Abstract
Superspreading and variants of concern (VOC) of the human pathogen severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are the main catalyzers of the coronavirus disease 2019 (COVID-19) pandemic. However, measuring their individual impact is challenging. By examining the largest database of SARS-CoV-2 genomes The Global Initiative on Sharing Avian Influenza Data [GISAID; n >1.2 million high-quality (HQ) sequences], we present evidence suggesting that superspreading has had a key role in the epidemiological predominance of VOC. There are clear signatures in the database compatible with large superspreading events (SSEs) coinciding chronologically with the worst epidemiological scenarios triggered by VOC. The data suggest that, without the randomness effect of the genetic drift facilitated by superspreading, new VOC of SARS-CoV-2 would have had more limited chance of success.
Keywords: SARS-CoV-2, superspreader, superspreader event, variant of concern, B.1.1.7, B.1.617.2
Superspreading and VOC
A superspreader (see Glossary) is an infected individual who is responsible for a disproportionally large number of secondary transmissions relative to the basic reproductive number (R 0 ). The role of superspreaders in infectious diseases has been known for more than 100 years [1] and has been well documented in past viral epidemic scenarios [2., 3., 4.]. Their importance in the SARS-CoV-2 pandemic was highlighted in the first wave of the pandemic [5,6], and further corroborated in later studies (e.g., [7., 8., 9., 10.]). SSEs are those that favor large-scale transmissions (massive events, indoor meetings, etc.). Many authors favor the use of the term ‘SSE’, thus highlighting the social circumstances and environment instead of the role of single superspreader individuals (despite many authors assuming their existence [10]). However, there is enough evidence to suggest that SSEs usually require the intervention of one or a few individuals contributing most of the transmissions [11., 12., 13., 14., 15., 16., 17.]; this would usually occur over a short period of time, when an infected person is shedding a very high viral load and contacting a high number of exposed individuals [13]. Nevertheless, our knowledge of the phenotypic characteristics of superspreader individuals is still in its infancy [18]. The rule that 20% of infected individuals cause 80% of the infections has been borne out by several COVID-19 studies [7,19]. In addition, several studies that used mathematical simulations highlight the importance of early SSEs as determinants of SARS-CoV-2 variant predominance [20,21].
At the same time, several genetic VOC of the virus have emerged over the past few months, triggering alerts from health authorities and governments worldwide. These variants are assumed to provide advantages to the virus in a given epidemiological environment (higher transmissibility), and some could also lead to higher severity and mortality [22]. Although their impact in the scientific literature and popular media is relatively new (since the start of 2021), these variants have probably existed since the beginning of the pandemic [23]; in part, the growing interest in VOC has been determined by their potential ability to evade the efficiency of emerging SARS-CoV-2 treatments and vaccines (which began to circulate in a few countries during late 2020). According to Public Health England (PHE; Technical briefing 18 from 9 July 2021)i, there are four current VOC: B.1.1.7 [Alpha (according to the WHO nomenclature) or VOC-2020/12/01 (according to the PHE) [24]], B.1.351 (Beta or VOC-202012/02 [25]), B.1.617.2 (Delta or VOC-202104/02 [26]), and P.1 (Gamma or VOC-202101/02 [27]).
Determinants of VOC predominance
Of the 1 493 747 genomes present in GISAID, most are labeled as HQ sequences (84%) and contain a lower proportion of ambiguities compared with low-quality (LQ) sequences (Table 1). There are >670 000 different HQ haplotypes (~77% of which were found only once), and >58 000 substitutions in the HQ sequences (~11 000 singletons) (Box 1 for the methods used to analyze the database).
Table 1.
Variation observed in SARS-CoV-2 sequence genomes recorded in GISAIDa
| All | LQ | HQ | S (HQ) | B.1.1.7 | B.1.1.7-S | B.1.351 | B.1.351-S | B.1.617.2 | B.1.617.2-S | P.1 | P.1-S | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | 1 493 747 | 240 458 | 1 253 289 | 1 253 289 | 511 492 | 511 492 | 4092 | 4092 | 4013 | 4013 | 10 538 | 10 538 |
| Ns | 69 643 | 52 253 | 45 082 | 6324 | 14 652 | 2041 | 127 | 31 | 535 | 91 | 368 | 47 |
| Indels | 13 579 | 5053 | 11 002 | 1361 | 2213 | 156 | 87 | 9 | 66 | 8 | 134 | 15 |
| MNPs | 8365 | 3352 | 5871 | 804 | 895 | 127 | 23 | 2 | 15 | 1 | 68 | 5 |
| DH | 822 117 | 186 180 | 670 833 | 98 310 | 227 496 | 21 145 | 2764 | 669 | 1924 | 486 | 6026 | 1082 |
| SH | 649 422 | 155 910 | 521 249 | 60 698 | 163 439 | 11 628 | 2248 | 435 | 1489 | 337 | 4887 | 694 |
| ST | 62 066 | 44 228 | 58 065 | 7833 | 36 866 | 5099 | 3587 | 548 | 2588 | 363 | 6028 | 825 |
| SST | 11 588 | 12 032 | 10 944 | 1468 | 7311 | 1002 | 1847 | 261 | 1429 | 197 | 2927 | 406 |
Abbreviations: DH, different haplotypes; MNPs, multi-nucleotide polymorphisms; Ns, ambiguities; SH, singleton haplotypes; SST, singleton substitutions; ST, substitutions. Values for VOC refer to HQ sequences.
Box 1. Methods used to analyze the GISAID database.
We downloaded 1 493 747 genomes deposited in the GISAID database [43]. All analyses were carried out with 1 253 289 labeled as HQ and using statistical and phylogenetic analysis procedures described previously [5,6,33,44]. The genome with GenBank identity code MN908947 was used as the reference sequence. Definition of the sequence motif of VOC is ambiguous and depends on the source (e.g., ‘genomes are assigned lineage to B.1.1.7 if they exhibit at least 5 of the 17 mutations inferred to have arisen on the phylogenetic branch immediately ancestral to the cluster’v). However, our definition is based on a sequence motif inferred from the most parsimonious phylogenetic tree of Figure 1 in the main text. A few haplotypes in GISAID could fall outside the VOC definition due to reversions and/or recombination, sequencing errors, and so on. To avoid conflicts in comparative studies, facilitate reproducibility of the analysis and a better inference of SSEs (e.g., inferring one-step haplotypes from the core of a SSE), we defined VOC by their most parsimonious sequence motifs.
BEAST v.2.6.2 [45] software was used to build the Bayesian tree and estimate the TMRCA of the A26063T (N501Y) mutation and other sublineages through a coalescent model with exponential growth and the reference sequence as an outgroup. We excluded indels and samples with evident errors in date of sampling from the inference, to avoid problems with the location of ancestral haplotypes. We used strict-clock, a rate of evolution of 0.80×10–3 (0.14×10–3–1.31×10–3) substitutions per site per year (s/s/y)vi and a Markov chain-Monte Carlo run of 300 000 000 steps sampling every 10 000 steps and 10% discarded as burn-in. We used Tracer (v. 1.7.1) [45] to explore distribution convergence. The Maximum clade credibility tree was visualized and edited using FigTree v.1.4.4vii. We additionally built median joining networks [46] using POPart software [47] to better visualize the star-like shape that is characteristic of a superspreader pattern of transmission, which is different to that generated by homogeneous and chain of transmissions. The methodology used in the present and previous studies [5,6,33] does not aim at capturing individual transmissions between infectors and infectees [that might be only captured (at least partially) by examining e.g., known contact tracing networks and intrahost variation], but to make visible the signatures left by SSEs on the global scale represented in GISAID.
We used R software to carry out the graphic representation of the data [48].
Alt-text: Box 1
A common feature of VOC is the accumulation of specific mutations in the surface spike protein [encoded by the spike (S) gene], which, according to different authors, might alter the way in which the virus interacts with the human angiotensin-converting enzyme 2 (ACE2), a receptor protein of the host cells in the lung and other tissues that constitute the main entrance to cell invasion. Approximately 13% of the substitutions in HQ sequences occurred within the S gene, and the four VOC show a comparable percentage of substitutions in this gene, ranging from ~14% to ~15% (B.1.1.7, ~14%; B.1.351, ~15%; B.1.617.2, ~14%; P.1, ~14%). The substitution A23063T, highlighted as one of the most remarkable mutations in several VOC, leads to the amino acid change N501Y in the S protein and might contribute to an increased infectivity of the virus by enhancing binding affinity to ACE2 (as of July 2021, the query ‘N501Y’ in PubMed yielded 149 items; e.g., [28])ii. Other mutations of interest in the S gene are: C23604A (P681H; adjacent to the furin cleavage site in the S protein), G23012 (E484K), and the deletion TACATG21765 (ΔH69/ΔV70 also in the S protein). It has also been claimed that the high transmissibility of B.1.1.7 can be explained by the large number of favorable mutations accumulated by this variant [24].
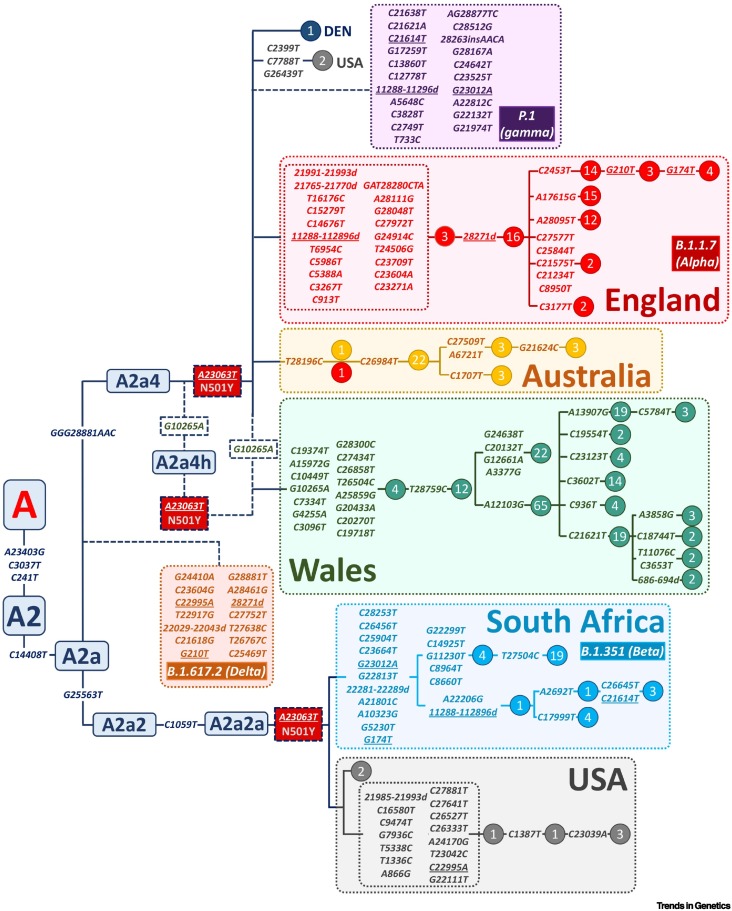
Figure 1 shows a maximum parsimony (MP) tree of the most important VOC, with special focus on B.1.1.7 because it was the first alarming one (and the first to be detected and popularized by the end of 2020), and is by far the best represented in GISAID of the four considered by the PHE (see later). The phylogenetic root of B.1.1.7 accumulated 21 mutational changes, 14 of which were non-synonymous substitutions. According to the maximum likelihood tree of Nextstrainiii, mutation A23063T, which belongs to the sequence motif of B.1.1.7, B.1.351, and P.1. occurred in the ancestral node on top of the A2a characteristic mutations (see the early clade nomenclature of [5]). Therefore, by retrieving genomes from the beginning of the pandemic to 31 October 2020, we captured most (if not all) the available ancestral sequences in GISAID of the main VOC up to the beginning of the most important outbreaks of B.1.1.7. The number of genomes available is scarce, namely, 67 for B.1.1.7 and 29 for B.1.351 (there are no representatives for the other VOC). All the VOC (B.1.1.7, B.1.351, B.1.617.2 and P.1) have a common ancestor in the node A2a, and diverged into three branches, namely A2a4 (B.1.1.7 and P.1), A2a2 > A2a2a (B.1.351), and another unnamed A2a-branch (B.1.617.2). A few interesting features can be inferred from a detailed MP and Bayesian phylogenies of the VOC (Figures 1 and 2A, Key figure), as discussed in this opinion.
Figure 1.
Maximum parsimony (MP) tree of genomes representing the ancestors of B.1.1.7 and B.1.351 [carriers of A23063T (N50Y) from 31 December 2019 to 31 October 2020 in the Global Initiative on Sharing Avian Influenza Data (GISAID) database] and their most closely related clades.
The phylogeny fits well with that inferred using a Bayesian approach (see Figure 2A in the main text). The motifs for the other two variants of interest (VOC) (P.1 and B.1.617.2) have been parsimoniously attached to the main tree skeleton. Lineage/haplogroup nomenclature for the ancestral nodes was taken from [5]. Phylogenetic reconstruction is blurred by the high evolutionary rate of the virus, which leads to recurrent mutations and recombination, both of which create homoplasy in the phylogenetic tree. Parallel mutations are underlined. Time to the most recent common ancestor (TMRCA) values were taken from the Bayesian tree in Figure 2A in the main text.
Figure 2.
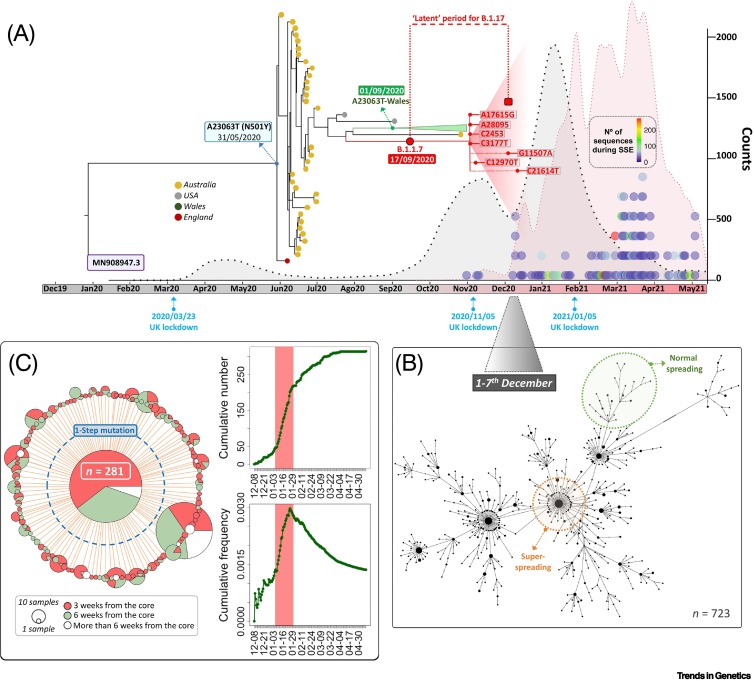
Key figure. Phylogeny, epidemiological context, and superspreader events (SSEs) for B.1.1.7.
(A) Bayesian phylogeny for B.1.1.7 genomes and phylogenetically related sequences until 31 October 2020. The large triangle indicates the point from which all B.1.1.7 sequences emerged. Attached to its node are the main subbranches and diagnostic mutations: almost all the important ones are present in the database until the end of the B.1.1.7 outbreak (coinciding with the fall of positive cases around March 2021). The red-broken line curve represents absolute counts of UK B.1.1.7 genomes, and the black-broken line curve the new positive cases (×0.035) from the beginning of the pandemic. The colored dot columns on the right represent candidate SSEs, with its own legend (right) indicating the number of representative genomes. (B) Network representing genomes circulating in the UK during 1–7 December 2020, showing the typical clustering pattern; by way of example, the orange-broken line shape indicates a section of the tree suggestive of superspreading, while the green section suggests normal spreading. (C) Median joining network of B.1.1.7 UK genomes representing one of the SSE candidates (mini-outbreak; mid-January 2021). Most of the genomes in the core (a proxy for primary transmission; PT) occurred in 3 weeks (red); in green are those occurring in the third to sixth weeks. The branches indicate one-step mutations from the core. The graphics on the left represent the mini-outbreak cumulative counts (top) for the core haplotype that increased very rapidly in only 5 days (red band), as well as its relative frequency with respect to the circulated viruses (bottom).
Time of the most recent common ancestor of A23063T and B.1.1.7
According to a Bayesian tree, the time of the most recent common ancestor (TMRCA) for A23063T within A2a4 (this mutation most likely appeared independently in the phylogeny of the virus) is 31 May 2020 [95% highest posterior density interval (HPD): 28 May 2020–4 June 2020; first instance A2a4+A23063T in GISAID sampled on 3 June 2020 in Australia (#480662)], approximately 3 months before the core of B.1.1.7 dated to 17 September 2020 (95% HPD TMRCA: 14 September 2020–19 September 2020; first instances in GISAID sampled on 20 September 2020 (#601443) and 21 September 2020 (#581117)] and approximately 5 months before the outbreak of B.1.1.7. Many of the subbranches deriving from the ancestral A23063T node extinguished, while its basal node carrying the problematic mutation A23063T passed unnoticed to GISAID for approximately 2 months (indicated as ‘latent’ period of B.1.1.7 in Figure 2A). Therefore, mutation A23063T alone could not fully explain the success of the descendent VOC, because this mutation circulated at inconspicuous frequencies for months without causing major outbreaks.
Unsuccessful A23063T phylogenetic branches
Several relevant phylogenetic branches emerging from an early A23063T node had locally moderate geographical success but most likely died out (or at least had no continuity in GISAID) in only a few weeks (e.g., an Australian branch characterized by mutation T28196C and a Welsh branch carrying 15 mutations on top of A23063T; Figure 1, Figure 2A).
Consistent rate accumulation of mutational changes over time
Mutations accumulate in the RNA of circulating viruses following an expected (or somehow acceleratediv) molecular clock, and a known substitution rate that is in the order of magnitude of ~10–3 (Box 2 ). Therefore, if we assume a consistent rate of accumulation of mutations in the genome of SARS-CoV-2 over time, the change that made B.1.1.7 more infectious is not obvious e.g., the last mutation(s) incorporated in its sequence motif (that triggered the outbreaks), complex epistatic interactions, and/or other nongenetic factors, such as genetic drift. There is evidence suggesting that important SSEs could have substantially enabled B.1.1.7 to succeed in many epidemiological contexts (see later).
Box 2. SARS-CoV-2 variation.
The range of SARS-CoV-2 substitution rates, as inferred from maximum likelihood trees, is ~0.8×10–3–0.542×10–3 substitutions per site per year (s/s/y) [5]vii. This rate is of the same order of magnitude as other RNA viruses. This allowed an estimation of the TMRCA of the SARS-CoV-2 to November 2019 [5] by fitting epidemiological data. Given that the RNA of the virus is ~30 000 base pairs (bp) long, this means that the genomes of the circulating viruses accumulate approximately one mutation every 2 weeks on average. According to this estimate, a SARS-CoV-2 circulating 18 months after the reference genome (sampled on 1 December 2019 [49]) would accumulate, on average, 36 substitutions with respect to this reference. The GISAID database explored in the present study contains more than 44 200 different substitutions, more than 7800 of which fall in the S gene and 70.8% of which (>5500/7800) are non-synonymous (see Table 1 in the main text). It is conceivable that some of these mutations could either facilitate or make more difficult the dispersion of the virus, while others could also help the virus escape the immune defenses of a previously vaccinated or infected host. A few of these variants have been concerning the scientific community over the past few months. However, it is likely that thousands of them have gone unnoticed, in part because many, despite having evolutionary advantages, could have died out by chance (e.g., occurring in people that never transmitted the virus or chains of transmissions that broke in only a few steps) and also because of the obvious limitations of the international sequencing efforts aimed at detecting these variants (mainly concentrated in Europe and North America; see main text). Genetic drift mediated by superspreading scenarios could have an important role in favoring the survival of both advantageous and disadvantageous SARS-CoV-2 variants and making them predominant in a short timeframe. From phylogenies and patterns of variation, it appears that the B.1.1.7 and B.1.617.2 variants (those represented in the analyzed GISAID database) could have emerged to a large extent, thanks to SSEs [6].
Alt-text: Box 2
Number of mutations accumulated in B.1.1.7 genomes
The core of B.1.1.7 comprises 21 mutational changes with respect to the reference genome (18 of which are substitutions; Figure 1), a number that is not far from expectation: given that the RNA of circulating SARS-CoV-2 incorporates approximately two substitutions per month according to its evolutionary rate, the expected number of substitutions that any SARS-CoV-2 would accumulate from December 2019 until mid-September 2020 would be, on average, ~17 to 18. There are in fact 4192 and 1308 sequences recorded in GISAID (and sampled before mid-September) accumulating >18 and >21 substitutions, respectively. In addition, the sequence motif of B.1.1.7 has 14 non-synonymous mutations; however, this is not unusual in GISAID either: there are 6145 non-synonymous substitutions recorded in the S gene (Box 2 and Table 1); and the accumulation of many of these mutations in the genome of a circulating SARS-CoV-2 could have occurred hundreds of times during the evolutionary pandemic history of this coronavirus; for example, there are >20 000 non-VOC sequences in GISAID having more non-synonymous substitutions in the S gene than in the B.1.1.7 sequence motif. In addition, a mutation does not need to be non-synonymous to alter the fitness of a microorganism [29].
Therefore, the argument that B.1.1.7 has accumulated an unusual number of mutations of concern (many of which are non-synonymous), that these could benefit its host transmissibility, and that this fact alone could explain large SARS-CoV-2 outbreaks, may be too simplistic and can be challenged. By way of mathematical modeling, Davies et al. [24] claimed altered transmission characteristics for this variant with respect to other coronaviruses circulating at the time because they ‘did not find substantial differences in social interactions between regions of high and low VOC-202012/01 (B.1.1.7) prevalence, as measured by Google mobility and social contact survey data from September to December’; this led the authors to disregard possible founder effects associated with the rapid spread of B.1.1.7. However, SSEs do not necessarily leave a discernible signature on social interactions and contact survey data. Nonetheless, SSEs were not considered as a variable to calibrate transmissibility of B.1.1.7 in the UK [24]. In addition, it is now well known that reduction in mobility is associated with low transmission rates [30., 31., 32.]; thus, many important outbreaks involving VOC (and non-VOC) occurred immediately after strict lockdown periods and/or after relaxing of measures to prevent COVID-19 (e.g., Christmas and summer holidays). By way of simulations aimed at measuring the impact of mobility restrictions in COVID-19, Lima et al. [31] concluded that ‘the superspreaders are responsible for most of the infection propagation and the impact of personal protective equipment in the spreading of the infection’. In this regard, little or no attention has been devoted to the role of superspreading to the estimate of R 0, even for the VOC [24], despite the many convergent lines of evidence signaling their role in the pandemic (see earlier).
Inferring superspreading from a genome database
Analyzing the details of SSEs would ideally need a careful examination of contact tracing networks and understanding the epidemiological circumstances occurring locally at the time of the event. In practice, this is only possible in a few cases (some of which are reported in the scientific literature [8,11]). There exists an alternative to detecting genome candidates responsible for SSEs by analyzing the signatures that this pattern of viral transmission can leave on the sequences stored in GISAID. Such an exercise was undertaken using the data stored during the early phase of the pandemic, and the results pointed to viral dispersion patterns compatible with the existence of superspreading on a worldwide scale [5]. This database recorded a high number of haplotypes that experienced a sudden increase in their frequency in a geographical location in a short time-period of a few days. The star-like phylogenies observed for these haplotypes and their one-mutational step-related haplotypes (Figure 2B,C), coupled with a substitution rate of SARS-CoV-2 on the order of 10–3 substitutions per site per year (s/s/y), and the incubation rate for the COVID-19 of 5 to 6 days (Box 3 ) would require the intervention of persons spreading the virus ‘more efficiently’ than others. This pattern was observed across the world and could explain a significant proportion of inferred secondary transmissions in the database. The results of this pioneering study were subsequently extended to a more restricted geographical scenario (Spain; [6]) and later to a much larger database [33].
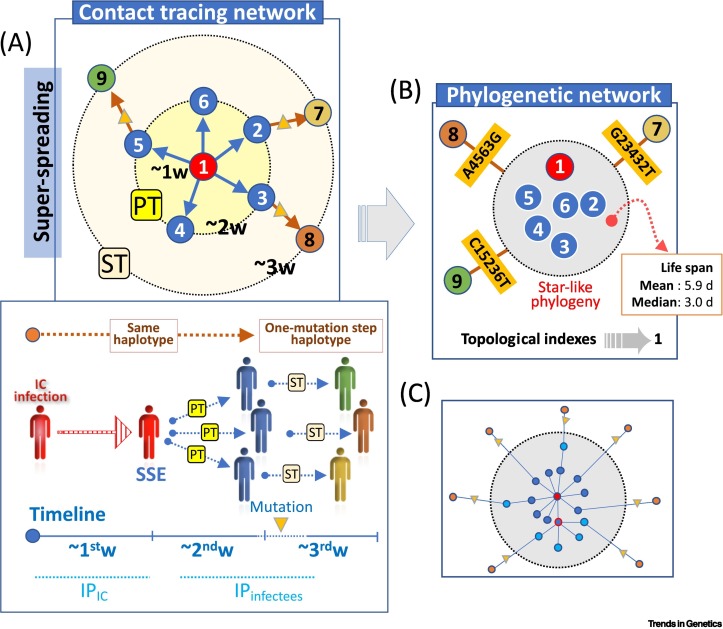
Box 3. Signature of SSEs in databases.
The mean incubation period (IP) of COVID-19 (time between exposure and symptom onset) is ~5 to 6 daysviii. On average, a person who is infected by another (primary transmissions; PT) has a low probability to contribute to a secondary transmission (ST) before elapsing enough time to incubate the virus (latency periodviii). Having evolutionary and transmission rates in mind (see Box 2 in the main text), it can be predicted how a phylogeny would look like in a SSE. A typical contact-tracing network (Figure IA) would begin in an index case (IC) (#1; red circle) that, once infected, will last about a week (w) to infect others (#2–6; dark-blue circles; ‘infectious period’); and about another week for these individuals to transmit the virus to others (#7–9; orange circles). The contact network finds a perfect parallelism in a phylogenetic network (Figure IB): #1–6 (blue circles) would, on average, tend to share the same viral haplotype (favored by narrow transmission bottlenecks and low levels of intrahost variation [50]), while the ones circulating during the third week and originated from ST (#7–9; other colored circles) would differ, on average, by one-step mutation from those circulating among PT. Star-like networks are typical signatures observed in GISAID genomes (see Figure 2B,C in the main text). Overall, if the same haplotype appears in an unusually large number of individuals in the same geographical place and in a short period of ~5 to 6 days, it is likely that several haplotypes have been transmitted by an IC or a few infectors. Topological indexes for these networks support superspreading [5,6,51]. In agreement with all these figures, the lifespan of identical genomes in GISAID has a mean of 5.9 days (median = 3.0 days).The reality might be more complex (Figure IC) due, for example, to interindividual variability in incubation period, transmission bottlenecks, diagnosis, and intervals of infectiousness, such that the root haplotype of a cluster might in fact reflect the overlapping of, for example, more than one superspreader [e.g., the IC and other primary infected individual (red-framed circle with dark-blue background)] and a few STs without mutations differing from the root (light-blue circles) coexisting with other STs differing by one-step mutations (triangles connecting orange circles); the fact that most identical genomes in the root occur in a short time period (deduced from sampling dates and assuming that these dates correlate well with short-term exposure [10]) points to a major role of superspreaders in this root haplotype (dark-blue circles) (Figure IC).
Figure I.
Unifying epidemiology and (phylo)genetics.
Alt-text: Box 3
There are obvious limitations to the procedure of inferring superspreading from a database [5,6]; in short: (i) genomic data alone do not allow the tracking of individual transmissions or, therefore, to estimate how many superspreaders may have contributed to the event (especially in massive social events); (ii) the database is not a random representation of circulating coronaviruses, and different countries contribute disproportionally to the database (see later); (iii) there are variable delays in data deposition, and potential errors in the records (e.g., with genomes and sampling dates [34]); (iv) sequences recorded in GISAID do not represent intrahost variation (but consensus genomes), and so on. However, this method, favored by a relatively high mutation rate (comparable with other RNA viruses) but relatively low transmission rate (Box 3), allows the evaluation of the superspreading phenomenon on a large scale, in which errors in the inferences are compensated by the ‘law of large numbers’ (estimates of evolutionary rates, incubation times, etc. only represent population averages and not interindividual variation). Theoretical expectations (Box 3) and different strands of evidence indicate that the procedure works satisfactorily. For instance, as recorded by Salas et al. [33], two SSEs reported in a skilled nursing facility and a business meeting in Boston [8] left a clear signature in GISAID [33]. In addition, the phylogenetic networks examined in a few of these SSEs [8,11,35] follow the expected pattern.
The typical phylogenetic pattern of SARS-CoV-2 genomes circulating in a population comprises star-like clusters caused by main outbreaks, probably due to a great extent by SSEs; and these clusters are interconnected by genome sequences that reflect the predominant chain of transmission patterns (e.g., Figure 2B and Box 3; Figure S13 in [6]).
The impact of SSEs in VOC dynamics
By searching for identical haplotypes appearing in a geographical region ≥20 times in a period of 5 days, we identified 765 candidates as potentially responsible for important SSEs worldwide (see Table S1 in the supplemental information online; hereafter ‘superspreader haplotypes’ or SSEh). These SSEh represented >93 000 (5.9%) of the database; by also considering the one-step mutations on top of SSEh, the number of genomes increased to >134 600 (7.7% of the database). Since one of the major contributors to GISAID is the UK, it is not surprising that ~31% (n = 237) of the SSEh were observed in this country. A total of 354 out of 765 SSEh (46.3%) fell within the B.1.1.7 definition, and 142 of these appeared in the UK (142/354 = 40.1%) (Figure 3).
Figure 3.
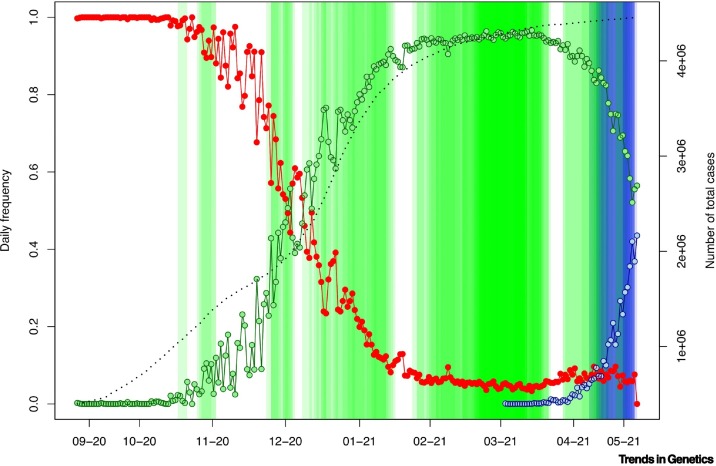
Frequency evolution of B.1.1.7 and B.1.617.2 in the UK and superspreader events (SSEs) occurring during the same period.
Note that the percentage of severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) cases sequenced by the UK increased gradually, especially from January and much more intensely since February 2021i. Therefore, we represent the proportion of B.1.1.7 (green) against the rest of the variants (red) and B.1.617.2 (blue), as well as the positive cases in UK for the same time period, as previously recordedix. SSEs are indicated in vertical green bars (the intensity of the color mirrors overlapping events). The fact that the number of detected SSE candidates increased in mid-March 2021 does not reflect the intensity of the pandemic in UK due to B.1.1.7 (improving from February 2021 onward), but rather the increased coverage of SARS-CoV-2 genome sequences by UK authorities and institutions. The black-broken line represents cumulative positive cases in the UK.
According to epidemiological data, the major outbreak in the UK originated from B.1.1.7 starting at the end of November 2020 onwardi (with a peak on 8 January 2021). The core haplotype of B.1.1.7 emerged for the first-time during mid-September 2020 (TMRCA, September 17, 2020; 95% HPD, 14 September 2020–19 September 2020), and its first appearance in the database corresponded to two genomes from the UK [#601443 (20 September 2020) and #581117 (21 September 2020)] (Figure 1, Figure 2A). However, the large outbreak of B.1.1.7 began at least 2 or 3 months later. Consistently, we detected important SSEh for B.1.1.7 starting in November 2020, become more intense in terms of their presence from December 2020 onward, coinciding with the peak of the pandemic in the UK (Figure 2A). We detected the initial four B.1.1.7 SSEh in UK, with a peak in March 2021 (44% of the events), which does not necessary reflect the epidemiological situation of the country (improving at that time) but which does coincide with the significant increase of sequencing efforts in the UKi, leading to a growing presence of B.1.1.7 in the database (Figure 2A). The large number of SSEs inferred from GISAID could have contributed to the exponential growth experienced by this variant in such a short time period, beyond the improved transmissibility attributed to this VOC.
We did not detect SSEs associated with B.1.351.2 or P.1, probably because these two VOC did not occur strongly in the UK and USA, the major contributors to the database during the period covered by the GISAID database used in the analysis (namely, until 18 May 2021). However, we detected important SSEs related to B.1.617.2 (n = 6), all of which appeared in the UK. The rapid replacement of the B.1.1.7 variant in the UK by the B.1.617.2 variant (first detected on 24 March 2021 [36]; Figure 3) could also be explained by the numerous SSEs occurring at the time of its exponential growth.
Concluding remarks
There are evolutionary processes that might have had a role in the epidemiological success of lineage B.1.1.7 and other VOC, and selective advantage has been invoked as the first choiceiv [24,36., 37., 38.]. However, evaluating the reasons behind the evolutionary advantage and success of VOC is far from simple, and many considerations should be taken into account before reaching definitive conclusions, including the potential role of recombination and the possible intervention of non-human hosts [39], among others. The convergent observation that a given VOC increases its predominance in several countries [36] did not fully consider the epidemiological situation of those countries and social behavior. For instance, B.1.617.2 replaced B.1.1.7 very quickly in the UK and other countries (e.g., Spain) in a more favorable epidemiological situation; for example, the UK Government initiated a national lockdown on 5 January 2021 to control the spread of B.1.1.7 and its incidence fell drastically over the next 2 months (Figure 2A); therefore, B.1.617.2 may have taken advantage of the depression of B.1.1.7 and easily filled in the ecological niche left by it; in such a scenario, genetic drift could have had a major role in helping B.1.617.2 gain territory in the UK. In this sense, the analysis undertaken in the present study also detected a large number of intense SSEs candidates that could have benefited the emergence of B.617.2 (Figure 3).
We contend that superspreading has a key role in the complex algorithm of the pandemic. The intervention of a mini outbreak in the transmission of the virus can change exponentially the dynamics of a SARS-CoV-2 variant in any epidemiological context. There are clear signatures suggesting the existence of numerous SSEs coinciding chronologically with the rise of VOC. At the same time, a large number of important SARS-CoV-2 epidemiological outbreaks have originated from variants not qualified as VOC but catalyzed by superspreading [e.g., worldwide [5]; Spain [6]; Boston [8]; Germany [40]; South Korea [17]; Georgia (USA)] [41]. Therefore, superspreading appears to be an omnipresent phenomenon in the pandemic. In this regard, it is noteworthy that most sequencing efforts are limited to a few countries (e.g., UK and USA have contributed 53.7% of the GISAID database) and continental regions (Europe and North America contribute 62.6% and 28.3% of the total database, respectively), implying that the variants responsible for important regional or even continental outbreaks (other VOC different to B.1.1.7., etc.) have never been characterized or recorded by GISAID.
Overall, this evidence leads to the conclusion that control of the pandemic requires a deep understanding of SSEs and of what makes a person a superspreader (see Outstanding questions). Individual and community prevention measurements that control the emergence of SSEs (adequate lockdown policies, vaccination, road traffic and social movement restrictions, use of face masks, indoor air quality improvement, etc.) could have helped prevent important outbreaks led by VOC, and the pandemic might have been mitigated more efficiently, independent of the circulating SARS-CoV-2 variant of the moment.
Outstanding questions.
What is the real impact of superspreading in the SARS-CoV-2 pandemic compared with other forces?
What are the phenotypic features that make a person a superspreader?
What are the determinants that make a VOC highly transmissible?
Alt-text: Outstanding questions
Acknowledgments
Acknowledgments
We gratefully acknowledge GISAID and hundreds of contributing laboratories for giving us access to the SAR-CoV-2 genome database. This study received support from the following projects: GePEM [Instituto de Salud Carlos III(ISCIII)/PI16/01478/Cofinanciado FEDER], DIAVIR (ISCIII/DTS19/00049/Cofinanciado FEDER; Proyecto de Desarrollo Tecnológico en Salud), Resvi-Omics (ISCIII/PI19/01039/Cofinanciado FEDER), BI-BACVIR [PRIS-3; Agencia de Conocimiento en Salud (ACIS) – Servicio Gallego de Salud (SERGAS) – Xunta de Galicia, Spain)], Programa Traslaciona Covid-19 (ACIS) and Axencia Galega de Innovación (GAIN; IN607B 2020/08 – Xunta de Galicia; Spain) awarded to A.S.; and projects ReSVinext (ISCIII/PI16/01569/Cofinanciado FEDER), Enterogen (ISCIII/PI19/01090/Cofinanciado FEDER), and GAIN (IN845D 2020/23 – Xunta de Galicia; Spain) awarded to F.M-T.
Declaration of interests
None declared by the authors.
Glossary
- Basic reproductive number (R0)
average number of secondary cases that a primary case will generate in a population assuming that nobody is either immune or vaccinated; usually, this value can only be obtained when measures to control the pandemic have been established (effective R or Re). A single mini-SSE involving only 30 primary SARS-CoV-2 infections and a R0 value of ~2.87 [42] might lead to an increase in infected individuals by more than one order of magnitude in only 1 month with respect to a normal spread.
- Mutation rate
generally expressed as the number of substitutions per site per replication cycle.
- Non-synonymous substitution
nucleotide mutation that alters the amino acid sequence of a protein.
- Phylogenetic (or evolutionary) tree
branching diagram showing the evolutionary relationships among species or intraspecific relationships.
- Substitution (evolutionary) rate
nucleotide substitutions per site per time unit (often estimated from phylogenetic trees).
- Superspreader
individual that infects an unusually large number of secondary cases. They usually have a higher viral shedding but the causes of their being a superspreader remain unknown.
- Superspreading event (SSE)
event in which a disproportionally large number of secondary cases relative to R0 occurs.
- Variant of concern (VOC)
genetic variant of the virus for which there is evidence of an increased transmissibility, impact on disease severity in terms of deaths, hospitalizations, diagnostic effectiveness, or effects on immunology generated by vaccination or natural infection, among others.
Footnotes
Supplemental information to this article can be found online at https://doi.org/10.1016/j.tig.2021.09.003.
Resources
iwww.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201iihttps://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/959438/Technical_Briefing_VOC_SH_NJL2_SH2.pdfiiihttps://nextstrain.orgivhttps://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563vhttps://virological.org/t/tracking-the-international-spread-of-sars-cov-2-lineages-b-1-1-7-and-b-1-351-501y-v2/592vihttp://virological.org/t/phylodynamic-analysis-90-genomes-12-feb-2020/356viihttps://nextstrain.org/ncov/globalviiiwww.who.intixhttps://ourworldindata.org/coronavirus/country/united-kingdom?country=GBR~OWID_WRLx https://virological.org/t/recombinant-sars-cov-2-genomes-involving-lineage-b-1-1-7-in-the-uk/658
Supplemental information
Characteristics of the top superspreader haplotype (SSEh) candidates detected in the GISAID database.
References
- 1.Mortimer P.P. Mr N the milker, and Dr Koch's concept of the healthy carrier. Lancet. 1999;353:1354–1356. doi: 10.1016/S0140-6736(98)11315-6. [DOI] [PubMed] [Google Scholar]
- 2.Wong G., et al. MERS, SARS, and ebola: The role of super-spreaders in infectious disease. Cell Host Microbe. 2015;18:398–401. doi: 10.1016/j.chom.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stein R.A. Super-spreaders in infectious diseases. Int. J. Infect. Dis. 2011;15:e510–e513. doi: 10.1016/j.ijid.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau M.S., et al. Spatial and temporal dynamics of superspreading events in the 2014-2015 West Africa Ebola epidemic. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2337–2342. doi: 10.1073/pnas.1614595114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gómez-Carballa A., et al. Mapping genome variation of SARS-CoV-2 worldwide highlights the impact of COVID-19 super-spreaders. Genome Res. 2020;30:1434–1448. doi: 10.1101/gr.266221.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gómez-Carballa A., et al. Phylogeography of SARS-CoV-2 pandemic in Spain: a story of multiple introductions, micro-geographic stratification, founder effects, and super-spreaders. Zool. Res. 2020;41:605–620. doi: 10.24272/j.issn.2095-8137.2020.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adam D.C., et al. Clustering and superspreading potential of SARS-CoV-2 infections in Hong Kong. Nat. Med. 2020;26:1714–1719. doi: 10.1038/s41591-020-1092-0. [DOI] [PubMed] [Google Scholar]
- 8.Lemieux J.E., et al. Phylogenetic analysis of SARS-CoV-2 in Boston highlights the impact of superspreading events. Science. 2020;371 doi: 10.1126/science.abe3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker A., et al. Genetic structure of SARS-CoV-2 reflects clonal superspreading and multiple independent introduction events, North-Rhine Westphalia, Germany, February and March 2020. Euro Surveill. 2020;25:2000746. doi: 10.2807/1560-7917.ES.2020.25.22.2000746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., et al. Secondary attack rate and superspreading events for SARS-CoV-2. Lancet. 2020;395 doi: 10.1016/S0140-6736(20)30462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekizuka T., et al. Haplotype networks of SARS-CoV-2 infections in the Diamond Princess cruise ship outbreak. Proc. Natl. Acad. Sci. U. S. A. 2020;117:20198–20201. doi: 10.1073/pnas.2006824117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sneppen K., et al. Overdispersion in COVID-19 increases the effectiveness of limiting nonrepetitive contacts for transmission control. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2016623118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goyal A., et al. Viral load and contact heterogeneity predict SARS-CoV-2 transmission and super-spreading events. eLife. 2021;10 doi: 10.7554/eLife.63537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei C., et al. A super-spreader of SARS-CoV-2 in incubation period among health-care workers. Respir. Res. 2020;21:327. doi: 10.1186/s12931-020-01592-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng V.C., et al. Nosocomial outbreak of COVID-19 by possible airborne transmission leading to a superspreading event. Clin. Infect Dis. 2021;73:e1356–e1364. doi: 10.1093/cid/ciab313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J., et al. COVID-19 outbreak associated with air conditioning in restaurant, Guangzhou, China, 2020. Emerg. Infect. Dis. 2020;26:1628–1631. doi: 10.3201/eid2607.200764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S., et al. Evaluation of COVID-19 epidemic outbreak caused by temporal contact-increase in South Korea. Int. J. Infect. Dis. 2020;96:454–457. doi: 10.1016/j.ijid.2020.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards D.A., et al. Exhaled aerosol increases with COVID-19 infection, age, and obesity. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2021830118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun K., et al. Transmission heterogeneities, kinetics, and controllability of SARS-CoV-2. Science. 2021;371 doi: 10.1126/science.abe2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goyal A., et al. Early super-spreader events are a likely determinant of novel SARS-CoV-2 variant predominance. medRxiv. 2021 doi: 10.1101/2021.03.23.21254185. Published online March 24, 2021. [DOI] [Google Scholar]
- 21.Kochanczyk M., et al. Super-spreading events initiated the exponential growth phase of COVID-19 with 0 higher than initially estimated. R. Soc. Open Sci. 2020;7:200786. doi: 10.1098/rsos.200786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davies N.G., et al. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plante J.A., et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies N.G., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tegally H., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- 26.Singh J., et al. SARS-CoV-2 variants of concern are emerging in India. Nat. Med. 2021;27:1131–1133. doi: 10.1038/s41591-021-01397-4. [DOI] [PubMed] [Google Scholar]
- 27.Faria N.R., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luan B., et al. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS Lett. 2021;595:1454–1461. doi: 10.1002/1873-3468.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kristofich J., et al. Synonymous mutations make dramatic contributions to fitness when growth is limited by a weak-link enzyme. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nouvellet P., et al. Reduction in mobility and COVID-19 transmission. Nat. Commun. 2021;12:1090. doi: 10.1038/s41467-021-21358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lima L.L., Atman A.P.F. Impact of mobility restriction in COVID-19 superspreading events using agent-based model. PLoS ONE. 2021;16 doi: 10.1371/journal.pone.0248708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glass D.H. European and US lockdowns and second waves during the COVID-19 pandemic. Math. Biosci. 2020;330:108472. doi: 10.1016/j.mbs.2020.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salas A., et al. Superspreading: the engine of the SARS-CoV-2 pandemic. Science. 2021 https://www.science.org/doi/full/10.1126/science.abe3261 Published online March 21, 2021. [Google Scholar]
- 34.Gozashti L., Corbett-Detig R. Shortcomings of SARS-CoV-2 genomic metadata. BMC Res. Notes. 2021;14:189. doi: 10.1186/s13104-021-05605-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballesteros N., et al. Deciphering the introduction and transmission of SARS-CoV-2 in the Colombian Amazon Basin. PLoS Negl. Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell F., et al. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill. 2021;26:2100509. doi: 10.2807/1560-7917.ES.2021.26.24.2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pascarella S., et al. SARS-CoV-2 B.1.617 Indian variants: are electrostatic potential changes responsible for a higher transmission rate? J. Med. Virol. 2021 doi: 10.1002/jmv.27210. Published online July 14, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volz E., et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J., et al. Potential transmission chains of variant B.1.1.7 and co-mutations of SARS-CoV-2. Cell Discov. 2021;7:44. doi: 10.1038/s41421-021-00282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Streeck H., et al. Infection fatality rate of SARS-CoV2 in a super-spreading event in Germany. Nat. Commun. 2020;11:5829. doi: 10.1038/s41467-020-19509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lau M.S.Y., et al. Characterizing superspreading events and age-specific infectiousness of SARS-CoV-2 transmission in Georgia, USA. Proc. Natl. Acad. Sci. U. S. A. 2020;117:22430–22435. doi: 10.1073/pnas.2011802117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Billah M.A., et al. Reproductive number of coronavirus: a systematic review and meta-analysis based on global level evidence. PLoS ONE. 2020;15 doi: 10.1371/journal.pone.0242128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pardo-Seco J., et al. Pitfalls of barcodes in the study of worldwide SARS-CoV-2 variation and phylodynamics. Zool. Res. 2021;42:87–93. doi: 10.24272/j.issn.2095-8137.2020.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drummond A.J., Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bandelt H.-J., et al. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 47.Leigh J.W., Bryant D. POPART: full-feature software for haplotype network construction. Methods Ecol. Evol. 2015;6:1110–1116. [Google Scholar]
- 48.R core Team . R Foundation for Statistical Computing; 2019. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 49.Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lythgoe K.A., et al. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372 doi: 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Colijn C., Gardy J. Phylogenetic tree shapes resolve disease transmission patterns. Evol. Med. Public Health. 2014;2014:96–108. doi: 10.1093/emph/eou018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of the top superspreader haplotype (SSEh) candidates detected in the GISAID database.