Abstract
In recent times, COVID-19, has a great impact on the healthcare sector and results in a wide range of respiratory illnesses. It is a type of Ribonucleic acid (RNA) virus, which affects humans as well as animals. Though several artificial intelligence-based COVID-19 diagnosis models have been presented in the literature, most of the works have not focused on the hyperparameter tuning process. Therefore, this paper proposes an intelligent COVID-19 diagnosis model using a barnacle mating optimization (BMO) algorithm with a cascaded recurrent neural network (CRNN) model, named BMO-CRNN. The proposed BMO-CRNN model aims to detect and classify the existence of COVID-19 from Chest X-ray images. Initially, pre-processing is applied to enhance the quality of the image. Next, the CRNN model is used for feature extraction, followed by hyperparameter tuning of CRNN via the BMO algorithm to improve the classification performance. The BMO algorithm determines the optimal values of the CRNN hyperparameters namely learning rate, batch size, activation function, and epoch count. The application of CRNN and hyperparameter tuning using the BMO algorithm shows the novelty of this work. A comprehensive simulation analysis is carried out to ensure the better performance of the BMO-CRNN model, and the experimental outcome is investigated using several performance metrics. The simulation results portrayed that the BMO-CRNN model has showcased optimal performance with an average sensitivity of 97.01%, specificity of 98.15%, accuracy of 97.31%, and F-measure of 97.73% compared to state-of-the-art methods.
Keywords: COVID-19, Evolutionary computing, Soft computing, Intelligent systems, Deep learning, Hyperparameter tuning, Decision making
1. Introduction
In recent times, Coronavirus Disease 2019 is referred to as COVID-19, which is considered an epidemic disease from the end of December 2019 in Wuhan, China. The name COVID-19 was dictated by the World Health Organization (WHO) as a novel and dangerous infection which comes under the class of Coronaviruses (CoV) and infectious viruses [1]. It results in severe cases in critical care respiratory conditions like severe acute respiratory syndrome (SARS-CoV) intended for shortness of breathing and finally death. Based on the survey reported by WHO, the risk evaluation of COVID-19 is higher all over the world [2]. Additionally, the maximum number of people was positive for COVID-19, and some people died due to COVID-19. Some other lung disorders are Viral and Bacterial pneumonia, leading to high mortality. Pneumonia diseases are caused due to the fungal infection of the lungs that is formed by pus and additional fluids filled in air sacs. However, bacterial pneumonia is critical; particularly, small kids are affected easily due to low immunity [3], [4], [5].
Real-Time Polymerase Chain Reaction (RT-PCR) is an effective tool for examining pneumonia diseases and Coronaviruses. But the RT-PCR samples accomplished maximum false-negative levels for COVID-19 positive cases [6]. Then, the radiological investigations are performed by applying Chest X-ray (CXR) as well as Computed Tomography (CT) scans to examine the health condition of patients. But the exposure to radiation results in high side effects. The CT scan provides a proficient model in screening, analysis, and progressive estimation of patients confirmed with COVID-19. The clinical studies have reported positive CXR models that pave the way for developing CT scans and reduce the medical overload of CT in COVID-19 [7]. The American College of Radiology suggested applying portable chest radiography to reduce the threats of Coronavirus disease [8]. Moreover, chest CT screening demands higher dose exposure and costlier treatments. Unlike other models, traditional X-ray machines are accessible and portable in clinics for robust scanning of lungs as 2D images [9]. Thus, CXR is initialized to diagnose COVID-19 cases to confirm the patients with highly suspected COVID-19 or pneumonia diseases [10], [11].
In computer-aided diagnosis (CAD), technologies are projected in the real-time solution for resolving the difficulties in Chest X-rays, and to help the radiologists in predicting the diseases from low-contrast X-ray images [12]. The CAD models are integrated units of computer models with currently developed image processing approaches for performing interventional operations. Recently, Artificial Intelligence is employed extensively for the advanced diagnostic function of CAD systems for different clinical applications like brain tumor classification or segmentation, reduced invasive aortic valve incorporation, and predicting pulmonary infections. Presently, Deep Learning (DL) models are part of a broader family of machine learning (ML) methods based on artificial neural network (ANN) with representation learning. It is applied for learning patterns and features from annotated data which is applicable for automated performance of a certain operation on earlier training like human sentiment classification as well as computer vision domains in surgery [13], [14], [15].
1.1. Previous works
Automatic investigations of COVID-19 and chest infections have been predicted under the application of clinical CT and X-ray imaging sectors. In recent times, [16] depicted that chest CT images and DL methodologies are significant in effectively identifying and segmenting COVID-19 disease. Therefore, it is concentrated to use CXR images for COVID19 positive patients and pneumonia diseases. Automated classification of lung diseases in X-ray images has been projected for TB screening, prediction of lung intensity as well as severe pneumonia infections. However, examining the COVID19 disease in Chest X-rays is still in the progressive stage and examined in peer-review published articles. For instance, some of the DL classification models, like the pre-trained InceptionV3 method, were applied to detect COVID-19 [17]. Moreover, the pneumonia diseases are examined using Convolutional Neural Networks (CNNs) with better classification accuracy as projected in [18]. Drop-weight-related Bayesian CNNs have been employed for validating the maximum correlation of uncertainty by accomplishing supreme prediction accuracy in detecting the COVID-19 from X-ray images [19].
In [20], pneumonia X-ray images with the help of 3 diverse DL methods are employed for COVID-19 diagnosis. Under the application of the ResNet model, the dataset is classified into several labels like Age, Gender, and so on. Moreover, Multi-Layer Perceptron (MLP) classifier is applied, which achieved maximum accuracy. Yadav and Jadhav [21] processed a classification approach under the application of pneumonia details where SVM classifier is used along with InceptionV3 and VGG-16 methodologies as DL modules. In this application, a dataset is classified into 3 types for enhancing the contrast as well as brightness zoom setting along with the augmentation model for an image with a dataset that has attained an optimal classification score. Abiyev and Ma’aitah [22] applied the Backpropagation Neural Network (BPNN), as well as Competitive Neural Network approaches for classifying the pneumonia information. By using pneumonia as well as healthy CXR images, some portions in the dataset are used as test data, showing better classification results.
Stephen et al. [23] developed a DL technology for classifying the pneumonia data from scratch for data training. The input size is 200 x 200 pixels, which is employed to determine the viabilities of classification with a sigmoid function. Consequently, the optimal success rate is achieved in pneumonia. Chouhan et al. [24] predicted the images of pneumonia by utilizing DL methods and 3 classes of the dataset. Initially, pre-processing is performed for noise elimination. Then, the augmentation mechanism is applied for all images and transfer learning is used for model training, resulting in effective classification accuracy. Islam et al. [25] developed a DL model using the integration of CNN and long short-term memory (LSTM) for COVID-19 detection and classification. Hussain et al. [26] employed DL with natural language processing tools to estimate the average sentiment, sentiment trend, and discussion topic related to COVID-19 vaccination. Melin et al. [27] proposed an ensemble neural network firefly algorithm for COVID-19 diagnosis.
Sujath et al. [28] projected a COVID-19 diagnosis model using linear regression, Multilayer perceptron, and Vector autoregression methods. A novel hybrid model is designed in [29] to predict COVID-19 using the integration of Statistical Neural Network models and the Non-linear Autoregressive Neural Network. Iwendi et al. [30] designed a fine-tuned Random Forest with AdaBoost technique for predicting the health conditions of COVID-19 patients. A CAD model for COVID-19 is presented in [31] using the fusion of CNN with statistical and textural features. Attallah et al. [32] presented an efficient CAD model to detect COVID-19 using CNNs and SVM models. El-bana et al. [33] presented a fine-tuned InceptionV3 deep model to detect COVID-19 using multi-modal learning. Some other COVID-19 diagnosis models are available in the literature [34], [35], [36]. Though several methods are available in the literature, there is still a need to improve the COVID-19 diagnostic performance. Besides, only a few works have concentrated on the parameter optimization of the DL based feature extraction techniques.
1.2. Paper contributions
This paper proposes an intelligent COVID-19 diagnosis model using a BMO algorithm with a CRNN model called BMO-CRNN. The proposed BMO-CRNN model intends to identify and categorize the existence of COVID-19 from Chest X-ray images. Firstly, image pre-processing is applied to eliminate the noise. Next, the CRNN technique is applied as feature extraction, and the BMO algorithm is used for the hyperparameter tuning of CRNN. The CRNN explores the redundant and complementary information and involves two recurrent neural network (RNN) layers. The former eliminates the redundant data, and the latter aims to learn the complementary data. Lastly, the SoftMax (SM) layer is utilized for classification purposes to categorize COVID-19 or non-COVID-19. A comprehensive simulation analysis is done to ensure the goodness of the BMO-CRNN technique using the Chest X-ray (CXR) dataset. The key contributions of the paper are summarized as follows.
-
•
An intelligent COVID-19 diagnostic method involving pre-processing, CRNN based feature extraction, and BMO based parameter optimization is proposed. To the best of our knowledge, the BMO-CRNN model has not been presented in the literature for this problem.
-
•
The design of CRNN based feature extraction involves a series of RNNs, which necessitate a minimal number of highly applicable parameters for classification, and a limited number of training samples is needed.
-
•
The parameter optimization of the CRNN model using the BMO algorithm using cross-validation helps improve the BMO-CRNN model’s diagnostic performance for unseen data. In addition, the use of the BMO algorithm for the parameter tuning of the CRNN model shows the novelty of this work.
-
•
Detailed experiments are conducted to validate the performance of the BMO-CRNN on the benchmark CXR dataset under various settings compared to SOTA methods.
The remaining portions of the paper are arranged as follows. Section 2 discusses the proposed BMO-CRNN model. Section 3 performs the simulation process, experiments, and discussion. At last, Section 4 concludes the paper.
2. Materials and methods
2.1. Overall architecture
The working principle involved in the BMO-CRNN model is depicted in Fig. 1, which comprises pre-processing, parameter tuning, feature extraction, and classification. Once the input image is pre-processed, the BMO algorithm determines the parameters of CRNN namely learning rate, batch size, activation function, and epoch count. When the parameters are identified, the feature extraction process is carried out. The major steps involved in the proposed model are given in Algorithm 1.
Fig. 1.
Overall architecture of the BMO-CRNN model.
|
Algorithm 1: Major Steps of the Proposed Model | |
|---|---|
| Input: Training CXR dataset | |
| 1. | Consider hyperparameters in CRNN as barnacles and assume the original impact of data prediction using CRNN. |
| 2. | Identify the number of barnacles and the hyperparameters of CRNN to be optimized. Compute the upper and lower boundary points of the optimization area. Then, create the barnacle population and identify the number of search iterations. |
| 3. | Substitute the hyperparameters of the CRNN respective to the points of barnacles. Partition the dataset into the training and testing part, afterward identify the subsequent series of training data by analysing the trend line. By comparing the predicted and actual data, the error can be determined. |
| 4. | Reiterate the above process until the termination criteria are fulfilled. |
| Output: Classified images (COVID-19 or Normal) | |
2.2. Gaussian Filtering (GF)
The input images from CXR dataset are pre-processed using the GF technique, which is a linear smoothing filter used for weight selection depending upon the structure of the Gaussian function. The GF technique is applied on the spatial or frequency domain as an effective low pass filtering technique, particularly for noise removal. The 1-D Gaussian function of zero means can be denoted as follows.
| (1) |
The parameter involved in the Gaussian distribution computes the width of the Gaussian functions. In the case of processing images, 2D discrete Gaussian function of zero mean namely smooth filter [37], and the respective function is expressed as follows.
| (2) |
2.3. BMO algorithm
The BMO method is a new bio-inspired optimization mechanism used for resolving the optimization issues. The BMO method is chosen over the other optimization algorithms due to the following reasons:
-
•
BMO algorithm balances the trade-off between exploitation and exploration for producing a new offspring toward a globally optimum solution.
-
•
BMO algorithm offers enhanced outcome over the other compared methods and attains global optimum, high exploration ability, and avoids local optima problem.
-
•
It is highly flexible and efficient over other algorithms.
This model is evolved based on the barnacle’s mating nature and related functions. Even though barnacles are hermaphrodites that carry male and female genitalia, the mating process is performed rarely, which applies extraordinarily long penises and finds a mate inside the striking distance [38]. The Barnacles ovaries are placed in a stalk and expand into the mantle, often expand into the thorax. Also, self-mating is carried out rarely in barnacles which do not leave the shells for mating. Barnacles can reproduce by sperm casting where male barnacles discharge the sperm to the water, and the female consumes and fertilizes the eggs is named sperm casting or self-mating. The lifecycle of BMO is shown in Fig. 2 [39].
Fig. 2.
Lifecycle of the BMO algorithm.
The different steps involved in the BMO algorithm are listed below, and the flowchart is shown in Fig. 3 [39]. The algorithm begins with the parameter initialization process. Then, the control variables are mapped to the individuals that exist in the population. Next, the population value is fixed, followed by selection and reproduction processes. Finally, the processes get iterated till the maximum number of iterations is reached.
Fig. 3.
Flowchart of the BMO algorithm.
Initialization: The estimation of control parameters and count of the population of barnacles are performed, and the sorting process is done to place the optimum solutions.
Selection process: This process is carried out randomly, but it is restricted to the barnacle’s penis length. Every barnacle has sperm and receives the sperm from another barnacle which is fertilized simultaneously where the female is fertilized using massive male barnacles.
Reproduction: The reproduction process is evolved from BMO that is varied then in evolutionary models. The BMO is an emphasizing one from the inheritance features of barnacles’ parents in developing an offspring related to the Hardy–Weinberg principle.
2.4. Cascaded Recurrent Neural Network (CRNN)
RNN [40] is a division of Artificial Neural Network (ANN), which is an extended version of the traditional feedforward neural network (FFNN) with loops and connections. In contrast with FFNN, the RNN is capable to compute the sequential input using a recurrent hidden state with the activation of the former step. Thus, the system represents the dynamic temporal behavior. Consider the sequence data , where implies the data in th time step, the RNN updates the recurrent hidden state using Eq. (3):
| (3) |
where denotes a non-linear function. Thus, RNN is composed of output . Ultimately, the data classification is performed by an output . In the classical RNN method, the update rule of the recurrent hidden state in (1) is executed as given below:
| (4) |
where and refer to the coefficient matrices for input and activation of recurrent hidden units. Assume is a sequence probability that is degraded as:
| (5) |
Followed by, the conditional probability distribution is developed using a recurrent network
| (6) |
where is gained from (3), (6). The hyperspectral pixel is treated as sequential data and a recurrent network is applied for modeling spectral sequence.
As the DL family is significant, RNNs show the best outcome in ML as well as computer vision operations. To overcome these problems, a model is developed with a sophisticated recurrent unit. LSTM is defined as the class of recurrent hidden units which is suitable for learning long-term series [41].
When compared with the LSTM unit, the gated recurrent unit (GRU) requires a smaller count of parameters that is highly applicable for classification, and a limited number of training samples is required [42]. Thus, GRU is selected as an essential component of RNN. The major components of GRU are 2 gating units which are employed for controlling the data flow within the unit. Rather than using the activation of hidden layer for band is expressed as,
| (7) |
where denotes the update gate which has been retrieved by
| (8) |
where means a sigmoid function, refers to a weight value, and depicts the weight vector. Likewise, is determined by,
| (9) |
where refers to an element-wise multiplication, and signifies the reset gate that is obtained from
| (10) |
In particular, the data sequence is classified into sub-sequences , where it is composed of different class labels. Followed by final sub-sequence , the length of alternate sub-sequences is , which refers to the closer integers less than or equal to . Therefore, the th sub-sequence , it is composed of given bands,
| (11) |
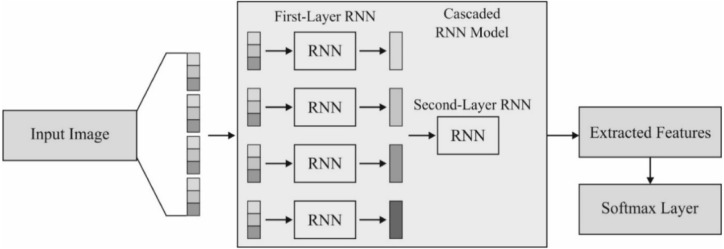
Additionally, each sub-sequence is fed into the first layer of RNNs, which has a similar architecture and distributes parameters for reducing their count. In the case of sub-sequence , every sample is composed of output from the GRU. The final feature representation for , is referred to be , where implies the size of the hidden layer in first layer RNN. Afterward, the are combined for generating sequence where length is . These sequences are induced into second layer RNN for learning complementary details. Likewise, the first layer RNNs applies the result of GRU finally as the learned feature . The classification result of can be attained by inducing the input into the resultant layer with equally sized candidate classes . Hence, 2-layer RNNs have massive weight parameters. Finally, selected as a loss function and apply the BPTT model for optimization. Fig. 4 shows the structure of the cascaded RNN model [42].
Fig. 4.
Structure of cascaded RNN model.
2.5. Classification using SoftMax layer
After the completion of the feature extraction process using BMO-CRNN, SM based classification is performed to identify the existence of COVID-19. It will map the input vectors from the -dimensional space into classes, as given in Eq. (12): labels, as defined below.
| (12) |
where represents the weight factors and K denotes the number of classes.
3. Experimental results analysis
This section validates the efficiency of the BMO-CRNN model on COVID-19 diagnostic process. The proposed model is simulated utilizing Python 3.6.5 tool along with few packages. The details related to the dataset, evaluation metrics, and comparative results analysis are made in the succeeding sections.
3.1. Dataset description
The CXR dataset is used for the classification of COVID-19 [45]. It contains two classes namely Normal and COVID-19, where a total of 27 images belong to the Normal class and 220 images are related to the COVID-19 class. Fig. 5 shows some test images from the CXR dataset.
Fig. 5.
Sample test images from CXR dataset (a) Normal (b) COVID-19.
3.2. Evaluation parameters
The classification results of the BMO-CRNN technique are examined using different evaluation parameters such as sensitivity, specificity, accuracy, and F-measure [46].
Sensitivity determines the proportion of positive samples which are properly identified, as defined in Eq. (13):
| (13) |
where TP, TN, FP, and FN represent true positive, true negative, false positive, and false negative, respectively.
Specificity determines the proportion of negative samples which are properly identified, as given in Eq. (14).
| (14) |
F-measure defines the harmonic mean of precision and sensitivity, which can be equated using Eq. (15):
| (15) |
Finally, accuracy determines the closeness of the measurements to a particular value, as defined in Eq. (16):
| (16) |
3.3. Results analysis
The convergence rate analysis of the BMO algorithms in terms of best cost attained under several iteration counts in Fig. 6. The figure displayed that the PSO algorithm has failed to showcase better convergence over the other methods. In addition, the GWO, ABC, and BFO algorithms have exhibited certainly increased convergence over PSO. However, the BMO algorithm has surpassed all the compared optimization algorithms and reached a minimum best score under varying iteration counts.
Fig. 6.
Convergence Curve of Different Optimization Algorithms.
To determine the classification performance of the BMO-CRNN model, a set of confusion matrices generated at the time of execution is demonstrated in Fig. 7. The confusion matrices are arranged under varying numbers of epochs ranging between 100–1000.
Fig. 7.
Confusion matrix of the BMO-CRNN model under varying epochs.
Table 1 and Fig. 8 inspect the efficiency of the BMO-CRNN technique against other methods. The proposed BMO-CRNN model has exhibited optimal classification results under varying epoch counts. For instance, on the epoch count of 100, the BMO-CRNN model has gained a maximum sensitivity of 96.78%, specificity of 97.54%, an accuracy of 96.92%, and an F-measure of 97.21%. Along with that, on the epoch count of 200, the BMO-CRNN model has got a maximum sensitivity of 96.82%, specificity of 97.89%, an accuracy of 96.98%, and an F-measure of 97.53%. At the same time, on the epoch count of 300, the BMO-CRNN model has attained a maximum sensitivity of 96.91%, specificity of 98.12%, an accuracy of 97.33%, and an F-measure of 97.87%. Furthermore, on the epoch count of 400, the BMO-CRNN model has resulted in a maximum sensitivity of 97.10%, specificity of 98.56%, accuracy of 97.54%, and an F-measure of 97.92%. Moreover, on the epoch count of 1000, the BMO-CRNN model has resulted in a maximum sensitivity of 98.38%, specificity of 98.82%, accuracy of 94.82%, and an F-measure of 99.09%. These values show that the BMO-CRNN technique has ensured the betterment under varying epoch counts.
Table 1.
Result analysis of the proposed BMO-CRNN model under varying epochs.
| No. of Epochs | Accuracy | Sensitivity | Specificity | F-measure |
|---|---|---|---|---|
| Epoch 100 | 97.98 | 99.09 | 88.89 | 98.87 |
| Epoch 200 | 99.19 | 99.55 | 96.30 | 99.55 |
| Epoch 300 | 98.79 | 99.09 | 96.30 | 99.32 |
| Epoch 400 | 98.79 | 99.09 | 96.30 | 99.32 |
| Epoch 500 | 99.19 | 99.55 | 96.30 | 99.55 |
| Epoch 600 | 98.79 | 99.09 | 96.30 | 99.32 |
| Epoch 700 | 99.19 | 99.55 | 96.30 | 99.55 |
| Epoch 800 | 96.76 | 97.27 | 92.59 | 98.17 |
| Epoch 900 | 97.17 | 97.73 | 92.59 | 98.40 |
| Epoch 1000 | 97.98 | 98.18 | 96.30 | 98.86 |
| Average | 98.38 | 98.82 | 94.82 | 99.09 |
Fig. 8.
Result analysis of the BMO-CRNN model under varying epochs.
Fig. 9 investigates the ROC analysis of the proposed BMO-CRNN model under varying epoch counts. The figure demonstrated that the BMO-CRNN model has resulted in a maximum ROC of 99.9370, 99.9899, 99.9684, 99.8822, 99.9892, 99.9389, 99.9846, 99.1193, 99.4028, and 99.7811 under epoch count of 100–1000 with intervals of 100.
Fig. 9.
ROC analysis of the BMO-CRNN model under varying epochs.
Table 2 and Fig. 10 investigate the performance of the BMO-CRNN model under five different folds, and the results demonstrated the enhanced classification results under each fold. For instance, with fold-1, the BMO-CRNN model has obtained an accuracy of 97.14%, sensitivity of 98.24%, specificity of 96.19%, and F-measure of 98.25%. Likewise, with fold-2, the BMO-CRNN model has attained the accuracy of 97.99%, sensitivity of 98.67%, specificity of 98.75%, and F-measure of 98.55%. Similarly, with fold-3, the BMO-CRNN model has accomplished accuracy of 98.18%, sensitivity of 98.21%, specificity of 96.63%, and F-measure of 98.09%. Moreover, with fold-4, the BMO-CRNN model has offered an accuracy of 98.94%, sensitivity of 98.80%, specificity of 98.93%, and F-measure of 99.58%. Furthermore, with fold-5, the BMO-CRNN model has resulted in an accuracy of 98.97%, sensitivity of 97.89%, specificity of 96.00%, and F-measure of 98.57%.
Table 2.
Result analysis of the proposed BMO-CRNN model under 5-Fold Cross Validation.
| No. of Folds | Accuracy | Sensitivity | Specificity | F-measure |
|---|---|---|---|---|
| Fold 1 | 97.14 | 98.24 | 96.19 | 98.25 |
| Fold 2 | 97.99 | 98.67 | 98.75 | 98.55 |
| Fold 3 | 98.18 | 98.21 | 96.63 | 98.09 |
| Fold 4 | 98.94 | 98.80 | 98.93 | 99.58 |
| Fold 5 | 98.97 | 97.89 | 96.00 | 98.57 |
| Average | 98.24 | 98.36 | 97.30 | 98.61 |
Fig. 10.
Result analysis of the BMO-CRNN technique under different folds.
4. Discussion
A detailed comparative study of the BMO-CRNN with other existing methods is performed in Table 3. Fig. 11 demonstrates the analysis of the BMO-CRNN with existing models in terms of sensitivity and specificity. The figure shows that the DT and CNN approaches have shown insignificant performance with the minimum sensitivity of 87% and 87.3%, respectively. Moreover, the ANFIS and K-NN models have exhibited somewhat enhanced outcomes with a nearer sensitivity of 88.48% and 89% correspondingly. In the same way, the DT and CoroNet models have exhibited moderate results with a sensitivity of 89.61% and 90% correspondingly. Likewise, the XGBoost, RNN, and CNNLSTM techniques exhibited near identical outcomes with the sensitivity of 92%, 92.04%, and 92.14%, respectively.
Table 3.
| Methods | Sens. | Spec. | Accuracy | F-measure |
|---|---|---|---|---|
| BMO-CRNN (Ours) | 98.38 | 98.82 | 94.82 | 99.09 |
| CoroNet | 90.00 | 92.14 | 90.21 | 91.00 |
| CNN | 87.73 | 86.97 | 87.36 | 89.65 |
| DTL | 89.61 | 92.03 | 90.75 | 90.43 |
| ANN | 93.78 | 91.76 | 86.00 | 91.34 |
| CNNLSTM | 92.14 | 91.98 | 84.16 | 90.01 |
| CNNRNN | 94.23 | 92.67 | 85.66 | 91.20 |
| LSTM | 93.42 | 92.64 | 86.66 | 91.89 |
| RNN | 92.04 | 90.87 | 84.16 | 90.61 |
| ANFIS | 88.48 | 87.74 | 88.11 | 89.04 |
| MLP | 93.00 | 87.23 | 93.13 | 93.00 |
| LR | 93.00 | 90.34 | 92.12 | 92.00 |
| XGBoost | 92.00 | 90.44 | 91.57 | 92.00 |
| K-NN | 89.00 | 90.65 | 88.91 | 89.00 |
| DT | 87.00 | 88.93 | 86.71 | 87.00 |
Fig. 11.
Comparative results analysis of the BMO-CRNN technique.
Similarly, the MLP and LR models have obtained exactly equivalent classifier results with a sensitivity of 93%. Concurrently, the LSTM, ANN, and CNNRNN models have obtained competitive results with the sensitivity of 93.42%, 93.78%, and 94.23%, respectively. At last, the proposed BMO-CRNN model has shown superior performance with a maximum sensitivity of 98.38%.
On examining the outcome in terms of specificity, the CNN and MLP techniques achieved ineffective performance with specificity of 86% and 87.23%. Simultaneously, the ANFIS and DT techniques have presented moderate results with identical specificity of 87.74% and 88.93%. On continuing with, the LR and XGBoost technologies have presented substantial outcomes with the specificity of 90.34% and 90.44%. Similarly, the LSTM, KNN, and ANN models have showcased closer outcomes with the specificity of 90.65%, 90.87%, and 91.76%, correspondingly. In the same way, the CNNLSTM and DTL methods have attained slightly better results with the specificity of 91.98% and 92.03%. Simultaneously, the CoroNet, LSTM, and CNNRNN techniques have secured competing outcomes with the specificity of 92.14%, 92.64%, and 92.67%, correspondingly. Finally, our BMO-CRNN framework has showcased supreme function with higher specificity of 98.82%.
Fig. 12 illustrates the results analysis of the BMO-CRNN with previous methods with respect to accuracy and F-measure. The figure depicted that the RNN and CNNLSTM methodologies have depicted inferior functions with lower accuracy of 84.16%. Concurrently, the CNNRNN and ANN technologies have presented moderate outcomes with identical accuracy of 85.66% and 86% correspondingly. Along with that, the LSTM and DT frameworks have depicted slight effects with the accuracy of 86.66% and 86.71%, respectively. Similarly, the CNN, ANFIS, and KNN techniques have been referred to as closer identical results with the accuracy of 87.36%, 88.11%, and 88.91%, correspondingly. Likewise, the CoroNet and DTL techniques have gained manageable classifier results with 90.21% and 90.75% accuracy. Meantime, the XGBoost, LR, and MLP methods have attained competing outcomes with the accuracy of 91.57%, 92.12%, and 93.13%, correspondingly. Consequently, the BMO-CRNN technique has showcased supreme function with optimal accuracy of 94.82%.
Fig. 12.
Accuracy and F-measure analysis of the BMO-CRNN technique.
Finally, on examining the F-measure analysis, it is prominent that the DT and KNN schemes have depicted worse performance with lower F-measure of 87% and 89%. Meantime, the ANFIS and CNN methods have presented moderate results with the nearby F-measure of 89.04% and 89.65%, correspondingly. In the same way, the CNNLSTM and DTL technologies have showcased slightly better results with the F-measure of 90.01% and 90.43%, correspondingly. Then, the RNN, CoroNet, and CNNRNN models have depicted closer identical results by offering F-measure of 90.61%, 91%, and 91.2%, correspondingly. In the same way, the ANN and LSTM approaches have reached substantial classifier outcomes with the F-measure of 91.34% and 91.89%. Meantime, the LR, XGBoost, and MLP approaches have gained competing outcomes with the F-measure of 92%, 92%, and 93%, correspondingly. Lastly, the BMO-CRNN method has showcased supreme outcomes with the best F-measure of 99.09%.
The detailed investigation of the results analysis of the BMO-CRNN model has demonstrated superior results with the maximum average sensitivity of 98.38%, specificity of 98.82%, the accuracy of 94.82%, and F-measure of 99.09%. Therefore, it can be employed as an appropriate tool to assist doctors in the COVID-19 diagnostic procedures.
5. Conclusion and future work
This paper has developed an intelligent COVID-19 detection model using the BMO-CRNN algorithm. The BMO-CRNN model comprises pre-processing, parameter tuning, feature extraction, and classification. Initially, image pre-processing techniques are used to remove the noise from the image. Besides, the CRNN model is used as feature extraction, and the hyperparameter tuning of CRNN is performed via the BMO algorithm for boosting the classification outcomes. The BMO algorithm determined the parameters of CRNN namely, learning rate, batch size, activation function, and epoch count. After the parameter’s identification process, the feature extraction was carried out using the CRNN model. Next, SM based classification was used to identify the existence of COVID-19. Finally, extensive experimentation was conducted to confirm the superior outcomes of the BMO-CRNN model using the CXR dataset. The obtained scores portrayed that the BMO-CRNN model has showcased optimal performance with an average accuracy of 94.82%. In the future, the diagnostic results of the BMO-CRNN model can be improved by the inclusion of the class attention layer in the CRNN model to capture the discriminative class-specific features.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
CRediT authorship contribution statement
K. Shankar: Conceptualization, Methodology, Software, Writing – original draft. Eswaran Perumal: Conceptualization, Methodology, Software, Writing – original draft. Vicente García Díaz: Conceptualization, Writing – review & editing, Project administration. Prayag Tiwari: Conceptualization, Writing – review & editing, Project administration. Deepak Gupta: Conceptualization, Writing – review & editing, Project administration. Abdul Khader Jilani Saudagar: Investigation, Validation, Writing – reviewing and editing, Supervision. Khan Muhammad: Investigation, Validation, Writing – reviewing and editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number 959.
Data Availability Statement
Data sharing is not applicable to this article as no new datasets were generated during the current study.
References
- 1.Paules C.I., Marston H.D., A.S. Fauci. Coronavirus infections—more than just the common cold. JAMA. 2020;323(8):707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 2.Demir F. DeepCoroNet: A deep LSTM approach for automated detection of COVID-19 cases from chest X-ray images. Appl. Soft Comput. 2021;103 doi: 10.1016/j.asoc.2021.107160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aslan M.F., Unlersen M.F., Sabanci K., Durdu A. CNN-based transfer learning–BiLSTM network: A novel approach for COVID-19 infection detection. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo L., Ren L., Yang S., Xiao M., Chang D., Yang F., Dela Cruz C.S., Wang Y., Wu C., Xiao Y., Zhang L. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ezzat D., Hassanien A.E., Ella H.A. An optimized deep learning architecture for the diagnosis of COVID-19 disease based on gravitational search optimization. Appl. Soft Comput. 2020 doi: 10.1016/j.asoc.2020.106742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wynants L., Van Calster B., Bonten M.M., Collins G.S., Debray T.P., De Vos M., Haller M.C., Heinze G., Moons K.G., Riley R.D., Schuit E. Prediction models for diagnosis and prognosis of covid-19 infection: Systematic review and critical appraisal. Bmj. 2020;369 doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nour M., Cömert Z., Polat K. A novel medical diagnosis model for COVID-19 infection detection based on deep features and Bayesian optimization. Appl. Soft Comput. 2020;97 doi: 10.1016/j.asoc.2020.106580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poggiali E., Dacrema A., Bastoni D., Tinelli V., Demichele E., Mateo Ramos P., Marcianò T., Silva M., Vercelli A., Magnacavallo A. Can lung US help critical care clinicians in the early diagnosis of novel coronavirus (COVID-19) pneumonia? Radiology. 2020;295(3):E6. doi: 10.1148/radiol.2020200847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marques G., Agarwal D., de la Torre Díez I. Automated medical diagnosis of COVID-19 through EfficientNet convolutional neural network. Appl. Soft Comput. 2020;96 doi: 10.1016/j.asoc.2020.106691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi F., Wang J., Shi J., Wu Z., Wang Q., Tang Z., He K., Shi Y., Shen D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19. IEEE Rev. Biomed. Eng. 2020 doi: 10.1109/RBME.2020.2987975. [DOI] [PubMed] [Google Scholar]
- 11.Sait U., KV G.L., Shivakumar S., Kumar T., Bhaumik R., Prajapati S., Bhalla K., Chakrapani A. A deep-learning based multimodal system for Covid-19 diagnosis using breathing sounds and chest X-ray images. Appl. Soft Comput. 2021 doi: 10.1016/j.asoc.2021.107522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin S., Wang B., Xu H., Luo C., Wei L., Zhao W., Hou X., Ma W., Xu Z., Zheng Z., Sun W. 2020. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system in four weeks. MedRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tiwari P., Uprety S., Dehdashti S., Hossain M.S. Terminformer: Unsupervised term mining and analysis in biomedical literature. Neural Comput. Appl. 2020:1–14. doi: 10.1007/s00521-020-05335-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherjee R., et al. IoT-cloud based healthcare model for COVID-19 detection: An enhanced k-Nearest Neighbour classifier based approach. Computing. 2021:1–21. [Google Scholar]
- 15.Madhavan M.V., et al. Res-CovNet: An internet of medical health things driven COVID-19 framework using transfer learning. Neural Comput. Appl. 2021:1–14. doi: 10.1007/s00521-021-06171-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shinde G.R., Kalamkar A.B., Mahalle P.N., Dey N., Chaki J., Hassanien A.E. Forecasting models for coronavirus disease (COVID19): A survey of the state-of-the-art. SN Comput. Sci. 2020;1(4):197. doi: 10.1007/s42979-020-00209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemdan E.E.-D., Shouman M.A., Karar M.E. 2020. COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in X-ray images. [Google Scholar]
- 18.Apostolopoulos I.D., Mpesiana T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020 doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghoshal B., Tucker A. 2020. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. [Google Scholar]
- 20.Baltruschat I.M., Nickisch H., Grass M., Knopp T., Saalbach A. Comparison of deep learning approaches for multi-label chest X-ray classification. Sci. Rep. 2019;9:6381. doi: 10.1038/s41598-019-42294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yadav S.S., Jadhav S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data. 2019;6:113. [Google Scholar]
- 22.Abiyev R.H., Ma’aitah M.K.S. Deep convolutional neural networks for chest diseases detection. J. Healthc. Eng. 2018;2018 doi: 10.1155/2018/4168538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephen O., Sain M., Maduh U.J., Jeong D.-U. An efficient deep learning approach to pneumonia classification in healthcare. J. Healthc. Eng. 2019;2019 doi: 10.1155/2019/4180949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chouhan V., Singh S.K., Khamparia A., Gupta D., Tiwari P., Moreira C., Damasevicius R., de Albuquerque V.H.C. A novel transfer learning based approach for pneumonia detection in chest X-ray images. Appl. Sci. 2020;10 [Google Scholar]
- 25.Islam M.Z., Islam M.M., Asraf A. A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hussain A., Tahir A., Hussain Z., Sheikh Z., Gogate M., Dashtipour K., Ali A., Sheikh A. 2020. Artificial intesslligence-enabled analysis of UK and US public attitudes on Facebook and Twitter towards COVID-19 vaccinations. medRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melin P., Sánchez D., Monica J.C., Castillo O. Optimization using the firefly algorithm of ensemble neural networks with type-2 fuzzy integration for COVID-19 time series prediction. Soft Comput. 2021:1–38. doi: 10.1007/s00500-020-05549-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Sujath R., Chatterjee J.M., Hassanien A.E. A machine learning forecasting model for COVID-19 pandemic in India. Stoch. Environ. Res. Risk Assess. 2020;34:959–972. doi: 10.1007/s00477-020-01827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhamodharavadhani S., Rathipriya R., Chatterjee J.M. Covid-19 mortality rate prediction for India using statistical neural network models. Front. Public Health. 2020;8 doi: 10.3389/fpubh.2020.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwendi C., Bashir A.K., Peshkar A., Sujatha R., Chatterjee J.M., Pasupuleti S., Mishra R., Pillai S., Jo O. COVID-19 patient health prediction using boosted random forest algorithm. Front. Public Health. 2020;8(357) doi: 10.3389/fpubh.2020.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ragab D.A., Attallah O. FUSI-CAD: Coronavirus (COVID-19) diagnosis based on the fusion of CNNs and handcrafted features. PeerJ Comput. Sci. 2020;6 doi: 10.7717/peerj-cs.306. e306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Attallah O., Ragab D.A., Sharkas M. MULTI-DEEP: A novel CAD system for coronavirus (COVID-19) diagnosis from CT images using multiple convolution neural networks. PeerJ. 2020;8 doi: 10.7717/peerj.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El-bana S., Al-Kabbany A., Sharkas M. A multi-task pipeline with specialized streams for classification and segmentation of infection manifestations in covid-19 scans. PeerJ Comput. Sci. 2020;6 doi: 10.7717/peerj-cs.303. e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J., Xie Y., Li Y., Shen C., Xia Y. 2020. Covid-19 screening on chest X-ray images using deep learning based anomaly detection. arXiv preprint arXiv:2003.12338. [Google Scholar]
- 35.Ko H., Chung H., Kang W.S., Kim K.W., Shin Y., Kang S.J., Lee J.H., Kim Y.J., Kim N.Y., Jung H., Lee J. COVID-19 pneumonia diagnosis using a simple 2D deep learning framework with a single chest CT Image: Model Development and Validation. J. Med. Internet Res. 2020;22(6) doi: 10.2196/19569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall L.O., Paul R., Goldgof D.B., Goldgof G.M. 2020. Finding covid-19 from chest X-rays using deep learning on a small dataset. arXiv preprint arXiv:2004.02060. [Google Scholar]
- 37.Wang M., Zheng S., Li X., Qin X. 2014 International Conference on Information Science, Electronics and Electrical Engineering, vol. 1. IEEE; 2014. A new image denoising method based on Gaussian filter; pp. 163–167. [Google Scholar]
- 38.Sulaiman M.H., Mustaffa Z., Saari M.M., Daniyal H. Barnacles mating optimizer: A new bio-inspired algorithm for solving engineering optimization problems. Eng. Appl. Artif. Intell. 2020;87 [Google Scholar]
- 39.https://transpireonline.blog/2020/02/03/a-new-optimization-algorithm-inspired-from-the-mating-behavior-of-barnacles-barnacles-mating-optimizer-algorithms-bmo/.
- 40.Mou L., Ghamisi P., Zhu X.X. Deep recurrent neural networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017;55(7):3639–3655. [Google Scholar]
- 41.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 42.Hang R., Liu Q., Hong D., Ghamisi P. Cascaded recurrent neural networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2019;57(8):5384–5394. [Google Scholar]
- 43.Shankar K., Perumal E., Tiwari P., Shorfuzzaman M., Gupta D. Deep learning and evolutionary intelligence with fusion-based feature extraction for detection of COVID-19 from chest X-ray images. Multimedia Syst. 2021:1–13. doi: 10.1007/s00530-021-00800-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shankar K., Perumal E. A novel hand-crafted with deep learning features based fusion model for COVID-19 diagnosis and classification using chest X-ray images. Complex Intell. Syst. 2020:1–17. doi: 10.1007/s40747-020-00216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.https://github.com/ieee8023/covid-chestxray-dataset.
- 46.Uthayakumar J., Vengattaraman T., Dhavachelvan P. Swarm intelligence based classification rule induction (CRI) framework for qualitative and quantitative approach: An application of bankruptcy prediction and credit risk analysis. J. King Saud Univ.-Comput. Inf. Sci. 2017 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new datasets were generated during the current study.