Abstract
Pridopidine is a selective Sigma-1 receptor (S1R) agonist in clinical development for Huntington disease (HD) and amyotrophic lateral sclerosis. S1R is a chaperone protein localized in mitochondria-associated endoplasmic reticulum (ER) membranes, a signaling platform that regulates Ca2+ signaling, reactive oxygen species (ROS) and mitochondrial fission. Here, we investigate the protective effects of pridopidine on various mitochondrial functions in human and mouse HD models. Pridopidine effects on mitochondrial dynamics were assessed in primary neurons from YAC128 HD mice expressing the mutant human HTT gene. We observe that pridopidine prevents the disruption of mitochondria-ER contact sites and improves the co-localization of inositol 1,4,5-trisphosphate receptor (IP3R) and its chaperone S1R with mitochondria in YAC128 neurons, leading to increased mitochondrial activity, elongation, and motility. Increased mitochondrial respiration is also observed in YAC128 neurons and in pridopidine-treated HD human neural stem cells (hNSCs). ROS levels were assessed after oxidative insult or S1R knockdown in pridopidine-treated YAC128 neurons, HD hNSCs, and human HD lymphoblasts. All HD models show increased ROS levels and deficient antioxidant response, which are efficiently rescued with pridopidine. Importantly, pridopidine treatment before H2O2-induced mitochondrial dysfunction and S1R presence are required for HD cytoprotection. YAC128 mice treated at early/pre-symptomatic age with pridopidine show significant improvement in motor coordination, indicating a delay in symptom onset. Additionally, in vivo pridopidine treatment reduces mitochondrial ROS levels by normalizing mitochondrial complex activity. In conclusion, S1R-mediated enhancement of mitochondrial function contributes to the neuroprotective effects of pridopidine, providing insight into its mechanism of action and therapeutic potential.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13311-021-01022-9.
Keywords: Huntington disease, Pridopidine, Sigma-1 receptor, Mitochondrial dysfunction, Oxidative stress
Introduction
Huntington disease (HD) is a neurodegenerative disorder caused by an expanded CAG trinucleotide repeat in the HTT gene that progressively affects motor and cognitive functions, ultimately leading to cell death. Mitochondrial dysfunction and oxidative stress are early, central mechanisms underlying neuronal death in neurodegenerative disorders including HD [1, 2]. Therefore, therapeutic targeting of mitochondria with small molecules may have the potential to mitigate the progression of these diseases.
Pridopidine is a highly selective and potent agonist of the Sigma-1 receptor (S1R) [3, 4]. The S1R is a molecular chaperone protein enriched in the mitochondria-associated endoplasmic reticulum (ER) membranes (MAM) and implicated in cell differentiation, oxidative signaling, neuroplasticity, and cognitive functions [5–7]. Pridopidine exerts neuroprotective effects in several models of neurodegenerative disorders including HD, Parkinson’s disease (PD), Alzheimer’s disease (AD), and amyotrophic lateral sclerosis (ALS), mediated by the activation of the S1R [8–13]. By its agonistic action at the S1R, pridopidine rescues mHTT-induced cell death in HD mouse neurons and human iPSCs [8], induces restoration of homeostatic synaptic plasticity in HD cortical neurons [14], and restores the impaired spine density and aberrant intracellular calcium (Ca2+) signaling in HD neurons [10]. In the 6-hydroxydopamine (6-OHDA) lesion mouse model for PD, pridopidine treatment induces neurorestoration of the nigrostriatal system and significantly increases the density of dopaminergic fiber and nigral dopamine cell bodies [15]. The neuroprotective effects of pridopidine are S1R mediated as they were abolished in S1R knockout mice [13].
The S1R plays a key role in the mitochondrial-associated membrane (MAM) domain of the ER and in mitochondrial function [16, 17]. MAM is a highly specialized region of the ER membrane with a lipid raft composition allowing it to interact with the outer mitochondrial membrane, which has key roles in various biological pathways, including mitochondrial fission, Ca2+ shuttling, phospholipid metabolism, autophagosome formation, and oxidative stress [18]. S1R deficiency induces MAM disruption, resulting in mis-localization of the ER Ca2+ channel inositol 1,4,5-trisphosphate (IP3) receptor (IP3R), mitochondrial dysfunction [19], and motor deficits [16].
The effects of pridopidine in ameliorating HD mitochondrial dysfunction have not been addressed until now. Here, we show that pridopidine restores mitochondrial functions, representing a mechanism by which pridopidine exerts its neuroprotective effects. In HD models, pridopidine treatment increases tethering between mitochondria and ER, improving mitochondrial elongation, movement, and respiration. We also observe that pridopidine, via activation of the S1R, rescues mitochondrial dysfunction induced by oxidative damage in both in vitro and ex vivo HD models, namely, in YAC128 transgenic mice, human HD lymphoblasts, and human HD neural stem cells (NSCs). Additionally, we show in vivo that early pridopidine treatment is effective in delaying onset of HD-related motor symptoms in the YAC128 HD mice.
Materials and Methods
Animals and Ethical Permits
Colonies of hemizygous YAC128 (line HD53; mHTT high expresser), homozygous YAC128 (line HD55; mHTT low expresser), and WT mice, all with FVB/N background, were housed in the housing Facility of the CNC, Coimbra, Portugal, and at UBC, Vancouver, Canada. Temperature was controlled (22–23 °C) and a 12-h light/dark cycle maintained. Food and water were available ad libitum. All mouse experiments were carried out in accordance with the guidelines of the Institutional Animal Care and Use of Committee and the European Community directive (2010/63/EU) and protocols approved by the Faculty of Medicine, University of Coimbra (ref ORBEA_189_2018/11042018) and UBC Committee (ref A16-0130) on Animal Care and the Canadian Council on Animal Care. All efforts were made to minimize animal suffering and to reduce the number of animals used.
Primary Neuronal Cultures
Primary cortical, striatal, and cortico-striatal co-cultures were generated from the offspring of crosses between wild-type (WT) mice (used as controls) or between homozygous YAC128 mice (line HD55) males or hemizygous YAC128 mice (line HD53) males and WT females from the same genetic background (FVB/N). Embryos from timed pregnant females were collected on gestational day E15.5–16.5.
For cortico-striatal co-cultures, brains were removed and transferred to Hibernate E medium (Thermo Fisher Sci., catalog no. A1247601) for up to 24 h. At this time, the samples from the remaining embryonic tissues were genotyped. The cortex and striatum were micro-dissected in ice-cold Hank’s balanced salt solution (HBSS; Gibco), then diced and pooled per genotype. The tissues were then dissociated with 0.05% trypsin-EDTA (Thermo Fisher Sci., catalog no. 25300054), which was then neutralized with 10% fetal bovine serum (FBS) in Neurobasal medium (Thermo Fisher Sci., catalog no. 21103049) and DNAse I treatment (153 U/ml). The dissociated tissues were centrifuged at 145×g for 5 min, and the pellet was then triturated with a pipette five to six times. Cells were seeded on poly-D-lysine-coated plates in Neurobasal media supplemented with 2% B27 (Thermo Fisher Sci., catalog no. 17504044), 100 U/ml penicillin-streptomycin (Gibco, catalog no. 15140122), and 2 mM L-glutamax (Thermo Fisher Sci., catalog no. 35050061). The cells were fed with one-third volume fresh medium every fifth day.
To obtain pure cortical and striatal neurons, tissue was microdissected in HBSS supplemented with 0.3% fatty acid free BSA (Sigma-Aldrich, catalog no. A8806). Tissue from embryos with the same genotype were pooled and digested with 0.003% trypsin in HBSS, then mechanically digested. Soybean (Sigma-Aldrich, catalog no. T9128) was used to block the trypsin. Neurons were plated at a density of either 130 × 103 cells/cm2 (high) or 85 × 103 cells/cm2 (low) on poly-D-lysine (0.1 mg/ml)-coated plates and maintained in Neurobasal medium supplemented with 2% B27, 1 mM glutamine, and 20 μg/ml gentamicin (Thermo Fisher Sci., catalog no. 15750060). Cultures were maintained at 37 °C in a humidified incubator with 5% CO2/95% air. After 3 days in vitro (DIV) half of the medium was replaced with fresh supplemented Neurobasal medium containing 5-fluoro-2′-deoxyuridine (5-FdU, 5 μM final concentration in the medium; Sigma-Aldrich, catalog no. F0503) to reduce dividing non-neuronal cells. At DIV 7 half of the medium was again replaced with fresh medium, and cells were used at DIV12. Striatal cultures used for single cell measurements displayed 82% of GABA (gamma-aminobutyric acid)-positive plus DARPP-32 (dopamine and cAMP regulated phosphoprotein of 32 kDa)-positive neurons [20].
Neuronal Transfection
Striatal neurons were transfected with pDsRed2-Mito Vector (MitoDsRed; Clontech, catalog no. 632421) at 8 DIV using the calcium phosphate precipitation method as previously described [21]. Briefly, plasmid was diluted in TE (in mM: 1 Tris-HCl pH 7.3, 1 mM EDTA), followed by the addition of CaCl2 (in mM: 2.5 CaCl2 in 10 HEPES, pH 7.2). The DNA solution was carefully added to 2× HEBS (in mM: 12 dextrose, 50 HEPES, 10 KCl, 280 NaCl and 1.5 Na2HPO4.2H2O, pH 7.2) while bubbling air through the solution. The mixture was then incubated for 25 min at room temperature. The precipitates were added dropwise to the coverslips in Neurobasal medium and incubated for 80 min at 37 °C. The DNA–Ca2+-phosphate precipitates were dissolved with freshly made dissolution medium (Neurobasal medium with 20 mM HEPES, pH 6.8) and incubated for 7 min at room temperature. The transfected neurons were then washed with Neurobasal medium and transferred back to their original dishes containing conditioned culture medium until DIV12.
Lymphoblast Cultures and Transfection
Lymphoblasts from a control subject (GM02174, CAG repeat 15/15) and an HD patient (NA04724, CAG repeat 67/15), obtained from the Coriell Institute, were grown in RPMI medium (Thermo Fisher Sci. 11875093) containing 10% FBS, 2 mM L-glutamax, and 100 U/ml penicillin-streptomycin. The lymphoblasts were passaged 1:3 every 5 to 6 days. For conducting experiments, the lymphoblasts were collected and centrifuged at 145×g for 5 min. The cell pellet was resuspended in fresh growth medium. Lymphoblasts between passage number 5 and 20 were used in each experiment.
S1R depletion was induced by treating with S1R-directed siRNA duplexes (Origene, cat no. SR426072). siRNA transfection was performed in antibiotic-free growth medium using the Polyplus JETPRIME transfection protocol as per manufacturer’s instructions (Polyplus). Forty-eight hours after transfection, the cells were subjected to pridopidine treatment and/or oxidative stress challenge. S1R knockdown efficiency was determined by immunoblot analysis.
Human Neural Stem Cell Culture
Neural stem cells (NSCs) were differentiated from heterozygous human-induced pluripotent stem cells (iPSC), HD4-iPSC, with a normal (19 CAG repeats) and an expanded allele (72 CAG repeats) generated by Park and colleagues [22], generously provided by Prof. George Daley (Harvard Medical School, Boston, Massachusetts, USA) and control AMS4-iPSC generated and characterized by Pereira de Almeida and collaborators [23] (Center for Neuroscience and Cell Biology, University of Coimbra, Portugal). iPSCs were maintained in Geltrex® (Thermo Fisher Sci., catalog no. A1413202) coated 6-well plates until 90% confluence, then the neural induction protocol was applied. Neural differentiation was based on dual SMAD inhibition with SB431542 (Lefty/Activin/transforming growth factor beta—TGFβ inhibitor; Tocris Bioscience, catalog no. 1614), dorsomorphin (bone morphogenetic protein—BMP inhibitor; Tebu-bio, catalog no. 04–0024), and XAV-939 (β-catenin-transcription inhibitor and axin stabilizing agent; Sigma-Aldrich, catalog no. X3004), as previously described [24–26]. Neural induction medium (N2 medium) consisted of a 1:1 mixture of DMEM/F12 (Thermo Fisher Sci., catalog no. 32500043) and Neurobasal, and supplemented with 1% N2 (100×) (Thermo Fisher Sci., catalog no. 17502048), 2 mM L-glutamine, 100 μM nonessential amino acids (Sigma-Aldrich, catalog no: M7145), 100 μM 2-mercaptoethanol (Sigma-Aldrich, catalog no. M6250), 1% penicillin/streptomycin (Thermo Fisher Sci., catalog no. 15140122), and 2% B27. Neural induction occurred between day 0 and days 12–15. From day 0 to day 5, cells were maintained in iPSC medium without FGF2 and incubated with 5 μM dorsomorphin and 10 μM SB431542. Medium was changed every other day. From day 5 to day 12, the medium was gradually replaced by 75% KnockOut Serum Replacement + 25% N2 medium, 50% iPSC + 50% N2 medium up to 100% N2 medium with 5 μM dorsomorphin, 10 μM SB431542, and 1 µM XAV939 [24–26]. Between days 12 and 15, fields full of rosettes became morphologically visible. To allow the cells to differentiate, cells were replated in Geltrex® coated 12-well plates. For detaching, 500 µl of 1× Accutase® (Thermo Fisher Sci., catalog no. A1110501) was added to the plate and incubated at 37 °C in 5% CO2, for 15–20 min. Accutase was diluted in DMEM/F12 medium pre-warmed to 37 °C. Cells were collected and spun for 3 min at 200×g at room temperature, and resuspended in 200 µl of media into 12-well plates. Cells were allowed to adhere for 30 min, and then 300 μL N2 medium, supplemented with 10 μM Y-27632 (Thermo Fisher Sci.), 10 ng/ml FGF2 (Tebu-bio, catalog no. 100-18B-A), and 10 ng/ml EGF (Tebu-bio, catalog no. AF-100–15-A), was added. Cells were incubated overnight at 37 °C, 5% CO2. Cells were passaged every 2–3 days for no more than 10–12 passages. Expression of the neural lineage marker proteins nestin and SOX2 was confirmed by immunocytochemistry upon each differentiation process (data not shown).
Pridopidine Incubations
Pridopidine was provided by Prilenia Therapeutics. A working stock concentration of 10 mM was prepared in sterile deionized water and stored at 4 °C for up to 2 weeks. Pridopidine incubations were done for 24 h in all cellular models used unless otherwise stated (Fig. 3h). Final working concentrations are described in figures and figure legends.
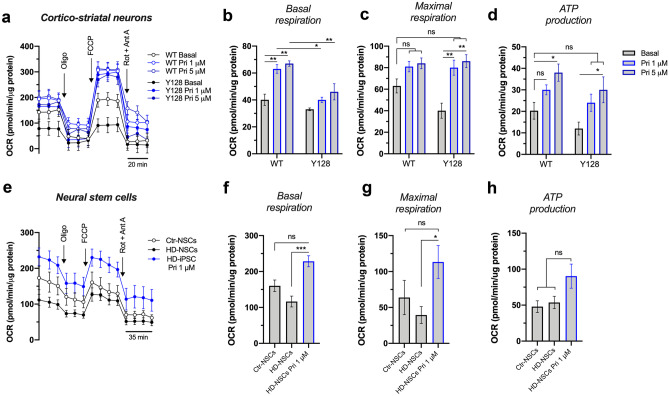
Fig. 3.
Pridopidine enhances mitochondrial respiration in HD cell models. a–d Oxygen consumption evaluated in WT and YAC128 cortical/striatal neurons treated with 1 and 5 µM pridopidine for 24 h using the Seahorse flux analyzer (three independent experiments). Quantification of b basal respiration, c maximal respiration, and d ATP production. e–h Oxygen consumption evaluated in neural stem cells (NSCs) treated with 1 µM pridopidine for 24 h using the Seahorse flux analyzer (three to four independent experiments). Quantification of f basal respiration, g maximal respiration, and h ATP production. Statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001 by Kruskal-Wallis test followed by Dunn multiple comparison test. ns non-significant
Immunocytochemistry and Co-localization Analysis
MitoDsRed-transfected striatal neurons were fixed with 4% paraformaldehyde (pre-warmed at 37 °C) for 20 min, permeabilized in 0.2% Triton X-100 in PBS for 2 min and blocked for 1 h at room temperature in 3% (w/v) BSA in PBS. IP3R3 antibody (1:1000 prepared in 3% (w/v) BSA in PBS; EMD Millipore, catalog no. AB9076) and S1R antibody (1:500 prepared in 3% (w/v) BSA in PBS; Santa Cruz Biotechnology, catalog no. sc-137075) were incubated overnight at 4 °C. Neurons were incubated with 4 μg/mL Hoechst 33342 (Thermo Fisher Sci., catalog no. H1399) for 20 min and mounted using Mowiol 40-88 (Sigma-Aldrich, catalog no. 324590). Confocal images were obtained as stacks, at 0.46-µm intervals along the z axis, using a Plan-Apochromat/1.4NA 63× lens on an Axio Observer.Z1 confocal microscope (Zeiss Microscopy, Germany) with Zeiss LSM 710 software. FIJI (ImageJ, National Institute of Health, USA) was used for image analysis. Z-stack images were normalized for background (rolling ball radius of 9 for mitochondria and 25 for IP3R3), and the FindFoci function was used to identify peak intensity regions in order to extract mitochondria-specific fluorescence [27]. To optimally resolve individual mitochondria, a threshold was applied followed by the Analyze Particles function to trace mitochondrial outlines. Mitochondria aspect ratio (the ratio between the major and minor axes of mitochondria) was used as an index of mitochondria length. For S1R and IP3R3 fluorescence, a threshold was set similarly to the one described above, and Integrated Density was obtained. S1R and IP3R3 Integrated Density was calculated inside of mitochondrial ROI selection to obtain co-localization with mitochondria.
Transmission Electron Microscopy and Analysis
Striatal neurons were washed and fixed in 2.5% (V/V) glutaraldehyde in 0.1 M phosphate buffer and pellet before sectioning. The ultrathin sections were prepared using Leica Ultracut UCT (Leica, Vienna, Austria) and contrasted with uranyl acetate and lead citrate. Sections were observed in a Tecnai 12 BioTWIN transmission electron microscope (FEI Company, Eindhoven, The Netherlands) at 100 kV. Digital images were taken using a magnification of 26,500× in a Veleta camera (Olympus Soft imaging Solutions, GmbH, Münster, Germany). All mitochondria of 6–7 different cells were snapped per condition and per independent experiment. Number of mitochondria-ER contacts sites (MERCS), MERCS length, ER and mitochondria profile areas, ER width, and mitochondria aspect ratio were quantified. Number of MERCS per mitochondria was obtained by dividing number of MERCS per number of mitochondria profiles. Percentage of mitochondria surface covered by ER was measured by dividing the MERCS length by mitochondria perimeter and multiplying by 100. MERCS were considered when the distance between ER and mitochondria < 40 nm.
Mitochondrial Movement Analysis
MitoDsRed-transfected striatal neurons were washed and incubated in Na+ medium (in mM: 140 NaCl, 5 KCl, 1 CaCl2, 1 MgCl2, 10 Glucose, 10 HEPES, pH 7.4) at 37 °C for mitochondrial movement studies. Neuronal projections were imaged every 5 s for a total of 145 frames, using a 63× objective with NA = 1.4, on a Carl Zeiss Axio Observed Z1 inverted confocal microscope using the CSU-X1M spinning disc technology (Zeiss, Jena, Germany). Mitochondrial movement analysis was performed using the Kymograph Macro [28] in FIJI. Briefly, histograms were matched to the first frame to correct fluorescence variations using Bleach Correction plugin developed by Miura and Rietdorf, and time lapse-dependent x-y drift was corrected by applying the TurboReg plugin. ROIs were designated using a segmented line following mitochondria trajectory across projections. Kymographs generated in a x-y dimension (distance vs time) were used to obtain the slope from which mitochondrial velocity was calculated.
Seahorse Oxygen Respirometry
Oxygen consumption rate (OCR) in WT and hemizygous YAC128 cortical/striatal co-cultures and NSCs was measured using Seahorse XFe-24/96 flux analyzers (Seahorse Bioscience, Billerica, MA, USA) following the manufacturer’s instructions. Cortical-striatal primary neurons were cultured in Seahorse XF96 V3 cell culture microplates (Agilent, catalog no. 101085-004) at a density of 20,000 cells/well. NSCs were seeded 30,000 cells/well onto an XF24 cell culture microplate (Agilent, catalog no. 102340–100) coated with Geltrex® and allowed to adhere for 24 h at 37 °C. Pridopidine (0.1, 1, and/or 5 µM) was added, when indicated in the graphs, 24 h before the experiment. The sensor cartridge plate was incubated with immersed sensors in a non-CO2 incubator at 37 °C for ~ 16 h (overnight). Prior to the experiments, cells were washed and incubated with XF assay medium (DMEM; Thermo Fisher Sci., catalog no. D5030) supplemented with glucose (20 mM for neurons; 17.5 mM for NSC), 1 mM pyruvate and 2 mM glutamine, pH = 7.4, at 37 °C. The cell medium was aspirated gently to avoid disturbing cell monolayers. Baseline measurements of OCR were sampled prior to sequential injection of mitochondrial complex V inhibitor oligomycin (1 µM; Sigma-Aldrich, catalog no. 75351), protonophore carbonyl cyanide-4-phenylhydrazone (FCCP) (0.5 µM for neurons and 0.3 µM for NSCs; Sigma-Aldrich, catalog no. C2920) and antimycin A (0.5 µM for neurons and 1 µM for NSCs; Sigma-Aldrich, catalog no. A8674) plus rotenone (0.5 µM for neurons and 1 µM for NSCs; Sigma-Aldrich, catalog no. R8875) to complete inhibit mitochondrial respiration. Accordingly, mitochondrial basal respiration, maximal respiration, and ATP production were automatically calculated and recorded by the Seahorse software. Data was normalized for protein levels.
Mitochondrial Membrane Potential
Mitochondrial membrane potential (Δψm) was assessed in cortical and striatal neurons using the positively changed fluorescent probe TMRM (tetramethylrhodamine methyl ester) (Thermo Fisher Sci., catalog no. T668) and in cortical/striatal co-cultures and lymphoblasts using an equivalent probe, TMRE (tetramethylrhodamine ethyl ester) (Abcam, catalog no. ab113852).
TMRM Assay: Cortical and striatal neurons previously treated, when indicated, with pridopidine (0.1 and 1 μM; 24 h) were incubated with 150 nM TMRM (quenching conditions) in Na+ medium for 30 min, at 37 °C. Under these conditions, retention of TMRM by mitochondria was studied to estimate changes in Δψm. Basal fluorescence (503-nm excitation and 525-nm emission) was recorded using a microplate reader Spectrofluorometer Gemini EM (Molecular Devices, USA) for 4 min, followed by the addition of 2.5 μM FCCP plus 2.5 μg/mL oligomycin to produce maximal mitochondrial depolarization and mitochondrial probe release. TMRM release was calculated based in the differences in fluorescence before and after addition of oligomycin/FCCP.
TMRE Assay: Primary neurons and lymphoblasts in suspension were cultured in 6-well plates. The cells were pretreated with/without pridopidine and hydrogen peroxide (H2O2) as per experimental condition followed by incubation with 25 nM TMRE diluted in complete cell culture media for 15 min at 37 °C. Primary neurons were detached from the plate using 0.05% trypsin-EDTA and collected in 10% FBS-containing PBS and centrifuged at 1000 rpm for 5 min. The lymphoblasts also were centrifuged at 1000 rpm for 5 min. The cell pellets were re-suspended in 1% FBS-containing PBS and subjected to FACS analysis using Fortessa Flow cytometer (BD Biosciences) with a PE filter (545 nm).
Measurement of Mitochondrial H2O2 Levels
Coverslip-plated cortical and striatal neurons were pre-treated with pridopidine (0.1 and 1 μM) for 24 h and incubated with the Mitochondria peroxy yellow 1 (MitoPY1) probe (8 μM; Tocris Bioscience, catalog no. 4428) in Na+ medium for 30 min at 37 °C. After incubation, MitoPY1 was washed out and neurons imaged at 1-min intervals for 30 min using LCI PlanNeofluar/1.3NA 63× lens on a Carl Zeiss Axio Observed Z1 inverted confocal microscope using the CSU-X1M spinning disc technology with Zen Black 2012 software (Zeiss, Jena, Germany). Fluorescence was recorded at 503-nm excitation and enhanced emission at 528 nm [29]. After basal reading for 10 min, neurons were stimulated with antimycin A (2 μM). Specific MitoPY1 fluorescence in mitochondria was confirmed after co-incubating cells with MitoTracker Deep Red (300 nM; Thermo Fisher Sci., catalog no. M22426). Fluorescence intensity at each time point was analyzed with FIJI using the time series analyzer plugin (v 3.0) (Balaji J. 2007).
NSCs were plated at 30,000 cells/well in 96-well assay plates coated with Geltex® for 24 h at 37 °C. Afterwards, NSCs were incubated for another 24 h with 1 μM pridopidine. Prior to acquisition, cells were washed with HBSS (in mM: 137.9 NaCl, 1.3 CaCl2, 0.5 MgCl2-6H2O 0.4 MgSO4-7H2O, 5.3 KCl, 0.4 KH2PO4, 4.2 NaHCO3, 0.3 Na2HPO4, 5.6 D-glucose, at pH 7.4) and incubated for 20 min with 10 μM MitoPY1 at 37 °C and 5% CO2. MitoPY1 fluorescence was obtained with a Microplate Spectrofluorometer Gemini EM (Molecular Devices, USA) using wavelength parameters described above. Basal levels were measured for 10–15 min followed by exposure to 3 μM myxothiazol (mitochondrial complex III inhibitor; Sigma-Aldrich, catalog no. T5580) and measured for an additional 30 min. The results were calculated as RFU per 30,000 cells.
In isolated mitochondria, H2O2 levels were measured using the Amplex Red-horseradish peroxidase method. Briefly, 5 µg of isolated mitochondria was resuspended in mitochondrial reaction buffer (MRB, in mM: 100 sucrose, 100 KCl, 2 KH2PO4, 5 HEPES, 0.01 EGTA, 3 succinate, 3 glutamate, 0.1 ADP-K, pH 7.4) supplemented with 10 μM Amplex Red reagent (Thermo Fisher Sci., catalog no. A12222) and freshly prepared 0.5 units/mL horseradish peroxidase (Sigma-Aldrich, catalog no. 77332). The homogenate was then dispensed into a 96-multiwell plate, and fluorescence was measured in a spectrofluorometer microplate reader by excitation at 570 nm and emission at 585 nm every 30 s, for 20 min. After 10 min of basal reading, mitochondria were challenged with antimycin A (2 µM). Results were analyzed as time-dependent changes in fluorescence.
CellROX Assay
H2O2 (9.8 M stock) was dissolved in cell culture medium prior to experimentation. Primary neurons and lymphoblasts cultured on PDL-coated plates were treated with H2O2 (0–1 mM) for up to 6 h. Oxidative stress was measured using the CellROX red reagent (Life Technologies, catalog no. C10422). Cells were treated with 5 μM CellROX red in complete medium, then incubated for 30 min. The cells were then washed with PBS and imaged on a Zeiss Axiovert inverted microscope (Zeiss, Jena, Germany) using a 10× objective. Exposure settings were maintained for all samples. Eight random fields were sampled, and the fluorescence intensity was measured using ImageJ software and normalized to the DAPI signal.
MTS Assay
Cell viability was assessed on lymphoblasts using the MTS assay, a measure of mitochondrial function, and cell survival. The assay was performed according to the manufacturer's instructions (Promega, catalog no. G1112). Control and HD lymphoblasts were treated with 5 μM pridopidine and 0.1 mM H2O2 as indicated in figure legends. In the day of experiment, MTS reagent was added to the culture media (1:10) and incubated at 37 °C for 4 h. Then, lymphoblasts were centrifuged at 145×g for 5 min and media was collected. The absorbance of the formazan dye produced by viable cells was quantified in the medium at OD = 490 nm.
Immunoblotting
Cells were homogenized in RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 0.5% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, 1 mM EDTA, 2 mM sodium orthovanadate, 50 mM sodium fluoride, 1 mM PMSF, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). The cell lysates were transferred to microtubes and incubated for 30 min on ice, then centrifuged at 20,800×g for 15 min at 4 °C. Proteins (25–50 μg cell lysate) were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride membrane, which was then blocked for 1 h at room temperature in Odyssey blocking buffer diluted 1:1 with PBS. The membranes were probed overnight at 4 °C with the relevant primary antibodies followed by three washes with 0.1% Tween in PBS (PBST), and re-probing with appropriate fluorescently tagged secondary antibodies. After three washes with PBST, the membranes were scanned and quantified using an Odyssey fluorescent scanner (LICOR Biosciences).
Antibodies used for immunoblotting analysis include anti-rabbit Nrf2 (Cell signal tech, #12721, 1:1000), anti-mouse S1R (Santa Cruz Biotech, SC-137075, 1:500), and anti-mouse actin (Sigma-Aldrich, A5441, 1:20,000).
Gene Expression Analysis by qRT-PCR
RNA extraction and qRT-PCR procedures were performed as previously described [30]. Briefly, RNA was extracted from cultured lymphoblasts using PureLink mini RNA extraction kit (Thermo Fisher Sci., catalog no. 12183018A). RNA (500 ng) was reverse transcribed using SuperScript III Reverse Transcriptase (Thermo Fisher Sci., catalog no. 18080093) according to manufacturer’s instructions. The qPCR was performed using the ABI Prism 7500 Sequence Detection System (Applied Biosystems). Each sample was run in triplicate. Gene expression was normalized to Rpl13a or UBC mRNA.
In Vivo Study Design
About 1.5-month-old WT and hemizygous YAC128 mice (males and females in equal proportion) were divided into four groups. One WT mice group and one YAC128 mice group received pridopidine treatment (30 mg/kg dissolved in a volume of 100 µL/25 g), whereas the two remaining groups received an equivalent volume of vehicle (sterile water) by oral gavage for 45 consecutive days, until 3 months of age. Mice were divided 4 animals per cage enriched in corn-husk nesting material and paper rolls, each cage representing one individual experiment, for a total of 9 animals per group. Pridopidine was dissolved in deionized sterile water. Animals were weighed every week and the volume of treatment adjusted accordingly. Mice were behaviorally tested in rotarod immediately before commencing treatment and on the day before finishing treatment. Tests were conducted blindly at a set time during the day. Mice were sacrificed 24 h after administration of the final pridopidine dose, and mitochondria isolated from the striatum.
Rotarod Analysis
Motor learning and coordination were assessed on a rotarod apparatus (Letica Scientific Instruments, Panlab, Barcelona, Spain). Mice were allowed to adapt to the behavior room for 2 h before the behavior studies. Procedures were consistent for all subjects and tests made at minimal noise levels. The training phase consisted of four trails per day (120 s each) at 1 h intervals, at a fixed speed of 14 rpm. In this test, mice must learn to run when placed on a constant rotating rod to prevent them from falling. Once the task is learned, the accelerating rotarod can be used to assess motor coordination and balance. The testing phase was carried out the following day on an accelerating rotarod from 4 to 40 rpm over 5 min and consisted of 3 trials, spaced 2 h apart. Rotarod scores are the average of 3 trials. Experiments were performed blinded for genotype and treatment.
Isolation of Functional Mitochondria
The striatum was dissected from mouse brains washed once in ice-cold mitochondria isolation buffer (MIB: in mM, 225 mannitol, 75 sucrose, 1 EGTA, 5 HEPES–KOH, pH 7.2). Striatal mitochondria were isolated using discontinuous Percoll density gradient centrifugation as previously described [31]. Briefly, striata were transferred to a 7-mL Dounce tissue grinder (Kontes Glass Co., Vineland, NJ, USA) and homogenized 8 times in 0.8 mL MIB supplemented with 1 mg/mL fatty acid free BSA using a pestle with 0.07–0.12-mm clearance, followed by another 8 strokes with a pestle with 0.02–0.056-mm clearance. The final homogenate was then centrifuged at 1100×g for 2 min, at 4 °C. The supernatant was collected and mixed with freshy made 80% Percoll (GE Healthcare, catalog no. 17-5445-02) prepared in mitochondrial dilution buffer (MDB: in mM, 1000 sucrose, 50 HEPES–KOH, 10 EGTA, pH 7.0), to create a 9.5% Percoll solution, which was further carefully layered on the top of freshly made 10% Percoll (diluted from 80% Percoll in MIB). The mitochondrial fraction was pelleted by centrifugation at 18,500×g for 10 min at 4 °C. The pellet was then resuspended in 1 mL of mitochondria washing buffer (MWB: in mM, 250 sucrose, 5 HEPES–KOH, 0.1 EGTA, pH 7.2) and centrifuged at 10,000×g for 5 min at 4 °C. Mitochondrial pellet was again resuspended in a small volume of ice-cold MWB, to create a concentrated mitochondria solution, and kept on ice for further analysis for a maximum of 2 h. Protein content of isolated mitochondria was quantified by Bio-Rad assay (Bio-Rad, catalog no. 5000006).
Mitochondrial Complex Activity
Five microgram of isolated mitochondria diluted in mitochondrial assay solution (MAS: in mM, 70 sucrose, 220 mannitol, 10 K2HPO4, 5 MgCl2, 1 EGTA, 2 HEPES–KOH) supplemented with 0.2% (w/v) fatty acid-free BSA, 10 mM pyruvate, 2 mM malate, and 4 µM FCCP were seeded in poly(ethyleneimine)-coated (1:15,000; Sigma-Aldrich, catalog no. 03880) XF24 seahorse plates by centrifugation at 2000×g for 18 min, at 4 °C [31, 32]. After centrifugation, the volume was adjusted to 450 µL and the Seahorse plate was equilibrated in a humidified CO2-free incubator at 37 °C for 10–12 min. Sequential electron flow through of the electron transport chain was evaluated after sequential injection of rotenone (2 μM; complex I inhibitor), succinate (10 mM; complex II substrate), antimycin A (4 μM; complex III inhibitor), and ascorbate/TMPD (N,N,N′,N′-tetramethyl p-phenylenediamine) (10 mM/100 μM; electron donors to cytochrome C/complex IV) [31, 32].
Statistical Analysis
Results are expressed as mean ± SEM (standard error of the mean) of the number of independent experiments or animals indicated in figure legends. Comparisons between multiple groups were performed by non-parametric one-way analysis of variance (ANOVA) using the Kruskal-Wallis test followed by Dunn multiple comparison test. Correction for multiple comparisons was done by two-way ANOVA followed by Tukey’s post hoc test. Comparison between two groups was performed by non-parametric Mann-Whitney test. The F-test was performed to analyze the interaction term. Outliers were detected using ROUT method (Q = 1%). Significance was accepted at p < 0.05. All analyses were performed using Prism software (GraphPad Version 8.0).
Results
Pridopidine Promotes Mitochondria-ER Tethering and Stimulates Mitochondrial Dynamics and Respiration in HD Neuronal Models
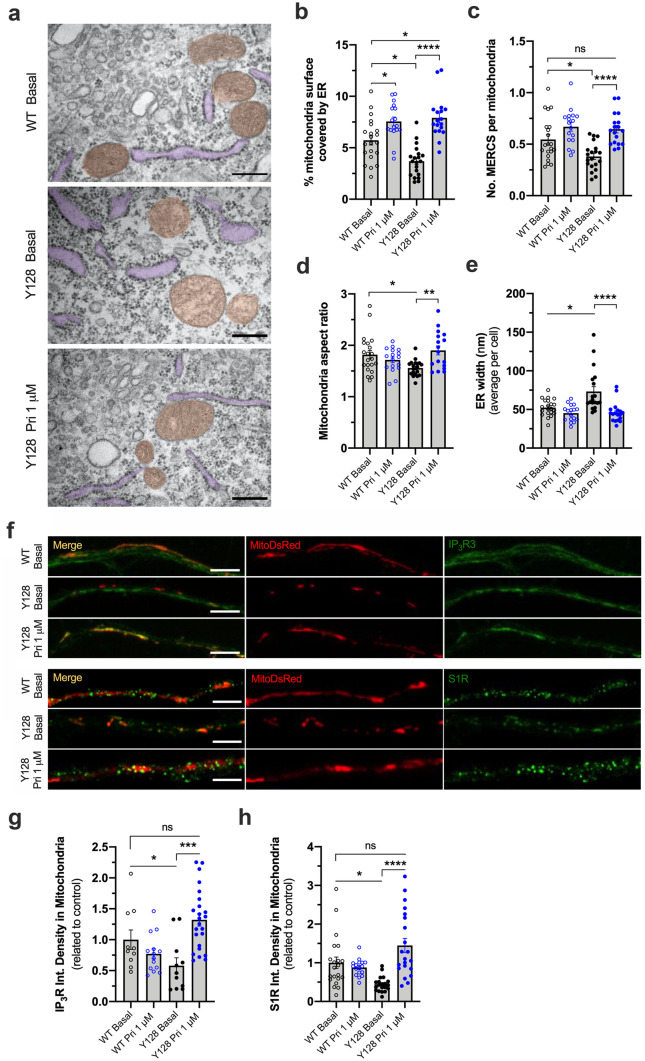
Mitochondria-ER tethering controls the amount of Ca2+ that is buffered into mitochondria, impacting the activity of Krebs cycle enzymes, mitochondrial respiration, and ROS production. These contacts are disrupted in several neurodegenerative disorders [33]. Using electron micrographs, we observe that YAC128 mouse striatal neurons show a decreased number of mitochondria-ER contact sites (MERCS) per mitochondria (p = 0.0274). The percent of mitochondrial surface covered by ER is also reduced compared with WT neurons (p = 0.0141) (Fig. 1a–c). Concordantly, co-localization of IP3R3 and S1R with mitochondria is reduced by ~ 40% in HD neurons (p = 0.049 for IP3R; p = 0.0008 for S1R) (Fig. 1f–h). An abnormally enlarged structure of the ER observed in YAC128 neurons (p = 0.0329) may account for these alterations in this signaling platform (Fig. 1a, e).
Fig. 1.
Pridopidine increases mitochondria-ER coupling and co-localization of IP3R and S1R with mitochondria in YAC128 neurons. a–e Electron micrographs were obtained from striatal neurons treated for 24 h with 1 μM pridopidine and b the % of mitochondria surface in contact with ER, c the number of MERCS per mitochondria, d the mitochondria aspect ratio, and e the ER width were quantified. In a, mitochondria and ER are highlighted in orange and purple, respectively. Each dot represents a measurement of a single cell from 3 independent primary cultures. Scale bar = 300 nm. f–h Representative images of YAC128 and WT striatal neurons treated for 24 h with 1 µM pridopidine and transfected with MitoDsRed plasmid. IP3R3 and S1R were labeled using specific antibodies and their co-localization with mitochondria was analyzed. Each data point represents an individual neurite analyzed from three independent cultures. Statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by Kruskal Wallis test followed by Dunn multiple comparison test. ns non-significant
Mitochondrial morphology is also impaired in HD. YAC128 neurons show a reduced aspect ratio (p = 0.0343) (Fig. 1d; supplementary Fig. 1A), indicating a higher proportion of fragmented mitochondria as a result of increased fission. No alterations in ER and mitochondria total area were observed (supplementary Fig. S1B, C).
Pridopidine treatment (1 μM) restores mitochondria-ER connectivity in YAC128 striatal neurons by increasing the number of MERCS per mitochondria and the percent of mitochondrial surface in contact with ER by 4.2% (p < 0.0001). Pridopidine additionally increases the proportion of elongated mitochondria in YAC128 striatal neurons to WT levels (p = 0.0044) (Fig. 1a–d; supplementary Fig. 1A). The increase in IP3R and S1R co-localization with mitochondria prompted by pridopidine treatment in YAC128 neurons (p = 0.0001 for IP3R; p < 0.0001 for S1R) further influences the reestablishment of MAM platform in this HD model (Fig. 1f–h). Interestingly, total S1R levels are reduced in YAC128 striatal neurons, and pridopidine treatment restores S1R levels to WT levels (supplementary Fig. S2A-C).
Furthermore, pridopidine restores the abnormal ER width observed in YAC128 neurons, leading to a more organized ER morphology (p < 0.0001) (Fig. 1e). In WT neurons, pridopidine also significantly increases the percent of mitochondrial surface in contact with ER by 1.8% (p = 0.0339) (Fig. 1b).
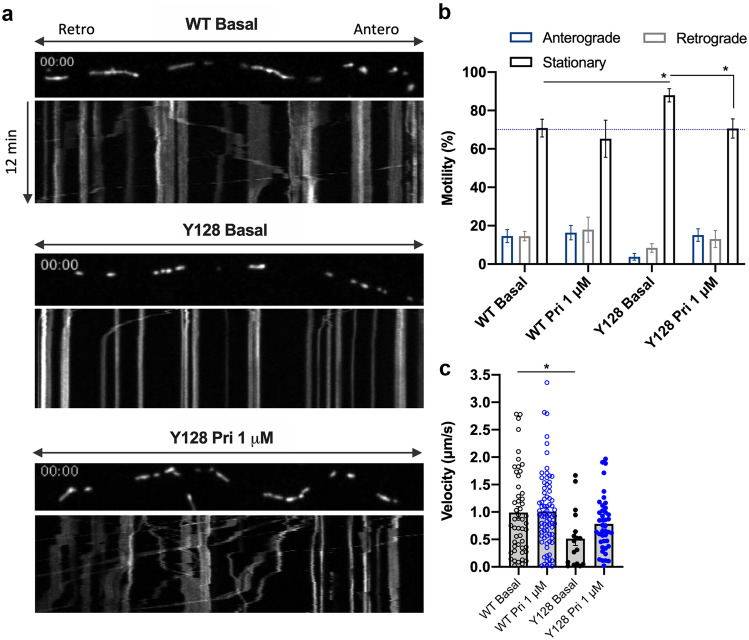
Mitochondrial fission is a means to enable mitochondria transport along the axon without affecting trafficking [34]. Therefore, we explored the possibility that the increased mitochondrial fission in HD neurons is a compensatory mechanism to improve mitochondrial trafficking. We observed that in YAC128 striatal neurons, mitochondrial anterograde transport is almost undetected, with approximately 90% of mitochondria in stationary phase (p = 0.0169) (Fig. 2a, b; supplementary video 1, 2). Moreover, motile mitochondria in YAC128 neurons move at half the velocity of wild-type neurons (p = 0.0218). Pridopidine treatment (1 μM) significantly increases the proportion of motile mitochondria (p = 0.0286), restablishing both anterograde and retrograde transport (Fig. 2a–b; supplementary video 3). These data indicate that pridopidine-induced mitochondria elongation does not negatively influence transport.
Fig. 2.
Pridopidine improves mitochondrial anterograde transport in YAC128 neurons. a Representative images of MitoDsRed-transfected mitochondria live tracked in spinning disk confocal with kymograph plotting distance over time. b Motility analysis of mitochondria by direction of transport. Two-way ANOVA indicates an interaction between the condition and the type of movement (F(6,87) = 3.943, p = 0.0016). c Quantification of velocity of mitochondrial trafficking from kymographs. Each data point represents the quantification of an individual kymograph obtained from individual neurites from four independent experiments. Statistical significance: *p < 0.05 by 2-way ANOVA followed by Tukey’s multiple comparison test. In c, *p < 0.05 by Kruskal-Wallis test followed by Dunn multiple comparison test
Indeed, rescue of mitochondrial dynamics by pridopidine treatment in YAC128 neurons may contribute to the enhanced mitochondrial maximal respiration (p = 0.0061 for 1 μM Pri, p = 0.002 for 5 μM Pri) and ATP production (p = 0.0358 for 5 μM Pri) (Fig. 3a–d). In WT neurons, pridopidine enhances mitochondrial basal respiration (p = 0.0058 for 1 μM Pri, p = 0.0016 for 5 μM Pri) and ATP production (p = 0.02 for 5 μM Pri) (Fig. 3b, d). Pridopidine also increases mitochondrial basal (p = 0.0043) and maximal respiration (p = 0.0177) in human iPSC-derived neural stem cells from a heterozygous HD patient (HD-NSCs) with a normal allele of 19 CAG repeats and an expanded allele retaining 72 CAG repeats (Fig. 3e–h).
Early Pridopidine Treatment Rescues Mitochondrial Function in HD Cells
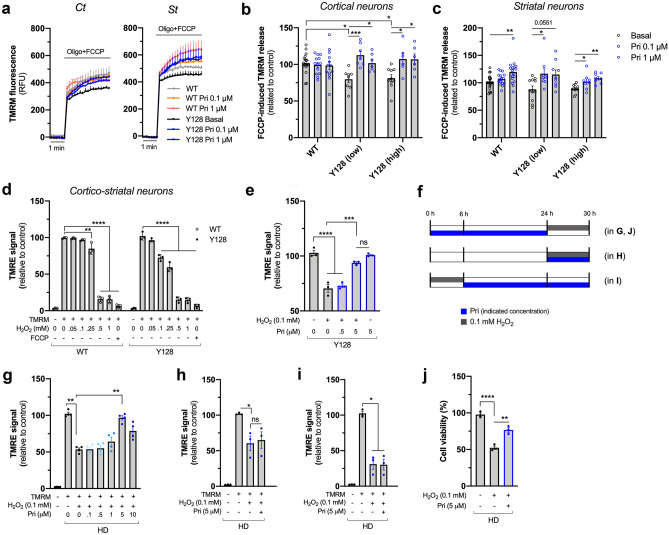
To gain further insight into the role of pridopidine in the regulation of mitochondrial function, mitochondrial membrane potential (Δψm) was evaluated as it directly affects ATP production. To this end, we employed two complementary methods: (i) tetramethylrhodamine methyl ester (TMRM) fluorescent probe was used under quenched conditions and its accumulation in mitochondria was assessed after mitochondrial depolarization with oligomycin plus FCCP and (ii) tetramethylrhodamine ethyl ester (TMRE) fluorescent probe accumulation in mitochondria was directly evaluated by flow cytometry.
Two previously described YAC128 mouse lines [35] that differ in the levels of mHTT expression, termed here as low or high YAC128, were used. As in the previous experiments, WT neurons were used as controls since ΔΨm is similar between WT and YAC transgenic mice expressing normal human full-length HTT retaining 18 glutamines (YAC18) [36]. In both lines, YAC128 cortical neurons exhibit lower mitochondrial retention of TMRM, observed by decreased fluorescence after complete mitochondrial depolarization with oligomycin and FCCP (p = 0.0193 vs Y128 low; p = 0.0222 vs Y128 high) (Fig. 4a, b), suggesting lower ΔΨm compared with wild-type cortical neurons. In striatal neurons, we observe a reduction which is not statistically significant (Fig. 4c). Remarkably, pridopidine (0.1 and 1 µM) significantly increases ΔΨm in YAC128 cortical and striatal neurons expressing both high and low levels of mHTT (YAC128 high: p = 0.015 for 0.1 µM Pri and p = 0.0201 for 1 µM Pri in Ct neurons; p = 0.0407 for 0.1 µM Pri and p = 0.0017 for 1 µM Pri in St neurons. YAC128 low: p = 0.0008 for 0.1 µM Pri, and p = 0.0303 for 1 µM Pri in Ct neurons; p = 0.0274 for 0.1 µM Pri in St neurons) (Fig. 4a–c). These results indicate that the effect of pridopidine on mitochondrial function is independent of mHTT expression levels. In WT striatal neurons, 1 µM pridopidine also increases ΔΨm (p = 0.0013) (Fig. 4c).
Fig. 4.
Early pridopidine treatment protects YAC128 neurons and HD lymphoblasts against H2O2-induced mitochondrial dysfunction. a–c Cortical (Ct, left panel) and striatal (St, right panel) WT and YAC128 neurons expressing low or high levels of mHTT were treated with pridopidine for 24 h, and TMRM was used to evaluate changes in ∆Ψm after depolarization with oligomycin plus FCCP (seven to ten independent cultures). b Quantification of TMRM release in cortical neurons. c Quantification of TMRM release in striatal neurons. d H2O2-dependent mitochondrial toxicity was established in WT and YAC128 cortical/striatal co-cultured neurons by incubating cells with increasing concentrations of H2O2, followed by staining with TMRE and analysis by flow cytometry (three independent cultures). e YAC128 neurons pre-treated with pridopidine for 24 h (at indicated concentrations) were incubated with 0.1 mM H2O2, and ∆Ψm was evaluated with TMRE by flow cytometry (three independent cultures). f Timeline of pridopidine incubations in lymphoblasts. Lymphoblasts were first treated with pridopidine for 24 h followed by an oxidative stimulus of 6 h with 0.1 mM H2O2 (in g, j); pridopidine and H2O2 incubations were performed simultaneously during 6 h before the experiments (in h); pridopidine treatment for 24 h was preceded by 0.1 mM H2O2 incubation for 6 h (in i). g HD human lymphoblasts were pre-treated with pridopidine for 24 h and then challenged with H2O2 for 6 h, and ∆Ψm was evaluated by quantifying the TMRE signal by flow cytometry. h Lymphoblasts were treated simultaneously with pridopidine and H2O2 and TMRE signal quantified. i Lymphoblasts challenged with H2O2 and then treated with pridopidine and TMRE signal quantified. j Cell viability was evaluated by the MTS assay in HD lymphoblasts treated or not with pridopidine for 24 h followed by 6 h incubation with H2O2 (three independent experiments). Statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by Kruskal-Wallis test followed by Dunn multiple comparison test. No statistical significance was observed between WT basal vs. YAC128 pridopidine treated conditions (in b and c). ns non-significant
To understand the efficacy of pridopidine treatment under a more potent cytotoxic/oxidative stimuli, we evaluated ΔΨm in response to increasing levels of hydrogen peroxide (H2O2) (Fig. 4d). Concentrations equal to or above 0.25 mM H2O2 affect mitochondrial function in wild-type neurons. YAC128 neurons are more sensitive to H2O2, showing a significant reduction already at 0.1 mM H2O2 (Fig. 4d). YAC128 neurons exposed to 0.1 mM H2O2 for 6 h show ~ 30% loss of ΔΨm (p < 0.0001). Pridopidine treatment completely reverts this H2O2-induced loss of ΔΨm (p = 0.001), showing a TMRM signal similar to YAC128 neurons unchallenged by H2O2 (comparing the two right bars) (Fig. 4e).
In order to test the protective effect of pridopidine in a human model, the previous experiment was replicated in lymphoblasts derived from HD patients, assessing multiple concentrations of pridopidine (0.1–10 µM). Three different protocols were established (Fig. 4f) to assess if pridopidine can exert a protective effect regardless of the timing of administration relative to the cytotoxic stimulus. Pridopidine was administered either 24 h before H2O2, together with H2O2 or 6 h after H2O2 challenge. The maximal protective effect of pridopidine was observed when pridopidine was administered before the cytotoxic stimulus of 0.1 mM H2O2, at a concentration of 5 μM (p = 0.0097). A higher dose of pridopidine (10 µM) is less efficacious and shows less restoration of membrane potential (p = 0.1389), indicating a bell-shaped dose response curve (Fig. 4g). Bell-shaped response curves are typical of S1R agonists and are demonstrated in numerous preclinical models [37–42]as well as in clinical trials [43–45]. The protective effect of pridopidine is only observed when pridopidine is administered prior to the cytotoxic stimuli (Fig. 4g). Simultaneous administration of pridopidine together with H2O2, or administration of pridopidine after H2O2 challenge, does not elicit the same protective effect (Fig. 4h, i). Importantly, the recovery of mitochondrial function observed when pridopidine treatment is administered prior to H2O2 stimuli is accompanied by a significant increase in cell viability of approximately 50% (p = 0.0016) (Fig. 4j).
Pridopidine Ameliorates Reactive Oxygen Species Production and Antioxidant Defense in HD Cell Models
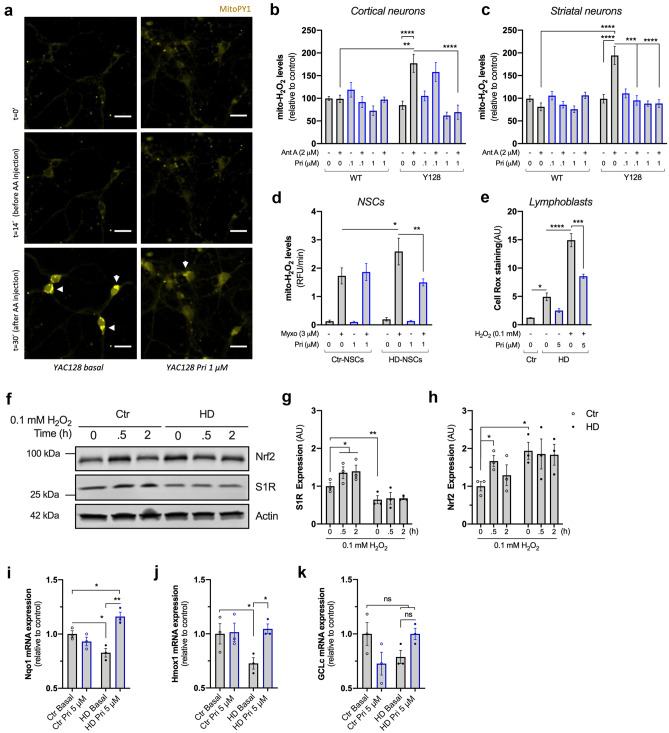
The higher susceptibility of HD mitochondria to H2O2 (Fig. 4d) may indicate that HD cells exhibit increased oxidative stress. To test this hypothesis, we assessed local flux of H2O2 with the fluorescent probe MitoPY1 that accumulates in mitochondria [29]. Antimycin A (Ant A)-stimulated cortical and striatal YAC128 neurons show a significant twofold increase in mitochondrial-driven H2O2 levels compared with WT neurons (p = 0.001 for Ct neurons; p < 0.0001 for St neurons) (Fig. 5a–c). Similarly, HD NSCs treated with the mitochondrial complex III inhibitor, myxothiazol (Myxo, 3 µM) show a large increase in mito-H2O2 levels compared with control NSCs (p = 0.0365) (Fig. 5d). HD lymphoblasts also present a threefold increase in CellROX fluorescence in response to H2O2 challenge (p = 0.0206) (Fig. 5e). Pridopidine efficiently decreases ROS levels in all three cellular HD models assessed. In both cortical (Fig. 5b) and striatal (Fig. 5c) neurons, pridopidine reverts the increased levels of mito-H2O2 prompted by Ant A (4B; p < 0.0001) (4C; p = 0.0005 for 0.1 µM Pri, p < 0.0001 for 1 µM Pri). In HD-NSCs, 1 µM pridopidine reduces (~ 42%) the observed increase in H2O2 levels induced by complex III inhibition (p = 0.0075) (Fig. 5d). In HD lymphoblasts, pridopidine treatment (5 µM) decreases ROS levels by 57% under H2O2-challenged conditions (p = 0.0001) (Fig. 5e).
Fig. 5.
Pridopidine reverts oxidative challenge-induced ROS production and impaired expression of Nrf2/ARE genes in HD cells. a Representative image of cortical and striatal YAC128 neurons treated or not with 1 μM pridopidine, and incubated with MitoPY1 fluorescence probe. Mitochondrial H2O2 levels were recorded before and after Antimycin A (Ant A, 2 μM), as indicated (considering ~ 20 cells/condition for St neurons and ~ 10 cells/condition for Ct neurons from four independent cultures). Scale bar = 30 μM. b Quantification of mitochondrial H2O2 levels in cortical neurons. c Quantification of mitochondrial H2O2 levels in striatal neurons. d Quantification of mitochondrial H2O2 in human NSCs treated with 1 μM pridopidine for 24 h in the presence or absence of myxothiazol (Myxo, 3 μM) as indicated in the graph (four independent experiments). e Quantification of ROS levels by CellROX in lymphoblasts treated with 0.1 mM H2O2 for 6 h followed by 5 μM pridopidine treatment for 24 h, where indicated (four independent experiments). f–h Control and HD lymphoblasts were challenged with 0.1 mM H2O2 for 0, 0.5, and 2 h and the total protein levels of S1R and Nrf2 were evaluated by western blotting. Representative images of the blots are depicted in f. Quantification of g S1R and h Nrf2 protein levels. Two-way ANOVA indicated in effect of genotype for both Nrf2 (F(1,12) = 6.987, p = 0.0214) and S1R (F(1,12) = 31.59, p < 0.001) expression. Data obtained from three independent experiments. i–k Control and HD lymphoblasts were treated with pridopidine (5 μM, 24 h), and expression of antioxidant genes Nqo1 (i), Hmox1 (j), and GCLc (k) were evaluated by qPCR (three independent experiments). Statistical significance: *p < 0.05, **p < 0.01, ****p < 0.0001 by 2-way ANOVA followed by Tukey’s multiple comparison test. In e, i–k, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by Kruskal-Wallis test followed by Dunn multiple comparison test. ns non-significant
Recent evidence indicates that the S1R modulates oxidative stress through activation of nuclear factor erythroid 2-related factor 2 (Nrf2) signaling and the downstream antioxidant response element (ARE) genes [46]. In WT lymphoblasts, both S1R and Nrf2 protein levels increase in response to 0.1 mM H2O2 challenge (p = 0.0256) (Fig. 5f–h). Interestingly, HD lymphoblasts exhibit lower levels of S1R and higher levels of Nrf2 under basal conditions (p = 0.0106 and p = 0.022, respectively) (Fig. 5f–h). An increase in Nrf2 levels suggests a compensatory mechanism in response to the observed decreased levels of S1R [47]. Reduced expression of S1R was previously described in other neurodegenerative disorders in which oxidative stress is involved [48, 49], strengthening the potential protective effect of pridopidine by activating the S1R.
HD lymphoblasts fail to respond to the oxidative stimulus showing no change in the expression of S1R and Nrf2 (Fig. 5f–h). No increase in Nrf2 suggests impaired expression of the downstream ARE genes and an impaired stress response in HD cells, further explaining the observed increase in ROS production (Fig. 5e). Therefore, we evaluated the expression of three Nrf2-ARE pathway-regulated genes: NAD(P)H dehydrogenase [quinone] 1 (Nqo1), heme oxygenase 1 (Hmox1), and glutamate-cysteine ligase catalytic subunit (GCLc) [50] (Fig. 5i–k). Two of the analyzed genes show significantly reduced mRNA expression in HD lymphoblasts (p = 0.0372 for Nqo1 and p = 0.0463 for Hmox1) and a tendency for reduced mRNA levels of GCLc (p = 0.0918). Pridopidine treatment upregulates the expression of Nqo1 (p = 0.0010) and Hmox1 (p = 0.0308) (Fig. 5i–j), further supporting pridopidine’s role in regulating oxidative stress.
The S1R Mediates the Beneficial Effects of Pridopidine on HD Mitochondria
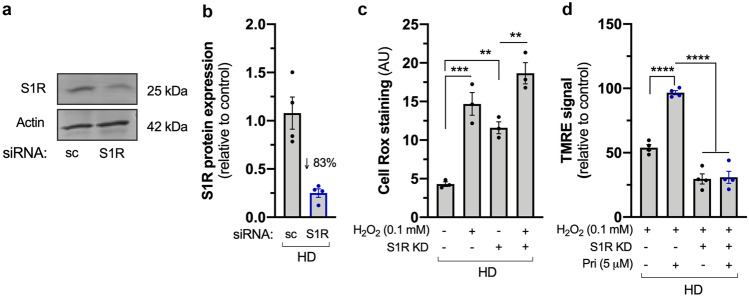
To further confirm that the effects of pridopidine are mediated by the S1R, we silenced the S1R in HD lymphoblasts, achieving approximately 83% reduction in protein levels (Fig. 6a, b). ΔΨm and oxidative stress were evaluated as before. S1R loss is sufficient to increase ROS levels 2.7-fold in HD lymphoblasts (p = 0.0063) (Fig. 6c), similar to the observed effect induced by H2O2 (p = 0.0005). Hence, the S1R plays an important role in the oxidative signaling response. Remarkably, the protective effect of pridopidine treatment on mitochondrial membrane potential upon H2O2 challenge is abolished in the absence of the S1R (p < 0.0001) (Fig. 6d), indicating that pridopidine effects on mitochondrial functions are S1R-dependent.
Fig. 6.
Pridopidine effects on mitochondrial function are mediated by the S1R. a Representative image and b quantification of western blot confirming S1R knockdown (~ 83%) in HD lymphoblasts. c Quantification of CellROX staining in HD lymphoblasts subjected to S1R siRNA and challenged with H2O2 (0.1 mM; 6 h) when indicated (n = 3). d Quantification of Δψm by TMRE signal in control and S1R-silenced HD lymphoblasts treated or not with H2O2 and pridopidine (5 μM, 24 h, n = 4). Statistical significance: **p < 0.01, ***p < 0.001, ****p < 0.0001 by Kruskal–Wallis test followed by Dunn multiple comparison test
Early Pridopidine Treatment in YAC128 Mice Delays Onset of Motor Deficits and Regularizes Mitochondrial Abnormalities
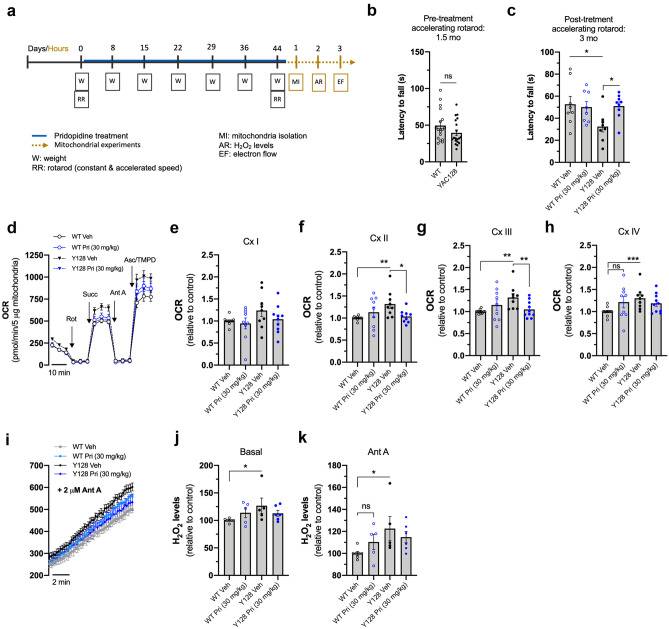
The encouraging findings in vitro led us to assess the neuroprotective effect of pridopidine in vivo. WT and YAC128 mice at 1.5 months of age (pre-symptomatic) were treated with either 30 mg/kg/day pridopidine or vehicle (sterile water) by oral gavage for 45 days (until 3 months of age; early symptomatic) (Fig. 7a). YAC128 mice exhibit motor deficits at 3 months of age in the rotarod performance test [51]; therefore, this test was applied before and after pridopidine treatment. Motor coordination score was measured after training as previously described [51], and the latency to fall was quantified in an accelerated rotarod from 5 to 40 rpm over 5 min. As expected, at 1.5 months (pre-treated) YAC128 mice display the same weight and motor coordination as wild-type mice (Fig. 7b; supplementary Fig. S3). At 3 months of age, vehicle-treated YAC128 mice show motor deficits compared with vehicle-treated wild-type mice, as observed by a reduced latency to fall during the accelerated rotarod test (p = 0.0471) (Fig. 7c). Conversely, HD mice treated with pridopidine exhibit a significant motor improvement in the accelerated rotarod, compared with vehicle-treated HD mice (p = 0.0270) (Fig. 7c), reaching a latency to fall similar to that of WT mice. This finding is in line with previous data showing pridopidine treatment improves motor function and motor learning [52].
Fig. 7.
Early pridopidine treatment normalizes mitochondrial complex activity and H2O2 production in isolated YAC128 striatal mitochondria. a Schematic representation of the in vivo/ex vivo experimental design: mice were treated with vehicle or pridopidine (blue line) for 45 days anticipated and followed by rotarod test (R); animal’s weight (W) was measured once a week during the time of treatment and pridopidine concentration adjusted. 24 h after treatment conclusion, striatum was dissected, and mitochondria isolated for functional analyses. Rotarod motor test was performed before (b) and after (c) the treatment and consisted of three trails on an accelerating rotarod from 5 to 40 rpm over 5 min; scores were averaged. (d) Electron flow measurement in striatal mitochondria isolated from vehicle- or pridopidine-treated WT and YAC128 mice using a Seahorse apparatus. Mitochondrial complex inhibitors and substrates, 2 μM rotenone, 10 mM succinate, 4 μM antimycin A, and 1 mM ascorbate/100 mM TMPD, were sequentially injected to calculate mitochondrial complex I (e), complex II (f), complex III (g), and complex IV (h) activities, respectively. i Mitochondrial levels of H2O2 were measured using Amplex Red probe, and the fluorescence was evaluated for 10 min before (j) and after (k) inhibition of mitochondrial complex III with antimycin A (2 μM). In i, XY lines show time-dependent changes in fluorescence after adding Ant A. Statistical significance: *p < 0.05, **p < 0.01, ***p < 0.001 by non-parametric Kruskal-Wallis test followed by Dunn multiple comparison test. ns non-significant
After behavioral analysis, functional mitochondria were isolated from the mice striatum. Mitochondrial complex activities in isolated mitochondria were evaluated by measuring the sequential electron flow through the electron transport chain using the Seahorse flux analyzer [31]. Sequential injections of rotenone, succinate, antimycin A, and ascorbate/TMPD induced individual stimulation or inhibition of mitochondrial complexes I, II, III, and IV, respectively, allowing the calculation of their activities (Fig. 7d–h). Striatal mitochondria from vehicle-treated YAC128 mice show significantly higher activity of complexes II (p = 0.0016) (Fig. 7f), III (p = 0.0021) (Fig. 7g), and IV (p = 0.0010) (Fig. 7h) compared with vehicle-treated wild-type mice, suggesting an early compensatory mechanism.
Mechanisms to increase mitochondrial complex activity can benefit proton leak and consequently regulate ROS production. Indeed, increased complex activities in YAC128 HD mouse striatal cells are accompanied by increased mitochondrial H2O2 production, as measured by resorfin fluorescence (Fig. 7i–k) before (Fig. 7j) and after (Fig. 7k) inhibition of complex III with antimycin A (p = 0.0276 in j; p = 0.0219 in k). These results further strengthen our in vitro data (Fig. 5), suggesting that abnormal mitochondrial ROS production along with abnormal complex activity may underlie the HD mitochondrial phenotype. In vivo pridopidine treatment in YAC128 mice normalizes the activity of mitochondrial complexes II and III (p = 0.0306 and p = 0.0084, respectively) (Fig. 7f, g). Pridopidine treatment also hinders the observed increase of H2O2 in HD mitochondria (Fig. 7i–k). Pridopidine treatment did not affect mitochondrial complex activities and H2O2 levels in wild-type mice.
Together, these data demonstrate neuroprotective effects of pridopidine in vivo in HD mice and suggests that regulation of oxidative stress is a key factor in the beneficial effect of pridopidine on mitochondrial function.
Discussion
The S1R is a Ca2+-sensitive ligand-operated receptor chaperone located at the MAM, regulating ER-mitochondria Ca2+ shuttling and cell survival [17]. Pridopidine, a high affinity and potent S1R agonist, demonstrates neuroprotective effects in numerous models of HD and other neurodegenerative disorders. We have evaluated the effect of pridopidine at 0.1–10 μM on mitochondrial function and oxidative status in HD models. This concentration range was chosen based on previous studies showing a beneficial effect of pridopidine at 0.1 and 1 μM, in a S1R-dependent manner [8, 10, 53]. At high concentrations (i.e., 10 μM), pridopidine is expected to show a bell-shaped response, a known feature of S1R agonists [54, 55].
In this study, we show that pridopidine improves mitochondria-ER interaction in HD primary neurons, further enhancing mitochondrial respiration and ATP production, mechanisms highly regulated by Ca2+ influx through the mitochondrial Ca2+ uniporter [56] (Fig. 8). This increase in bioenergetics also supports mitochondrial anterograde transport in neurons. The effect of pridopidine is not confined to the central nervous system or in vitro systems. Indeed, pridopidine effectively reduces ROS levels via the S1R in HD neural cells, human lymphoblasts, and in adult YAC128 mouse mitochondria (Fig. 8). Moreover, we show that in YAC128 HD mice, pridopidine treatment restores abnormal mitochondrial complex activity and H2O2 production and maintains motor capabilities in vivo.
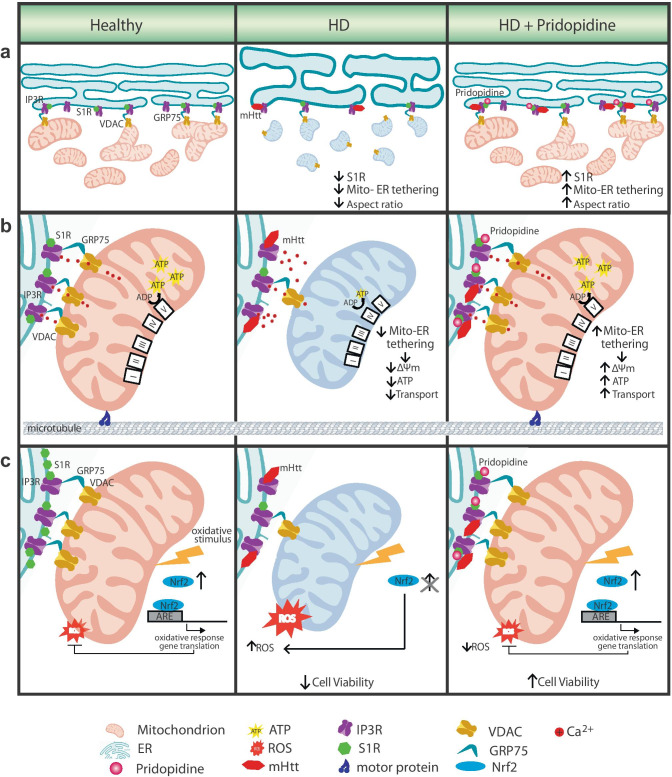
Fig. 8.
S1R agonist pridopidine restores multiple mitochondrial processes disrupted in HD cells. a Mitochondria-ER tethering is essential for mitochondrial health. In healthy cells, mitochondria-ER tethering is facilitated via S1R stabilization of the tethering complex consisting of IP3R, GRP75 and VDAC [16]. These contacts are also important to maintain the balance between mitochondrial fission and fusion (left). In HD cells, mHTT binds to the IP3R at the ER membrane [75], which may disrupt these contacts and lead to a reduction in mitochondria-ER tethering, and influence mitochondrial fission (fragmented, round mitochondria). ER also presents a swollen structure (center). Pridopidine activation of the S1R increases mitochondria-ER tethering and restores mitochondrial and ER morphology (right). b Mitochondria-ER tethering regulates Ca2+ flux to the mitochondria via the IP3R (left) [76]. As a result of the disruption in mitochondria and ER communication in HD cells, mitochondria bioenergetics is compromised, as observed by decreases in Δψm and ATP production (center); concomitantly, a reduction in bidirectional mitochondrial transport is observed. Pridopidine treatment increases mitochondrial membrane potential, ATP production, and mitochondrial trafficking (right). c In healthy cells oxidative insult (i.e. H2O2) enhances Nrf2 levels, leading to upregulation of ARE genes as a defense mechanism (left). This response, necessary for cell survival, is abrogated in HD cells (center). Pridopidine treatment restores the Nrf2-ARE response to oxidative stress and rescues cell viability (right).
Pridopidine Effect on Mitochondria-ER Crosstalk: Linking Bioenergetics and Movement
The role of MERCS is gaining interest in neurobiology research as it constitutes key signaling hubs that regulate several mechanisms implicated in neurodegeneration. Interestingly, mitochondria-ER coupling dynamics vary substantially in neurodegenerative disorders. Abnormally increased coupling is described in models of familial and sporadic forms of Alzheimer´s disease [57–59] as well as in demented patients with high ventricular levels of Aβ42 [60]. Reduced mitochondria-ER coupling is observed in Parkinson’s disease models with mutant α-synuclein and was also recently described in a HD transgenic model expressing exon 1 of the HTT gene carrying 115 CAG repeats [61, 62]. In all cases, Ca2+ exchange between the two organelles is affected, disrupting mitochondrial ATP production.
Pharmacological treatments targeted at reinforcing ER-mitochondria crosstalk prevent axonal degeneration [49] and rescue locomotor deficits in HD models [63]. A functional interaction between mitochondria and ER also stimulates neuronal plasticity and enhances dendritic spine density [64]. The role of the S1R in maintaining the integrity and functionality of MERCS is evident, as inhibition or loss of S1R function reduces contacts between mitochondria and ER in motor neurons both in vitro and in vivo [16]. This dysfunctional interplay is a potential cause for impaired bioenergetics and mitochondrial transport towards the synapse. S1R chaperones the IP3R at these sites [17], and its mis-localization from the MAM causes an overall decrease in O2 consumption [65].
Impaired ER-mitochondria connectivity extensively disturbs mitochondrial respiration and ATP production [33]. In our HD YAC128 neurons, we observe a significant reduction in the mitochondrial-ER connectivity. Moreover, we detected abnormal ER and mitochondrial morphology in YAC128 neurons. In fact, inhibition of wild-type HTT expression drastically alters the structure of the ER network [66], but whether the distorted ER network can influence MERCS remains unknown. We also observe a reduction in the total levels of S1R in HD neurons and lymphoblasts, as well as its co-localization with mitochondria, further supporting the role of the S1R in the regulation of MERCS (Fig. 8a). Pridopidine treatment significantly restores the mitochondria-ER juxtaposition, as well as mitochondrial and ER structure in YAC128 HD neurons, to WT levels. Pridopidine restoration of mitochondrial-ER coupling may contribute to the previously reported protective effects of pridopidine on synaptic plasticity [14] and spine density in HD models [10], and for increases in mitochondrial bioenergetics and cell survival demonstrated in this study (Fig. 8b). This effect is mediated via activation of the S1R as deletion of S1R expression abolishes the protective effects of pridopidine.
Impaired mitochondria-ER connectivity also influences mitochondrial transport. S1R deficiency disrupts mitochondrial axonal transport [16], which is in accordance with our data, while activation of the S1R enhances mitochondrial movement [67]. Remarkably, after pridopidine treatment we observe a significant decrease in the number of stationary mitochondria in YAC128, that were equally redirected to anterograde and retrograde transport, similar to what was observed in non-treated WT neurons. Similar results are described with another S1R agonist, the (+)SKF-10047, in primary neurons derived from 3xTg-Alzhemer’s disease and non-Tg mice [67]. Pridopidine also enhances mitochondria velocity in HD YAC128 neurons. In SOD1G93A co-cultures, a model of ALS pathology that shows disrupted ER-mitochondria connectivity [68], pridopidine rescues mitochondrial axonal transport, mediated by S1R. Although it is not yet elucidated how MERCS affect mitochondrial movement and velocity, recent proteomic reports show that Miro1, a Ca2+ sensor GTPase that regulates mitochondrial transport, is present in MAMs and may regulate these contacts [69, 70].
Pridopidine Mediates Regulation of Redox Status and Cell Survival Through S1R
Increased production of ROS and changes in redox homeostasis are major contributors to neuronal death. Mitochondria are the primary source of ROS since about 1% of O2 consumed generates superoxide anion due to the leakage of electrons at complexes I and III. Our group has previously shown that striatal cells expressing mHTT exhibit enhanced mitochondrial ROS levels, which was related to altered activities of antioxidant defense systems and decreased response to oxidant stressors due to impaired Nrf2-ARE transcriptional pathway [71].
The role of the S1R in the regulation of ROS production is evident by the observation that genetic deletion of the S1R, or its pharmacological inhibition, leads to increased ROS levels. On the other hand, S1R overexpression decreases ROS levels via the activation of Nrf2-ARE regulated genes (Nqo1, Hmox1, and GCLc) [72]. Similar data is obtained in this study using different HD models. YAC128 primary neurons, human HD lymphoblasts, and human HD NSCs all show increased susceptibility to oxidative stimulus as measured by high ROS production. Additionally, mitochondria isolated from YAC128 neurons also show increased ROS production after complex III inhibition, which may be related to increased activity of mitochondrial complexes. In fact, such an increase in complexes activity can be an early compensatory event to avoid ATP depletion. At later stages, no changes in complexes activities or respiratory activity are described for this model [73].
In HD lymphoblasts, the abnormal increase in ROS production is associated with low levels of S1R and an impaired response to oxidative stress. Following H2O2 exposure, WT lymphoblasts show an increase in Nrf2 as a defense mechanism. This defense mechanism is impaired in HD lymphoblasts as evident by their inability to increase Nrf2 levels and an improper response to the oxidative stress. Cells’ inability to increase Nrf2 hampers their downstream protective transcriptional increase of ARE-regulated genes, leading to cell death. Similar increases in ROS levels are observed in S1R knockout hepatocytes and in COS-7 cells treated with the S1R antagonists haloperidol, BD 1047, and BD 1063. On the other hand, S1R overexpression decreases the levels of ROS two-fold, via activation of the Nrf2-ARE-regulated genes [72]. Pridopidine treatment significantly enhances the transcription of Nrf2-ARE-antioxidant defense genes, further reducing ROS levels and increasing cell survival. This effect is mediated by activation of the S1R as genetic deletion of the S1R in HD lymphoblasts abolishes the protective effect of pridopidine (Fig. 8c).
The observed multifaceted rescue of mitochondrial function by pridopidine at the cellular level translates to a beneficial effect in vivo. Treatment of pre-symptomatic YAC128 HD mice with pridopidine significantly improves their performance in the rotarod test, implying a delay in the onset of motor symptoms. This improved performance is likely to result from maintenance of neuronal function.
An important observation from this study is the effect of timing: pridopidine was most efficacious for restoring Δψm when administered prior to oxidative challenge. This observation is additionally enforced by the maintenance of motor function in HD mice when they were treated at early, pre-symptomatic stages with pridopidine. These data highlight the potential utility of pridopidine administration at early stages of HD and correlate with observations from the PRIDE-HD clinical trial in which pridopidine was most efficacious in maintaining functional capacity in early HD patients [74].
Conclusions
Modulation of S1R function has significant therapeutic potential due to its involvement in mitochondria-ER tethering and multiple downstream effects (Fig. 8a, b). In this study, we show that S1R activation by pridopidine rescues multiple mitochondrial functions, potentially contributing to the overall neuroprotective effect of pridopidine observed in preclinical and clinical studies. The cellular deficits addressed in this study are early events in the pathophysiology of HD and are consistent with the concept of early treatment aimed at achieving maximal beneficial effects.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors wish to acknowledge George Q. Daley (Harvard Medical School, Boston, MA, USA) and Luís Pereira de Almeida (Center for Neuroscience and Cell Biology, University of Coimbra, Coimbra, Portugal) for providing the HD and control iPSCs, respectively, from which NSCs were generated. We also thank I. Luísa Ferreira (Center for Neuroscience and Cell Biology, University of Coimbra, Coimbra, Portugal) for setting up the studies in isolated mitochondria from mouse brain using the Seahorse apparatus. The authors also acknowledge Lars Haag and Eva Idsund Jonsson from Electron Microscopy Unit (EMil), at Karolinska Institutet (KI), Stockholm, for processing TEM samples.
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Abbreviations
- S1R
Sigma-1 receptor
- HD
Huntington disease
- ER
Endoplasmic reticulum
- MAM
Mitochondria-associated membrane
- MERCS
Mitochondria-ER contact sites
- PD
Parkinson’s disease
- AD
Alzheimer’s disease
- ALS
Amyotrophic Lateral Sclerosis
- ROS
Reactive oxygen species
- iPSC
Induced pluripotent stem cells
- 6-OHDA
6-Hydroxydopamine
- IP3
Inositol 1,4,5-trisphosphate
- IP3R
IP3 receptor
- NSC
Neural stem cell
- Htt
Huntingtin
- mHtt
Mutant huntingtin
- WT
Wild-type
- TMRM
Tetramethylrhodamine methyl ester
- TMRE
Tetramethylrhodamine ethyl ester
- FCCP
Carbonyl cyanide-4-phenylhydrazone
- Ant A
Antimycin A
- Myxo
Myxothiazol
- Nrf2
Nuclear factor erythroid 2-related factor 2
- ARE
Antioxidant response element
- Nqo1
NAD(P)H dehydrogenase [quinone] 1
- Hmox1
Heme oxygenase 1
- GCLc
Glutamate-cysteine ligase catalytic subunit
Author Contribution
LN, PL, MG, MH, and ACR designed the research. LN and PL performed most of the research and analysis. CL performed the research in NSCs. SM performed and analyzed in vivo/ex vivo data. CM performed some experiments in primary neurons. PC cultured and collected neurons for TEM and performed ICC for S1R. MA provided conditions and funding to perform TEM experiments. LN, NGE, MG, and ACR wrote the manuscript with the agreement of the others. All authors approved the final manuscript.
Funding
This work was supported by Prilenia Therapeutics Ltd (transferred rights from Teva Pharmaceutical Industries Ltd). ACR also acknowledges financial support from ‘Fundação Luso-Americana para o Desenvolvimento’ (FLAD)—Life Science 2020, Portugal. CNC is supported by the European Regional Development Fund (ERDF), through Centro 2020 Regional Operational Programme: project CENTRO-01-0145-FEDER-000012-HealthyAging2020, the COMPETE 2020-Operational Programme for Competitiveness and Internationalization and Portuguese national funds via FCT—Fundação para a Ciência e a Tecnologia, I.P.: project POCI-01-0145-FEDER-007440 and UIDB/04539/2020.
Data Availability
Data and materials described in the present manuscript will be available upon adequate request.
Declarations
Ethics Approval
All mouse experiments were carried out in accordance with the guidelines of the Institutional Animal Care and Use of Committee and the European Community directive (2010/63/EU) and protocols approved by the Faculty of Medicine, University of Coimbra (ref: ORBEA_189_2018/11042018) and UBC Committee (ref A16-0130) on Animal Care and the Canadian Council on Animal Care. All efforts were made to minimize animal suffering and to reduce the number of animals used.
Consent for Publication
All authors read and agreed to publish the present manuscript in its current format.
Conflict of Interest
MG and MH were employed by Teva Pharmaceutical Industries Ltd (transferred rights to Prilenia Therapeutics Ltd) during the time this study was conducted. LN and CM received a post-doctoral and a research grant, respectively, financed by Teva Pharmaceutical during part of the study. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Michael R. Hayden, and A. Cristina Rego equally contributed to this work.
Contributor Information
Luana Naia, Email: luana.naia@ki.se.
Philip Ly, Email: philiply@mail.ubc.ca.
Sandra I. Mota, Email: sandraisab.mota@gmail.com
Carla Lopes, Email: carlanmlopes@hotmail.com.
Carina Maranga, Email: carinamaranga@hotmail.com.
Patrícia Coelho, Email: patriciardcoelho1212@gmail.com.
Noga Gershoni-Emek, Email: noga.gershoni@prilenia.com.
Maria Ankarcrona, Email: maria.ankarcrona@ki.se.
Michal Geva, Email: Michal.Geva@prilenia.com.
Michael R. Hayden, Email: Michael.Hayden@prilenia.com
A. Cristina Rego, Email: acrego@cnc.uc.pt, Email: a.cristina.rego@gmail.com.
References
- 1.Kawamata H, Manfredi G. Proteinopathies and OXPHOS dysfunction in neurodegenerative diseases. J Cell Biol [Internet]. 2017 Dec 4;216(12):3917–29. Available from: 10.1083/jcb.201709172 [DOI] [PMC free article] [PubMed]
- 2.Panov A V., Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002 Aug;5(8):731–6. Available from: https://www.nature.com/articles/nn884 [DOI] [PubMed]
- 3.Sahlholm K, Sijbesma JWA, Maas B, Kwizera C, Marcellino D, Ramakrishnan NK, et al. Pridopidine selectively occupies sigma-1 rather than dopamine D2 receptors at behaviorally active doses. Psychopharmacology (Berl) [Internet]. 2015 Sep;232(18):3443–53. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26159455 [DOI] [PMC free article] [PubMed]
- 4.Johnston TH, Geva M, Steiner L, Orbach A, Papapetropoulos S, Savola J-M, et al. Pridopidine, a clinic-ready compound, reduces 3,4-dihydroxyphenylalanine-induced dyskinesia in Parkinsonian macaques. Mov Disord [Internet]. 2018 Dec 21; Available from: 10.1002/mds.27565 [DOI] [PubMed]
- 5.Francardo V, Bez F, Wieloch T, Nissbrandt H, Ruscher K, Cenci MA. Pharmacological stimulation of sigma-1 receptors has neurorestorative effects in experimental parkinsonism. Brain [Internet]. 2014 Jul;137(7):1998–2014. Available from: 10.1093/brain/awu107 [DOI] [PubMed]
- 6.Hyrskyluoto A, Pulli I, Törnqvist K, Huu Ho T, Korhonen L, Lindholm D. Sigma-1 receptor agonist PRE084 is protective against mutant huntingtin-induced cell degeneration: involvement of calpastatin and the NF-κB pathway. Cell Death Dis [Internet]. 2013 May 23;4(5):e646–e646. Available from: http://www.nature.com/articles/cddis2013170 [DOI] [PMC free article] [PubMed]
- 7.Vagnerova K, Hurn PD, Bhardwaj A, Kirsch JR. Sigma 1 Receptor Agonists Act as Neuroprotective Drugs Through Inhibition of Inducible Nitric Oxide Synthase. Anesth Analg [Internet]. 2006 Aug;103(2):430–4. Available from: https://insights.ovid.com/crossref?an=00000539-200608000-00029 [DOI] [PubMed]
- 8.Eddings CR, Arbez N, Akimov S, Geva M, Hayden MR, Ross CA. Pridopidine protects neurons from mutant-huntingtin toxicity via the sigma-1 receptor. Neurobiol Dis [Internet]. 2019 Sep;129:118–29. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0969996118306120 [DOI] [PMC free article] [PubMed]
- 9.Kusko R, Dreymann J, Ross J, Cha Y, Escalante-Chong R, Garcia-Miralles M, et al. Large-scale transcriptomic analysis reveals that pridopidine reverses aberrant gene expression and activates neuroprotective pathways in the YAC128 HD mouse. Mol Neurodegener [Internet]. 2018 Dec 21;13(1):25. Available from: 10.1186/s13024-018-0259-3 [DOI] [PMC free article] [PubMed]
- 10.Ryskamp D, Wu J, Geva M, Kusko R, Grossman I, Hayden M, et al. The sigma-1 receptor mediates the beneficial effects of pridopidine in a mouse model of Huntington disease. Neurobiol Dis [Internet]. 2017 Jan;97:46–59. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0969996116302479 [DOI] [PMC free article] [PubMed]
- 11.Ryskamp D, Wu L, Wu J, Kim D, Rammes G, Geva M, et al. Pridopidine stabilizes mushroom spines in mouse models of Alzheimer’s disease by acting on the sigma-1 receptor. Neurobiol Dis [Internet]. 2019 Apr;124:489–504; Available from: https://www.sciencedirect.com/science/article/pii/S0969996118305953 [DOI] [PMC free article] [PubMed]
- 12.Ionescu A, Gradus T, Altman T, Maimon R, Saraf Avraham N, Geva M, et al. Targeting the Sigma-1 Receptor via Pridopidine Ameliorates Central Features of ALS Pathology in a SOD1G93A Model. Cell Death Dis [Internet]. 2019 Mar 1;10(3):210. Available from: http://www.nature.com/articles/s41419-019-1451-2 [DOI] [PMC free article] [PubMed]
- 13.Francardo V, Geva M, Bez F, Denis Q, Steiner L, Hayden MR, et al. Pridopidine Induces Functional Neurorestoration Via the Sigma-1 Receptor in a Mouse Model of Parkinson’s Disease. Neurotherapeutics. 2019 Apr;16(2):465–479. Available from: 10.1007/s13311-018-00699-9 [DOI] [PMC free article] [PubMed]
- 14.Smith-Dijak AI, Nassrallah WB, Zhang LYJ, Geva M, Hayden MR, Raymond LA. Impairment and Restoration of Homeostatic Plasticity in Cultured Cortical Neurons From a Mouse Model of Huntington Disease. Front Cell Neuroscience [Internet]. 2019 May 16;13:209. Available from: 10.3389/fncel.2019.00209/full [DOI] [PMC free article] [PubMed]
- 15.Francardo V, Geva M, Bez F, Denis Q, Steiner L, Hayden MR, et al. Pridopidine Induces Functional Neurorestoration Via the Sigma-1 Receptor in a Mouse Model of Parkinson’s Disease. Neurotherapeutics [Internet]. 2019 Apr 12;16(2):465–79. Available from: 10.1007/s13311-018-00699-9 [DOI] [PMC free article] [PubMed]
- 16.Bernard-Marissal N, Médard J-J, Azzedine H, Chrast R. Dysfunction in endoplasmic reticulum-mitochondria crosstalk underlies SIGMAR1 loss of function mediated motor neuron degeneration. Brain [Internet]. 2015 Apr;138(Pt 4):875–90. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25678561 [DOI] [PubMed]
- 17.Hayashi T, Su TP. Sigma-1 Receptor Chaperones at the ER- Mitochondrion Interface Regulate Ca2+ Signaling and Cell Survival. Cell. 2007;131(3):596–610. doi: 10.1016/j.cell.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 18.Paillusson S, Stoica R, Gomez-Suaga P, Lau DHW, Mueller S, Miller T, et al. There’s Something Wrong with my MAM; the ER–Mitochondria Axis and Neurodegenerative Diseases. Trends Neurosci [Internet]. 2016 Mar;39(3):146–57. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0166223616000217 [DOI] [PMC free article] [PubMed]
- 19.Watanabe S, Ilieva H, Tamada H, Nomura H, Komine O, Endo F, et al. Mitochondria‐associated membrane collapse is a common pathomechanism in SIGMAR1 ‐ and SOD1 ‐linked ALS. EMBO Mol Med [Internet]. 2016 Dec;8(12):1421–37. Available from: 10.15252/emmm.201606403 [DOI] [PMC free article] [PubMed]
- 20.Naia L, Rego AC. Isolation and Maintenance of Striatal Neurons. Bio-Protocol [Internet]. 2018;8(8). Available from: https://bio-protocol.org/e2823 [DOI] [PMC free article] [PubMed]
- 21.Fu Z-X, Tan X, Fang H, Lau P-M, Wang X, Cheng H, et al. Dendritic mitoflash as a putative signal for stabilizing long-term synaptic plasticity. Nat Commun [Internet]. 2017 Dec 26;8(1):31. Available from: http://www.nature.com/articles/s41467-017-00043-3 [DOI] [PMC free article] [PubMed]
- 22.Park I-H, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-Specific Induced Pluripotent Stem Cells. Cell [Internet]. 2008 Sep;134(5):877–86. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0092867408010015 [DOI] [PMC free article] [PubMed]
- 23.Onofre I, Mendonça N, Lopes S, Nobre R, de Melo JB, Carreira IM, et al. Fibroblasts of Machado Joseph Disease patients reveal autophagy impairment. Sci Rep [Internet]. 2016 Sep 22;6(1):28220. Available from: http://www.nature.com/articles/srep28220 [DOI] [PMC free article] [PubMed]
- 24.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol [Internet]. 2009 Mar 1;27(3):275–80. Available from: http://www.nature.com/articles/nbt.1529 [DOI] [PMC free article] [PubMed]
- 25.Delli Carri A, Onorati M, Castiglioni V, Faedo A, Camnasio S, Toselli M, et al. Human Pluripotent Stem Cell Differentiation into Authentic Striatal Projection Neurons. Stem Cell Rev Reports [Internet]. 2013 Aug 27;9(4):461–74. Available from: 10.1007/s12015-013-9441-8 [DOI] [PubMed]
- 26.Nicoleau C, Varela C, Bonnefond C, Maury Y, Bugi A, Aubry L, et al. Embryonic stem cells neural differentiation qualifies the role of Wnt/β-Catenin signals in human telencephalic specification and regionalization. Stem Cells [Internet]. 2013 Sep;31(9):1763–74. Available from: 10.1002/stem.1462 [DOI] [PubMed]
- 27.Herbert AD, Carr AM, Hoffmann E. FindFoci: A Focus Detection Algorithm with Automated Parameter Training That Closely Matches Human Assignments, Reduces Human Inconsistencies and Increases Speed of Analysis. Lichten M, editor. PLoS One [Internet]. 2014 Dec 5;9(12):e114749. Available from: 10.1371/journal.pone.0114749 [DOI] [PMC free article] [PubMed]
- 28.Rietdorf J, A S. Multi Kymograph [Internet]. 2008. Available from: http://fiji.sc/Multi_Kymograph
- 29.Dickinson BC, Lin VS, Chang CJ. Preparation and use of MitoPY1 for imaging hydrogen peroxide in mitochondria of live cells. Nat Protoc [Internet]. 2013 Jun;8(6):1249–59. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23722262 [DOI] [PMC free article] [PubMed]
- 30.Ehrnhoefer DE, Southwell AL, Sivasubramanian M, Qiu X, Villanueva EB, Xie Y, et al. HACE1 is essential for astrocyte mitochondrial function and influences Huntington disease phenotypes in vivo. Hum Mol Genet [Internet]. 2018 Jan 15;27(2):239–53. Available from: https://academic.oup.com/hmg/article/27/2/239/4600049 [DOI] [PMC free article] [PubMed]
- 31.Ferreira IL, Carmo C, Naia L, I. Mota S, Cristina Rego A. Assessing Mitochondrial Function in In Vitro and Ex Vivo Models of Huntington’s Disease. In: Precious S V., et al., editors. Huntington’s Disease, Methods in Molecular Biology [Internet]. Springer; 2018. p. 415–42. Available from: 10.1007/978-1-4939-7825-0_19 [DOI] [PubMed]
- 32.Rogers GW, Brand MD, Petrosyan S, Ashok D, Elorza AA, Ferrick DA, et al. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PLoS One [Internet]. 2011;6(7):e21746. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21799747 [DOI] [PMC free article] [PubMed]
- 33.Rossi A, Pizzo P, Filadi R. Calcium, mitochondria and cell metabolism: A functional triangle in bioenergetics. Biochim Biophys Acta - Mol Cell Res [Internet]. 2019 Jul;1866(7):1068–78. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0167488918304798 [DOI] [PubMed]
- 34.Lewis TL, Kwon S-K, Lee A, Shaw R, Polleux F. MFF-dependent mitochondrial fission regulates presynaptic release and axon branching by limiting axonal mitochondria size. Nat Commun [Internet]. 2018 Dec 27;9(1):5008. Available from: http://www.nature.com/articles/s41467-018-07416-2 [DOI] [PMC free article] [PubMed]
- 35.Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y, et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet [Internet]. 2003 Jul 1;12(13):1555–67. Available from: http://www.ncbi.nlm.nih.gov/pubmed/12812983 [DOI] [PubMed]
- 36.Panov A V, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci [Internet]. 2002;5(8):731–6. Available from: http://www.nature.com/neuro/journal/v5/n8/pdf/nn884.pdf [DOI] [PubMed]
- 37.Lucas G, Rymar V V, Sadikot AF, Debonnel G. Further evidence for an antidepressant potential of the selective σ1 agonist SA 4503: Electrophysiological, morphological and behavioural studies. Int J Neuropsychopharmacol. 2008; [DOI] [PubMed]
- 38.Hayashi T, Maurice T, Su TP. Ca2+ signaling via σ1-receptors: Novel regulatory mechanism affecting intracellular Ca2+ concentration. J Pharmacol Exp Ther. 2000; [PubMed]
- 39.Monnet FP, Debonnel G, Fournier A, De Montigny C. Neuropeptide Y potentiates the N-methyl-D-aspartate response in the CA3 dorsal hippocampus. II. Involvement of a subtype of sigma receptor. J Pharmacol Exp Ther. 1992; [PubMed]
- 40.Hong W, Nuwayhid SJ, Werling LL. Modulation of bradykinin-induced calcium changes in SH-SY5Y cells by neurosteroids and sigma receptor ligands via a shared mechanism. Synapse. 2004; [DOI] [PubMed]
- 41.Matsuno K, Nakata K, Kobayashi T, Mita S, Okamoto K, Senda T. SA4503, a novel cognitive enhancer, with σ1 receptor agonistic properties. Behav Brain Res. 2002;83(1–2):221–224. doi: 10.1016/s0166-4328(97)86074-3. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi T, Matsuno K, Nakata K, Mita S. Enhancement of acetylcholine release by SA4503, a novel sigma 1 receptor agonist, in the rat brain. J Pharmacol Exp Ther. 1996;279(1):106–113. [PubMed] [Google Scholar]
- 43.Rousseaux CG, Greene SF. Sigma receptors [σ Rs]: Biology in normal and diseased states. J Recept Signal Transduct. 2016;36(4):327–388. doi: 10.3109/10799893.2015.1015737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Urani A, Romieu P, Roman FJ, Yamada K, Noda Y, Kamei H, et al. Enhanced antidepressant efficacy of σ1 receptor agonists in rats after chronic intracerebroventricular infusion of β-amyloid-(1–40) protein. Eur J Pharmacol. 2004; [DOI] [PubMed]
- 45.Pande AC, Genève J, Scherrer B, Smith F, Leadbetter RA, de Meynard C. A placebo-controlled trial of igmesine in the treatment of major depression. Eur Neuropsychopharmacol. 1999;
- 46.Su T-P, Su T-C, Nakamura Y, Tsai S-Y. The Sigma-1 Receptor as a Pluripotent Modulator in Living Systems. Trends Pharmacol Sci. 2016;37(4):262–278. doi: 10.1016/j.tips.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weng T-Y, Hung DT, Su T-P, Tsai S-YA. Loss of Sigma-1 Receptor Chaperone Promotes Astrocytosis and Enhances the Nrf2 Antioxidant Defense. Oxid Med Cell Longev [Internet]. 2017;2017:1–14. Available from: https://www.hindawi.com/journals/omcl/2017/4582135/ [DOI] [PMC free article] [PubMed]
- 48.Mishina M, Ohyama M, Ishii K, Kitamura S, Kimura Y, Oda K, et al. Low density of sigma1 receptors in early Alzheimer’s disease. Ann Nucl Med [Internet]. 2008 Apr 23;22(3):151–6. Available from: 10.1007/s12149-007-0094-z [DOI] [PubMed]
- 49.Bernard-Marissal N, van Hameren G, Juneja M, Pellegrino C, Louhivuori L, Bartesaghi L, et al. Altered interplay between endoplasmic reticulum and mitochondria in Charcot–Marie–Tooth type 2A neuropathy. Proc Natl Acad Sci [Internet]. 2019 Feb 5;116(6):2328–37. Available from: 10.1073/pnas.1810932116 [DOI] [PMC free article] [PubMed]
- 50.Nguyen T, Sherratt PJ, Pickett CB. Regulatory mechanisms controlling gene expression mediated by the antioxidante response element. Annu Rev Pharmacol Toxicol [Internet]. 2003 Apr;43(1):233–60. Available from: 10.1146/annurev.pharmtox.43.100901.140229 [DOI] [PubMed]
- 51.Van Raamsdonk JM, Pearson J, Slow EJ, Hossain SM, Leavitt BR, Hayden MR. Cognitive dysfunction precedes neuropathology and motor abnormalities in the YAC128 mouse model of Huntington’s disease. J Neurosci [Internet]. 2005 Apr 20;25(16):4169–80. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15843620 [DOI] [PMC free article] [PubMed]
- 52.Garcia-Miralles M, Geva M, Tan JY, Yusof NABM, Cha Y, Kusko R, et al. Early pridopidine treatment improves behavioral and transcriptional deficits in YAC128 Huntington disease mice. JCI Insight [Internet]. 2017 Dec 7;2(23). Available from: https://insight.jci.org/articles/view/95665 [DOI] [PMC free article] [PubMed]
- 53.Geva M, Kusko R, Soares H, Fowler KD, Birnberg T, Barash S, et al. Pridopidine activates neuroprotective pathways impaired in Huntington Disease. Hum Mol Genet [Internet]. 2016 Jul 27;[Epub ahead of print]. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27466197 [DOI] [PMC free article] [PubMed]
- 54.Brimson JM, Brimson S, Chomchoei C, Tencomnao T. Using sigma-ligands as part of a multi-receptor approach to target diseases of the brain. Expert Opinion on Therapeutic Targets. 2020. [DOI] [PubMed]
- 55.Maurice T. Bi-phasic dose response in the preclinical and clinical developments of sigma-1 receptor ligands for the treatment of neurodegenerative disorders. Expert Opin Drug Discov. 2020;in press. [DOI] [PubMed]
- 56.Niescier RF, Hong K, Park D, Min K-T. MCU Interacts with Miro1 to Modulate Mitochondrial Functions in Neurons. J Neurosci [Internet]. 2018 May 16;38(20):4666–77. Available from: 10.1523/JNEUROSCI.0504-18.2018 [DOI] [PMC free article] [PubMed]
- 57.Area-Gomez E, del Carmen Lara Castillo M, Tambini MD, Guardia-Laguarta C, de Groof AJC, Madra M, et al. Upregulated function of mitochondria-associated ER membranes in Alzheimer disease. EMBO J [Internet]. 2012 Nov 5;31(21):4106–23. Available from: 10.1038/emboj.2012.202 [DOI] [PMC free article] [PubMed]
- 58.Hedskog L, Pinho CM, Filadi R, Ronnback A, Hertwig L, Wiehager B, et al. Modulation of the endoplasmic reticulum-mitochondria interface in Alzheimer’s disease and related models. Proc Natl Acad Sci [Internet]. 2013 May 7;110(19):7916–21. Available from: 10.1073/pnas.1300677110 [DOI] [PMC free article] [PubMed]
- 59.Leal NS, Dentoni G, Schreiner B, Naia L, Piras A, Graff C, et al. Amyloid β-Peptide Increases Mitochondria-Endoplasmic Reticulum Contact Altering Mitochondrial Function and Autophagosome Formation in Alzheimer’s Disease-Related Models. Cells [Internet]. 2020 Nov 28;9(12):2552. Available from: https://www.mdpi.com/2073-4409/9/12/2552 [DOI] [PMC free article] [PubMed]
- 60.Leal NS, Dentoni G, Schreiner B, Kämäräinen O-P, Partanen N, Herukka S-K, et al. Alterations in mitochondria-endoplasmic reticulum connectivity in human brain biopsies from idiopathic normal pressure hydrocephalus patients. Acta Neuropathol Commun [Internet]. 2018 Dec 1;6(1):102. Available from: 10.1186/s40478-018-0605-2 [DOI] [PMC free article] [PubMed]
- 61.Paillusson S, Gomez-Suaga P, Stoica R, Little D, Gissen P, Devine MJ, et al. α-Synuclein binds to the ER–mitochondria tethering protein VAPB to disrupt Ca2+ homeostasis and mitochondrial ATP production. Acta Neuropathol [Internet]. 2017 Jul 23;134(1):129–49. Available from: 10.1007/s00401-017-1704-z [DOI] [PMC free article] [PubMed]
- 62.Cherubini M, Lopez-Molina L, Gines S. Mitochondrial fission in Huntington’s disease mouse striatum disrupts ER-mitochondria contacts leading to disturbances in Ca2+ efflux and Reactive Oxygen Species (ROS) homeostasis. Neurobiol Dis [Internet]. 2020 Mar;136:104741. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0969996120300164 [DOI] [PubMed]
- 63.Basso V, Marchesan E, Peggion C, Chakraborty J, von Stockum S, Giacomello M, et al. Regulation of ER-mitochondria contacts by Parkin via Mfn2. Pharmacol Res [Internet]. 2018 Dec;138:43–56. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1043661818303219 [DOI] [PubMed]
- 64.Gómez-Suaga P, Pérez-Nievas BG, Glennon EB, Lau DHW, Paillusson S, Mórotz GM, et al. The VAPB-PTPIP51 endoplasmic reticulum-mitochondria tethering proteins are present in neuronal synapses and regulate synaptic activity. Acta Neuropathol Commun [Internet]. 2019 Dec 6;7(1):35. Available from: 10.1186/s40478-019-0688-4 [DOI] [PMC free article] [PubMed]
- 65.Díaz-Vegas AR, Cordova A, Valladares D, Llanos P, Hidalgo C, Gherardi G, et al. Mitochondrial Calcium Increase Induced by RyR1 and IP3R Channel Activation After Membrane Depolarization Regulates Skeletal Muscle Metabolism. Front Physiol [Internet]. 2018 Jun 25;9. Available from: 10.3389/fphys.2018.00791/full [DOI] [PMC free article] [PubMed]
- 66.Omi K, Hachiya NS, Tokunaga K, Kaneko K. siRNA-mediated inhibition of endogenous Huntington disease gene expression induces an aberrant configuration of the ER network in vitro. Biochem Biophys Res Commun [Internet]. 2005 Dec;338(2):1229–35. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0006291X05023338 [DOI] [PubMed]
- 67.Cavendish JZ, Sarkar SN, Colantonio MA, Quintana DD, Ahmed N, White BA, et al. Mitochondrial Movement and Number Deficits in Embryonic Cortical Neurons from 3xTg-AD Mice. J Alzheimer’s Dis [Internet]. 2019 Jul 2;70(1):139–51. Available from: 10.3233/JAD-190143 [DOI] [PMC free article] [PubMed]
- 68.Watanabe S, Ilieva H, Tamada H, Nomura H, Komine O, Endo F, et al. Mitochondria-associated membrane collapse is a common pathomechanism in SIGMAR1- and SOD1-linked ALS. EMBO Mol Med [Internet]. 2016;8(12):1421–37. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27821430 [DOI] [PMC free article] [PubMed]
- 69.Wang X, Wen Y, Dong J, Cao C, Yuan S. Systematic In-Depth Proteomic Analysis of Mitochondria-Associated Endoplasmic Reticulum Membranes in Mouse and Human Testes. Proteomics [Internet]. 2018 Jul;18(14):1700478. Available from: 10.1002/pmic.201700478 [DOI] [PubMed]
- 70.Lee K-S, Huh S, Lee S, Wu Z, Kim A-K, Kang H-Y, et al. Altered ER–mitochondria contact impacts mitochondria calcium homeostasis and contributes to neurodegeneration in vivo in disease models. Proc Natl Acad Sci [Internet]. 2018 Sep 18;115(38):E8844–53. Available from: 10.1073/pnas.1721136115 [DOI] [PMC free article] [PubMed]
- 71.Ribeiro M, Rosenstock TR, Oliveira AM, Oliveira CR, Rego AC. Insulin and IGF-1 improve mitochondrial function in a PI-3K/Akt-dependent manner and reduce mitochondrial generation of reactive oxygen species in Huntington’s disease knock-in striatal cells. Free Radic Biol Med [Internet]. 2014 Sep;74:129–44. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24992836 [DOI] [PubMed]
- 72.Pal A, Fontanilla D, Gopalakrishnan A, Chae Y-K, Markley JL, Ruoho AE. The sigma-1 receptor protects against cellular oxidative stress and activates antioxidant response elements. Eur J Pharmacol [Internet]. 2012 May;682(1–3):12–20. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0014299912001008 [DOI] [PMC free article] [PubMed]
- 73.Hamilton J, Pellman JJ, Brustovetsky T, Harris RA, Brustovetsky N. Oxidative metabolism in YAC128 mouse model of Huntington's disease. Hum Mol Genet. [Internet] 2015 Sep 1;24(17):4862–78. Available from: https://academic.oup.com/hmg/article/24/17/4862/647965 [DOI] [PMC free article] [PubMed]
- 74.Reilmann R, Olanow CW, Leinonen M, McGarry A, Geva M, Squitieri F, et al. Novel PET data and analysis of early HD from Pride-HD. In: Huntington Study Group Annual Meeting. 2019.
- 75.Tang T, Tu H, Chan EYW, Maximov A, Wang Z, Wellington CL, et al. Huntingtin and Huntingtin-Associated Protein 1 Influence Neuronal Calcium Signaling Mediated by Inositol- ( 1 , 4 , 5 ) Triphosphate Receptor Type 1. Neuron. 2003;39:227–39. Available from: 10.1016/S0896-6273(03)00366-0 [DOI] [PMC free article] [PubMed]
- 76.Naia L, Ferreira IL, Ferreiro E, Rego AC. Mitochondrial Ca2+handling in Huntington’s and Alzheimer’s diseases – Role of ER-mitochondria crosstalk. Biochem Biophys Res Commun. 2017;483(4). Available from: https://www.sciencedirect.com/science/article/pii/S0006291X16312517 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and materials described in the present manuscript will be available upon adequate request.