Abstract
Cilia are highly specialized organelles that extend from the cell membrane and function as cellular signaling hubs. Thus, cilia formation and the trafficking of signaling molecules into cilia are essential cellular processes. TULP3 and Tubby (TUB) are members of the tubby-like protein (TULP) family that regulate the ciliary trafficking of G-protein coupled receptors, but the functions of the remaining TULPs (i.e., TULP1 and TULP2) remain unclear. Herein, we explore whether these four structurally similar TULPs share a molecular function in ciliary protein trafficking. We found that TULP3 and TUB, but not TULP1 or TULP2, can rescue the defective cilia formation observed in TULP3-knockout (KO) hTERT RPE-1 cells. TULP3 and TUB also fully rescue the defective ciliary localization of ARL13B, INPP5E, and GPR161 in TULP3 KO RPE-1 cells, while TULP1 and TULP2 only mediate partial rescues. Furthermore, loss of TULP3 results in abnormal IFT140 localization, which can be fully rescued by TUB and partially rescued by TULP1 and TULP2. TUB’s capacity for binding IFT-A is essential for its role in cilia formation and ciliary protein trafficking in RPE-1 cells, whereas its capacity for PIP2 binding is required for proper cilia length and IFT140 localization. Finally, chimeric TULP1 containing the IFT-A binding domain of TULP3 fully rescues ciliary protein trafficking, but not cilia formation. Together, these two TULP domains play distinct roles in ciliary protein trafficking but are insufficient for cilia formation in RPE-1 cells. In addition, TULP1 and TULP2 play other unknown molecular roles that should be addressed in the future.
Keywords: cilia, cilia formation, ciliary trafficking, RPE1, TULP
INTRODUCTION
Primary cilia are microtubule-based sensory organelles present on the surface of most mammalian cell types. Although long thought dispensable, cilia are now recognized to have essential roles in various cellular processes, including developmental signaling and adult homeostasis (Drummond, 2012; Fliegauf et al., 2007). Disruptions of ciliary function lead to ciliopathies, such as the developmental disorders polycystic kidney disease, retinitis pigmentosa, Bardet-Biedl syndrome, and Joubert syndrome (Ansley et al., 2003; Green et al., 1989; Hildebrandt et al., 2011; Liu et al., 2002; Pazour et al., 2000; Valente et al., 2006).
Primary cilia receive many extracellular signals, including Hedgehog, Wnt, and Notch (Ezratty et al., 2011; Huangfu et al., 2003; Oishi et al., 2006; Rohatgi et al., 2007). Therefore, to ensure proper function, the ciliary membrane houses many transmembrane signaling molecules such as platelet-derived growth factor receptors (PDGF), Smoothened (Smo), transient receptor potential (TRP) channels, and G protein-coupled receptors (GPCRs) (Christensen et al., 2007; Colbert et al., 1997; Corbit et al., 2005; Hilgendorf et al., 2016; Mukhopadhyay et al., 2013; Rohatgi et al., 2007). Together, these receptors modulate a number of critical developmental signaling pathways, but our understanding of the mechanisms by which these membrane proteins are targeted to cilia and concentrated there remains sparse.
Intraflagellar transport (IFT) is a well-characterized, conserved system that controls ciliary protein trafficking. Its specialized transport machinery comprises two discrete multi-protein complexes—IFT-A and IFT-B (Cole et al., 1998; Rosenbaum and Witman, 2002). The IFT-B complex, in cooperation with kinesin-2, mediates anterograde transport towards the ciliary tip, while the IFT-A complex, along with dynein, mediates retrograde transport back towards the ciliary base (Ishikawa and Marshall, 2011; Scholey, 2003; Taschner et al., 2012). IFT is thought to play a critical role in establishing and sustaining proper cilia formation and function (Rosenbaum and Witman, 2002; Scholey, 2003). Still, although many membrane proteins depend on IFT proteins for ciliary trafficking (Crouse et al., 2014; Keady et al., 2011; Nachury et al., 2010), it remains unclear whether the IFT complexes regulate this process directly themselves or whether other regulatory mechanisms are also required.
Mammalian TULP3 reportedly plays a key role in the ciliary trafficking of several membrane proteins, including the rhodopsin family GPCRs Sstr3, Mchr1, Npy2r, GPR161, as well as the polycystins (Badgandi et al., 2017; Mukhopadhyay et al., 2010; 2013). TULP3 captures ciliary membrane proteins in a phosphoinositide 4,5-bisphosphate (PI(4,5)P2)-dependent mechanism, presumably by binding to the PI(4,5)P2-binding domain, and then moves together with its cargo into the cilia via IFT-A binding (Badgandi et al., 2017). Tubby (TUB), the founding member of the tubby-like protein (TULP) family, is also required for the ciliary trafficking of Sstr3, Mchr1, and Npy2r (Loktev and Jackson, 2013; Sun et al., 2012). These molecular functions seem to be conserved throughout the animal kingdom, as the Drosophila tubby homolog dTULP and the C. elegans homolog tub-1 are also involved in the ciliary trafficking of membrane receptors (DiTirro et al., 2019; Park et al., 2013).
Tubby mice show maturity-onset obesity with neurosensory deficits (Kleyn et al., 1996; Noben-Trauth et al., 1996), both common ciliopathy traits. Although the particular cell types and GPCRs that cause the obesity phenotype of tubby mice have not yet been identified, the phenotype is almost certainly linked to TUB’s ciliary functions. On the other hand, TULP3 was originally identified as a negative regulator of Hedgehog signaling (Norman et al., 2009), and the TULP3-dependent nature of ciliary GPCR trafficking has been clearly implicated in the underlying mechanism (Mukhopadhyay et al., 2010).
While some of the molecular functions of mammalian TULPs, especially of TULP3 and TUB, have been identified, those of TULP1 and TULP2 have not. Whereas the loss of TULP1 function leads to retinal degeneration in both humans and mice (Hagstrom et al., 1999; Ikeda et al., 2000), there are not yet any functional data available for TULP2. Because TULP1 and TULP2 each have either the conserved IFT-A-binding domain or the membrane phosphoinositide-binding domains of the TULPs, and because these domains are required for the ciliary trafficking of GPCRs, there is a good chance TULP1 and TULP2 share some of the molecular functions of TULP3 and TUB.
Here, we report the results of our investigation into the ciliary functions of the four TULPs—TULP1, TULP2, TULP3, and TUB—in hTERT RPE-1 (RPE1) cells. We found that, despite significant amino acid-level similarity, these four proteins play separate roles in primary cilia assembly and trafficking. The functions of TULP3 and TUB are largely analogous with respect to cilia formation and ciliary protein trafficking. In contrast, although TULP1 and TULP2 show limited capacity to control ciliary protein trafficking, the addition of an IFT-A binding domain increases it. Together, while TULP3 and TUB are critical ciliary membrane trafficking regulators, TULP1 and TULP2 must play different molecular roles that should be investigated in the future.
MATERIALS AND METHODS
Cell culture
Both RPE1 wild-type (CRL-4000; American Type Cell Collection [ATCC], USA) and TULP3 knockout (KO) cells were cultured in DMEM/F-12 (11320082; Thermo Fisher Scientific, USA) supplemented with 10% fetal bovine serum (FBS) (10099141; Thermo Fisher Scientific) and 1% Penicillin/Streptomycin (P/S) (15-140-122; Thermo Fisher Scientific). Cells were grown on coverslips in 12-well culture plates at a density of 0.5 × 106 cells/well and cultured in a CO2 incubator. Cells were shifted from 10% to 0.5% serum 24 h after transfection to induce ciliation and fixed 72 h post-transfection. Plasmid transfections were carried out with a NEPA21 Electroporator (NEPA GENE, Japan) at a pulse voltage of 150 V and pulse length of 5 ms.
RNA isolation and quantitative RT-PCR
Total RNA was extracted using the RNeasy mini kit (74104; Qiagen, Germany). Complementary DNA (cDNA) was synthesized from total RNA with the RevertAid Reverse Transcriptase (EP0442; Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. To assess gene expression, cDNAs were amplified using the SensiFast SYBR HiRox Kit (92020; Meridian Bioscience, USA). Each reaction was performed in triplicate. A comprehensive list of PCR primers appears in Supplementary Table S1. Amplification conditions were as follows: 2 min at 95°C for polymerase activation, 40 cycles at 95°C denaturation for 5 s, 60°C annealing for 10 s, and 72°C extension for 20 s. The cycle threshold (Ct) values for the target genes and GAPDH were measured and calculated with the 7500 fast system sequence detection software (Applied Biosystems, USA).
Immunoblot analysis
Cells were washed with phosphate-buffered saline (PBS), lysed on ice in RIPA buffer (BRI-9001; TIB Molbiol, Germany) containing protease inhibitors, and centrifuged at 15,000 × g for 20 min. Samples were then mixed with a loading buffer containing 2-mercaptoethanol (1610747; Bio-Rad, USA) and incubated at 95°C for 5 min. Each protein sample was then resolved via SDS-PAGE using 8% polyacrylamide gels and electrophoretically transferred to a PVDF membrane (IPVH00010; Millipore Sigma, USA). The membranes were then incubated with blocking buffer (5% skimmed milk in Tris-buffered saline [TBS] containing 0.1% Tween 20; TBS-T) for 1 h at room temperature. Primary antibodies specific to TULP3 (13637-1-AP; Proteintech, USA) at 1:1,000 and TUB (17928-1-AP; Proteintech) at 1:1,000 were diluted in blocking buffer and incubated with the membranes overnight at 4°C on a rocking platform. The following day, the membranes were washed four times at 10 min intervals with TBS-T. HRP-conjugated-anti-Rabbit IgG at 1:5,000 (A32731; Sigma-Aldrich, USA) was diluted in blocking buffer and incubated with the membranes as a secondary antibody for 1 h at room temperature on a rocking platform. The membranes were then rinsed in TBS-T four times at 10 min intervals before being visualized with an ECL detection reagent (RPN2232; GE Healthcare, USA).
Establishment of the TULP3 KO RPE1 cell line using CRISPR/Cas9
A single-guide RNA (sgRNA) sequence (5’-AGAAATGATGAAGATGCGAC-3’) targeting exon 1 of the TULP3 gene was designed using the CRISPR/Cas9 design program (Sentmanat et al., 2018). Forward and reverse oligonucleotides matching the sgRNA sequence were ordered, annealed, and cloned into the pSpCas9 (BB)-2A-Puro (PX459) V2.0 vector (Addgene plasmid #62988) (Ran et al., 2013). RPE1 cells were transfected with the PX459 vector containing the sgRNA-encoding cassette by electroporation. Cells were subcultured at a ratio of 1:6 in media for 72 h and maintained until the formation of distinct colonies. Individual clones were isolated and expanded. Genomic DNA from single clones was extracted and analyzed for TULP3 mutations by sequencing. Lines with biallelic TULP3 disruption were selected, and the absence of TULP3 protein expression in these TULP3 KO RPE1 cells was confirmed with immunoblotting.
Plasmids
Mouse Tulp1, Tulp2, Tulp3, and TUB cDNAs were cloned into the pIRES2-DsRed2 vector (Clontech plasmid #632420), which expresses both the gene of interest and the dsRed gene. TUB PIP2- and IFT-A-mutant cDNAs were cloned into the pLVX-EF1α-IRES-ZsGreen vector (Clontech plasmid #631982), which expresses both the gene of interest and the ZsGreen gene. To examine the subcellular localization of TULPs in RPE1 cells, mouse Tulp1, Tulp2, Tulp3, TUB, and Tulp1IFT-A(+) cDNAs were cloned into the pCMV-Tag3B vector (Agilent Technologies plasmid #211173), which makes it possible to visualize the expression of a gene of interest using an anti-Myc antibody.
Immunofluorescence analysis
Samples were fixed with 4% paraformaldehyde in 1× PBS at room temperature for 10 min. After three washes with PBS, cells were permeabilized with 0.2% Triton X-100 (PBS-T) for 30 min and blocked with 5% goat serum in PBS for 1 h at room temperature. For TULP3 staining, cells were blocked in PBS containing 0.1% Triton X-100 and 2% donkey serum. Samples were subsequently incubated in a blocking solution containing primary antibodies at 4°C overnight. The following day, coverslips were rinsed three times with PBS, incubated with a blocking solution containing secondary antibodies at room temperature for 1 h. They were then rinsed twice with PBS, mounted in Vectashield (Vector Laboratories, USA), and examined using a Zeiss LSM700 confocal microscope (Germany) with a 60× plan-apochromat oil-immersion objective. When comparing the expression and localization of ciliary proteins between groups, all samples were prepared simultaneously, and the resulting confocal images were obtained under the same conditions. We only examined DsRed-positive RPE1 cells to exclude any non-transfected cells.
The primary antibodies used for immunofluorescence were diluted as follows: mouse anti-α-acetylated tubulin, 1:1,000 (T6793; Millipore Sigma); mouse anti-γ-acetylated tubulin, 1:800 (T6557; Sigma-Aldrich); chicken anti-mCherry, 1:700 (ab205402; Abcam, UK); rabbit anti-TULP3, 1:100 (13637-1-AP; Proteintech), 1:100; rabbit anti-ARL13B, 1:400 (17711-1-AP; Proteintech); rabbit anti-GPR161, 1:400 (13398-1-AP; Proteintech); rabbit anti-INPP5E, 1:400 (17797-1-AP; Proteintech); rabbit anti-IFT140, 1:300 (17460-1-AP; Proteintech); rabbit anti-c-Myc 1:500 (C3956; Sigma-Aldrich), 1:500; rabbit anti-IFT88, 1:300 (13967-1-AP; Proteintech). The secondary antibodies used were diluted as follows: Alexa Fluor 488, 568-conjugated anti-mouse IgG, 1:400 (A11029 and A11031; Invitrogen, USA); Alexa Fluor 568-conjugated anti-rabbit IgG, 1:400 (A11011; Invitrogen), Alexa 633-conjugated anti-rabbit IgG, 1:400 (A21070; Invitrogen); Alexa Fluor 555-conjugated anti-chicken IgY, 1:500 (A32932; Invitrogen). Control and non-transfected TULP3 KO cells were also counterstained with TO-PRO3 at 1:1,000 (1878895; Invitrogen).
Data analysis
All data are presented as mean ± SEM and based on results obtained from at least three independent experiments. All statistical analyses were carried out using GraphPad Prism 9 (GraphPad Software, USA). One-way ANOVAs with Tukey’s post-hoc tests were used for multiple comparisons. Pearson’s χ2 test was used for comparisons of ciliary IFT140 localization.
RESULTS
Generating a TULP-free RPE1 cell line
To investigate the role of TULPs in ciliary trafficking, we decided to utilize hTERT RPE-1 (RPE1) cells because they produce clear cilia and are commonly used to study cilia formation and ciliary protein trafficking. We first asked which TULPs are expressed in RPE1 cells. We found via quantitative RT-PCR that while TULP3 is abundant in RPE1 cells, the other TULPs are almost undetectable (Supplementary Fig. S1A). We further confirmed the presence of TULP3 and the absence of TUB in RPE1 cells via immunoblot (Supplementary Fig. S1B). In addition, we found that endogenous TULP3 is localized to cilia with a slight enrichment in the ciliary base (Supplementary Fig. S1E). Based on these preliminary results, we wanted to generate a TULP-free RPE1 cell line so we could re-introduce the TULPs and identify any shared ciliary functions. We therefore attempted to generate TULP3-knockout RPE1 (TULP3 KO RPE1) cells using the CRISPR/Cas9 system (Ran et al., 2013). We obtained one TULP3 KO RPE1 cell line by targeting the first exon of TULP3, leading to a frameshift mutation in the distal end of the second exon (Supplementary Fig. S1C). We then confirmed the successful knockout of TULP3 using immunocytochemistry and western blot analysis (Supplementary Figs. S1D and S1E).
Effect of TULPs in cilia formation in RPE1 cells
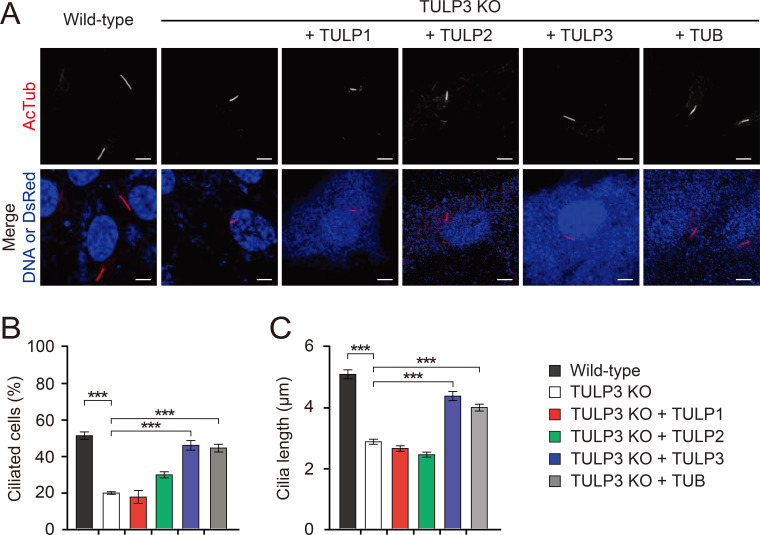
Consistent with previous results (Han et al., 2019), we found TULP3 KO RPE1 cilia were significantly disrupted compared to wild-type RPE1 cilia (Fig. 1). When we compared the cilia of serum-starved wild-type and TULP3 KO RPE1 cells, we found that while 50% of wild-type RPE1 cells assembled a cilium, only 20% of TULP3 KO RPE1 cells assembled one (Fig. 1B). In addition, the primary cilia of TULP3 KO RPE1 cells averaged only half the length of wild-type RPE1 cilia (Fig. 1C). This suggests TULP3 is critical for proper cilia formation in RPE1 cells (Fig. 1C). We then found that re-introduction of Tulp3 into TULP3 KO RPE1 cells by transfection fully rescued their cilia defects, confirming the specificity of TULP3’s role in cilia formation. When we introduced other TULPs into TULP-free RPE1 cells, we found that TUB, but not Tulp1 or Tulp2, fully rescued cilia formation and length (Figs. 1B and 1C). To exclude the possibility that mislocalization of TULP1 and TULP2 in TULP3 KO RPE1 cells attributes to their inability to rescue defective cilia formation, we examined the intracellular localization of TULP1 and TULP2. TULP1 was mainly localized in the nucleus as previously reported (He et al., 2000), and TULP2 was enriched in cilia (Supplementary Fig. S2). Their subcellular localization was not dependent on TULP3, indicating that TULP1 and TULP2 did not play a role in cilia formation.
Fig. 1. Effects of TULPs on ciliogenesis in RPE1 cells.
(A) Wild-type RPE1 cells and TULP3 KO RPE1 cells, transfected with the indicated TULPs. Cells were stained with an antibody specific to the axonemal marker Ac-tubulin. Transfected cells were identified with DsRed staining. Non-transfected cells were identified with the nuclear stain To-Pro3. Scale bars = 5 μm. (B and C) Quantification of ciliated cells (B) and cilia length (C). Data are presented as mean ± SEM from three independent experiments. More than 300 cells were counted for each genotype for the ciliary formation experiment and more than 100 were counted for each genotype for the ciliary length experiment. One-way ANOVAs with Tukey’s post-hoc tests. ***P < 0.001.
Effect of TULPs on ciliary membrane protein trafficking in RPE1 cells
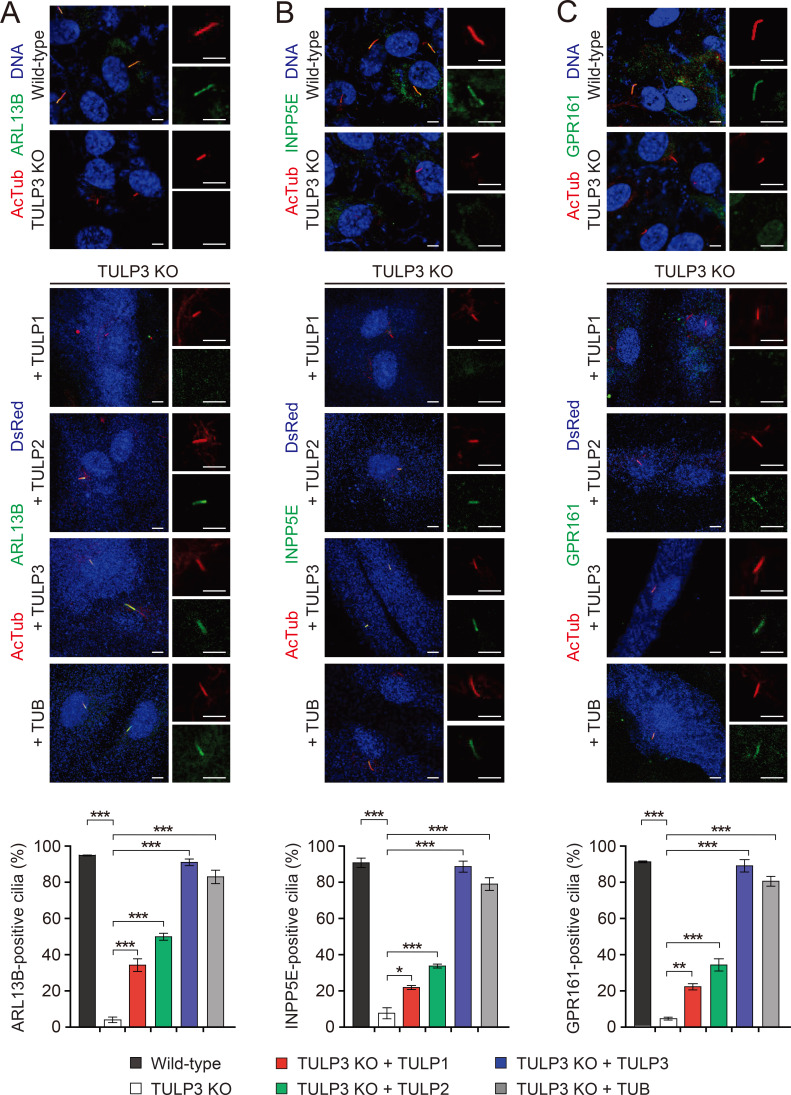
TULP3 regulates the localization of several ciliary membrane-associated proteins, as well as a subset of GPCRs (Badgandi et al., 2017; Han et al., 2019; Mukhopadhyay et al., 2010; 2013). Indeed, consistent with previous reports (Han et al., 2019), we found that TULP3 KO RPE1 cells showed impaired ciliary localization of ARL13B, INPP5E, and GPR161, and that TULP3 expression rescued these trafficking defects (Fig. 2). We therefore proceeded to explore further whether the other TULPs could also rescue this impaired ciliary protein localization. While the expression of TUB induced a nearly full rescue of ciliary protein localization, Tulp1 and Tulp2 induced only a partial rescue. Specifically, compared to that facilitated by TULP3, expression of TULP1 and TULP2 in TULP3 KO RPE1 cells facilitated approximately 25% and 40%, respectively, of the ciliary localization of ARL13B, INPP5E, and GPR161 (Fig. 2). These data suggest TULP1 and TULP2 may be additional modulators of ciliary protein trafficking, though to a lesser extent than TUB and TULP3.
Fig. 2. Effects of TULPs on ciliary trafficking of ARL13B, INPP5E, and GPR161 in RPE1 cells.
(A-C) Wild-type RPE1 cells and TULP3 KO RPE1 cells, transfected with the indicated TULPs. Cells were stained with antibodies specific for the axonemal marker Ac-tubulin, as well as ARL13B (A), INPP5E (B), or GPR161 (C). Transfected cells were identified with DsRed staining. Non-transfected cells were identified with the nuclear stain To-Pro3. Enlarged images of single cilia displayed on the right. Scale bars = 5 μm. A quantification of ciliated cells expressing each ciliary protein is shown below. Data are presented as mean ± SEM from three independent experiments. More than 100 ciliated cells were counted for each condition. One-way ANOVAs with Tukey’s post-hoc tests. *P < 0.05, **P < 0.01, ***P < 0.001.
Effect of TULPs on IFT subunit trafficking in RPE1 cells
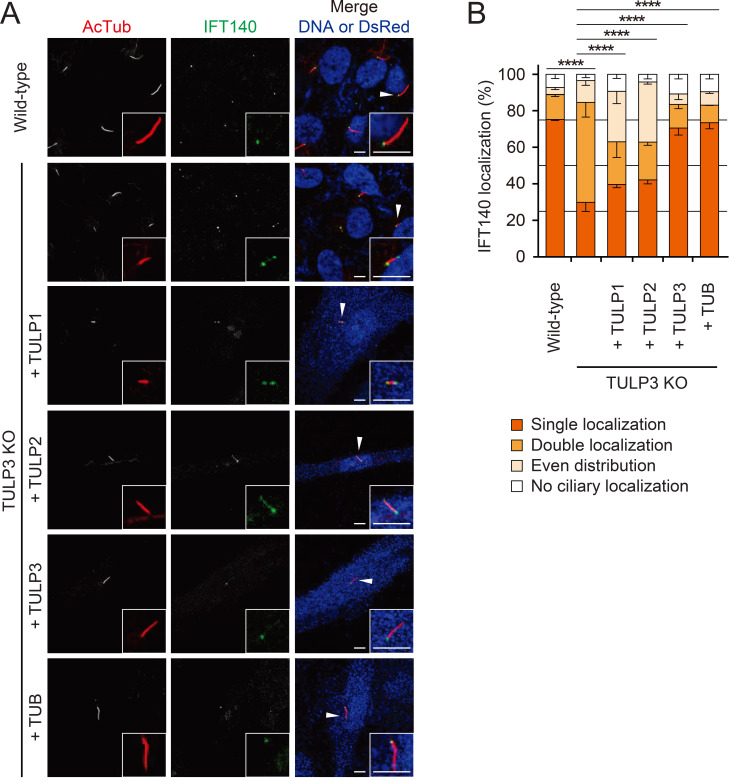
The core IFT-A complex is essential for the ciliary localization of TULP3, as TULP3 localization is lost upon ablation of the core IFT-A subunits IFT140, IFT122, or WDR19 (Mukhopadhyay et al., 2010; Qin et al., 2011). Because TULP3 acts as an adaptor between the core IFT-A complex and ciliary membrane cargo (Badgandi et al., 2017), we asked whether the loss of TULP3 affects the ciliary localization of the core IFT-A subunits. Specifically, we examined IFT140 localization in RPE1 cells because the phosphoinositide-dependent localization of TULP3 also affects the ciliary localization of IFT140 (Garcia-Gonzalo et al., 2015). In wild-type RPE1 cells, IFT140 is primarily localized to a singular focus at the base of the cilium, with some occasional faint staining at the ciliary tip (Fig. 3, Supplementary Fig. S3C). Upon depletion of TULP3, however, IFT140 staining was either localized in two foci, one at the ciliary base and one at the tip, or it was uniformly distributed across the entire cilium (Fig. 3). This suggests TULP3 plays an integral role in the trafficking of IFT140. Upon the expression of TUB and TULP3 in TULP3 KO RPE1 cells, approximately 70% of the cells showed a reversion of IFT140 staining to the singular focus observed in wild-type RPE1 cells (Fig. 3). Furthermore, expression of TULP1 and TULP2 also rescued IFT140 localization back to the ciliary base, though to a lesser degree than TUB and TULP3 (Fig. 3). In contrast, TULP3 depletion did not affect the ciliary distribution of IFT88, indicating that TULPs only control the trafficking of IFT-A (Supplementary Figs. S3A and S3B).
Fig. 3. Effects of TULPs on ciliary trafficking of IFT140 in RPE1 cells.
(A) Wild-type RPE1 cells and TULP3 KO RPE1 cells, transfected with the indicated TULPs. Cells were stained with antibodies specific for the axonemal marker Ac-tubulin and IFT140. Transfected cells were identified with DsRed staining. Non-transfected cells were identified with the nuclear stain To-Pro3. Insets, enlarged images of the cilia marked with arrowheads. Scale bars = 5 μm. (B) Quantification of IFT140 localization. Data are presented as mean ± SEM from three independent experiments. Total counted cells are 100-150 for each condition. Pearson’s χ2 test. ****P < 0.0001.
Tubby’s IFT-A and PIP2-binding domains contribute differentially to ciliary protein trafficking
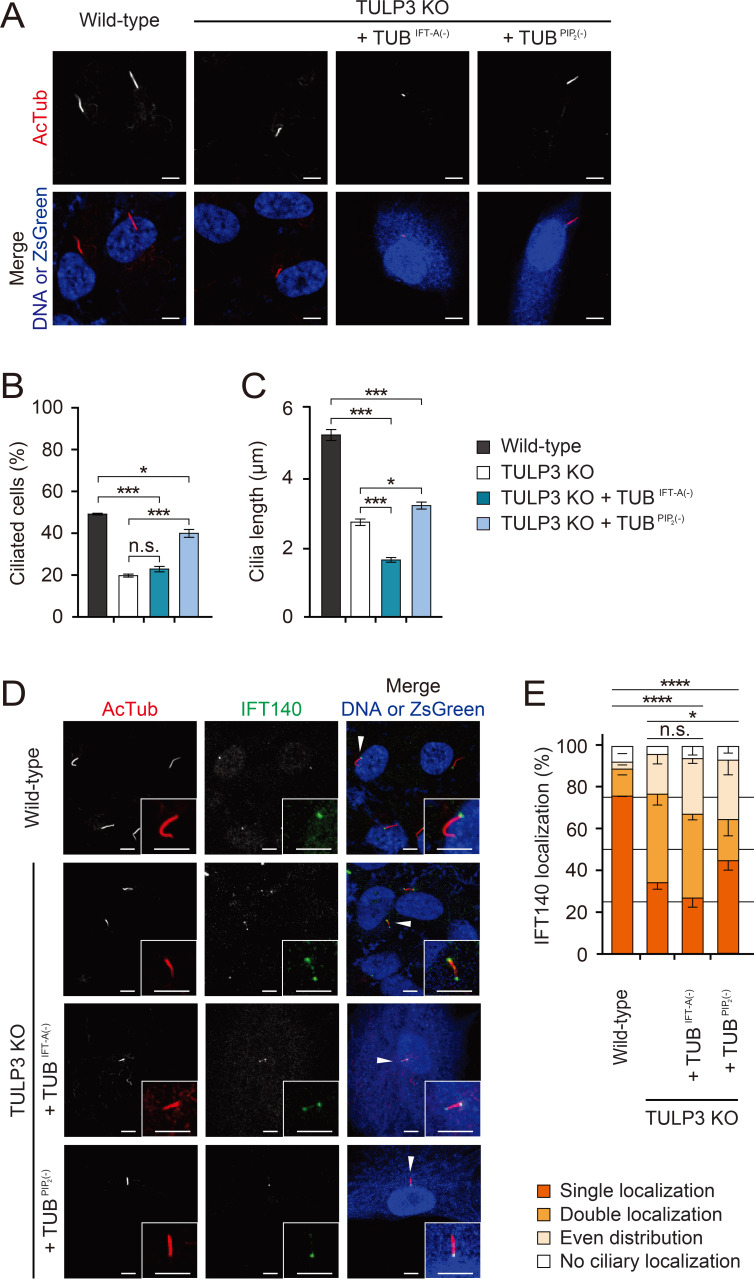
Previous studies have shown that TULP’s N-terminal IFT-A binding domain and C-terminal PIP2 binding domain are crucial for proper regulation of ciliary membrane protein trafficking (Mukhopadhyay et al., 2010; Park et al., 2013). We decided to evaluate the contribution of each of these domains in the TULPs to cilia formation and IFT-A trafficking in RPE1 cells. When we expressed a mutant form of TUB defective in IFT-A binding (TUBIFT-A(–)) in TULP3 KO RPE1 cells, not only did it fail to rescue cilia formation, but it also induced the formation of even shorter cilia than those produced by control TULP3 KO RPE1 cells. This was reminiscent of the dominant negative effect mutant TULP3 defective in IFT-A binding has on GPCR ciliary trafficking (Mukhopadhyay et al., 2010) (Figs. 4A-4C). In contrast, a mutant form of TUB defective in PIP2 binding (TUBPIP2(–)) did partially rescue cilia formation and cilia length (Figs. 4A-4C). Furthermore, while TUBIFT-A(–) did not rescue IFT140 localization, TUBPIP2(–) permitted a partial rescue of IFT140 localization (Figs. 4D and 4E). These data indicate that while both domains of TUB are vital for both cilia formation and IFT-A trafficking, IFT-A binding is more important in RPE1 cells. Consistent with these results, TUBPIP2(–) permitted a near total rescue of the ciliary localization of ARL13B, INPP5E, and GPR161, but TUBIFT-A(–) did not (Supplementary Fig. S4). These data indicate that TUB’s IFT-A binding domain is essential for the trafficking of ciliary membrane proteins.
Fig. 4. Fig. 4. Differential contributions of the IFT-A and PIP2-binding domains of TUB to ciliary formation and IFT140 trafficking.
(A) Wild-type RPE1 cells and TULP3 KO RPE1 cells, transfected with the indicated forms of TUB. TUBIFT-A(–), a mutant TUB that does not bind IFT-A; TUBPIP2(–), a mutant TUB that does not bind PIP2. Cells were stained with an antibody specific for the axonemal marker Ac-tubulin. Transfected cells were identified with either DsRed or ZsGreen. Non-transfected cells were identified with the nuclear marker To-Pro3. Scale bars = 5 μm. (B and C) Quantification of ciliated cells (B) and cilia length (C). Data are presented as mean ± SEM from three independent experiments. More than 300 cells were counted for each genotype for the ciliary formation experiment and more than 90 were counted for each genotype for the ciliary length experiment. One-way ANOVAs with Tukey’s post-hoc tests. (D) Wild-type RPE1 cells and TULP3 KO RPE1 cells, transfected with the indicated forms of TUB. Cells were stained with antibodies specific for the axonemal marker Ac-tubulin and IFT140. Transfected cells were identified with ZsGreen. Non-transfected cells were identified with the nuclear marker To-Pro3. Insets, enlarged images of the cilia marked with arrowheads. Scale bars = 5 μm. (E) Quantification of IFT140 localization. Data are presented as mean ± SEM from three independent experiments. Total counted cells are 90-150 for each condition. Pearson’s χ2 test. *P < 0.05, ***P < 0.001, ****P < 0.0001. n.s., not significant.
Enhancement of TULP1 IFT-A binding improved its regulation of cilia protein trafficking
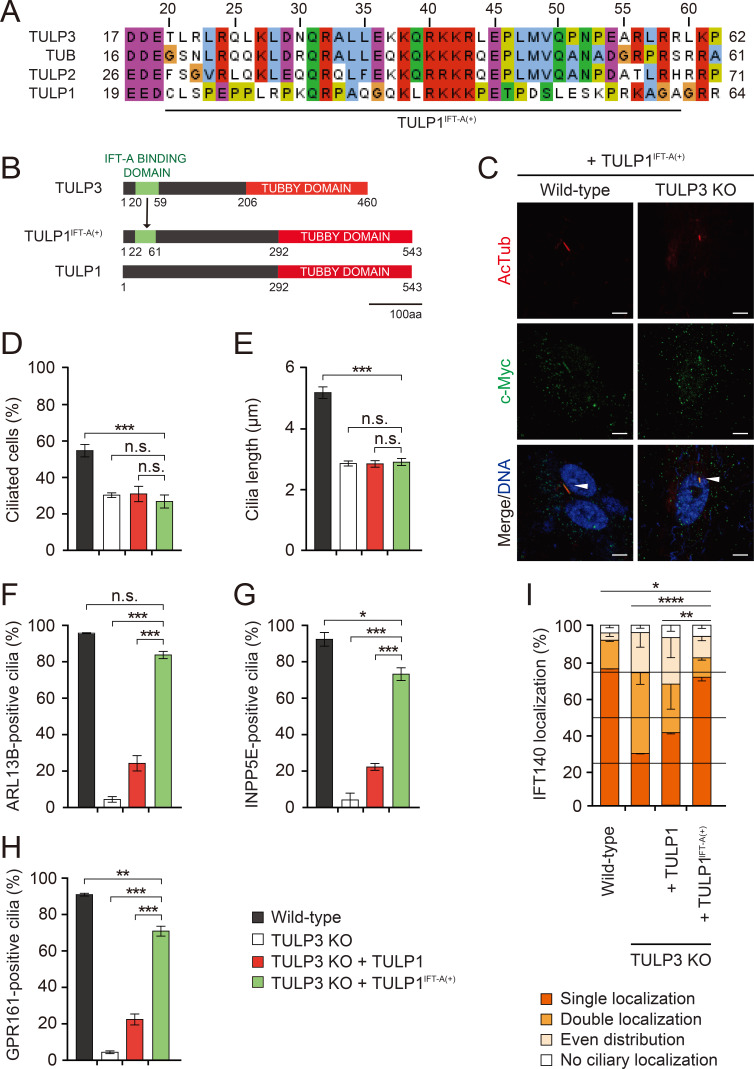
The IFT-A binding domain of the TULPs seems to be essential for cilia formation and ciliary protein trafficking. While TULP1 contains a conserved Tubby domain, it does not bind to the core IFT-A complex. It contains a far less conserved IFT-A binding domain than the other TULPs that also failed to interact with IFT-A in an in vitro protein interaction assay (Fig. 5A) (Mukhopadhyay and Jackson, 2011; Mukhopadhyay et al., 2010). We reasoned that adding IFT-A binding to TULP1 may allow it to fully rescue cilia formation and ciliary protein trafficking in TULP3 KO RPE1 cells. To test this hypothesis, we substituted the putative IFT-binding domain of TULP3 (20-59 amino acids of TULP3) for the corresponding 40-amino-acid stretch in TULP1 (22-61 amino acids of TULP1). Hereafter, we refer to this chimeric protein as TULP1IFT-A(+) (Fig. 5B). In contrast to the nuclear localization of TULP1, TULP1IFT-A(+) was mainly localized in the cilia. This implicates the IFT-A binding domain in dictating the cilia localization of TULP1IFT-A(+) (Fig. 5C). Next, we examined the effect of TULP1IFT-A(+)on cilia formation and ciliary protein trafficking. We found that expression of TULP1IFT-A(+) in TULP3 KO RPE1 cells had no effect on the formation of their cilia or on their ciliary length defects (Figs. 5D and 5E), but almost completely rescued the localization of ARL13B, INPP5E, GPR161, and IFT140 (Figs. 5F-5I). This suggests TULP1’s inability to control ciliary protein trafficking was due to its inability to bind IFT-A.
Fig. 5. Fig. 5. IFT-A binding in the TULPs is essential for the proper ciliary trafficking and localization of IFT140.
(A) Multiple sequence alignment of the putative IFT-A binding domain from the TULPs. (B) Schematic depicting the formation of TULP1IFT-A(+). The putative IFT-A binding domain of TULP3 (amino acids 20-59) was used to replace the corresponding 40 amino acid sequence in TULP1. (C) Immunocytochemical analysis of TULP1 and TULP1IFT-A(+) in wild-type and TULP3 KO RPE1 cells. Cells were stained with antibodies specific for the axonemal marker Ac-tubulin and c-Myc for TULP1 and TULP1IFT-A(+). Nuclei were stained with To-Pro3. TULP1IFT-A(+)-positive cilia are marked with arrowheads. Scale bars = 5 μm. (D and E) Quantification of ciliated cells (D) and cilia length (E). Data are presented as mean ± SEM from three independent experiments. More than 300 cells were counted for each genotype for the ciliary formation experiment and more than 100 were counted for each genotype for the ciliary length experiment. One-way ANOVAs with Tukey’s post-hoc tests. ***P < 0.001. (F-H) Quantification of ciliated cells expressing each ciliary protein. Data are presented as mean ± SEM from three independent experiments. More than 100 cells were counted for each condition. One-way ANOVAs with Tukey’s post-hoc tests. *P < 0.05, **P < 0.01, ***P < 0.001. (I) Quantification of IFT140 localization. Data are presented as mean ± SEM from three independent experiments. Total counted cells are 90-150 for each condition. Pearson’s χ2 test. *P < 0.05, **P < 0.01, ****P < 0.0001. n.s., not significant.
DISCUSSION
Here, we have shown that TUB and TULP3, but not TULP1 or TULP2, support cilia formation and protein trafficking in RPE1 cells. The main characteristic distinguishing the TULPs with respect to this phenomenon is their capacity to bind to the IFT-A complex, as a chimeric TULP1 harboring the ITF-A binding domain of TULP3 rescued ciliary protein trafficking just as TULP3 did.
Although previous studies found that TULP3 did not affect ciliogenesis (Mukhopadhyay et al., 2010), we found complete knockout of TULP3 in RPE1 cells using CRISPR/Cas9 downregulated cilia formation. This finding is consistent with another study that used TULP3 KO RPE1 cells (Han et al., 2019). We expect that this discrepancy is due to an incomplete knockdown of TULP3 via RNA interference. It is noteworthy, however, that the cilia of Tulp3-/- mouse embryonic fibroblasts (MEFs) are comparable to those of wild-type MEFs (Norman et al., 2009). This could be due to differential requirements of TULP3 for ciliogenesis in each cell type, which would mean TULP3 is a non-universal regulator of ciliogenesis. It is also possible that TUB plays a redundant role in Tulp3–/– MEFs.
Although we found that IFT140 localization depended on TULP3 and TUB (Fig. 3), a previous study found that TULP3 knockdown did not affect IFT140 localization (Mukhopadhyay et al., 2010), perhaps due to incomplete TULP3 depletion. However, it is unclear whether the mislocalization of IFT140 is due to the inability of INPP5E to localize in cilia in TULP3 KO RPE1 cells because INPP5E affects both the ciliary localization and level of the IFT-A subunits IFT140 and IFT139 (Garcia-Gonzalo et al., 2015). Based on our findings, the interaction between IFT140 and TULP3 is more important, because we found that expression of TUB lacking its PIP2-binding domain in TULP3 KO cells still rescued INPP5E localization (Supplementary Fig. S4B), but not IFT140 localization (Figs. 4D and 4E).
Although chimeric TULP1 containing the IFT-A binding domain of TULP3 fully rescued the ciliary trafficking of IFT-A and other proteins, it did not rescue the defective cilia formation of TULP3 KO RPE1 cells. In addition, TULP2, which contains both IFT-A and PIP2-binding domains, was localized to cilia, despite lacking any ciliary rescue ability. This data suggest the IFT-A and PIP2-binding domains of TUB and TULP3 are not the only domains required for cilia formation and ciliary protein trafficking. Future studies will be necessary to determine which additional unknown domains of TULP3 and TUB contribute to proper cilia formation in RPE1 cells.
In conclusion, we have demonstrated that TULP3 and TUB have a similar capacity to regulate cilia formation and ciliary protein trafficking in RPE1 cells. It is likely that more functions of TULP3 and TUB will be revealed in the future, and it will be intriguing to explore the molecular functions of TULP1 and TULP2, both of which may not have anything to do with cilia.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by National Research Foundation of Korea (NRF) Grants funded by the Korean Government (NRF-2016R1A5A2008630 and NRF-2018R1A2B3001668).
Footnotes
AUTHOR CONTRIBUTIONS
J.J.H. and K.E.K. conducted most of the experiments. S.Y.P. performed plasmid transfections. J.B. and J.J.H. analyzed the data. J.T.S. and S.J.M. designed and supervised the project and wrote the paper.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Ansley S.J., Badano J.L., Blacque O.E., Hill J., Hoskins B.E., Leitch C.C., Kim J.C., Ross A.J., Eichers E.R., Teslovich T.M., et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- Badgandi H.B., Hwang S.H., Shimada I.S., Loriot E., Mukhopadhyay S. Tubby family proteins are adapters for ciliary trafficking of integral membrane proteins. J. Cell Biol. 2017;216:743–760. doi: 10.1083/jcb.201607095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S.T., Pedersen L.B., Schneider L., Satir P. Sensory cilia and integration of signal transduction in human health and disease. Traffic. 2007;8:97–109. doi: 10.1111/j.1600-0854.2006.00516.x. [DOI] [PubMed] [Google Scholar]
- Colbert H.A., Smith T.L., Bargmann C.I. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans . J. Neurosci. 1997;17:8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D.G., Diener D.R., Himelblau A.L., Beech P.L., Fuster J.C., Rosenbaum J.L. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit K.C., Aanstad P., Singla V., Norman A.R., Stainier D.Y., Reiter J.F. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- Crouse J.A., Lopes V.S., SanAgustin J.T., Keady B.T., Williams D.S., Pazour G.J. Distinct functions for IFT140 and IFT20 in opsin transport. Cytoskeleton (Hoboken) 2014;71:302–310. doi: 10.1002/cm.21173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTirro D., Philbrook A., Rubino K., Sengupta P. The Caenorhabditis elegans Tubby homolog dynamically modulates olfactory cilia membrane morphogenesis and phospholipid composition. Elife. 2019;8:e48789. doi: 10.7554/eLife.48789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond I.A. Cilia functions in development. Curr. Opin. Cell Biol. 2012;24:24–30. doi: 10.1016/j.ceb.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty E.J., Stokes N., Chai S., Shah A.S., Williams S.E., Fuchs E. A role for the primary cilium in Notch signaling and epidermal differentiation during skin development. Cell. 2011;145:1129–1141. doi: 10.1016/j.cell.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., Omran H. When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalo F.R., Phua S.C., Roberson E.C., Garcia G., 3rd, Abedin M., 3rd, Schurmans S., 3rd, Inoue T., 3rd, Reiter J.F., 3rd Phosphoinositides regulate ciliary protein trafficking to modulate hedgehog signaling. Dev. Cell. 2015;34:400–409. doi: 10.1016/j.devcel.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J.S., Parfrey P.S., Harnett J.D., Farid N.R., Cramer B.C., Johnson G., Heath O., McManamon P.J., O'Leary E., Pryse-Phillips W. The cardinal manifestations of Bardet-Biedl syndrome, a form of Laurence-Moon-Biedl syndrome. N. Engl. J. Med. 1989;321:1002–1009. doi: 10.1056/NEJM198910123211503. [DOI] [PubMed] [Google Scholar]
- Hagstrom S.A., Duyao M., North M.A., Li T. Retinal degeneration in tulp1−/− mice: vesicular accumulation in the interphotoreceptor matrix. Invest. Ophthalmol. Vis. Sci. 1999;40:2795–2802. doi: 10.1093/hmg/9.2.155. [DOI] [PubMed] [Google Scholar]
- Han S., Miyoshi K., Shikada S., Amano G., Wang Y., Yoshimura T., Katayama T. TULP3 is required for localization of membrane-associated proteins ARL13B and INPP5E to primary cilia. Biochem. Biophys. Res. Commun. 2019;509:227–234. doi: 10.1016/j.bbrc.2018.12.109. [DOI] [PubMed] [Google Scholar]
- He W., Ikeda S., Bronson R.T., Yan G., Nishina P.M., North M.A., Naggert J.K. GFP-tagged expression and immunohistochemical studies to determine the subcellular localization of the tubby gene family members. Brain Res. Mol. Brain Res. 2000;81:109–117. doi: 10.1016/s0169-328x(00)00164-9. [DOI] [PubMed] [Google Scholar]
- Hildebrandt F., Benzing T., Katsanis N. Ciliopathies. N. Engl. J. Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgendorf K.I., Johnson C.T., Jackson P.K. The primary cilium as a cellular receiver: organizing ciliary GPCR signaling. Curr. Opin. Cell Biol. 2016;39:84–92. doi: 10.1016/j.ceb.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Liu A., Rakeman A.S., Murcia N.S., Niswander L., Anderson K.V. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Ikeda S., Shiva N., Ikeda A., Smith R.S., Nusinowitz S., Yan G., Lin T.R., Chu S., Heckenlively J.R., North M.A., et al. Retinal degeneration but not obesity is observed in null mutants of the tubby-like protein 1 gene. Hum. Mol. Genet. 2000;9:155–163. doi: 10.1093/hmg/9.2.155. [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Marshall W.F. Ciliogenesis: building the cell's antenna. Nat. Rev. Mol. Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Keady B.T., Le Y.Z., Pazour G.J. IFT20 is required for opsin trafficking and photoreceptor outer segment development. Mol. Biol. Cell. 2011;22:921–930. doi: 10.1091/mbc.E10-09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleyn P.W., Fan W., Kovats S.G., Lee J.J., Pulido J.C., Wu Y., Berkemeier L.R., Misumi D.J., Holmgren L., Charlat O., et al. Identification and characterization of the mouse obesity gene tubby: a member of a novel gene family. Cell. 1996;85:281–290. doi: 10.1016/s0092-8674(00)81104-6. [DOI] [PubMed] [Google Scholar]
- Liu Q., Zhou J., Daiger S.P., Farber D.B., Heckenlively J.R., Smith J.E., Sullivan L.S., Zuo J., Milam A.H., Pierce E.A. Identification and subcellular localization of the RP1 protein in human and mouse photoreceptors. Invest. Ophthalmol. Vis. Sci. 2002;43:22–32. [PMC free article] [PubMed] [Google Scholar]
- Loktev A.V., Jackson P.K. Neuropeptide Y family receptors traffic via the Bardet-Biedl syndrome pathway to signal in neuronal primary cilia. Cell Rep. 2013;5:1316–1329. doi: 10.1016/j.celrep.2013.11.011. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Jackson P.K. The tubby family proteins. Genome Biol. 2011;12:225. doi: 10.1186/gb-2011-12-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Wen X., Chih B., Nelson C.D., Lane W.S., Scales S.J., Jackson P.K. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 2010;24:2180–2193. doi: 10.1101/gad.1966210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Wen X., Ratti N., Loktev A., Rangell L., Scales S.J., Jackson P.K. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 2013;152:210–223. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- Nachury M.V., Seeley E.S., Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu. Rev. Cell Dev. Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noben-Trauth K., Naggert J.K., North M.A., Nishina P.M. A candidate gene for the mouse mutation tubby. Nature. 1996;380:534–538. doi: 10.1038/380534a0. [DOI] [PubMed] [Google Scholar]
- Norman R.X., Ko H.W., Huang V., Eun C.M., Abler L.L., Zhang Z., Sun X., Eggenschwiler J.T. Tubby-like protein 3 (TULP3) regulates patterning in the mouse embryo through inhibition of Hedgehog signaling. Hum. Mol. Genet. 2009;18:1740–1754. doi: 10.1093/hmg/ddp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi I., Kawakami Y., Raya A., Callol-Massot C., Izpisúa Belmonte J.C. Regulation of primary cilia formation and left-right patterning in zebrafish by a noncanonical Wnt signaling mediator, duboraya . Nat. Genet. 2006;38:1316–1322. doi: 10.1038/ng1892. [DOI] [PubMed] [Google Scholar]
- Park J., Lee J., Shim J., Han W., Lee J., Bae Y.C., Chung Y.D., Kim C.H., Moon S.J. dTULP, the Drosophila melanogaster homolog of tubby, regulates transient receptor potential channel localization in cilia. PLoS Genet. 2013;9:e1003814. doi: 10.1371/journal.pgen.1003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Dickert B.L., Vucica Y., Seeley E.S., Rosenbaum J.L., Witman G.B., Cole D.G. Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene Tg737, are required for assembly of cilia and flagella. J. Cell Biol. 2000;151:709–718. doi: 10.1083/jcb.151.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Lin Y., Norman R.X., Ko H.W., Eggenschwiler J.T. Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1456–1461. doi: 10.1073/pnas.1011410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Milenkovic L., Scott M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Rosenbaum J.L., Witman G.B. Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Scholey J.M. Intraflagellar transport. Annu. Rev. Cell Dev. Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- Sentmanat M.F., Peters S.T., Florian C.P., Connelly J.P., Pruett-Miller S.M. A survey of validation strategies for CRISPR-Cas9 editing. Sci. Rep. 2018;8:1–8. doi: 10.1038/s41598-018-19441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Haley J., Bulgakov O.V., Cai X., McGinnis J., Li T. Tubby is required for trafficking G protein-coupled receptors to neuronal cilia. Cilia. 2012;1:21. doi: 10.1186/2046-2530-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M., Bhogaraju S., Lorentzen E. Architecture and function of IFT complex proteins in ciliogenesis. Differentiation. 2012;83:S12–S22. doi: 10.1016/j.diff.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente E.M., Silhavy J.L., Brancati F., Barrano G., Krishnaswami S.R., Castori M., Lancaster M.A., Boltshauser E., Boccone L., Al-Gazali L., et al. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.