Abstract
Many anti-severe acute respiratory syndrome coronavirus 2 (anti-SARS-CoV-2) neutralizing antibodies target the angiotensin-converting enzyme 2 (ACE2) binding site on viral spike receptor-binding domains (RBDs). Potent antibodies recognize exposed variable epitopes, often rendering them ineffective against other sarbecoviruses and SARS-CoV-2 variants. Class 4 anti-RBD antibodies against a less-exposed, but more-conserved, cryptic epitope could recognize newly emergent zoonotic sarbecoviruses and variants, but they usually show only weak neutralization potencies. Here, we characterize two class 4 anti-RBD antibodies derived from coronavirus disease 2019 (COVID-19) donors that exhibit breadth and potent neutralization of zoonotic coronaviruses and SARS-CoV-2 variants. C118-RBD and C022-RBD structures reveal orientations that extend from the cryptic epitope to occlude ACE2 binding and CDRH3-RBD main-chain H-bond interactions that extend an RBD β sheet, thus reducing sensitivity to RBD side-chain changes. A C118-spike trimer structure reveals rotated RBDs that allow access to the cryptic epitope and the potential for intra-spike crosslinking to increase avidity. These studies facilitate vaccine design and illustrate potential advantages of class 4 RBD-binding antibody therapeutics.
Keywords: cryo-EM, coronavirus, neutralizing antibody, receptor-binding domain, sarbecovirus, SARS-CoV-2, Spike trimer: structural biology, virology: X-ray crystallography
Graphical abstract

Jette et al. characterize antibodies derived from convalescent COVID-19 donors that broadly recognize sarbecoviruses and neutralize ACE2-tropic strains, including all SARS-CoV-2 variants of concern. Structures reveal binding to a highly conserved RBD epitope using long CDRH3 loops, with an orientation that inhibits ACE2 binding to the RBD and allows IgG avidity.
Introduction
The current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic is a crisis of immediate global concern, but two other zoonotic betacoronaviruses, SARS-CoV and MERS-CoV (Middle East respiratory syndrome), also resulted in epidemics within the last 20 years (de Wit et al., 2016). All three viruses likely originated in bats (Li et al., 2005; Zhou et al., 2021), with SARS-CoV and MERS-CoV having adapted to intermediary animal hosts, most likely palm civets (Song et al., 2005) and dromedary camels (Haagmans et al., 2014), respectively, prior to infection of humans. Serological surveys of people living near caves where bats carry diverse coronaviruses suggest direct transmission of SARS-CoV-like viruses (Wang et al., 2018), raising the possibility of future outbreaks resulting from human infection with SARS-like betacoronaviruses (sarbecoviruses).
Coronaviruses encode a trimeric spike glycoprotein (S) that serves as the machinery for fusing the viral and host cell membranes (Fung and Liu, 2019). The first step in fusion is contact of S with a host receptor. The receptor-binding domains (RBDs) at the apex of the S trimers of SARS-CoV-2, SARS-CoV, HCoV-NL63, and some animal coronaviruses utilize angiotensin-converting enzyme 2 (ACE2) as their receptor (Hoffmann et al., 2020; Li et al., 2003; Zhou et al., 2020b). RBDs can adopt either “down” or “up” conformations, with ACE2 binding to RBDs only possible in an “up” conformation (Kirchdoerfer et al., 2016; Li et al., 2019; Walls et al., 2016, 2020; Wrapp et al., 2020; Yuan et al., 2017). A phylogenetic tree of the relationship between coronavirus S protein RBDs shows that sarbecovirus RBDs form a separate branch (Figure 1 A).
Figure 1.
C118 and C022 show diverse binding and neutralization of sarbecoviruses
(A) Sarbecovirus (lineage B) phylogenetic tree classified based on RBD sequence conservation.
(B) Left: cartoon rendering of SARS-CoV-2 S trimer (PDB: 6VYB) showing location of “up” RBD (surface, orange and purple). Right: amino acid sequence conservation of 12 RBDs calculated as described (Landau et al., 2005) plotted on a surface representation of a SARS-CoV-2 RBD structure (PDB: 7BZ5). Primary RBD epitopes for the indicated representatives from defined classes of RBD-binding antibodies (class 1–4) (Barnes et al., 2020a) are indicated as yellow dotted lines (PDB: 7K90, 6W41, 7JX3, and 7K8M). C022 epitope indicated as blue dotted line.
(C) Comparison of binding of the indicated monoclonal IgGs to a panel of sarbecovirus RBDs from ELISA data shown as area under the curve (AUC) values. Data presented are mean AUC values from two independent experiments. IOMA IgG is an anti-HIV-1 antibody serving as a negative control (Gristick et al., 2016).
(D) Neutralization IC50 values for the indicated IgGs against SARS-CoV-2 (D614G version of the original variant (GenBank: NC_045512)), SARS-CoV-2 variants of concern, and other ACE2-tropic sarbecovirus pseudoviruses. Geomean = geometric mean IC50 in which IC50 values >50,000 ng/mL were entered as 50000 ng/mL for the calculation. IC50 values are means of 2–7 independent experiments.
SD, standard deviation. See also Figures S1 and S2.
Consistent with their obligate role in viral entry, sarbecovirus S trimers are the primary targets of neutralizing antibodies (Brouwer et al., 2020; Cao et al., 2020; Fung and Liu, 2019; Kreer et al., 2020; Liu et al., 2020b; Robbiani et al., 2020; Rogers et al., 2020; Seydoux et al., 2020; Shi et al., 2020; Zost et al., 2020b), with many focusing on the RBD (Barnes et al., 2020a, 2020b; Brouwer et al., 2020; Cao et al., 2020; Kreer et al., 2020; Liu et al., 2020b; Pinto et al., 2020; Robbiani et al., 2020; Rogers et al., 2020; Seydoux et al., 2020; Zost et al., 2020a). Structural analysis of the binding epitopes of anti-SARS-CoV-2 RBD antibodies enabled their classification into four initial categories: class 1, derived from VH3-53/VH3-63 germlines and including a short heavy chain complementarity determining region 3 (CDRH3) that binds an epitope overlapping with the ACE2 binding site and recognizes only “up” RBDs; class 2, whose epitope also overlaps with the ACE2 binding site, but which can bind to both “up” and “down” RBD conformations; class 3, which bind to the opposite side of “up” and “down” RBDs adjacent to an N-glycan attached to residue N343; and class 4, which are often weakly neutralizing antibodies that target a cryptic epitope facing the interior of the spike protein on “up” RBDs (Barnes et al., 2020a) (Figure S1; Video S1).
The video shows soluble SARS-CoV-2 spike (PDB: 7BZ5) and the ‘down’ and ‘up’ conformations of the RBD domain. Next, the sequence conservation of 12 sarbecovirus RBDs is mapped onto the surface of the RBD domain. Finally, The C118-SARS-CoV RBD and C022-SARS-CoV-2 RBD structures are shown.
Potent anti-SARS-CoV-2 neutralizing antibodies are typically class 1 or class 2 anti-RBD antibodies that block the ACE2 binding site (Barnes et al., 2020a; Dejnirattisai et al., 2021; Huang et al., 2021; Liu et al., 2020b; Piccoli et al., 2020; Tortorici et al., 2020). Because class 1 and class 2 RBD epitopes are not well conserved (Figure 1B), antibodies in these classes are unlikely to strongly cross-react across sarbecovirus RBDs. However, an in vitro-selected variant of an ACE2 blocking antibody isolated from a SARS-infected survivor exhibited increased cross-reactive properties, showing neutralization of SARS-CoV-2 and other betacoronaviruses (Rappazzo et al., 2021). In general, however, as isolated from infected donors, class 3 and class 4 RBD-binding antibodies are better prospects for neutralizing across multiple strains and thereby potentially protecting against emergent sarbecoviruses. Indeed, S309, a class 3 anti-RBD antibody isolated from a SARS-CoV-infected donor, demonstrated cross-reactive neutralization of SARS-CoV-2 (Pinto et al., 2020). Furthermore, reports of class 4 human antibodies that exhibit cross-reactive binding and neutralization among sarbecoviruses (Liu et al., 2020a; Starr et al., 2021a; Tortorici et al., 2021) suggest that further investigation of antibodies from COVID-19 convalescent donors could lead to discoveries of potent and broadly cross-reactive class 4 antibodies that recognize the highly conserved, “cryptic” RBD epitope.
Here, we investigated C118 and C022, two class 4 human antibodies isolated from COVID-19 donors (Robbiani et al., 2020) that show breadth of binding and neutralization across sarbecoviruses and SARS-CoV-2 variants of concern. We report crystal structures of C118 complexed with SARS RBD and C022 complexed with SARS-CoV-2 RBD, which revealed interactions with a conserved portion of the RBD in common with interactions of previously described cross-reactive but more weakly neutralizing class 4 antibodies, e.g., CR3022 (Huo et al., 2020; Yuan et al., 2020a, 2020b), S304/S2A4 (Piccoli et al., 2020), and EY6A (Zhou et al., 2020a). Unlike these class 4 anti-RBD antibodies, C118 and C022 also occlude portions of the ACE2 binding site to facilitate more potent neutralization. A single-particle cryoelectron microscopy (cryo-EM) structure of a C118-S trimer complex demonstrated binding of C118 to an intact trimer, revealing an S configuration with increased separation between the RBDs than found in class 1–3 Fab-S or ACE2-S trimer structures and revealing the potential for intra-spike crosslinking. These results define a cross-reactive class 4-like epitope on sarbecovirus RBDs that can be targeted in vaccine design and illustrate a mechanism by which the cryptic RBD epitope can be accessed on intact CoV S trimers.
Results
C022 and C118 IgGs recognize and neutralize diverse sarbecoviruses, including SARS-CoV-2 variants
From a survey to identify cross-reactive monoclonal antibodies isolated from SARS-CoV-2-infected donors from the New York area (Robbiani et al., 2020), we found antibodies isolated from different donors, C118 (VH3-30/VL4-69 encoded) and C022 (VH4-39/VK1-5 encoded), that recognized a diverse panel of 12 sarbecovirus RBDs spanning clades 1, 1/2, 2, and 3 (Figure 1). As evaluated by enzyme-linked immunosorbent assay (ELISA), C118 bound to RBDs from all sarbecoviruses tested, and C022 bound to all but two RBDs, similar to the class 4 anti-RBD antibody CR3022 (Figure 1C). By comparison, the cross-reactive class 3 anti-SARS RBD antibody S309 (Pinto et al., 2020) recognized half of the set of sarbecovirus RBDs, and C144, a more potent SARS-CoV-2 class 2 neutralizing antibody (Robbiani et al., 2020), bound to the SARS-CoV-2 RBD, but not to RBDs from the other 11 sarbecovirus strains (Figure 1C).
To further define the C022 and C118 antibody epitopes, we evaluated binding of C118 and C022 to a panel of RBDs with mutations chosen from circulating variants that conferred resistance to one or more classes of anti-RBD antibodies (Li et al., 2020; Starr et al., 2021b; Weisblum et al., 2020). We also assessed binding to RBD substitutions identified in the B.1.1.7 and B.1.351 SARS-CoV-2 variants of concern (Rambaut et al., 2020; Tegally et al., 2020) and to mutations in the MA10 mouse-adapted SARS-CoV-2 virus (Leist et al., 2020). Relative to wild-type (WT) RBD, C118, C022, CR3022, and S309 demonstrated a similar binding profile with respect to the RBD substitutions tested and exhibited a broader range of binding to the RBD mutants than did the more potent class 2 C144 antibody (Figure 1C; Figure S2A). Collectively, the ELISA binding data suggested that C022 and C118 recognize a highly conserved epitope and are therefore likely to be class 4 anti-RBD antibodies.
We next measured neutralization potencies using an in vitro pseudovirus-based assay that quantitatively correlates with authentic virus neutralization (Schmidt et al., 2020) to evaluate SARS-CoV-2, SARS-CoV-2 RBD mutants, SARS-CoV-2 variants (Annavajhala et al., 2021; Faria et al., 2021; Rambaut et al., 2020; Tegally et al., 2020; Voloch et al., 2020; West et al., 2021; Zhang et al., 2021), and sarbecovirus strains known to infect human ACE2-expressing target cells (SARS-CoV-2, SARS-CoV, WIV1, SHC104, WIV16, Pangolin GD, and Pangolin GX) (Figure 1D; Figures S2B–S2D). Against a panel of SARS-CoV-2-pseudotyped viruses harboring single amino acid RBD substitutions, C118 and C022 neutralized all viruses with potencies similar to “WT” SARS-CoV-2, consistent with the results obtained in ELISA binding assays (S gene with D614 residue; GenBank: NC_045512) (Figure S2). For comparisons with SARS-CoV-2 variants of concern, the S gene we used to make “WT” SARS-CoV-2 pseudovirus included the D614G substitution in the context of the Wuhan-Hu-1 spike (Korber et al., 2020), resulting in a 2- to 4-fold reduction in 50% inhibitory concentration (IC50) values for C022 and C118 antibodies (Figure 1D).
We found that C118 and C022 IgGs neutralized all four SARS-CoV-2 variants and all ACE2-tropic sarbecoviruses with IC50 values <1 μg/mL, with the exception of C118, which inhibited SARS-CoV-pseudotyped viruses with an IC50 = 3.9 μg/mL (Figure 1D; Figures S2B–S2D). By contrast, the class 4 anti-RBD antibody CR3022 showed weak or no neutralization against the majority of pseudoviruses tested, with the exception of SARS-CoV (IC50 ∼1.1 μg/mL) and WIV1 (IC50 ∼0.6 μg/mL). The SARS-CoV-2-derived antibody COVA1-16 IgG neutralized SARS-CoV-2 variants of concern with IC50 values similar to C118 and C022 IgG, but it showed weak neutralization for WIV1 (11.3 μg/mL) and SHC014 (20.2 μg/mL) and no neutralization for SARS-CoV (>50 μg/mL), which is consistent with previously published studies (Liu et al., 2020a, 2020b, 2021; Brouwer et al., 2020). The class 3 S309 antibody showed strong neutralization potencies (IC50 values between 16 and 120 ng/mL) against all viruses with the exceptions of the B.1.1.7 SARS-CoV-2 variant of concern and SHC014. The class 2 anti-RBD antibody C144 was highly potent against SARS-CoV-2 and the B.1.1.7 and B.1.429 variants (IC50 values between 1 and 2 ng/mL) but did not neutralize the other SARS-CoV-2 variants or sarbecoviruses. Taken together, of the IgGs evaluated C118, and C022 exhibited the greatest breadth of sarbecovirus neutralization (Figure 1D; Figure S2), consistent with their broad cross-reactive binding profile demonstrated by ELISA (Figure 1C; Figure S2A).
Crystal structures of C022-RBD and C118-RBD reveal class 4 RBD interactions and conservation of epitope residues
To understand the mechanism underlying the breadth of neutralization of C022 and C118, we solved structures of complexes between C118 Fab bound to SARS-CoV RBD and C022 bound to SARS-CoV-2 RBD to resolutions of 2.7 and 3.2 Å, respectively, chosen based on which complexes formed well-ordered crystals (Figures 2A and 2B; Table S1).
Figure 2.
Crystal structures of C022 and C118 Fabs bound to RBDs reveal class 4-like RBD binding
(A and B) Cartoon renderings of crystal structures of (A) C0118 Fab complexed with SARS-CoV RBD and (B) C022 Fab complexed with SARS-CoV-2 RBD. Dashed circle shows location of SARS-CoV N357RBD residue, with the inset showing the N357RBD asparagine and glycan modeled based on the SARS-CoV spike-S230 structure (PDB: 6NB6).
(C and D) CDR loops and RBD epitope residues of (C) C118 Fab and (D) C022 Fab overlaid on RBDs represented as gray surfaces with stick representations of epitope residues. Framework region residues, which account for some of the contacts for both antibodies, are not shown in the right panels.
(E) Comparison of Fab poses for binding to an RBD-ACE2 complex. C118 Fab (blue), C022 Fab (red), CR3022 Fab (PDB: 6W41; orange), and EY6A Fab (PDB: 6CZC pink) modeled onto an ACE2-RBD structure (PDB: 6M0J; RBD shown as a gray surface and ACE2 shown as a green cartoon).
(F) Fab and ACE2 competition experiment by surface plasmon resonance. ACE2-Fc was immobilized on a chip, and then complexes of SARS-CoV-2 with either C118, C022, CR3022, or C144 Fab flowed over. A binding event indicated no competition for RBD binding between ACE2 and the corresponding Fab.
See also Figures S1 and S2 and Table S1.
The C118-RBD and C022-RBD structures showed that both Fabs recognize an epitope that is highly conserved among sarbecoviruses at the base of the RBD (Figure 1B), which is exposed only in “up” RBD conformations as first described for the class 4 RBD-binding antibodies CR3022 (Huo et al., 2020; Yuan et al., 2020a, 2020b) and EY6A (Zhou et al., 2020a). C022 and C118 use four of six complementarity determining region (CDR) loops to interact with an epitope that extends toward the RBD ridge near the ACE2 binding site and, in the case of C022, includes an overlapping interacting residue (K417RBD) (Figures 2C and 2D). In both structures, CDRH3 loops, CDRL2 loops, and portions of FWRL3 mediate the majority of RBD contacts and establish extensive polar and van der Waals interactions with RBD residues (Figures 2C and 2D), accounting for 71% of epitope buried surface area (BSA) on the RBD for the C022-RBD and C118-RBD structures, respectively (Table S2). No contacts were made in either complex with the N343RBD N-glycan (SARS-CoV-2 S numbering). SARS-CoV contains an additional potential N-linked glycosylation site at N357RBD (SARS-CoV S numbering), which, if glycosylated, would not be contacted by C118, a favorable feature for cross-reactive recognition given that this potential N-linked glycosylation site is conserved in all S protein sequences except for SARS-CoV-2 (Figure 2A).
Overlaying the RBDs of our Fab-RBD structures with the RBD of the ACE2-RBD structure (PDB: 6M0J) showed that the binding poses of both C118 and C022 placed the VL domain of each Fab in a position that would clash with concurrent ACE2 binding, in contrast with the CR3022 and EY6A binding poses (Figure 2E). This binding orientation would sterically prevent RBD-ACE2 interactions, as has been suggested for other class 4 anti-RBD antibodies (Liu et al., 2020a; Piccoli et al., 2020). To verify direct competition with ACE2, we conducted competition binding experiments using surface plasmon resonance (SPR). In the first setup, sACE2-Fc was coupled to a biosensor chip, and a Fab-RBD complex was injected. RBD binding to immobilized sACE2-Fc was blocked in the presence of C118, C022, and C144 Fabs, which contrasts binding in the presence of CR3022 Fab (Figure 2F). Similar results were observed when SARS-CoV-2 RBD was coupled to a biosensor chip, an RBD-binding IgG was injected, and then soluble ACE2 was injected over the RBD-IgG complex (Figure S2E). Taken together, these results suggest that C118 and C022 block ACE2 binding to RBD and utilize a primary neutralization mechanism that prevents spike attachment to host cell ACE2 receptors.
Features of C118 and C022 recognition of the class 4 epitope
Class 4 RBD-binding antibodies contact a common epitope at the base of the RBD that is distant from the ACE2 binding site (Figure 3 A). The epitopes of three class 4 antibodies, C118, C022, and COVA1-16, also include a patch reaching toward the ridge on the left side of the RBD as depicted in Figure 3A.
Figure 3.
The C118 and C022 epitopes include a conserved RBD helix
(A) Epitopes for ACE2 and monoclonal antibodies (mAbs) calculated from analyses of structures of RBD or S trimer complexes (human antibodies isolated from COVID-19 patients are C118, C022, COVA1-16, EY6A, and S2A4). RBDs shown are derived from SARS-CoV-2 except for the C118 panel, which is SARS-CoV RBD.
(B) Alignment of sequences for sarbecovirus RBDs (residue numbering for SARS-CoV-2 RBD). Secondary structure for SARS-CoV-2 RBD shown above alignment. Dots designate binding or neutralization for C118 (blue), C022 (red), or CR3022 (orange) for each strain. Diamonds designate RBD epitope residues for C118 binding to SARS-CoV (blue) and C022 (red) or CR3022 (orange) binding to SARS-CoV-2. Left boxes show binding by ELISA or neutralization of pseudovirus for each antibody for each strain; data for COVA1-16 are from Liu et al. (2020a). Circles show binding or neutralization, blank spaces designate not tested, and dashes designate no binding or neutralization. Shadings in the sequence alignment indicate conserved portions of epitope (green). Colored boxes show differing portion of epitope covering the α4 helix and following loop (pink).
(C) Cartoon representation of SARS-CoV-2 RBD (gray) showing overlapping antibody-interacting residues (green) as sticks in epitopes for C118, C022, COVA1-16, and CR3022 (corresponding to green shading in B).
(D) Cartoon representation of SARS-CoV-2 RBD (gray) showing α4 helix and following (sticks, pink) that differ in their contacts with C118, C022, COVA1-16, and CR3022 (pink shading in B).
(E) Cartoon representation of RBDs showing α4 region of RBD and C118 (left) or C022 (right) interacting loops with interacting Fab residues in light blue (C118) and light pink (C022).
See also Figures S3 and S4.
To compare the C118 and C022 epitopes with epitopes of other class 4 anti-RBD antibodies, we analyzed RBD residues contacted by C118, C022, COVA1-16, and CR3022 on aligned sequences of sarbecovirus RBDs (Figure 3B). Sequence conservation among sarbecoviruses at the C022 and C118 epitopes involves a majority of residues that are strictly conserved or conservatively substituted between SARS-CoV-2 and other RBDs (Figure 3B), likely explaining the broad cross-reactivity observed for these antibodies (Figure 1C). Comparison of the C118 and C022 epitopes showed a majority of recognized RBD residues are shared between the two antibodies (70% of C118 epitope also contacted by C022) (Figure 3B). CR3022 contacted a similar number of residues as C118 and C022, including the conserved patch at the RBD base (Figures 3B and 3C); however, a region from 404RBD to 417RBD that comprises an unstructured loop and the α4 helix above an internal RBD β sheet contained only a single CR3022 contact residue (R408RBD) and was not contacted by antibodies EY6A, S2A4, and S304, whereas C118, C022, and COVA1-16 showed contacts with this region (Figures 3B and 3D).
The α4 helix is proximal to the ACE2 receptor-binding motif and has less sequence conservation across the 12 sarbecoviruses (Figure 3B). To accommodate binding in this region, C118 uses insertions in its FWRL3 (54B-56LC) to form a β strand adjacent to the α4 helix, establishing both side-chain and backbone interactions (Figure 3E, left panel). C022 showed similar binding in this region but used non-contiguous CDRH1, CDRH3, and CDRL2 loops (Figure 3E, right panel). C022 contacts were located more to the C-terminal end of the α4 helix than the C118 contacts and encompassed the disordered RBD loop that includes the ACE2-interacting residue K417RBD (K404RBD in SARS-CoV) (Lan et al., 2020) (Figure 3E, right panel). Additionally, C022 buried more surface area on RBD in this region than C118 (323 versus 150 Å2). Four of eight and five of nine RBD contacts for C118 and C022, respectively, were fully conserved among sarbecoviruses (Figure 3B), suggesting that interactions in this region may be possible with other sarbecoviruses. In particular, the conserved residue R408RBD (R395RBD in SARS-CoV) was contacted by both antibodies and alone was responsible for 94 and 95 Å2 of BSA buried on the RBDs for C118 and C022, respectively. Despite both C118 and C022 engaging the α4 helix and residue R408RBD, mutations at this position known to affect class 1 and class 4 anti-RBD antibodies (Greaney et al., 2021) had no effect on these antibodies (Figure S2A). Overall, engagement of the α4 helix region provided 16% (C118) and 36% (C022) of the BSA buried on RBD and extended their epitopes past the cryptic epitope to bind adjacent to or overlapping with the ACE2 binding site.
Shared features of the C022 and COVA1-16 class 4 anti-RBD antibodies
The C022 epitope on RBD closely resembles the epitope of COVA1-16 (Figures 3A and 3B), a class 4 antibody isolated from a SARS-CoV-2 convalescent donor derived from VH1-46/VK1-33 V-gene segments (Brouwer et al., 2020) (Figure S3). Yet, COVA1-16 showed weak neutralization (>10 μg/mL) against WIV1-CoV, SHC014, and SARS-CoV pseudoviruses, which contrasts the potent C022 neutralization (Figure 1D). After superimposing the RBDs from crystal structures of SARS-CoV-2 RBD complexed with COVA1-16 (PDB: 7JMW) and C022 (this study), the VH-VL domains of the bound Fabs were related by a root mean square deviation (RMSD) of 1.3 Å (235 Cα atoms), with the majority of conformational differences occurring in the CDRH1 and CDRH2 loops (Figure S4A). Despite being derived from different V gene segments (which would affect their VH gene segment-encoded CDRH1 and CDRH2 loops), C022 and COVA1-16 recognized similar epitopes, contacting a common set of 23 RBD residues that include interactions with the RBD α4 helix (Figure 3B).
Although C022 and COVA1-16 share a generally similar mode of binding, there are differences in interactions of residues encoded within their different VH gene segments (i.e., their CDRH1 and CDRH2 loops) (Figure S4B). For example, the C022 contact with T430RBD was part of an extensive clasp made by an interaction between the C022 CDRH1 residue R33HC with backbone carbonyls of D427RBD, D428RBD, and F429RBD and with the side chain of T430RBD (Figure S4C). Two of the same RBD residues (D427RBD and F429RBD) interacted with an arginine from COVA1-16, but this arginine (R100BHC) is located at the base of the CDRH3 loop rather than within CDRH1, as is the case with C022 R33HC. The larger separation distance from the RBD of COVA1-16 R100BHC allowed it to form a side-chain-backbone H-bond with D427RBD similar to a side-chain-backbone H-bond involving C022 R33HC and D428RBD, but precluded interactions with D428RBD and T430RBD (Figure S4D). In addition, the COVA1-16 CDRH1 was shorter than the C022 CDRH1 (7 versus 9 residues) (Figure S3A) and was shifted away from the RBD relative to the C022 CDRH1. These differences, in addition to fewer LC interactions by COVA1-16, resulted in less total BSA for COVA1-16 relative to C022 (1,607 versus 1,875 Å2, respectively) despite similar contributions from CDRH3 loops (Table S2).
Interactions with RBD main-chain atoms facilitate recognition of diverse RBDs
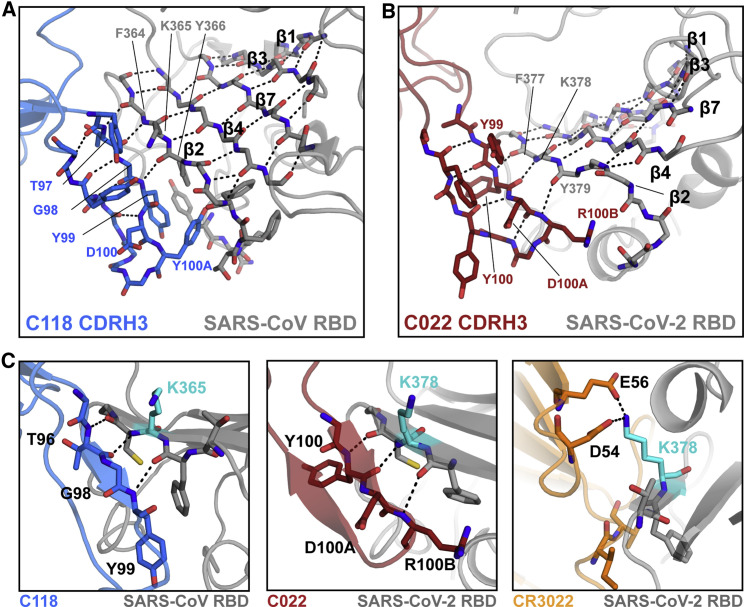
The paratopes of both C118 and C022 were dominated by their long CDRH3 loops (20 and 21 residues, respectively) (Figures 4A and 4B; Figure S3), which make up approximately half of the BSAs of each paratope (461 of 1,020 Å2 for C118 and 537 of 969 Å2 for C022) (Table S2). The C118 and C022 CDRH3s comprise two anti-parallel β strands that extend a largely internal RBD β sheet (β strands β1–β4 and β7) through main-chain H-bonds between the RBD β2 strand (377–379RBD) and the first CDRH3 β strand (CDRH3 residues 97–99 [C118] or 100-100B [C022]) (Figures 4A and 4B). A similar feature is also seen in the structure of the COVA1-16-RBD complex (Liu et al., 2020a), which shares a nearly identical CDRH3 sequence with C022 (Figure S4E).
Figure 4.
C118 and C022 Fabs primarily use their CDRH3s for main-chain backbone contacts with the RBD β2 strand
(A) Close-up cartoon showing β-hairpin formed by C118 CDRH3 (blue sticks) and β sheet formation with SARS-CoV RBD (gray cartoon with sticks). H-bonds shown as black dashed lines.
(B) Close-up cartoon showing β-hairpin formed by C022 CDRH3 (red sticks) and β sheet formation with SARS-CoV-2 RBD (gray cartoon with sticks). H-bonds shown as black dashed lines.
(C) Cartoon and stick representation of C118-RBD (left), C022-RBD (middle), and CR3022-RBD (right) showing distinct interactions with residue K365SARS/K378SARS2 (cyan).
See also Table S2.
C118 and C022 form extensive backbone interactions with RBD, with 10 and 9 H-bonds formed with the backbone of RBD, respectively. Extensive backbone interactions in the C118 and C022 epitopes could contribute to their breadth of binding and neutralization across sarbecoviruses, because backbone interactions would facilitate binding despite side-chain substitutions, which are rare across the RBD sequences listed (Figure 3B), but could occur in other CoV RBDs. For example, the backbone H-bonds between the CDRH3s of C118 and C022 with the RBD β2 strand allow for binding despite substitution at position K378RBD (K365RBD in SARS-CoV) (Figure S2A; Figure 4C). By contrast, the class 4 antibody CR3022 uses side-chain interactions (potential electrostatic interactions between D54HC and E56HC and K378RBD); thus, CR3022 is sensitive to mutation at K378NRBD (Figure S2A). This is consistent with CR3022 not binding to Rf1-CoV RBD (Figure 1C), which contains an asparagine at the equivalent position to SARS-CoV-2 K378RBD (Figure 3B), whereas C118 and C022 binding to Rf1-CoV RBD was not affected. Overall, main-chain H-bond interactions likely reduce sensitivity to RBD side-chain substitutions, making antibodies such as C118 and C022 more tolerant to differences between sarbecovirus strains or variants.
C118-S cryo-EM structure shows increased S trimer opening
On an S trimer, the class 4 cryptic epitope is at the base of the RBD, where it faces toward the center of the trimer (Barnes et al., 2020a; Huo et al., 2020; Yuan et al., 2020b). The epitope is buried in the closed, prefusion S conformation and interacts with portions of the spike S2 subunit and neighboring “down” RBDs. Compared with class 2 or class 3 anti-RBD antibodies that recognize their epitopes in “up” or “down” RBD conformations (Barnes et al., 2020a), the class 4 epitope is less accessible and requires two “up” RBDs for antibody binding (Piccoli et al., 2020). Additionally, class 4 antibody binding may also require RBD rotation to prevent steric clashes with neighboring “up” RBDs, as observed for the complexes of S trimer with EY6A, S2A4, and S304 (Piccoli et al., 2020; Zhou et al., 2020a).
Given the similar binding poses of C118 and C022 antibodies, which bind with a more acute angle with respect to the RBD than EY6A or CR3022 (Figure 2E), and the increased breadth and potency of C118 and C022 relative to other class 4 anti-RBD antibodies (Figure 1D), we sought to understand the requirements for epitope recognition on a S trimer. Thus, we solved a single-particle cryo-EM structure of C118 Fabs bound to SARS-CoV-2 S 6P trimers (Hsieh et al., 2020), finding two distinct states defined by RBDs adopting various rotational conformations (Figures 5A and 5B; Figure S5), as well as C118 Fab bound to dissociated S1 subunit protomers (Figure S5B). For the state 1 C118-S trimer complex structure solved to 3.4 Å, we subsequently used symmetry expansion and local refinement to generate a 3.7-Å map of the C118 VHVL-RBD interface (Figure S5B–S5E).
Figure 5.
Cryo-EM structure of C118-S complex shows binding to cryptic epitope and the potential for intra-spike crosslinking
(A) 3.4 Å cryo-EM density for the C118-S trimer complex (state 1). Side view (left panel) illustrates orientation with respect to the viral membrane (dashed line). Top view (right panel) shows symmetric binding at the trimer apex with C118 HC (blue) oriented in the interior.
(B) 4.4 Å cryo-EM density for the C118-S trimer complex (state 2). Top view illustrates asymmetry of complex as a result of RBD rotation in one protomer.
(C) Composite model of an open SARS-CoV-2 trimer bound by class 4 Fabs, C118 (this paper, blue), EY6A (PDB: 6ZDH, pink), and S2A4 (PDB: 7JVC, brown), and the class 4 anti-SARS antibodies S304 (PDB: 7JW0, green) and H014 (PDB: 7CAK, yellow).
(D) Comparison of S trimer openness by measurements of Cα distances for D428RBD between adjacent “up” RBDs in S trimers complexed with the class 1 antibody S2E12 (PDB: 7K43, gray), soluble ACE2 (PDB: 7KMS, green), and the class 4 antibodies C118 (this study, blue) and EY6A (PDB: 6ZDH, pink).
(E) Prediction of potential intra-spike avidity effects by measurement of Cα distances between the C termini of adjacent CH1 domains for the mAb-S trimer complexes described in (C). Measurements were used to evaluate the potential for intra-spike crosslinking by an IgG binding to a single spike trimer as described previously (Barnes et al., 2020a). For the H014-S complex, the CH1-CL domains were rigid body fit into the cryo-EM density (EMD-30333) prior to measurements.
(F) IC50 values and molar neutralization ratios (MNRs) were defined as: (IC50 Fab or bispecific IgG (nM)/IC50 IgG (nM)) (Klein and Bjorkman, 2010) for C118 and C022. IC50 values shown for the IgGs are from Figure 1D. IC50 values for all assays against SARS-CoV-2 and SARS-CoV are means of two to seven independent experiments. Two MNRs are presented in the MNRs (bispecific/Fab) column: the MNR was calculated using a bispecific IgG versus the bivalent IgG (left), and the MNR was calculated using a Fab versus the bivalent IgG (right). Neutralization results with MNRs ≤ 5 are indicated as not demonstrating avidity effects (−), MNRs > 10 are indicated as demonstrating minimal avidity (+), results with one MNR > 50 are indicated as moderate avidity (++), and MNRs demonstrating strong avidity effects (one MNR > 700) are indicated as +++.
The C118 pose with respect to the RBD observed in the C118-SARS-CoV-2 S structure was similar to the C118-SARS-CoV RBD crystal structure (Figure S5F), demonstrating consistent recognition of the antibody epitope on both SARS-CoV-2 and SARS-CoV RBDs. Furthermore, the C118 binding pose was oriented higher on the RBD relative to other class 4 anti-RBD antibodies (Figure 5C) and was consistent with SPR competition data that suggested C118 would sterically hinder ACE2 binding to the same protomer (Figure S2E).
Despite differences in binding poses relative to other class 4 antibodies (Figure 5C), C118 binding also resulted in RBD conformations displaced farther from the trimer center relative to S2E12 (a class 1 anti-RBD neutralizing antibody) (Tortorici et al., 2020) and ACE2 (Yan et al., 2020) (Figure 5D). On average, class 4 anti-RBD antibody binding resulted in an ∼15- to 20-Å displacement of the RBD relative to ACE2-bound conformations, which likely results in destabilization of the spike trimer. Indeed, S1 shedding induced by class 4 antibodies has been described as a possible neutralization mechanism (Huo et al., 2020; Piccoli et al., 2020; Wec et al., 2020). The presence of C118-S1 protomer classes in our cryo-EM data suggested that C118 also induces shedding (Figure S5), but the role S1 shedding and premature S-triggering plays in C118-mediated neutralization requires further investigation.
C118 and C022 neutralization of sarbecoviruses demonstrates differential effects of avidity enhancement
Neutralization of SARS-CoV-2 and SARS-CoV by COVA1-16 was found to be mediated by avidity effects based on potent neutralization by the bivalent COVA1-16 IgG, but not the monovalent Fab (Liu et al., 2020a). To evaluate whether intra-spike crosslinking, one source of avidity enhancement for bivalent antibodies, was possible for C118 or C022 IgGs, we examined the C118-S trimer structure to ask whether the positioning of two Fabs on adjacent RBDs would be compatible with binding by a single IgG. As previously described for other anti-RBD IgGs, we compared the distance between residues near the C termini of adjacent Fab CH1 domains to analogous distances in crystal structures of intact IgGs, setting a cutoff of ≤65 Å as potentially allowing a single IgG to include both Fabs (Barnes et al., 2020a). The measured distance for the C termini of adjacent Fab CH1 domains in the symmetric State 1 C118-S trimer structure was 41 Å (Figure 5E), suggesting that intra-spike crosslinking would be possible for C118 IgGs bound to spike trimers. The asymmetric State 2 C118-S structure included distances of 50, 89, and 92 Å (Figure 5E), also allowing intra-spike crosslinking between one combination of two bound RBDs, as well as the potential for inter-spike crosslinking between adjacent spikes on the virion surface. In comparison, no other class 4 anti-RBD Fab-S trimer structures showed measured distances that would be compatible with intra-spike crosslinking (Figure 5E); thus, any potential avidity effects for those IgGs could occur only via inter-spike crosslinking.
To further evaluate whether avidity could also facilitate cross-reactive neutralization by the C118 and C022 antibodies, we compared neutralization of SARS-CoV-2, SARS-CoV, WIV1, and SHC014 by the bivalent C118 and C022 IgGs and by two monovalent forms of each antibody: a 50-kDa Fab and an IgG size-matched bispecific IgG containing only one relevant Fab. The bispecific IgGs included one C118 or C022 RBD-binding Fab and a second non-RBD-binding Fab derived from the HIV-1 antibody 3BNC117 (Scheid et al., 2011). To interpret neutralization results, we calculated molar neutralization ratios (MNRs) defined as: (IC50 Fab or bispecific IgG (nM)/IC50 IgG (nM)) (Klein and Bjorkman, 2010). In the absence of avidity effects resulting from either crosslinking within a spike trimer (intra-spike crosslinking) or cross-linking between adjacent spike trimers (inter-spike crosslinking), an MNR would be 2.0, which accounts for twice as many relevant Fabs in a bivalent IgG compared with its monovalent forms.
Using pseudotyped SARS-CoV-2, SARS-CoV, WIV1, and SHC014, we derived neutralization potencies of the bivalent IgG, monovalent bispecific IgG, and Fab forms of C118 and C022 and then calculated MNRs for the bivalent IgG to bispecific IgG comparison (bispecific MNR) and for the bivalent IgG to Fab comparison (Fab MNR) (Figure 5F). Comparisons between the Fab and bispecific IgG forms of monovalent antibody allowed evaluation of potential steric affects that could increase neutralization potencies for larger IgGs compared with smaller Fabs. With the exception of the low MNRs derived from the IgG comparison with the bispecific and Fab forms of C118 against SARS-CoV-2 (MNRs of 5 and 3, respectively), we found mostly high MNRs ranging from the lowest values of 13 and 33 for the MNRs for C118 against SARS-CoV (where 11 is a minimal estimate because the C118 bispecific was non-neutralizing) to the highest values of 708 and 1,444 for the C022 bispecific and Fab MNRs against SHC014. Four of the bispecific to Fab MNR comparisons showed a 2-fold or higher Fab MNR than the comparable bispecific MNR, suggesting that at least some of the increased potencies of the bivalent IgGs compared with their counterpart Fabs resulted from steric effects. However, six of the eight monovalent to bivalent comparisons exhibited MNRs well over 70, suggestive of strong avidity effects. By contrast, mean MNRs derived for broadly neutralizing anti-HIV-1 Env antibodies are ≤10 (Wang et al., 2017), consistent with the low spike density on HIV-1 virions that largely prevents inter-spike crosslinking, and the architecture of the HIV-1 Env trimer, which prohibits intra-spike crosslinking for all known HIV-1 broadly neutralizing antibodies (Klein and Bjorkman, 2010). Taken together with the analysis of the C118-S trimer structure, the observed avidity effects for C118 IgGs binding to WIV1 and SHC014 and for the related C022 IgGs binding to the four viruses tested could arise from intra-spike and inter-spike crosslinking.
The question as to why C118 exhibits little or no avidity effects for neutralization of SARS-CoV-2 and SARS-CoV is difficult to address because the same IgG showed strong avidity effects against WIV1 and SHC014, and C022, which binds similarly to C118, showed avidity effects in neutralization of all four pseudoviruses. These results could derive from different binding characteristics for C118 to the SARS-CoV-2 and SARS-CoV RBDs compared with C118 and C022 interactions with the other sarbecoviruses evaluated. Indeed, simulations of avidity effects demonstrated that some combinations of IgG concentration and antigen-binding affinity and kinetic constants showed no advantages of bivalent versus monovalent binding (Klein, 2009; Klein and Bjorkman, 2010). Thus, the effects of avidity are a complicated function of concentration and binding constants that preclude predictions in the absence of experimental data.
Discussion
Concerns about coronaviruses having spillover potential, as well as the increasing prevalence of SARS-CoV-2 variants, necessitates identification of cross-reactive antibodies. Antibodies elicited against infectious viruses for which there are multiple circulating variants, either within an individual or the population, often show a trade-off between potency and breadth (Corti et al., 2010; Wagh et al., 2016). In the case of antibody responses against SARS-CoV-2, the cause of the current global pandemic, many strongly neutralizing antibodies have been isolated that block ACE2 receptor interactions (Barnes et al., 2020a; Dejnirattisai et al., 2021; Huang et al., 2021; Liu et al., 2020b; Piccoli et al., 2020). However, the ACE2-binding region of the RBD also tends to accumulate amino acid changes, as evidenced by substitutions identified in the current SARS-CoV-2 variants of concern (Annavajhala et al., 2021; Faria et al., 2021; Rambaut et al., 2020; Tegally et al., 2020; Voloch et al., 2020; West et al., 2021; Zhang et al., 2021), thus reducing the potential efficacies of vaccines and monoclonal antibody therapies. Recent studies suggest that antibodies against the S2 subunit offer the potential of greater cross-reactivity across coronaviruses, but these antibodies generally lack strong neutralization potency (Sauer et al., 2021; Shah et al., 2021; Wang et al., 2021).
The class 4 RBD-binding epitope, which is more conserved than the class 1 and class 2 RBD epitopes, represents a plausible target for the elicitation of antibodies with broad cross-reactive recognition across sarbecoviruses. Indeed, some recently described class 4 antibodies (e.g., CR3022, H014, COVA1-16, EY6A, ADI-56046) neutralize two or more sarbecovirus strains and/or can bind RBDs from multiple sarbecoviruses (Liu et al., 2020a). However, although many class 4 antibodies show some cross-reactivity, they generally exhibit decreased potencies against heterologous sarbecovirus strains. For example, the SARS-CoV-derived CR3022 antibody does not potently neutralize SARS-COV-2 (Huo et al., 2020), and the SARS-CoV-2-derived COVA1-16 antibody does not potently neutralize SARS-CoV, WIV1, or SHC014 (Liu et al., 2020a, 2020b).
Here we characterized two antibodies, C118 and C022, derived from different COVID-19 convalescent donors (Robbiani et al., 2020), which show breadth of and potent neutralization against sarbecoviruses of all three clades. The structural similarity of RBD binding poses between C022 and COVA1-16 (Liu et al., 2020a), which was derived from yet a third COVID-19 convalescent donor (Brouwer et al., 2020), suggests that these sorts of cross-reactive antibodies are commonly elicited by natural infection and that their epitope represents an attractive target for immunogen design. Of particular importance for the current pandemic, circulating variants of concern or variants of interest did not confer resistance to the C118 and C022 antibodies. In addition, C118 and C022 antibodies were not affected by naturally occurring RBD mutations that undermine the activity of several antibodies approved for therapeutic use (Hoffmann et al., 2021; Starr et al., 2021b).
Analysis of our C118-RBD and C022-RBD complex structures revealed key details of cross-reactive recognition and broad sarbecovirus neutralization. First, C118 and C022 utilize long CDRH3s to facilitate interactions with the cryptic RBD epitope at the base of the RBD. In contrast with less potent class 4 antibodies such as CR3022 (Huo et al., 2020; Yuan et al., 2020a, 2020b) and EY6A (Zhou et al., 2020a) that also contact this region, the longer CDRH3 provides the opportunity to target a highly conserved patch of residues across sarbecoviruses with an orientation that extends the epitope upward to the ACE2 binding site, a structural feature shared with COVA1-16 (Liu et al., 2020a). Second, the aforementioned binding poses of C118, C022, and COVA1-16, as well as overlap of the C022 epitope with the edge of the ACE2 binding site, suggested competition with ACE2 as part of their neutralization mechanisms. Indeed, competition experiments reported here for C118 and C022 and by others for COVA1-16 (Liu et al., 2020a) demonstrated competition with ACE2 for SARS-CoV-2 RBD binding. Third, C118 and C022 formed many interactions with backbone atoms of RBD residues, adding a second level of buffering against viral escape because amino acid substitutions at these positions are less likely to abrogate antibody binding. Finally, the demonstration that C118 and C022 bivalency increased potency of neutralization against some of the viruses evaluated showed the potential for these antibodies to utilize avidity effects for neutralization of sarbecoviruses. Given the requirement for two “up” RBDs on a S trimer for class 4 antibody binding, bivalent binding within a single S trimer would be possible. Thus, we suggest that intra-spike crosslinking would be an advantage for neutralization of sarbecoviruses, where avidity effects likely play a role.
In conclusion, class 4 antibodies that access the cryptic RBD epitope and compete with ACE2 binding are important for understanding cross-reactivity of human SARS-CoV-2 antibody responses elicited by natural sarbecovirus infection. We suggest that potent class 4 anti-RBD antibodies could be used therapeutically to avoid resistance to SARS-CoV-2 variants of concern, perhaps after in vitro selection to further improve their potencies. Structural characterization of these antibodies could also be used to inform future vaccine design efforts to produce immunogens that preferentially elicit C118 and C022-like cross-reactive antibodies by blocking RBD epitopes recognized by class 1 and class 2 antibodies, which typically are not cross-reactive to other sarbecoviruses.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat Anti-Human IgG(H+L)-HRP | SouthernBiotech | Cat# 2015-05; RRID:AB_2795588 |
| Goat Anti-Human IgG-HRP | SouthernBiotech | Cat# 2040-05; RRID:AB_2795644 |
| Bacterial and virus strains | ||
| SARS-CoV-2 pseudotyped reporter virus | Robbiani et al., 2020 | https://www.nature.com/articles/s41586-020-2456-9 |
| SARS-CoV pseudotyped reporter virus | Robbiani et al., 2020 | https://www.nature.com/articles/s41586-020-2456-9 |
| WIV1-CoV pseudotyped reporter virus | Cohen et al., 2021 | https://www.science.org/lookup/doi/10.1126/science.abf6840 |
| SCH014-CoV pseudotyped reporter virus | Cohen et al., 2021 | https://www.science.org/lookup/doi/10.1126/science.abf6840 |
| SARS-CoV-2 B.1.1.7 pseudotyped reporter virus | Scheid et al., 2021 | https://linkinghub.elsevier.com/retrieve/pii/S0092867421005353 |
| SARS-CoV-2 B.1.351 pseudotyped reporter virus | Scheid et al., 2021 | https://linkinghub.elsevier.com/retrieve/pii/S0092867421005353 |
| SARS-CoV-2 B.1.429 pseudotyped reporter virus | Bjorkman lab (this paper) | N/A |
| SARS-CoV-2 B.1.526 pseudotyped reporter virus | Bjorkman lab (this paper) | N/A |
| E. coli DH5 Alpha | Zymo Research | Cat# T3009 |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s Modified Eagle Medium (DMEM) | GIBCO | Cat# 11960-044 |
| Fetal bovine serum (FBS) | Sigma-Aldrich | Cat# F4135 |
| Gentamicin solution | Sigma-Aldrich | Cat# G1397 |
| CAS:1405-41-0 | ||
| Blasticidin S HCl | GIBCO | Cat# A1113902 |
| CAS:3513-03-9 | ||
| Expi293 Expression Medium | GIBCO | Cat# A1435102 |
| Expi293 Expression System Kit | GIBCO | Cat# A14635 |
| LB Broth (Miller) | Sigma-Aldrich | Cat# L3522 |
| 1-Step Ultra TMB-ELISA Substrate Solution | Thermo Scientific | Cat# 34029 |
| HBS-EP+ Buffer 20x | Teknova | Cat# H8022 |
| Critical commercial assays | ||
| Luciferase Cell Culture Lysis 5X Reagent | Promega | Cat# E1531 |
| Nano-Glo Luciferase Assay System | Promega | Cat# N1110 |
| Deposited data | ||
| C118 Fab/SARS-CoV-2 S 2P state 1 coordinates | This paper | PDB: 7RKV |
| C118 Fab/SARS-CoV-2 S 2P state 1 CryoEM map | This paper | EMDB: 24504 |
| C118 Fab/SARS-CoV-2 S 2P state 2 CryoEM map | This paper | EMDB: 24505 |
| C118 Fab/SARS-CoV RBD crystal structure | This paper | PDB: 7RKS |
| C022 Fab/SARS-CoV-2 RBD crystal structure | This paper | PDB: 7RKU |
| Experimental models: Cell lines | ||
| HEK293T cells | Pear et al., 1993 | Cat# CCLV-RIE 1018 |
| RRID:CVCL_0063 | ||
| HEK293TAce2 cells | BEI | Cat# NR-52511 |
| Expi293F cells |
GIBCO |
Cat# A14527 |
| RRID:CVCL_D615 | ||
| Recombinant DNA | ||
| SARS-CoV-2 S (residues 16-1206) | Pamela J. Bjorkman, California Institute of Technology (This paper) | GenBank: MN985325.1 |
| SARS-CoV2-Strunc B.1.429 | Pamela J. Bjorkman, California Institute of Technology (This paper) | GenBank QQM19141 residues 1-1252 |
| pTwist-CMV BetaGlobin-SARS-CoV-2 S RBD (residues 331-524) | Barnes et al., 2020b | https://linkinghub.elsevier.com/retrieve/pii/S0092867420307571 |
| pTwist-CMV BetaGlobin-SARS-CoV S RBD (residues 318-510) | Barnes et al., 2020b | https://linkinghub.elsevier.com/retrieve/pii/S0092867420307571 |
| CR3022 Fab HC, CR3022 IgG HC, CR3022 LC | Yuan et al., 2020b | https://www.science.org/lookup/doi/10.1126/science.abb7269 |
| COVA1-16 IgG HC, COVA1-16 LC | Liu et al., 2020a | https://linkinghub.elsevier.com/retrieve/pii/S1074761320304647 |
| C118 Fab HC, C118 IgG HC, C118 LC | Robbiani et al., 2020 | https://www.nature.com/articles/s41586-020-2456-9 |
| C022 Fab HC, C022 IgG HC, C022 LC | Robbiani et al., 2020 | https://www.nature.com/articles/s41586-020-2456-9 |
| C144 Fab HC, C144 IgG HC, C144 LC | Robbiani et al., 2020 | https://www.nature.com/articles/s41586-020-2456-9 |
| S309 IgG HC, S309 LC | Pinto et al., 2020 | https://www.nature.com/articles/s41586-020-2349-y |
| See Table S4 for additional Recombinant DNA Reagents | ||
| Software and algorithms | ||
| GISAID | Shu and McCauley, 2017 | https://www.gisaid.org RRID:SCR_018251 |
| Clustal Omega | Sievers et al., 2011 | https://www.ebi.ac.uk/Tools/msa/clustalo/RRID: SCR_001591 |
| MUSCLE | Edgar, 2004 | https://www.ebi.ac.uk/Tools/msa/muscle/RRID:SCR_011812 |
| PhyML 3.0 | Guindon et al., 2010 | http://www.atgc-montpellier.fr/phyml/RRID: SCR_014629 |
| PRESTO | http://www.atgc-montpellier.fr/presto/ | |
| Gen5 | BioTek | https://www.biotek.com/products/software-robotics-software/gen5-microplate-reader-and-imager-software/RRID:SCR_017317 |
| Prism 9 | GraphPad | https://www.graphpad.com/scientific-software/prism/RRID:SCR_002798 |
| SerialEM 3.7 | Mastronarde, 2005 | https://bio3d.colorado.edu/SerialEM/RRID:SCR_017293 |
| cryoSPARC 3.1 | Punjani et al., 2017 |
https://cryosparc.com RRID:SCR_016501 |
| UCSF Chimera | Pettersen et al., 2004 | http://plato.cgl.ucsf.edu/chimera/RRID:SCR_004097 |
| Goddard et al., 2007 | ||
| XDS | Kabsch, 2010 | https://xds.mr.mpg.de/ |
| RRID:SCR_015652 | ||
| PHASER | McCoy et al., 2007 | https://phenix-online.org/documentation/reference/phaser.html |
| RRID:SCR_014219 | ||
| Phenix | Adams et al., 2010 | https://phenix-online.org/RRID:SCR_014224 |
| Coot | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/RRID:SCR_014222 |
| AIMLESS | Winn et al., 2011 | https://www.ccp4.ac.uk/html/aimless.html |
| RRID:SCR_015747 | ||
| MolProbity | Chen et al., 2010 | http://molprobity.biochem.duke.edu |
| RRID:SCR_014226 | ||
| PyMOL 2.3.5 | Schrodinger, Inc. | https://pymol.org/2/RRID:SCR_000305 |
| ConSurf Database | Landau et al., 2005 | https://consurf.tau.ac.il |
| RRID:SCR_002320 | ||
| Biacore T200 software | Cytiva | N/A |
| Other | ||
| Pierce Streptavidin Coated Plates, Clear, 96-Well | Thermo Scientific | Cat# 15125 |
| HisTrap FF | GE Healthcare Life Sciences | Cat# 17-5255-01 |
| HisTrap HP | GE Healthcare Life Sciences | Cat# 17-5248-02 |
| HiLoad 16/600 Superdex 200 pg | GE Healthcare Life Sciences | Cat# 28-9893-35 |
| Superose 6 Increase 10/300 GL | GE Healthcare Life Sciences | Cat# 29-0915-96 |
| HiTrap MabSelect SuRe column, 5 mL | GE Healthcare Life Sciences | Cat# 11-0034-95 |
| HiTrap MabSelect SuRe column, 1 mL | GE Healthcare Life Sciences | Cat# 11-0034-93 |
| Superdex 200 Increase 10/300 GL | GE Healthcare Life Sciences | Cat# 28-9909-44 |
| Amicon Ultra-15 Centrifugal Filter Devices | Millipore | Cat# UFC903096 |
| PD-10 Desalting Columns | GE Healthcare Life Sciences | Cat# 17-0851-01 |
| 300 Mesh UltrAuFoil® Holey Gold Films, R 1.2/1.3 | Electron Microscopy Sciences | Cat# Q350AR13A |
| Series S Sensor Chip CM5 | GE Healthcare Life Sciences | Cat# BR-1005-30 |
Resource availability
Lead contact
All requests for further information or requests for resources and reagents should be directed to the Lead Contact, Pamela Bjorkman (bjorkman@caltech.edu).
Materials availability
All expression plasmids generated in this study for CoV proteins, CoV pseudoviruses, human Fabs and IgGs are available upon request.
Experimental model and subject details
Cell lines
Cells for pseudovirus production (HEK293T) were cultured at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Sigma-Aldrich) and 5 μg/ml Gentamicin (Sigma-Aldrich).
Target cells for pseudovirus neutralization experiments (HEK293TACE2) were generated as described (Robbiani et al., 2020) and cultured at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Sigma-Aldrich), 5 μg/ml gentamicin (Sigma-Aldrich), and 5μg/mL Blasticidin (GIBCO).
Expi293F cells (GIBCO) for protein expression were maintained at 37°C and 8% CO2 in Expi293 expression medium (GIBCO), transfected using an Expi293 Expression System Kit (GIBCO) and maintained under shaking at 130 rpm. All cell lines were female and were not specifically authenticated.
Bacteria
E. coli DH5 Alpha (Zymo Research) used for propagation of expression plasmids were cultured with shaking at 250 rpm at 37°C in LB broth (Sigma-Aldrich).
Viruses
To generate pseudotyped viral stocks, HEK293T cells were transfected with pNL4-3ΔEnv-nanoluc and pSARS-CoV2-Strunc (Robbiani et al., 2020) using polyethylenimine, leading to production of HIV-1-based pseudovirions carrying the SARS-CoV-2 S protein at the surface. Eight hours after transfection, cells were washed twice with phosphate buffered saline (PBS) and fresh media was added. Supernatants containing pseudovirus were harvested 48 hours post transfection, filtered and stored at −80°C. Infectivity of pseudoviruses was determined by titration on 293TACE2 cells.
Method details
Phylogenetic trees
Sequence alignments of RBDs were made with Clustal Omega (Sievers et al., 2011). Phylogenetic trees were calculated from amino acid alignments using PhyML 3.0 (Guindon et al., 2010) and visualized with PRESTO (http://www.atgc-montpellier.fr/presto).
Graphical abstract
Portions of the graphical abstract were produced using https://biorender.com/. The remaining parts were made using Adobe Illustrator.
Protein Expression
Fabs and IgGs were expressed and purified as previously described (Scharf et al., 2015; Schoofs et al., 2019; Liu et al., 2020a) and stored at 4°C. Bispecific IgGs (C118 or C022 plus 3BNC117, a non-coronavirus binding HIV-1 antibody (Scheid et al., 2011)) were produced by co-transfection of two heavy chain and two light chain genes that included knobs-into-holes mutations in IgG Fc and a domain cross-over in the 3BNC117 Fab to prevent incorrect light chain pairing (Schaefer et al., 2011). Antibody CDR lengths were determined using the IMGT definitions (Lefranc et al., 2015; Lefranc et al., 2009).
The following C-terminally 6xHis-tagged RBD proteins were transfected and expressed as described previously (Cohen et al., 2021): SARS-CoV-2 RBD (residues 328-533), SARS-CoV-2 RBD mutants (residues 319-541), SARS RBD (residues 318-510), SHC014 RBD (residues 307-524), WIV-1 RBD (residues 307-528), RaTG13 RBD (residues 319-541), Rs4081 RBD (residues 310-515), Yun11 RBD (residues 310-515), Rf1 RBD (residues 310-515), RmYN02 RBD (298-503), BM-4831 RBD (residues 310-530), BtKY72 RBD (residues 309-530). A trimeric SARS-CoV-2 ectodomain (residues 16-1206 of the early SARS-CoV-2 GenBank MN985325.1 sequence isolate with 6P (Hsieh et al., 2020) stabilizing mutations, a mutated furin cleavage site between S1 and S2, a C-terminal TEV site, foldon trimerization motif, octa-His tag, and AviTag) was expressed as described (Barnes et al., 2020a; Barnes et al., 2020b). A gene encoding a 6xHis-tagged soluble human ACE2 construct (residues 1-615) was purchased from Addgene (Catalog # 149268) and expressed and purified as described (Chan et al., 2020).
SARS-CoV-2 S trimer, RBDs, and soluble ACE2 were purified by Nickel-NTA and size-exclusion chromatography using a Superdex 200 column (GE Life Sciences) as described (Barnes et al., 2020a; Cohen et al., 2021). Peak fractions were identified by SDS-PAGE, and those containing S trimer, monomeric RBDs, or soluble ACE2 were pooled, concentrated, and stored at 4°C (RBDs) or flash frozen in nitrogen and stored at −80°C (S trimer) until use.
ELISAs
Purified RBD at 10 μg/ml in 0.1 M NaHCO3 pH 9.8 was coated onto Nunc® MaxiSorp 384-well plates (Sigma) and stored overnight at 4°C. The following day, plates were blocked with 3% bovine serum albumin (BSA) in TBS-T Buffer (TBS + 0.1% Tween20) for 1hr at room temperature. Blocking solution was removed from the plates, purified IgGs at 50 μg/mL were serially diluted by 4-fold with TBS-T/3% BSA and added to plates for 3 hr at room temperature. Plates were washed with TBS-T and then incubated with 1:15,000 dilution of secondary HRP-conjugated goat anti-human IgG for 45 minutes at room temperature (Southern Biotech). Plates were washed again with TBS-T and developed using SuperSignal ELISA Femto Maximum Sensitivity Substrate (ThermoFisher) and read at 425 nm. ELISAs were done in duplicate, and curves were plotted and integrated to obtain the area under the curve (AUC) using Graphpad Prism v9.1.0.
Neutralization assays
SARS-CoV-2, SARS-CoV-2 variants of concern (Annavajhala et al., 2021; Faria et al., 2021; Rambaut et al., 2020; Tegally et al., 2020; Voloch et al., 2020; West et al., 2021; Zhang et al., 2021), SARS-CoV, WIV1, and SHC014 pseudoviruses based on HIV-1 lentiviral particles were prepared as described (Cohen et al., 2021; Crawford et al., 2020; Robbiani et al., 2020) using genes encoding S protein sequences with cytoplasmic tail deletions: 21 amino acid deletions for SARS-CoV-2, SARS-CoV-2 variants of concern, WIV1, and SHC014 and a 19 amino acid deletion for SARS-CoV. Plasmids expressing the spike protein found in the bat (Rinolophus Sinicus) coronavirus bCoV-WIV16 as well as the pangolin (Manis javanica) coronaviruses from Guandong, China (pCoV-GD) and Guanxi, China (pCoV-GX) have been described previously and are based on ALK02457 (Genebank), Pangolin_CoV_EPI_ISL_410721(Gisaid) and Pangolin_CoV_EPI_ISL_410542 (Gisaid) (Muecksch et al., 2021).
Relative to the SARS-CoV-2 spike gene (Wuhan-Hu-1 Spike Glycoprotein Gene, D614G mutant, designated as ‘wt’ in Figure 1D), the SARS-CoV-2 variants of concern included the D614G mutation and the following other substitutions: B.1.351: L18F, D80A, D215G, del242-244, R246I, K417N, E484K, N501Y, A701V; B.1.1.7: del69-70, del144, N501Y, A570D, P681H, T716I, S982A, D1118H; B.1.429: S13I, W152C, L452R, and B.1.526: L5F, T95I, D253G, E484K, A701V. For neutralization assays presented in Figure 1D, four-fold dilutions of purified IgGs (starting concentrations of 50 μg/mL) were incubated with a pseudotyped virus for 1 hour at 37°C. Cells were washed twice with phosphate-buffered saline (PBS) and lysed with Luciferase Cell Culture Lysis 5x reagent (Promega) after incubation with 293TACE2 target cells for 48 hours at 37°C. NanoLuc Luciferase activity in lysates was measured using the Nano-Glo Luciferase Assay System (Promega). Relative luminescence units (RLUs) were normalized to values derived from cells infected with pseudotyped virus in the absence of IgG. Half-maximal inhibitory concentrations (IC50 values) were determined using 4- or 5-parameter nonlinear regression in AntibodyDatabase (West et al., 2013).
Relative to the SARS-CoV-2 spike gene (Wuhan-Hu-1; NC_045512, D614 sequence designated as ‘wt’ in Figure S2), a panel of plasmids expressing RBD mutant SARS-CoV-2 S proteins in the context of pSARS-CoV-2-SΔ19 have been described previously (Muecksch et al., 2021; Robbiani et al., 2020; Schmidt et al., 2020; Weisblum et al., 2020). The E484K substitution was constructed in the context of a pSARS-CoV-2-SΔ19 variant with a mutation in the furin cleavage site (R683G) to increase infectivity (Muecksch et al., 2021). The IC50 values of this pseudotype (E484K/R683G) was compared to a wild-type SARS-CoV-2 S sequence carrying R683G in the subsequent analyses. For neutralization assays presented in Figure S2, monoclonal antibodies were four-fold serially diluted and incubated with SARS-CoV-2 pseudotyped HIV-1 reporter virus for 1 h at 37 °C (final starting concentration of 2.5 μg/ml). The antibody and pseudotyped virus mixture was added to HT1080ACE2.cl 14 cells (Schmidt et al., 2020). After 48 h, cells were washed with PBS and lysed with Luciferase Cell Culture Lysis 5 × reagent (Promega). Nanoluc luciferase activity in cell lysates was measured using the Nano-Glo Luciferase Assay System (Promega) and the Glomax Navigator (Promega). Relative luminescence units were normalized to those derived from cells infected with SARS-CoV-2 pseudovirus in the absence of monoclonal antibodies. The 50% inhibitory concentration (IC50) was determined using 4-parameter nonlinear regression (least-squares regression method without weighting; constraints: top = 1, bottom = 0) (GraphPad Prism).
SPR-based ACE2 binding competition experiments
Surface Plasmon Resonance (SPR) experiments were done using a Biacore T200 instrument (GE Healthcare). For Figure 2F, purified ACE2-Fc was conjugated to each of the four flow cells using primary amine chemistry at pH 4.5 (Biacore manual) to a CM5 chip (GE Healthcare) to a response level of ∼1000 resonance units (RUs). C118 Fab-SARS-CoV-2 RBD, C022 Fab-SARS-CoV-2 RBD, C144 Fab-SARS-CoV-2 RBD, and CR3022 Fab-SARS-CoV-2 RBD complexes were formed in HBS-EP+ buffer (150mM sodium chloride, 10mM HEPES, 3mM EDTA, 0.05% Tween-20, pH 7.6) by incubating 10uM Fab with 1uM RBD for 2 hours at room temperature. Complexes were injected on the ACE2-Fc-CM5 chip for a contact time of 300 s at 30μL/min and a dissociation time of 30 s in HBS-EP+ buffer.
For Figure S2E, purified SARS CoV-2 RBD was conjugated to each of the four flow cells using primary amine chemistry at pH 4.5 (Biacore manual) to a CM5 chip (GE Healthcare) to a response level of ∼700 RUs. C118, C022, C144, and CR3022 IgG (1000nM) in buffer HBS-EP+ (150mM sodium chloride, 10mM HEPES, 3mM EDTA, 0.05% Tween-20, pH 7.6) were each injected on the RBD-CM5 chip for a contact time of 600 s at 30μL/min. A second injection of soluble ACE2 at 250nM was injected over the immobilized RBD-Fab at 30μL/min for a contact time of 300 s and dissociation time of 30 s in HBS-EP+ buffer. Data were analyzed and plotted using Prism 9 (Graphpad).
X-ray crystallography
Fab-RBD complexes were assembled by incubating an RBD with a 1.5x molar excess of Fab for 1 hr on ice followed by size exclusion chromatography on an S200 10/300 increase column (GE Life Sciences). Fractions containing complex were pooled and concentrated to 8mg/mL. Crystallization trials using commercially-available screens (Hampton Research) were performed at room temperature using the sitting drop vapor diffusion method by mixing equal volumes of a Fab-RBD complex and reservoir using a TTP LabTech Mosquito instrument. Crystals were obtained for C118 Fab-SARS RBD in 0.2M sodium fluoride, 20% w/v polyethylene glycol 3,350 and for C022 Fab-SARS-CoV-2 RBD in 0.05M ammonium sulfate, 0.05M Bis-Tris, 30% v/v pentaerythritol ethoxylate (15/4 EO/OH). Crystals were cryoprotected by adding glycerol directly to drops to a final concentration of 20% and then looped and cryopreserved in liquid nitrogen.
X-ray diffraction data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 12-2 on a Pilatus 6M pixel detector (Dectris). Data from single crystals were indexed and integrated in XDS (Kabsch, 2010) and merged using AIMLESS in CCP4 (Winn et al., 2011) (Table S1). The C022-RBD structure was solved by molecular replacement in PHASER (McCoy et al., 2007) using unmodified RBD coordinates (PDB 7K8M) and coordinates from C102 Fab (PDB 7K8M) after trimming heavy chain and light chain variable domains using Sculptor (Bunkóczi and Read, 2011) as search models. Coordinates were refined with phenix.refine from the PHENIX package ver. 1.17.1 (Adams et al., 2010) and cycles of manual building in Coot (ver 0.8.9.1) (Emsley et al., 2010) (Table S1).
Cryo-EM Sample Preparation
C118 Fab-S trimer complex was assembled by incubating purified SARS-CoV-2 S trimer at a 1.2:1 molar excess of purified Fab per S protomer at RT for 30 min. 17 uL of complex was mixed with 0.8uL of a 0.5% w/v F-octylmaltoside solution (Anatrace) and then 3μL were immediately applied to a 300 mesh, 1.2/1.3 AuUltraFoil grid (Electron Microscopy Sciences) that had been freshly glow discharged for 1 min at 20mA using a PELCO easiGLOW (Ted Pella). The grid was blotted for 3.5 s with Whatman No. 1 filter paper at 22°C and 100% humidity then vitrified in 100% liquid ethane using a Mark IV Vitrobot (FEI) and stored under liquid nitrogen.
Cryo-EM data collection and processing
Single-particle cryo-EM data were collected for the C118-S trimer complex as previously described (Barnes et al., 2020a). Briefly, for the C118-S trimer complex, micrographs were collected on a Talos Arctica transmission electron microscope (Thermo Fisher) operating at 200 kV using a 3x3 beam image shift pattern with SerialEM automated data collection software (Mastronarde, 2005). Movies were obtained on a Gatan K3 Summit direct electron detector operating in counting mode at a nominal magnification of 45,000x (super-resolution 0.4345 Å/pixel) using a defocus range of −0.7 to −2.0 μm. Movies were collected with an 3.6 s exposure time with a rate of 13.5 e-/pix/s, which resulted in a total dose of ∼60 e-/Å2 over 40 frames. The 2,970 movies were patch motion corrected for beam-induced motion including dose-weighting within cryoSPARC v3.1 (Punjani et al., 2017) after binning super resolution movies by 2 (0.869 Å/pixel). The non-dose-weighted images were used to estimate CTF parameters using Patch CTF in cryoSPARC, and micrographs with poor CTF fits and signs of crystalline ice were discarded, leaving 2,487 micrographs. Particles were picked in a reference-free manner using Gaussian blob picker in cryoSPARC (Punjani et al., 2017). An initial 923,707 particle stack was extracted, binned x4 (3.48 Å/pixel), and subjected to ab initio volume generation (4 classes) and subsequent heterogeneous refinement. The 3D classes that showed features for a Fab-S trimer complex were 2D classified to identify class averages corresponding to intact S-trimer complexes with well-defined structural features. This routine resulted in a new particle stack of 110,789 particles, which were unbinned (0.836 Å/pixel) and re-extracted using a 432 box size. Particles were then moved to Relion v3.1 (Zivanov et al., 2018), for further 3D classification (k = 6)., which revealed two distinct states of the C118-S trimer complex.
Particles from state 1 (53,728 particles) and state 2 (31,422 particles) were separately refined using non-uniform 3D refinement imposing either C3 or C1 symmetry in cryoSPARC, respectively, to final resolutions of 3.4 Å and 4.5 Å according to the gold-standard FSC (Bell et al., 2016), respectively. To improve features at the C118-RBD interface, particles from State 1 were symmetry expanded and classified for a focused, non-uniform 3D local refinement in cryoSPARC. A soft mask was generated around the C118 VHVL – RBD domains (5-pixel extension, 10-pixel soft cosine edge) for local refinements. These efforts resulted in a modest improvement in the RBD-C118 Fab interface (Figure S5B), with an overall resolution of 3.7 Å according to the gold-standard FSC.
Cryo-EM Structure Modeling and Refinement
Initial coordinates were generated by rigid-body docking reference structures into cryo-EM density using UCSF Chimera (Goddard et al., 2007). The following coordinates were used: SARS-CoV-2 S 6P trimer: PDB 7K4N (mutated to include 6P mutations), PDB 7BZ5, and C118 Fab variable domains: this study. These initial models were then refined into cryo-EM maps using one round of rigid body refinement, morphing and real space refinement in Phenix (Adams et al., 2010). Sequence-updated models were built manually in Coot (Emsley et al., 2010) and then refined using iterative rounds of refinement in Coot and Phenix (Adams et al., 2010). Glycans were modeled at possible N-linked glycosylation sites (PNGSs) in Coot using ‘blurred’ maps processed with a variety of B-factors (Terwilliger et al., 2018). Validation of model coordinates was performed using MolProbity (Chen et al., 2010) and is reported in Table S3.
Structure Analyses
Interacting residues were determined using PDBePISA (Krissinel and Henrick, 2007) for the C118 and C022 epitopes using the following criteria: Potential H-bonds were assigned using a distance of < 3.6A ° and an A-D-H angle of > 90°, and the maximum distance allowed for a van der Waals interaction was 4.0A °. H-bonds assigned for the C022-RBD complex should be considered tentative due to the relatively low resolution of the structure (3.2Å). Epitope patches for other antibodies in Figure 4A were defined as residues containing an atom within 4Å of the partner protein as determined in PyMOL (Schrödinger, 2011). Buried surface areas (BSAs) were determined with PDBePISA (Krissinel and Henrick, 2007) using a 1.4A ° probe. Structure figures were made using PyMOL ver. 2.3.5 (Schrödinger, 2011) or UCSF Chimera ver. 1.14 (Goddard et al., 2018). Fab-RBD-ACE2 complex figures (Figure 2E) were made by aligning RBD Cα atoms of Fab-RBD (this study and PDBs 6W41 and 6ZCZ) and RBD-ACE2 structures (PDB 6M0J). As density at position N357RBD for our C118-SARS RBD structure precluded building of the glycan, it was modeled (Figure 2A) by aligning Cα atoms of residues 353-371 of SARS-CoV spike-S230 structure (PDB 6NB6, chain E) and overlaying the glycan at N357RBD from the SARS-CoV spike on the RBD model of the C118-RBD crystal structure. Sequence alignments were done using the MUSCLE server (https://www.ebi.ac.uk/Tools/msa/muscle/) (Edgar, 2004). Secondary structure was defined as described in (Huo et al., 2020).
To predict whether intra-spike crosslinking by a single IgG binding to a spike trimer might be possible, we measured the distance between residue 222HC Cα atoms in the CH1 domains of adjacent Fabs in Fab-S structures as previously described (Barnes et al., 2020a). This distance was compared to analogous distances in crystal structures of intact IgGs (42Å, PDB 1HZH; 48Å, PDB 1IGY; 52Å, PDB 1IGT). We accounted for potential influences of crystal packing in intact IgG structures, flexibility in the VH-VL/CH1-CL elbow bend angle, and uncertainties in CH1-CL domain placement in Fab-S cryo-EM structures, by setting a cut-off of ≤ 65Å for this measured distance as potentially allowing for a single IgG to include both Fabs when binding a spike trimer.
Quantification and statistical analysis
All software used for structural analysis and processing is listed in the Key resources table. The half-maximal inhibitory concentrations for monoclonal antibodies (IC50s) reported in Figure 1 were determined by normalizing relative luminescence units (RLUs) to values derived from SARS-CoV-2 pseudovirus infected cells in the absence of IgG antibody followed by 4- or 5-parameter nonlinear regressions analyzed using AntibodyDatabase (West et al., 2013). IC50s reported in Figure S2 were determined by normalizing RLUs to values derived from cells infected with SARS-CoV-2 pseudovirus in the absence of antibodies followed by a 4-parameter nonlinear regression (least-squares regression method without weighting) using GraphPad Prism. Numbers of replicates and experiments and statistical tests for each experiment are indicated in in the respective figure legends and STAR Methods.
Acknowledgments
We thank J. Vielmetter, P. Hoffman, and the Protein Expression Center in the Beckman Institute at Caltech for expression assistance and K. Dam for assistance with soluble ACE2 purification. Electron microscopy was performed in the Caltech Cryo-EM Center with assistance from S. Chen and A. Malyutin. We thank the Gordon and Betty Moore and Beckman Foundations for gifts to Caltech to support the Molecular Observatory. We thank J. Kaiser, Director of the Molecular Observatory at Caltech, and beamline staff C. Smith and S. Russi at SSRL for data collection assistance. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by the National Institutes of Health (NIH), National Institute of General Medical Sciences (NIGMS) (P30GM133894). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. This work was supported by the NIH (P01-AI138938-S1 to P.J.B. and M.C.N.; R01AI078788 to T.H.; and R01AI640511 to P.D.B.), the Caltech Merkin Institute for Translational Research (P.J.B.), and a George Mason University Fast Grant (to P.J.B.). F.M. is supported by the Bulgari Women & Science Fellowship in COVID-19 Research. C.O.B was supported by the Hanna Gray Fellowship Program from the Howard Hughes Medical Institute and the Postdoctoral Enrichment Program from the Burroughs Wellcome Fund. M.C.N. is a Howard Hughes Medical Institute (HHMI) investigator.
Author contributions
C.A.J., A.A.C., P.J.B., and C.O.B. conceived and designed experiments. Proteins were produced and characterized by A.A.C., K.E.H.-T., C.O.B., and C.A.J. Binding and neutralization studies were done by A.A.C. and F.M. with assistance from P.N.P.G., Y.E.L., and F.S. SPR binding competition experiments were done by C.A.J. with assistance from J.R.K. Structural studies were performed by C.A.J. with assistance from C.O.B. Structure analysis was done by C.A.J. with assistance from C.O.B. and A.A.C. Sequence analysis was done by A.P.W. The paper was written by C.A.J., P.J.B., and C.O.B. with assistance from A.A.C., T.H., M.C.N., P.D.B., and other authors.
Declaration of interests
The Rockefeller University has filed provisional patent applications in connection with this work on which M.C.N. (US patent 63/021,387) is listed as an inventor.
Published: September 8, 2021; corrected online: December 15, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109760.
Supporting Citations
The following reference appears in the supplemental information: Kane et al. (2016).
Supplemental information
Data and code availability
-
•
Atomic models of C118 Fab complexed with SARS-CoV RBD and C022 Fab complexed with SARS-CoV-2 RBD have been deposited in the Protein Data Bank (PDB) (https://www.rcsb.org/) under accession codes 7RKS and 7RKU, respectively. The atomic model and cryo-EM maps generated for the C118 Fab–SARS-CoV-2 S complex have been deposited at the PDB (https://www.rcsb.org/) and the Electron Microscopy Databank (EMDB) (http://www.emdataresource.org/) under accession codes 7RKV (state 1 coordinates), EMD-24504 (state 1) and EMD-24505 (state 2). All models and maps are publicly available as of the date of publication.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annavajhala M.K., Mohri H., Zucker J.E., Sheng Z., Wang P., Gomez-Simmonds A., Ho D.D., Uhlemann A.-C. A Novel SARS-CoV-2 Variant of Concern, B.1.526, Identified in New York. medRxiv. 2021 doi: 10.1101/2021.02.23.21252259. [DOI] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J.M., Chen M., Baldwin P.R., Ludtke S.J. High resolution single particle refinement in EMAN2.1. Methods. 2016;100:25–34. doi: 10.1016/j.ymeth.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunkóczi G., Read R.J. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr. D Biol. Crystallogr. 2011;67:303–312. doi: 10.1107/S0907444910051218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.A., Gnanapragasam P.N.P., Lee Y.E., Hoffman P.R., Ou S., Kakutani L.M., Keeffe J.R., Wu H.-J., Howarth M., West A.P., et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735–741. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Suguitan A.L., Jr., Pinna D., Silacci C., Fernandez-Rodriguez B.M., Vanzetta F., Santos C., Luke C.J., Torres-Velez F.J., Temperton N.J., et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., et al. Protocol and Reagents for Pseudotyping Lentiviral Particles with SARS-CoV-2 Spike Protein for Neutralization Assays. Viruses. 2020;12:513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Zhou D., Ginn H.M., Duyvesteyn H.M.E., Supasa P., Case J.B., Zhao Y., Walter T.S., Mentzer A.J., Liu C., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184:2183–2200.e22. doi: 10.1016/j.cell.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Claro I.M., Candido D., Moyses Franco L.A., Andrade P.S., Coletti T.M., Silva C.A.M., Sales F.C., Manuli E.R., Agular R.S., et al. 2021. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological.com.https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 [Google Scholar]
- Fung T.S., Liu D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Ferrin T.E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29:463–476.e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristick H.B., von Boehmer L., West A.P., Jr., Schamber M., Gazumyan A., Golijanin J., Seaman M.S., Fätkenheuer G., Klein F., Nussenzweig M.C., Bjorkman P.J. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat. Struct. Mol. Biol. 2016;23:906–915. doi: 10.1038/nsmb.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Haagmans B.L., Al Dhahiry S.H.S., Reusken C.B.E.M., Raj V.S., Galiano M., Myers R., Godeke G.-J., Jonges M., Farag E., Diab A., et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect. Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.-L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.-C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.A., Tan T.K., Chen T.H., Huang C.G., Harvey R., Hussain S., Chen C.P., Harding A., Gilbert-Jaramillo J., Liu X., et al. Breadth and function of antibody response to acute SARS-CoV-2 infection in humans. PLoS Pathog. 2021;17:e1009352. doi: 10.1371/journal.ppat.1009352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., et al. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host Microbe. 2020;28:445–454.e6. doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M., Zang T.M., Rihn S.J., Zhang F., Kueck T., Alim M., Schoggins J., Rice C.M., Wilson S.J., Bieniasz P.D. Identification of Interferon-Stimulated Genes with Antiretroviral Activity. Cell Host Microbe. 2016;20:392–405. doi: 10.1016/j.chom.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., Corbett K.S., Graham B.S., McLellan J.S., Ward A.B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein J.S. California Institute of Technology; 2009. Investigations in the design and characterization of HIV-1 neutralizing molecules; p. 166. [Google Scholar]
- Klein J.S., Bjorkman P.J. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 2010;6:e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., et al. Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell. 2020;182:843–854.e12. doi: 10.1016/j.cell.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M.P., Giudicelli V., Ginestoux C., Jabado-Michaloud J., Folch G., Bellahcene F., Wu Y., Gemrot E., Brochet X., Lane J., et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M.P., Giudicelli V., Duroux P., Jabado-Michaloud J., Folch G., Aouinti S., Carillon E., Duvergey H., Houles A., Paysan-Lafosse T., et al. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 2015;43:D413–D422. doi: 10.1093/nar/gku1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist S.R., Dinnon K.H., 3rd, Schäfer A., Tse L.V., Okuda K., Hou Y.J., West A., Edwards C.E., Sanders W., Fritch E.J., et al. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell. 2020;183:1070–1085.e12. doi: 10.1016/j.cell.2020.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li Z., Tomlinson A.C., Wong A.H., Zhou D., Desforges M., Talbot P.J., Benlekbir S., Rubinstein J.L., Rini J.M. The human coronavirus HCoV-229E S-protein structure and receptor binding. eLife. 2019;8:e51230. doi: 10.7554/eLife.51230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wu N.C., Yuan M., Bangaru S., Torres J.L., Caniels T.G., van Schooten J., Zhu X., Lee C.D., Brouwer P.J.M., et al. Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. Immunity. 2020;53:1272–1280.e5. doi: 10.1016/j.immuni.2020.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu H., Yuan M., Huang D., Bangaru S., Zhao F., Lee C.D., Peng L., Barman S., Zhu X., Nemazee D., et al. A combination of cross-neutralizing antibodies synergizes to prevent SARS-CoV-2 and SARS-CoV pseudovirus infection. Cell Host Microbe. 2021;29:806–818.e6. doi: 10.1016/j.chom.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muecksch F., Weisblum Y., Barnes C.O., Schmidt F., Schaefer-Babajew D., Lorenzi J.C.C., Flyak A.I., DeLaitsch A.T., Huey-Tubman K.E., Hou S., et al. Development of potency, breadth and resilience to viral escape mutations in SARS-CoV-2 neutralizing antibodies. bioRxiv. 2021 doi: 10.1101/2021.03.07.434227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear W.S., Nolan G.P., Scott M.L., Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Piccoli L., Park Y.-J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–1042.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E., on behalf of COVID-19 Genomics Consortium UK (CoG-UK) 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological.org.https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science. 2021;371:823–829. doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Rapid isolation of potent SARS-CoV-2 neutralizing antibodies and protection in a small animal model. bioRxiv. 2020 doi: 10.1101/2020.05.11.088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M.M., Tortorici M.A., Park Y.-J., Walls A.C., Homad L., Acton O., Bowen J., Wang C., Xiong X., de van der Schueren W., et al. Structural basis for broad coronavirus neutralization. bioRxiv. 2021 doi: 10.1101/2020.12.29.424482. [DOI] [PubMed] [Google Scholar]
- Schaefer W., Regula J.T., Bähner M., Schanzer J., Croasdale R., Dürr H., Gassner C., Georges G., Kettenberger H., Imhof-Jung S., et al. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc. Natl. Acad. Sci. USA. 2011;108:11187–11192. doi: 10.1073/pnas.1019002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L., Wang H., Gao H., Chen S., McDowall A.W., Bjorkman P.J. Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env. Cell. 2015;162:1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid J.F., Mouquet H., Ueberheide B., Diskin R., Klein F., Oliveira T.Y., Pietzsch J., Fenyo D., Abadir A., Velinzon K., et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid J.F., Barnes C.O., Eraslan B., Hudak A., Keeffe J.R., Cosimi L.A., Brown E.M., Muecksch F., Weisblum Y., Zhang S., et al. B cell genomics behind cross-neutralization of SARS-CoV-2 variants and SARS-CoV. Cell. 2021;184:3205–3221.e24. doi: 10.1016/j.cell.2021.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.-H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C., et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217:e20201181. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoofs T., Barnes C.O., Suh-Toma N., Golijanin J., Schommers P., Gruell H., West A.P., Jr., Bach F., Lee Y.E., Nogueira L., et al. Broad and Potent Neutralizing Antibodies Recognize the Silent Face of the HIV Envelope. Immunity. 2019;50:1513–1529.e9. doi: 10.1016/j.immuni.2019.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger L. The PyMOL Molecular Graphics System; 2011. The PyMOL Molecular Graphics System. [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.H., Feng J., et al. Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity. 2020;53:98–105.e5. doi: 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P., Canziani G.A., Carter E.P., Chaiken I. The Case for S2: The Potential Benefits of the S2 Subunit of the SARS-CoV-2 Spike Protein as an Immunogen in Fighting the COVID-19 Pandemic. Front. Immunol. 2021;12:637651. doi: 10.3389/fimmu.2021.637651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.D., Tu C.C., Zhang G.W., Wang S.Y., Zheng K., Lei L.C., Chen Q.X., Gao Y.W., Zhou H.Q., Xiang H., et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Czudnochowski N., Zatta F., Park Y.-J., Liu Z., Addetia A., Pinto D., Beltramello M., Hernandez P., Greaney A.J., et al. Antibodies to the SARS-CoV-2 receptor-binding domain that maximize breadth and resistance to viral escape. bioRxiv. 2021 doi: 10.1101/2021.04.06.438709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- Terwilliger T.C., Adams P.D., Afonine P.V., Sobolev O.V. A fully automatic method yielding initial models from high-resolution cryo-electron microscopy maps. Nat. Methods. 2018;15:905–908. doi: 10.1038/s41592-018-0173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Czudnochowski N., Starr T.N., Marzi R., Walls A.C., Zatta F., Bowen J.E., Jaconi S., Di Iulio J., Wang Z., et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature. 2021;597:103–108. doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloch C.M., da Silva F.,R., Jr., de Almeida L.G.P., Cardoso C.C., Brustolini O.J., Gerber A.L., Guimarães A.P.d.C., Mariani D., da Costa R.M., Ferreira O.C., Jr., et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. medRxiv. 2020 doi: 10.1101/2020.12.23.20248598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagh K., Bhattacharya T., Williamson C., Robles A., Bayne M., Garrity J., Rist M., Rademeyer C., Yoon H., Lapedes A., et al. Optimal Combinations of Broadly Neutralizing Antibodies for Prevention and Treatment of HIV-1 Clade C Infection. PLoS Pathog. 2016;12:e1005520. doi: 10.1371/journal.ppat.1005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Tortorici M.A., Bosch B.J., Frenz B., Rottier P.J.M., DiMaio F., Rey F.A., Veesler D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Gristick H.B., Scharf L., West A.P., Galimidi R.P., Seaman M.S., Freund N.T., Nussenzweig M.C., Bjorkman P.J. Asymmetric recognition of HIV-1 Envelope trimer by V1V2 loop-targeting antibodies. eLife. 2017;6:e27389. doi: 10.7554/eLife.27389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Li S.-Y., Yang X.-L., Huang H.-M., Zhang Y.-J., Guo H., Luo C.-M., Miller M., Zhu G., Chmura A.A., et al. Serological Evidence of Bat SARS-Related Coronavirus Infection in Humans, China. Virol. Sin. 2018;33:104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., van Haperen R., Gutiérrez-Álvarez J., Li W., Okba N.M.A., Albulescu I., Widjaja I., van Dieren B., Fernandez-Delgado R., Sola I., et al. A conserved immunogenic and vulnerable site on the coronavirus spike protein delineated by cross-reactive monoclonal antibodies. Nat. Commun. 2021;12:1715. doi: 10.1038/s41467-021-21968-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C.C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P., Jr., Scharf L., Horwitz J., Klein F., Nussenzweig M.C., Bjorkman P.J. Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proc. Natl. Acad. Sci. USA. 2013;110:10598–10603. doi: 10.1073/pnas.1309215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West A.P., Barnes C.O., Yang Z., Bjorkman P.J. SARS-CoV-2 lineage B.1.526 emerging in the New York region detected by software utility created to query the spike mutational landscape. bioRxiv. 2021 doi: 10.1101/2021.02.14.431043. [DOI] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA. 2021;325:1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Duyvesteyn H.M.E., Chen C.-P., Huang C.-G., Chen T.-H., Shih S.-R., Lin Y.-C., Cheng C.-Y., Cheng S.-H., Huang Y.-C., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Ji J., Chen X., Bi Y., Li J., Hu T., Song H., Chen Y., Cui M., Zhang Y., et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. bioRxiv. 2021 doi: 10.1101/2021.03.08.434390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The video shows soluble SARS-CoV-2 spike (PDB: 7BZ5) and the ‘down’ and ‘up’ conformations of the RBD domain. Next, the sequence conservation of 12 sarbecovirus RBDs is mapped onto the surface of the RBD domain. Finally, The C118-SARS-CoV RBD and C022-SARS-CoV-2 RBD structures are shown.
Data Availability Statement
-
•
Atomic models of C118 Fab complexed with SARS-CoV RBD and C022 Fab complexed with SARS-CoV-2 RBD have been deposited in the Protein Data Bank (PDB) (https://www.rcsb.org/) under accession codes 7RKS and 7RKU, respectively. The atomic model and cryo-EM maps generated for the C118 Fab–SARS-CoV-2 S complex have been deposited at the PDB (https://www.rcsb.org/) and the Electron Microscopy Databank (EMDB) (http://www.emdataresource.org/) under accession codes 7RKV (state 1 coordinates), EMD-24504 (state 1) and EMD-24505 (state 2). All models and maps are publicly available as of the date of publication.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.