in Raman reporters. Current efforts are focused on optimizing SERS probe synthesis for high uniformity and reproducibility, and advancing instrumentations for higher sensitivity, portability, and compactness.
PLASMON-ENHANCED FLUORESCENCE (PEF) DETECTION
PEF signal amplification on metal nanostructures has been conducted intensively in recent decades.105,106 The interaction between localized electromagnetic field and fluorophores, if they are sufficiently close but not in quenching region, can enhance fluorescence emission intensity significantly, which can be up to more than 103 times in single-molecule levels.107
Meanwhile, fluorescence detection is one of the most common biosensing methods because of its simplicity and versatility so that it can be potentially applied for high-throughput clinical applications. Especially compared to conventional protein detection techniques, such as western blotting and enzyme-linked immunosorbent assay (ELISA), fluorescence-based protein detection has advantages that require small sample amounts, shorter assay time, and fewer processing steps. However, fluorescence-based EV sensing has been challenging especially for EV protein detection due to their small sizes and weak signals. As a tool for transcending the limitation of current protein detection, PEF based protein detection has been widely applied for various clinical applications, e.g., T. gondii, cytomegalovirus, rubella, type 1 diabetes and hypertensive heart disease.70,108,109 Moreover, a PEF detection platform was also developed to monitor the expression of cardiac troponin I and creatine kinase-MB in serum for the prognosis of myocardial infarction, demonstrating a signal enhancement of 130-fold for near-infrared fluorescence detection.71
Recently, broadening the concept of PEF-based circulating tumor cell72 and protein detections, a PEF detection platform based on periodic nanoholes was reported for multiplexed EV marker profiling.57 An Au chip with nanohole arrays achieved fluorescence intensity enhancements by the factor of 23 and 9 from Cy3 and Cy5 streptavidin monolayer. The plasmonic resonance wavelength of nanohole structure (λ = 667 nm) induces the enhancement due to the large spectral overlap with a Cy5 channel (λexcitation = 649 and λemission = 666 nm). They further investigated the PEF effect from EVs captured on glass and nPLEX chip using a DOPA-based bioadhesive layer. ~10 times more EVs were detected on nPLEX, and the mean fluorescence intensity of nPLEX was 8.6 times higher than glass. Most importantly, the single EV analysis technology allowed for obtaining a much richer information set and discerning the heterogeneity for tumor marker on individual vesicles. The PEF signal application is an elegant approach to improve the detection sensitivity through intensity enhancement. However, as discussed before, plasmonic resonances can only enhance fluorescence in the wavelengths that overlap with plasmonic resonances, usually in the red and near-infrared regions, limiting the level of multiplexing in EV molecular profiling. Arranging a rare marker in the PEF channel or multi-modal multiplexing by combining PEF and SERS probes could be potentially employed to expand the level of multiplexing. Additionally, as a general limitation of fluorescence-based EV detection, a universal fluorescence labeling method with simple steps is still under study.
SURFACE FUNCTIONALIZATION
Since metallic substrates/nanoparticles are mainly used in plasmonic platforms, a few surface chemistry methods are commonly used to capture EVs, their proteins, and miRNAs, as discussed below.
Non-specific binding: EVs can be physically adsorbed on the metallic substrates nonspecifically.36,38,44-47,95,96,98,99 This approach is mostly used to characterize EVs using SERS, where the close proximity of target molecules to the metallic surface is critical for sensitive detection. The metallic surface can be functionalized with positively charged compounds, such as cysteamine, to improve the capture efficiency of negatively charged EVs.97
Immunocapture: Antibodies have been widely used to capture target EVs and their transmembrane proteins with high specificity and sensitivity. Antibodies can be conjugated on metallic substrates/nanoparticles through physical adsorption.30,66,110 The most common approach, however, is using thiol compounds (e.g., 11-mercaptoundecanoic acid, thiol-polyethylene glycol, etc.) to form a carboxyl-terminated self-assembly monolayer (SAM), which are then activated by N-(3-dimethylami-nopropyl)-N-ethylcarbodiimide (EDC) - N-hydroxysuccinimide (NHS) reaction to covalently bind antibodies onto the SAM.24,29,31,32,34,37,39,50,56,88,92 Alternatively, streptavidin or biotinylated SAM can also be prepared, and antibody conjugation is achieved by biotin-avidin chemistry.25,26,33,40,54,60,64,65,89,111 Another approach used in commercial Biacore chip (GE Healthcare) is carboxymethylated dextran deposition through standard amine coupling.59,62,63 Although antibodies demonstrate high specificity and sensitivity, they commonly suffer from high cost, limited shelf life, temperature-sensitivity, and low chemical and pH resistance.
Aptamers (peptides/single-stranded DNA/synthetic RNA): Aptamers (small synthetic molecules that have binding affinity to specific targets) are selected through in vitro Systematic Evolution of Ligands by Exponential Enrichment (SELEX) process. Aptamers are superior in terms of lower production cost, higher chemical and temperature stabilities, and higher reproducibility.112 The surface chemistries used to bind aptamer onto metallic surfaces/nanoparticles include physical adsorption,27,28,49,93 thiolated peptides,100 and avidin-biotin chemistry.51,91,94,101 However, compared to antibodies, limited availability for various target molecules and cross-reactivity might hinder their practical applications.
Complementary oligonucleotides: Thiolated oligonucleotides are used to functionalize metallic substrates/nanoparticles and capture EV miRNA targets.42,48 Alternatively, surface carboxylation can be used and then activated by EDC for DNA conjugation.55 In addition, cholesterol-labeled DNA anchors can be prepared to incorporate DNA into the EV membrane through hydrophobic interaction.53,101 Through such an approach, SERS probes can be bound onto target EVs.
BRIDGING BENCH TO BEDSIDE
For every clinical profiling and diagnostic success story, there are invariably many failed prior attempts, often cited as the “translational valley of death”.113 According to the National Institute of Health, 80-90% of research projects failed to reach testing in humans, and the rate is even worse for new drugs.114 Such concerns also apply to EV bioengineering efforts, not from ineffective technologies, but insufficient synergies between hardware and clinical realities. Scientific development of EV technologies can, at times, lend itself to an over-reliance on the platforms, notably their limit of detection and precision under controlled environmental and biological conditions. Yet translational challenges often extend into reliable testing performance across diverse clinical specimen types of varying quality (e.g., pre-analytical handling, degradation, matrix effects). EVs are found in all biofluids, but the EV contents are vastly different depending on their origins. For example, urine contains EVs mostly from the urogenital tract (kidney, bladder, prostate),115 and plasma/serum is the preferred biofluid for systemic conditions.116 This makes it desirable to have a technology that can analyze EVs from different types of biofluids, both from a matrix composition perspective as well as from a volume perspective, since some biofluids may require a much higher volume to be processed to achieve a sufficient number of specific EVs.117 The matrix effects in biofluids such as plasma vs urine vs cerebrospinal fluid (CSF) are very different, and reaction conditions that work for one biofluid may not be optimal for another.118 For example, EV recovery rates in CSF are inferior to plasma/serum even with twenty times higher starting volumes.118 Utilizing a technology that is flexible enough to accommodate these variables and allow for optimization is desirable. Although there are thousands of biomarkers proposed each year in peer-reviewed articles, unfortunately, only very few of those end up as clinical biomarkers.119 There are many challenges on the way, but some biomarkers fail when applied in the correct “intended use population” and some are not robust enough when sample matrix variation effects are taken into consideration.
Moreover, to better leverage EVs' dynamic and circulating nature, serial analyses become paramount to increase performance. Serial sampling also makes it possible to utilize biomarkers that cannot be used in tissues where serial sampling is not possible or practical. Biomarkers with large heterogeneity in the population may not be useful for a single time-point analysis, but an EV-based “liquid biopsy” that can be repeated allows for each individual to create their own baseline, circumventing some of these problems.120 To this end, cumbersome and time demanding approaches pose significant hurdles to adoption, particularly when not aligned with clinical workflows. The field’s increased attention towards single EV analyses magnifies such challenges. Specifically, bioengineering solutions are pressured to take advantage of the shift away from bulk EV readouts and make the process easily available to clinical laboratories. Instead of increasing assay complexity and turnaround times to meet such challenges, leaner and faster tactics are encouraged where clinical labs can efficiently scale throughput to meet the demand of the test with an automated or semi-automated workflow. The other type of testing that is increasing in demand is the point-of-care setting where typically the test throughput is lower, but the instrument is simple and can rapidly return the test result.121 SPR based detection approaches fit really well for both types of testing, where detector chips can be manufactured for parallel processing of many patient samples, but are yet cheap and simple enough to produce single-patient chips that can be read quickly on a small instrument footprint.
CLINICAL APPLICATIONS
The following sections summarize and detail some clinical instances where SPR was used for EV analysis.
Cancer
Most studies to date have used SPR platforms as a proof-of-principle to detect cancers. These studies include detecting multiple myeloma-derived exosomes using colloidal Au nanoparticles with SPR biosensor chip,23 lung cancer diagnosis using exosomal epidermal growth factor receptor (EGFR) and programmed death-ligand 1 (PD-L1) as biomarkers,25 breast cancer molecular profiling using exosome-specific markers (CD9, CD63) and cancer-specific marker (HER2),24,26 high-throughput screening for non-small cell lung cancer (NSCLC) in clinical specimens,30 ovarian cancer analysis,32 and pancreatic cancer.89
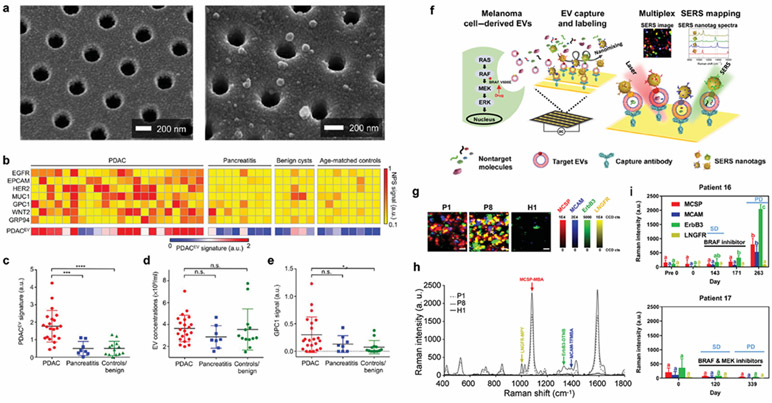
Im et al. developed the nPLEX system based on transmission SPR through periodic nanohole arrays (Figure 5).32 They showed that the levels of EpCAM and CD24 were elevated in exosomes from ovarian cancer cells. Based on exosome analysis in ascites from 30 patients (non-cancer, n = 10) and ovarian cancer (n = 20), they demonstrated 97% detection accuracy. The group also presented a next-generation nPLEX system (Figure 8a) to analyze pancreatic ductal adenocarcinoma (PDAC)-derived EVs in higher throughput.89 They defined a PDACEV signature marker set comprising EGFR, EpCAM, MUC1, GPC1, and WNT2 that showed an overall accuracy of 84% with 86% sensitivity and 81% specificity in differentiating PDAC from pancreatitis, benign, and control patient groups (Figure 8b). The PDACEV signature was highly elevated in PDAC-derived EVs (Figure 8c), whereas total EV concentrations (Figure 8d) and a single marker of GPC1 (Figure 8e) demonstrated a low detection power.
Figure 8. EV molecular profiling for cancer diagnosis.
a Multiparametric plasma EV profiling using periodic nanoholes for the diagnosis of pancreatic malignancy. Scanning electron micrographs show the periodically arranged nanopore array and EVs captured on the surface. b Heatmap analysis of EV markers. The PDACEV signature is defined as a combined marker panel of EGFR, EPCAM, MUC1, GPC1, and WNT2. c The established PDACEV signature signals, d EV concentrations, and e single GPC1 signal as measured for plasma EVs collected from 22 PDAC patients, 8 with pancreatitis, 5 with benign cystic tumors, and 8 age-matched controls. Reprinted with permission from ref. 89. Copyright 2017 American Association for the Advancement of Science. f Tracking extracellular vesicle phenotypes for melanoma monitoring. EVs from melanoma cells are captured, and SERS nanotags are attached. The characterization of EV phenotypes is performed by SERS mapping (MCSP-MBA, red; MCAM-TFMBA, blue; ErbB3-DTNB, green; LNGFR-MPY, yellow). g Representative false-color SERS spectral images and h corresponding average SERS spectra from patient and normal samples. i Monitoring EV phenotypic evolution of patients 16 and 17 during targeted therapies. Patient 16 was treated with dabrafenib. Stable disease (SD) on day 143 and developed progressive disease (PD) after cessation of treatment (day 263) were shown. Patient 17 received the combination treatment of dabrafenib and trametinib. SD on day 120 and PD at the third visit (day 339) were shown. Reprinted with permission from ref. 54. Copyright 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC) http://creativecommons.org/licenses/by-nc/4.0/.
Another nanoplasmonic platform used for cancer diagnosis is based on plasmonic-enhanced laser desorption/ionization mass spectrometry (LDI-MS).38,122-124 Plasmonic-enhanced LDI-MS offers rapid (in seconds) detection of metabolic fingerprints with low sample volume, which potentially could be used to investigate disease prognosis125 such as medulloblastoma diagnosis and radiotherapy evaluation.126 Sun et al. demonstrated the in vitro profiling of small molecules in exosomes on the metabolic level, showing its potential in early lung cancer diagnosis.38 Alternatively, breast cancer-derived exosome detection was performed using colorimetric nanoplasmonic platform,94 simplifying the system required for detection.
SERS platforms identified distinctive Raman spectrum signatures and applied principal component analysis for cancer diagnosis, such as early pancreatic cancer diagnosis,43 exosomal phosphoprotein detection in cancer-derived exosomes,47 liver cancer diagnosis,53 etc. Molecular profiling of circulating miRNAs in serum or plasma has emerged as a potential diagnostic and prognostic option in cancer.127 Pang et al. demonstrated dual-SERS nanoprobes for pancreatic cancer-specific exosomal miRNA detection (Figure 8f-g).55 They could successfully quantify miR-10b in exosome and residual plasma of blood samples (Figure 8h) from patients of PDAC (n = 5), chronic pancreatitis (CP, n = 5), and normal controls (NC, n = 5).
Various SERS nanoprobes, coupled with different Raman reporters, were also developed to label specific subgroups of exosomes for cancer diagnosis. It was demonstrated that aptamers for PSMA, HER2, and AFP have affinities to exosomes from prostate cancer, breast cancer, and hepatocellular carcinoma, respectively.51 Antibodies were also used to detect pancreatic cancer-derived exosomes52 and breast cancer-derived exosomes.103
Tracking patient treatment responses and drug resistance in real-time by using biomarkers in EVs is a promising way to improve cancer therapeutics. Wang et al. introduced a multiplex EV phenotype analyzer chip for direct EV phenotyping in plasma, allowing multiplex biomarker detection by simultaneously labeling target EVs with AuNPs conjugated with target-specific antibodies and Raman reporters.54 They suggested four biomarkers - melanoma chondroitin sulfate proteoglycan (MCSP), melanoma cell adhesion molecule (MCAM), low affinity nerve growth factor receptor (LNGFR), and receptor tyrosine protein kinase (ErbB3), which are correlated with melanoma treatment or progression.128 By evaluating the profiles of these proteins, they detected cancer-specific EV phenotypes from plasma sample of melanoma patients (n = 11) and healthy people (n = 12) as well as changes of plasma EV phenotypes during treatment in melanoma patients (n = 8) undergoing molecular targeted therapies, offering potentials to guide personalized cancer therapy.
Neurodegenerative disease
Since exosomes have the capability to cross the blood-brain barrier, molecular profiling of central nervous system (CNS)-derived exosomes in blood could potentially discover important biomarkers of neurodegeneration such as Alzheimer’s disease (AD) and Parkinson’s disease.
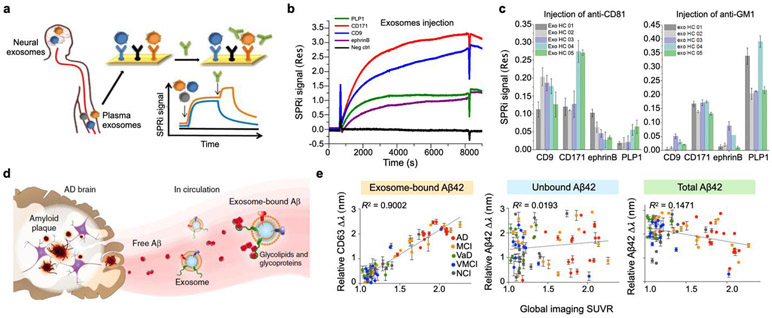
SPR imaging was employed to simultaneously detect and characterize CNS-derived exosomes in patient plasma29 (Figure 9a). Purified exosomes were captured on the SPR substrate using capture antibodies: anti-CD9 (generic marker), anti-PLP1 (oligodendrocyte marker), anti-CD171, and anti-ephrinB (neuronal markers) (Figure 9b). Captured exosomes were then characterized with antibodies targeting CD81 (generic marker) and GM1 (ganglioside marker). The results showed that the abundance of CD81 in different subpopulations of exosomes are inhomogeneous. Although CD81 is considered a generic exosomal marker, CD171-positive exosomes contained a larger amount of CD81 (Figure 9c). This shows that molecular profiling of different subpopulations of exosomes is important due to their heterogeneity in molecular levels, which might be useful for disease diagnosis. Similarly, GM1 was high in CD171 and PLP-1 positive exosomes, reflecting their neuronal and oligodendrocyte origin. The presence of GM1 has been associated with various neurodegenerative diseases.129,130 For instance, gangliosides accelerate α-synuclein aggregation in neurons, causing the spread of Parkinson’s disease.131
Figure 9. Subtyping of brain-derived exosomes reflecting brain diseases.
a Detection and characterization of different brain-derived subpopulations of plasma exosomes by SPR imaging. b Sensorgram of exosome detection on the SPRi chip with spots of different antibodies. c SPRi intensities related to the injections of anti-CD81 and anti-GM1 in sequence for exosomes derived from blood plasma of five healthy subjects. Reprinted with permission from ref. 29. Copyright 2018 American Chemical Society. d Aβ proteins, the main component of amyloid plaques found in AD brain pathology, are released into the extracellular space. Through their surface glycoproteins and glycolipids, exosomes can associate with the released Aβ proteins. e Subtyping of circulating exosome-bound amyloid β using nanoplasmonics with periodic nanoholes. Correlations of different populations of circulating Aβ42 with global average PET brain imaging. When correlated to the global imaging data of brain amyloid plaque, the exosome-bound Aβ42 measurements demonstrated the best correlation. Reprinted with permission from ref. 34. Copyright 2019 Springer Nature. Creative Common CC-BY.
AD, in particular, is severe dementia associated with a progressive loss of memory and cognitive functions. Current AD diagnosis through neuropsychological assessments is subjective, and most cases are detected in the late-stage. Although different molecular assays are being developed for early-stage detection, these approaches are either invasive or expensive.132,133 Blood analysis is an ideal candidate, but pathological AD molecules (e.g., amyloid β, Aβ) are in low concentrations in blood that are difficult to be detected by conventional approaches such as ELISA. On the other hand, exosomes in the blood not only carry pathological AD proteins, but their markers are also enriched in human brain amyloid plaques. Therefore, plasma exosome analysis could potentially characterize AD non-invasively for early-stage diagnosis.
Amplified plasmonic exosome (APEX) was presented recently to analyze exosomes in blood samples (Figure 5f).34 The periodic nanohole array was first functionalized with antibodies against exosomal membrane proteins (Aβ42, APP, α-syn, CHL1, IRS-1, NCAM, Tau). The APEX platform was used to measure unbound Aβ42 and exosome-bound Aβ42 from blood samples of AD patients (Figure 9d). Exosome-bound Aβ42 in native plasma was enriched using Aβ42 antibodies, and the relative amount of CD63 was measured. Total circulating Aβ42 was measured using both Aβ42 antibodies for enrichment and detection. Finally, unbound Aβ42 was measured by first removing large particles (>50 nm, filtration) in the plasma and then detecting unbound Aβ42 using the APEX platform.
A total of 84 patients was recruited for the study: 17 patients with AD; 18 patients with mild cognitive impairment, 16 healthy control with no cognitive impairment, 9 controls with vascular dementia, 12 controls with vascular mild cognitive impairment and 12 controls with acute stroke. Brain amyloid plaque load was determined using PET brain images. The concentration of exosome-bound Aβ42 had the best correlation to PET imaging (R2 = 0.9002, Figure 9e), suggesting that exosomes are strongly associated with prefibrillar Aβ42 aggregates that can readily form fibrils. Interestingly, the results showed a stronger correlation to the brain plaque load in the cingulate region (early AD-affected region). In terms of specificity, the concentration of exosome-bound Aβ42 could differentiate patients with AD and mild cognitive impairment (P < 0.01), as well as with other controls (P < 0.0001, Student’s t-test).
The APEX platform provides sensitive blood-based measurements of circulating exosomes directly from native plasma samples with multiplexing capabilities. In addition to CD63 for exosome identification, additional markers could be used as well to identify other subpopulations for combinatorial analysis to determine various stages of AD and provide guidance in disease-modifying therapies.
Cardiovascular disease
In the US, 647,000 people die from cardiovascular diseases every year, equating to 25% of total death in the US.134 While several well-advanced biomarkers exist, detecting and monitoring different cardiovascular diseases through EV analysis have some niche applications. For example, in one study, it was shown that the detection of EVs positive for ICAM-1 could be a prognostic indicator of coronary heart disease.59 In this study, Human umbilical vein endothelial cells (HUVECs) were used as an in vitro model of patient vascular endothelial cells. To mimic EVs derived coronary heart disease patients, HUVECs were stimulated with TNF-α, which induces an inflammatory stress response. Anti-ICAM-1 was selected as a diagnostic marker due to better binding affinity and discrimination ability between unstimulated and TNF-α-stimulated EVs. In SPR analysis with patient-derived EVs, The ICAM-1 levels in the coronary heart disease patient samples (n = 10) were significantly higher than that of healthy control (n = 6, p = 0.007). Moreover, the level of ICAM-1-positive EV was not changed after the TNF-α stimulation in the patient cohort (p = 0.86), but much more EVs were detected after TNF-α stimulation in the healthy cohort (p = 0.07).
Others
Besides clinical applications reviewed above, EVs are also potentially important biomarkers in other diseases. In the study of pathogen infection, it is discovered that viruses are transmitted via vesicles as populations of viral particles and this type of transmission enhances their infection efficiency.135,136 For instance, exosomes in the plasma of hepatitis B patients were shown to contain nucleic acids and proteins of hepatitis B virus.137 It was found that exosomes could shuttle hepatitis B viruses into uninfected cells as efficiently as free-virus infection, serving as important regulators of hepatitis B virus transmission. Recently, Elrashdy et al. proposed that one of the potential mechanisms for the relapse of the COVID-19 infection could be a cellular transport pathway associated with the release of the SARS-CoV-2-loaded EVs, whereby such the “Trojan horse” strategy represents possible explanation for the re-appearance of the viral RNA in the recovered COVID-19 patients.138 On the other hand, miRNA profiling of bovine-milk derived exosomes could identify potential biomarkers for early mastitis detection due to bacterial infection.139 In addition, EVs were found to be promising biomarkers for diabetes,140 kidney injury, renal diseases,141 and traumatic brain injury.142 EV characterization and molecular profiling using plasmonic platforms can be further explored in various areas of clinical diagnosis, theranostics, and monitoring.
FUTURE PERSPECTIVE
In this review, we have shown that SPR represents a number of different technologies to sensitively analyze EVs. The research and clinical implications are detailed as such methods would allow us to better understand the role of EVs in health and disease. However, at the same time, there are ample opportunities to further develop plasmonic sensing approaches. The current research and clinical needs are summarized below:
Single EV analysis. Most of plasmonic sensing platforms, although superior to conventional methods, still requires a certain number of EVs, measuring bulk properties from an ensemble of vesicles. Analyzing single EVs could reveal distinctive molecular profiles of cell-specific EVs, which will further promote clinical use of these vesicles and allow us to construct a comprehensive EV atlas per different biological parameters (e.g., cellular origin, cell state). For instance, in cancer, single EV analysis could be extremely valuable in studying EV biogenesis, tumor heterogeneity, rare tumor subtypes, phenotypic changes occurring during therapy, and normal host EV variations that occur concomitantly with tumoral changes.2,15 Several plasmonic platforms have demonstrated proof-of-concept for single EV analysis,31,39,57 showing that plasmonic technology has high potential in developing clinical systems for single EV analysis.
Multiplex measurements to improve specificity. Multiplex measurements are critical for EV subpopulation characterization or multiple parameters analysis to improve the specificity in disease diagnosis. For example, Yang et al. showed that the specificity of identifying PDAC increased from 52% (using a single biomarker GPC1) to 81% by using a panel of 5 biomarkers.89 SPR imaging and periodic nanoholes have demonstrated high multiplexing capability and are suitable to investigate complex clinical problems. Multiple capture antibodies/aptamers can be patterned using a microspotter or microfluidic technology, while multiplexed profiling analysis can be performed with the help of machine-learning classification.
Better signal amplification strategies for rare targets and markers. Since mutant nucleic acids and proteins are rare in EV, better amplification strategies are needed to increase the sensitivity of the plasmonic response and detect such rare targets and markers using plasmonic technology. Signal amplification strategies in plasmonics include surface design with nanostructures,42,48 sandwich assays with detection probes,48,52,53 chemical deposition/reduction processes,28,34,55 and plasmon-enhanced fluorescence detection.57 Other amplification strategies can be explored to further improve the sensitivity of the plasmonic platforms targeting rare RNAs or mutant proteins in EVs.
Commercialization of technologies. Although there are hundreds of papers on SPR, up to now, plasmonic technology has yet to be commercialized clinically. Commercial SPR systems, such as Biacore, are still primarily used in research to measure real-time binding kinetics and search for promising biomarkers. Much effort is needed in designing a robust plasmonic system that can be used in clinical settings. Exosome Diagnostics, Inc. is currently spearheading such an effort, developing cost-effective sensor chips and automated signal readers. Commercial SPR products will be one of the most promising in vitro diagnostic instruments and contribute to the world’s second-largest sales market and production base.143
Identification of clinically relevant biomarkers. Hundreds of EV biomarkers have been identified, but, unfortunately, less than a handful have proven clinically useful for disease diagnosis and monitoring. This is evident by the fact that the research effort is still focused on platform development and improving sensitivity, LOD, measurement precision, and robustness. In recent years, more researchers have been focusing on identifying clinically useful biomarkers using plasmonic platforms.32,34,89 More translational research is needed to facilitate the adaptation of plasmonic technology in clinical applications. In that effort, it is also critical that the establishment of a biomarker signature is done in the real intended use population instead of more generic studies where patients and controls are chosen due to availability rather than clinically appropriate samples taken at the time-point where it is intended to be used.
Prospective clinical trials. With the successful identification of clinically useful biomarkers and a well-defined clinical decision that the biomarker is trying to solve, prospective clinical trials using plasmonic technology for disease diagnosis and monitoring are essential. These clinical trials have to be well controlled with the collaborative effort of clinical investigators and research staffs such that the deployment of plasmonic technology in real clinical settings can be achieved efficiently. The intended use population needs to be well defined for the clinical question, and the population need to have the right distribution of patients reflecting the real-world scenario since this is where many biomarkers fail.
In conclusion, SPR is a very promising technique for EV detection, proteomics, and miRNA profiling with great clinical translation potentials. While this review focused on the application of plasmonic platforms on EV detection and analysis, it is important to note that plasmonic platforms are targeting nano-sized particles. Biological particles that fall in the same size range of EVs can also be detected and characterized by plasmonic platforms, such as lysosomes, viruses, and bacteria.124 For instance, plasmonic technologies were demonstrated to detect SARS-CoV-2 virus using nanoplasmonic Au nanoprobe or nanoisland substrate, down to pM sensitivity,144-146 demonstrating the use of plasmonic sensors in other nano-sized biological entities and expanding their uses in a broad range of biomedical applications.
Funding:
This work was supported in part by U.S. NIH Grants R00CA201248 (H.I.), R21CA217662 (H.I.), R01CA229777 (H.L.), U01CA233360 (H.L., C.M.C.), R21DA049577 (H.L.), R01CA204019 (R.W.), R21CA236561 (R.W.), US DOD-W81XWH1910199 (H.L.), DOD-W81XWH1910194 (H.L), Fundamental Research Program (PNK 6070) of the Korean Institute of Materials Science, and Singapore National Research Foundation Competitive Research Programme NRFCRP13-2014-01 (A.Q.L.).
VOCABULARY
- Extracellular vesicles
Micro- and nano-sized particles that are released naturally by cells and carry molecules such as proteins, nucleic acids, lipids, etc., originated from the cells.
- Surface plasmon resonance
A physical phenomenon whereby free photons on metal surfaces are converted into localized charge-density oscillations (known as surface plasmons) under the stimulation of light.
- Nanoplasmonics
The physical phenomenon of surface plasmon resonance occurs on metal surfaces with nanostructures or nanoprobes.
- Surface-enhanced Raman spectroscopy
A physical technique used to enhance the Raman scattering using rough metal surfaces or nanostructures.
- Plasmonic-enhanced fluorescence
A physical technique used to enhance the fluorescence intensity of fluorophores through plasmonic coupling interactions.
- Clinical translational research
Research that transfers laboratory discoveries to clinical studies, accelerating the adoption of new technologies or techniques in hospital settings.
- Diagnostics
Techniques and approaches used to identify illnesses, symptoms and diseases.
Footnotes
Competing interest: J.S. is the Chief Scientific Officer of Exosome Diagnostics Inc. Exosome Diagnostics Inc. licensed a patent invented by H.I., H.L. and R.W. from MGH. H.I. and H.L. serve as consultants for Exosome Diagnostics, Inc. None of these activities are related to the manuscript.
REFERENCES
- 1.Mathieu M; Martin-Jaular L; Lavieu G; Théry C Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat Cell Biol 2019, 21, 9–17. [DOI] [PubMed] [Google Scholar]
- 2.Fraser K; Jo A; Giedt J; Vinegoni C; Yang KS; Peruzzi P; Chiocca EA; Breakefield XO; Lee H; Weissleder R Characterization of Single Microvesicles in Plasma from Glioblastoma Patients. Neuro Oncol 2019, 21, 606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verma M; Lam TK; Hebert E; Divi RL Extracellular Vesicles: Potential Applications in Cancer Diagnosis, Prognosis, and Epidemiology. BMC Clin Pathol 2015, 15, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X; Yuan X; Shi H; Wu L; Qian H; Xu W Exosomes in Cancer: Small Particle, Big Player. J Hematol Oncol 2015, 8, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skog J; Würdinger T; van Rijn S; Meijer DH; Gainche L; Sena-Esteves M; Curry WT; Carter BS; Krichevsky AM; Breakefield XO Glioblastoma Microvesicles Transport RNA and Proteins That Promote Tumour Growth and Provide Diagnostic Biomarkers. Nat Cell Biol 2008, 10, 1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKiernan J; Donovan MJ; O’Neill V; Bentink S; Noerholm M; Belzer S; Skog J; Kattan MW; Partin A; Andriole G; Brown G; Wei JT; Thompson IM; Carroll P A Novel Urine Exosome Gene Expression Assay to Predict High-Grade Prostate Cancer at Initial Biopsy. JAMA Oncol 2016, 2, 882–889. [DOI] [PubMed] [Google Scholar]
- 7.Ailawadi S; Wang X; Gu H; Fan GC Pathologic Function and Therapeutic Potential of Exosomes in Cardiovascular Disease. Biochim Biophys Acta 2015, 1852, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibrahim A; Marbán E Exosomes: Fundamental Biology and Roles in Cardiovascular Physiology. Annu Rev Physiol 2016, 78, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Properzi F; Logozzi M; Fais S Exosomes: The Future of Biomarkers in Medicine. Biomark Med 2013, 7, 769–778. [DOI] [PubMed] [Google Scholar]
- 10.Saman S; Kim W; Raya M; Visnick Y; Miro S; Saman S; Jackson B; McKee AC; Alvarez VE; Lee NC; Hall GF Exosome-Associated Tau Is Secreted in Tauopathy Models and Is Selectively Phosphorylated in Cerebrospinal Fluid in Early Alzheimer Disease. J Biol Chem 2012, 287, 3842–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuhrmann G; Neuer AL; Herrmann IK Extracellular Vesicles - A Promising Avenue for the Detection and Treatment of Infectious Diseases. Eur J Pharm Biopharm 2017, 118, 56–61. [DOI] [PubMed] [Google Scholar]
- 12.Silverman JM; Reiner NE Exosomes and Other Microvesicles in Infection Biology: Organelles with Unanticipated Phenotypes. Cell Microbiol 2011, 13, 1–9. [DOI] [PubMed] [Google Scholar]
- 13.Wang B; Xing D; Zhu Y; Dong S; Zhao B The State of Exosomes Research: A Global Visualized Analysis. Biomed Res Int 2019, 2019, 1495130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei Z; Batagov AO; Schinelli S; Wang J; Wang Y; El Fatimy R; Rabinovsky R; Balaj L; Chen CC; Hochberg F; Carter B; Breakefield XO; Krichevsky AM Coding and Noncoding Landscape of Extracellular RNA Released by Human Glioma Stem Cells. Nat Commun 2017, 8, 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee K; Fraser K; Ghaddar B; Yang K; Kim E; Balaj L; Chiocca EA; Breakefield XO; Lee H; Weissleder R Multiplexed Profiling of Single Extracellular Vesicles. ACS Nano 2018, 12, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liedberg B; Nylander C; Lundström I Biosensing with Surface Plasmon Resonance--How It All Started. Biosens Bioelectron 1995, 10, i–ix. [DOI] [PubMed] [Google Scholar]
- 17.Yadav SP; Bergqvist S; Doyle ML; Neubert TA; Yamniuk AP MIRG Survey 2011: Snapshot of Rapidly Evolving Label-Free Technologies Used for Characterizing Molecular Interactions. J Biomol Tech 2012, 23, 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brockman JM; Nelson BP; Corn RM Surface Plasmon Resonance Imaging Measurements of Ultrathin Organic Films. Annu Rev Phys Chem 2000, 51, 41–63. [DOI] [PubMed] [Google Scholar]
- 19.Lamprecht B; Krenn JR; Schider G; Ditlbacher H; Salerno M; Félidj N; Leitner A; Aussenegg FR; Weeber JC Surface Plasmon Propagation in Microscale Metal Stripes. Appl Phys Lett 2001, 79, 51–53. [Google Scholar]
- 20.Knoll W Interfaces and Thin Films as Seen by Bound Electromagnetic Waves. Annu Rev Phys Chem 1998, 49, 569–638. [DOI] [PubMed] [Google Scholar]
- 21.Im H; Lesuffleur A; Lindquist NC; Oh S-H Plasmonic Nanoholes in a Multichannel Microarray Format for Parallel Kinetic Assays and Differential Sensing. Anal Chem 2009, 81, 2854–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh P SPR Biosensors: Historical Perspectives and Current Challenges. Sens Actuators B Chem 2016, 229, 110–130. [Google Scholar]
- 23.Di Noto G; Bugatti A; Zendrini A; Mazzoldi EL; Montanelli A; Caimi L; Rusnati M; Ricotta D; Bergese P Merging Colloidal Nanoplasmonics and Surface Plasmon Resonance Spectroscopy for Enhanced Profiling of Multiple Myeloma-Derived Exosomes. Biosens Bioelectron 2016, 77, 518–524. [DOI] [PubMed] [Google Scholar]
- 24.Grasso L; Wyss R; Weidenauer L; Thampi A; Demurtas D; Prudent M; Lion N; Vogel H Molecular Screening of Cancer-Derived Exosomes by Surface Plasmon Resonance Spectroscopy. Anal Bioanal Chem 2015, 407, 5425–5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C; Zeng X; An Z; Yang Y; Eisenbaum M; Gu X; Jornet JM; Dy GK; Reid ME; Gan Q; Wu Y Sensitive Detection of Exosomal Proteins via a Compact Surface Plasmon Resonance Biosensor for Cancer Diagnosis. ACS Sens 2018, 3, 1471–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sina AA; Vaidyanathan R; Dey S; Carrascosa LG; Shiddiky MJ; Trau M Real Time and Label Free Profiling of Clinically Relevant Exosomes. Sci Rep 2016, 6, 30460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q; Zou L; Yang X; Liu X; Nie W; Zheng Y; Cheng Q; Wang K Direct Quantification of Cancerous Exosomes via Surface Plasmon Resonance with Dual Gold Nanoparticle-Assisted Signal Amplification. Biosens Bioelectron 2019, 135, 129–136. [DOI] [PubMed] [Google Scholar]
- 28.Liao G; Liu X; Yang X; Wang Q; Geng X; Zou L; Liu Y; Li S; Zheng Y; Wang K Surface Plasmon Resonance Assay for Exosomes Based on Aptamer Recognition and Polydopamine-Functionalized Gold Nanoparticles for Signal Amplification. Mikrochim Acta 2020, 187, 251. [DOI] [PubMed] [Google Scholar]
- 29.Picciolini S; Gualerzi A; Vanna R; Sguassero A; Gramatica F; Bedoni M; Masserini M; Morasso C Detection and Characterization of Different Brain-Derived Subpopulations of Plasma Exosomes by Surface Plasmon Resonance Imaging. Anal Chem 2018, 90, 8873–8880. [DOI] [PubMed] [Google Scholar]
- 30.Fan Y; Duan X; Zhao M; Wei X; Wu J; Chen W; Liu P; Cheng W; Cheng Q; Ding S High-Sensitive and Multiplex Biosensing Assay of NSCLC-Derived Exosomes via Different Recognition Sites Based on SPRi Array. Biosens Bioelectron 2020, 154, 112066. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y; Zhai C; Zeng Q; Khan AL; Yu H Multifunctional Detection of Extracellular Vesicles with Surface Plasmon Resonance Microscopy. Anal Chem 2020, 92, 4884–4890. [DOI] [PubMed] [Google Scholar]
- 32.Im H; Shao H; Park YI; Peterson VM; Castro CM; Weissleder R; Lee H Label-Free Detection and Molecular Profiling of Exosomes with a Nano-Plasmonic Sensor. Nat Biotechnol 2014, 32, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park J; Im H; Hong S; Castro CM; Weissleder R; Lee H Analyses of Intravesicular Exosomal Proteins Using a Nano-Plasmonic System. ACS Photonics 2018, 5, 487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim CZJ; Zhang Y; Chen Y; Zhao H; Stephenson MC; Ho NRY; Chen Y; Chung J; Reilhac A; Loh TP; Chen CLH; Shao H Subtyping of Circulating Exosome-Bound Amyloid Β Reflects Brain Plaque Deposition. Nat Commun 2019, 10, 1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu S; Li H; Yang M; Pang SW Highly Sensitive Detection of Exosomes by 3D Plasmonic Photonic Crystal Biosensor. Nanoscale 2018, 10, 19927–19936. [DOI] [PubMed] [Google Scholar]
- 36.Thakur A; Qiu G; Ng SP; Guan J; Yue J; Lee Y; Wu CL Direct Detection of Two Different Tumor-Derived Extracellular Vesicles By SAM-AuNIs LSPR Biosensor. Biosens Bioelectron 2017, 94, 400–407. [DOI] [PubMed] [Google Scholar]
- 37.Lv X; Geng Z; Su Y; Fan Z; Wang S; Fang W; Chen H Label-Free Exosome Detection Based on a Low-Cost Plasmonic Biosensor Array Integrated with Microfluidics. Langmuir 2019, 35, 9816–9824. [DOI] [PubMed] [Google Scholar]
- 38.Sun X; Huang L; Zhang R; Xu W; Huang J; Gurav DD; Vedarethinam V; Chen R; Lou J; Wang Q; Wan J; Qian K Metabolic Fingerprinting on a Plasmonic Gold Chip for Mass Spectrometry Based in Vitro Diagnostics. ACS Cent Sci 2018, 4, 223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raghu D; Christodoulides JA; Christophersen M; Liu JL; Anderson GP; Robitaille M; Byers JM; Raphael MP Nanoplasmonic Pillars Engineered for Single Exosome Detection. PLoS One 2018, 13, e0202773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zeng X; Yang Y; Zhang N; Ji D; Gu X; Jornet J; Wu Y; Gan Q Plasmonic Interferometer Array Biochip as a New Mobile Medical Device for Cancer Detection. IEEE J Sel Top Quantum Electron 2019, 25, 7201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang K; Liu F; Fan J; Sun D; Liu C; Lyon CJ; Bernard DW; Li Y; Yokoi K; Katz MH; Koay EJ; Zhao Z; Hu Y Nanoplasmonic Quantification of Tumor-Derived Extracellular Vesicles in Plasma Microsamples for Diagnosis and Treatment Monitoring. Nat Biomed Eng 2017, 1, 0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joshi GK; Deitz-McElyea S; Liyanage T; Lawrence K; Mali S; Sardar R; Korc M Label-Free Nanoplasmonic-Based Short Noncoding RNA Sensing at Attomolar Concentrations Allows for Quantitative and Highly Specific Assay of MicroRNA-10b in Biological Fluids and Circulating Exosomes. ACS Nano 2015, 9, 11075–11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carmicheal J; Hayashi C; Huang X; Liu L; Lu Y; Krasnoslobodtsev A; Lushnikov A; Kshirsagar PG; Patel A; Jain M Label-Free Characterization of Exosome via Surface Enhanced Raman Spectroscopy for the Early Detection of Pancreatic Cancer. Nanomedicine 2019, 16, 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sivashanmugan K; Huang W-L; Lin C-H; Liao J-D; Lin C-C; Su W-C; Wen T-C Bimetallic Nanoplasmonic Gap-Mode SERS Substrate for Lung Normal and Cancer-Derived Exosomes Detection. J Taiwan Inst Chem Eng 2017, 80, 149–155. [Google Scholar]
- 45.Avella-Oliver M; Puchades R; Wachsmann-Hogiu S; Maquieira A Label-Free SERS Analysis of Proteins and Exosomes with Large-Scale Substrates from Recordable Compact Disks. Sens Actuators B Chem 2017, 252, 657–662. [Google Scholar]
- 46.Lee C; Carney RP; Hazari S; Smith ZJ; Knudson A; Robertson CS; Lam KS; Wachsmann-Hogiu S 3D Plasmonic Nanobowl Platform for the Study of Exosomes in Solution. Nanoscale 2015, 7, 9290–9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong S; Wang Y; Liu Z; Zhang W; Yi K; Zhang X; Zhang X; Jiang C; Yang S; Wang F Beehive-Inspired Macroporous SERS Probe for Cancer Detection through Capturing and Analyzing Exosomes in Plasma. ACS Appl Mater Interfaces 2020, 12, 5136–5146. [DOI] [PubMed] [Google Scholar]
- 48.Lee JU; Kim WH; Lee HS; Park KH; Sim SJ Quantitative and Specific Detection of Exosomal miRNAs for Accurate Diagnosis of Breast Cancer Using a Surface-Enhanced Raman Scattering Sensor Based on Plasmonic Head-flocked Gold Nanopillars. Small 2019, 15, e1804968. [DOI] [PubMed] [Google Scholar]
- 49.Wang Z; Zong S; Wang Y; Li N; Li L; Lu J; Wang Z; Chen B; Cui Y Screening and Multiple Detection of Cancer Exosomes Using an SERS-Based Method. Nanoscale 2018, 10, 9053–9062. [DOI] [PubMed] [Google Scholar]
- 50.Pang Y; Shi J; Yang X; Wang C; Sun Z; Xiao R Personalized Detection of Circling Exosomal PD-L1 Based on Fe3O4@TiO2 Isolation and SERS Immunoassay. Biosens Bioelectron 2020, 148, 111800. [DOI] [PubMed] [Google Scholar]
- 51.Ning C-F; Wang L; Tian Y-F; Yin B-C; Ye B-C Multiple and Sensitive SERS Detection of Cancer-Related Exosomes Based on Gold–Silver Bimetallic Nanotrepangs. Analyst 2020, 145, 2795–2804. [DOI] [PubMed] [Google Scholar]
- 52.Li TD; Zhang R; Chen H; Huang ZP; Ye X; Wang H; Deng AM; Kong JL An Ultrasensitive Polydopamine Bi-Functionalized SERS Immunoassay for Exosome-Based Diagnosis and Classification of Pancreatic Cancer. Chem Sci 2018, 9, 5372–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tian Y-F; Ning C-F; He F; Yin B-C; Ye B-C Highly Sensitive Detection of Exosomes by SERS Using Gold Nanostar@ Raman Reporter@ Nanoshell Structures Modified with a Bivalent Cholesterol-Labeled DNA Anchor. Analyst 2018, 143, 4915–4922. [DOI] [PubMed] [Google Scholar]
- 54.Wang J; Wuethrich A; Sina AA; Lane RE; Lin LL; Wang Y; Cebon J; Behren A; Trau M Tracking Extracellular Vesicle Phenotypic Changes Enables Treatment Monitoring in Melanoma. Sci Adv 2020, 6, eaax3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pang Y; Wang C; Lu L; Wang C; Sun Z; Xiao R Dual-SERS Biosensor for One-Step Detection of MicroRNAs in Exosome and Residual Plasma of Blood Samples for Diagnosing Pancreatic Cancer. Biosens Bioelectron 2019, 130, 204–213. [DOI] [PubMed] [Google Scholar]
- 56.Reiner AT; Fossati S; Dostalek J Biosensor Platform for Parallel Surface Plasmon-Enhanced Epifluorescence and Surface Plasmon Resonance Detection. Sens Actuators B Chem 2018, 257, 594–601. [Google Scholar]
- 57.Min J; Son T; Hong JS; Cheah PS; Wegemann A; Murlidharan K; Weissleder R; Lee H; Im H Plasmon-Enhanced Biosensing for Multiplexed Profiling of Extracellular Vesicles. Adv Biosyst 2020, e2000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kretschmann E; Raether H Radiative Decay of Non Radiative Surface Plasmons Excited by Light. Zeitschrift für Naturforschung A 1968, 23, 2135–2136. [Google Scholar]
- 59.Hosseinkhani B; van den Akker N; D’Haen J; Gagliardi M; Struys T; Lambrichts I; Waltenberger J; Nelissen I; Hooyberghs J; Molin DGM; Michiels L Direct Detection of Nano-Scale Extracellular Vesicles Derived from Inflammation-Triggered Endothelial Cells Using Surface Plasmon Resonance. Nanomedicine 2017, 13, 1663–1671. [DOI] [PubMed] [Google Scholar]
- 60.Sina AA; Vaidyanathan R; Wuethrich A; Carrascosa LG; Trau M Label-Free Detection of Exosomes Using a Surface Plasmon Resonance Biosensor. Anal Bioanal Chem 2019, 411, 1311–1318. [DOI] [PubMed] [Google Scholar]
- 61.Carney RP; Hazari S; Rojalin T; Knudson A; Gao T; Tang Y; Liu R; Viitala T; Yliperttula M; Lam KS Targeting Tumor‐Associated Exosomes with Integrin‐Binding Peptides. Adv Biosyst 2017, 1, 1600038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Desplantes R; Lévêque C; Muller B; Lotierzo M; Ferracci G; Popoff M; Seagar M; Mamoun R; El Far O Affinity Biosensors Using Recombinant Native Membrane Proteins Displayed on Exosomes: Application to Botulinum Neurotoxin B Receptor. Sci Rep 2017, 7, 1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Skalska J; Oliveira FD; Figueira TN; Mello ÉO; Gomes VM; McNaughton-Smith G; Castanho MARB; Gaspar D Plant Defensin PvD1 Modulates the Membrane Composition of Breast Tumour-Derived Exosomes. Nanoscale 2019, 11, 23366–23381. [DOI] [PubMed] [Google Scholar]
- 64.Rupert DLM; Shelke GV; Emilsson G; Claudio V; Block S; Lässer C; Dahlin A; Lötvall JO; Bally M; Zhdanov VP; Höök F Dual-Wavelength Surface Plasmon Resonance for Determining the Size and Concentration of Sub-Populations of Extracellular Vesicles. Anal Chem 2016, 88, 9980–9988. [DOI] [PubMed] [Google Scholar]
- 65.Rupert DL; Lässer C; Eldh M; Block S; Zhdanov VP; Lotvall JO; Bally M; Höök F Determination of Exosome Concentration in Solution Using Surface Plasmon Resonance Spectroscopy. Anal Chem 2014, 86, 5929–5936. [DOI] [PubMed] [Google Scholar]
- 66.Zhu L; Wang K; Cui J; Liu H; Bu X; Ma H; Wang W; Gong H; Lausted C; Hood L; Yang G; Hu Z Label-Free Quantitative Detection of Tumor-Derived Exosomes through Surface Plasmon Resonance Imaging. Anal Chem 2014, 86, 8857–8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li JF; Li CY; Aroca RF Plasmon-enhanced Fluorescence Spectroscopy. Chem Soc Rev 2017, 46, 3962–3979. [DOI] [PubMed] [Google Scholar]
- 68.Bauch M; Toma K; Toma M; Zhang Q; Dostalek J Plasmon-Enhanced Fluorescence Biosensors: A Review. Plasmonics 2014, 9, 781–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang B; Pinsky BA; Ananta JS; Zhao S; Arulkumar S; Wan H; Sahoo MK; Abeynayake J; Waggoner JJ; Hopes C; Tang M; Dai H Diagnosis of Zika Virus Infection on a Nanotechnology Platform. Nat Med 2017, 23, 548–550. [DOI] [PubMed] [Google Scholar]
- 70.Li X; Kuznetsova T; Cauwenberghs N; Wheeler M; Maecker H; Wu JC; Haddad F; Dai H Autoantibody Profiling on a Plasmonic Nano-Gold Chip for the Early Detection of Hypertensive Heart Disease. Proc Natl Acad Sci U S A 2017, 114, 7089–7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu W; Wang L; Zhang R; Sun X; Huang L; Su H; Wei X; Chen CC; Lou J; Dai H; Qian K Diagnosis and Prognosis of Myocardial Infarction on a Plasmonic Chip. Nat Commun 2020, 11, 1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang R; Le B; Xu W; Guo K; Sun X; Su H; Huang L; Huang J; Shen T; Liao T; Liang Y; Zhang JXJ; Dai H; Qian K Magnetic “Squashing” of Circulating Tumor Cells on Plasmonic Substrates for Ultrasensitive NIR Fluorescence Detection. Small Methods 2019, 3, 1800474. [Google Scholar]
- 73.Park S; Xiao X; Min J; Mun C; Jung HS; Giannini V; Weissleder R; Maier SA; Im H; Kim D Self‐Assembly of Nanoparticle‐Spiked Pillar Arrays for Plasmonic Biosensing. Adv Funct Mater 2019, 29, 1904257. [Google Scholar]
- 74.Luan J; Seth A; Gupta R; Wang Z; Rathi P; Cao S; Gholami Derami H; Tang R; Xu B; Achilefu S; Morrissey JJ; Singamaneni S Ultrabright Fluorescent Nanoscale Labels for the Femtomolar Detection of Analytes with Standard Bioassays. Nat Biomed Eng 2020, 4, 518–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Battaglia TM; Masson JF; Sierks MR; Beaudoin SP; Rogers J; Foster KN; Holloway GA; Booksh KS Quantification of Cytokines Involved in Wound Healing Using Surface Plasmon Resonance. Anal Chem 2005, 77, 7016–7023. [DOI] [PubMed] [Google Scholar]
- 76.Lyon LA; Musick MD; Smith PC; Reiss BD; Peña DJ; Natan MJ Surface Plasmon Resonance of Colloidal Au-modified Gold Films. Sens Actuators B Chem 1999, 54, 118–124. [Google Scholar]
- 77.Mitchell JS; Wu Y; Cook CJ; Main L Sensitivity Enhancement of Surface Plasmon Resonance Biosensing of Small Molecules. Anal Biochem 2005, 343, 125–135. [DOI] [PubMed] [Google Scholar]
- 78.Law WC; Yong KT; Baev A; Prasad PN Sensitivity Improved Surface Plasmon Resonance Biosensor for Cancer Biomarker Detection Based on Plasmonic Enhancement. ACS Nano 2011, 5, 4858–4864. [DOI] [PubMed] [Google Scholar]
- 79.He L; Musick MD; Nicewarner SR; Salinas FG; Benkovic SJ; Natan MJ; Keating CD Colloidal Au-Enhanced Surface Plasmon Resonance for Ultrasensitive Detection of DNA Hybridization. J Am Chem Soc 2000, 122, 9071–9077. [Google Scholar]
- 80.Rothenhäusler B; Knoll W Surface–Plasmon Microscopy. Nature 1988, 332, 615–617. [Google Scholar]
- 81.Wong CL; Olivo M Surface Plasmon Resonance Imaging Sensors: A Review. Plasmonics 2014, 9, 809–824. [Google Scholar]
- 82.Thiel AJ; Frutos AG; Jordan CE; Corn RM; Smith LM In Situ Surface Plasmon Resonance Imaging Detection of DNA Hybridization to Oligonucleotide Arrays on Gold Surfaces. Anal Chem 1997, 69, 4948–4956. [Google Scholar]
- 83.Wegner GJ; Wark AW; Lee HJ; Codner E; Saeki T; Fang S; Corn RM Real-Time Surface Plasmon Resonance Imaging Measurements for the Multiplexed Determination of Protein Adsorption/Desorption Kinetics and Surface Enzymatic Reactions on Peptide Microarrays. Anal Chem 2004, 76, 5677–5684. [DOI] [PubMed] [Google Scholar]
- 84.Zeng Y; Hu R; Wang L; Gu D; He J; Wu S-Y; Ho H-P; Li X; Qu J; Gao BZ; Shao Y Recent Advances in Surface Plasmon Resonance Imaging: Detection Speed, Sensitivity, and Portability. Nanophotonics 2017, 6, 1017–1030. [Google Scholar]
- 85.Giebel K; Bechinger C; Herminghaus S; Riedel M; Leiderer P; Weiland U; Bastmeyer M Imaging of Cell/substrate Contacts of Living Cells with Surface Plasmon Resonance Microscopy. Biophys J 1999, 76, 509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang S; Shan X; Patel U; Huang X; Lu J; Li J; Tao N Label-Free Imaging, Detection, and Mass Measurement of Single Viruses by Surface Plasmon Resonance. Proc Natl Acad Sci U S A 2010, 107, 16028–16032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yu H; Shan X; Wang S; Chen H; Tao N Molecular Scale Origin of Surface Plasmon Resonance Biosensors. Anal Chem 2014, 86, 8992–8997. [DOI] [PubMed] [Google Scholar]
- 88.Yang Y; Shen G; Wang H; Li H; Zhang T; Tao N; Ding X; Yu H Interferometric Plasmonic Imaging and Detection of Single Exosomes. Proc Natl Acad Sci U S A 2018, 115, 10275–10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang KS; Im H; Hong S; Pergolini I; Del Castillo AF; Wang R; Clardy S; Huang CH; Pille C; Ferrone S; Yang R; Castro CM; Lee H; Del Castillo CF; Weissleder R Multiparametric Plasma EV Profiling Facilitates Diagnosis of Pancreatic Malignancy. Sci Transl Med 2017, 9, eaal3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Qiu G; Ng SP; Wu CM Differential Phase-Detecting Localized Surface Plasmon Resonance Sensor with Self-Assembly Gold Nano-Islands. Opt Lett 2015, 40, 1924–1927. [DOI] [PubMed] [Google Scholar]
- 91.Bathini S; Raju D; Badilescu S; Kumar A; Ouellette RJ; Ghosh A; Packirisamy M Nano-Bio Interactions of Extracellular Vesicles with Gold Nanoislands for Early Cancer Diagnosis. Research (Wash D C) 2018, 2018, 3917986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Raju D; Bathini S; Badilescu S; Ouellette RJ; Ghosh A; Packirisamy M LSPR Detection of Extracellular Vesicles Using a Silver-PDMS Nano-Composite Platform Suitable for Sensor Networks. Enterp Inf Syst 2020, 14, 532–541. [Google Scholar]
- 93.Jiang Y; Shi M; Liu Y; Wan S; Cui C; Zhang L; Tan W Aptamer/AuNP Biosensor for Colorimetric Profiling of Exosomal Proteins. Angew Chem Int Ed Engl 2017, 56, 11916–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Y; Wang D; Yue S; Lu Y; Yang C; Fang J; Xu Z Sensitive Multicolor Visual Detection of Exosomes via Dual Signal Amplification Strategy of Enzyme-Catalyzed Metallization of Au Nanorods and Hybridization Chain Reaction. ACS Sens 2019, 4, 3210–3218. [DOI] [PubMed] [Google Scholar]
- 95.Tirinato L; Gentile F; Di Mascolo D; Coluccio ML; Das GLC; Pullano SAP,G; Francardi M; Accardo A; De Angelis F; Candeloro P; Di Fabrizio E SERS Analysis on Exosomes Using Super-Hydrophobic Surfaces. Microelectron Eng 2012, 97, 337–340. [Google Scholar]
- 96.Park J; Hwang M; Choi B; Jeong H; Jung JH; Kim HK; Hong S; Park JH; Choi Y Exosome Classification by Pattern Analysis of Surface-Enhanced Raman Spectroscopy Data for Lung Cancer Diagnosis. Anal Chem 2017, 89, 6695–6701. [DOI] [PubMed] [Google Scholar]
- 97.Shin H; Jeong H; Park J; Hong S; Choi Y Correlation between Cancerous Exosomes and Protein Markers Based on Surface-Enhanced Raman Spectroscopy (SERS) and Principal Component Analysis (PCA). ACS Sens 2018, 3, 2637–2643. [DOI] [PubMed] [Google Scholar]
- 98.Yan Z; Dutta S; Liu Z; Yu X; Mesgarzadeh N; Ji F; Bitan G; Xie Y-H A Label-Free Platform for Identification of Exosomes from Different Sources. ACS Sens 2019, 4, 488–497. [DOI] [PubMed] [Google Scholar]
- 99.Ferreira N; Marques A; Águas H; Bandarenka H; Martins R; Bodo C; Costa-Silva B; Fortunato E Label-Free Nanosensing Platform for Breast Cancer Exosome Profiling. ACS Sens 2019, 4, 2073–2083. [DOI] [PubMed] [Google Scholar]
- 100.Lee C; Carney R; Lam K; Chan JW SERS Analysis of Selectively Captured Exosomes Using an Integrin‐Specific Peptide Ligand. J Raman Spectrosc 2017, 48, 1771–1776. [Google Scholar]
- 101.Zhang X; Liu C; Pei Y; Song W; Zhang S Preparation of a Novel Raman Probe and Its Application in the Detection of Circulating Tumor Cells and Exosomes. ACS Appl Mater Interfaces 2019, 11, 28671–28680. [DOI] [PubMed] [Google Scholar]
- 102.Chen H; Luo C; Yang M; Li J; Ma P; Zhang X Intracellular Uptake of and Sensing with SERS-Active Hybrid Exosomes: Insight into a Role of Metal Nanoparticles. Nanomedicine (Lond) 2020, 15, 913–926. [DOI] [PubMed] [Google Scholar]
- 103.Kwizera EA; O’Connor R; Vinduska V; Williams M; Butch ER; Snyder SE; Chen X; Huang X Molecular Detection and Analysis of Exosomes Using Surface-Enhanced Raman Scattering Gold Nanorods and a Miniaturized Device. Theranostics 2018, 8, 2722–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zong S; Wang L; Chen C; Lu J; Zhu D; Zhang Y; Wang Z; Cui Y Facile Detection of Tumor-Derived Exosomes Using Magnetic Nanobeads and SERS Nanoprobes. Anal Methods 2016, 8, 5001–5008. [Google Scholar]
- 105.Knoblauch R; Geddes CD Review of Advances in Metal-Enhanced Fluorescence. In Reviews in Plasmonics 2017; Geddes CD, Ed.; Springer, Cham: Switzerland, 2019; pp 253–283. [Google Scholar]
- 106.Geddes CD; Lakowicz JR Editorial: Metal-Enhanced Fluorescence. J Fluoresc 2002, 12, 121–129. [Google Scholar]
- 107.Kinkhabwala A; Yu Z; Fan S; Avlasevich Y; Müllen K; Moerner WE Large Single-Molecule Fluorescence Enhancements Produced by a Bowtie Nanoantenna. Nat Photon 2009, 3, 654–657. [Google Scholar]
- 108.Tabakman SM; Lau L; Robinson JT; Price J; Sherlock SP; Wang H; Zhang B; Chen Z; Tangsombatvisit S; Jarrell JA; Utz PJ; Dai H Plasmonic Substrates for Multiplexed Protein Microarrays with Femtomolar Sensitivity and Broad Dynamic Range. Nat Commun 2011, 2, 466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wan H; Merriman C; Atkinson MA; Wasserfall CH; Mcgrail KM; Liang Y; Fu D; Dai H Proteoliposome-Based Full-Length ZnT8 Self-Antigen for Type 1 Diabetes Diagnosis on a Plasmonic Platform. Proc Natl Acad Sci U S A 2017, 114, 10196–10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qiu G; Thakur A; Xu C; Ng S-P; Lee Y; Wu C-ML Detection of Glioma-Derived Exosomes with the Biotinylated Antibody-Functionalized Titanium Nitride Plasmonic Biosensor. Adv Funct Mater 2019, 29, 1806761. [Google Scholar]
- 111.Shu S; Yang Y; Allen CL; Hurley E; Tung KH; Minderman H; Wu Y; Ernstoff MS Purity and Yield of Melanoma Exosomes Are Dependent on Isolation Method. J Extracell Vesicles 2020, 9, 1692401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou J; Rossi J Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat Rev Drug Discov 2017, 16, 181–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Butler D Translational Research: Crossing the Valley of Death. Nature 2008, 453, 840–842. [DOI] [PubMed] [Google Scholar]
- 114.Seyhan AA Lost in Translation: The Valley of Death across Preclinical and Clinical Divide–Identification of Problems and Overcoming Obstacles. Transl Med Commun 2019, 4, 1–19. [Google Scholar]
- 115.Street JM; Koritzinsky EH; Glispie DM; Yuen PS T. Urine Exosome Isolation and Characterization. Methods Mol Biol 2017, 1641, 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Keller S; Ridinger J; Rupp AK; Janssen JW; Altevogt P Body Fluid Derived Exosomes as a Novel Template for Clinical Diagnostics. J Transl Med 2011, 9, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Momen-Heravi F; Balaj L; Alian S; Trachtenberg AJ; Hochberg FH; Skog J; Kuo WP Impact of Biofluid Viscosity on Size and Sedimentation Efficiency of the Isolated Microvesicles. Front Physiol 2012, 3, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Soares Martins T; Catita J; Martins Rosa I; A B da Cruz E Silva O; Henriques AG Exosome Isolation from Distinct Biofluids Using Precipitation and Column-Based Approaches. PLoS One 2018, 13, e0198820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.LeBleu VS; Kalluri R Exosomes as a Multicomponent Biomarker Platform in Cancer. Trends Cancer 2020, 6, 767–774. [DOI] [PubMed] [Google Scholar]
- 120.Giannopoulou L; Zavridou M; Kasimir-Bauer S; Lianidou ES Liquid Biopsy in Ovarian Cancer: The Potential of Circulating miRNAs and Exosomes. Transl Res 2019, 205, 77–91. [DOI] [PubMed] [Google Scholar]
- 121.Jeong S; Park J; Pathania D; Castro CM; Weissleder R; Lee H Integrated Magneto-Electrochemical Sensor for Exosome Analysis. ACS Nano 2016, 10, 1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shu W; Wang Y; Liu C; Li R; Pei C; Lou W; Lin S; Di W; Wan J Construction of a Plasmonic Chip for Metabolic Analysis in Cervical Cancer Screening and Evaluation. Small Methods 2020, 4, 1900469. [Google Scholar]
- 123.Yang J; Wang R; Huang L; Zhang M; Niu J; Bao C; Shen N; Dai M; Guo Q; Wang Q; Wang Q; Fu Q; Qian K Urine Metabolic Fingerprints Encode Subtypes of Kidney Diseases. Angew Chem Int Ed Engl 2020, 132, 1720–1727. [DOI] [PubMed] [Google Scholar]
- 124.Vedarethinam V; Huang L; Xu W; Zhang R; Gurav DD; Sun X; Yang J; Chen R; Qian K Detection and Inhibition of Bacteria on a Dual-Functional Silver Platform. Small 2019, 15, e1803051. [DOI] [PubMed] [Google Scholar]
- 125.Samarah LZ; Vertes A Mass Spectrometry Imaging Based on Laser Desorption Ionization from Inorganic and Nanophotonic Platforms. View 2020, 20200063. [Google Scholar]
- 126.Cao J; Shi X; Gurav DD; Huang L; Su H; Li K; Niu J; Zhang M; Wang Q; Jiang M; Qian K Metabolic Fingerprinting on Synthetic Alloys for Medulloblastoma Diagnosis and Radiotherapy Evaluation. Adv Mater 2020, 32, e2000906. [DOI] [PubMed] [Google Scholar]
- 127.Hayes J; Peruzzi PP; Lawler S MicroRNAs in Cancer: Biomarkers, Functions and Therapy. Trends Mol Med 2014, 20, 460–469. [DOI] [PubMed] [Google Scholar]
- 128.Tsao SC; Wang J; Wang Y; Behren A; Cebon J; Trau M Characterising the Phenotypic Evolution of Circulating Tumour Cells during Treatment. Nat Commun 2018, 9, 1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gaspar R; Pallbo J; Weininger U; Linse S; Sparr E Ganglioside Lipids Accelerate A-Synuclein Amyloid Formation. Biochim Biophys Acta Proteins Proteom 2018, 1866, 1062–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Marconi S; De Toni L; Lovato L; Tedeschi E; Gaetti L; Acler M; Bonetti B Expression of Gangliosides on Glial and Neuronal Cells in Normal and Pathological Adult Human Brain. J Neuroimmunol 2005, 170, 115–121. [DOI] [PubMed] [Google Scholar]
- 131.Rushworth JV; Hooper NM Lipid Rafts: Linking Alzheimer’s Amyloid-Β Production, Aggregation, and Toxicity at Neuronal Membranes. J Alzheimer’s Dis 2011, 2011, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lleó A; Cavedo E; Parnetti L; Vanderstichele H; Herukka SK; Andreasen N; Ghidoni R; Lewczuk P; Jeromin A; Winblad B; Tsolaki M; Mroczko B; Visser PJ; Santana I; Svenningsson P; Blennow K; Aarsland D; Molinuevo JL; Zetterberg H; Mollenhauer B Cerebrospinal Fluid Biomarkers in Trials for Alzheimer and Parkinson Diseases. Nat Rev Neurol 2015, 11, 41–55. [DOI] [PubMed] [Google Scholar]
- 133.Villemagne VL; Doré V; Burnham SC; Masters CL; Rowe CC Imaging Tau and Amyloid-Β Proteinopathies in Alzheimer Disease and Other Conditions. Nat Rev Neurol 2018, 14, 225–236. [DOI] [PubMed] [Google Scholar]
- 134.Benjamin EJ; Muntner P; Alonso A; Bittencourt MS; Callaway CW; Carson AP; Chamberlain AM; Chang AR; Cheng S; Das SR; Delling FN; Djousse L; Elkind MSV; Ferguson JF; Fornage M; Jordan LC; Khan SS; Kissela BM; Knutson KL; Kwan TW et al. Heart Disease and Stroke Statistics-2019 Update: A Report from the American Heart Association. Circulation 2019, 139, e56–e528. [DOI] [PubMed] [Google Scholar]
- 135.Chen YH; Du W; Hagemeijer MC; Takvorian PM; Pau C; Cali A; Brantner CA; Stempinski ES; Connelly PS; Ma HC; Jiang P; Wimmer E; Altan-Bonnet G; Altan-Bonnet N Phosphatidylserine Vesicles Enable Efficient En Bloc Transmission of Enteroviruses. Cell 2015, 160, 619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Altan-Bonnet N; Chen YH Intercellular Transmission of Viral Populations with Vesicles. J Virol 2015, 89, 12242–12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang Y; Han Q; Hou Z; Zhang C; Tian Z; Zhang J Exosomes Mediate Hepatitis B Virus (HBV) Transmission and NK-Cell Dysfunction. Cell Mol Immunol 2017, 14, 465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Elrashdy F; Aljaddawi AA; Redwan EM; Uversky VN On the Potential Role of Exosomes in the Covid-19 Reinfection/Reactivation Opportunity. J Biomol Struct Dyn 2020, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cai M; He H; Jia X; Chen S; Wang J; Shi Y; Liu B; Xiao W; Lai S Genome-Wide MicroRNA Profiling of Bovine Milk-Derived Exosomes Infected with Staphylococcus Aureus. Cell Stress Chaperones 2018, 23, 663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang H; Liu J; Qu D; Wang L; Wong CM; Lau CW; Huang Y; Wang YF; Huang H; Xia Y; Xiang L; Cai Z; Liu P; Wei Y; Yao X; Ma RCW; Huang Y Serum Exosomes Mediate Delivery of Arginase 1 as a Novel Mechanism for Endothelial Dysfunction in Diabetes. Proc Natl Acad Sci U S A 2018, 115, E6927–E6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.van Balkom BW; Pisitkun T; Verhaar MC; Knepper MA Exosomes and the Kidney: Prospects for Diagnosis and Therapy of Renal Diseases. Kidney Int 2011, 80, 1138–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Taylor DD; Gercel-Taylor C Exosome Platform for Diagnosis and Monitoring of Traumatic Brain Injury. Philos Trans R Soc Lond B Biol Sci 2014, 369, 20130503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Song H; Zhu Y The in Vitro Diagnostics Industry in China. View 2020, 1, e5. [Google Scholar]
- 144.Liu R; Fu A; Deng Z; Li Y; Liu T Promising Methods for Detection of Novel Coronavirus SARS-CoV-2. View 2020, 1, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Moitra P; Alafeef M; Dighe K; Frieman MB; Pan D Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Qiu G; Gai Z; Tao Y; Schmitt J; Kullak-Ublick GA; Wang J Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. [DOI] [PubMed] [Google Scholar]