As part of a larger investigation of a 1997 tuberculosis outbreak at a homeless shelter in upstate New York, it became necessary to provide DNA typing data from a nonviable strain of Mycobacterium tuberculosis. Patient A, initially diagnosed with tuberculosis in 1992, was believed to be the source of infection in this outbreak. We wished to know whether the 1997 M. tuberculosis isolate from patient A represented a reactivation of his 1992 infection or later reinfection by a different strain. The only 1992 samples from patient A were 6-year-old LJ agar slants. Several attempts to recover this strain so that sufficient amounts of DNA could be obtained for IS6110-based restriction fragment length polymorphism analysis (RFLP) (3) were unsuccessful. In response to this, we instead utilized the PCR-based spoligotyping method (2) based on analysis of spacers in the direct-repeat (DR) region of M. tuberculosis complex organisms.
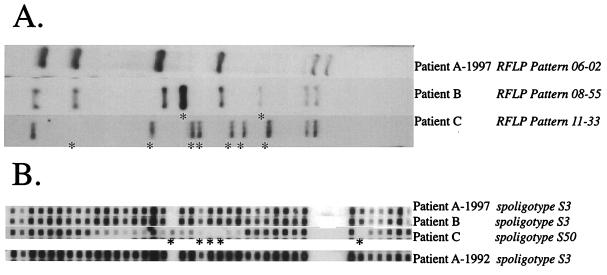
By using IS6110-based RFLP, patient A and six other individuals were shown to be infected during the 1997 outbreak with a strain of M. tuberculosis exhibiting a 6-banded fingerprint (06-02) as shown in Fig. 1A (top lane). The strain isolated from another patient (patient B) produced an 8-banded pattern (08-55) having six bands in common with the 06-02 pattern.
FIG. 1.
Comparison of results from M. tuberculosis DNA typing methods used in this study. Fingerprint patterns were compared with Bio Image Whole Band Analyzer and Matching software package version 3.4 from Genomic Solutions (Ann Arbor, Mich.). (A), IS6110-based RFLP results; (B), spoligotyping results. When included, the year indicates when the patient sample was taken. Fingerprint designations are listed in italics following the patient identifier. Asterisks in both panels highlight the differences from the 1997 isolate from patient A.
Spoligotyping was performed on all the 1997 isolates from individuals involved in the outbreak, and from a sample of nonviable cells from the 1992 culture of patient A, by using a commercially available spoligotyping kit (Isogen, Maarssen, The Netherlands). Spoligotyping of the 06-02 1997 strain from patient A produced the hybridization pattern shown in Fig. 1B. The six other isolates with the 06-02 fingerprint also exhibited the S3 spoligotype (data not shown). The 08-55 strain (Fig. 1B, patient B) produced the same spoligotyping pattern as the 06-02 strains, strengthening the argument that patient B was involved in the pattern of transmission from patient A. The 09-67 strain, which was not believed to be similar enough to the 06-02 strain to have been derived from a recent transmission, also showed a different spoligotyping pattern (Fig. 1B, patient C). Although the 1992 isolate from patient A was unable to be fingerprinted by IS6110 due to nonviability, there was sufficient DNA for a PCR to yield a spoligotyping pattern. The 1992 specimen from patient A gave a hybridization pattern identical to that observed for the 06-02 and 08-55 strains (Fig. 1B). This lends support to the hypothesis that patient A had not been cured of his 1992 infection and was the probable source of infection not only for patient B but also for the other six patients involved in this outbreak.
IS6110-based fingerprints vary in stability for reasons that are still unclear (1). Even serial isolates from the same patient may vary in the number of copies of IS6110 they contain (4). Because spoligotyping is based on a genomic target with a slower molecular clock than IS6110 transposition (3) it can compensate for that variability in some cases, and in this study it supported the inclusion of patient B but not patient C in the pattern of transmission from patient A.
Spoligotyping is rapidly becoming an accepted standard technique in molecular epidemiology of M. tuberculosis complex organisms due to its flexibility and low cost. The small size of the DR region makes it an attractive target for PCR-based analysis of even partially degraded DNA from nonviable cells. With spoligotyping, the opportunity to analyze samples from strain collections that may no longer be viable will help provide a longer time frame for transmission studies.
We acknowledge the contributions of Max Salfinger and the Clinical Mycobacteriology Laboratory of the Wadsworth Center.
This research was supported in part by the Centers for Disease Control and Prevention, National Tuberculosis Genotyping and Surveillance Network cooperative agreement, and by a grant from the Pittsfield Massachusetts Anti-tuberculosis Association.
REFERENCES
- 1.Dale J W, Tang T H, Wall S, Zainuddin Z F, Plikaytis B. Conservation of IS6110 sequence in strains of Mycobacterium tuberculosis with single and multiple copies. Tubercle Lung Dis. 1997;78:225–227. doi: 10.1016/s0962-8479(97)90002-2. [DOI] [PubMed] [Google Scholar]
- 2.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Embden J D, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeh R W, Ponce de Leon A, Agasino C B, Hahn J A, Daley C L, Hopewell P C, Small P M. Stability of Mycobacterium tuberculosis DNA genotypes. J Infect Dis. 1998;177:1107–1111. doi: 10.1086/517406. [DOI] [PubMed] [Google Scholar]