Abstract
Background
Bloodstream infections (BSIs) are an emerging cause of significant morbidity and mortality in severe Coronavirus disease 2019 (COVID-19). We aimed to assess the prevalence, clinical profile and outcome of BSIs in critically ill COVID-19 patients.
Methods
This was a single-centre retrospective study conducted at a tertiary care hospital in Western India. All patients (age > 18 years) with reverse-transcription polymerase chain reaction (RT-PCR) confirmed COVID-19 admitted in the intensive care unit (ICU) were included. Hospital electronic records were searched for demographic data, time of bloodstream infection since admission, clinical profile, antimicrobial resistance pattern and clinical outcome of all patients who developed BSIs.
Results
Out of 750 patients admitted in COVID ICU, 8.5% developed secondary BSIs. All severe COVID-19 patients who developed BSIs succumbed to illness. A significant proportion of BSIs were Gram-negative pathogens (53/64, 82.8%). Acinetobacter baumannii was the commonest isolate, followed by Klebsiella pneumoniae (32.8% and 21.9%, respectively). Multidrug-resistance organisms (MDRO) were found in 57.8% of the cases. The majority of MDRO belonged to K. pneumoniae and Enterococcus groups. The proportion of Gram-negative bacteria resistant to carbapenems was 47.2% (25/53). On multivariate analysis, raised total leukocyte counts, mechanical ventilation and presence of comorbidities were significantly associated with the incidence of BSIs.
Conclusion
We found a significant prevalence of Acinetobacter baumannii in COVID-19 associated BSIs. The presence of comorbidities raised leukocyte counts and mechanical ventilation should alarm clinicians for possible BSIs. The timely initiation of empirical antibiotics and rapid de-escalation is vital to improve the outcome. At the same time, strict compliance of infection control practices should be accomplished to reduce the occurrence of MDRO.
Keywords: COVID-19, Pneumonia, ICU, Antimicrobial resistance, BSIs, Acinetobacter baumannii
Background
The COVID-19 pandemic is an ongoing public health crisis causing the death of more than three million people worldwide at the end of May 2021 [1]. Critical COVID-19 is reported in around 5% of the cases, which requires intensive care admission [2]. The case-fatality rate is highly variable (1.39–14%) depending on the demography of infection, clinical standard, and epidemic wave dynamics [3]. During the course of hospitalization, it is difficult to predict the secondary bacterial infections, which warrant the use of empirical antimicrobials in patients with severe COVID-19. Recent reports show conflicting results regarding the prevalence of secondary bacterial infections (ranges from 14.3 to 67.7%), particularly bloodstream infections [4–8]. In addition, previous reports have shown prolonged hospital stay, morbidity, and mortality (odds ratio = 3.31, 95% CI 1.82–5.99) in the presence of bacterial superinfections [9, 10]. There is a scarcity of data from India regarding the BSIs and their impact on mortality in COVID-19. Knowing the local epidemiology and the impact of BSIs is paramount in order to apply prompt management and guide an empirical antimicrobial therapy when clinically appropriate. We aim to assess the prevalence, clinical profile, risk factors, frequency and distribution of microorganisms, antimicrobial susceptibility, and clinical outcome in severe COVID-19 with BSIs.
Methods
This retrospective observational study was carried out at a tertiary care centre in western Rajasthan, India, in a dedicated COVID-ICU referral centre. From July 2020 to December 2021, all patients with confirmed COVID-19 (RT-PCR positive on nasopharyngeal or oropharyngeal swab) who were admitted to COVID-19 ICU were included. Only patients who developed BSIs 48 h after the hospital admission were included. Patients who already had BSIs at the time of current admission were excluded. The study was approved by the institutional ethical committee (reference no -AIIMS/IEC/2020/3174). Bloodstream infections were defined by the presence of viable bacterial or fungal microorganisms in the bloodstream (demonstrated by the positivity of one or more blood cultures) that have elicited an inflammatory response characterized by the alteration of clinical, laboratory, and hemodynamic parameters [11]. For skin contaminants (like coagulase-negative Staphylococcus), at least two consecutive blood cultures for the same pathogen were considered as BSIs. Isolation of the same microorganisms from the bloodstream within 14 days was not considered a novel event and excluded from the analysis. Blood cultures were usually sent in patients with persisting fever (> 38.3 C), raised leukocyte counts, clinical signs of new septic foci, and clinical deterioration after initial improvement. Regarding central venous access, we have tried to minimize it in a view of possible central line-associated bloodstream infections (CLABSI). The central line was inserted only in cases with mechanical ventilation and difficult peripheral access.
All electronic health records were searched, and the following clinical data were extracted: Demographic parameters (age, gender), risk factors for hospital-acquired BSIs like indwelling catheters, mechanical ventilation, use of immunosuppressants (corticosteroids or Tocilizumab for primary disease), comorbidities (end-stage renal disease, chronic liver disease, or malignancy), use of antibiotics, the timing of blood culture drawn, duration of ICU stay, and outcome of patients. Haematological and biochemical parameters were collected, including haemoglobin, total leukocyte count, platelet count, c-reactive protein (CRP), procalcitonin, and lactate dehydrogenase (LDH). Quick sequential organ failure assessment (q-SOFA) score at baseline (at the time of ICU admission) was collected for each patient. Septic shock was defined with persisting hypotension requiring vasopressors to maintain MAP ≥ 65 mm Hg and having a serum lactate level > 2 mmol/L (18 mg/dL) despite adequate volume resuscitation [12].
Identification of BSIs and antimicrobial susceptibility pattern
Eight to ten ml of blood samples were collected aseptically and were inoculated into the BACTEC (BD, Biomerieux, France) bottles. The bottles were then loaded onto the automated system and incubated for 5 days minimum. Upon flagging positive, the bottles were taken out of the instrument, and the time to detection (TTD) was noted. All those bottles which did not flag positive within five days were considered sterile. The positively flagged bottles were subjected to Gram’s stain and culture onto blood agar and MacConkey agar. After 16–18 h of incubation at 37 °C, the cultures were read for the growth of colonies and the Gram’s stain and catalase tests were performed for further work-up. Identification of microorganisms in the majority of the cases was made by an automated VITEK-2 system. Antibiotic susceptibility testing was performed by Kirby Bauer’s disk diffusion method. The zone of inhibition for different antibiotics is compared to the criteria set by the Clinical and Laboratory Standards Institute (CLSI) [13]. Multidrug-resistant organism (MDRO) was defined as acquired non-susceptibility to at least one agent in three or more antimicrobial categories [14]. CR-BSIs cases were defined as a carbapenem-nonsusceptible and extended-spectrum cephalosporin-resistant (e.g. ceftriaxone, ceftazidime, ceftizoxime, and cefotaxime) isolates recovered from bloodstream [15].
Statistical analysis
Data analysis was performed using SPSS software, version 20.0 (IBM Corp, Armonk, NY). Descriptive data were summarized and tabulated with continuous variables in the form of mean ± standard deviation, Median (inter quartile range) and categorical data in the form of percentages or frequencies. Linear regression model was used to highlight the association between day of culture positivity (duration from the day of hospitalization to blood culture positivity) and mortality. Coefficient of determination (R square) was calculated to show the percentage of variation in mortality attributed to day of culture positivity. Univariate analysis was performed to identify the association between various factors and incidence of BSIs. We then conducted a multivariate analysis of variables that were found to be significantly associated with Covid-19 with a similar threshold of significance (p-value < 0.05).
Results
During the study period of 6 months (July 2020–December 2021), a total of 750 patients with RT-PCR confirmed COVID-19 were admitted to ICU. Overall, 8.5% of patients (64/750) developed BSIs. Demographic and clinical characteristics of all patients (BSIs and non-BSIs) are summarized in Table 1. The median age of all patients with BSIs was 65 years (IQR 54–70). The demographic features (age, gender) did not differ significantly between BSIs and non-BSIs patients (Table 1). The median qSOFA score was 2. Significant prevalence of comorbidities was found in BSIs patients compared to non-BSIs (70% vs 39.7%). Among the comorbidities, hypertension, diabetes, ischemic heart disease, and end-stage renal disease were common in BSIs patients (50%, 43.8%, 10.9%, 9.4%, respectively). Only 26.6% of the patients had central venous access in the BSIs group. However, it did not differ significantly compared to non-BSIs patients (p-value = 0.31). Mechanical ventilation was used in 68.8% of patients with BSIs compared to 13.6% of non-BSIs patients (p-value = 0.001).
Table 1.
Demographic, clinical and laboratory data of COVID-19 ICU patients (BSIs and non-BSIs group)
| Parameters | All patients (n = 750) | Patients with BSIs (n = 64) | Non-BSIs (n = 686) | p value |
|---|---|---|---|---|
| Age (median) | 62 years (IQR 51–72) | 65 years (IQR 54–70) | 60 (IQR 48–69) | 0.31 |
| (Mean ± SD) | 60 ± 17.71 years | 61.32 ± 13.74 years | 59.13 ± 16.88 years | |
| Male (%) | 562 (75) | 42 (65.6) | 487 (71) | 0.37 |
| q SOFA Score (median) | 2 | |||
| Comorbidities and risk factors (%) | ||||
| Comorbidities# | 317 (42.3) | 44 (70) | 273 (39.7) | 0.01 |
| Hypertension | 183 (24.4) | 32 (50) | 151 (22) | |
| Diabetes mellitus | 207 (27.6) | 28 (43.8) | 179 (26) | |
| Ischemic heart diseases | 67 (8.9) | 7 (10.9) | 60 (8.7) | |
| Immunosuppressants use | 64 (8.5) | 9 (10.9) | 55 (8) | |
| CKD/ESRD$ | 40 (5.3) | 6 (9.4) | 34 (5) | |
| Hypothyroidism | 47 (6.3) | 5 (7.8) | 42 (6.1) | |
| Stroke | 41(5.5) | 4 (6.4) | 37 (5.4) | |
| COPD | 29 (3.9) | 2 (3.2) | 27 (3.9) | |
| Rheumatoid arthritis | 23 (3.1) | 2 (3.2) | 21 (3.0) | |
| Chronic liver disease | 29 (3.9) | 1 (1.6) | 28 (4.1) | |
| Invasive devices (insitu), (%) | ||||
| Foleys Catheter | 485 (64.7) | 46 (71.9) | 439 (64) | 0.20 |
| Endotracheal intubation | 137 (18.3) | 44 (68.8) | 93 (13.6) | 0.001 |
| Arterial line | 282 (37.6) | 21 (32.8) | 261 (38) | 0.24 |
| Central venous line | 251 (33.5) | 17 (26.6) | 234 (34.1) | 0.31 |
| Laboratory parameters, median (IQR) | ||||
| Hb (g/dl) | 11.1 [8.8–12.9] | 10 [8.25–12] | 11. 3 [8.45–13.1] | 0.51 |
| Total leukocyte counts (103/μl) | 10.7 [7.73–15.91] | 16.5 [11.2–22.8] | 10.7 [7.73–15.91] | 0.001 |
| Platelet count (103/μl) | 247 [161–350] | 198 [111–341] | 273 [198–362] | 0.10 |
| CRP (mg/L) | 100 [40.2–145.7] | 128 [97–171] | 96.1 [39.1–139.8] | 0.06 |
| Procalcitonin (ng/ml) | 0.92 [0.145–6.99] | 5 [1.75–15.5] | 0.19 [0.09–1.12] | 0.02 |
| AST (IU/L) | 48 [30–129] | 44 [29–134] | 49 [31–135] | 0.73 |
| ALT (IU/L) | 37 [22–89] | 42 [26–92] | 36 [21–89] | 0.60 |
| Blood urea nitrogen (mg/dl) | 48.5 [31–99.25] | 100 [56–130] | 35 [22–62] | 0.002 |
| Creatinine (mg/dl) | 1.1 [0.84–1.8] | 2 [1–3] | 1.04 [0.84–1.5] | 0.01 |
| Bacterial microorganism identified in blood-stream | ||||
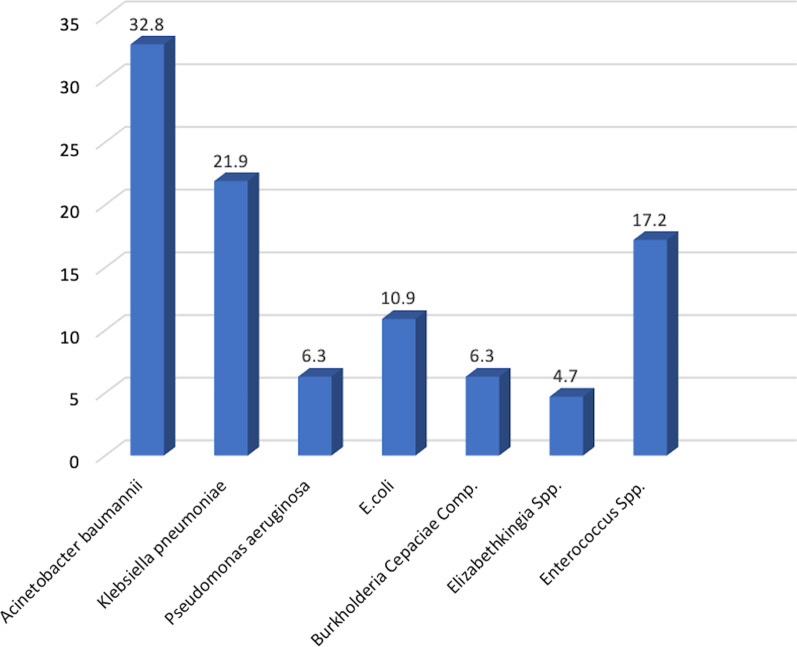
| Acinetobacter baumannii | 21 (32.8) | |||
| Klebsiella pneumoniae | 14 (21.9) | |||
| Enterococcus spp. | 11 (17.2) | |||
| Escherichia coli | 7 (10.9) | |||
| Burkholderia cepaciae complex | 4 (6.3) | |||
| Pseudomonas aeruginosa | 4 (6.3) | |||
| Elizabethkingia spp. | 3 (4.7) |
q SOFA Quick Sequential Organ Failure Assessment; CKD/ESRD chronic kidney disease/End stage renal disease; COPD chronic obstructive pulmonary disease; Hb hemoglobin; CRP C reactive protein; ALT alanine aminotransferase; AST aspartate aminotransferase
#Some patients had more than one comorbid state
$Out of 40 patients, 14 patients needed hemodialysis support
Corticosteroids were used in all patients, including non-BSIs patients (as a part of the standard treatment regimen of severe COVID-19). Seven patients received tocilizumab for COVID-19 pneumonia and developed BSIs. Tocilizumab use was not associated with incidence of BSIs (12.3% vs 8.2% in tocilizumab vs non-tocilizumab group, p-value = 0.29). Two patients were renal transplant recipients and receiving tacrolimus during hospitalization. Piperacillin-tazobactam (n = 42) was the most common empirical antibiotic used, followed by Levofloxacin (n = 7) and Ceftriaxone (n = 5). Subsequently, antibiotics were changed based on the blood culture sensitivity report and clinical improvement.
Biochemical parameters showed significant elevated CRP and procalcitonin levels in BSIs patients on admission (median CRP 128 mg/L and procalcitonin 5 ng/ml, Table 1). The median time from onset of symptoms to ICU admission was 5 days (IQR 3–7), and the median time from ICU admission to the first BSI episode was 8 days (IQR 4–11.8). The overall mortality rate was 100% (64) in the BSIs group and 33.8% (232) in the non-BSIs group, which was statistically significant (p-value < 0.0001). The patients who died in the hospital spent a median of 11.5 days (IQR 7.25–16 days). Out of 64 patients with BSIs, 43 developed septic shock, and three patients had features of septic encephalopathy. We analyzed the role of BSIs and sepsis in the clinical outcome of COVID-19 patients admitted to ICU. Linear regression analysis showed a positive correlation (coefficient of determination, R2 = 74.2%) between time to positivity of blood cultures and time to death (from the day of hospitalization) after adjusting confounding factors like age and sex (Fig. 1). Univariate analysis showed a significant association of procalcitonin level, leukocytosis and presence of comorbidities with BSIs (Table 2). Further multivariate analysis revealed comorbidities, mechanical ventilation, and raised leukocyte counts as an independent predictor of BSIs (Table 2).
Fig. 1.
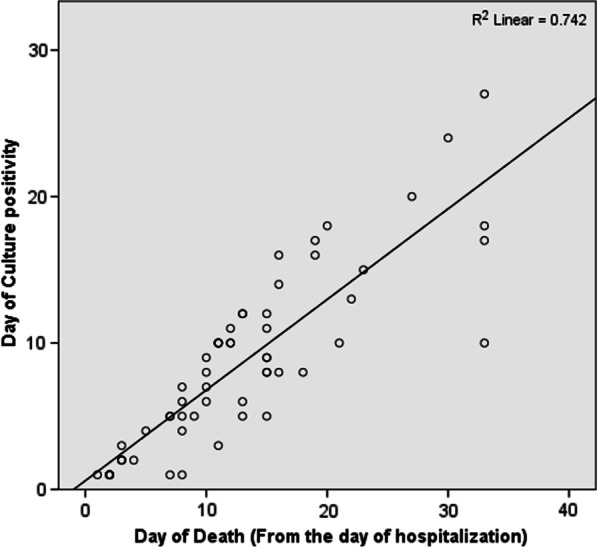
Linear regression graph showing positive correlation between day of culture positivity and day of mortality
Table 2.
Different variables and their associations with the incidence of BSIs in Covid-19 ICU patients in the univariate and multivariate analysis
| Characteristics | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|
| OR (95% CI) | p value | OR (95% CI) | p value | |
| Age (> 60 years) | 1.0 (0.98–1.2) | 0.36 | ||
| Gender (Male) | 0.78 (0.36–1.7) | 0.53 | ||
| CRP (> 6 mg/L) | 1.3 (0.97–1.8) | 0.31 | ||
| Procalcitonin (> 0.05 ng/ml) | 1.8 (0.89–2.7) | 0.04 | 1.5 (0.78–2.2) | 0.12 |
| Comorbiditiesa | 3.2 (1.6–6.3) | 0.003 | 2.9 (1.6–5.8) | 0.042 |
| Fever | 1.7 (0.86–3.3) | 0.129 | ||
| TLC (> 14 × 103/μl) | 4.2 (2.8–8.9) | 0.006 | 3.7 (2.6–7.10) | 0.01 |
| Immunomodulatory drugs (Tocilizumab) | 1.4 (0.54–2.1) | 0.29 | ||
| Mechanical Ventilation | 9.0 (4.7–17.4) | < 0.001 | 4.1 (2.9–10.4) | 0.001 |
CRP C reactive protein, OR odds ratio, CI confidence interval
aPatient with comorbid conditions which could be single or combination of more than one disease, refer to Table 1
Characteristics of microorganisms and antimicrobial resistance profile associated with BSIs
All of the BSIs were monomicrobial in this study (Table 1). The majority of the isolates were Gram-negative microorganisms (53/64, 82.8%). Among Gram-positive microorganisms, all isolates were from the Enterococcus group (11/64, 17.2%). The most common Gram-negative bacteria (GNB) were Acinetobacter baumannii (32.8%, 21/64), and Klebsiella pneumoniae (21.9%, 14/64). The remaining Gram-negative microorganisms (17.2%) were Burkholderia cepaciae complex, Pseudomonas aeruginosa, and Elizabethkingia spp. (Fig. 2). None of the patients had polymicrobial BSIs (isolation of > 1 microorganism from the bloodstream) in this study. Out of sixty-four patients, 57.8% patients were infected with MDRO. The majority of MDRO belonged to K. pneumoniae and Enterococcus spp. The incidence of Carbapenems resistance Gram-negative bacteria (CR-GNB) was 47.2% (25/53).
Fig. 2.
Frequency and distribution of pathogenic organism isolated from COVID-19 ICU patients (%)
Among isolated pathogens, the highest resistance for Acinetobacter baumannii was observed with ceftriaxone (76.2%), and piperacillin-tazobactam (76.2%, Table 3). Klebsiella pneumoniae isolates showed the highest resistance rate against aztreonam (85.7%). All E. coli isolates were resistant to ceftriaxone (100%). Among Gram-positive organisms, Enterococcus spp. showed maximum resistance for erythromycin, and ampicillin (90.9%, and 81.8%, respectively), (Table 4).
Table 3.
Antibiotic resistance pattern of predominant Gram-negative blood stream isolates in Covid-19 ICU patients (%)
| Antibiotic |
Acinetobacter baumannii (n = 21) |
Klebsiella pneumoniae (n = 14) | Psuedomonas aeruginosa (n = 4) | Escherichia coli (n = 7) | Burkholderia cepacia complex (n = 4) | Elizabethkingia spp. (n = 3) |
|---|---|---|---|---|---|---|
| Ceftriaxone | 76.2 | 71.4 | NA | 100 | NA | NA |
| Cefepime | 52.4 | 78.6 | 0 | 85.7 | NA | 66.6 |
| Ceftazidime | NA | NA | NA | NA | 50 | NA |
| Piperacillin-Tazobactam | 76.2 | 78.6 | 0 | 71.4 | NA | 33.3 |
| Meropenem | 33.3 | 21.4 | 0 | 14.3 | 75 | 100 |
| Colistin | 9.5 | 7.1 | 0 | 14.3 | 50 | 33.3 |
| Ciprofloxacin/Levofloxacin | 66.7 | 78.6 | NA | 85.7 | 50 | NA |
| Cefoperazone/sulbactam | NA | 71.4 | 0 | 57.1 | NA | NA |
| Amikacin | 52.4 | 57.1 | 0 | 57.1 | NA | 66.6 |
| Tigecycline | 0 | 0 | NA | 0 | NA | NA |
| Aztreonam | NA | 85.7 | 50 | 71.4 | NA | 66.6 |
| Imipenem | 9.5 | NA | 0 | 28.6 | NA | NA |
| Ertapenem | 23.8 | 71.4 | NA | NA | NA | NA |
Table 4.
Antibiotic resistance pattern of predominant Gram-positive blood stream isolates in Covid-19 ICU patients (%)
| Antibiotic | Enterococcus spp. (N = 11) |
|---|---|
| Ampicillin | 81.8 |
| Tigecycline | 0 |
| Ciprofloxacin | 81.8 |
| Tetracycline | 54.5 |
| Erythromycin | 90.9 |
| Linezolid | 0 |
| Teicoplanin | 18.1 |
| Vancomycin | 18.1 |
Discussion
The emerging data regarding the secondary bacterial infections in COVID-19 are scarce and conflicting, even more so with BSIs. In this study, we found the overall prevalence of BSIs in ICU patients to be 8.5%. A recent study from India found the prevalence of secondary infections to be 3.6% [16]. However, in their report, both hospital-acquired and community-acquired cases were taken. In addition, apart from BSIs, respiratory specimens were also analyzed, which could be contaminants or colonizers. Another report by Khurana et al. showed a 13% prevalence of secondary infections [17]. Contrary to previous studies, Lai et al. described the prevalence of BSIs up to 50% among non-survivors severe COVID-19 pneumonia patients [18]. Further large prospective studies are needed to determine the precise prevalence of BSIs in severe COVID-19 and its implications on the outcome of patients.
Interestingly, we found a 100% mortality rate in severe COVID-19 pneumonia patients with BSIs. Previous studies have reported 21–68% mortality in this group of patients [4, 5, 16, 19]. The disparity in mortality rate could be due to various factors, first of all the severity of the COVID-19 illness and the need for ICU admission. In previous studies, other sources of superinfection were analyzed e.g.: respiratory, urinary tract, and local tissue samples with a higher possibility of contaminations. Furthermore, the follow-up period was in some studies was brief or incomplete. Comorbidity also played an important role in mortality, with nearly half of the patients in this study having diabetes and hypertension. Few studies also showed high COVID-19 associated mortality in the male gender [20, 21]. However, our report did not find a similar result. Finally, we found sepsis as a determining factor in clinical outcomes, with a temporal relationship between sepsis/septic shock and mortality.
In this report, we found significantly elevated CRP and procalcitonin levels in patients who developed BSIs. However, the utility of inflammatory markers for predicting BSIs and empirical initiation of antibiotics in these patients remains debatable. Few studies reported the poor correlation of serum procalcitonin and CRP levels with bacterial coinfections, especially in the setting of immunomodulatory/immunosuppressive therapy [5, 19, 22]. Kreitmann et al. reported that median CRP and procalcitonin did not differ significantly in patients with or without bacterial coinfections (median procalcitonin, 0.4 vs 0.72 ng/ml, CRP, 182 vs 159 mg/L) [23]. Although procalcitonin levels were significantly raised in the BSIs group, we did not find an independent association between the incidence of BSIs and procalcitonin and other inflammatory markers (CRP). The inflammatory markers should be interpreted cautiously in COVID-19 before the initiation of empirical antibiotics.
Gram-negative microorganisms were predominant BSIs in this report. Similar observations were described by a multicentric study from India with a predominance of Gram-negative pathogens (78%) [16]. Conversely, Elabaddi et al. and a few other studies reported the increased prevalence of Gram-positive microorganisms, particularly Staphylococcus aureus (prevalence varies from 44 to 79.6%) in COVID-19 ICU patients [5, 19]. This heterogenicity in prevalence and distribution of microorganisms may attribute to different patient settings, the number of patients on mechanical ventilation, duration of hospital stays and follow-up and isolation of the pathogen from other specimens (like respiratory, urine and pus samples) in addition to BSIs. In this study, we observed the relatively high proportion of Enterococcus isolates as BSI, and the majority were multidrug-resistant (81.8% MDRO). Bonazetti et al. proposed a theory that SARS-CoV-2 mediated disruption of the gut barrier and bacterial translocation could trigger increased BSIs, especially Enterococci spp. [4]. Genotypic analysis was not available at our Centre, which would have provided some insight into this occurrence. Conflicting evidences are emerging regarding the prevalence of MDRO in the COVID-19 pandemic. Few reports showed a decreased prevalence of MDRO due to effective implementation of infection control practices; in contrast, there are studies that showed a high transmission of MDRO due to prolonged ICU stay and use of multiple antibiotics in COVID-19 [24–26].
In this study, we also observed the high prevalence of A. baumannii (32.8%). However, previous reports described A. baumanni in < 1% of the cases [4, 27, 28]. The other worrying aspect was the high proportion of CR-GNB in these isolates (nearly 50%). This increase in prevalence and resistance could be explained by prolonged ICU stay, mechanical ventilation, central venous catheter use, and inadvertent use of carbapenem with antimicrobial selection pressure, which are the usual triggers [29, 30]. Moreover, inadequate compliance with infection control procedures (like hand hygiene, disinfection of hospital equipment and environment) is another vital factor contributing to multidrug resistance organism infections.
The appropriateness of the use of empirical antimicrobials in severe COVID-19 patients is a matter of intense debate. According to a recent systematic review, the prevalence of empirical antibiotic use in COVID-19 patients was 74% [31]. Which was lower than our study (> 90% in our report); however, the pool of the patients in that review also included the mild to moderate COVID-19 patients. Despite the increasing use of empirical antibiotics, there is a lack of data favouring their use. Chedid et al. did not found a significant difference in antibiotic use among survivors and non-survivors COVID-19 patients [31]. Furthermore, exposure to antimicrobials increases the risk of drug resistance. According to a recent study, the use of combination antimicrobials was found to be associated with secondary infections [32]. Future prospective studies are needed to validate the aforementioned points before reaching the consensus regarding empirical antibiotic use in COVID-19.
This study has a number of limitations; first, this was a retrospective analysis which probably led to selection bias and inclusion of severe patients, which may have impacted the clinical outcome. Secondly, data were from a single centre, which may preclude its generalized applicability. All patients received corticosteroids and other immunosuppressants resulting in a high prevalence of secondary bacterial infections. In addition, the information about prognostic markers (e.g. SOFA, APACHE score) and central line days (central line insertion/1000 catheter days) was also lacking. Lastly, the lack of prospective design precluded the genotypic analysis of MDRO (A. baumannii), which is vital in view of patient cross-contamination in ICU settings.
Conclusions
BSIs in severe Covid-19 are associated with poor outcome. We report a high prevalence of Acinetobacter baumannii BSIs in the ICU setting. The presence of comorbidities and leukocytosis should alert the clinicians for possible BSIs. At the same time, clinicians should be prudent while interpreting the results of CRP and procalcitonin. We encourage implementing Antimicrobial stewardship and infection control practices, which could reduce the secondary bacterial infections in COVID-19 illness. Further prospective studies are warranted to determine the precise burden of secondary bacterial infections and their impact on mortality and morbidity in COVID-19.
Acknowledgements
We thank all healthcare workers who took part in patients management.
Abbreviations
- BSIs
Bloodstream infections
- Covid-19
Corona virus disease
- RT-PCR
Reverse-transcription polymerase chain reaction
- ICU
Intensive care unit
- MDRO
Multidrug-resistance microorganisms
- CLABSI
Central line-associated blood stream infections
- CRP
C-reactive protein
- LDH
Lactate dehydrogenase
- q-SOFA
Quick sequential organ failure assessment
- ESRD
End stage renal disease
- GNB
Gram-negative bacteria
- TGF-β
Transforming growth factor-β
- CR-GNB
Carbapenems resistance Gram-negative bacteria
Authors' contributions
DK, DSM, NP, and VN developed the study conception and design. VT, NM, GKB, AS undertook data curation. DK, DSM, ND performed data analysis and interpretation. DSM, NK, NP, and VN drafted manuscript. PB, MKG, SM, ND conducted critical revision of the manuscript for important intellectual content. SM, MKG, and ND provided administrative, technical or material support. DSM is the guarantor of the study. The corresponding author attests that all listed authors meet the ICMJE criteria for authorship and that no other meeting the criteria have been omitted. All authors read and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
This study was approved by the institutional ethical committee of All India Institutes of Medical Sciences Jodhpur, Rajasthan (Reference No—AIIMS/IEC/2020/3174). The study was conducted in accordance with ethical principles for medical research involving human subjects as outline in the Declaration of Helsinki. Written informed consent was obtained from all participants and parents/legally authorized representative of minor participants involved in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Naveenraj Palanisamy, Email: naveendoc22@gmail.com.
Nakka Vihari, Email: nakkavihari13@gmail.com.
Durga Shankar Meena, Email: dsmims14@gmail.com.
Deepak Kumar, Email: deepak1007sharma@gmail.com.
Naresh Midha, Email: midhanaresh@gmail.com.
Vibhor Tak, Email: takv@aiimsjodhpur.edu.in.
Ankur Sharma, Email: sharmaank@aiimsjodhpur.edu.
Gopal Krishana Bohra, Email: gopalbohra17@gmail.com.
Nikhil Kothari, Email: kotharin@aiimsjodhpur.edu.in.
Naveen Dutt, Email: duttn@aiimsjodhpur.edu.in.
Pradeep Kumar Bhatia, Email: bhatiapk@aiimsjodhpur.edu.in.
Mahendra Kumar Garg, Email: mkgargs@gmail.com.
Sanjeev Misra, Email: misralko@gmail.com.
References
- 1.WHO COVID-19 Dashboard. Geneva: World Health Organization, 2020. Available online: https://covid19.who.int/. Accessed 8 June 2021.
- 2.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 3.Jain VK, Iyengar K, Vaish A, Vaishya R. Differential mortality in COVID-19 patients from India and western countries. Diabetes Metab Syndr. 2020;14:1037–1041. doi: 10.1016/j.dsx.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonazzetti C, Morena V, Giacomelli A, Oreni L, Casalini G, Galimberti LR, et al. Unexpectedly high frequency of Enterococcal BSIs in coronavirus disease 2019 patients admitted to an Italian ICU: an observational study. Crit Care Med. 2021;49:e31–e40. doi: 10.1097/CCM.0000000000004748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giacobbe DR, Battaglini D, Ball L, Brunetti I, Bruzzone B, Codda G, et al. BSIs in critically ill patients with COVID-19. Eur J Clin Invest. 2020;50:e13319. doi: 10.1111/eci.13319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langford BJ, So M, Raybardhan S, Leung V, Westwood D, MacFadden DR, et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26:1622–1629. doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaillancourt M, Jorth P. The unrecognized threat of secondary bacterial infections with COVID-19. mBio. 2020;11:e01806–20. doi: 10.1128/mBio.01806-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ripa M, Galli L, Poli A, Oltolini C, Spagnuolo V, Mastrangelo A, COVID-BioB Study Group, et al. Secondary infections in patients hospitalized with COVID-19: incidence and predictive factors. Clin Microbiol Infect. 2021;27:451–57. [DOI] [PMC free article] [PubMed]
- 9.Musuuza JS, Watson L, Parmasad V, Putman-Buehler N, Christensen L, Safdar N. Prevalence and outcomes of co-infection and superinfection with SARS-CoV-2 and other pathogens: a systematic review and meta-analysis. PLoS ONE. 2021;16:e0251170. doi: 10.1371/journal.pone.0251170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buehler PK, Zinkernagel AS, Hofmaenner DA, Wendel Garcia PD, Acevedo CT, Gómez-Mejia A, et al. Bacterial pulmonary superinfections are associated with longer duration of ventilation in critically ill COVID-19 patients. Cell Rep Med. 2021;2:100229. doi: 10.1016/j.xcrm.2021.100229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plachouras D, Kärki T, Hansen S, Hopkins S, Lyytikäinen O, Moro ML, Reilly J, Zarb P, Zingg W, Kinross P, Weist K, Monnet DL, Suetens C, The Point Prevalence Survey Study Group. Antimicrobial use in European acute care hospitals: results from the second point prevalence survey (PPS) of healthcare-associated infections and antimicrobial use, 2016 to 2017. Euro Surveill. 2018;23(46):1800393. [DOI] [PMC free article] [PubMed]
- 12.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA. 2016;315(8):801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphries RM, Ambler J, Mitchell SL, Castanheira M, Dingle T, Hindler JA, CLSI Methods Development and Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing, et al. CLSI methods development and standardization working group best practices for evaluation of antimicrobial susceptibility tests. J Clin Microbiol. 2018;56:e01934–17. [DOI] [PMC free article] [PubMed]
- 14.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 15.Guh AY, Bulens SN, Mu Y, Jacob JT, Reno J, Scott J, et al. Epidemiology of Carbapenem-resistant Enterobacteriaceae in 7 US Communities, 2012–2013. JAMA. 2015;314(14):1479–1487. doi: 10.1001/jama.2015.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vijay S, Bansal N, Rao BK, Veeraraghavan B, Rodrigues C, Wattal C. Secondary infections in hospitalized COVID-19 patients: Indian experience. Infect Drug Resist. 2021;14:1893–1903. doi: 10.2147/IDR.S299774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khurana S, Singh P, Sharad N, Kiro VV, Rastogi N, Lathwal A. Profile of co-infections & secondary infections in COVID-19 patients at a dedicated COVID-19 facility of a tertiary care Indian hospital: implication on antimicrobial resistance. Indian J Med Microbiol. 2021;39:147–153. doi: 10.1016/j.ijmmb.2020.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai CC, Wang CY, Hsueh PR. Co-infections among patients with COVID-19: the need for combination therapy with non-anti-SARS-coV-2 agents? J Microbiol Immunol Infect. 2020;53:505–512. doi: 10.1016/j.jmii.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elabbadi A, Turpin M, Gerotziafas GT, Teulier M, Voiriot G, Fartoukh M. Bacterial coinfection in critically ill COVID-19 patients with severe pneumonia. Infection. 2021;3:1–4. doi: 10.1007/s15010-020-01553-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin JM, Bai P, He W, Wu F, Liu XF, Han DM. Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health. 2020;8:152. doi: 10.3389/fpubh.2020.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yanez ND, Weiss NS, Romand JA, Treggiari MM. COVID-19 mortality risk for older men and women. BMC Public Health. 2020;20:1742. doi: 10.1186/s12889-020-09826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bari SF, Khan A, Lawson T. C reactive protein may not be reliable as a marker of severe bacterial infection in patients receiving tocilizumab. BMJ Case Rep. 2013;2013:bcr2013010423. [DOI] [PMC free article] [PubMed]
- 23.Kreitmann L, Monard C, Dauwalder O, Simon M, Argaud L. Early bacterial co-infection in ARDS related to COVID-19. Intensive Care Med. 2020;46:1787–1789. doi: 10.1007/s00134-020-06165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole J, Barnard E. The impact of the COVID-19 pandemic on healthcare acquired infections with multidrug resistant organisms. Am J Infect Control. 2021;49:653–654. doi: 10.1016/j.ajic.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donà D, Di Chiara C, Sharland M. Multi-drug-resistant infections in the COVID-19 era: a framework for considering the potential impact. J Hosp Infect. 2020;106:198–199. doi: 10.1016/j.jhin.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baiou A, Elbuzidi AA, Bakdach D, Zaqout A, Alarbi KM, Bintaher AA. Clinical characteristics and risk factors for the isolation of multi-drug-resistant Gram-negative bacteria from critically ill patients with COVID-19. J Hosp Infect. 2021;110:165–171. doi: 10.1016/j.jhin.2021.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. 2008;46:1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 29.Montrucchio G, Corcione S, Sales G, Curtoni A, De Rosa FG, Brazzi L. Carbapenem-resistant Klebsiella pneumoniae in ICU-admitted COVID-19 patients: keep an eye on the ball. J Glob Antimicrob Resist. 2020;23:398–400. doi: 10.1016/j.jgar.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pascale R, Bussini L, Gaibani P, Bovo F, Fornaro G, Lombardo D. Carbapenem resistant bacteria in Intensive Care Unit during COVID-19 pandemic: multicenter before-after cross sectional study. Infect Control Hosp Epidemiol. 2021 doi: 10.1017/ice.2021.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chedid M, Waked R, Haddad E, Chetata N, Saliba G, Choucair J. Antibiotics in treatment of COVID-19 complications: a review of frequency, indications, and efficacy. J Infect Public Health. 2021;14:570–76. doi: 10.1016/j.jiph.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Y, Li W, Wang Z, Chen H, Tian L, Liu D. Nosocomial infection among patients with COVID-19: a retrospective data analysis of 918 cases from a single center in Wuhan. China Infect Control Hosp Epidemiol. 2020;41:982–983. doi: 10.1017/ice.2020.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.