Abstract
The human lung cell A549 is susceptible to infection with a number of respiratory viruses. However, A549 cells are resistant to Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2) infection in conventional submerged culture, and this would appear to be due to low expression levels of the SARS-CoV-2 entry receptor: angiotensin-converting enzyme-2 (ACE2). Here, we examined SARS-CoV-2 susceptibility to A549 cells after adaptation to air-liquid interface (ALI) culture. A549 cells in ALI culture yielded a layer of mucus on their apical surface, exhibited decreased expression levels of the proliferation marker KI-67 and intriguingly became susceptible to SARS-CoV-2 infection. We found that A549 cells increased the endogenous expression levels of ACE2 and TMPRSS2 following adaptation to ALI culture conditions. Camostat, a TMPRSS2 inhibitor, reduced SARS-CoV-2 infection in ALI-cultured A549 cells. These findings indicate that ALI culture switches the phenotype of A549 cells from resistance to susceptibility to SARS-CoV-2 infection through upregulation of ACE2 and TMPRSS2.
Keywords: SARS-CoV-2, A549 cells, Air-liquid interface, ACE2, TMPRSS2, Viral entry
1. Introduction
Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2) causes severe respiratory diseases in humans. The virus spike (S) protein is a glycoprotein localized on SARS-CoV-2 virion surface and triggers viral entry and membrane fusion in target cells through the interaction with host factors: entry receptors and proteases [1]. To date, two type I transmembrane proteins have been identified as entry receptors for SARS-CoV-2: angiotensin-converting enzyme-2 (ACE2) and neuropilin-1 (NRP1) [[2], [3], [4]]. In particular, the interaction between ACE2 and S protein has attracted attention, because emerging SARS-CoV-2 variants possess amino acid substitutions in the receptor binding domain of S protein which can increase the binding affinity to human ACE2 [5]. In addition, the exogenous expression of ACE2 confers SARS-CoV-2 susceptibility to cells which are normally resistant to infection [6,7]; therefore, ACE2 is a major determinant of SARS-CoV-2 susceptibility. On the other hand, certain host proteases proteolytically activate (or prime) S protein for membrane fusion. For example, TMPRSS2 is a host serine protease which act as a priming enzyme of SARS-CoV-2 S protein. TMPRSS2 mediates direct fusion of SARS-CoV-2 virion on the plasma membrane and facilitates SARS-CoV-2 entry [2,8]. Collectively, cells expressing both ACE2 and TMPRSS2 are highly permissive to SARS-CoV-2 entry.
Airway epithelial cells are positive for ACE2/TMPRSS2 and are primary targets of in vivo infection of SARS-CoV-2 [9,10]. Primary human airway epithelial cells can be cultured and used for basic research studies and antiviral development [[11], [12], [13]]. In experiments, primary airway epithelial cells are differentiated and maintained under an air-liquid interface under (ALI) culture condition, which allows cells to grow on microporous membranes in culture inserts with air exposure and culture medium through the basolateral side. It has been reported that 96% of expressed genes represent similar expression patterns between brushed biopsy of nasal epithelial cells and those cultured under ALI conditions [14]. In comparison with conventional submerged culture condition where cells are grown on a plastic surface with overlayed culture medium, primary airway epithelial cells in ALI culture recapitulate the transcriptional profile of in vivo airway epithelia [15]. The ALI culture method is a more physiologically relevant way for infection of primary airway epithelial cells and provides an appropriate ex vivo respiratory model.
A549 and Calu-3 cells are human lung-derived epithelial cells and widely used for studies of virus infection. In contrast to Calu-3 cells, only limited entry and growth of SARS-CoV-2 occurs in A549 cells [7]. The decreased susceptibility of A549 cells to SARS-CoV-2 infection was attributed to the low expression level of ACE2 and the retinoic acid-inducible gene-I (RIG-I)-mediated antiviral response [1,16]. These studies were performed by inoculation of SARS-CoV-2 to cells under submerged culture conditions. In contrast, these cell lines can be cultured and differentiated under ALI conditions. ALI-cultured A549 cells are used for cell-based toxicity assays by inhalation exposure of exhausts, nanoparticles and chemical compounds [[17], [18], [19]]. A549 cells exert different expression profiles of genes involved in cellular processes such as those in the cell cycle, DNA damage repair and transporters between in submerged and ALI cultures [20,21]. In the present study, we compared the susceptibility of A549 cells to SARS-CoV-2 infection under these culture conditions.
2. Materials and methods
2.1. Cells
Calu-3 cells (ATCC, Manassas, VA) were maintained in Eagle's Minimum Essential Medium (MEM) supplemented with 10% fetal bovine serum (FBS). A549 (RIKEN BRC, Tsukuba, Japan) and Vero-TMPRSS2 [22] cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% FBS. All cells were incubated at 37 °C with 5% CO2. Generation of human ACE2-and TMPRSS2-expressing cells were described previously [22]. A549 cells stably expressing ACE2 and TMPRSS2 were selected in the presence of puromycin (for ACE2) or blasticidin S (for TMPRSS2).
2.2. Viruses
SARS-CoV-2 WK-521 strain (EPI_ISL_408667) were provided by Dr. Saijyo and Dr. Shimojima (National Institute of Infectious Diseases, Tokyo, Japan). The original stock was prepared by inoculation of Vero-TMPRSS2 cells and titrated by plaque assay as previously described [22].
2.3. Immunoblotting
Cells were lysed in lysis buffer (1% NP-40; 20 mM Tris-HCl, pH 7.5; 150 mM NaCl; and 5 mM EDTA) supplemented with cOmplete ULTRA protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN). Cell lysate was analyzed by SDS-PAGE and immunoblotting using anti-ACE2 (4355T, Cell Signaling Technology, Danvers, MA) or anti-TMPRSS2 (ab92323, Abcam, Cambridge, UK) antibodies. HRP-conjugated anti-β-actin antibody (PM053-7, MBL, Tokyo, Japan) was used to detect the loading control. Immune complexes were detected using HRP-conjugated anti-rabbit IgG secondary antibody and the Immobilon Western Chemiluminescent HRP Substrate (Millipore; Merck, Darmstadt, Germany).
2.4. A549 cell culture in the air-liquid interface (ALI)
Millicell culture inserts for 24-well plate with a 0.4 μm pore size polyethylene terephthalate (PET) membrane and 0.3 cm2 culture area (Millipore; Merck) were coated with type I collagen. A549 cells were seeded onto the insert at 100,000 cells and cultured in DMEM supplemented with 10% FBS for 24 h. Culture medium in the apical and basal chambers were then replaced with PneumaCult ALI medium (STEMCELL Technologies, Vancouver, Canada). On day 3 after seeding, the cells were subjected to air-lift by removing medium from both the apical and basal chambers but supplying fresh PneumaCult ALI medium only in the basal chamber. Cells were maintained in ALI for at least an additional 21 days. The culture medium in the basal chamber was changed every 3 days and the apical area was washed with the culture medium every 7 days.
2.5. Cell staining
A549 cells in the culture insert were fixed with 10% phosphate-buffered formalin before ALI culture or after ALI culture for 21 days. Clipped cells with PET membranes were embedded in paraffin, sectioned at 4 μm-thickness and mounted on Platinum PRO micro slide glass (Matsunami Glass, Osaka, Japan). Slides were stained with hematoxylin and eosin (H&E) and periodic acid-schiff (PAS) staining (MUTO Pure Chemicals, Tokyo, Japan).
2.6. Multi-cycle growth of SARS-CoV-2
For assays in submerged culture, cells in 24-well plates were inoculated with SARS-CoV-2 at a multiplicity of infection (MOI) of 1 (Fig. 1 A), 10 (Fig. 1A–C) or 0.1 (Fig. 1G and H) for 1 h. Cells were then washed with PBS to remove unabsorbed viruses and cultured in fresh medium with 2% FBS. The culture supernatants for virus titration were harvested at the indicated time points. For assays in ALI culture, ALI-cultured A549 cells were infected at the apical surface with SARS-CoV-2 at 50,000 plaque forming unit (pfu) in 100 μl of culture medium. After 1 h of incubation, the apical area of cells was washed twice with PBS, and then cells were maintained under ALI culture conditions. At the indicated time points, the mucus fluids containing progeny viruses in the apical area were suspended by incubation with 200 μl of culture medium for 30 min. The harvested fluids were subjected to virus titration.
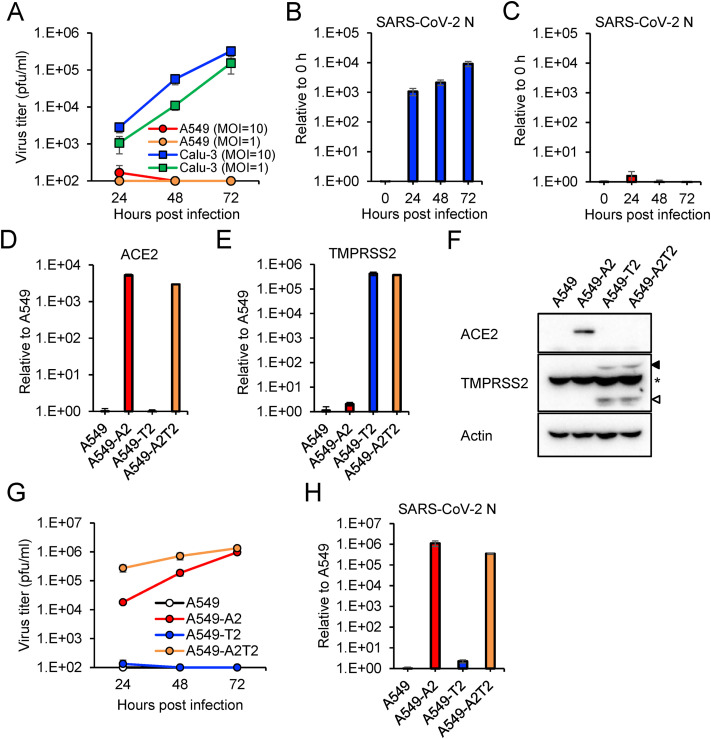
Fig. 1.
Infectivity of SARS-CoV-2 in A549 cells under submerged culture condition
(A) Growth curves of SARS-CoV-2 in A549 and Calu-3 cells. Viral titers in the culture supernatant were determined using a plaque assay. (B and C) SARS-CoV-2 RNA levels in Calu-3 cells (B) and A549 cells (C) were quantified by qRT-PCR and were normalized to the expression levels of β-actin. (D and E) Expression levels of exogenously transduced ACE2 (D) and TMPRSS2 (E) were quantified by qRT-PCR and normalized to the expression levels of β-actin. (F) Expression of exogenously transduced proteins was detected by Western Blotting with antibodies specific to the proteins indicated on the left. The full-length and cleaved forms of TMPRSS2 are indicated by close and open arrowheads, respectively. Asterisks indicate nonspecific bands that cross-reacted with anti-TMPRSS2 antibody. (G) Growth curves of SARS-CoV-2 in A549 cells expressing ACE2 and TMPRSS2. Viral titers in the culture supernatant were determined using a plaque assay. (H) The levels of SARS-CoV-2 N gene at 72 hpi were quantified by qRT-PCR and normalized to the expression levels of β-actin. The values in the graphs are expressed as the mean ± SD of triplicate samples.
2.7. Quantitative reverse-transcription PCR (qRT-PCR)
Total RNA was extracted from cells with TRIzol (Invitrogen; Themo Fisher Scientific, Waltham, MA) and Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA). The RNA samples were analyzed by qRT-PCR with Thunderbird Probe One-step Probe qRT-PCR Kit (Toyobo, Osaka, Japan). Probe and primers targeting SARS-CoV-2 nucleocapsid (N) gene were used for viral RNA quantification and described previously as N2 set [23]. The following Taqman Gene Expression Assays (Applied Biosystems; Thermo Fisher Scientific) consisting of predesigned probe and primers were used for host gene expression analysis: KI-67 (Hs01032443_m1), ACE2 (Hs01085333_m1), NRP1 (Hs00826128_m1), Cathepsin L (Hs00964650_m1), TMPRSS2 (Hs00237175_m1) and β-actin (Hs99999903_m1). Negative samples were included in the analysis with Ct values set at the limit of detection value (Ct = 50). RNA levels were normalized to β-actin and calculated by the ΔΔCt method.
2.8. Virus infection assay with the TMPRSS2 inhibitor, camostat mesylate
ALI-cultured A549 cells were infected at the apical surface with SARS-CoV-2 at 250,000 pfu in the presence of 50 μM camostat mesylate (FUJIFILM Wako Pure chemical, Osaka, Japan) for 2 h. The apical area of cells was washed three times with PBS and then cells were maintained under ALI culture for 4 h. At 6 h post-infection (hpi), cells were mixed with TRIzol reagent and subjected to RNA extraction and qRT-PCR assay as described above.
3. Results
3.1. The susceptibility of A549 cells to SARS-CoV-2 infection
A549 and Calu-3 cells were cultured in conventional submerged condition and inoculated with SARS-CoV-2 at different MOIs. The infectious virus titers and virus RNA levels were increased in Calu-3 cells infected with SARS-CoV-2 in a time-dependent manner (Fig. 1A and B). In contrast, SARS-CoV-2 replication was barely detectable in A549 cells even at a high MOI using virus titration and qRT-PCR assays (Fig. 1A and C). It has been reported that exogenous expression of ACE2 renders A549 cells susceptible to SARS-CoV-2 infection [[24], [25], [26]]. We also generated A549 cells with overexpression of ACE2 and/or TMPRSS2 (A549-A2, -T2 and -A2T2 cells). The enhanced expression of the transduced genes was validated by qRT-PCR and immunoblotting (Fig. 1D–F). Immunoblotting analysis revealed that the full-length of ACE2 protein was present in the lysate of A549-A2 but not in that of A549-A2T2 (Fig. 1F), consistent with a previous report that ACE2 is processed by TMPRSS2 but maintains its receptor activity for SARS-CoV [27]. SARS-CoV-2 replication was observed exclusively in ACE2-overexpressing A549 cells (A549-A2 and A549-A2T2 cells) (Fig. 1G and H). The virus titers in A549-A2T2 cells were higher than those in A549-A2 at 24 and 48 hpi (Fig. 1G), indicating that TMPRSS2 facilitates SARS-CoV-2 infection in the presence of ACE2 in A549 cells.
3.2. The susceptibility of A549 cells under ALI-culture to SARS-CoV-2 infection
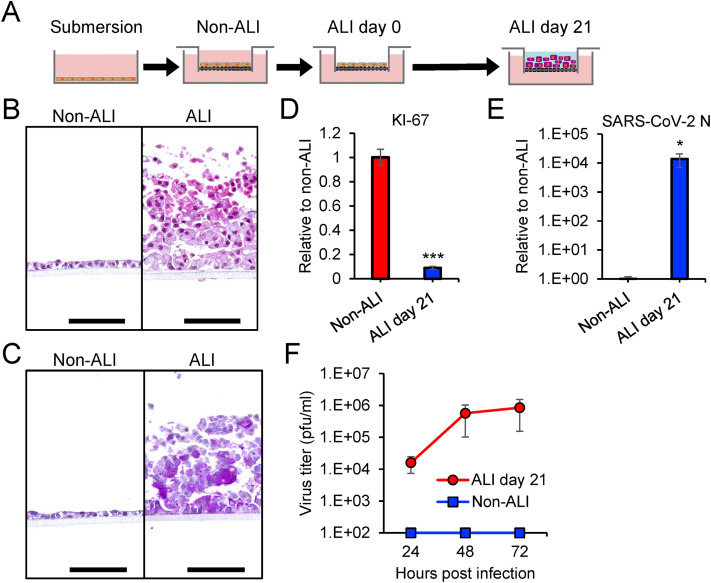
Primary human nasal and bronchial epithelial cells can be differentiated under ALI culture conditions. The differentiated epithelial cells are susceptible to SARS-CoV-2 infection and used as an ex vivo model to study SARS-CoV-2 infection in the human airway [[11], [12], [13]]. We examined the impact of ALI culture on the susceptibility of A549 cells to SARS-CoV-2 infection. A549 cells were seeded onto culture inserts and fed commercial differentiation medium only in the basal chamber for ALI culture (Fig. 2 A). A549 cells formed a monolayer in the insert under submerged culture with maintenance medium of DMEM containing 10% FBS (non-ALI), while the cells increased in number with a multilayered structure in ALI culture (Fig. 2B). A549 cells in ALI culture yielded a layer of mucus which was synthesized by the cells and detected by PAS staining (Fig. 2C). The expression levels of the proliferation marker KI-67 was markedly decreased in A549 cells in ALI culture (Fig. 2D) as previously reported [18]. SARS-CoV-2 was inoculated onto the apical surface of A549 cells adapted to ALI culture (ALI day 21). A549 cells under submerged culture (non-ALI) was used as a control. Viral replication and progeny viruses were observed only in A549 cells under ALI conditions (Fig. 2E and F), indicating that ALI culture confers susceptibility of SARS-CoV-2 to infection of A549 cells.
Fig. 2.
Infectivity of SARS-CoV-2 in A549 cells under air-liquid interface culture condition
(A) Schematic representation of the ALI culture system. A549 cells were seeded onto a culture insert with growth medium (non-ALI condition). The medium at the apical chamber was then removed to start the ALI culture (ALI day 0). Cells were maintained for another 21 days by supplying ALI medium to the basal chamber (ALI day 21). (B and C) A549 cells in non-ALI and ALI day 21 were examined by hematoxylin and eosin (H&E) (B) and periodic acid-schiff (PAS) staining (C). Mucus in cells displayed purple color after PAS staining. Scale bars = 100 μm. (D and E) Relative levels of KI-67 (D) and SARS-CoV-2 N (E) were measured by qRT-PCR and normalized to the expression levels of β-actin. (F) Growth curves of SARS-CoV-2 in A549 cells in non-ALI and ALI culture conditions. The mucus fluids in the apical area were suspended in culture medium and subjected to viral titration using a plaque assay. The values in the graphs are expressed as the mean ± SD of triplicate samples. ∗p < 0.05, ∗∗∗p < 0.001, two-tailed Student's t-test. . (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Upregulated expression of ACE2 and TMPRSS2 in A549 cells in ALI-culture
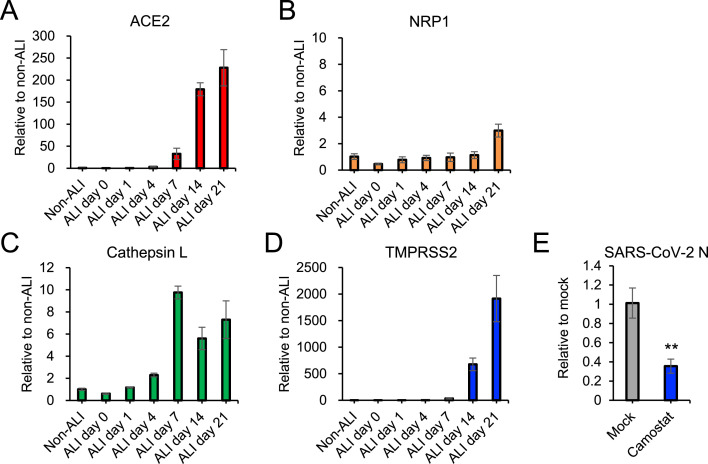
ALI-culture reportedly upregulates the expression of ACE2 in primary human tracheobronchial epithelial cells [28]. Therefore, we compared the expression levels of ACE2 between A549 cells in submerged non-ALI and ALI cultures. After 21 days of ALI culture, the ACE2 level was markedly elevated more than 200-fold above non-ALI culture (Fig. 3 A). In contrast, a consistent expression level of another SARS-CoV-2 entry receptor, NRP1, was observed in A549 cells through the culture period (from day 0 to day 21, Fig. 3B). We also examined the expression levels of host proteases, cathepsin and TMPRSS2, which are responsible for SARS-CoV-2 S protein priming in membrane fusion. The increased expression of both proteases was observed in ALI-cultured A549 cells (Fig. 3C and D). Notably, TMPRSS2 expression levels were more than three orders of magnitude higher after 21 days of ALI culture (Fig. 3D). In order to investigate the impact of increased expression of TMPRSS2 on SARS-CoV-2 entry, we examined the inhibitory effect of camostat, a TMPRSS2 inhibitor, on SARS-CoV-2 entry into the cells. Camostat inhibited viral entry into ALI-cultured A549 cells (Fig. 3E), indicating that TMPRSS2 facilitates SARS-CoV-2 entry in ALI-cultured A549 cells.
Fig. 3.
Gene expression levels of host factors involved in SARS-CoV-2 entry
(A-D) Relative expression levels of ACE2 (A), NRP1 (B), cathepsin L (C) and TMPRSS2 (D) in A549 cells during the adaptation to ALI culture were measured by qRT-PCR. β-actin mRNA levels were used as the reference control. (E) ALI-cultured A549 cells were infected with SARS-CoV-2 in the presence of 50 μM camostat. Viral RNA levels at 6 hpi were measured by qRT-PCR and normalized to the expression levels of β-actin. The values in the graphs are expressed as the mean ± SD of triplicate samples. ∗∗p < 0.01, two-tailed Student's t-test.
4. Discussion
ALI culture conditions allow the differentiation of primary airway epithelial cells under the similar condition as the physiological state. Human lung-derived cells A549 and Calu-3 can also be differentiated under ALI culture and are used as models for toxicological and pharmacological studies with exhausts, nanoparticles and chemical compounds [[17], [18], [19]]. Here, we applied ALI-cultured A549 cells for experiments with SARS-CoV-2. In contrast to A549 cells in submerged culture, A549 cells in ALI-culture permitted SARS-CoV-2 infection and yielded sufficient level of progeny virus without exogenous gene expression. These results indicated that ALI-culture switches the phenotype of A549 cells from resistance to susceptibility to SARS-CoV-2 infection.
A549 is a human lung carcinoma-derived cell line and widely used in virological studies. This cell line showed limited endogenous ACE2 expression level and was non-permissive to SARS-CoV-2 infection under submerged culture conditions (Fig. 1F–H) [7]. Our study and other studies demonstrated that exogenous expression of human ACE2 mediates SARS-CoV-2 entry and renders A549 cells permissive for SARS-CoV-2 infection [[24], [25], [26]]. Upregulation of endogenous ACE2 level is the possible primary cause of the susceptibility of ALI-cultured A549 cells to SARS-CoV-2 infection. ALI culture also increased the expression level of TMPRSS2 in A549 cells, which activates the S protein and facilitates cellular entry of SARS-CoV-2. Since camostat, a TMPRSS2 inhibitor, reduced SARS-CoV-2 entry into ALI-cultured A549 cells, TMPRSS2 plays a role in SARS-CoV-2 infection in the cells.
One limitation of our study is that we could not infer the mechanism of ACE2 and TMPRSS2 upregulations under ALI culture conditions. In contrast to ACE2 and TMPRSS2, the level of another SARS-CoV-2 receptor NRP1 was consistent between cells in submerged- and ALI-culture. ACE2 expression is reportedly upregulated by ALI culture in primary human tracheobronchial epithelial cells [28]. These findings indicate that ALI culture increases the expression levels of specific host factors involved in SARS-CoV-2 infection and this event is not specific for the A549 cell line. Qiao et al. have currently proposed that the androgen receptor enhances expression levels of both ACE2 and TMPRSS2 in lung epithelial cells through the binding to their promoter regions [29]. Investigation of the relationship between androgen receptor and ACE2/TMPRSS2 expression event in A549 cells may provide insights into the mechanism of ACE2 and TMPRSS2 upregulations in ALI culture.
In conclusion, this study demonstrated that ALI culture confers SARS-CoV-2 susceptibility to A549 cells. Unlike primary human airway cells, ALI-cultured A549 cells are free from donor variation and established easily. ALI-cultured A549 cells thus could be used as an alternative model to primary human airway cells for investigation of the inhaled exposure of SARS-CoV-2 and the evaluation of antivirals against SARS-CoV-2.
Declaration of competing interest
Kentaro Uemura, Shinsuke Toba, Takao Sanaki and Akihiko Sato are employees of Shionogi & Co., Ltd. Other authors declare no competing interests.
Acknowledgments
We thank Drs. Saijyo, Shimojima and Ito at National Institute of Infectious Diseases, Japan for providing SARS-CoV-2 WK-521 strain. This work was supported by the Japan Agency for Medical Research and Development (AMED), Japan (PJ21fk0108104, JP21wm0125008, PJ20fk0108509), Japan Science and Technology Agency (JST) Moonshot R&D, Japan (JPMJMS2025), the World-leading Innovative and Smart Education (WISE) Program (1801) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and the fund from the Atlantic Philanthropies Director/Employee Designated Gift Program.
References
- 1.Murgolo N., Therien A.G., Howell B., Klein D., et al. SARS-CoV-2 tropism, entry, replication, and propagation: considerations for drug discovery and development. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020 doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daly J.L., Simonetti B., Klein K., Chen K.E., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020 doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peacock T.P., Penrice-Randal R., Hiscox J.A., Barclay W.S. SARS-CoV-2 one year on: evidence for ongoing viral adaptation. J. Gen. Virol. 2021;102 doi: 10.1099/jgv.0.001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uemura K., Sasaki M., Sanaki T., Toba S., Y, et al. MRC5 cells engineered to express ACE2 serve as a model system for the discovery of antivirals targeting SARS-CoV-2. Sci. Rep. 2021;11:5376. doi: 10.1038/s41598-021-84882-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saccon E., Chen X., Mikaeloff F., Rodriguez J.E., et al. Cell-type-resolved quantitative proteomics map of interferon response against SARS-CoV-2. iScience. 2021;24:102420. doi: 10.1016/j.isci.2021.102420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuyama S., Nao N., Shirato K., Kawase M., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muus C., Luecken M.D., Eraslan G., Sikkema L., et al. Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nat. Med. 2021;27:546–559. doi: 10.1038/s41591-020-01227-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rendeiro A.F., Ravichandran H., Bram Y., Chandar V., et al. The spatial landscape of lung pathology during COVID-19 progression. Nature. 2021;593:564–569. doi: 10.1038/s41586-021-03475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao S., Ning K., Kuz C.A., Vorhies K., et al. Long-term modeling of SARS-CoV-2 infection of in vitro cultured polarized human airway epithelium. mBio. 2020;11 doi: 10.1128/mBio.02852-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pizzorno A., Padey B., Julien T., Trouillet-Assant S., et al. Characterization and treatment of SARS-CoV-2 in nasal and bronchial human airway epithelia. Cell Rep Med. 2020;1:100059. doi: 10.1016/j.xcrm.2020.100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Outlaw V.K., Bovier F.T., Mears M.C., Cajimat M.N., et al. Inhibition of coronavirus entry in vitro and ex vivo by a lipid-conjugated peptide derived from the SARS-CoV-2 spike glycoprotein HRC domain. mBio. 2020;11 doi: 10.1128/mBio.01935-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh B., Park B., Bhowmik D., Nishida K., et al. Strong correlation between air-liquid interface cultures and in vivo transcriptomics of nasal brush biopsy. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;318:L1056–L1062. doi: 10.1152/ajplung.00050.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pezzulo A.A., Starner T.D., Scheetz T.E., Traver G.L., et al. The air-liquid interface and use of primary cell cultures are important to recapitulate the transcriptional profile of in vivo airway epithelia. Am. J. Physiol. Lung Cell Mol. Physiol. 2011;300:L25–L31. doi: 10.1152/ajplung.00256.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada T., Sato S., Sotoyama Y., Orba Y., et al. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021 doi: 10.1038/s41590-021-00942-0. [DOI] [PubMed] [Google Scholar]
- 17.Barraud C., Corbière C., Pottier I., Estace E., et al. Impact of after-treatment devices and biofuels on diesel exhausts genotoxicity in A549 cells exposed at air-liquid interface. Toxicol. Vitro. 2017;45:426–433. doi: 10.1016/j.tiv.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 18.Movia D., Bazou D., Volkov Y., Prina-Mello A. Multilayered Cultures of NSCLC cells grown at the Air-Liquid Interface allow the efficacy testing of inhaled anti-cancer drugs. Sci. Rep. 2018;8:12920. doi: 10.1038/s41598-018-31332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He R.W., Gerlofs-Nijland M.E., Boere J., Fokkens P., et al. Comparative toxicity of ultrafine particles around a major airport in human bronchial epithelial (Calu-3) cell model at the air-liquid interface. Toxicol. Vitro. 2020;68:104950. doi: 10.1016/j.tiv.2020.104950. [DOI] [PubMed] [Google Scholar]
- 20.Öhlinger K., Kolesnik T., Meindl C., Gallé B., et al. Air-liquid interface culture changes surface properties of A549 cells. Toxicol. Vitro. 2019;60:369–382. doi: 10.1016/j.tiv.2019.06.014. [DOI] [PubMed] [Google Scholar]
- 21.Wu J., Wang Y., Liu G., Jia Y., et al. Characterization of air-liquid interface culture of A549 alveolar epithelial cells. Braz. J. Med. Biol. Res. 2017;51 doi: 10.1590/1414-431X20176950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki M., Uemura K., Sato A., Toba S., et al. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirato K., Nao N., Katano H., Takayama I., et al. Development of genetic diagnostic methods for detection for novel coronavirus 2019(nCoV-2019) in Japan. Jpn. J. Infect. Dis. 2020;73:304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 24.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045. doi: 10.1016/j.cell.2020.04.026. e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Vries M., Mohamed A.S., Prescott R.A., Valero-Jimenez A.M., et al. A comparative analysis of SARS-CoV-2 antivirals characterizes 3CLpro inhibitor PF-00835231 as a potential new treatment for COVID-19. J. Virol. 2021 doi: 10.1128/JVI.01819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y., Hu G., Wang Y., Ren W., et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., et al. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88:1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia H.P., Look D.C., Shi L., Hickey M., et al. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao Y., Wang X.M., Mannan R., Pitchiaya S., et al. Targeting transcriptional regulation of SARS-CoV-2 entry factors. Proc. Natl. Acad. Sci. U. S. A. 2020 doi: 10.1073/pnas.2021450118. [DOI] [PMC free article] [PubMed] [Google Scholar]