Abstract
The COVID-19 (coronavirus disease 2019) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to loss of human life in millions and devastating socio-economic consequences worldwide. The disease has created urgent needs for intervention strategies to control the crisis and meeting these needs requires a deep understanding of the structure-function relationships of viral proteins and relevant host factors. The trimeric spike (S) protein of the virus decorates the viral surface and is an important target for development of diagnostics, therapeutics and vaccines. Rapid progress in the structural biology of SARS-CoV-2 S protein has been made since the early stage of the pandemic, advancing our knowledge on the viral entry process considerably. In this review, we summarize our latest understanding of the structure of the SARS-CoV-2 S protein and discuss the implications for vaccines and therapeutics.
Current Opinion in Virology 2021, 50:173–182
This review comes from a themed issue on Virus structure and expression
Edited by José R Castón and Adam Zlotnic
For complete overview about the section, refer “Virus structure and expression (2022)”
Available online 8th September 2021
https://doi.org/10.1016/j.coviro.2021.08.010
1879-6257/© 2021 Elsevier B.V. All rights reserved.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the COVID-19 (coronavirus disease 2019) pandemic [1••], and its infection has led to millions of lives lost and devastating socio-economic consequences throughout the globe. There are urgent needs for innovative vaccine and therapeutic strategies to control this unprecedented crisis, as well as potential future needs if it becomes seasonal with continuous emergence of new variants. A deep understanding of the structure-function relationships of viral proteins and relevant host factors will be required in order to meet these needs. Coronaviruses (CoVs) are enveloped positive-stranded RNA viruses that enter a host cell by fusion of its envelope lipid bilayer with the target cell membrane. This first critical step of viral infection is catalyzed by its trimeric spike (S) protein, which decorates the virion surface as a major antigen and induces neutralizing antibody responses. The protein is therefore an important target for development of diagnostics, therapeutics and vaccines. Remarkable progress in the structural biology of SARS-CoV-2 S protein has been made since the initial outbreak of the virus [2], substantially advancing our molecular understanding of the viral entry process. Here we summarize our current knowledge on the structure of the SARS-CoV-2 S protein and discuss the implications for vaccines and therapeutics.
Overall structure of SARS-CoV-2 S protein
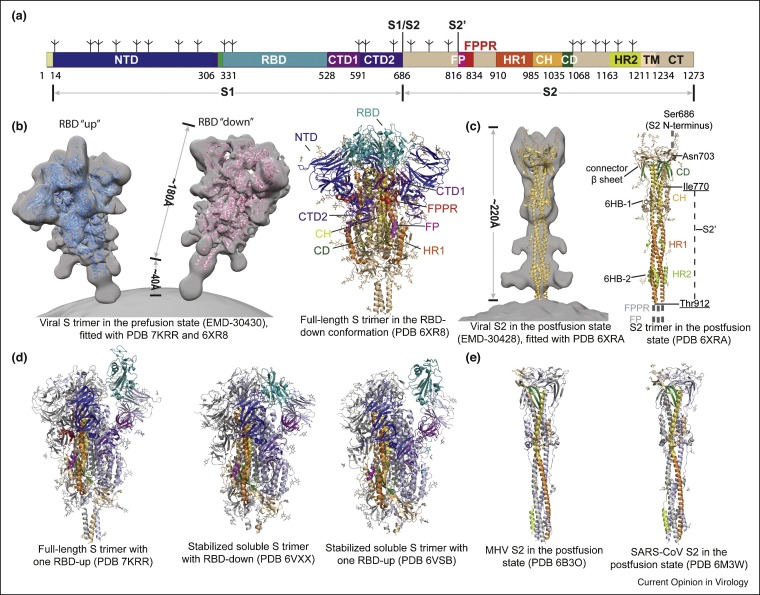
The SARS-CoV-2 spike glycoprotein is a type I membrane protein (Figure 1 a), which forms a trimer, anchored to the viral membrane by its transmembrane segment, while decorating the virion surface with it large ectodomain (Figure 1b). It binds to the receptor angiotensin-converting enzyme 2 (ACE2) on a host cell and undergo large structural rearrangements to promote membrane fusion [1••,3•]. The protein is heavily glycosylated with each protomer containing 22 N-linked glycosylation sites [4,5]. The full-length S protein of the Wuhan-Hu-1 strain from the initial outbreak has 1273 amino acid residues, including a N-terminus signal peptide, a receptor-binding fragment S1 and a fusion fragment S2. S1 can be further divided into N-terminal domain (NTD), receptor-binding domain (RBD) and C-terminal domains (CTD1 and CTD2), while S2 includes fusion peptide (FP), fusion-peptide proximal region (FPPR), heptad repeat 1 (HR1), central helix (CH), connector domain (CD), heptad repeat 2 (HR2), transmembrane segment (TM) and the cytoplasmic tail (CT), depicted in Figure 1a.
Figure 1.
Distinct conformational states of the SARS-CoV-2 spike protein.
(a) Schematic representation of the SARS-CoV-2 spike protein organization. Segments of S1 and S2 include: NTD, N-terminal domain; RBD, receptor-binding domain; CTD1, C-terminal domain 1; CTD2, C-terminal domain 2; S1/S2, S1/S2 cleavage site; S2′, S2′ cleavage site; FP, fusion peptide; FPPR, fusion peptide proximal region; HR1, heptad repeat 1; CH, central helix region; CD, connector domain; HR2, heptad repeat 2; TM, transmembrane anchor; CT, cytoplasmic tail; and tree-like symbols for glycans. (b) Left: viral SARS-CoV-2 S trimer in the prefusion conformation (EMD-30430; Ref. [15•]), fitted with the structures of purified proteins (PDB ID: 7KRR and 6XR8; Refs. [13••,50••]). Right: cryo-EM structure of the full-length S trimer in the RBD-down conformation (PDB ID: 6XR8). (c) Left: viral SARS-CoV-2 S2 trimer in the postfusion conformation (EMD-30428; Ref. [15•]), fitted with the structure of the purified protein (PDB ID: 6XRA; Ref. [13••]). Right: cryo-EM structure of the full-length S2 trimer in the postfusion conformation (PDB ID: 6XRA). (d) Additional structures of coronavirus S proteins, including the full-length SARS-CoV-2 S trimer carrying G614 in the one RBD-up conformation (PDB ID: 7KRR), the stabilized soluble SARS-CoV-2 S trimer in the RBD-down conformation (PDB ID: 6VXX; Ref. [7•]), the stabilized soluble SARS-CoV-2 S trimer in the one RBD-up conformation (PDB ID: 6VSB; Ref. [6••]). (e) MHV (mouse hepatitis virus) S2 in the postfusion state (PDB ID: 6B3O; Ref. [25]), and SARS-CoV S2 in the postfusion state (PDB ID: 6M3W; Ref. [24]).
Structures of S protein fragments derived from the Wuhan-Hu-1 strain, including the S ectodomain stabilized in its prefusion conformation [6••,7•], RBD-ACE2 complexes [8••,9•,10,11], and segments of S2 in the postfusion state [12], were determined within the first several months of the pandemic. Soon after, structures of detergent-solubilized, full-length S proteins in both prefusion and postfusion conformations [13••,14], as well as those of the intact S trimer on the virion surface, studied by cryo-electron tomography [15•,16••,17•,18••], were also reported (Figure 1b and c). Overall, the SARS-CoV-2 S structure shows many similarities to those of other coronavirus spike proteins [19, 20, 21, 22, 23]. In the prefusion structure, the S1 fragment, adopting a ‘V’ shaped architecture with the NTD at one arm and the RBD, CTD1 and CTD2 at the other (also see Figure 2 a), which wrap around the central helical bundle formed by the prefusion S2 fragment, projecting the N-terminal end of HR1 toward the viral membrane. Three RBDs form the apex of the S trimer, sampling two distinct conformations — ‘up’ representing a receptor-accessible state and ‘down’ representing a receptor-inaccessible state (Figure 1b). The three NTDs are located at the periphery of the trimer, each making contacts with the RBD from the adjacent protomer. The CTD1 and CTD2 pack underneath the RBD against S2 and between the two neighboring NTDs, indicating they could modulate these domains and play important roles in the structural rearrangements required for membrane fusion.
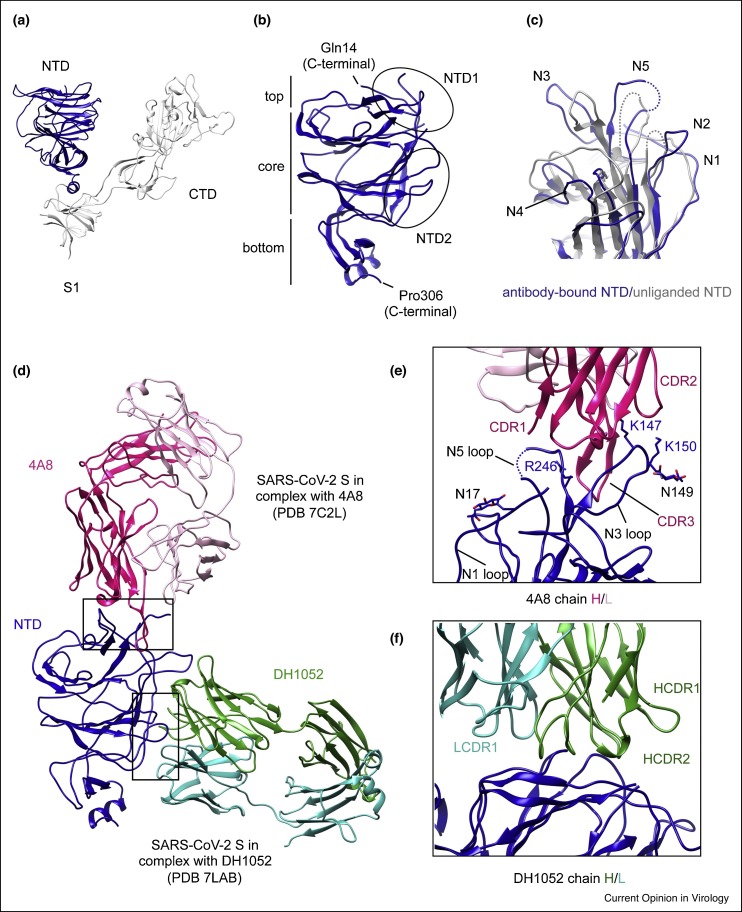
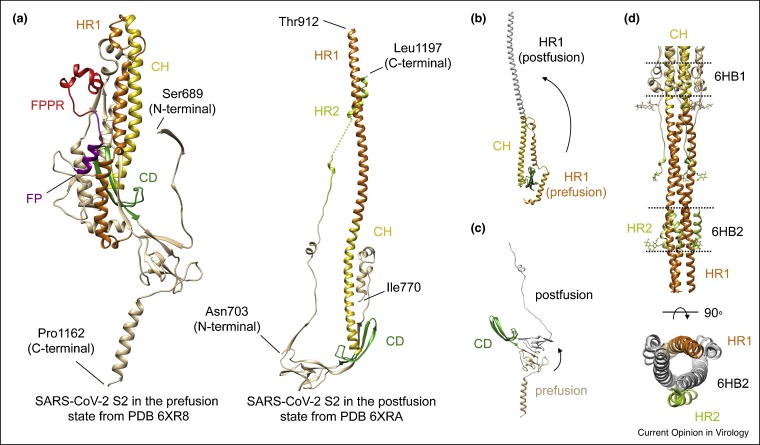
Figure 2.
Structures of NTD and its antibody complexes.
(a) Cryo-EM structure of S1 fragment from the full-length SARS-CoV-2 S trimer (PDB ID: 6XR8), with the NTD highlighted in blue and the rest of S1 in gray. (b) Close-up view of the NTD in the SARS-CoV-2 S protein. (c) The NTD (in blue) from its complex with 4A8 is superposed with the domain from the full-length S trimer in gray, showing shifts of the five surface loops (N1–N5). (e) and (f) Close-up view of the binding interface for the NTD-4A8 and NTD-DH1205 complexes with contacting residues in the NTD highlighted in sticks. (d) Superposition of the structures of the NTD in complex with antibody 4A8 Fab (PDB ID: 7C2L; Ref. [28••]) and DH1052 Fab (PDB ID: 7LAB; Ref. [32]), as indicated. Heavy and light chains of 4A8 are colored in red and pink, respectively, and those of DH1052 are in green and cyan, respectively.
In the postfusion conformation, S1 dissociates as a monomer, while S2 adopts a rigid, baseball-bat-like shape (∼220 Å long), and the HR1 flips over to form a continuous long helix together with the CH, which is further surrounded by short helices and β-sheets at the distal end of the membrane (Figure 1c and e). The connector domain (CD), together with a segment (residues 718–729) in the S1/S2–S2′ fragment, form a three-stranded β sheet, and residues 1127–1135 join the connector β sheet to expand it into four strands. Another segment (residues 737–769) in the S1/S2–S2′ fragment makes up three helical regions locked by two disulfide bonds that pack against the groove of the CH part of the coiled coil to form a short, six-helix bundle structure (6HB-1). The N-terminal region of HR2 adopts a one-turn helical conformation and also packs against the groove of the HR1 coiled coil; the C-terminal region of HR2 forms a longer helix that makes up the second six-helix bundle structure with the rest of the HR1 coiled coil (6HB-2) [13••,24,25].
N-terminal domain
At the periphery of the spike (Figure 1b) [13••], the NTD projects away from the threefold axis, and can be divided into the top, core and bottom regions (Figure 2b). The core structure has a galectin-like antiparallel β-sandwich fold, formed by one six-stranded β-sheet and the other with seven strands. The top region has two antiparallel β strands connected by a short loop, while the bottom region is primarily made up of two short β sheets and a helix. The overall structure of the NTD is decorated by eight N-linked glycans and similar to that of the S proteins from Middle East respiratory syndrome coronavirus (MERS-CoV) [26] and bovine coronavirus [27].
The exact function of the NTD in SARS-CoV-2 S remains unknown, although NTDs of other coronaviruses have been shown to recognize sugars upon initial attachment or specific protein receptors, or play a role in the prefusion-to-postfusion transition [27]. Nonetheless, NTD-targeted neutralizing antibodies (nAbs), with a potency at the nM level, have been isolated from SARS-CoV-2 infected patients [28••], suggesting a functionally critical role of this domain. High-resolution structures of the S protein in complex with NTD-directed neutralizing antibodies (4A8, FC05, CM25, 4-18, S2M28, and DH1205) have been determined [28••,29•,30,31•,32], showing that these antibodies primarily bind to two glycan-free surfaces of the domain, designated NTD-1 and NTD-2 regions, respectively (Figure 2b; Ref. [33]). Most antibodies target the NTD-1 region, which is thus named the NTD-1-antigenic supersite. It is located at the edge of the NTD top-core region, including five surface loops: N1 (residues 14–26), N2 (residues 67–79), N3 (residues 141–156), N4 (residues 177–186), N5 (residues 246–260) (Figure 2c), and a β-hairpin structure near N3, surrounded by four N-linked glycans (Asn17, Asn74, Asn122 and Asn149). These loops reconfigure upon binding to various antibodies (Figure 2c).
In the S-4A8 complex structure (Figure 2d) [28••], the third complementarity determining region (CDR3) of the 4A8 heavy chain inserts to a cleft formed by the N3 β-hairpin/loop and N5 loop, while the CDR1 and CDR2 interact with the tips of the two loops. Moreover, the glycan at Asn149 is very close to the interface and may also contribute to antibody binding (Figure 2e). Other antibodies, such as S2M28, 4-18, DH1050, CM25, FC05, 12C9 [33], also use their CDR1-3 to contact the N3 and N5 loops, but some interact with the nearby N1 loop or the glycan at Asn17 as well. Despite the differences in approaching angles among these antibodies, their interface with the NTD-1 is highly conserved. Up till now, NTD-2 is recognized by non-neutralizing antibodies, such as by DH1052 and 81D6 [33]. The CDR loops of both heavy and light chains in DH1052 interact with the surface formed by residues spanning 27–32, 59–62 and 211–218 in the NTD (Figure 2f), with possible involvement of the glycan at Asn603 of the CTD-2. Not surprisingly, the newly emerged SARS-CoV-2 variants of concerns, including Alpha (lineage B.1.1.7), Beta (B.1.351), Gamma (B.1.1.28) and Delta (B.1.617.2), all have mutations and/or deletions within the NTD-1-supersite, rendering resistance to neutralization by NTD-directed antibodies [34,35].
Receptor binding domain
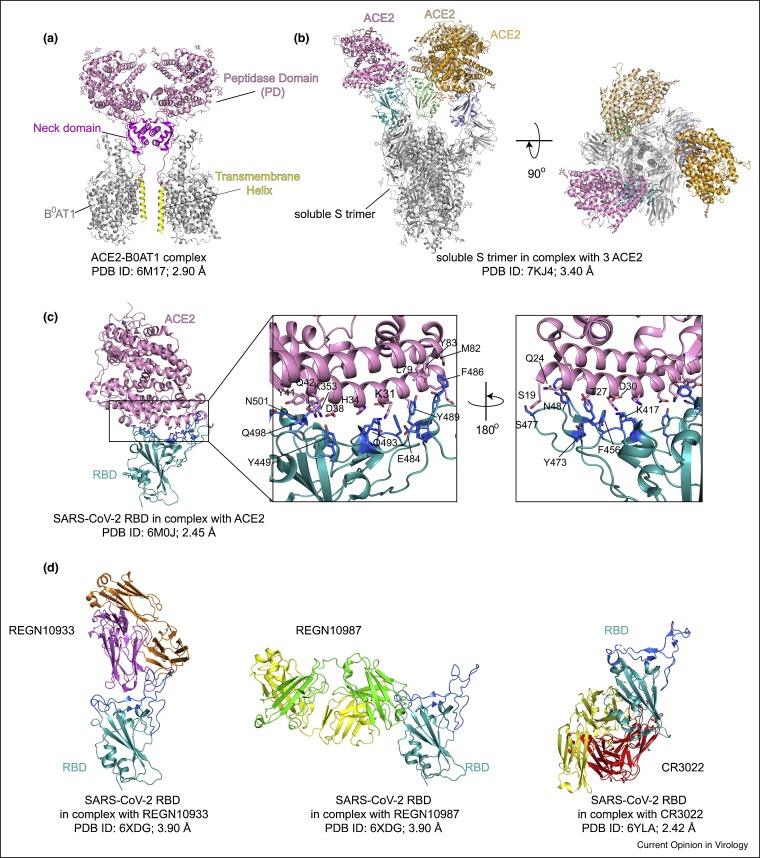
The RBD contains two subdomains — a five-stranded antiparallel β sheet connected by short helices and loops, and an extended loop, named receptor binding motif (RBM) [8••,9•,36]. In the host cell, ACE2 is an important component of renin-angiotensin system (RAS) and catalyzed the hydrolysis of angiotensin II to angiotensin 1–7 [9•]. The full-length human ACE2 is also a chaperone of the amino acid transporter B°AT1 and forms a homodimer mediated by its neck domain in the presence of B°AT1 (Figure 3 a) [10]. Cryo-EM structures of the soluble uncleaved S protein in complex with monomeric ACE2 show that the S trimer can bind one, two or three ACE2s in the RBD-up conformations (Figure 3b) [37,38]. The crystal structure of the SARS-CoV-2 RBD-ACE2 complex reveals a similar structure to the SARS-CoV RBD-ACE2 complex [8••,9•]. A gently concave outer surface of the extended RBM interacts with the N-terminal helix of the claw-like peptidase domain (PD) of ACE2 (Figure 3c) [8••,9•,10]. Hydrogen bonds and salt bridges between a series of polar residues, such as K417, E484, N487 and N501 of the RBD and D30, K31, H34, Y41 and K353 of the ACE2, dominate the RBD-ACE2 interaction (Figure 3c) [8••,9•,10]. Additional hydrophobic interactions between F486 of the RBD and L79, M82 and Y83 of the ACE2 also contribute to the receptor engagement (Figure 3c) [8••,9•,10]. Mutations of the key residues, such as N501Y, K417N and E484K, have been identified in the fast-spreading variants of concern, leading to enhanced affinity for ACE2 and immune evasion [39,40].
Figure 3.
Structures of ACE2, ACE2-S complexes and RBD-antibody complexes.
(a) Cryo-EM structure of the full-length ACE2 in complex with BoAT1 (PDB ID: 6M17; Ref. [10]), with the peptidase domain (PD) of ACE2 in pink, its neck domain in magenta, the transmembrane helix in yellow, and BoAT1 in gray. (b) The side view (left) and the top view (right) of cryo-EM structure of the soluble S trimer complexed with three ACE2s (PDB ID: 7KJ4; Ref. [37]). (c) Left, the crystal structure of SARS-CoV-2 RBD in complex with ACE2 (PDB ID: 6M0J; Ref. [8••]), with ACE2 in pink and the RBD in cyan. Middle and right, close-up views of the binding interface with contacting residues from the N-terminal helix of the ACE2 and the RBM of the RBD shown in sticks. (d) Left, cryo-EM structure of the RBD in complex with antibody REGN10933 (PDB ID: 6XDG; Ref. [43••]). Middle, cryo-EM structure of the RBD in complex with antibody REGN10987 (PDB ID: 6XDG; Ref. [43••]). Right, crystal structure of the RBD in complex with antibody CR3022 (PDB ID: 6YLA; Ref. [45]). The RBD is shown in cyan and heavy and light chains of the antibodies in various colors. The RBM is highlighted in dark blue.
The RBD is a dominant target of nAbs elicited by either natural infection or vaccination, confirming its pivotal role during infection [41,42]. The RBD-directed nAbs can recognize multiple distinct epitopes, showing great potencies at the pM-nM level in vitro neutralization assays (Figure 3d) [42]. The nAbs that target the ACE2-binding-site, such as REGN10933, C144 and S2H14, directly compete for ACE2 association [41,42,43••,44]. Those recognizing the non-ACE2-binding-site, such as REGN10987 and C135, probably prevent ACE2 binding either by clashing with ACE2 or by blocking the transition of the RBD from the ‘down’ to the ‘up’ conformation [42,43••,44]. Other nAbs against the so-called ‘cryptic supersite’, such as CR3022 and S304, can destabilize the S trimer and induce S1 dissociation [41,42,45]. Although the great potency of this class of antibodies makes them promising therapeutic agents, emergence of resistant variants could limit their clinical applications for treating the COVID-19.
A recombinant human ACE2, named APN01, is under evaluation as a treatment for COVID-19 in a phase 2 clinical trial, based on the favorable results from a previous phase 1 trial [46], and evidence that the protein blocks SARS-CoV-2 infection effectively in vitro [47]. Other ACE2-based fusion inhibitors have been developed with optimized binding and potency comparable to those of the nAbs [37,48,49]. The ACE2 constructs with multivalency, such as the dimeric protein sACE22.v2.4-IgG1 carrying the mutation T27Y/L79T/N330Y and the trimeric protein ACE2-foldon T27W, can inhibit the viral infection with a potency 1000-fold and 1700-fold greater than that of the monomeric soluble ACE2 with the wildtype sequence [37,48]. Substitution of T27 with an aromatic residue appears to further stabilize the binding interface through non-polar interactions with residues Y489, F456 and Y473 of the RBD (Figure 3c). In addition, a series of miniproteins, created using computer-generated scaffolds to mimic the N-terminal helix of ACE2, can bind the RBD and inhibit viral infection at a concentration below the nM level [49]. These ACE2-derived inhibitors may show even greater potency to those SARS-CoV-2 variants that have gained increased receptor binding than the Wuhan-Hu-1 virus. Nonetheless, pharmacokinetics, in vivo efficacy and safety profile of these new designs still require further validation.
C-terminal domains
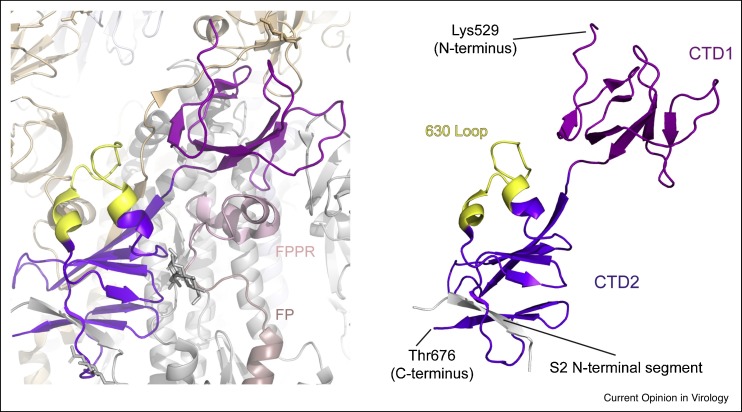
The C-terminal domains (CTDs) are formed primarily by β-structures of segments from S1, as well as the N-terminal segment of S2 adjacent to the furin cleavage site (Figure 4 ). CTD1 contains two antiparallel β-sheets, with two strands and four strands, respectively. CTD2 also has two β-sheets: a four-stranded one and another four-stranded one that includes a strand from the S2 subunit [6••,7•,13••,50••]. In the RBD-down conformation of the S trimer, a structural element in the CTD2, named the ‘630 loop’, becomes well-ordered in the G614 variant while disordered in the Wuhan-Hu-1 strain [13••,50••]. When structured, the 630 loop inserts into a gap between the NTD and CTD1 of the same protomer, stabilizing the CTD2 structure. It is also located in the vicinity of the S1/S2 boundary as well as the FPPR of a neighboring protomer [50••]. The FPPR and the 630 loop help retain the RBDs in the down conformation but move out of their positions when the adjacent RBD flips up. Thus, the CTDs, together with the FPPR and the 630 loop are key components of the S fusion machinery that modulate the fusogenic structural rearrangements of S protein.
Figure 4.
Structures of CTDs.
Structures of CTDs form the full-length S trimer (PDB ID: 7KRQ; Ref. [50••]) are shown, with CTD1 in magenta, CTD2 in purple, the 630-loop in yellow, and the β-strand in the CTD2 from S2 subunit in gray.
S2 structure
In the prefusion conformation [13••], three S2 subunits tightly pack around a central three-stranded coiled-coil of ∼140 Å long, formed by CH (Figure 1b). Portion of the HR1 together with another segment of S2 (residues 758–784) adopt α-helical conformation and assemble into a nine helix-bundle with the central coiled-coil, forming the most rigid part of the entire S trimer. The CD region links CH and the C-terminal HR2 through a linker region (Figure 5 a). The FP forms a short helix and tucks in a pocket formed by two neighboring S protomers. The structured FPPR clashes with the CTD1 if the RBD moves up and thus appears to help clamp the prefusion S trimer in the closed, RBD-down conformation. It has also been suggested to function as a pH-dependent switch domain that modulates the RBD position [38]. The remaining HR2, TM and CT segments are disordered in the most S trimer structures, but show low-resolution density in the cryo-ET reconstructions that can be tilted away from the threefold axis of the trimer with an angle from 17° to 60° [16••].
Figure 5.
Structures and proposed conformational changes of SARS-CoV S2.
(a) Close-up view of S2 in the prefusion (left) and postfusion (right) conformations from PDB ID: 6XR8 and 6XRA, with the fusion peptide (FP) highlighted in purple, the FPPR in red, central helix (CH) in gold, connector domain (CD) in green, HR1 in orange and HR2 in green. (b) Proposed structural transition of the HR1 from the prefusion to postfusion conformation. (c) Proposed conformational change of the HR2. (d). Six-helix bundle structures in the postfusion S2 with HR1 in orange and HR2 in green.
In the postfusion conformation [13••,24], the HR1 and CH form a continuous α-helix and three copies of them assemble into a long central three-stranded coiled-coil of ∼180 Å (Figure 5a). Two proline substitutions at the boundary between the HR1 and CH to prevent formation of the postfusion helix have been introduced to stabilize the prefusion conformation and such a design has been used for structural studies and the first-generation vaccines [6••,7•,51]. Part of the HR2 folds into α-helix and packs against the groove between two HR1-CH helices to form a six-helix bundle structure, reminiscent of the postfusion organization of other viral fusion proteins [52,53]. The CD remains unchanged from the prefusion conformation, as a three-stranded β-sheet covering the C-terminal end of HR1-CH helices. Comparison of the prefusion and postfusion conformations of S suggests that HR1 undergoes large rearrangements to form a coiled-coil, translocating its N-terminal end by a large distance to project the FP towards the target cell membrane (Figure 5b). In addition, the HR2 and the TM at its C-terminal end must fold back to pack along the groove of the HR1-CH coiled-coil to form the postfusion six-helical bundle (Figure 5c). These refolding events effectively bring the viral and target cell membranes close together, ultimately leading to membrane fusion (Figure 5d). Interestingly, five N-linked glycans decorate the postfusion S2 surface along the long axis with a regular spacing and may protect the S2 from the host immune responses.
Implications for vaccines and therapeutics
The SARS-CoV-2 S protein is the key component of almost all the first-generation COVID-19 vaccines [51]. Based on structural studies, concerns have been raised that the inactivated-virus vaccines or those used the wildtype sequence of the Wuhan-Hu-1 strain may have too many postfusion spikes and induce mainly non-neutralizing antibodies [13••,18••]. Indeed, these vaccines have induced lower levels of neutralizing antibody responses than other S constructs containing stabilization modifications to prevent conformational changes [54]. Additional studies have identified the G614 S trimer as a possible superior immunogen candidate [50••,55], as it is naturally constrained in a prefusion state presenting both the RBD-down and RBD-up conformations with great stability. Moreover, the global spread of SARS-CoV-2 and the consequently vast number of replication events make emergence of new variants inevitable, and substantially increases the genetic diversity of the virus, which will bring much greater challenges for vaccine development than it was at the beginning of the pandemic. Indeed, genetic diversity is also the major hurdle for development or optimization of vaccines against several other human pathogens, such HIV-1, hepatitis C virus and influenza virus [56, 57, 58]. If SARS-CoV-2 becomes seasonal, structure-based innovative strategies will likely be needed for developing next-generation vaccines designed to elicit broadly neutralizing antibody responses. Likewise, high-resolution structural information has been instrumental for creating peptide-based and ACE2-based fusion inhibitors [12,37,49,59], it will undoubtedly continue to serve as a foundation for rational design of antiviral therapeutics to fight against the pandemic.
Conclusion
Tremendous progress in the structural biology of SARS-CoV-2 spike protein has been made since the initial outbreak of the virus. The structural knowledge not only fills the major gap in our understanding of the viral entry process, but also provides a solid foundation for development and optimization of vaccines and therapeutics against the current and future pandemics of coronaviruses.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We thank all the former and current members of the Chen laboratory at Boston Children’s Hospital for their contributions to our research. Our work was supported by N.I.H. grants AI147884 (to B.C.), AI141002 (to B.C.), AI127193 (to B.C. and James Chou), a Fast grant by Emergent Ventures (to B.C.) and a COVID-19 Award by Massachusetts Consortium on Pathogen Readiness (MassCPR; to B.C.).
References
- 1••.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper first reported the identification and characterization of SARS-CoV-2 (named 2019-nCoV at the time), which caused an epidemic of acute respiratory syndrome in humans in Wuhan, China. It shows that SARS-CoV-2 shares 79.6% sequence identity to SARS-CoV and 96% identity to a bat coronavirus. It has also confirmed that the new virus uses the same cell entry receptor—angiotensin converting enzyme II (ACE2)—as does SARS-CoV.
- 2.Barcena M., Barnes C.O., Beck M., Bjorkman P.J., Canard B., Gao G.F., Gao Y., Hilgenfeld R., Hummer G., Patwardhan A., et al. Structural biology in the fight against COVID-19. Nat Struct Mol Biol. 2021;28:2–7. doi: 10.1038/s41594-020-00544-8. [DOI] [PubMed] [Google Scholar]
- 3•.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper has demonstrated that SARS-CoV-2 uses the SARS-CoV receptor ACE2 for entry and the serine protease TMPRSS2 for S protein priming.
- 4.Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H., et al. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601. doi: 10.1016/j.chom.2020.08.004. e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6••.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first cryo-EM structure of a stabilized soluble SARS-CoV-2 (or 2019-nCoV) spike protein trimer in the prefusion conformation was reported. The paper has also shown that the predominant state of the trimer has one of the three receptor-binding domains (RBDs) rotated up in a receptor-accessible conformation.
- 7•.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;183:1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that the receptor-binding domains of SARS-CoV-2 S and SARS-CoV S bind with similar affinities to human ACE2, and that the furin cleavage site at the boundary between the S1/S2 subunits is processed during biogenesis. Cryo-EM structures of the SARS-CoV-2 S ectodomain trimer and inhibition SARS-CoV-2 S mediated entry by SARS-CoV S murine polyclonal antibodies have also been described.
- 8••.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]; The high-resolution crystal structure of the SARS-CoV-2 RBD–ACE2 complex has revealed detailed interactions at the binding interface. There is a high degree of similarity between binding of SARS-CoV-2 and SARS-CoV with ACE2, suggesting convergent evolution of the RBDs for improved binding with the receptor.
- 9•.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reported another crystal structure of the RBD of SARS-CoV-2 S in complex with ACE2. Comparison with the SARS-CoV RBD has revealed that the ACE2-binding site of the SARS-CoV-2 RBD has a more compact conformation and that several residue changes in the SARS-CoV-2 RBD stabilize two virus-binding hotspots at the RBD–ACE2 interface. Moreover, the bat coronavirus RaTG13 also uses human ACE2 as its receptor, providing evidence for the potential animal-to-human transmission of SARS-CoV-2.
- 10.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia S., Liu M., Wang C., Xu W., Lan Q., Feng S., Qi F., Bao L., Du L., Liu S., et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30:343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cryo–EM structures derived from a preparation of the full-length S protein, representing its prefusion and postfusion conformations have been reported. The paper has shown that the spontaneous transition to the postfusion state is independent of target cells and that the prefusion trimer has three RBDs clamped down by a segment adjacent to the fusion peptide. In addition, the postfusion structure is strategically decorated by N-linked glycans, suggesting possible protective roles against host immune responses and harsh external conditions.
- 14.Bangaru S., Ozorowski G., Turner H.L., Antanasijevic A., Huang D., Wang X., Torres J.L., Diedrich J.K., Tian J.H., Portnoff A.D., et al. Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. Science. 2020;370:1089–1094. doi: 10.1126/science.abe1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Yao H., Song Y., Chen Y., Wu N., Xu J., Sun C., Zhang J., Weng T., Zhang Z., Wu Z., et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738. doi: 10.1016/j.cell.2020.09.018. e713. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is a study on the molecular assembly of the authentic SARS-CoV-2 using cryoelectron tomography and subtomogram averaging. Native structures of the S proteins in prefusion and postfusion conformations, as well as the ribonucleoproteins and their higher-order assemblies have been structurally characterized, revealing the architecture of the virus in exceptional details.
- 16••.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, cryo-electron microscopy and tomography have been used to image intact SARS-CoV-2 virions and determine the high-resolution structure, conformational flexibility and distribution of S trimers in situ on the virion surface.
- 17•.Turonova B., Sikora M., Schurmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., von Bulow S., Gecht M., Bagola K., Horner C., et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, cryo–electron tomography, subtomogram averaging and molecular dynamics simulations were used to structurally analyze SARS-CoV-2 spike (S) protein in situ. It shows that the stalk region of S contains three hinges, giving the head unexpected orientational freedom.
- 18••.Liu C., Mendonca L., Yang Y., Gao Y., Shen C., Liu J., Ni T., Ju B., Liu C., Tang X., et al. The architecture of inactivated SARS-CoV-2 with postfusion spikes revealed by cryo-EM and cryo-ET. Structure. 2020;28:1218–1224. doi: 10.1016/j.str.2020.10.001. e1214. [DOI] [PMC free article] [PubMed] [Google Scholar]; An inactivated SARS-CoV-2 preparation of the original strain has been genetically and structurally characterized. It shows that the virus particles are roughly spherical or moderately pleiomorphic and that most spikes appear nail shaped, thus resembling a postfusion state.
- 19.Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8 doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Z., Tomlinson A.C., Wong A.H., Zhou D., Desforges M., Talbot P.J., Benlekbir S., Rubinstein J.L., Rini J.M. The human coronavirus HCoV-229E S-protein structure and receptor binding. eLife. 2019;8 doi: 10.7554/eLife.51230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan X., Cao D., Kong L., Zhang X. Cryo-EM analysis of the post-fusion structure of the SARS-CoV spike glycoprotein. Nat Commun. 2020;11 doi: 10.1038/s41467-020-17371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.J., Rey F.A., Veesler D. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci U S A. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou H., Chen Y., Zhang S., Niu P., Qin K., Jia W., Huang B., Zhang S., Lan J., Zhang L., et al. Structural definition of a neutralization epitope on the N-terminal domain of MERS-CoV spike glycoprotein. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10897-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng G., Xu L., Lin Y.L., Chen L., Pasquarella J.R., Holmes K.V., Li F. Crystal structure of bovine coronavirus spike protein lectin domain. J Biol Chem. 2012;287:41931–41938. doi: 10.1074/jbc.M112.418210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reported isolation and characterization of monoclonal antibodies (mAbs) from 10 convalescent COVID-19 patients. The antibody 4A8 is a potent neutralizing antibody that targets the N-terminal domain (NTD) of the S protein, suggesting that the NTD may be a promising target for therapeutic mAbs against COVID-19.
- 29•.Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J., et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021 doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this paper, structures for seven potent NTD-directed neutralizing antibodies in complex with SARS-CoV-2 spike or isolated NTD have been determined by either cryo-EM or X-ray crystallography. The structures revealed that all seven antibodies target a common surface, which appear to be a single supersite, outside of the RBD.
- 30.Voss W.N., Hou Y.J., Johnson N.V., Delidakis G., Kim J.E., Javanmardi K., Horton A.P., Bartzoka F., Paresi C.J., Tanno Y., et al. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes. Science. 2021 doi: 10.1126/science.abg5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347. doi: 10.1016/j.cell.2021.03.028. e2316. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work described a large number of human monoclonal Abs (mAbs) derived from memory B cells, targeting the SARS-CoV-2 S N-terminal domain (NTD) with a subset neutralizing the virus ultrapotently. A supersite recognized by all known NTD-specific neutralizing mAbs has been defined, and these mAbs inhibit cell-to-cell fusion, activate effector functions, and protect hamsters from the viral challenge.
- 32.Li D., Edwards R.J., Manne K., Martinez D.R., Schafer A., Alam S.M., Wiehe K., Lu X., Parks R., Sutherland L.L., et al. The functions of SARS-CoV-2 neutralizing and infection-enhancing antibodies in vitro and in mice and nonhuman primates. bioRxiv. 2021 [Google Scholar]
- 33.Tong P., Gautam A., Windsor I., Travers M., Chen Y., Garcia N., Whiteman N.B., McKay L.G.A., Lelis F.J.N., Habibi S., et al. Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike. bioRxiv. 2021 doi: 10.1016/j.cell.2021.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 36.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 37.Xiao T., Lu J., Zhang J., Johnson R.I., McKay L.G.A., Storm N., Lavine C.L., Peng H., Cai Y., Rits-Volloch S., et al. A trimeric human angiotensin-converting enzyme 2 as an anti-SARS-CoV-2 agent. Nat Struct Mol Biol. 2021;28:202–209. doi: 10.1038/s41594-020-00549-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou T., Tsybovsky Y., Gorman J., Rapp M., Cerutti G., Chuang G.Y., Katsamba P.S., Sampson J.M., Schon A., Bimela J., et al. Cryo-EM structures of SARS-CoV-2 spike without and with ACE2 reveal a pH-dependent switch to mediate endosomal positioning of receptor-binding domains. Cell Host Microbe. 2020;28:867–879. doi: 10.1016/j.chom.2020.11.004. e865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gobeil S.M., Janowska K., McDowell S., Mansouri K., Parks R., Stalls V., Kopp M.F., Manne K., Saunders K., Edwards R.J., et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation and antigenicity. bioRxiv. 2021 doi: 10.1126/science.abi6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai Y., Zhang J., Xiao T., Lavine C.L., Rawson S., Peng H., Zhu H., Anand K., Tong P., Gautam A., et al. Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1126/science.abi9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042. doi: 10.1016/j.cell.2020.09.037. e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finkelstein M.T., Mermelstein A.G., Parker Miller E., Seth P.C., Stancofski E.D., Fera D. Structural analysis of neutralizing epitopes of the SARS-CoV-2 spike to guide therapy and vaccine design strategies. Viruses. 2021;13 doi: 10.3390/v13010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]; A series of neutralizing antibodies generated from humanized mice and convalescent patients against the SARS-CoV-2 Spike protein were characterized in binding, neutralization and structure to identify pairs of highly potent antibodies ideal for cocktail antibody therapy, which may enhance the antiviral potency and minimize the chance of virus escape.
- 44.Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., et al. Neutralization of SARS-CoV-2 by destruction of the prefusion spike. Cell Host Microbe. 2020;28:445–454.e446. doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haschke M., Schuster M., Poglitsch M., Loibner H., Salzberg M., Bruggisser M., Penninger J., Krahenbuhl S. Pharmacokinetics and pharmacodynamics of recombinant human angiotensin-converting enzyme 2 in healthy human subjects. Clin Pharmacokinet. 2013;52:783–792. doi: 10.1007/s40262-013-0072-7. [DOI] [PubMed] [Google Scholar]
- 47.Monteil V., Kwon H., Prado P., Hagelkruys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado Del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. e907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao L., Goreshnik I., Coventry B., Case J.B., Miller L., Kozodoy L., Chen R.E., Carter L., Walls A.C., Park Y.J., et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020;370:426–431. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Zhang J., Cai Y., Xiao T., Lu J., Peng H., Sterling S.M., Walsh R.M., Jr., Rits-Volloch S., Zhu H., Woosley A.N., et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science. 2021;372:525–530. doi: 10.1126/science.abf2303. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cryo-EM structures of a full-length G614 S trimer have been determined and the trimer adopts three distinct prefusion conformations that differ primarily by the position of one receptor-binding domain. A loop disordered in the D614 S trimer wedges between domains within a protomer in the G614 spike. This added interaction appears to prevent premature dissociation of the G614 trimer effectively increasing the number of functional spikes and enhancing infectivity and to modulate structural rearrangements for membrane fusion.
- 51.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chan D.C., Fass D., Berger J.M., Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 53.Weissenhorn W., Dessen A., Harrison S.C., Skehel J.J., Wiley D.C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 54.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 55.Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., Rangarajan E.S., Pan A., Vanderheiden A., Suthar M.S., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11 doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burton D.R. Advancing an HIV vaccine; advancing vaccinology. Nat Rev Immunol. 2019;19:77–78. doi: 10.1038/s41577-018-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duncan J.D., Urbanowicz R.A., Tarr A.W., Ball J.K. Hepatitis C virus vaccine: challenges and prospects. Vaccines (Basel) 2020;8 doi: 10.3390/vaccines8010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei C.J., Crank M.C., Shiver J., Graham B.S., Mascola J.R., Nabel G.J. Next-generation influenza vaccines: opportunities and challenges. Nat Rev Drug Discov. 2020;19:239–252. doi: 10.1038/s41573-019-0056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]