Abstract
The coronavirus disease 2019(COVID-19) is recognized as systemic inflammatory response syndrome. It was demonstrated that a rapid increase of cytokines in the serum of COVID-19 patients is associated with the severity of disease. However, the mechanisms of the cytokine release are not clear. By using immunofluorescence staining we found that the number of CD11b positive immune cells including macrophages in the spleens of died COVID-19 patients, was significantly higher than that of the control patients. The incidence of apoptosis as measured by two apoptotic markers, TUNEL and cleaved caspase-3, in COVID-19 patients’ spleen cells is higher than that in control patients. By double immunostaining CD11b or CD68 and SARS-CoV-2 spike protein, it was found that up to 67% of these immune cells were positive for spike protein, suggesting that viral infection might be associated with apoptosis in these cells. Besides, we also stained the autophagy-related molecules (p-Akt、p62 and BCL-2) in spleen tissues, the results showed that the number of positive cells was significantly higher in COVID-19 group. And compared with non-COVID-19 patients, autophagy may be inhibited in COVID-19 patients. Our research suggest that SARS-CoV-2 may result in a higher rate of apoptosis and a lower rate of autophagy of immune cells in the spleen of COVID-19 patients. These discoveries may increase our understanding of the pathogenesis of COVID-19.
Keywords: Apoptosis, Autophagy, COVID-19, Spleen, Cytokines
1. Introduction
The coronavirus disease 2019(COVID-19) pandemic has resulted in more than 110 million diagnosed patients and about twenty-four million deaths worldwide since December 2019. The three large-scale outbreaks of coronary virus (CoVs) diseases over the past two decades include severe acute respiratory syndrome (SARS), Middle Eastern respiratory syndrome (MERS), and COVID-19 [1], [2], [3]. These diseases have three common features: (1) They were all caused by CoVs; (2) They exhibited a rapid increase of cytokines in the serum (also called cytokine storm); (3) They could result in life-threatening systemic inflammation, ARDS, and Multiple Organ Dysfunction Syndrome(MODS).
SARS-CoV-2 uses its spike protein(S protein) to bind with the angiotensin-converting enzyme 2 (ACE2) receptor on target cells. After entry, the virus rapidly replicates within host cells and accentuates the inflammatory response. Viral infection can also cause apoptosis or other types of cell death of infected cells. Morphologically, there are three classifications of cell death occur under physiological conditions: apoptosis, autophagic cell death and necrosis [4]. The apoptosis or execution of infected cells has been considered as an important defensive response to control infection and regulates the immune and inflammatory processes [5]. In addition, cell death that depends on autophagy is characterizes by cytoplasmic vacuolization, autophagosome formation and clearance of material via the lysosome [6]. Through these functions to ensure proper cell corpse engulfment and degradation.During SARS-CoV-2 infection, multiple cytokines were found to be elevated in the serum including interleukin-1β, interleukin-6, IP-10, TNF, interferon-γ, macrophage inflammatory protein (MIP) 1α and 1β, and VEGF [7]. However, the precise mechanisms through which cytokine storms occurred in COVID-19 patients are still not fully known.
The spleen, the largest lymph organ, contains about 25% of the body’s lymph tissue and plays an important role in innate/acquired immune responses against various pathogens. Multiple cell types such as neutrophils, monocytes, macrophages, and natural killer (NK) cells were found in the spleen and may be involved in the control of pathogens [8]. Among these immune cells,several studies indicate that macrophages play a critical role in the pathological consequences of COVID-19. Immunostaining of postmortem tissue from six patients who had died from COVID-19 revealed that ACE2 expresses on macrophages in spleens and lymph nodes and viral nucleocaspid protein (NP) can be found in ACE2 + cells, CD169 + macrophages [9]. Whatsmore, Liao [10] and Hibah Shaath [11] both reported the presence of proinflammatory monocyte-derived macrophages in the bronchoalveolar lavage(BAL) from patients with severe COVID-9,Chen further confirmed that macrophage cluster-1 is one of the marker of severe COVID-19. Based on the above discoveries, we focused on macrophages in this study.
Macrophages, whose marker is CD68, have a variety of functions including the removal of cell debris, regulation of the function of neighboring cells, and the secretion of cytokines [12]. CD11b is the marker of inflammatory cells including monocytes, macrophages, and nature killer (NK) cells. We aim to evaluate the apoptosis and autophagy of immune cells in spleen tissue obtained from patients who died from COVID-19. Our findings indicate that apoptosis and autophagy of immune cells in the spleen may contribute to the occurrence of cytokine storms in COVID-19 patients. To our knowledge, this is the first report that defines the spleen as an important source of virus replication and cytokine production in COVID-19 patients.
2. Materials and methods
2.1. Patients and controls
Spleen tissue samples were obtained by autopsy from three patients who died from COVID-19 and compared them with an age-matched group of two non-COVID-19 patients. The study was approved by the Medical Ethical Committee of the Wuhan Infectious Diseases Hospital (KY-2020–15.01). All data were collected from Zhongnan Hospital of Wuhan University. Tissues were used for hematoxylin and eosin staining and immunohistochemical staining.
2.2. Antibodies and reagents
We used the following antibodies:anti-CD11b(1:500, ab8878, Abcam), anti-CD68(1:500, GB14043, Servicebio), anti-cleaved caspase-3 (Asp175) (5A1E) (1:400, #9664, Cell Signaling Technology, USA), anti-SARS-CoV-2 spike glycoprotein antibody-Coronavirus(1:500, ab272504, Abcam), anti-phospho Akt(1:800, 13038T, Cell Signaling Technology), anti-SQSTM1/p62(1:250, 23214S, Cell Signaling Technology), anti-BCL-2(1:200, sc-492, Santa Cruz Biotechnology), Alexa Fluor 647 goat anti-rat IgG(1:500, ab150159, Abcam), Alexa Fluor 488 goat anti-rabbit (1:500, ab150077, Abcam), Alexa Fluor 647 Goat anti-mouse (1:500, ab150115, Abcam), DAPI (Beyotime Institute of Biotechnology, China).
2.3. Immunofluorescence staining
The tissue slices were incubated with goat serum for blocking for 45 min. The slices were then incubated with primary antibodies at 4 ℃ overnight. After washing three times with phosphate-buffered saline(PBS) for 10 min each time, the samples were incubated with species-specific secondary antibodies separately for 60 min at room temperature. They have then washed with PBS again and incubated with DAPI for nucleus counter-staining and use a fluorescence microscope to obtain pictures.
2.4. TUNEL staining in spleen tissue
We carried out the TUNEL assay through the One Step TUNEL Apoptosis Assay Kit(Beyotime, Institute of Biotechnology, China). The spleen slices were deparaffinized in xylene and rehydrated through graded alcohol, and then treated with proteinase K for 20 min at room temperature. The slices were washed with PBS three times. We prepared the TUNEL reaction mixture by the instructions and then applied the liquid to the slices. They were put in a humidified atmosphere in the dark for 1 h at 37 ℃. Afterward, the tissue was washed with PBS three times again. The FITC labeled TUNEL-positive cells were imaged under inverted fluorescence microscopy.
2.5. Hematoxylin and eosin staining
The slices were stained for hematoxylin and eosin staining kit (Beyotime) following the manufacturer’s instructions.
2.6. Confocal microscopy
Histological immunofluorescence staining was visualized on a Leica SP8 STED laser confocal microscopy system.
2.7. Statistical analyses
All data analyses were performed in SPSS 23.0 software (SPSS Inc., Chicago, IL, USA). P < 0.05 was considered to indicate a statistically significant difference. Graphs were constructed using GraphPad Prism version 6(Graphpad Software, San Diego, CA, USA).
3. Results
3.1. HE staining findings
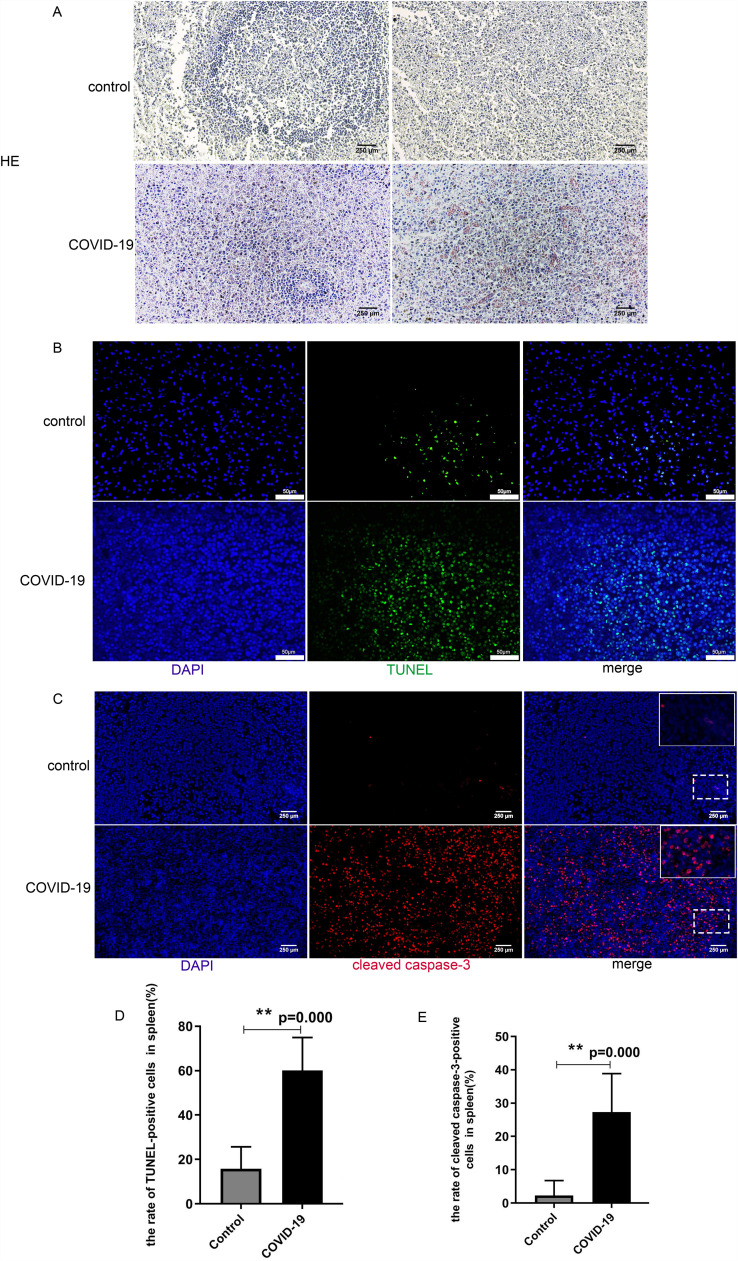
Compared to non-COVID-19 patients, all the spleens from COVID-19 patients showed that white pulp decreased, while red pulp was congested. Besides, the spleen tissues of COVID-19 patients were disorganized and the number of cells was increased microscopically ( Fig. 1A).
Fig. 1.
Increased apoptosis of spleen cells in COVID-19 patients. A. H&E staining of spleens from non-COVID-19 control (upper panel) and COVID-19 patients(lower panel). Spleens from COVID-19 patient 1 showing increased red pulp, white pulp decreased. Spleen from COVID-19 patient 2 showing mild red pulp infarction. B.Representative pictures showed TUNNEL-positive cells in the spleen tissues from non-COVID-19 control (upper panel) and COVID-19 patients(lower panel). The merged pictures of DAPI (blue) with TUNEL (green)-positive cells in the spleen were shown; C. Representative pictures showed cleaved caspase 3-positive cells in the spleen tissues from non-COVID-19 control (upper panel) and COVID-19 patients(lower panel). The merged pictures of DAPI (blue) with cleaved caspase-3 (red)-positive cells in the spleen were shown. D.Percentage of TUNEL-positive cells in the spleens of the control and COVID-19 group (N = 5, **P = 0.00). E. Percentage of cleaved caspase-3-positive cells in the spleens of the control and COVID-19 group (N = 5, **P = 0.00). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Apoptosis and cell death in spleen tissue
To examine apoptotic cells in spleen tissue, we stained the tissue slides by TUNEL assay. There were 15.8% TUNEL positive cells in the spleens of control patients compared to 60.1% TUNEL positive cells in the spleens of COVID-19 patients(Fig. 1B, D). These results implied that the incidence of apoptosis in COVID-19 patients' spleen cells is significantly higher than in non-COVID-19 patients(P = 0.00).
Caspase-3 has long been regarded as the hallmark of apoptosis.Caspase-3 initiates apoptosis through cleavage at specific sites and cleaved caspase-3 is a marker of caspase-dependent apoptosis and cell death. Therefore, we examined cleaved caspase-3 to further evaluate the death of spleen cells. The results confirmed that the percentage of cells positive with cleaved caspase-3 was significantly elevated in COVID-19 patients compared to those of the non-COVID-19 control group (Fig. 1C, E).
3.3. Immune cells including macrophages underwent cell death
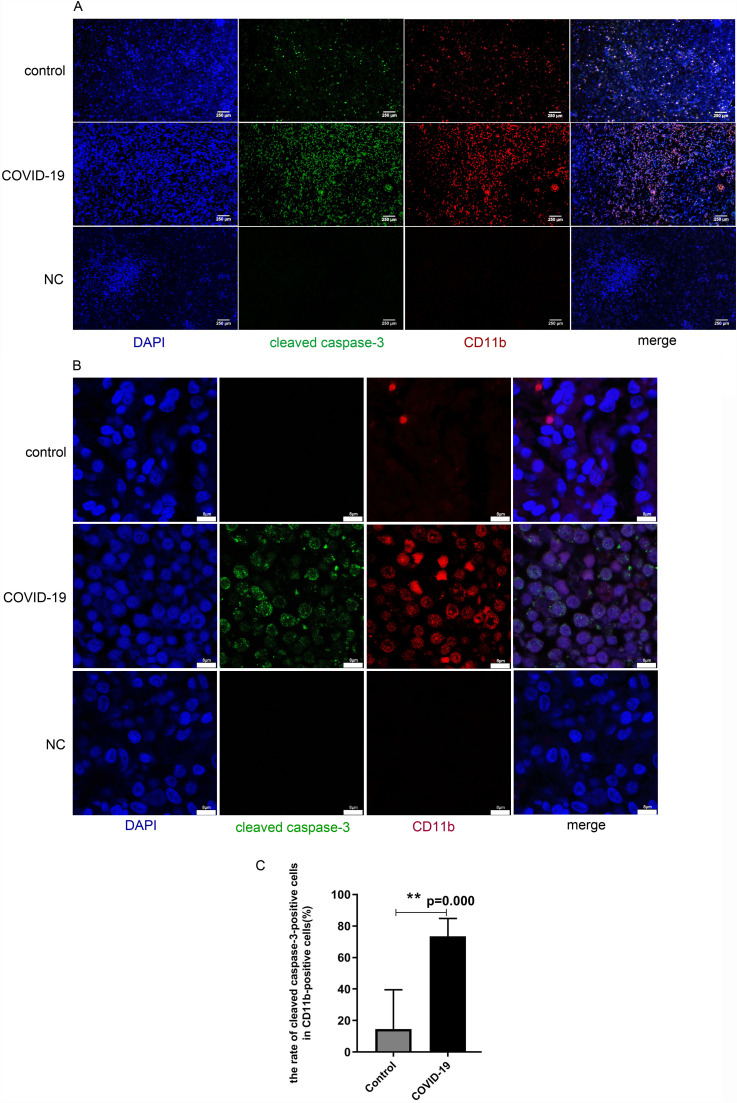
To further illustrate the cell types that underwent cell death in the spleen tissue, we co-stained the tissue sections with both antibodies against CD11b and antibodies against cleaved caspase-3. Cells positive for both CD11b and cleaved caspase-3 were considered dead immune cells ( Fig. 2A). Confocal images of the slides confirmed the result (Fig. 2B). The data showed that in COVID-19 patients, the number of dead immune cells in the spleen tissues was significantly higher than in the control group(P = 0.00)(Fig. 2C).
Fig. 2.
Increased apoptosis of CD11b positive immune cells in tissues from spleens of COVID-19 patients. Tissues from non-COVID-19 control patients were shown at the upper panels, Tissues from COVID-19 patients were shown at the middle panels, Negative control (NC: incubate with blocking solution without primary antibody) were shown at the bottom panels. A. Representative pictures were immunofluorescence co-stained with antibodies against cleaved caspase-3 (green) and CD11b (red). B. Confocal images of cleaved caspase-3 (green) and CD11b (red), respectively. C.Percentage of apoptotic cells in the CD11b-positive cells in tissues from for the control and COVID-19 group respectively (N = 5, **P = 0.00). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
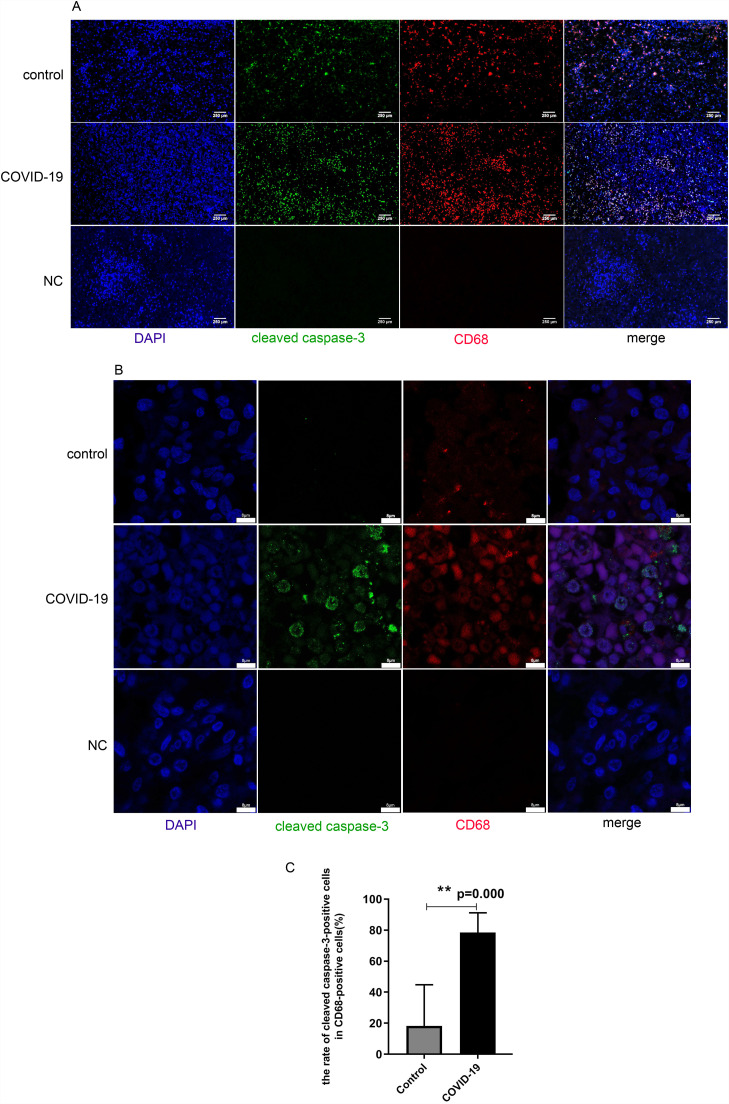
Furthermore, sections were also stained with both antibodies against CD68 and antibodies against cleaved caspase-3 to ascertain macrophages that underwent death. Cells positive for both CD68 and cleaved caspase-3 were considered as dead macrophages ( Fig. 3A). Confocal images of the slides confirmed the result (Fig. 3B). The data showed that in COVID-19 patients, the number of dead macrophages in the spleen tissues was significantly higher than in the control group (P = 0.00)(Fig. 3C).
Fig. 3.
Increased apoptosis of macrophages in tissues from spleens of COVID-19 patients. Tissues from non-COVID-19 control patients were shown at the upper panels, Tissues from COVID-19 patients were shown at the middle panels, Negative control (NC: incubate with blocking solution without primary antibody) were shown at the bottom panels. A. Representative pictures of immunofluorescence co-stained with antibodies against cleaved caspase 3 (green) and CD68 (red), respectively. Merged pictures represent the co-localization of red, green, and blue staining; B. Confocal images of cleaved caspase-3 (green) and CD68(red) by co-immunostaining. C. Percentage of apoptotic cells in the CD68-positive cells from tissues of the control and COVID-19 group. (N = 5, **P = 0.00). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
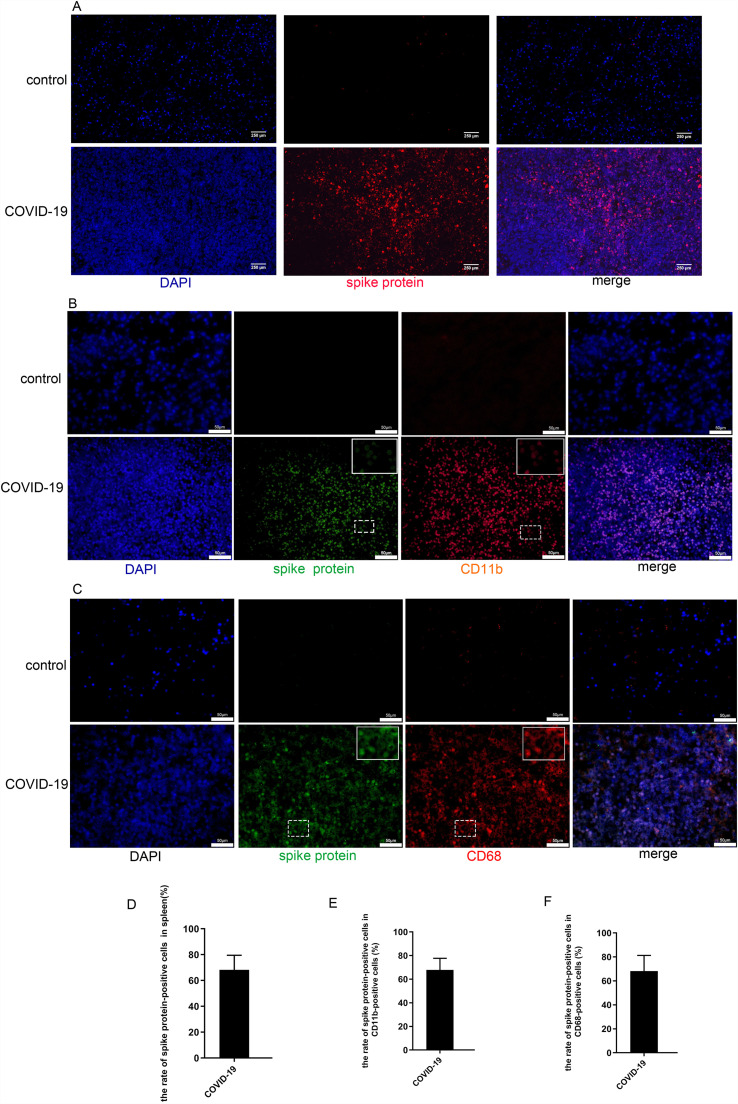
3.4. SARS-CoV-2 spike protein was expressed in immune cells
Since SARS-CoV-2 enters host cells by binding its S protein to ACE2 on the host cell’s membrane [13], [14], [15]. We have found that SARS-CoV-2 S protein was expressed in 68% of cells in spleen tissues of COVID-19 patients but not in that of controls ( Fig. 4A, D). We then sought to explore whether SARS-CoV-2 could attack immune cells and macrophages. We performed double immunostaining of CD11b and SARS-CoV-2 S protein, as well as CD68 and SARS-CoV-2 S protein in the two groups. The data presented showed that SARS-CoV-2 S protein and CD11b co-expression were readily detectable in these tissues from COVID-19 patients(Fig. 4B). The SARS-CoV-2 S protein-positive rate among CD11b positive cells was up to 67%(Fig. 4E). In addition, cells of spleen tissue from COVID-19 patients were also successively co-stained with SARS-CoV-2 S protein and CD68 antibodies(Fig. 4C). The SARS-CoV-2 S protein-positive rate among CD68 positive cells was up to 68.1%(Fig. 4F). In contrast, spleen tissue of non-COVID-19 control patients stained with little anti-CD68/anti-CD11b antibodies while no SARS-CoV-2 Spike proteins were detected in control patients. These results indicated SARS-CoV-2 may infect immune cells including macrophages during infection of SARS-CoV-2.
Fig. 4.
SARS-CoV-2 Spike protein was expressed in immune cells. A. Representative pictures of immunofluorescence co-staining of SARS-CoV-2 Spike protein (red) in tissues from COVID-19 patients(upper panel). B. Representative pictures of immunofluorescence staining of SARS-CoV-2 Spike protein (green) and CD11b (red) in COVID-19 spleen tissues(upper panel). C. Representative pictures of immunofluorescence co-staining of SARS-CoV-2 Spike protein (green) and CD68 (red). D.E.F. Percentages of SARS-CoV-2 Spike protein-positive cells in total(D), CD68 positive(E), CD11b positive (F) cells respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
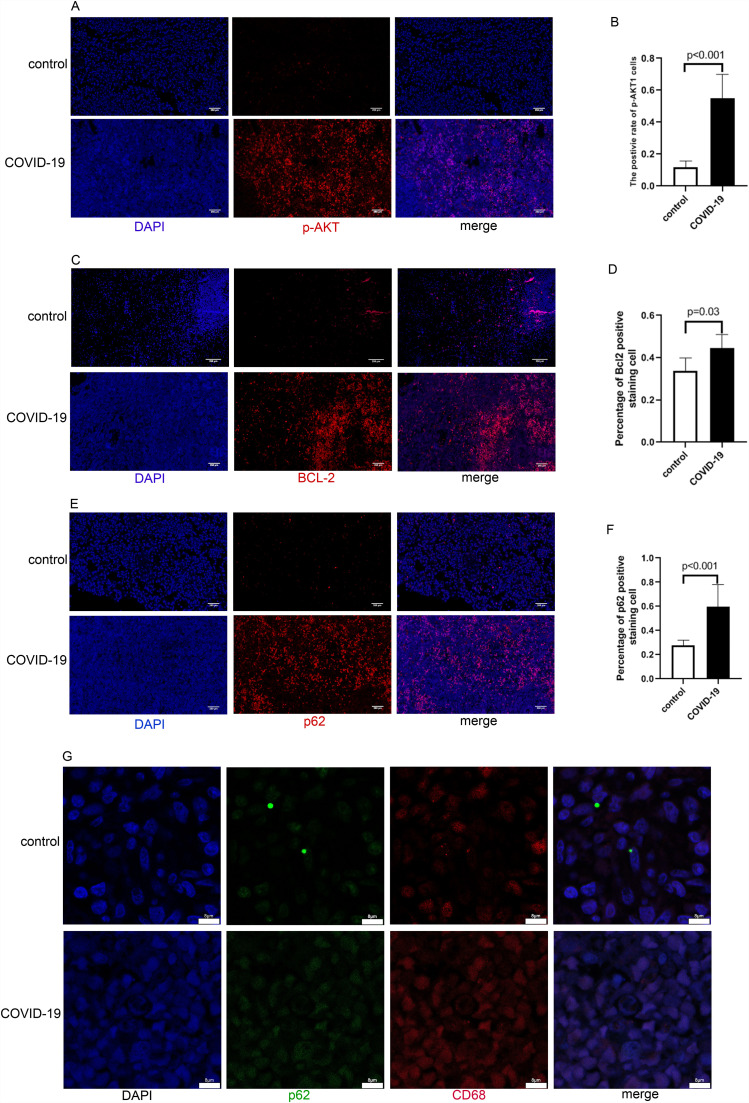
3.5. Autophagy in spleen tissue
Autophagy-related molecules, including p-Akt、p62 and BCL-2 were stained to examine positive cells in spleen tissue. We found that in COVID-19 patients, the number of staining positive cells in the spleen tissues was significantly higher than in the control group ( Fig. 5A-F). The above three molecules regulate autophagy through different signaling pathways and p62 was found to be accumulated in many cell types with autophagy deficiency [16], [17]. Their increased expression facilitate autophagic degradation, suggest that autophagy may be inhibited in COVID-19 patients. Then, we co-stained the tissue sections with p62 against CD68 to further illustrate the cell types that underwent autophagy in the spleen tissue. Confocal laser microscopy showed that CD68 and p62 were co-localized in COVID-19 patients(Fig. 5G). The result indicated that the autophagy of macrophages may be inhibited in COVID-19 patients.
Fig. 5.
Autophagy-related molecules of spleen cells in COVID-19 patients. Tissues from non-COVID-19 control patients were shown at the upper panels, Tissues from COVID-19 patients were shown at the lower panels. A. Representative pictures showed p-AKT-positive cells in the spleens of the control and COVID-19 group. B. Percentage of p-AKT-positive cells in the spleens of two groups (N = 5, P < 0.001). C. Representative pictures showed BCL-2-positive cells in the spleens of the control and COVID-19 group. D. Percentage of BCL-2-positive cells in the spleens of two groups (N = 5, P = 0.03). E. Representative pictures showed p62-positive cells in the spleens of the control and COVID-19 group.F.Percentage of p62-positive cells in the spleens of two groups (N = 5, P < 0.001). G. Confocal images of p62 (green) and CD68 (red), respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Reports of elevated cytokine levels and beneficial effects of immunosuppressant agents in COVID-19 patients suggest that the pathogenesis of COVID-19 may be related to cytokine storms [18], [19]. Studies also showed that cytokine storm was associated with poor outcomes [20]. However, previous studies of COVID-19 have done little on the role of the spleen in the pathogenesis of cytokine storm although the spleen is the largest secondary lymphoid organ in the body and as such hosts a wide range of immunological functions [12]. In this study, we have found that (1) COVID-19 patients have a higher rate of apoptotic and dead cells in spleen tissue than that of non-COVID-19 control patients; (2) there were more immune cells including macrophages in COVID-19 patients compared to the control group within the spleen tissue, indicating that infiltrating immune cells may play an important role in the pathogenesis of COVID-19; (3) the TUNEL and cleaved caspase-3 positive immune cells including macrophages in COVID-19 patients were also significantly higher than that in the control group respectively; (4) the high rate of S protein in immune cells suggests that SARS-CoV-2 replicates within these immune cells; (5)the number of autophagy-related molecules of spleen tissues in COVID-19 patients was significantly higher than in the control group. In addition, the lower blood oxygen and possible thrombosis in the spleen reported in COVID-19 patients [21] may contribute to the massive death of immune cells. Therefore, these data suggest that SARS-CoV-2 may result in a higher rate of death and a lower rate of autophagy of immune cells in the spleen of COVID-19 patients.
Previous studies demonstrated that beta coronavirus infection of monocytes, macrophages, and dendritic cells can trigger the secretion of IL-6 and other inflammatory cytokines [18]. IL-6, an important mediator of the acute inflammatory response, correlates with respiratory failure, ARDS, and adverse clinical outcomes [22], [23], [24]. The increased systemic cytokine production results in a systemic "cytokine storm" [25]. On one hand, the death of macrophages and other inflammatory immune cells in the spleen observed may be one of the mechanisms that induce cytokine storm; on the other hand, loss of these immune cells will disrupt the immune system and aggravate the damage to the host by the virus.
Apoptosis and pyroptosis are two types of programmed cell death and apoptosis is characterized by morphological changes, including cell shrinkage, nuclear condensation, and plasma membrane blebbing [26]. Apoptosis plays a critical role in host defense against viral infections by eliminating infected cells. Meanwhile, most of the apoptotic cells undergo secondary necrosis which will lead to increased membrane permeability. Subsequently, the cellular contents which contain various cytokines will be released into the blood [27], [28]. The spleen contains lymphocytes and many types of leukocytes, including neutrophils, monocytes, dendritic cells (DCs), and macrophages [29] ( Fig. 6). During infection, the significant increase of CD11b or CD68 positive cells in the spleen tissue suggests many immune cells were infiltrated. Our results clearly showed increased apoptosis and death of these immune cells in the spleen of COVID-19 patients, suggesting their crucial role in pathogenesis (Fig. 5). ACE2 is widely distributed and expressed in different tissues and organs such as the lungs, liver, intestine, spleen, brain, muscle, pituitary, and skin in the human body [20], [30]. It is so far unclear why the spleen serves as a vital target of the SARS-COV-2 virus and exhibits massive apoptosis within the tissue cells. However, the results at least explain partly why spleens from COVID-19 patients are smaller than normal as discovered by clinicians and pathologists [21]. It was demonstrated that ACE2-expressing cells include AT1 cells, airway epithelial cells, fibroblasts, endothelial cells, and macrophages [31]. By binding with ACE2 with its S protein, SARS COV-2 may target these organs or cells to replicate within them. Consistently, we find that the virus’ S protein was widely expressed in the immune cells including macrophages in the spleen, suggesting that the virus is replicating in these cells and causing apoptosis. However, further studies are needed to prove this hypothesis.
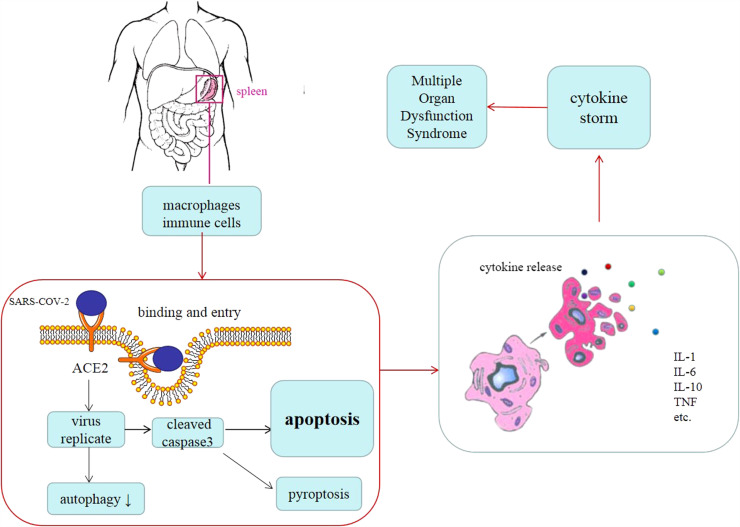
Fig. 6.
Diagram of proposed mechanisms of “cytokine storm” in COVID-19 patients. After infection, SARS-COV-2 viruses attack and replicate in the immune cells in the spleen. As a result, massive apoptosis and autophagy take place within the spleen. A large number of various cytokines are released from apoptotic immune cells and CD68 positive macrophages into the blood vessels and cause multiple organ dysfunction syndromes including ARDS.
In terms of the relationship between inflammation and autophagy, autophagy can inhibit the inflammatory response and the activation of inflammasomes through classical and non-classical pathways. On the other hand, inflammation can improve the level of autophagy and promote the formation of autophagosomes [32]. Along these lines, it was demonstrated recently that autophagy activation can inhibit MERS-CoV infection.In turns, MERS-CoV infection can block autophagy at the autolysosome formation stage. Such an effect would benefit the viral replication in several ways [33]. Based on the above, we suspect that autophagy generally antagonize coronavirus replication. In addition, studies have shown that in macrophages cells, the inhibition of autophagy leads to higher levels of both IL-1β and IL-23, and these in turn stimulate T lymphocytes to secrete IL-17, IFN-γ and IL-22 [34]. In our study,we found that autophagy may be inhibited in COVID-19 patients, suggesting that the replication of SARS-COV-2 may be enhanced in spleen.Moreover, the release of inflammatory cytokines caused by autophagy inhibition of macrophages may aggravate the cytokine storm. All of these may have contributed to the COVID-19 patient's death. As above, further research is needed to confirm our hypothesis.
In conclusion, the spleen might not only be one of the organs with severe inflammatory loci but also a reservoir of SARS-CoV-2 and a major source of the cytokines released into the blood. Our study may shed new light on further understanding of.pathogenesis in COVID-19 patients.
CRediT authorship contribution statement
Haiqin Ping, Hengning Ke: Conceptualization, Methodology. Haiqin Ping, Kai Zhang, Xin Tong: Data curation, Writing – original draft. Yunyun Wang, Lu Zhiyan: Visualization, Investigation. Xien Gui, Xinghuan Wang, Liang Liu: Supervision. Haiqin Ping: Software, Validation. Hengning Ke, Zhaojun Chen, Caiyun Cai: Writing – review & editing.
Conflict of interest
There are no conflicts of interest to declare.
Acknowledgments
The authors would like to thank William Ke for his editing of the manuscript. This work was funded by the National Natural Science Foundation of China, China (31760263) to Dr. Hengning Ke and the Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences, China (2020-PT320-004).
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J., Liang Z., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of coronavirus disease 2019. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41(12):1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green D.R., Llambi F. Cell death signaling. Cold Spring Harb. Perspect. Biol. 2015;7(12) doi: 10.1101/cshperspect.a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roulston A., Marcellus R.C., Branton P.E. Viruses and apoptosis. Annu Rev. Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 6.Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., Alnemri E.S., Altucci L., Amelio I., Andrews D.W., Annicchiarico-Petruzzelli M., Antonov A.V., Arama E., Baehrecke E.H., Barlev N.A., Bazan N.G., Bernassola F., Bertrand M., Bianchi K., Blagosklonny M.V., Blomgren K., Borner C., Boya P., Brenner C., Campanella M., Candi E., Carmona-Gutierrez D., Cecconi F., Chan F.K., Chandel N.S., Cheng E.H., Chipuk J.E., Cidlowski J.A., Ciechanover A., Cohen G.M., Conrad M., Cubillos-Ruiz J.R., Czabotar P.E., D’Angiolella V., Dawson T.M., Dawson V.L., De Laurenzi V., De Maria R., Debatin K.M., DeBerardinis R.J., Deshmukh M., Di Daniele N., Di Virgilio F., Dixit V.M., Dixon S.J., Duckett C.S., Dynlacht B.D., El-Deiry W.S., Elrod J.W., Fimia G.M., Fulda S., García-Sáez A.J., Garg A.D., Garrido C., Gavathiotis E., Golstein P., Gottlieb E., Green D.R., Greene L.A., Gronemeyer H., Gross A., Hajnoczky G., Hardwick J.M., Harris I.S., Hengartner M.O., Hetz C., Ichijo H., Jäättelä M., Joseph B., Jost P.J., Juin P.P., Kaiser W.J., Karin M., Kaufmann T., Kepp O., Kimchi A., Kitsis R.N., Klionsky D.J., Knight R.A., Kumar S., Lee S.W., Lemasters J.J., Levine B., Linkermann A., Lipton S.A., Lockshin R.A., López-Otín C., Lowe S.W., Luedde T., Lugli E., MacFarlane M., Madeo F., Malewicz M., Malorni W., Manic G., Marine J.C., Martin S.J., Martinou J.C., Medema J.P., Mehlen P., Meier P., Melino S., Miao E.A., Molkentin J.D., Moll U.M., Muñoz-Pinedo C., Nagata S., Nuñez G., Oberst A., Oren M., Overholtzer M., Pagano M., Panaretakis T., Pasparakis M., Penninger J.M., Pereira D.M., Pervaiz S., Peter M.E., Piacentini M., Pinton P., Prehn J., Puthalakath H., Rabinovich G.A., Rehm M., Rizzuto R., Rodrigues C., Rubinsztein D.C., Rudel T., Ryan K.M., Sayan E., Scorrano L., Shao F., Shi Y., Silke J., Simon H.U., Sistigu A., Stockwell B.R., Strasser A., Szabadkai G., Tait S., Tang D., Tavernarakis N., Thorburn A., Tsujimoto Y., Turk B. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25(3):486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Z., Cai T., Fan L., Lou K., Hua X., Huang Z., Gao G. Clinical value of immune-inflammatory parameters to assess the severity of coronavirus disease 2019. Int. J. Infect. Dis. 2020;95:332–339. doi: 10.1016/j.ijid.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto D., Chow A., Noizat C., Teo P., Beasley M.B., Leboeuf M., Becker C.D., See P., Price J., Lucas D., Greter M., Mortha A., Boyer S.W., Forsberg E.C., Tanaka M., van Rooijen N., García-Sastre A., Stanley E.R., Ginhoux F., Frenette P.S., Merad M. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeqing Feng, Bo Diao, Rongshuai Wang, et al. The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) directly decimates human spleens and lymph nodes. Preprint at medRxiv. https://doi.org/10.1101/2020.03.27.20045427 (2020).
- 10.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 11.Shaath H., Vishnubalaji R., Elkord E., Alajez N.M. Single-cell transcriptome analysis highlights a role for neutrophils and inflammatory macrophages in the pathogenesis of severe COVID-19. Cells. 2020;9(11):2374. doi: 10.3390/cells9112374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis S.M., Williams A., Eisenbarth S.C. Structure and function of the immune system in the spleen. Sci. Immunol. 2019;4(33):eaau6085. doi: 10.1126/sciimmunol.aau6085. eaau6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63(3):457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wrapp D., Wang N., Corbett K.S., et al. Cryo-EM structure of the 2019-nCoV spikeintheprefusion conformation. Science. 2020;13(367):1260–1263. doi: 10.1126/science.abb2507. 6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartlett B.J., Isakson P., Lewerenz J., Sanchez H., Kotzebue R.W., Cumming R.C., Harris G.L., Nezis I.P., Schubert D.R., Simonsen A., Finley K.D. p62, Ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy. 2011;7(6):572–583. doi: 10.4161/auto.7.6.14943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nogalska A., Terracciano C., D’Agostino C., King Engel W., Askanas V. p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion-body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta Neuropathol. 2009;118(3):407–413. doi: 10.1007/s00401-009-0564-6. [DOI] [PubMed] [Google Scholar]
- 18.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368(6490):473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 19.The RECOVERY Collaborative Group. Dexamethasone in hospitalized patients with COVID-19—preliminary report. N Engl J Med DOI: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed]
- 20.Huang K.-J., Su I.-J., Theron M., Wu Y.C., Lai S.K., Liu C.C., Lei H.Y. An interferon-gamma-related cytokine storm in SARS patients. J. Med Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ren L., Liu Q., Wang R., Chen R., Ao Q., Wang X., Zhang J., Deng F., Feng Y., Wang G., Zhou Y., Li L., Liu L. Clinicopathologic features of COVID-19: a case report and value of forensic autopsy in studying SARS-CoV-2 infection. Am. J. Forensic Med. Pathol. 2021;42(2):164–169. doi: 10.1097/PAF.0000000000000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130(5):2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruan Q., Yang K., Wang W., Jiang L., Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46:846–848. doi: 10.1007/s00134-020-05991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang S., Tanaka T., Narazaki M., Kishimoto T. Targeting interleukin-6 signaling in clinic. Immunity. 2019;50(4):1007–1023. doi: 10.1016/j.immuni.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 25.Fajgenbaum D.C., June C.H. Cytokine storm. N. Engl. J. Med. 2020;383(23):2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galluzzi L., Vitale I., Abrams J.M., Alnemri E.S., Baehrecke E.H., Blagosklonny M.V., Dawson T.M., Dawson V.L., El-Deiry W.S., Fulda S., Gottlieb E., Green D.R., Hengartner M.O., Kepp O., Knight R.A., Kumar S., Lipton S.A., Lu X., Madeo F., Malorni W., Mehlen P., Nuñez G., Peter M.E., Piacentini M., Rubinsztein D.C., Shi Y., Simon H.U., Vandenabeele P., White E., Yuan J., Zhivotovsky B., Melino G., Kroemer G. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19(1):107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majno G., Joris I. Apoptosis, oncosis, and necrosis. An overview of cell death. Am. J. Pathol. 1995;146:3–15. [PMC free article] [PubMed] [Google Scholar]
- 28.Rock K.L., Kono H. The inflammatory response to cell death. Annu Rev. Pathol. 2008;3:99–126. doi: 10.1146/annurev.pathmechdis.3.121806.151456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nolte M.A., ’t Hoen E.N.M., van Stijn A., Kraal G., Mebius R.E. Isolation of the intact white pulp.Quantitative and qualitative analysis of the cellular composition of the splenic compartments. Eur. J. Immunol. 2000;30(2):626–634. doi: 10.1002/1521-4141(200002)30:2<626::AID-IMMU626>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 30.Han T., Kang J., Li G., Ge J., Gu J. Analysis of 2019-nCoV receptor ACE2 expression in different tissues and its significance study. Ann. Transl. Med. 2020;8(17):1077. doi: 10.21037/atm-20-4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;202(5):756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mo Y., Sun Y.Y., Liu K.Y. Autophagy and inflammation in ischemic stroke. Neural Regen. Res. 2020;15(8):1388–1396. doi: 10.4103/1673-5374.274331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gassen N.C., Niemeyer D., Muth D., Corman V.M., Martinelli S., Gassen A., Hafner K., Papies J., Mösbauer K., Zellner A., Zannas A.S., Herrmann A., Holsboer F., Brack-Werner R., Boshart M., Müller-Myhsok B., Drosten C., Müller M.A., Rein T. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-coronavirus infection. Nat. Commun. 2019;10(1):5770. doi: 10.1038/s41467-019-13659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peral de Castro C., Jones S.A., Ní Cheallaigh C., Hearnden C.A., Williams L., Winter J., Lavelle E.C., Mills K.H.G., Harris J. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J. Immunol. 2012;189(8):4144–4153. doi: 10.4049/jimmunol.1201946. [DOI] [PubMed] [Google Scholar]