Abstract
Cyclase-associated protein 2 (CAP2) has been addressed as a candidate biomarker in various cancer types. Previously, we have shown that CAP2 is expressed during multi-step hepatocarcinogenesis; however, its underlying mechanisms in liver cancer cells are not fully elucidated yet. Here, we demonstrated that endoplasmic reticulum (ER) stress induced CAP2 expression, and which promoted migration and invasion of liver cancer cells. We also found that the ER stress-induced CAP2 expression is mediated through activation of protein kinase C epsilon (PKCε) and the promotor binding of activating transcription factor 2 (ATF2). In addition, we further demonstrated that CAP2 expression promoted epithelial-mesenchymal transition (EMT) through activation of Rac1 and ERK. In conclusion, we suggest that ER stress induces CAP2 expression promoting EMT in liver cancer cells. Our results shed light on the novel functions of CAP2 in the metastatic process of liver cancer cells.
Keywords: activating transcription factor 2, cyclase-associated protein 2, endoplasmic reticulum-stress, epithelial-mesenchymal transition, extracellular signal-regulated kinase, hepatocellular catcinoma, protein kinase C epsilon
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most malignant tumors harboring huge molecular heterogeneity (Woo et al., 2017). Development of HCC is progressed stepwisely from liver cirrhosis, dysplastic nodules, and to early and progressed HCCs (Ojima et al., 2016). Previously, by performing gene expression profiling of the multi-step hepatocarcinogenesis, we identified potential driver genes that drive the multi-step progression of liver cancer (Jee et al., 2019). Among them, Cyclase-associated protein 2 (CAP2) was found as one of candidate drivers which showed stepwise expression during malignant conversion of pre-cancerous dysplastic lesions to early HCC.
CAP2 protein is highly conserved between yeast and mammal and plays a role in mediating the dynamics of actin polymerization by binding to adenylyl cyclase and binding to G-actin (Kosmas et al., 2015). There are two homologs of CAP1 and CAP2; CAP1 is expressed ubiquitously, whereas CAP2 is expressed only in brain, heart, skeletal muscle, and skin (Kosmas et al., 2015). CAP2 has also been suggested to express in various types of cancers including thyroid, kidney, bladder, and liver cancers. In particular, consistent with our finding, CAP2 is expressed in 70%-100% of early HCCs, but only in 5%-10% precancerous lesions (Sakamoto, 2009). CAP2 expression was generally increased as HCC evolves from early to advanced stages (Fu et al., 2015; Ojima et al., 2016; Sakamoto et al., 2008; Shibata et al., 2006). Moreover, CAP2 expression was significantly associated with overall survival and disease-free survival of HCC patients (Fu et al., 2015). It also has been shown that CAP2 could promote invasion of HCC cells (Effendi et al., 2013). These studies and ours consistently support that CAP2 expression play an important role in liver cancers. However, underlying mechanisms of CAP2 expression and its functions in liver cancer cells are not fully elucidated yet. With this concern, in this study, we aimed to investigate the functional roles of CAP2 in liver cancer progression.
In addition, we have suggested that endoplasmic reticulum (ER) is increased in dysplastic nodules of liver, and which may contribute to the oncogene expression that drives malignant conversion of the pre-cancerous lesions (Jee et al., 2019). ER stress causes unfolded protein responses (UPR) that trigger homeostatic recovery or cell death (Madden et al., 2019; Urra et al., 2016). Enhanced UPR apparently mediates the paradoxical microenvironment in cancers, giving rise to aggressive behaviors of cancer cells (Yadav et al., 2014). In this study, we could demonstrate that the ER stress induces CAP2 expression in HCC cells, which may provide new insights on the functions of CAP2 in HCC progression.
MATERIALS AND METHODS
Cells, antibodies, and other reagents
Human liver cancer cells of SNU423 (catalog No. KCLB00423) and Huh7 cells (catalog No. KCLB60104) were purchased from Korean Cell Line Bank (KCLB, Korea) and cultured in DMEM (catalog No. 11965084; Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS) (catalog No. 12483-020; Invitrogen), 100 U/ml penicillin, and 100 µg/ml streptomycin (catalog No. 15140122; Invitrogen). Human liver cell (THLE-2 cell) was purchased from ATCC (catalog No. CRL-2706; ATCC, USA) and cultured in BEGM supplemented with growth factors (catalog No. CC3170; Lonza, USA). Mycoplasma tests were performed regularly with polymerase chain reaction (PCR) analysis (e-Myco; COSMO GENETECH, Korea). Authentication of the cell lines were described in Supplementary Information 1-3.
Anti-ERK (1:1,000, catalog No. 9102), anti-phospho-ERK (T202/Y204) (1:1,000, catalog No. 9101), anti-AKT (1:1,000, catalog No. 9272), anti-phospho-AKT (S473) (1:1,000, catalog No. 4060), anti-phospho-SAPK/JNK (T183/Y185) (1:1,000, catalog No. 4668), anti-p38 (1:1,000, catalog No. 9212), anti-phopho-p38 (T180/Y182) (1:1,000, catalog No. 4511), anti-PKCε (1:1,000, catalog No. 2683), anti-phospho-threonine (1:1,000, catalog No. 9386), anti-vimentin (1:1,000, catalog No. 5741), anti-E-cadherin (1:1,000, catalog No. 3195), and anti-ATF2 (1:1,000, catalog No. 35031), anti-mouse IgG, HRP-linked antibody (1:1,000, catalog No. 7076), anti-rabbit IgG, HRP-linked antibody (1:1,000, catalog No. 7074), and Dylight594 Phalloidin (1:20, catalog No. 12877) were purchased from Cell Signaling Biotechnology (USA). Anti-phospho-PKCε (S729) (1:1,000, catalog No. ab63387), anti-N-cadherin (1:1,000, catalog No. ab76057), and anti-snail (1:1,000, catalog No. ab180714) antibodies were from Abcam (UK), anti-CAP2 (1:1,000, catalog No. sc-100916), anti-SAPK/JNK (1:1,000, catalog No. sc-7345), and anti-β-actin (1:1,000, catalog No. sc-47778) antibodies were from Santa Cruz Biotechnology (USA), and anti-Rac1 antibody (1:1,000, catalog No. PA1-091), donkey anti-mouse IgG (H+L) antibody, alexa fluor 488 (1:500, catalog No. R37114), and donkey anti-rabbit IgG (H+L) antibody, alexa fluor 594 (1:500, catalog No. R37119) was from Invitrogen. SB203580 (catalog No. S8307), GF109203X (catalog No. G2911), thapsigargin (catalog No. T9033), tunicamycin (catalog No. T7765), SP600125 (catalog No. S5567), and dithiothreitol (DTT) (catalog No. D9779) were from Sigma-Aldrich (USA), U0126 (catalog No. 662005) was from MERK (Germany), and NSC23766 (catalog No. 2161) was from TOCRIS (USA).
Gene expression constructs and lentiviral vector transfection
Lentiviral constructs expressing CAP2 shRNAs were purchased from Dharmacon (USA). The CAP2 (NM_001363533.1), Rac1-wild type (Rac1-WT, NM_006908.5)-HA cDNA constructs were cloned into pCDH-CMV-MCS-EF1-Puro using Infusion HD cloning kit (catalog No. 011614; Clontech, Japan), a lentiviral vector for cDNA expression (catalog No. CD510-1; System Biosciences, USA). Rac1-Q61L-HA and Rac1-N17-HA were constructed by site-directed mutagenesis using Rac1-WT as a template. The constructs were used to make stable cell lines as described (Yoon et al., 2019). The targets of CAP2 shRNAs and the PCR primers were summarized in Supplementary Table S1 and S2. The human CAP2 promoter sequence was amplified from SNU423 genomic DNA. The 2.5 kb-length clone (–2,500/+100 bp) and truncated derivatives (–2,000/+100, –1,500/+100, –1,000/+100, –500/+100) were obtained using the following. All promoter constructs were cloned into the pGL3-basic luciferase reporter (catalog No. E1751; Promega, USA) vector using Infusion HD cloning kit (Clontech) and verified by DNA sequencing analysis. The candidate ATF2 binding sequences (-2185ATGACGTTAA-2176) of pGL3-CAP2 (–2.5 kb) were mutated using site-directed mutagenesis and verified by DNA sequencing. The putative transcription factor binding sites were analyzed by the PROMO computer program (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) (Farre et al., 2003).
siRNA-mediated knockdown
Knockdown experiments for ATF2 or PKCε were performed using 100 nM of negative control siRNA (siNeg, catalog No. sc-37007; Santa Cruz Biotechnology), siATF2 (catalog No. sc-29205; Santa Cruz Biotechnology) or siPKCε (catalog No. sc-36251; Santa Cruz Biotechnology). The siRNAs were transfected into liver cancer cells using Lipofectamine 3000 (catalog No. L3000015; Invitrogen). After 48 h of transfection, the cells were harvested for further study.
Luciferase reporter assay
Huh7 cells were plated in 6-well plates and transfected on the following day using Lipofectamine 3000 (Invitrogen). To investigate the CAP2 promoter region, the cells were co-transfected with pGL3-basic vector (Promega) or various reporter constructs with a deletion or point mutant of CAP2 promoter, and pTurbo-GFP plasmid (catalog No. SHC003; Sigma-Aldrich) for normalization of transfection efficiency. After 24 h of transfection, the cells were treated with thapsigargin (100 nM, catalog No. T9033; Sigma-Aldrich) for 24 h and analyzed the luciferase expression using real-time reverse transcription-PCR (qRT-PCR). The relative luciferase expressions were calculated as a fold of induction compared with the pGL3-basic vector control or an unstimulated vector control. All luciferase expression data were normalized with that of GFP mRNA.
Chromatin immunoprecipitation (ChIP) assay
Huh7 cells were grown overnight in 150-mm dishes to ~70% confluency, and the cells were treated with or without 100 nM thapsigargin (Sigma-Aldrich) for 24 h. At this time, cells were cross-linked with formaldehyde, harvested, and chromatin immunoprecipitations were performed. ChIP assay was performed using a SimpleChIP Enzymatic Chromatin IP kit (catalog No. 9003) from Cell Signaling Technology.
Immunoprecipitation was performed using a polyclonal antibody against ATF2 (catalog No. 35031; Cell Signaling Technology). As a positive control, an antibody of Histone H3 (catalog No. 4620; Cell Signaling Technology) was used to precipitate the histone-bound RPL30 promoter (catalog No. 7014; Cell Signaling Technology). The recovered DNA was analyzed by PCR with primers flanking the putative transcription factor binding sites as indicated. As a negative control, we used normal rabbit IgG (catalog No. 2729; Cell Signaling Technology). The primers used for PCR of the ChIP fragents are shown in Supplementary Table S2.
Rac1 GTPase activity assay
Rac1 activity was measured by using a Rac Activation Assays kit (catalog No. STA-401-1; Cell Biolabs, USA). Briefly, the cell lysates were incubated with agarose beads coupled to the p21-binding domain (PBD) of p21-activated protein kinase (PAK). The amount of bound form Rac1 was measured by western blot analysis using an anti-Rac1 antibody.
Real-time qPCR
Cells were harvested and total RNAs were isolated using an RNeasy kit (catalog No. 74104; Qiagen, Netherlands). The PrimeScript RT kit (catalog No. RR037A; Takara, Japan) was used to reverse transcribe the 1 μg total RNA into cDNA. PCR was done using a CFX96 Real Time PCR Detection System (Bio-Rad) with IQ SYBR Green Supermix (catalog No. BR170-8882; Bio-Rad). Analysis of each sample was performed at least three times for each experiment, and the data were presented as the average values of 2-ΔΔCT ± SD. The sequences of primers are shown in Supplementary Table S2.
Immunoprecipitation and Western blots
For immunoprecipitation, 500 µg of proteins were incubated overnight with anti-ATF2 antibody (catalog No. 35031, 1:100; Cell Signaling Technology), subsequently with protein A/G agarose beads (catalog No. sc-2003; Santa Cruz Biotechnology) for 1 h. The beads were washed five times from which immuno-precipitates were extracted with 2 × SDS sample buffer.
For preparing total cell lysates, cells were lysed in lysis buffer (20 mM Tris-HCl [pH 8.0], 1% Triton X-100, 2 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride), incubated on ice for 20 min, and centrifuged for 20 min to remove cell debris. A total of 20 μg of whole-cell lysate was used in SDS-polyacrylamide gel electrophoresis. The proteins were then electro-transferred to a nitrocellulose membrane and incubated overnight with antibodies at 4°C. Then, the membranes were incubated with peroxidase-conjugated secondary antibodies (anti-mouse IgG, HRP-linked antibody, catalog No. 7076, 1:1,000 [Cell Signaling Technology]; anti-rabbit IgG, HRP-linked antibody, catalog No. 7074, 1:1,000 [Cell Signaling Technology]) for 1 h at room temperature, and the signal was detected using an enhanced chemiluminescence detection kit (catalog No. 34580; PIERCE, USA).
Confocal microscopy
To demonstrate the localization of proteins, cells were grown on Lab-Tek four-well glass chamber slides (catalog No. C6807; NUNC, USA). After 24 h incubation, cells were fixed and permeabilized with cold methanol for 5 min, washed with phosphate-buffered saline (PBS), and incubated with primary antibody (1:100) and secondary antibody conjugates (1:500). Images were collected on a laser scanning confocal microscope LSM710 (Carl Zeiss, Germany) equipped with argon (488 nm) and krypton (568 nm) lasers, using a 40 × water immersion objective lens. Images were processed with ZEN 2009 light edition (Carl Zeiss).
Cell proliferation
Cells (5 × 103) were split into 96-well plates and incubated in media containing 10% FBS for 48 h. Cell proliferation was measured by WST-1 assay (catalog No. 05015944001; Roche, Korea). Each experiment was performed in four replicates at least three times.
Cell migration/invasion assay
Cell migration/invasion assay were performed in 24-well modified Boyden chamber (8-μm pore size, catalog No. 3422 Costar; Corning Life Sciences, USA). For invasion assay, the Transwell filter inserts were coated with collagen type I (catalog No. A10644-01; Invitrogen). In the migration assay, 1 × 105 cells in 0.2 ml of serum-free DMEM were seeded into the upper chamber insert. The lower chamber was filled with 0.6 ml DMEM/10% FBS. After 6 h, cells at the membrane undersurface were fixed, stained. The images were captured by light microscope (magnification, ×5). For spectrophotometric analysis, the crystal violet-stained cells were solubilized with 200 μl 10% acetic acid (v/v, catalog No. 414; Duksan Pure Chemicals, Korea) for 15 min. The absorbance at 570 nm of the extracts (50 µl) was measured with by a spectrophotometer (96-well plate reader).
Wound healing assay
Cell motility was measured by wound healing assay. Cells (8 × 105) were split into 60 mm dishes and incubated in media containing 10% FBS for 24 h. Cells were cultured until a monolayer form, then scratched with 200 µl tip. The cells were changed to media containing 1% FBS and incubated for 24 h or 48 h. Images were captured using bright field and 4× magnification. The wound width was measured as the average distance between edges of the gap. The scale bar was equivalent in all images and all experiments were performed at least three times for reproducible results.
Statistical analysis
The data were presented as mean ± SEM. Statistical analysis was performed using the SigmaPlot 12.5 software. Differences were assessed by paired Student’s t-test. All the data were derived from at least three independent experiments.
RESULTS
CAP2 induces migration and invasion but not proliferation of liver cancer cells
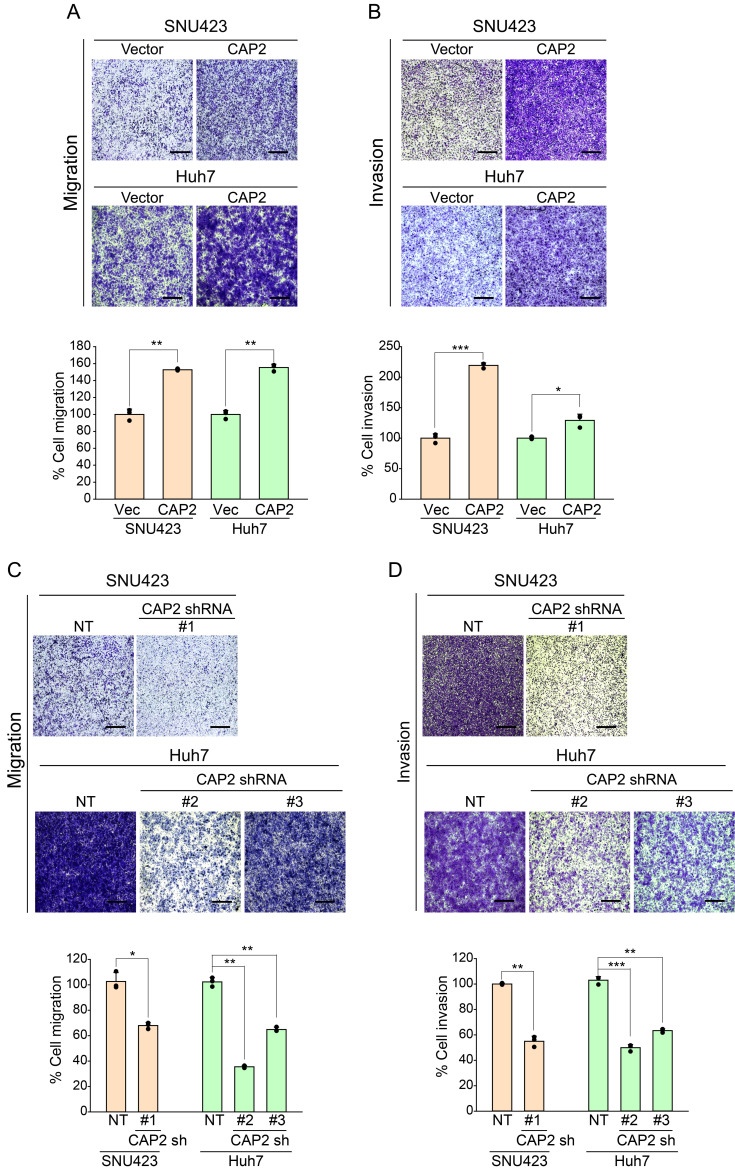
We established the CAP2 overexpression systems using SNU423 (SNU423-CAP2) and Huh7 cells (Huh7-CAP2) (Supplementary Figs. S1A and S1B). The CAP2-overexpressing cells showed higher migration and invasion properties than the Control vector-cells (SNU423-Vector and Huh7-Vector; Fig. 1A and 1B, Supplementary Fig. S2). In addition, we could demonstrate that the shRNA-mediated knockdown of CAP2 (SNU423-CAP2 sh and Huh7-CAP2 sh) significantly suppressed the migration and invasion of liver cancer cells (Fig. 1C and 1D, Supplementary Figs. S1C and S2). However, CAP2 expression did not affect the liver cancer cell proliferation, indicating that CAP2 functions to promote metastasis but not proliferation of the liver cancer cells (Supplementary Fig. S3).
Fig. 1. CAP2 increases migration and invasion of liver cancer cells.
Migration (A and C) and invasion (B and D) were measured in Transwell migration/invasion assays. Migrated/invaded cells were fixed with 10% formaldehyde and stained with 1% crystal violet. NT, non-targeting control. Images were captured under a light microscope (magnification, ×5). Scale bars = 500 μm. The data are presented as mean ± SEM for triplicate determinations. *P < 0.05, **P < 0.01, and ***P < 0.001.
ER stress induces CAP2 expression through promoter binding ATF2
As we have predicted that CAP2 expression is induced by ER stress from the transcriptome data analyses, we examined the effects of ER stress modulators on CAP2 expression. We could demonstrate that the treatment ER stress inducers such as thapsigargin, tunicamycin or DTT can induce the expression of CAP2 mRNAs and proteins (Fig. 2A and 2B). Induction of ER stress was confirmed by examining the expression of an ER stress marker GRP78 (Supplementary Fig. S4). We also demonstrated that the thapsigargin treatment could promote invasion of the cancer cells, and which was abolished by knockdown of CAP2 expression (Supplementary Fig. S5). These findings suggest that CAP2 plays a critical role in the ER stress-mediated invasion of liver cancer cells.
Fig. 2. ER stress induces CAP2 expression via promoter binding of ATF2.
(A) SNU423 and Huh7 cells are treated with/without thapsigargin (Thap; 50 nM and 100 nM, respectively), tunicamycin (Tuni; 50 ng/ml), or DTT (1 mM) for 24 h. Con, control. Expression of CAP2 mRNA is measured by qRT-PCR and normalized to β-actin mRNA. Data represent mean ± SEM of 4 independent experiments. (B) SNU423 and Huh7 cells are treated with thapsigargin (50 nM or 100 nM, respectively) for indicated times. Cell lysates are examined by western blotting with the indicated antibodies. CAP2 is indicated by asterisks (*). (C) Schematic representation of the CAP2 luciferase reporters. ChIP primer sets are shown. (D and E) Huh7 cells are transiently co-transfected with pTurbo-GFP and deleted constructs (D) or ATF2 binding site mutated construct (ATF2-Mut) (E) of the CAP2 promoter. After treatment with 100 nM thapsigargin for 24 h, total RNAs are extracted. Luciferase expression is monitored by qRT-PCR; Luciferase expression levels are normalized by that of GFP mRNA (right). Data represent mean ± SEM of the three independent experiments. (F) Cross-linked chromatin preparation from control and thapsigargin (100 nM, 24 h) treated cells are immunoprecipitated with anti-ATF2, anti-Histone H3 (H3), or normal rabbit IgG (IgG) antibodies. The ATF2 binding sites on the immunoprecipitated DNA are determined by RT-PCR using their corresponding primers. Amplificons of the input chromatin (input) prior to immunoprecipitation were served as controls for each chromatin extraction and PCR amplification. Chromatin immunoprecipitation using a Histone H3 antibody is served as a positive control (P.C) and a non-specific antibody (normal rabbit IgG) is served as a negative control (N.C). (G and H) Huh7 cells are transiently transfected with siNegative control (siNeg) or siATF2. After 24 h, thapsigargin (100 nM) is treated for 24 h and the CAP2 expression levels are measured by qRT-PCR (G) and western blotting (H). CAP2 is indicated by asterisk (*). The data represent mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01, and ***P < 0.001.
In addition, we compared the expression of CAP2 in normal liver cells (THLE-2 cell) and HCC cells (SNU423 and Huh7 cells), which revealed no significant difference between them. However, unlike the HCC cells, THLE-2 cells did not induce CAP2 expression by thapsigargin treatment (Supplementary Fig. S6), which may indicate the cancer cell-specific function of CAP2.
To further investigate the underlying molecular mechanisms of the ER stress-mediated CAP2 expression, we cloned the 5`-flanking region of the CAP2 gene using the template genomic DNA of SNU423. The sequence of this 5`-flanking region encompassed residues –2.5 kb to +100 bp relative to the putative transcription start site. The promoter activity of these regions was measured by generating a set of serial deletion constructs by fusing the truncated forms of the 5`-flanking region to the 5`-end of a luciferase reporter gene in the pGL3-basic vector (Fig. 2C). Huh7 cells were transiently transfected with these reporter constructs and then treated with 100 nM thapsigargin. We found that the construct containing the –2.5 kb to +100 bp fragment showed the highest promoter activity under ER stress condition. Deletion of the 5`-region could abolish the promoter activity (Fig. 2D). These results suggest that the –2.5 kb to –2.0 kb region contains the binding site for cis-responsive element which is required for the CAP2 expression by ER stress.
Next, we computationally predicted the binding of ATF2 (activating transcription factor 2) to this promotor site by using PROMO software. To validate this prediction result, we performed a site-directed mutagenesis experiment, which revealed that the mutations at the ATF2 binding site significantly reduced the promoter activity (Fig. 2E). In addition, we further confirmed the binding of ATF2 with CAP2 promoter region by performing a chromatin immunoprecipitation (ChIP) experiment. We could observe the enriched immunoprecipitated chromatins in the PCR products using CAP2 promoter-specific primers (ChIP-1 primer and ChIP-2 primer). Moreover, thapsigargin treatment could enhance the binding of ATF2 and CAP2 promoter (Fig. 2F). As a negative control, PCR reactions were performed with the primers (ChIP-2 primers) for the 2.5 kb downstream region of the ATF2 binding site of CAP2, which showed no immunoprecipitation. We also demonstrated that knockdown of ATF2 suppressed the thapsigargin-induced CAP2 expression in liver cancer cells (Supplementary Fig. S7, Fig. 2G and 2H). Taken together these results, we suggest that ER stress-induced CAP2 expression is mediated through the promoter binding of ATF2.
PKCε is involved in ER stress-mediated ATF2 activation
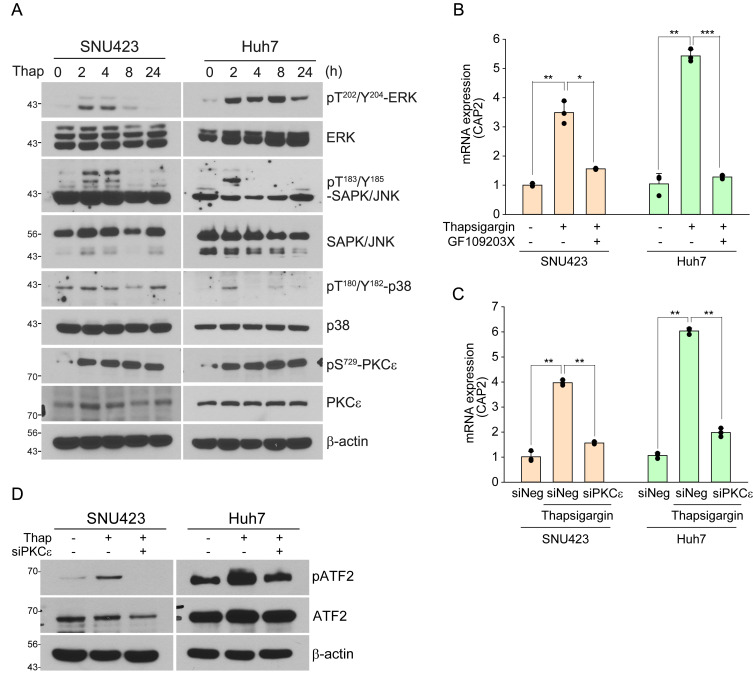
ATF2 is activated by several stress-activated protein kinases such as ERK1, JNK, p38, and protein kinase epsilon (PKCε) in response to a variety of stimuli (Lau and Ronai, 2012). Thus, we examined these protein kinases in the ER stress-induced liver cancer cells, which revealed that the thapsigargin treatment could activate ERK, JNK, p38, and PKCε (Fig. 3A). We also evaluated the effects of the inhibitors of stress-activated protein kinases. The PKCε inhibitor GF109203X reduced stress-induced CAP2 transcription but did not the other inhibitors of ERK1 (U0126, 20 μM), JNK (SP600125, 25 μM), and p38 (SB203580, 10 μM) (Fig. 3B, Supplementary Fig. S8). These results may indicate that only PKCε is involved in the ER stress-mediated ATF2 activation. We further confirmed this finding by evaluating the effect of siRNA-mediated knockdown of PKCε (siPKCε), which could suppress the thapsigargin-mediated CAP2 expression (Supplementary Fig. S9, Fig. 3C). Treatment of siPKCε could also suppress the ATF2 activation by thapsigargin treatment (Fig. 3D). Thus, we suggest that PKCε is involved in ER stress-mediated ATF2 activation.
Fig. 3. PKCε is involved in the ER stress-induced ATF2 activation.
(A) SNU423 and Huh7 cells are treated with thapsigargin (Thap; 50 nM and 100 nM, respectively) for the indicated times and cell lysates are examined by western blotting with the indicated antibodies. (B) SNU423 and Huh7 cells are treated with thapsigargin (50 nM and 100 nM, respectively) alone or with thapsigargin plus the PKCε inhibitor GF109203X (GF, 20 μM) for 24 h. Relative expression of CAP2 mRNA is measured by qRT-PCR. Data represent mean ± SEM of 3 independent experiments. (C and D) SNU423 and Huh7 cells are transiently transfected with siNegative control (siNeg) or siPKC ε and after 24 h, thapsigargin (100 nM) is treated for 24 h. CAP2 expression is measured by qRT-PCR. Data represent mean ± SEM of three independent experiments (C). ATF2 activation is monitored by western blotting with the indicated antibodies (D). *P < 0.05, **P < 0.01, and ***P < 0.001.
CAP2 induces EMT in liver cancer cells via Rac1-ERK activation
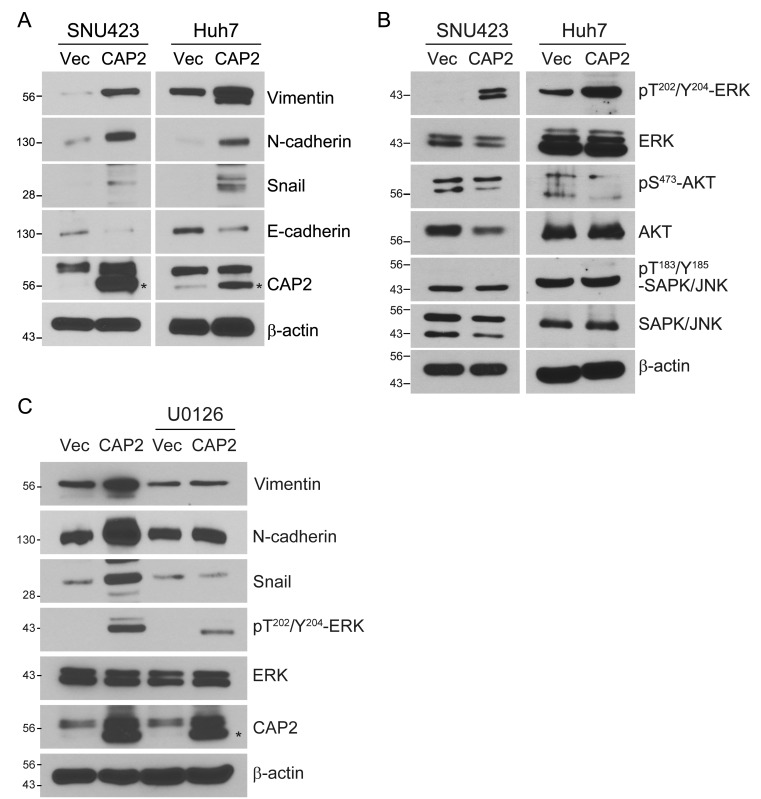
As EMT has been addressed to induce the migration and invasion of cancer cells, we evaluated whether CAP2 expression can induce EMT by examining the expression of the EMT markers such as the suppression of E-cadherin and cytokeratin and the expression of vimentin, N-cadherin, twist, snail, and slug (Ahmed et al., 2020; Zeisberg and Neilson, 2009). We could observe that the CAP2-overexpressing cells induced EMT, showing higher expression of vimentin, N-cadherin, and snail, but lower expression of E-cadherin (Fig. 4A). Moreover, we also found that the CAP2-overexpressing cells enhanced the expression of the active form ERK (Fig. 4B). Treatment of an ERK inhibitor (U0126, 20 μM) suppressed the expression levels of vimentin, N-cadherin, and snail (Fig. 4C). These results strongly suggest that CAP2 can induce EMT in liver cancer cells, and which is mediated by activation of ERK.
Fig. 4. CAP2 induces EMT in liver cancer cells via ERK activation.
(A and B) SNU423-Vector, SNU423-CAP2, Huh7-Vector, and Huh7-CAP2 cells are subjected to Western blotting with the indicated antibodies. (C) SNU423-Vector and SNU423-CAP2 cells are treated for 24 h with or without the ERK inhibitor U0126 (20 μM), followed by Western blotting with the indicated antibodies. CAP2 is indicated by asterisks (*).
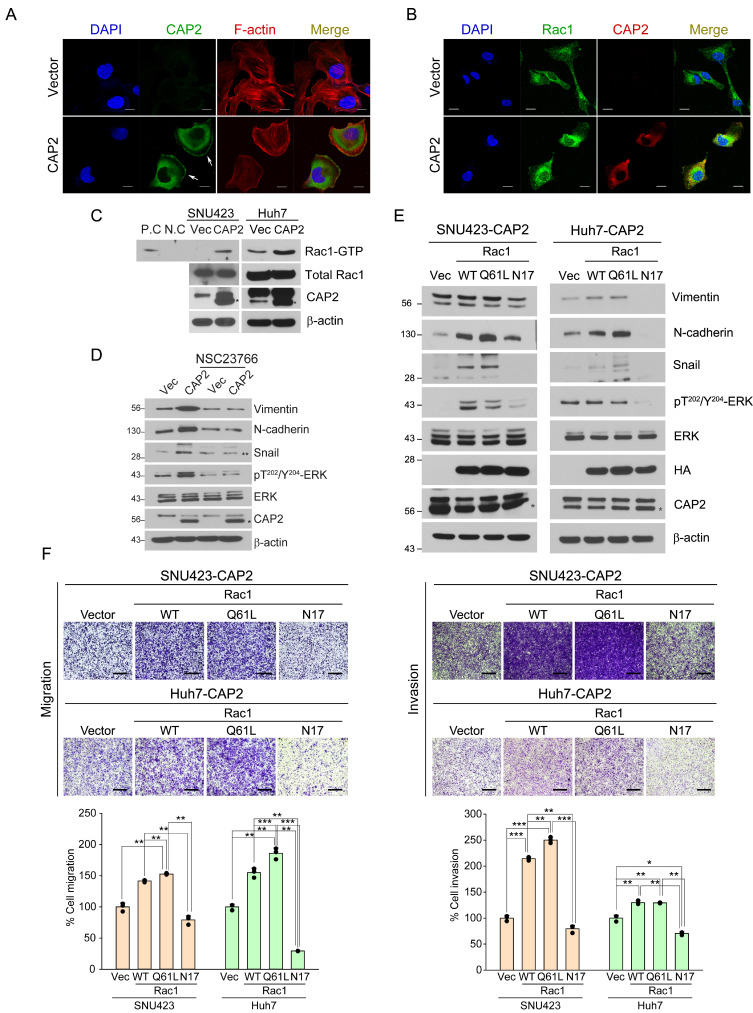
In addition, CAP2 has been addressed to play an important role in actin dynamics (Kosmas et al., 2015). Indeed, in the CAP2-overexpressing cells, we could observe the localized expression of CAP2 protein in the membrane lamellipodia and ruffles, which are the characteristic feature of the actively migrating cells with EMT (Fig. 5A). Moreover, Rac1 is suggested to regulate these membrane features by converting F-actin filaments into G-actin monomers, and which is mediated by activation of p21-activated kinase (PAK)-cofilin signaling pathway (Bid et al., 2013). In fact, Rac1-ERK pathway has been suggested to play important roles in cell migration and invasion (Ebi et al., 2013; Jiang et al., 2014; Lin et al., 2018; Pang et al., 2020). With this concern, we could observe that the formation of F-actin filaments was decreased by the overexpression of CAP2 in liver cancer cells (Fig. 5A). Moreover, CAP2 and Rac1 proteins were co-localized at membrane lamellipodia and ruffles, which may imply the involvement of Rac1 in the CAP2-mediated EMT (Fig. 5B). We could validate the interaction of CAP2 and Rac1 by a pull-down experiment using the GST linked to the Rac1 effector PAK (Fig. 5C). Also, we could demonstrate that the treatment of a Rac1 inhibitor (NSC23766, 50 μM) suppressed the ERK activity and the expression of EMT biomarkers such as vimentin, N-cadherin, and snail (Fig. 5D). We further validated these findings by generating the CAP2-expresing cells with different forms of Rac1, i.e., Rac1 wild-type HA (Rac1-WT), dominant-positive mutant Rac1-Q61L-HA (Rac1-Q61L), dominant-negative mutant Rac1-N17-HA (Rac1-N17), or control vector (Vector) (Fig. 5E). As expected, the CAP2-expressing cells with Rac1-WT and Rac1-Q61L exhibited the increased protein levels of vimentin, N-cadherin, and snail. By contrast, the CAP2-expressing cells with Rac1-N17 exhibited the decreased levels of the ERK activity as well as those EMT proteins.
Fig. 5. Rac1 is involved in CAP2-mediated ERK activation and EMT.
(A and B) SNU423-Vector and SNU423-CAP2 cells are stained with the indicated fluorescent antibodies. Nuclei are counter-stained with DAPI. Images are captured under a laser scanning confocal microscope (×40). White arrows indicate membrane ruffles. Scale bars = 10 μm. (C) Total cell lysates prepared from the indicated cells are incubated with agarose beads coupled to PAK PBD. Bound Rac1 is detected with Western blotting using a Rac1 Ab (upper panel). The amount of total Rac1 is also measured (lower panel). P.C, positive control; N.C, negative control. Asterisks (*) indicate CAP2. (D) SNU423-Vector and SNU423-CAP2 cells were treated for 24 h with NSC23766 (50 μM) and subjected to western blotting with the indicated antibodies. Asterisk (*) indicates CAP2, double asterisk (**) indicates Snail. (E) SNU423-CAP2 and Huh7-CAP2 cells stably expressing Vector (Vec), Rac1-WT, Rac1-Q61L, or Rac1-N17 are subjected to Western blotting with the indicated antibodies. Asterisks (*) indicate CAP2. (F) Migration and invasion of the indicated cells were measured in Transwell assays. Migrated/invaded cells are fixed with 10% formaldehyde and stained with 1% crystal violet. WT, wild type. Images are captured under a light microscope (magnification, ×5). Scale bars = 500 μm. Data are presented as mean ± SEM for triplicate experiments. *P < 0.05, **P < 0.01, and ***P < 0.001.
Furthermore, the Rac1-WT or the Rac1-Q61L-expressing cells showed the increased migration and invasion, whereas the Rac1-N17-expressing cells showed the reduced migration and invasion compared to those of the Vector cells (Fig. 5F). Treatment of an ERK inhibitor U0126 (20 μM) did not affect the ER stress-induced CAP2 expression, indicating that ERK is a downstream effector of CAP2 (Supplementary Fig. S10). Taken together these results, we suggest that CAP2 induces EMT via Rac1 and ERK activation.
DISCUSSION
In this study, we demonstrated that ER stress induces CAP2 expression, and which facilitated EMT promoting migration and invasion of liver cancer cells. We also demonstrated that PKCε-ATF2 signaling is involved in ER stress-induced CAP2 expression.
Cancer cells can survive in micro-environmental conditions that cause ER-stress. The ability to tolerate constant ER stress improves cancer cell survival, angiogenesis, metastatic ability, drug resistance, and immune suppression (Cubillos-Ruiz et al., 2017). ER stress promotes UPR that controls the balance between cell survival and cell death, playing critical roles in cancer cell adaptation to stress and cancer development (Sisinni et al., 2019; Wang and Kaufman, 2014). We found that CAP2 is one of downstream effector molecules that respond to ER stress in liver cancer cells. Moreover, we successfully demonstrated that the CAP2 expression was localized in the lamellipodia and ruffles of liver cancer cells indicating EMT (see Figs. 2 and 5A). We also suggested that Rac1-ERK signaling is involved in the CAP2-mediated EMT. Rac1 is a GTP-binding protein which has been known to regulate actin cytoskeleton reorganization to form cell surface extensions (lamellipodia/ruffles) through ERK activation (Bright et al., 2018; De et al., 2019; Ebi et al., 2013; Jiang et al., 2014; Lin et al., 2018; Pang et al., 2020). ERKs act as critical regulators of cell proliferation by promoting cell cycle progression. However, in some systems, hyperactivation of ERKs by the expression of the activated forms of Ras, Raf, or MEK1 causes cell cycle arrest, promoting mesenchymal transition with increased cell migration and invasion (Meloche and Pouyssegur, 2007; Murphy and Blenis, 2006; Shin et al., 2019). Consistently, we could demonstrate that the activation of CAP2 induced EMT but not proliferation of liver cancer cells, and which was mediated through Rac1 and ERK activation (see Figs. 4 and 5). These results may suggest that the metastasis of cancer cells might be controlled by stage-specific fine modulation of ERK activity, although further mechanism studies are required.
In conclusion, although our study was limited to in vitro experiments, we could successfully demonstrate that ER stress-mediated CAP2 expression plays a critical role in the metastatic process of liver cancer cells. We suggest that targeting CAP2 and its downstream pathways can be a new therapeutic strategy for treatment of HCC patients.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by grants from the National Research Foundation of Korea (NRF), funded by the Korean government (MSIP) (NRF-2017R1E1A1A01074733, NRF-2017M3A9B6061509, NRF-2017M3C9A6047620, NRF-2019R1A5A2026045, and NRF-2019R1I1A1A01057206).
Footnotes
AUTHOR CONTRIBUTIONS
S.Y. performed experiments and wrote the manuscript. B.S. performed experiments. H.G.W. contributed to overall study design, wrote the manuscript, and directed the study.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ahmed M.B., Islam S.U., Sonn J.K., Lee Y.S. PRP4 kinase domain loss nullifies drug resistance and epithelial-mesenchymal transition in human colorectal carcinoma cells. Mol. Cells. 2020;43:662–670. doi: 10.14348/molcells.2020.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bid H.K., Roberts R.D., Manchanda P.K., Houghton P.J. RAC1: an emerging therapeutic option for targeting cancer angiogenesis and metastasis. Mol. Cancer Ther. 2013;12:1925–1934. doi: 10.1158/1535-7163.MCT-13-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright M.D., Clarke P.A., Workman P., Davies F.E. Oncogenic RAC1 and NRAS drive resistance to endoplasmic reticulum stress through MEK/ERK signalling. Cell. Signal. 2018;44:127–137. doi: 10.1016/j.cellsig.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz J.R., Bettigole S.E., Glimcher L.H. Tumorigenic and immunosuppressive effects of endoplasmic reticulum stress in cancer. Cell. 2017;168:692–706. doi: 10.1016/j.cell.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De P., Aske J.C., Dey N. RAC1 takes the lead in solid tumors. Cells. 2019;8:382. doi: 10.3390/cells8050382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebi H., Costa C., Faber A.C., Nishtala M., Kotani H., Juric D., Della Pelle P., Song Y., Yano S., Mino-Kenudson M., et al. PI3K regulates MEK/ERK signaling in breast cancer via the Rac-GEF, P-Rex1. Proc. Natl. Acad. Sci. U. S. A. 2013;110:21124–21129. doi: 10.1073/pnas.1314124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effendi K., Yamazaki K., Mori T., Masugi Y., Makino S., Sakamoto M. Involvement of hepatocellular carcinoma biomarker, cyclase-associated protein 2 in zebrafish body development and cancer progression. Exp. Cell Res. 2013;319:35–44. doi: 10.1016/j.yexcr.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Farre D., Roset R., Huerta M., Adsuara J.E., Rosello L., Alba M.M., Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Li M., Wu D.C., Liu L.L., Chen S.L., Yun J.P. Increased expression of CAP2 indicates poor prognosis in hepatocellular carcinoma. Transl. Oncol. 2015;8:400–406. doi: 10.1016/j.tranon.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee B.A., Choi J.H., Rhee H., Yoon S., Kwon S.M., Nahm J.H., Yoo J.E., Jeon Y., Choi G.H., Woo H.G., et al. Dynamics of genomic, epigenomic, and transcriptomic aberrations during stepwise hepatocarcinogenesis. Cancer Res. 2019;79:5500–5512. doi: 10.1158/0008-5472.CAN-19-0991. [DOI] [PubMed] [Google Scholar]
- Jiang Q.H., Wang A.X., Chen Y. Radixin enhances colon cancer cell invasion by increasing MMP-7 production via Rac1-ERK pathway. ScientificWorldJournal. 2014;2014:340271. doi: 10.1155/2014/340271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmas K., Eskandarnaz A., Khorsandi A.B., Kumar A., Ranjan R., Eming S.A., Noegel A.A., Peche V.S. CAP2 is a regulator of the actin cytoskeleton and its absence changes infiltration of inflammatory cells and contraction of wounds. Eur. J. Cell Biol. 2015;94:32–45. doi: 10.1016/j.ejcb.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Lau E., Ronai Z.A. ATF2 - at the crossroad of nuclear and cytosolic functions. J. Cell Sci. 2012;125:2815–2824. doi: 10.1242/jcs.095000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C.H., Shih C.H., Lin Y.C., Yang Y.L., Chen B.C. MEKK1, JNK, and SMAD3 mediate CXCL12-stimulated connective tissue growth factor expression in human lung fibroblasts. J. Biomed. Sci. 2018;25:19. doi: 10.1186/s12929-018-0421-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden E., Logue S.E., Healy S.J., Manie S., Samali A. The role of the unfolded protein response in cancer progression: from oncogenesis to chemoresistance. Biol. Cell. 2019;111:1–17. doi: 10.1111/boc.201800050. [DOI] [PubMed] [Google Scholar]
- Meloche S., Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26:3227–3239. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- Murphy L.O., Blenis J. MAPK signal specificity: the right place at the right time. Trends Biochem. Sci. 2006;31:268–275. doi: 10.1016/j.tibs.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Ojima H., Masugi Y., Tsujikawa H., Emoto K., Fujii-Nishimura Y., Hatano M., Kawaida M., Itano O., Kitagawa Y., Sakamoto M. Early hepatocellular carcinoma with high-grade atypia in small vaguely nodular lesions. Cancer Sci. 2016;107:543–550. doi: 10.1111/cas.12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Zhao Y., Wang Y., Wang X., Wang R., Liu N., Li P., Ji M., Ye J., Sun T., et al. TNFAIP8 promotes AML chemoresistance by activating ERK signaling pathway through interaction with Rac1. J. Exp. Clin. Cancer Res. 2020;39:158. doi: 10.1186/s13046-020-01658-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M. Early HCC: diagnosis and molecular markers. J. Gastroenterol. 2009;44(Suppl 19) doi: 10.1007/s00535-008-2245-y. [DOI] [PubMed] [Google Scholar]
- Sakamoto M., Mori T., Masugi Y., Effendi K., Rie I., Du W. Candidate molecular markers for histological diagnosis of early hepatocellular carcinoma. Intervirology. 2008;51(Suppl 1) doi: 10.1159/000122603. [DOI] [PubMed] [Google Scholar]
- Shibata R., Mori T., Du W., Chuma M., Gotoh M., Shimazu M., Ueda M., Hirohashi S., Sakamoto M. Overexpression of cyclase-associated protein 2 in multistage hepatocarcinogenesis. Clin. Cancer Res. 2006;12:5363–5368. doi: 10.1158/1078-0432.CCR-05-2245. [DOI] [PubMed] [Google Scholar]
- Shin S., Buel G.R., Nagiec M.J., Han M.J., Roux P.P., Blenis J., Yoon S.O. ERK2 regulates epithelial-to-mesenchymal plasticity through DOCK10-dependent Rac1/FoxO1 activation. Proc. Natl. Acad. Sci. U. S. A. 2019;116:2967–2976. doi: 10.1073/pnas.1811923116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisinni L., Pietrafesa M., Lepore S., Maddalena F., Condelli V., Esposito F., Landriscina M. Endoplasmic reticulum stress and unfolded protein response in breast cancer: the balance between apoptosis and autophagy and its role in drug resistance. Int. J. Mol. Sci. 2019;20:857. doi: 10.3390/ijms20040857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urra H., Dufey E., Avril T., Chevet E., Hetz C. Endoplasmic reticulum stress and the hallmarks of cancer. Trends Cancer. 2016;2:252–262. doi: 10.1016/j.trecan.2016.03.007. [DOI] [PubMed] [Google Scholar]
- Wang M., Kaufman R.J. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer. 2014;14:581–597. doi: 10.1038/nrc3800. [DOI] [PubMed] [Google Scholar]
- Woo H.G., Choi J.H., Yoon S., Jee B.A., Cho E.J., Lee J.H., Yu S.J., Yoon J.H., Yi N.J., Lee K.W., et al. Integrative analysis of genomic and epigenomic regulation of the transcriptome in liver cancer. Nat. Commun. 2017;8:839. doi: 10.1038/s41467-017-00991-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R.K., Chae S.W., Kim H.R., Chae H.J. Endoplasmic reticulum stress and cancer. J. Cancer Prev. 2014;19:75–88. doi: 10.15430/JCP.2014.19.2.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Choi J.H., Kim S.J., Lee E.J., Shah M., Choi S., Woo H.G. EPHB6 mutation induces cell adhesion-mediated paclitaxel resistance via EPHA2 and CDH11 expression. Exp. Mol. Med. 2019;51:1–12. doi: 10.1038/s12276-019-0261-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg M., Neilson E.G. Biomarkers for epithelial-mesenchymal transitions. J. Clin. Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.