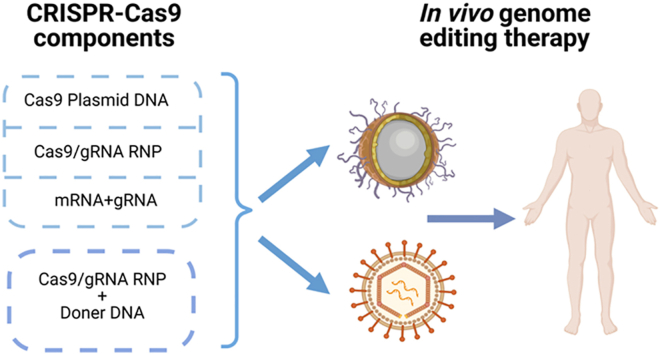
Abstract
Within less than a decade since its inception, CRISPR-Cas9-based genome editing has been rapidly advanced to human clinical trials in multiple disease areas. Although it is highly anticipated that this revolutionary technology will bring novel therapeutic modalities to many diseases by precisely manipulating cellular DNA sequences, the low efficiency of in vivo delivery must be enhanced before its therapeutic potential can be fully realized. Here we discuss the most recent progress of in vivo delivery of CRISPR-Cas9 systems, highlight innovative viral and non-viral delivery technologies, emphasize outstanding delivery challenges, and provide the most updated perspectives.
KEY WORDS: CRISPR, Cas9, Genome editing therapy, LNP, Nanoparticle, AAV, Lentivirus, In vivo
Graphical abstract
CRISPR-Cas9 genome editing technology has the potential to provide life changing cures to many patients. Safe and efficient delivery vectors are the key to the broad clinical success.
1. Introduction
Previous genome editing technologies, such as zinc-finger nucleases and transcription activator-like effector nucleases, both of which require an individually designed protein molecule to recognize each distinct target gene, are labor intensive, costly, and time consuming. Discovery of the clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR-associated protein 9 (Cas9) system added a robust tool to the genome editing toolbox. It dramatically lowered the bar of conducting genome editing experiments, making it one of the most useful techniques in biomedical research1, 2, 3. Additionally, it has shown unprecedented potential in revolutionizing treatment of human diseases4, 5, 6. CRISPR-Cas9 therapeutics hold apparent advantages over traditional gene therapies, such as gene replacement therapy, RNA interference (RNAi) therapy, and antisense therapy, in that the repair on a mutated gene or the insertion of a missing gene is permanently and precisely ironed to the host genome after a single genome editing event. Due to its high accuracy of target gene recognition, CRISPR-Cas9 averts activation of oncogenes and mutagenesis. In addition, it eliminates repeated dosing in certain applications, although repeated administration of the CRISPR-Cas9 system has improved genome editing efficiency in some preclinical studies. CRISPR-Cas9 therapeutics remind everyone with ever stronger signal that the era of precision medicine is coming.
Although some diseases are very likely to quickly benefit from CRISPR-Cas9 technology through ex vivo manner, the more profound clinical success of CRISPR-Cas9 therapeutics can only be accomplished if the system can be directly administered to patients. However, safe and effective in vivo delivery systems for the components of CRISPR-Cas9 therapeutics are still yet to mature (Fig. 1). This article summarizes the most recent progress of in vivo delivery of CRISPR-Cas9 systems for genome editing, highlights innovative delivery technologies using viral and non-viral vectors, emphasizes outstanding challenges to in vivo delivery faced by the CRISPR-Cas9 genome editing community, and provides the most updated perspectives.
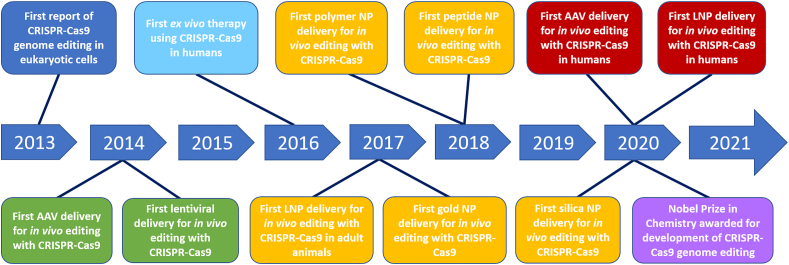
Figure 1.
Milestones of in vivo delivery of CRISPR-Cas9 therapeutics. Key developmental milestones for in vivo delivery of CRISPR-Cas9. AAV: adeno-associated virus, NP: nanoparticle, LNP: lipid nanoparticle. CRISPR: clustered regularly interspaced short palindromic repeats, Cas9: CRISPR-associated protein 9.
2. The biology of CRISPR-Cas9 mediated genome editing in mammalian cells
Though CRISPR-Cas systems have bacterial origins from species such as, but not limited to, Streptococcus pyogenes, Staphylococcus aureus, and Streptococcus thermophilus, their utility reaches beyond the bacterial immune system, and can be used for genome editing in mammalian cells2,7. CRISPR-Cas systems can be differentiated based on the number of Cas proteins involved in their mechanism. A type I system uses multiple Cas proteins while a type II system uses only one Cas protein8,9. Because of this contrast, type II CRISPR-Cas systems are more desirable for genome engineering as its components can be produced with greater ease. Type II CRISPR-Cas systems involve multiple key components for genome engineering, including a Cas protein, CRISPR RNA (crRNA), and trans-activating crRNA (tracrRNA)8,10, 11, 12. Cas9 is an endonuclease with two nuclease domains. These domains are the HNH, which cleaves complementary DNA strands, and RuvC, which cleaves non-complementary DNA strands2,8.
Mammalian genome editing with CRISPR-Cas9 was simplified by engineering a single guide RNA (sgRNA), which takes the place of crRNA and tracrRNA components in the CRISPR-Cas system13. The sgRNA is essential for precise gene editing as its 5ʹ-terminal 20-nucleotide sequence interacts with a target sequence of host DNA via Watson and Crick base pairing rules, while its 3ʹ duplex structure allows binding to Cas9 proteins8. Before being able to perform a double strand break (DSB), Cas9 proteins must locate the target DNA sequence by recognizing the protospacer adjacent motif (PAM), which is a short sequence of nucleotides directly adjacent to the target DNA sequence8,11,14. Once a Cas9 protein binds to the target strand of DNA, it undergoes a conformational change, allowing for a targeted DSB to occur 3–4 nucleotides upstream of the PAM8,11,15.
Following a DSB, there are multiple fates for the broken chromosomal DNA. Primary DSB repair mechanisms include non-homologous end joining (NHEJ) or homology directed repair (HDR) pathways (Fig. 2). NHEJ involves direct ligation of the two broken chromosomal DNA strands to one another and is the main cellular DSB repair mechanism. This process is error-prone due to random insertions or deletions (indels) of nucleotides to assist in ligation, which can cause gene disruption via frameshift or nonsense mutations16. Beyond gene disruption, NHEJ can also be used to induce significant deletions when carried out with two different sgRNAs. Gene disruption or deletion via NHEJ is desirable when treating diseases whose pathogenesis is characterized by an over-expression of a certain protein, such as transthyretin amyloidosis. HDR pathways require the presence of an exogenous donor DNA template that is homologous to the target region of chromosomal DNA, allowing for gene insertion following DSB. HDR's applications rest in targeted integration of transgene. Gene editing with HDR is the desired manner for the treatment of diseases caused by missing an entire gene, such as X-linked retinitis pigmentosa, hemophilia A or B, and phenylketonuria. When compared, NHEJ is more efficient and can occur at any phase of the cell cycle. It is utilized to edit DNA more frequently than HDR, which is less efficient, especially in post-mitotic cells, such as neurons, retinal pigment epithelial (RPE) cells, cardiac myocytes and mature muscle cells. These two repair mechanisms are most used by researchers as they allow for a straightforward approach to gene disruption, deletion, correction, or insertion.
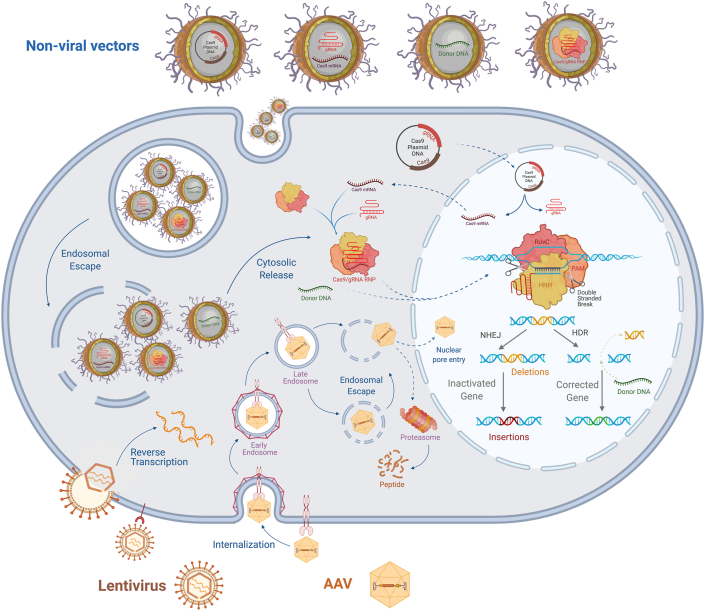
Figure 2.
CRISPR-Cas9 genome editing through viral or non-viral delivery. Representative depiction of mechanisms and strategies involved in CRISPR-Cas9 delivery with both viral and non-viral vectors. AAV and lentivirus both bind to cell surface receptors prior to cellular infection. Following cellular internalization, AAVs have the capacity to escape the endosomes and transport across the nuclear membrane prior to uncoating, though the capsid degradation mediated by proteasome can also occur in the cytoplasm. Following lentiviral cell membrane fusion is uncoating and release of its RNA contents, which then undergo reverse transcription to form complementary DNA. Non-viral vectors offer the advantage of carrying various forms of CRISPR-Cas9 cargoes including plasmid DNA, RNA, donor DNA, and RNP. Cellular entry of non-viral vectors is accomplished via endocytosis which requires the NP to escape these endosomes in order to carry out its intended genome editing. Following endosomal escape and cytosolic release, the cargo carried by a non-viral NP must travel to varying sites, such as the nucleus for transcription and/or cytoplasm for translation. Once necessary transcription and translation steps have taken place with nucleic acid delivery approaches, a RNP is formed and can translocate across the nuclear membrane for targeted genome editing. RNPs work to perform targeted DSBs by PAM- and sgRNA-mediated recognition of a specific sequence of chromosomal DNA. Once this recognition occurs, the Cas9 nuclease can perform a DSB utilizing its two nuclease domains the HNH and RuvC which cleave complementary and non-complementary DNA strands, respectively. Following a DSB, there are multiple fates for genome editing such as, but not limited to, NHEJ and HDR. NHEJ is utilized for genomic disruption or deletion, while HDR is utilized for gene correction, but requires the administration of an exogenous donor DNA template. AAV: adeno-associated virus, NP: nanoparticle, CRISPR: clustered regularly interspaced short palindromic repeats, Cas9: CRISPR-associated protein 9, RNP: ribonucleoprotein complexes, PAM: protospacer adjacent motif, DSB: double-stranded break, NHEJ: non-homologous end joining, HDR: homology-directed repair.
Alternative DSB repair mechanisms include homology-mediated end joining (HMEJ), microhomology-mediated end joining (MMEJ), and homology-independent targeted integration (HITI). MMEJ's mechanism includes 5–25 bp microhomologous arm sequences that align between both strands of the broken DNA, allowing for end joining once the nucleotide sequences directly adjacent to the original break and microhomologies are cleaved17,18. MMEJ can be utilized as a mechanism for gene insertion that requires shorter homology arms than HDR18,19. Additionally, HMEJ works in a similar mechanism to MMEJ, but instead with 800 bp homologous arm sequences to assist with DNA repair or transgene insertion19. Lastly, HITI requires administration of an exogenous donor DNA template for insertion, however unlike HDR, HITI does not require chromosomal DNA homology for insertion, and instead utilizes a three sgRNA system to cleave chromosomal and donor DNA prior to insertion20.
3. Selecting the right cargoes for CRISPR-Cas9-based gene editing therapy
Although Cas9 protein, sgRNA, and donor DNA template (if for HDR) must present at the nucleus of target cells for genome editing therapy, they are not necessarily the cargo originally loaded into the delivery systems for administration. To enable CRISPR-Cas9-based genome editing in vivo, there are multiple choices in terms of cargoes. Direct delivery of Cas9/sgRNA ribonucleoprotein (RNP) complexes is the most straightforward choice, due to the high genome editing efficiency, rapid onset of action, short resident time in the target cell, and reduced off-target effects. Alternatively, both the Cas9 protein and sgRNA may be replaced by other forms of entities, such as plasmid DNA, mRNA, or a viral genome inside a viral vector. In this case, Cas9 proteins and sgRNAs are produced on site in the target cells to form RNPs for genome editing. In addition, when HDR is the desired approach, even the donor DNA sequence can be delivered by way of plasmid DNA or viral genome. Depending on the disease, desired dosing regimen, route of administration, and delivery system, researchers have a variety of options to deliver DNA, RNA and/or protein payloads to the target cells (Fig. 2). There are four options for Cas9 enzymes: protein, plasmid DNA, mRNA, or viral genome, three options for sgRNA: RNA, plasmid DNA, or viral genome, and three options for DNA donor template: DNA oligos, plasmid DNA, or viral genome. Mathematically, there are twelve options (by multiplying four by 3) for NHEJ-mediated genome disruption or deletion, and thirty-six options (by multiplying four by three by 3) for HDR-mediated gene insertion. In this section, we discuss the options available for each component, analyze advantages and disadvantages of the options, and provide general guidance for consideration when making selections.
3.1. Cas9
Purified Streptococcus pyogenes Cas9 (SpCas9) protein is a positively charged molecule with a molecular weight of ∼163 kDa. The large molecule size represents a problem for efficient entry into both the cell and cell nucleus. In addition, its bacterial origin makes the Cas9 protein immunogenic in humans, which may decrease genome editing efficiency and increase the risk of a genome editing therapy21. Pre-existing anti-Cas9 antibodies have been detected in humans due to the frequent interaction with bacteria in the past thousands of years22. Furthermore, the purification process is still costly and time consuming. Moreover, it requires significant work to ensure the purity, and maintain stability and biological activity of Cas9 protein during the entire purification, formulation, shipping, and storage process. Using mRNA or plasmid DNA encoding Cas9 can avoid problems related to protein production and purification while reducing the cost. If mRNA or plasmid DNA are successfully delivered, Cas9 can be produced in target cells where gene manipulation is expected to occur. However, intracellular delivery is difficult for both naked mRNA and plasmid DNA as they are unstable and unable to enter target cells by themselves. When a comparison is made between them, mRNAs are more fragile and expensive. However, mRNAs do not pose the risk of insertional mutagenesis on chromosomal DNA. What's more, mRNAs do not need to enter cell nucleus and can produce Cas9 protein at cytosolic ribosomal sites as quickly as ∼6 h post intracellular delivery. This is a significant advantage over plasmid DNA which must be delivered into the cell nucleus to initiate Cas9 expression. In addition, mRNA-mediated Cas9 expression is more transient compared to plasmid DNA-mediated Cas9 transgene expression in target cells, further minimizing the exposure of the cellular genome to Cas9 and reducing the risk of off-target effects. The fourth option to deliver Cas9 is viral gene delivery. Cas9 genes can be engineered into the recombinant genome of viral vectors, such as adeno-associated viruses (AAVs) or lentiviruses, which are proficient in producing Cas9 in target cells. However, it has been shown that long-term Cas9 production in target cells led to increased off-target effects and immunogenicity21,23,24. Viral vectors also pose the risk of random gene integration into the host genome. Although both SpCas9 and Staphylococcus aureus Cas9 (SaCas9) genes can be delivered by AAVs, it is important to note that the genes of some novel Cas nucleases are too long to be packaged in a single AAV vectors. Finally, mRNA, plasmid DNA, and viral genome all require additional time to produce Cas9 enzymes in target cells, resulting in delayed action. The delay may decrease genome editing efficiency when an sgRNA is delivered concurrently, because sgRNAs may be degraded in the cell before Cas9 protein is made.
3.2. sgRNA
An sgRNA is a negatively charged macromolecule with a molecular weight of ∼31 kDa. Due to the ease of synthesis, sgRNA is a popular cargo choice. Alternatively, plasmid DNA and viral genome offer appealing advantages by expressing sgRNA right in target cells. Plasmid DNA takes additional time to transcribe sgRNA in the cell, causing a delayed expression of sgRNA. If Cas9 nuclease is co-delivered along with plasmid DNA encoding sgRNA, Cas9 nucleases may not survive in the cell by the time when sgRNA is expressed. This timing issue will lead to a decreased genome editing efficiency. However, if a plasmid DNA is delivered to express both Cas9 and sgRNA in target cells, the timing issue is generally not a concern. If sgRNA is expressed by viral vectors such as AAVs, sgRNA may be produced in target cells for a prolonged period of time. This may provide convenience and flexibility for Cas9 delivery as Cas9 protein is short-lived in cells.
3.3. DNA donor template
The selection of a donor DNA template applies to HDR-based and HITI-based genome editing. Exogenous DNA template can be delivered in the form of plasmid DNA, usually when the insertion is a long DNA sequence. Alternatively, the DNA donor can be delivered as a synthetic single-stranded DNA oligonucleotide when the insertion sequence is short. Although a plasmid DNA is more stable than a short DNA oligonucleotide, its intranuclear delivery efficiency is lower due to the larger size. Moreover, when a plasmid DNA is delivered, it is important to make sure that there is no CRISPR targeting site in the plasmid DNA. Otherwise, the HDR-edited gene locus will be cut by Cas9 protein, resulting in a failed chromosomal DNA repair. AAV vectors can also deliver the DNA donor template25, 26, 27, or simultaneously deliver both Cas9 and the donor DNA28. In the latter case, it helps increase gene editing efficiency because a single AAV vector gets two genome editing components into the same target cell.
4. Current vectors for in vivo delivery of CRISPR-Cas9 therapeutics
CRISPR-Cas9 system can be efficiently delivered into mammalian cells by different ways such as using delivery vectors or physical methods. Delivery vectors will be discussed in depth in this section (Table 1). However, physical methods, including microinjection and electroporation, are more applicable to ex vivo CRISPR-Cas9 based genome editing therapy. Thus, they are not discussed in this review. We suggest interested readers refer to the cited review articles covering physical methods29, 30, 31, 32.
Table 1.
Viral and non-viral vectors for in vivo delivery of CRISPR-Cas9 therapeutics.
| Delivery system | Payload option | Packaging capacity/loading capacity | Advantage | Disadvantage | Progress |
|---|---|---|---|---|---|
| Adeno-associated virus | Single-stranded DNA viral genome | ∼ 4.7 kb | High transduction efficiency Transduces dividing and non-dividing cells in various tissues Very low immunogenicity Clinically proven safe and efficient |
Limited packaging capacity Preexisting neutralizing antibodies Inflammation Long-term Cas9 expression may increase off-target effects High cost Difficult large-scale production |
Clinical stage |
| Lentivirus | Single-stranded RNA viral genome | ∼ 10 kb | High transduction efficiency Transduces dividing and non-dividing cells in various tissues Large packaging capacity Very low immunogenicity |
Complicated packaging construct High cost Difficult large-scale production |
Clinical stage |
| Lipid nanoparticles | RNP plasmid DNA encoding Cas9 and/or sgRNA Cas9 mRNA sgRNA Donor DNA |
Lack of report in most studies | High biocompatibility Minimal immunogenicity Relatively easy for large-scale production Low toxicity Temporal release of CRISPR-Cas9 may reduce off-target effects All-in-one delivery |
Limited delivery efficiency | Clinical stage |
| Polymer nanoparticles | RNP plasmid DNA encodinzg Cas9 and/or sgRNA Cas9 mRNA sgRNA Donor DNA |
Lack of report in most studies | Minimal immunogenicity Relatively easy for large-scale production Temporal release of CRISPR-Cas9 may reduce off-target effects All-in-one delivery |
Limited delivery efficiency Variable biocompatibility and toxicity Limited delivery efficiency |
Preclinical stage |
| Peptide nanoparticles | RNP plasmid DNA encoding Cas9 and/or sgRNA Cas9 mRNA sgRNA Donor DNA |
Lack of report in most studies | Relatively easy for large-scale production Temporal release of CRISPR-Cas9 may reduce off-target effects All-in-one delivery |
Limited delivery efficiency In vivo degradation Potential immunogenicity from foreign peptides |
Preclinical stage |
| Inorganic nanoparticles | RNP plasmid DNA encoding Cas9 and/or sgRNA Cas9 mRNA sgRNA Donor DNA |
5%–10% (w/w) loading efficiency by silica nanoparticles. Lack of report in other studies | High biocompatibility Minimal immunogenicity Relatively easy for large-scale production Temporal release of CRISPR-Cas9 may reduce off-target effects All-in-one delivery |
Limited delivery efficiency | Preclinical stage |
4.1. Current viral vectors for in vivo delivery of CRISPR-Cas9 system
Viruses are natural experts for in vivo gene delivery33. Recombinant viral vectors have been in leading positions for human gene therapy due to high efficacy and long-term transgene expression. Among many viral vectors, AAVs and lentiviruses are the most widely used vectors in clinical studies, dominating in vivo and ex vivo gene therapy clinical trials, respectively34, 35, 36. They have been playing critical roles as the delivery vectors in pioneering genome editing therapies (Fig. 3). Although adenoviruses, retroviruses, and herpes simplex viruses are also investigated for in vivo genome editing, their applications are limited by different weaknesses.
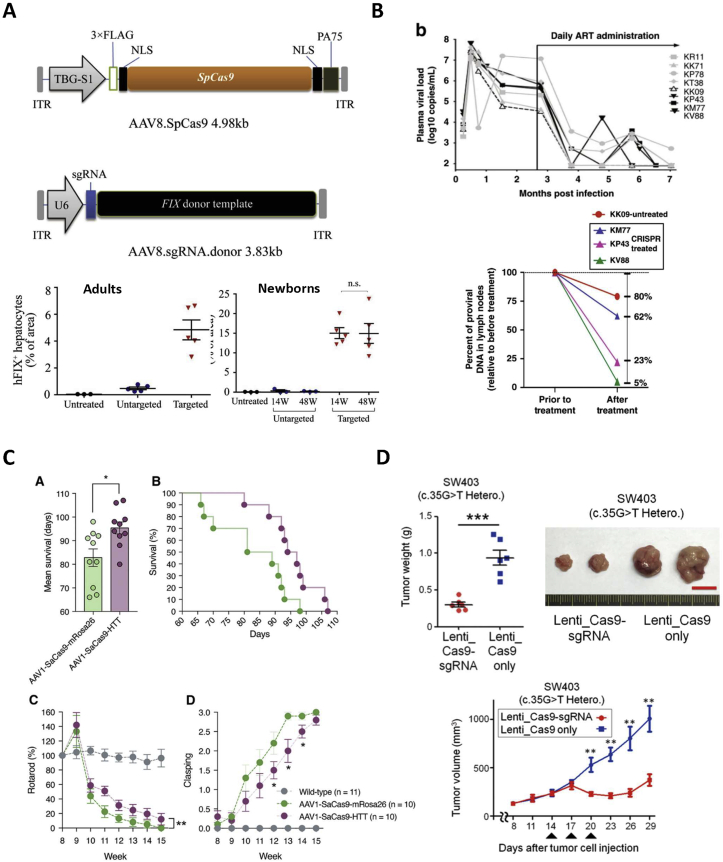
Figure 3.
Viral vector-mediated in vivo therapeutic genome editing. (A) Schematic representation of dual AAV8 strategy for site-specific gene insertion in hepatocytes treating Hemophilia B52. Copyright Elsevier 2020. Under the permission of the Creative Commons Attribution-Non Commercial-No Derivatives License 4.0 (CC BY-NC-ND 4.0) (https://creativecommons.org/licenses/by-nc-nd/4.0/). (B) AAV9 for systemic delivery of CRISPR-Cas9 for deletion of SIV proviral DNA in ART treated rhesus macaques158. Copyright Springer Nature 2020. Under the permission of the Creative Commons Attribution 4.0 international license (CC BY 4.0) international license (CC BY 4.0). (https://creativecommons.org/licenses/by/4.0/) (C) AAV1 for systemic delivery of CRISPR-Cas9 for disruption of mutant HTT in medium sized spiny neurons to treat Huntington’s disease144. Copyright Elsevier 2019. Under the permission of the Creative Commons Attribution-Non Commercial-No Derivatives License 4.0 (CC BY-NC-ND 4.0) (https://creativecommons.org/licenses/by-nc-nd/4.0/). (D) Systemic lentiviral delivery of CRISPR-Cas9 for KRAS disruption to suppress colorectal adenocarcinoma growth72. Copyright Cold Spring Harbor Laboratory Press 2018. Under the permission of the Creative Commons Attribution-Non Commercial License 4.0 (CC BY-NC 4.0) (https://creativecommons.org/licenses/by-nc/4.0/).
4.1.1. AAV
AAVs are very efficient vectors for in vivo gene delivery and gene editing as they can infect both dividing and non-dividing cells in varying tissues, mediate long-term and robust transgene expression, induce minimal immunogenicity, and have no known human diseases associated with their infection37, 38, 39. More than 250 AAV serotypes have been discovered and many of them have exhibited robust ability to deliver genes to various organs and cell types in vivo. Natural single stranded AAVs (ssAAVs) are composed of an icosahedral protein capsid that encapsulates a single-stranded DNA genome about 4.7 kb in length. Engineered self-complimentary AAVs (scAAVs) carry a double-stranded DNA genome, which reduces its capacity to carry transgenes40. Recombinant AAV genomes do not contain any gene encoding viral replication proteins, which makes space for transgenes, or expressing Cas9 nuclease, sgRNA, and in some cases donor DNA template41. Clinical success of AAV-based gene therapies has been well illustrated by regulatory approvals of Zolgensma®, Luxturna®, and Glybera®36.
Though AAVs represent an excellent platform for gene therapy, they still possess weaknesses when used for in vivo delivery of CRISPR-Cas9. One significant drawback is the packaging limitation, which complicates all-in-one delivery as SpCas9 itself is about 4.2 kb in size. Since recombinant AAVs must also contain regulatory elements essential for gene expression, such as the promoter region and polyadenylation signal, the small packaging capacity creates significant inconvenience and disqualifies AAVs for many applications where large gene sequences must be delivered. Solutions to the packaging limit challenge include using a truncated SpCas9, which is smaller in size but offers reduced genome editing efficiencies, or using SaCas9, which is about 3.1 kb in size and shows comparable genome editing efficiencies42,43. An alternative method is a dual-AAV strategy that splits the components of the transgene cassette into two independent viruses that are then co-infected44,45. Utilizing dual AAVs is especially valuable for HDR-based gene correction and insertion as a donor DNA template must also be included with Cas9 and sgRNA. Another significant problem faced by AAVs is the pre-existing neutralizing antibodies in patients from prior AAV infection, which requires serological testing before one can be treated38. AAV's long-term transgene expression can be seen as a merit for sustained gene therapy, however, it can also be a risk as continuous expression of Cas9 nucleases may cause significant off-target effects46. Lastly, large-scale production of AAV is still difficult47, 48, 49.
Since the first report of AAV-based CRISPR-Cas9 delivery for in vivo genome editing, it has made exciting progress in numerous disease models, including blood disorders50, 51, 52, metabolic liver diseases44,53, 54, 55, muscular diseases56, 57, 58, and ocular diseases59,60. In 2020, in a landmark clinical trial sponsored by Editas Medicines (NCT03872479), AAV became the very first CRISPR-Cas9 delivery vector for direct injection into a patient of Leber congenital amaurosis type 10.
4.1.2. Lentivirus
Lentivirus is an enveloped RNA virus with a single stranded genome that belongs to the retrovirus family61, 62, 63, 64, 65. Unlike other retroviruses, lentiviruses and can infect both dividing and non-dividing cells. Lentiviruses, such as HIV-1, are more complex than retroviruses as they contain an additional six genes; tat, rev, vpr, vpu, nef, and vif that are involved in viral replication and pathogenesis. However, some of these accessory genes involved in the virulence of lentiviruses can be removed to decrease their immunogenicity66,67. Typically, lentiviruses integrate their genomes into the host genome, which can significantly prolong transgene expression time, but this may increase the risk for adverse effects. Researchers have modified lentiviral integrase DNA to induce mutations leading to prevention of proviral integration, forming what is known as integration defective lentiviruses (IDLV)71.
Benefits of using lentiviruses for in vivo gene therapy include its 10 kb carrying capacity and prolonged transgene expression66,68. These attributes allow for decreased challenges with packaging limit when compared with AAV. Disadvantages of in vivo utilization include the potential for oncogenicity, insertional mutagenesis, and supra-physiological levels of RNA and/or proteins due to prolonged transgene expression time, however, these are reduced when using IDLVs69. Additionally, the inclusion of multiple regulatory genes involved in replication may further complicate the packaging construct69. Though they have promise as an in vivo delivery system, lentiviral vectors are an excellent option for ex vivo gene editing as they offer efficient cellular transduction in numerous cell types and shorter culture times needed for cell transfection70. These attributes make ex vivo editing with lentiviruses desirable due to their relative ease of use, which lessens challenges associated with large scale production needed for clinical utility.
Preclinical studies have demonstrated the utility of lentiviral vectors for both systemic and localized in vivo delivery of Cas9 and sgRNA for therapeutic gene disruption in wet age-related macular degeneration (AMD) and cancer models71, 72, 73. When used for ex vivo transduction, lentivirus has proved superior to other vectors in clinical studies. Two lentiviral vector-based ex vivo gene therapeutics, Kymriah® and Zynteglo®, have been approved for use by the US Food and Drug Administration (FDA) and European Medicines Agency (EMA), respectively36.
4.2. Non-viral vectors for in vivo CRISPR-Cas9 delivery
Many types of non-viral vectors used for protein, gene and RNAi delivery were adopted or modified for in vivo CRISPR-Cas9 delivery. These include liposomes, lipid nanoparticles (LNPs), polymeric nanoparticles, peptide nanoparticles, and inorganic nanoparticles (Fig. 4). They possess multiple attractive advantages, such as minimal immunogenicity, low cost in large scale production, the capability to deliver all components of a CRISPR-Cas9 system within one vector (all-in-one delivery), and the flexibility to deliver various cargoes (RNP, plasmid DNA, mRNA, sgRNA, donor DNA)32,74. Multiple non-viral vectors are evaluated for delivery of genes or RNAi in clinical studies.
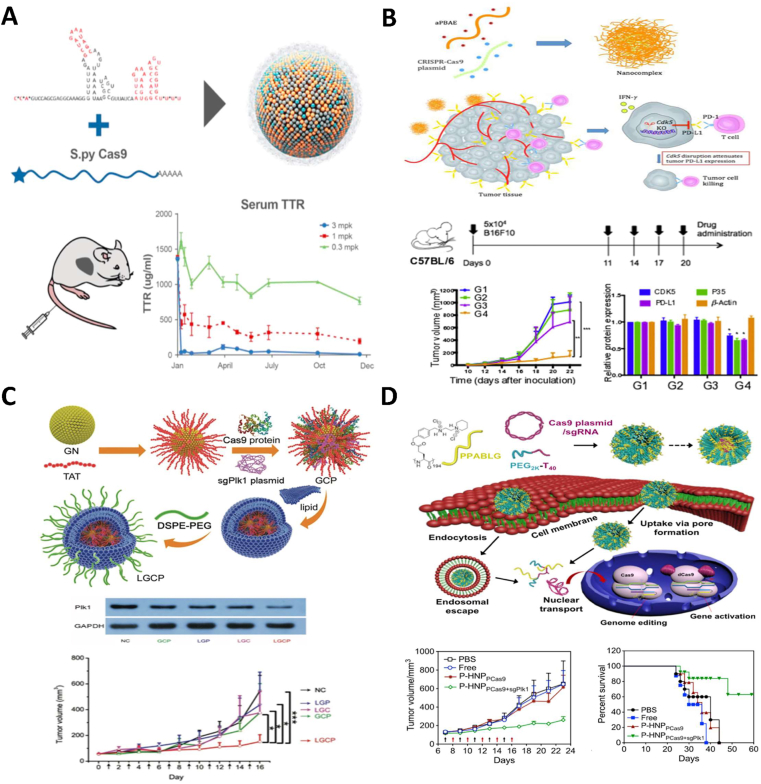
Figure 4.
Non-viral vector-mediated in vivo therapeutic genome editing. (A) Biodegradable LNP encapsulated Cas9 mRNA and sgRNA for TTR disruption in hepatocytes and prolonged reduction of serum TTR in a transthyretin amyloidosis model90. Copyright Cell Press 2018. Under the permission of the Creative Commons Attribution-Non Commercial-No Derivatives License 4.0 (CC BY-NC-ND 4.0) (https://creativecommons.org/licenses/by-nc-nd/4.0/). (B) Polymer nanocomplex for Cas9 plasmid DNA delivery for CDK5 disruption treating PD-L1-expressing melanoma and triple-negative breast cancer132. Copyright Elsevier 2020. Under the permission of the Creative Commons Attribution-Non Commercial-No Derivatives License 4.0 (CC BY-NC-ND 4.0) (https://creativecommons.org/licenses/by-nc-nd/4.0/). (C) Lipid encapsulated gold NP delivery of sgRNA plasmids and Cas9 protein for Plk1 disruption and melanoma suppression117. Copyright John Wiley and Sons 2017. Under the permission of the Creative Commons Attribution 4.0 international license (CC BY 4.0) (https://creativecommons.org/licenses/by/4.0/). (D) Helical polypeptide-based delivery of Cas9/sgRNA expressing plasmids for Plk1 disruption in HeLa cell tumors111. Copyright PNAS 2018. Under the permission of the Creative Commons Attribution-Non Commercial-No Derivatives License 4.0 (CC BY-NC-ND 4.0) (https://creativecommons.org/licenses/by-nc-nd/4.0/).
4.2.1. Lipid-based nanocarriers
Liposomes have been used for drug delivery for multiple decades75. Liposomes are characterized by a hydrophobic lipid bilayer and an inner aqueous pool76. In recent years, LNPs joined liposomes to address the challenge of safe and efficient gene delivery. LNPs are structurally distinct from liposomes, as the continuous lipid bilayer and large inner aqueous pool are absent77. However, both types of nanocarriers are primarily composed of natural phospholipids or their derivatives, cholesterol, cationic or ionizable lipids, and polyethylene glycol (PEG)-lipids. To condense and protect the negatively charged DNA or RNA cargoes, permanently charged cationic lipids, such as 1,2-di-O-octadecenyl-3-trimethylammonium propane (DOTMA), 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and dimethyldioctadecylammonium (DDAB), were commonly used in early versions of the nanoparticles for the delivery of plasmid DNA and RNA molecules (e.g., siRNAs, miRNAs and mRNAs). In the past decade or so, these permanently charged lipids were gradually surpassed and replaced by ionizable cationic lipids, such as ALC-0315 and SM-102, which demonstrated superior transfection efficiency and reduced cytotoxicity for mRNA delivery78.
With the surge of preclinical and clinical studies of genome editing therapies, a great amount of knowledge accumulated for DNA and RNA delivery was quickly translated into the delivery of CRISPR-Cas979. Although cationic liposomes were the very first type of non-viral vector for traditional human gene replacement therapy80,81, their application in gene therapy is still in the clinical investigation stage. In comparison, LNP-based Onpattro® has already received regulatory approval. Very recently, two LNP-based mRNA vaccines were authorized for emergency use in the United States to fight against COVID-19 pandemic82,83. LNPs have been used in many preclinical studies for CRISPR-Cas9 delivery in the past several years84, 85, 86, 87, 88, 89. The most promising application is in the genome editing of liver cells. Several groups reported that upon intravenous administration, LNPs efficiently deliver the payloads to liver cells due to the strong liver tropism induced by low-density lipoprotein (LDL) receptors on liver cells77,86,90,91. Recently, Siegwart and colleagues reported a novel modifiable LNP platform called selective organ targeting (SORT) LNP, by adding a fifth lipid component to the established LNP formulation. SORT LNPs delivered different CRISPR cargoes, including RNPs and Cas9 mRNA/sgRNAs, for efficient genome editing in the liver, lungs, and spleen after intravenous administration92. Although this strategy is very promising to expand the usefulness of LNPs, the most possible reason why SORT LNPs preferentially accumulated in the liver and spleen is because they are easily opsonized and captured by the reticuloendothelial systems. In addition, selective delivery to the lungs may result from the positive surface charge of intravascular SORT LNPs. Therefore, to achieve more success in other tissues, such as the brain, heart and kidney, stealth SORT LNPs will be needed. Nonetheless, LNP-delivered CRISPR-Cas9 therapeutics has made rapid progress. In 2020, Intellia Therapeutics’ NTLA-2001, a CRISPR-Cas9 gene editing system delivered by LNP, was injected into a patient in a human clinical trial (NCT04601051) for the treatment of transthyretin amyloidosis.
4.2.2. Polymer-based nanoparticles
Polymer nanoparticles have gained popularity as delivery systems due to their numerous strengths including controlled drug release, payload protection, minimal immunogenicity, biodegradability, and improved bioavailability of active components. Polymer-based delivery vectors have been used in clinic for small molecular weight drugs and protein/peptide drugs for many years. The safety and effectiveness of polymeric nanoparticles in gene therapy have been investigated in multiple clinical trials93. Commonly used polymers for gene and protein delivery include poly (lactic-co-glycolic-acid) (PLGA), poly (β-amino ester) (PBAE), polyethylenimine (PEI), cyclodextrin (CD) derivatives, and poly (ethylene-glycol) (PEG). Though these nanoparticles hold great promise as delivery systems, they also provide the challenge of variable biocompatibility and toxicity.
Polymer nanoparticles have shown utility as in vivo delivery systems for CRISPR-Cas9 payloads. Chen et al.94 coated pre-assembled RNPs by a thin glutathione-cleavable covalently crosslinked polymer layer and yielded a very fine nanocapsule with a diameter of only ∼25 nm. In an Ai14 mouse strain, this polymeric nanocapsule induced efficient genome editing in retinal pigment epithelium cells following subretinal injection, and skeletal muscle after intramuscular injection. In another study, Wan et al.95 skillfully leveraged the amides and oxyanions that specifically present in Cas9 proteins and complexed RNPs with low-molecular-weight PEI modified CD through hydrogen bonding and salt bridge effects. The resulting stable nanoparticle efficiently enabled genomic disruption of mutant KRAS in a murine tumor model. Other polymer-based or hybrid delivery systems have also shown success in delivering CRISPR-Cas9 components in preclinical studies, including solid-tumor malignancies96, 97, 98, and disease states characterized by over-activation of the immune system, such as septic shock99, peritonitis99, allograft rejection100, glioma101, and type two diabetes mellitus99,100,102,103.
4.2.3. Peptide nanoparticles
Peptides have attracted much attention in CRISPR-Cas9 delivery because they can be used as not only backbone materials, but also surface modifying molecules of nanoparticles. Several types of peptides have been explored for self-assembly of nanoparticles. Peptide nanoparticles aid in payload protection, cellular targeting, intracellular delivery, and endosomal escape104, 105, 106. Though peptide-based delivery mechanisms have multiple positive aspects, they are still faced with multiple challenges, such as immunogenicity, reduced activity due to protease degradation, and potential toxicities caused by foreign peptides.
Peptide nanoparticles and conjugates have been used successfully in delivering CRISPR-Cas9 components in preclinical studies for genome editing107, 108, 109, 110, 111. Recently, He and colleagues reported a novel self-assembled peptide nanoparticle which selectively delivered CRISPR-Cas9 components to the mitochondria of cancer cells for mitochondrial DNA editing112. This work greatly expanded the potential applications of CRISPR-Cas9 to mitochondrial DNA editing. Though these preclinical models shown positive therapeutic effects, peptide-based delivery systems have yet to enter clinical trials involving CRISPR-Cas9 therapeutics.
4.2.4. Inorganic nanoparticles
Gold nanoparticles have been used to aid in the delivery of imaging agents, nucleic acids and proteins. Gold is an attractive option for nanoparticle formulation as it is relatively biocompatible, can permeate multiple cell types, and can be conjugated with other components, such as cell penetrating peptides, nucleic acids, lipids and/or polymers to increase delivery efficiency113,114. Although these nanoparticles are considered relatively non-immunogenic, their toxicity profile is largely dependent on physicochemical properties such as size, shape, charge, and surface modifications115,116. Recently, gold nanoparticles have been used in preclinical studies as a medium for conjugation of Cas9-sgRNA RNPs or Cas9 protein and sgRNA-encoding plasmids for in vivo genome editing via local administration117,118.
Silica nanoparticles are another noteworthy delivery system, which have shown the capability to successfully delivery CRISPR-Cas9 to multiple organs and cell types for genome editing. These delivery systems have seen an introduction into CRISPR delivery as they are very versatile, biocompatible, and can be modified in multiple ways based on delivery strategy119. Some of this versatility is seen in alterations of particle size as well as chemical modifications which allow for better encapsulation and/or conjugation of CRISPR-Cas9 components or other materials involved in delivery, such as lipids or polymers. When utilized for CRISPR delivery in preclinical studies, these nanoparticles are commonly mesoporous and formulated with tetraethyl orthosilicate (TEOS) as well as conjugated with some form of lipid or organic compound. Multiple preclinical applications demonstrated the potential of silica nanoparticles for in vivo delivery of CRISPR-Cas9 to treat murine models of hypercholesterolemia120, hepatocellular carcinoma121, pulmonary adenocarcinoma122, and melanoma123 as well as proof-of-concept editing in retinal pigment epithelium124 and brain neurons125.
5. CRISPR-Cas9 mediated in vivo genome editing therapy
Many diseases require safe and efficient in vivo delivery of CRISPR-Cas9 components in order to enable accurate genome editing in target cells and tissues. So far, multiple viral vectors and non-viral vectors have demonstrated the usefulness for CRISPR-Cas9-mediated genome editing in various tissues in preclinical studies.
5.1. Genome editing therapy in liver
The liver is a popular target for therapeutic gene editing as it involves in metabolism and the production of numerous essential proteins. CRISPR-Cas9 has been administered systemically with both viral and non-viral vectors for therapeutic gene editing in the liver to treat conditions such as hemophilia, familial hypercholesterolemia, phenylketonuria, and ornithine transcarbamylase deficiency.
Hemophilia B is an X-linked condition that results from dysfunction or deficiency of coagulation factor IX (FIX). Recently, Wang et al.52 utilized a dual AAV8 delivery system to intravenously deliver SpCas9 and sgRNA genomes as well as codon-optimized partial human FIX for HDR at the albumin locus in hepatocytes of hemophilia B-induced mice52. CRISPR-Cas9 treated adult and neonatal hemophilia B-induced mice showed a site-specific gene integration with an on-target indel percentage of 21%–34% in adults and 26.1%–40.1% in neonates as well as an HDR percentage of 3.0%–4.6% in adults and 8.2%–16.1% in neonates. Both adult and neonatal mice exhibited a significant increase in FIX protein and FIX activity when compared to control treated mice, and these results were sustained until the study was completed 48 weeks post-injection (Fig. 3A).
Familial hypercholesterolemia can be related to overexpression of proprotein convertase subtilisin kexin 9 (PCSK9), as it regulates the degradation of LDL receptors in hepatocytes in response to blood cholesterol levels126. This disease is inherited in an autosomal dominant pattern and leads to increased levels of LDL in the bloodstream, causing atherosclerosis and thromboembolic events127. Liu et al.88 utilized bioreducible LNPs formulated with cholesterol, DOPE, and DSPE-PEG2000 to deliver Cas9 mRNA and sgRNA targeting PCSK9 for NHEJ-mediated disruption in mice hepatocytes via single intravenous injection. CRISPR-Cas9 therapy led to a 20% decrease in serum PCSK9 when compared to untreated mice. Treated mice also showed no significant changes in aspartate aminotransferase (AST), alanine aminotransferase (ALT), and total bilirubin when compared to mice treated with phosphate buffered saline.
Phenylketonuria is a metabolic liver disease inherited in an autosomal recessive manner in which the body has a deficiency of the enzyme phenylalanine hydroxylase (PAH), leading to decreased metabolism of l-phenylalanine and hyperphenylalaninemia128. Those untreated with this condition develop neurologic complications due to hyperphenylalaninemia. Villiger et al.54 demonstrated effective administration of a dual AAV8 delivery system via intravenous injection to phenylketonuria-induced mice. AAVs with genomes encoding a SaCas9 base editor and a sgRNA targeting PAH in hepatocytes induced an on-target indel percentage of up to 10.5% and gene correction of up to 63% in hepatocytes. PAH correction led to a significant increase in PAH enzyme activity, a decrease in blood phenylalanine levels, and maintenance of mouse body weight and fur pigmentation. Following AAV8 therapy, mice showed no significant off-target indels at ten predicted loci as well as no indication of DNA damage or cell proliferation.
Ornithine transcarbamylase deficiency (OTCD) is inherited in an X-linked recessive manner and affects key components of the urea cycle causing supra-physiological levels of ammonia to accumulate, which can lead to neurological complications and mortality129,130. Wang et al.55 showed the utility of gene correction with dual AAV8 delivery of genomes expressing SaCas9, sgRNA targeting OTC in hepatocytes, and donor DNA template via intravenous injection for the treatment of OTCD in mice. CRISPR-Cas9 therapy induced an on-target indel percentage that ranged from 22% to 38% and an HDR efficiency of 2%–10%. Genome editing effects were seen at three and eight weeks when liver samples were harvested for analysis. CRISPR-treated mice showed restoration of OTC activity up to 79% of wild type as well as significantly increased survival time and lower levels of ammonia, which were also comparable to levels in wild type mice.
These studies showed encouraging results for therapeutic genome editing in the liver. Studies conducted by both Wang et al.52 and Villiger et al.54 demonstrated the ability for prolonged therapeutic effects of up to 48 and 26 weeks, respectively. Alternatively, the studies conducted by Liu et al.88 and Wang et al.55 lacked off-target indel data and a long-term follow-up period, leaving the long-term efficacy and toxicity of each therapy to be further investigated.
5.2. Genome editing therapy in tumors
CRISPR-Cas9 can serve as a novel treatment method for various malignancies as NHEJ-mediated gene disruption or deletion is a significant tool in blocking tumor growth mechanisms and/or immune resistance. Systemic injection is typically the preferred route of administration because many cancers are metastatic and present with tumor involvement in multiple organs. However, CRISPR-Cas9 components can also be delivered via intratumoral injection to treat localized cancer. CRISPR-Cas9 therapeutics have showed success as a treatment in preclinical malignancy models, including triple negative breast cancer, nasopharyngeal carcinoma, and melanoma.
Triple-negative breast cancer (TNBC) is an aggressive form of breast cancer that is highly metastatic. Guo et al.97 have demonstrated the ability of a tumor targeting nanolipogel (tNLG) delivery system for deleting oncogene lipocalin 2 (LCN2). The tNLG delivery system is a non-cationic, lipid–polymer hybrid, composed of DOPC, and DSPE-PEG-COOH as well as alginate. TNLGs are covalently conjugated with an antibody targeting intracellular adhesion molecule 1 (ICAM1) to maximize editing efficiency and specificity. This robust delivery system encapsulated three plasmids expressing Cas9 and sgRNAs targeting differing DNA sequences of LCN2 for NHEJ-mediated deletion and was administered via repeated intravenous injections. TNLG therapy induced an on-target genome editing efficiency of 80%, which led to significant decreases in LCN2 expression as well as tumor volume and mass when compared to control-treated mice. Additionally, this therapy was only associated with minimal systemic toxicity.
CRISPR-Cas9 genome editing therapy was also combined with photodynamic therapy for cancer treatment. Deng et al.131 co-delivered CRISPR-Cas9 RNP with photosensitizer chlorin e6 to a xenograft nasopharyngeal carcinoma mouse model using a near infrared (NIR) light and reducing environment-responsive polymeric nanoparticle. This delivery system was administered via repeated intravenous injection and encapsulated SpCas9/sgRNA RNP targeting nuclear factor erythroid 2-related factor 2 (NRF2), an antioxidant regulator, for NHEJ-mediated disruption in CNE2 tumor cells. Upon administration, NIR light was exposed to the tumor location, which destabilized lysosomal membranes and allowed the entry of Cas9/sgRNA RNP into the cytoplasm. This therapy induced an on-target indel percentage of 26.7%–32.28% with off-target percentages up to 4.1% in the tumor and liver tissues. Treatment with Cas9/sgRNA, chlorin e6, and NIR (T-CC/NIR) led to a decreased expression of NRF2, hypoxia-inducible factor 1α (HIF1α), and vascular endothelial growth factor-A (VEGF-A). Additionally, T-CC/NIR-treated mice showed an increased survival time by 80% as well as significantly decreasing tumor volume when compared to controls. CRISPR-treated mice had similar tissue structure in major organs and body weight fluctuations when compared to untreated mice.
The over-expression of cyclin dependent kinase 5 (CDK5) in melanoma and TNBC is involved in angiogenesis, apoptosis, and immune checkpoint regulation of tumor cells132. In a recent study, Deng et al.98 used a poly (beta-amino-ester) copolymer nanoparticle to administer a SpCas9/sgRNA expressing plasmid targeting CDK5 for NHEJ-mediated disruption. CDK5 disruption has exhibited the ability to decrease levels of programmed-death ligand-1 (PD-L1) which activates its receptor, programmed cell death protein (PD-1), on T cells, leading to decreased T cell activation and increased tumor cell proliferation133,134. In both models, CRISPR therapy led to significant reductions in the expression of CDK5, PD-L1, and P35, as well as decreased tumor volume, weight, and lung metastasis (TNBC model). CDK5 disruption led to greater inhibition of tumor growth than the administration of anti-PD-L1 antibodies, and in the melanoma model, prolonged lifespan to 40 days compared to 26 days in PBS-treated mice. CRISPR-treated mice experienced no severe systemic toxicity (Fig. 4).
None of the preclinical malignancy studies reported long-term data after genome editing treatment, which is most likely due to the severity and progression of disease states. Also, some studies lacked indel data which complicates conclusions on their potential efficacy and safety. Interestingly, from antibody conjugation, to immune checkpoint inhibition, to NIR exposure, each study utilized unique tactics to improve genome editing effects and specificity. Creative approaches such as these may prove to be a key component for the future clinical application of in vivo CRISPR therapies.
5.3. Genome editing therapy in muscle
Skeletal and cardiac muscle have been targeted for CRISPR-based gene editing using both systemic and local injection strategies. Treatment with both non-viral and viral delivery systems has shown promise in conditions such as DMD and catecholaminergic polymorphic ventricular tachycardia (CPVT).
DMD is a disease typically inherited in an X-linked recessive manner resulting from mutations in the DMD gene that encodes the protein dystrophin5. Loss-of-function mutations in DMD causes muscle weakness and wasting that can be fatal as dystrophin plays a crucial role in muscle structure and function by stabilizing the sarcolemma135. Zhang et al.56 reported successful rescue of DMD mice through in vivo genome editing by using a dual AAV delivery strategy, with ssAAV-packaged SpCas9 and ssAAV- or scAAV-packaged sgRNA. In mice with exon 44 of DMD deleted, intraperitoneal injection of AAV9 targeting exon 45 of DMD in skeletal and cardiac myocytes resulted in exon skipping or reframing therapy and caused dose-dependent on-target indel percentages of up to 30%. On-target indels were significantly higher in tibialis anterior (TA) and triceps when treated with scAAVs package sgRNA. CRISPR-treated mice experienced significant restoration of wild type dystrophin in TA, triceps, diaphragm, and heart. Treated mice also showed a significant increase in specific force from extensor digitorum longus and soleus muscles, as well as increased maximal tetanic force when compared to untreated mice, with scAAVs more so than ssAAVs. Genome editing effects persisted for 4 weeks following AAV delivery when mice were sacrificed for analysis.
CRISPR-Cas9 mediated genome editing in muscular tissues was also achieved using extracellular vesicles (EVs). Gee et al.136 loaded RNP into an extracellular nanovesicle called NanoMEDIC (nanomembrane-derived extracellular vesicles for the delivery of macromolecular cargo), and demonstrated successful genome editing in cultured cells and in mdx mice. Efficient loading of RNP into EVs in producer cells was achieved through a chemical-induced dimerization process. In addition, an expression vector containing two lentiviral vector elements, including HIV-1 Tat, was used to express sgRNA. Moreover, self-cleaving ribozymes were flanked to sgRNA to facilitate its release in target cells. Despite the elegant yet complicated design, NanoMEDIC encapsulated SpCas9/sgRNA RNPs enabled exon skipping in muscular cells in a luciferase mouse model for up to 160 days, indicating the potential for in vivo DMD treatment. Although this proof-of-concept study did not report off-target editing or adverse event data, Tat may be included in EVs, which represents a potential risk factor to targeted cells.
CPVT is a disease typically caused by a missense mutation in the RYR2 gene that encodes the ryanodine receptor 2 (RYR2), which is found on the sarcoplasmic reticulum (SR) in cardiomyocytes within the heart. This mutation is inherited in an autosomal dominant pattern and leads to incorrect expression of a protein subunit involved in the regulation of calcium release from RYR2, increasing intracellular calcium concentration in the sarcomere137. Pan et al.138 utilized AAV9 for NHEJ-mediated gene disruption in a CPVT mice model with an RYR2 mutation. Mice were treated via subcutaneous injection of AAV9 expressing both SaCas9 and sgRNA targeting exon eight of RYR2 for disruption in cardiomyocytes. CRISPR-Cas9 therapy led to an on-target indel percentage of 9.91%–12.69% with no significant off-target indels occurring. RYR2 disruption led to significant decreases in RYR2 expression and calcium release from SR in cardiomyocytes when compared to control-treated mice. Following adrenergic stimulation, 71% of the control AAV9-treated mice experienced arrhythmia, while arrhythmia was not seen in any of the CRISPR-treated mice. CRISPR-treated mice also showed minimal systemic toxicity.
Of the preclinical studies targeting muscle, none included a significant long-term follow up and some did not report off-target indel data. Though the results from each showed encouraging possibilities for CRISPR therapies targeting muscles, the lack of long-term data and missing indel data will require more studies to be completed to validate the long-term efficacy and safety of these therapies.
5.4. Genome editing therapy in the central nervous system (CNS)
Local injection of CRISPR-Cas9 into the CNS has enabled gene editing for the treatment of multiple devastating neurodegenerative conditions. Both non-viral and viral vectors have been used in treatment of conditions such as Alzheimer's disease, Huntington's disease, and amyotrophic lateral sclerosis.
Alzheimer's disease is inherited in an autosomal dominant pattern and is characterized by accumulation of amyloid beta plaque and neurofibrillary tangles in the brain, which can lead to synaptic dysfunction, hippocampal neuron loss, and memory impairment139,140. For amyloid beta formation to occur, amyloid precursor protein must first be processed by beta secretase 1 (BACE1), which represents a potential yet difficult therapeutic target141,142. In an effort to ameliorate familial Alzheimer's disease in mice, Park et al.109 developed a simple but effective amphiphilic peptide nanoparticle to deliver Cas9/sgRNA RNP targeting BACE1 for NHEJ-mediated disruption via intrahippocampal injection. The cationic R7L10 peptide successfully delivered RNP to post-mitotic neuronal cells in mice, with an on-target indel percentage of roughly 45% and without significant off-target indels in nine predicted loci. BACE1 disruption caused a significant reduction in BACE1 expression, percent area of total amyloid beta plaque, and amyloid beta 42 accumulation and secretion. Two intrahippocampal injections showed significantly decreased levels of BACE1 and percent area of amyloid beta when compared to mice treated with a single injection. Treated mice also showed a significant improvement in spatial working memory and associative learning when compared to untreated mice. This therapy demonstrated relative safety, causing minimal increases in inflammation, reactive microglia, blood urea nitrogen, and cleaved caspase 3. Therapeutic and toxic effects of this therapy were measured up until 12 weeks post-injection.
Huntington's disease (HD) is caused by a mutation to the huntingtin (HTT) gene causing expression of an altered HTT protein that is neurotoxic and is commonly inherited in an autosomal dominant pattern143. Ekman et al.144 reported the utility of NHEJ-mediated HTT disruption in treating Huntingtonʹs disease in mice. Mice were given a single intra-striatal injection of AAV1 expressing SaCas9 and sgRNA targeting HTT in medium sized spiny neurons of the brain. This therapy induced an on-target indel percentage of up to 15% with no significant off-target indels occurring at ten predicted loci. HTT disruption led to a significant decrease in HTT protein and mutant HTT proteins, as well as a 15% increase in mean survival when compared to control-treated mice. Additionally, mice with reduced levels of HTT protein showed improved motor function and decreased hind limb clasping. CRISPR-treated mice showed no significant difference in body weight when compared to control-treated mice, as both declined over the course of the 15-week study. Body weight decline is an indicator of HD onset and researchers in this study attributed this outcome in both groups to disease onset prior to AAV-therapy. Genome editing effects were seen for 15 weeks post-injection when all mice eventually died (Fig. 3C).
Amyotrophic lateral sclerosis (ALS) typically causes motor neurons loss and presents with symptoms including muscle atrophy, paralysis, and eventual respiratory dysfunction4,145. ALS has multiple inheritance patterns and its pathogenesis can be attributed to mutations in superoxide dismutase 1 (SOD1), causing a malfunctioning protein which increases the oxidative stress and induces progressive motor neuron loss146. Lim et al.147 have demonstrated the potential of using AAV9 expressing SpCas9 and sgRNA targeting mutant SOD1 to induce nonsense mutations in murine astrocytes of the CNS. Intrathecal injection of the AAV9-based CRISPR therapy led to an on-target editing efficiency of about 1.2% in the cervical spinal cord with 4% off-target editing in one of ten predicted loci, though it occurred in a non-protein coding region. SOD1 editing resulted in a significant decrease in SOD1 reactive inclusions in ventral white matter and anterior grey matter when compared to control-treated mice. Additionally, SOD1 editing led to increases in mean survival time and disease duration as well as a higher rotarod percentage, increased motor function, and greater hind limb grip strength when compared to control-treated mice. CRISPR-treated mice showed no signs of discomfort or toxicity throughout the study, indicating the potential safety profile of this therapy. The duration of genome editing effects in this study varied based on survival times which ranged from 128.6 to 150.2 days.
All highlighted CNS genome editing studies utilized a local administration technique and showed positive therapeutic outcomes related to each specific disease state. However, these effects were not sustained for more than 6 months. Though these studies have offered promising results, direct local administration enables genome editing in limited areas surrounding the injection sites. Thus, neurological diseases affecting large areas of CNS would require multiple invasive local injections, which increases surgical costs and risk for potential infections. So far, proof-of concept studies illustrating the safety and efficacy of treating neurological diseases by systemic administration of CRISPR-Cas9 system have yet to be reported.
5.5. Genome editing therapy in eyes
CRISPR-Cas9 has found utility for treatment of numerous ocular conditions. Local delivery is desired in this case as a much smaller dose is required. In addition, local injection reduces the potential for off-target editing or adverse events. Delivery with both non-viral and viral delivery systems has showed promise in treating conditions such as X-linked juvenile retinoschisis and age-related macular degeneration.
X-linked juvenile retinoschisis is caused by a mutation in retinoschisin 1 (RS1), causing decreased expression by foveal and macular cone photoreceptors as well as bipolar cells148. RS1 helps preserve the structure and function of the retina via cell adhesion. Decreased RS1 level can lead to splitting in the macula or surrounding areas of retina, causing reduced signal transduction and progressive vision loss149. Chou and colleagues developed a modified polyamidoamine (PAMAM) dendrimer nanoparticle delivery system to encapsulate donor RS1 DNA template plasmids as well as plasmids expressing Cas9/sgRNA targeting RS1150. This dendrimer nanoparticle delivery system was administered via single intravitreal injection for RS1 gene insertion via HITI. RS1 gene knock-in was seen at post-injection day 18 and was determined using green fluorescent protein (GFP) signaling. Knock-in persisted until post-injection Day 30 when mice were sacrificed. Analysis of mice eyes showed no histological abnormalities as well as GFP+ ganglion cells in the retina.
Ling et al.71 reported that a single subretinal injection of IDLV vectors successfully enabled genome editing and reduced choroidal neovascularization (CNV) in a murine model of wet age-related macular degeneration. This lentivirus-based therapy delivered SpCas9 mRNA and an sgRNA expression cassette targeting VEGFA in retinal pigment epithelial cells for NHEJ-mediated disruption. CRISPR-treated mice showed an on-target indel percentage of up to 44% and no significant off-target indels detected in seven predicted loci. VEGFA disruption led to a significant reduction of both VEGFA expression and CNV area when compared to PBS-treated mice. Seven days following gene editing therapy, mice showed no significant CD3+ T cell infiltration into retinas or RPE as well as no significant difference in anti-Cas9 IgG when compared to PBS-treated mice. The minimal immunogenicity of this therapy may be due to the route of administration as the eye is considered an immune-privileged organ. Genome editing and therapeutic effects persisted for two weeks post-injection when mice were sacrificed for analysis.
For ocular diseases caused by loss-of-function mutations in larger genes, traditional gene replacement therapy or NHEJ-based genome editing therapy cannot use AAV due to its limitation in cargo capacity. Even though dual AAV strategy achieved success in several preclinical studies44,55, the absolute genome editing efficiency in post-mitotic neuronal cells is very low. One of the reasons is that dual AAV strategy does not get every component in target cells at the same time. To improve the genome editing efficiency, Nishiguchi et al.151 reported a single AAV strategy to enable MMEJ in a murine model of retinal dystrophy. MMEJ utilizes smaller homology arms (about 20 bp) in the packaging construct to allow more room for CRISPR components and donor DNA, to correct vision-impairing mutations152,153. In this study, a single AAV2/8 vector was used to deliver SaCas9, sgRNAs, and donor DNA for mutation replacement. Gnat 1IRD2/IRD2 Pde6ccpfl1/cpfl1 mice, who are blind due to mutations causing rod and cone cell loss154, were treated via subretinal injection with sgRNAs targeting Gnat1 in rod cells for correction of the IRD2 mutation. Successful mutation correction efficiency was between 10% and 11% at 1- and 3-months post-injection, which is much higher than dual AAV strategy. No significant off-target indels were detected at 14 predicted loci. Treated mice showed an increase in rod cell activity as well as improvements in light sensitivity and visual acuity.
Though not all preclinical studies demonstrating ocular gene editing provided a long-term follow up, these studies showed positive effects. Utilization of HITI and MMEJ provides an interesting contrast to many other preclinical studies that opted for more traditional editing strategies. In the case of MMEJ, this may be a great option to allow for all-in-one packaging of key CRISPR components to bypass the packaging limit challenges seen with AAV delivery.
5.6. Genome editing therapy in inner ear
CRISPR has shown its capability as a genome editing tool in the inner ear. Preclinical studies have demonstrated the feasibility of local delivery into the inner ear using both viral and non-viral vectors. Though inner ear editing is not as widely established as editing in other organs, localized otic CRISPR-Cas9 administration is significant for the treatment of hearing loss disorders to preserve physiological function and improve hearing.
Liu's group initially explored the feasibility of CRISPR-Cas9-mediated genome editing in the inner ear in a transgenic GFP mouse model79. In this proof-of-concept study, cationic lipid transfection reagent Lipofectamine RNAiMAX or Lipofectamine 2000 successfully delivered CRISPR-Cas9 RNP and enabled inner ear genome editing. This paved the way for the following therapeutic studies. In a recent study, the same group demonstrated that cationic liposome-mediated inner ear delivery of CRISPR-Cas9 RNP successfully ameliorated autosomal dominant hearing loss using a Beethoven mouse model where mutant transmembrane channel-like protein 1 (TMC1) causes hearing loss155. SpCas9 and sgRNA targeting TMC1 were complexed with Lipofectamine 2000 for delivery via single intracochlear injection. Five days post injection, mice showed an on-target indel percentage ranging from 0.3% to 2.25%, causing successful NHEJ-mediated disruption of TMC1 with no significant off-target indels. TMC1 disruption led to significant preservation of both inner and outer hair cells and significant hearing improvements at eight weeks post-injection. However, mice experienced a decrease in inner hair cell sensory transduction as well as outer hair cell damage following genome editing therapy, possibly due to the well-known cytotoxicity of Lipofectamine 2000.
More recently, György et al.156 showed one time treatment of AAV-mediated CRISPR-Cas9 genome editing prevented autosomal dominant hearing loss in Beethoven mice for up to one year. A single intracochlear injection of AAV2 expressing SaCas9 and sgRNA targeting TMC1 for NHEJ-mediated disruption induced on-target indels ranging from 0.2% (7 days post-injection) to 2.2% (55 days post-injection). Treated mice showed improvements in auditory brainstem response (ABR) for up to 52 weeks post-injection, with one of the treated mice demonstrated an ABR that was indistinguishable from wild type. Twenty-four weeks post injection, treated mice showed preservation of both inner and outer hair cells while untreated mice showed a significant loss in both cell types. This therapy was specific to mutant TMC1 with only 0.0075% indels found at wild type alleles. Though the prevention of hearing loss in this study is encouraging, AAV2-treated mice showed the consequence of reduced inner and outer hair cell mechanosensitivity.
These representative studies demonstrated the feasibility, efficacy, and relative safety of inner ear editing with CRISPR, while György et al.156 took it a step further in showing positive therapeutic effects for a prolonged period of time. Though there are not many preclinical models for genome editing in the inner ear, these studies provide an excellent foundation to build on for future inner ear genome editing therapies.
5.7. Genome editing therapy in the immune system
Targeting various cells of the immune system for gene editing has significantly increased the therapeutic versatility of CRISPR-Cas9. Systemic administration of CRISPR for immune modulation is desirable in treating many different conditions such as type two diabetes mellitus and allograft rejection.
Wang's group103 developed a cationic lipid-assisted nanoparticle (CLAN) delivery system for therapeutic genome editing in the immune system of murine models of type II diabetes mellitus (T2DM). CRISPR-Cas9 delivery was accomplished using intravenous injection of CLAN encapsulated Cas9 mRNA and sgRNA, or Cas9/sgRNA encoding plasmids. CLANs were prepared using PEG-PLGA and cationic lipid N,N-bis(2-hydroxyethyl)-N-methyl-N-(2-cholesteryloxycarbonylaminoethyl) ammonium bromide (BHEM-Chol). Treatment of T2DM included CLANs encapsulating SpCas9, sgRNA, and CD68 promoter encoding plasmids that targeted netrin 1 (NTN1) in macrophages and monocytes for NHEJ-mediated deletion. CRISPR therapy induced 19.6% on-target indels with less than 0.5% indels in five potential off-target loci. NTN1 deletion helped to modulate T2DM-induced inflammatory response by significantly decreasing serum levels of tumor necrosis factor-α (TNF-α), interleukin-6 (IL-6), and adipose tissue levels of netrin-1 and CD11b+F4/80+ macrophages. This immune modulation successfully decreased blood glucose and increased insulin sensitivity in T2DM-induced mice with minimal systemic toxicity, however these effects were only reported for 16 days post-treatment.
In another study, CLAN encapsulated Cas9 mRNA and sgRNA was tested to ablate cluster of differentiation cell surface receptor 40 (CD40) in dendritic cells (DCs) to induce acute allograft tolerance100. NHEJ-mediated disruption of CD40 in mice decreased allograft rejection, prolonged graft survival time, and showed more intact tissue alignment with less graft damage when compared to Rapamycin-treated and untreated mice. It is important to note that not all DCs showed CD40 knockout. Also, all edited DCs eventually were replaced by new ones. The fact that all mice had undergone complete graft rejection after 12 days indicated that there are a number of obstacles to overcome before one can specifically modulate the immune system function by CRSIPR-Cas9-based genome editing.
5.8. Genome editing therapy in HIV/SIV treatment
Lastly, CRISPR has shown promise as an adjunct therapy in the treatment of human immunodeficiency virus (HIV) via systemic injection. Dash et al.157 showed encouraging data related to the use of CRISPR-Cas9 along with long-acting slow-effective release antiretroviral therapy (LASER ART) in treating murine models of HIV. AAV9 expressing SaCas9 and sgRNA targeting HIV1 was given via single intravenous injection for systemic NHEJ-mediated gene deletion. CRISPR-Cas9 therapy induced an on-target indel percentage ranging from 60% to 80% with no detectable off-target indels in 100 predicted loci. Neither HIV1 deletion nor LASER ART therapies alone provided encouraging prospects for long term suppression of HIV1 viral load. When used in combination, CRISPR and LASER ART showed encouraging outcomes as a potential solution for long term viral suppression while further increasing CD4+ T cells. Combination therapy significantly increased CD4+ T cells and decreased HIV1 viral load in multiple organs when compared to untreated HIV-induced mice. Additionally, 39% of mice receiving CRISPR/ART combination therapy showed no viral rebound and four of these mice had complete elimination of HIV1 DNA. The study was terminated for specimen analysis at week 14, leaving only five weeks to observe the outcomes following CRISPR-Cas9 therapy.
Mancuso et al.158 furthered the application of CRISPR-Cas9 for in vivo therapeutic genome editing by treating simian immunodeficiency virus (SIV) infected rhesus macaques. Macaques were treated via intravenous injection of AAV9 expressing SaCas9 and sgRNAs targeting the Gag and LTR regions of the SIV viral genome for systemic NHEJ-mediated gene deletion in addition to ART. Treated macaques showed an on-target excision efficiency ranging from 37% to 92% in blood as well as varying levels in other tissues. SIV viral DNA excision led to a 38%–95% decline in proviral DNA found in lymph nodes compared to 20% in a macaque treated with ART alone. Macaques treated with CRISPR and ART did not show any adverse effects following AAV9 inoculation, though no quantification of off-target editing was reported. All genome editing results were determined at 3 weeks following CRISPR therapy when animals were sacrificed for analysis (Fig. 3B).
Eliminating any immunodeficiency virus from hosts is certainly an ambitious attempt, but neither study provided prolonged follow up. Regardless of perceived clinical feasibility, Mancuso et al.158 showed success in a non-human primate model, which brought in vivo CRISPR-Cas9 therapies one step closer to clinical application.
6. Challenges to in vivo delivery of CRISPR/Cas9 therapeutics
A number of challenges need to be addressed before CRISPR-Cas9 therapeutics can be widely applied in the clinic. These challenges can be classified by several aspects; however, we only discuss delivery-related challenges in this section. Readers are encouraged to read excellent review articles discussing challenges related to the biology1,159,160, ethics161,162, and affordability3,163 of CRISPR therapeutics.
To enable in vivo genome editing using CRISPR-Cas9 system, NHEJ-based gene disruption or deletion requires both Cas9 nucleases and sgRNAs, while HDR-based gene insertion requires Cas9, sgRNA, and exogenous donor DNA, both with proper timing in target cells. Although the knowledge and experience accumulated in protein and gene delivery are extremely helpful, it is clear that in vivo delivery of CRISPR-Cas9 is far more difficult than delivery of proteins and genes alone. For protein and gene delivery, only one therapeutic component, either protein, or DNA/RNA must be delivered to some target cells. While for CRISPR-Cas9 therapeutics, two or three functional components must present in the same nucleus of a target cell at the same time. This poses additional challenges not only to the delivery efficiency but also to the logistics of the delivery task.
6.1. The in vivo instability of CRISPR-Cas9 therapeutics, which includes biochemistry-related or immunogenicity-related instability
The intrinsic biochemical properties of protein and nucleic acid molecules results in poor in vivo stability of Cas9 protein, plasmid DNA, mRNA, sgRNA, as well as exogenous donor DNA. Cas9 protein can be degraded through protease-mediated degradation and hydrolysis in cells and tissues. In addition, when injected into blood, its positive charge can easily recruit negatively charged components of the blood and immune system, leading to fast clearance. The poor in vivo stability of DNA/RNA has been well acknowledged in gene and RNAi delivery164. Widespread nucleases in blood and interstitial fluid can quickly degrade DNA/RNA if they are present freely in the body. Therefore, the cargoes must be well protected by delivery vectors.
Since exogenous plasmid DNA, RNA, and protein molecules are all immunogenic, when they are administered in vivo, they may elicit host immune responses. The immunogenicity of plasmid DNA can be reduced by removing CpG motifs165,166. In addition, various judicious chemical modifications have decreased the immunogenicity and increased the stability of RNA molecules167. The immunogenicity of commonly used Cas9 nucleases poses a different challenge, because they are obtained from Staphylococcus aureus and Streptococcus pyogenes, which are frequently encountered by most humans21. Pre-existing anti-Cas9 antibodies have been discovered in humans22. One potential solution that may help overcome the challenge is to engineer bacterial Cas9 nucleases by removing their epitopes. Another possible strategy is to look for other suitable CRISPR nucleases. There are enormous efforts on discovering novel CRISPR nucleases, some of them have demonstrated superior genome editing efficiency and accuracy168. Nonetheless, the components of the CRISPR-Cas9 system must be fully encapsulated within the delivery vectors to maintain the stability and avoid triggering a host immune response during in vivo transportation.
6.2. The biodistribution of delivery vectors to the tissue of interest following systemic injection
Intravascular AAVs are known to transduce cells in multiple organs and tissues, with limited levels of tissue tropism shown by differing serotypes. Several strategies are being explored to improve the tissue tropism of AAVs, such as direct evolution, discovering novel AAVs from ancient species and creating novel AAVs by engineering capsid proteins37. Non-viral vectors greater than several hundred nanometers in diameter are easily blocked by capillary beds in the lungs following intravenous injection, while nanoparticles less than 100 nm are most successful for systemic delivery. Cationic ingredients are frequently used to condense negatively charged DNA and RNA molecules, conferring positive charge to delivery vectors. Following intravenous injection, cationic vectors quickly bind to negatively charged blood components and form large complexes. These complexes are usually blocked in the lungs, leading to rapid clearance from blood circulation and decreased distribution to target tissues. This may be beneficial for pulmonary-targeted therapies, but the positive surface charge must be stably masked when targeting other tissues. Non-viral vectors in blood are also promptly recognized as foreign particles by the host immune system, leading to fast clearance from the bloodstream and decreased distribution to target tissues. PEGylation is a widely used strategy to help shield the surface charge and protect the non-viral vectors from opsonization. It also improves the storage stability of nanoparticles by preventing aggregation. However, the density of PEG chains on nanoparticles should be optimized because too many PEG molecules can inhibit cellular uptake169, 170, 171. An additional consideration is that PEG itself is immunogenic. The host immune system can produce anti-PEG IgM, which results in accelerated blood clearance of PEGylated vectors during repeated administration172,173. To avoid the immunogenicity issue of PEG, it has been shown that endogenous macromolecules may be used as alternative surface masking molecules174.
6.3. Cell type-specific delivery
Since DNAs and RNAs are negatively charged hydrophilic macromolecules, they are not able to permeate across the hydrophobic and negatively charged cell membrane. Although viral and non-viral vectors can protect them from degradation in the bloodstream and in tissues, neither shows the desired capability of cell type-specific delivery following intravascular administration. Efforts to achieve cell type-specific gene delivery by non-viral vectors include surface modification by various targeting moieties such as small molecule ligands, aptamers, peptides, and antibodies175,176. For diseases that require systemic administration of CRISPR-Cas9 systems, cell type-specific delivery is ideal to avoid delivery-mediated off-target effects. However, it is worth noting that the clinical success of a genome editing therapy may be achieved for some diseases before a perfect cell type-specific delivery vector is available. This is because repairing the genome of a certain portion of target cells in the desired tissue by local administration of CRISPR-Cas9 therapeutics may bring sufficient clinical benefits to patients while keep the safety risk low. Several preclinical studies highlighted earlier have shown promising therapeutic outcomes after local injection of CRPSPR-Cas9 systems, such as in the inner ear155, muscle118 and brain109. Even with these advances, the widespread success of local administration of CRISPR-Cas9 genome editing therapy depends on the disease state, severity, properties of the protein encoded by the target gene, and the desired tissue.
6.4. Efficient intracellular release of CRISPR-Cas9 cargoes
Receptor-mediated endocytosis directs CRISPR delivery vectors into early endosomes, which will evolve into late endosomes, and eventually lysosomes, where the low pH environment and digestive enzymes can destroy CRISPR-Cas9 cargoes. Releasing the cargoes into the cytosol before late endosomes/lysosomes are formed is a critical step. It is not a concern for viral vectors, but it is very challenging for non-viral vectors. Different strategies of endosomal escape have been tested, such as cationic lipids-induced instability of the endosome membrane177 and PEI polymer-induced “proton sponge” effect178. However, endosomal release efficiency is still very low179. It was reported that membrane fusion peptides and cell penetrating peptides have demonstrated the ability to break endosomal/lysosomal membranes180,181, which may help resolve this issue. Alternatively, direct cytosolic delivery of nanoparticles through membrane fusion bypassed the endosome trap and increased cargo availability in the cytoplasm174.
6.5. Efficient nuclear entry
For most genome editing applications, the cell nucleus is where CRISPR-Cas9 components perform their function. Dividing cells break up their nuclear membrane during each cell cycle, which allows exogenous large molecules, including Cas9 nucleases, plasmid DNA, mRNA, sgRNA, and donor DNA template, to enter the cell nucleus. However, for non-dividing cells, such as neurons, myocytes, and adipocytes, the nuclear membrane is a formidable barrier. Although it has been demonstrated that nuclear localization sequences could increase the nuclear entry of plasmid DNA180,182, more evidence from in vivo studies is needed. Many preclinical and clinical data demonstrated successful genome editing using the CRISPR-Cas9 system in both dividing and non-dividing cells. However, the mechanism for Cas9 nucleases to enter cell nuclei is still elusive. Illustrating the sequential process will not only solve a piece of the puzzle regarding CRISPR-Cas9-mediated genome editing in mammalian cells, but also shed light on the design of next generation delivery systems.
7. Conclusion and future perspectives
CRISPR-Cas9-mediated genome editing has been recognized as one of the most revolutionary technologies in the 21st century. The precise and permanent repair on the genome of patients’ cells will enable a complete cure to numerous inherited and acquired diseases, ranging from cancer, metabolic diseases, nervous system diseases, and infectious diseases. The number of registered clinical trials has been rapidly growing in the past several years (Clinicaltrials.gov). However, the majority of clinical trials are conducted by ex vivo delivery74, and only a small number of them use in vivo delivery (Table 2). It clearly shows that safe and effective in vivo delivery is the most significant hurdle to widespread clinical success of CRISPR-Cas9 therapeutics. Because the biological nature and mechanism of CRISPR-Cas9 is much more sophisticated than a single gene, RNA, or protein, no existing gene or protein delivery system can be directly adapted for the CRISPR-Cas9 system. Although the experience and lessons learned for protein and gene delivery have been very valuable, CRISPR-Cas9 components need significantly upgraded and innovative in vivo delivery systems. Both viral vectors and non-viral vectors have made remarkable progress for in vivo delivery of CRISPR-Cas9 therapeutics. However, only a small number of studies reported long-term efficacy and benefit of CRISPR-Cas9 genome editing therapies, which should be essential for all genome editing studies. In addition, few non-viral delivery studies reported the loading efficiency of CRISPR-Cas9 cargoes, which makes it difficult to evaluate the potential of the vectors for future development. Nonetheless, appropriate delivery vectors should be designed according to the choice of CRISPR-Cas9 cargoes as discussed earlier, in addition to the route of administration and disease. We are optimistic that different delivery systems will be proven effective for in vivo CRISPR-Cas9-based genome editing therapies.
Table 2.
Clinical trials of in vivo CRISPR-Cas9 therapeutics.
| Disease | Target gene | Cargo | Vector | Route of administration | Phase | Status | Sponsor | ClinicalTrials.gov identifier |
|---|---|---|---|---|---|---|---|---|
| Refractory herpetic viral keratitis | Herpes simplex virus type I | CRISPR-Cas9 mRNA | Lentivirus | Corneal injection | Phase 1/2 | Recruiting | Shanghai BDgene Co., Ltd. | NCT04560790 |
| Human papillomavirus-related malignant neoplasm | HPV16 and HPV18 E6/E7 | CRISPR-Cas9 plasmid | Gel | Topical application | Phase 1 | Unknown | First Affiliated Hospital, Sun Yat-sen University | NCT03057912 |
| Hereditary transthyretin amyloidosis | Transthyretin | CRISPR/Cas9 system | LNP | Intravenous administration | Phase 1 | Recruiting | Intellia Therapeutics | NCT04601051 |
| Leber congenital amaurosis 10 | Centrosomal protein 290 | CRISPR/Cas9 system | AAV | Subretinal injection | Phase 2 | Recruiting | Editas Medicine, Inc./Allergan | NCT03872479 |
Completely addressing the in vivo delivery challenges requires collaborative efforts from multiple disciplines. Cas9 nuclease engineering might help overcome several challenges, such as reducing its immunogenicity and facilitating endosomal escape as a membrane fusion protein. A Cas9 fusion protein with multiple functions, such as nucleic acids binding, cellular entry, endosomal escaping and nucleus entry, might dramatically change the design of next generation non-viral vectors for CRISPR-Cas9 delivery. Additionally, a clear illustration of biological processes involved in CRISPR-Cas9 genome editing, such as the intranuclear translocation of Cas9 proteins, will help improve the genome editing efficiency. Novel delivery systems which can carry all components of CRISPR-Cas9 in a single vector will most likely win the competition. Cell type-specific systemic delivery remains a challenge in the future and requires novel biomaterials to address the issue. Novel functional lipids, polymers, and peptides as well as viral vectors will continue to enrich the delivery arsenal. In parallel to the development of systemic delivery vectors, local administration should be explored for diseases which have been traditionally considered to be treated by systemic administration. Future efforts should also include efficient delivery of CRISPR-Cas9 to mitochondria for genome editing because many diseases are caused by faulty mitochondrial DNA183. The advancement of in vivo CRISPR-Cas9 delivery systems may also dramatically alter the landscape of ex vivo genome editing. Ex vivo genome editing therapy poses a myriad of challenges to cell extraction, culture, manipulation, freezing, and transportation, which are strictly regulated by regulatory authorities. Therefore, it is very possible that when highly specific in vivo delivery technologies are available for clinical genome editing therapy, the need for ex vivo genome editing may dramatically reduce, or even vanish.
Special attention must be given to the specificity of in vivo genome editing, which is critical to safe applications of CRISPR-Cas9 therapeutics in the clinic. The consequence of genome modification at off-target sites could be devastating because permanent genome editing at the incorrect sites may deactivate tumor suppressor genes or accidently activate oncogenes, both of which may lead to the development of cancer. In recent years, multiple techniques became available to detect off-target genome editing184, 185, 186, 187, 188, 189, 190, 191, 192. In the meantime, tremendous efforts have been made to reduce the off-target effects, including engineering Cas9 nucleases such as dimeric Cas9 nucleases193,194, high-fidelity Cas9195, enhanced specificity Cas9196, hyper-accurate Cas9197, Cas9 variants with expanded PAM compatibilities198, Cas9 fused with artificial inhibitory domains199. Additional efforts were made toward engineering sgRNA molecules, such as truncated sgRNA200 and paired sgRNAs201. As discussed earlier, innovative targeted delivery vectors play vital roles in reducing the delivery-mediated off-target effects. It is important to note that smart non-viral delivery systems may play greater roles even beyond delivery itself, such as reducing off-target genome editing. Stimuli-responsive non-viral delivery systems are promising to enable spatiotemporal genome editing by releasing or activating CRISPR-Cas9 components in target cells only when endogenous or exogenous signals trigger the release202. Off-target genome mutations at unintended sites may be significantly reduced through transient exposure of the cellular genome to CRISPR-Cas9 system. For example, Chen et al.203 demonstrated that optogenetic regulation of in vivo genome editing minimized the off-target effects by using cationic polymer-coated Au nanorods and second near infrared light. It is anticipated that the combination of advanced delivery technologies and molecular engineering technologies will work synergistically to minimize the off-target effects and pave a safe way toward clinical applications.
CRISPR-Cas9-mediated genome editing has shown unprecedented power and promise in medicine. Although the life-changing cures to many diseases seem to be just around the corner, safe and efficient delivery is half the game. Through integration of the knowledge accumulated in delivery science and vector engineering, together with continued collaborative efforts on the development of in vivo delivery systems, CRISPR-Cas9 therapeutics will soon realize their full potential.
Acknowledgments
Matthew Behr is thankful to the Summer Undergraduate Research Fellowship (SURF) Program at MCPHS University for financial support (USA). Hongwei Zhang acknowledges the internal funding from the School of Pharmacy – Boston, MCPHS University. The authors are grateful to Jieyu Fu for help with creating illustrations. The illustrations were created with BioRender.com.
Author contributions
Bing Xu and Hongwei Zhang conceptualized and designed this article. Matthew Behr, Jing Zhou and Hongwei Zhang wrote and revised the manuscript. Bing Xu provided critical comments and edited the manuscript. All authors have read and approved the final article.
Conflicts of interest
Hongwei Zhang is an inventor of an AAV gene therapy patent which is active in the US, EU and Japan. Other authors have no conflicts of interest to declare.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
References
- 1.Cox D.B., Platt R.J., Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 3.Doudna J.A. The promise and challenge of therapeutic genome editing. Nature. 2020;578:229–236. doi: 10.1038/s41586-020-1978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardiman O., van den Berg L.H. The beginning of genomic therapies for ALS. N Engl J Med. 2020;383:180–181. doi: 10.1056/NEJMe2012930. [DOI] [PubMed] [Google Scholar]
- 5.Verhaart I.E.C., Aartsma-Rus A. Therapeutic developments for Duchenne muscular dystrophy. Nat Rev Neurol. 2019;15:373–386. doi: 10.1038/s41582-019-0203-3. [DOI] [PubMed] [Google Scholar]
- 6.Wang D., Zhang F., Gao G. CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell. 2020;181:136–150. doi: 10.1016/j.cell.2020.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komor A.C., Badran A.H., Liu D.R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;169:559. doi: 10.1016/j.cell.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Oost J., Westra E.R., Jackson R.N., Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasiunas G., Barrangou R., Horvath P., Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2 doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sternberg S.H., LaFrance B., Kaplan M., Doudna J.A. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015;527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McVey M., Lee S.E. MMEJ repair of double-strand breaks (directors cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakuma T., Nakade S., Sakane Y., Suzuki K.T., Yamamoto T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat Protoc. 2016;11:118–133. doi: 10.1038/nprot.2015.140. [DOI] [PubMed] [Google Scholar]
- 19.Yao X., Wang X., Hu X., Liu Z., Liu J., Zhou H. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017;27:801–814. doi: 10.1038/cr.2017.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crudele J.M., Chamberlain J.S. Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat Commun. 2018;9:3497. doi: 10.1038/s41467-018-05843-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Charlesworth C.T., Deshpande P.S., Dever D.P., Camarena J., Lemgart V.T., Cromer M.K. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lattanzi A., Meneghini V., Pavani G., Amor F., Ramadier S., Felix T. Optimization of CRISPR/Cas9 delivery to human hematopoietic stem and progenitor cells for therapeutic genomic rearrangements. Mol Ther. 2019;27:137–150. doi: 10.1016/j.ymthe.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Senturk S., Shirole N.H., Nowak D.G., Corbo V., Pal D., Vaughan A. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nat Commun. 2017;8:14370. doi: 10.1038/ncomms14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaj T., Staahl B.T., Rodrigues G.M.C., Limsirichai P., Ekman F.K., Doudna J.A. Targeted gene knock-in by homology-directed genome editing using Cas9 ribonucleoprotein and AAV donor delivery. Nucleic Acids Res. 2017;45:e98. doi: 10.1093/nar/gkx154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S., Sun S., Moonen D., Lee C., Lee A.Y., Schaffer D.V. CRISPR-READI: efficient generation of knockin mice by CRISPR RNP electroporation and AAV donor infection. Cell Rep. 2019;27 doi: 10.1016/j.celrep.2019.05.103. 3780-3789.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bak R.O., Porteus M.H. CRISPR-mediated integration of large gene cassettes using AAV donor vectors. Cell Rep. 2017;20:750–756. doi: 10.1016/j.celrep.2017.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishiyama J., Mikuni T., Yasuda R. Virus-mediated genome editing via homology-directed repair in mitotic and postmitotic cells in mammalian brain. Neuron. 2017;96 doi: 10.1016/j.neuron.2017.10.004. 755-768.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H.X., Li M., Lee C.M., Chakraborty S., Kim H.W., Bao G. CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery. Chem Rev. 2017;117:9874–9906. doi: 10.1021/acs.chemrev.6b00799. [DOI] [PubMed] [Google Scholar]
- 30.Glass Z., Lee M., Li Y., Xu Q. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2018;36:173–185. doi: 10.1016/j.tibtech.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wan T., Ping Y. Delivery of genome-editing biomacromolecules for treatment of lung genetic disorders. Adv Drug Deliv Rev. 2021;168:196–216. doi: 10.1016/j.addr.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Yin H., Kauffman K.J., Anderson D.G. Delivery technologies for genome editing. Nat Rev Drug Discov. 2017;16:387–399. doi: 10.1038/nrd.2016.280. [DOI] [PubMed] [Google Scholar]
- 33.Kay M.A., Glorioso J.C., Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- 34.High K.A., Roncarolo M.G. Gene therapy. N Engl J Med. 2019;381:455–464. doi: 10.1056/NEJMra1706910. [DOI] [PubMed] [Google Scholar]
- 35.Anguela X.M., High K.A. Entering the modern era of gene therapy. Annu Rev Med. 2019;70:273–288. doi: 10.1146/annurev-med-012017-043332. [DOI] [PubMed] [Google Scholar]
- 36.High K.A. Turning genes into medicines—what have we learned from gene therapy drug development in the past decade? Nat Commun. 2020;11:5821. doi: 10.1038/s41467-020-19507-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat Rev Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hastie E., Samulski R.J. Adeno-associated virus at 50: a golden anniversary of discovery, research, and gene therapy success—a personal perspective. Hum Gene Ther. 2015;26:257–265. doi: 10.1089/hum.2015.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flotte T.R. Birth of a new therapeutic platform: 47 years of adeno-associated virus biology from virus discovery to licensed gene therapy. Mol Ther. 2013;21:1976–1981. doi: 10.1038/mt.2013.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weitzman M.D., Linden R.M. Adeno-associated virus biology. Methods Mol Biol. 2011;807:1–23. doi: 10.1007/978-1-61779-370-7_1. [DOI] [PubMed] [Google Scholar]
- 41.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat Rev Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 42.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Friedland A.E., Baral R., Singhal P., Loveluck K., Shen S., Sanchez M. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y., Wang L., Bell P., McMenamin D., He Z., White J. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hung S.S., Chrysostomou V., Li F., Lim J.K., Wang J.H., Powell J.E. AAV-mediated CRISPR/Cas gene editing of retinal cells in vivo. Invest Ophthalmol Vis Sci. 2016;57:3470–3476. doi: 10.1167/iovs.16-19316. [DOI] [PubMed] [Google Scholar]
- 46.Vakulskas C.A., Behlke M.A. Evaluation and reduction of CRISPR off-target cleavage events. Nucleic Acid Therapeut. 2019;29:167–174. doi: 10.1089/nat.2019.0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang H., Xie J., Xie Q., Wilson J.M., Gao G. Adenovirus-adeno-associated virus hybrid for large-scale recombinant adeno-associated virus production. Hum Gene Ther. 2009;20:922–929. doi: 10.1089/hum.2009.125. [DOI] [PubMed] [Google Scholar]
- 48.Grieger J.C., Samulski R.J. Adeno-associated virus vectorology, manufacturing, and clinical applications. Methods Enzymol. 2012;507:229–254. doi: 10.1016/B978-0-12-386509-0.00012-0. [DOI] [PubMed] [Google Scholar]
- 49.Qu W., Wang M., Wu Y., Xu R. Scalable downstream strategies for purification of recombinant adeno- associated virus vectors in light of the properties. Curr Pharmaceut Biotechnol. 2015;16:684–695. doi: 10.2174/1389201016666150505122228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J.P., Cheng X.X., Zhao M., Li G.H., Xu J., Zhang F. Curing hemophilia A by NHEJ-mediated ectopic F8 insertion in the mouse. Genome Biol. 2019;20:276. doi: 10.1186/s13059-019-1907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen H., Shi M., Gilam A., Zheng Q., Zhang Y., Afrikanova I. Hemophilia A ameliorated in mice by CRISPR-based in vivo genome editing of human Factor VIII. Sci Rep. 2019;9:16838. doi: 10.1038/s41598-019-53198-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Q., Zhong X., Li Q., Su J., Liu Y., Mo L. CRISPR-Cas9-mediated in vivo gene integration at the albumin locus recovers hemostasis in neonatal and adult hemophilia B mice. Mol Ther Methods Clin Dev. 2020;18:520–531. doi: 10.1016/j.omtm.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richards D.Y., Winn S.R., Dudley S., Nygaard S., Mighell T.L., Grompe M. AAV-mediated CRISPR/Cas9 gene editing in murine phenylketonuria. Mol Ther Methods Clin Dev. 2020;17:234–245. doi: 10.1016/j.omtm.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villiger L., Grisch-Chan H.M., Lindsay H., Ringnalda F., Pogliano C.B., Allegri G. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 55.Wang L., Yang Y., Breton C., Bell P., Li M., Zhang J. A mutation-independent CRISPR-Cas9-mediated gene targeting approach to treat a murine model of ornithine transcarbamylase deficiency. Sci Adv. 2020;6 doi: 10.1126/sciadv.aax5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Y., Li H., Min Y.L., Sanchez-Ortiz E., Huang J., Mireault A.A. Enhanced CRISPR-Cas9 correction of Duchenne muscular dystrophy in mice by a self-complementary AAV delivery system. Sci Adv. 2020;6 doi: 10.1126/sciadv.aay6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu L., Lau Y.S., Gao Y., Li H., Han R. Life-Long AAV-mediated CRISPR genome editing in dystrophic heart improves cardiomyopathy without causing serious lesions in MDX mice. Mol Ther. 2019;27:1407–1414. doi: 10.1016/j.ymthe.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koo T., Lu-Nguyen N.B., Malerba A., Kim E., Kim D., Cappellari O. Functional rescue of dystrophin deficiency in mice caused by frameshift mutations using Campylobacter jejuni Cas9. Mol Ther. 2018;26:1529–1538. doi: 10.1016/j.ymthe.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu S., Du J., Chen N., Jia R., Zhang J., Liu X. In vivo CRISPR/Cas9-Mediated genome editing mitigates photoreceptor degeneration in a mouse model of X-linked retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2020;61:31. doi: 10.1167/iovs.61.4.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moreno A.M., Fu X., Zhu J., Katrekar D., Shih Y.V., Marlett J. In situ gene therapy via AAV-CRISPR-Cas9-mediated targeted gene regulation. Mol Ther. 2018;26:1818–1827. doi: 10.1016/j.ymthe.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinberg J.B., Matthews T.J., Cullen B.R., Malim M.H. Productive human immunodeficiency virus type 1 (HIV-1) infection of nonproliferating human monocytes. J Exp Med. 1991;174:1477–1482. doi: 10.1084/jem.174.6.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Naldini L. Lentiviruses as gene transfer agents for delivery to non-dividing cells. Curr Opin Biotechnol. 1998;9:457–463. doi: 10.1016/s0958-1669(98)80029-3. [DOI] [PubMed] [Google Scholar]
- 63.Naldini L., Blomer U., Gallay P., Ory D., Mulligan R., Gage F.H. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 64.Miyoshi H., Smith K.A., Mosier D.E., Verma I.M., Torbett B.E. Transduction of human CD34+ cells that mediate long-term engraftment of NOD/SCID mice by HIV vectors. Science. 1999;283:682–686. doi: 10.1126/science.283.5402.682. [DOI] [PubMed] [Google Scholar]
- 65.Dull T., Zufferey R., Kelly M., Mandel R.J., Nguyen M., Trono D. A third-generation lentivirus vector with a conditional packaging system. J Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Naldini L., Trono D., Verma I.M. Lentiviral vectors, two decades later. Science. 2016;353:1101–1102. doi: 10.1126/science.aah6192. [DOI] [PubMed] [Google Scholar]
- 67.Consiglio A., Quattrini A., Martino S., Bensadoun J.C., Dolcetta D., Trojani A. In vivo gene therapy of metachromatic leukodystrophy by lentiviral vectors: correction of neuropathology and protection against learning impairments in affected mice. Nat Med. 2001;7:310–316. doi: 10.1038/85454. [DOI] [PubMed] [Google Scholar]
- 68.Gentner B., Schira G., Giustacchini A., Amendola M., Brown B.D., Ponzoni M. Stable knockdown of microRNA in vivo by lentiviral vectors. Nat Methods. 2009;6:63–66. doi: 10.1038/nmeth.1277. [DOI] [PubMed] [Google Scholar]
- 69.Kay M.A. State-of-the-art gene-based therapies: the road ahead. Nat Rev Genet. 2011;12:316–328. doi: 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- 70.Naldini L. Ex vivo gene transfer and correction for cell-based therapies. Nat Rev Genet. 2011;12:301–315. doi: 10.1038/nrg2985. [DOI] [PubMed] [Google Scholar]
- 71.Ling S., Yang S., Hu X., Yin D., Dai Y., Qian X. Lentiviral delivery of co-packaged Cas9 mRNA and a VEGFa-targeting guide RNA prevents wet age-related macular degeneration in mice. Nat Biomed Eng. 2021;5:144–156. doi: 10.1038/s41551-020-00656-y. [DOI] [PubMed] [Google Scholar]
- 72.Kim W., Lee S., Kim H.S., Song M., Cha Y.H., Kim Y.H. Targeting mutant KRAS with CRISPR-Cas9 controls tumor growth. Genome Res. 2018;28:374–382. doi: 10.1101/gr.223891.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen S.H., Hsieh Y.Y., Tzeng H.E., Lin C.Y., Hsu K.W., Chiang Y.S. ABL genomic editing sufficiently abolishes oncogenesis of human chronic myeloid leukemia cells in vitro and in vivo. Cancers (Basel) 2020;12 doi: 10.3390/cancers12061399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wei T., Cheng Q., Farbiak L., Anderson D.G., Langer R., Siegwart D.J. Delivery of tissue-targeted scalpels: opportunities and challenges for in vivo CRISPR/Cas-based genome editing. ACS Nano. 2020;14:9243–9262. doi: 10.1021/acsnano.0c04707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Allen T.M., Cullis P.R. Drug delivery systems: entering the mainstream. Science. 2004;303:1818–1822. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- 76.Lasic D.D. The mechanism of vesicle formation. Biochem J. 1988;256:1–11. doi: 10.1042/bj2560001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Witzigmann D., Kulkarni J.A., Leung J., Chen S., Cullis P.R., van der Meel R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv Drug Deliv Rev. 2020;159:344–363. doi: 10.1016/j.addr.2020.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S. Safety and efficacy of the BNT162b2 mRNA COVID-19 vaccine. N Engl J Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zuris J.A., Thompson D.B., Shu Y., Guilinger J.P., Bessen J.L., Hu J.H. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Leone P., Janson C.G., McPhee S.J., During M.J. Global CNS gene transfer for a childhood neurogenetic enzyme deficiency: canavan disease. Curr Opin Mol Ther. 1999;1:487–492. [PubMed] [Google Scholar]
- 81.Leone P., Janson C.G., Bilaniuk L., Wang Z., Sorgi F., Huang L. Aspartoacylase gene transfer to the mammalian central nervous system with therapeutic implications for Canavan disease. Ann Neurol. 2000;48:27–38. doi: 10.1002/1531-8249(200007)48:1<27::aid-ana6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 82.US Food and Drug Administration. Pfizer-BioNTech covid-19 vaccine FDA EAU letter of authorization. Available from: https://www.fda.gov/media/144412/download.
- 83.US Food and Drug Administration. Moderna COVID-19 vaccine EUA letter of authorization. Available from: https://www.fda.gov/media/144636/download.
- 84.Rosenblum D., Gutkin A., Kedmi R., Ramishetti S., Veiga N., Jacobi A.M. CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci Adv. 2020;6 doi: 10.1126/sciadv.abc9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qiu M., Glass Z., Chen J., Haas M., Jin X., Zhao X. Lipid nanoparticle-mediated codelivery of Cas9 mRNA and single-guide RNA achieves liver-specific in vivo genome editing of Angptl3. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2020401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei T., Cheng Q., Min Y.L., Olson E.N., Siegwart D.J. Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat Commun. 2020;11:3232. doi: 10.1038/s41467-020-17029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Patel S., Ashwanikumar N., Robinson E., Xia Y., Mihai C., Griffith J.P., 3rd Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat Commun. 2020;11:983. doi: 10.1038/s41467-020-14527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu J., Chang J., Jiang Y., Meng X., Sun T., Mao L. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv Mater. 2019;31 doi: 10.1002/adma.201902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang M., Zuris J.A., Meng F., Rees H., Sun S., Deng P. Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc Natl Acad Sci U S A. 2016;113:2868–2873. doi: 10.1073/pnas.1520244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Finn J.D., Smith A.R., Patel M.C., Shaw L., Youniss M.R., van Heteren J. A Single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 91.Bottger R., Pauli G., Chao P.H., Al Fayez N., Hohenwarter L., Li S.D. Lipid-based nanoparticle technologies for liver targeting. Adv Drug Deliv Rev. 2020;154–155:79–101. doi: 10.1016/j.addr.2020.06.017. [DOI] [PubMed] [Google Scholar]
- 92.Cheng Q., Wei T., Farbiak L., Johnson L.T., Dilliard S.A., Siegwart D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat Nanotechnol. 2020;15:313–320. doi: 10.1038/s41565-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Anselmo A.C., Mitragotri S. Nanoparticles in the clinic: an update. Bioeng Transl Med. 2019;4 doi: 10.1002/btm2.10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen G., Abdeen A.A., Wang Y., Shahi P.K., Robertson S., Xie R. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat Nanotechnol. 2019;14:974–980. doi: 10.1038/s41565-019-0539-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wan T., Chen Y., Pan Q., Xu X., Kang Y., Gao X. Genome editing of mutant KRAS through supramolecular polymer-mediated delivery of Cas9 ribonucleoprotein for colorectal cancer therapy. J Control Release. 2020;322:236–247. doi: 10.1016/j.jconrel.2020.03.015. [DOI] [PubMed] [Google Scholar]
- 96.Li L., Song L., Liu X., Yang X., Li X., He T. Artificial virus delivers CRISPR-Cas9 system for genome editing of cells in mice. ACS Nano. 2017;11:95–111. doi: 10.1021/acsnano.6b04261. [DOI] [PubMed] [Google Scholar]
- 97.Guo P., Yang J., Huang J., Auguste D.T., Moses M.A. Therapeutic genome editing of triple-negative breast tumors using a noncationic and deformable nanolipogel. Proc Natl Acad Sci U S A. 2019;116:18295–18303. doi: 10.1073/pnas.1904697116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Deng H., Tan S., Gao X., Zou C., Xu C., Tu K. Cdk 5 knocking out mediated by CRISPR-Cas9 genome editing for PD-L1 attenuation and enhanced antitumor immunity. Acta Pharm Sin B. 2020;10:358–373. doi: 10.1016/j.apsb.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu C., Lu Z., Luo Y., Liu Y., Cao Z., Shen S. Targeting of NLRP3 inflammasome with gene editing for the amelioration of inflammatory diseases. Nat Commun. 2018;9:4092. doi: 10.1038/s41467-018-06522-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Y., Shen S., Zhao G., Xu C.F., Zhang H.B., Luo Y.L. In situ repurposing of dendritic cells with CRISPR/Cas9-based nanomedicine to induce transplant tolerance. Biomaterials. 2019;217:119302. doi: 10.1016/j.biomaterials.2019.119302. [DOI] [PubMed] [Google Scholar]
- 101.Rui Y., Wilson D.R., Choi J., Varanasi M., Sanders K., Karlsson J. Carboxylated branched poly(beta-amino ester) nanoparticles enable robust cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci Adv. 2019;5 doi: 10.1126/sciadv.aay3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu Y., Cao Z.T., Xu C.F., Lu Z.D., Luo Y.L., Wang J. Optimization of lipid-assisted nanoparticle for disturbing neutrophils-related inflammation. Biomaterials. 2018;172:92–104. doi: 10.1016/j.biomaterials.2018.04.052. [DOI] [PubMed] [Google Scholar]
- 103.Luo Y.L., Xu C.F., Li H.J., Cao Z.T., Liu J., Wang J.L. Macrophage-specific in vivo gene editing using cationic lipid-assisted polymeric nanoparticles. ACS Nano. 2018;12:994–1005. doi: 10.1021/acsnano.7b07874. [DOI] [PubMed] [Google Scholar]
- 104.Spicer C.D., Jumeaux C., Gupta B., Stevens M.M. Peptide and protein nanoparticle conjugates: versatile platforms for biomedical applications. Chem Soc Rev. 2018;47:3574–3620. doi: 10.1039/c7cs00877e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fan T., Yu X., Shen B., Sun L. Peptide self-assembled nanostructures for drug delivery applications. J Nanomater. 2017;2017:4562474. [Google Scholar]
- 106.Nakase I., Kobayashi S., Futaki S. Endosome-disruptive peptides for improving cytosolic delivery of bioactive macromolecules. Pept Sci. 2010;94:763–770. doi: 10.1002/bip.21487. [DOI] [PubMed] [Google Scholar]
- 107.Krishnamurthy S., Wohlford-Lenane C., Kandimalla S., Sartre G., Meyerholz D.K., Theberge V. Engineered amphiphilic peptides enable delivery of proteins and CRISPR-associated nucleases to airway epithelia. Nat Commun. 2019;10:4906. doi: 10.1038/s41467-019-12922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shen Y., Cohen J.L., Nicoloro S.M., Kelly M., Yenilmez B., Henriques F. CRISPR-delivery particles targeting nuclear receptor-interacting protein 1 (NRIP1) in adipose cells to enhance energy expenditure. J Biol Chem. 2018;293:17291–17305. doi: 10.1074/jbc.RA118.004554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Park H., Oh J., Shim G., Cho B., Chang Y., Kim S. In vivo neuronal gene editing via CRISPR-Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer's disease. Nat Neurosci. 2019;22:524–528. doi: 10.1038/s41593-019-0352-0. [DOI] [PubMed] [Google Scholar]
- 110.Lostalé-Seijo I., Louzao I., Juanes M., Montenegro J. Peptide/Cas9 nanostructures for ribonucleoprotein cell membrane transport and gene edition. Chem Sci. 2017;8:7923–7931. doi: 10.1039/c7sc03918b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang H.X., Song Z., Lao Y.H., Xu X., Gong J., Cheng D. Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc Natl Acad Sci U S A. 2018;115:4903–4908. doi: 10.1073/pnas.1712963115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.He H., Lin X., Wu D., Wang J., Guo J., Green D.R. Enzymatic noncovalent synthesis for mitochondrial genetic engineering of cancer cells. Cell Reports Physical Science. 2020;1 doi: 10.1016/j.xcrp.2020.100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ding Y., Jiang Z., Saha K., Kim C.S., Kim S.T., Landis R.F. Gold nanoparticles for nucleic acid delivery. Mol Ther. 2014;22:1075–1083. doi: 10.1038/mt.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Giljohann D.A., Seferos D.S., Daniel W.L., Massich M.D., Patel P.C., Mirkin C.A. Gold nanoparticles for biology and medicine. Angew Chem Int Ed Engl. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dykman L.A., Khlebtsov N.G. Immunological properties of gold nanoparticles. Chem Sci. 2017;8:1719–1735. doi: 10.1039/c6sc03631g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ahmad S., Zamry A.A., Tan H.T., Wong K.K., Lim J., Mohamud R. Targeting dendritic cells through gold nanoparticles: a review on the cellular uptake and subsequent immunological properties. Mol Immunol. 2017;91:123–133. doi: 10.1016/j.molimm.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 117.Wang P., Zhang L., Xie Y., Wang N., Tang R., Zheng W. Genome editing for cancer therapy: delivery of Cas9 protein/sgRNA Plasmid via a gold nanocluster/lipid core-shell nanocarrier. 2017;4:1700175. doi: 10.1002/advs.201700175. Adv Sci (Weinh) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lee K., Conboy M., Park H.M., Jiang F., Kim H.J., Dewitt M.A. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat Biomed Eng. 2017;1:889–901. doi: 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou Y., Quan G., Wu Q., Zhang X., Niu B., Wu B. Mesoporous silica nanoparticles for drug and gene delivery. Acta Pharm Sin B. 2018;8:165–177. doi: 10.1016/j.apsb.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gong J., Wang H.X., Lao Y.H., Hu H., Vatan N., Guo J. A versatile nonviral delivery system for multiplex gene-editing in the liver. Adv Mater. 2020;32:2003537. doi: 10.1002/adma.202003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang B.C., Luo B.Y., Zou J.J., Wu P.Y., Jiang J.L., Le J.Q. Co-delivery of sorafenib and CRISPR/Cas9 based on targeted core-shell hollow mesoporous organosilica nanoparticles for synergistic HCC therapy. ACS Appl Mater Interfac. 2020;12:57362–57372. doi: 10.1021/acsami.0c17660. [DOI] [PubMed] [Google Scholar]
- 122.Pan Y., Yang J., Luan X., Liu X., Li X., Yang J. Near-infrared upconversion-activated CRISPR-Cas9 system: a remote-controlled gene editing platform. Sci Adv. 2019;5 doi: 10.1126/sciadv.aav7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu Q., Wang C., Zheng Y., Zhao Y., Wang Y., Hao J. Virus-like nanoparticle as a co-delivery system to enhance efficacy of CRISPR/Cas9-based cancer immunotherapy. Biomaterials. 2020;258:120275. doi: 10.1016/j.biomaterials.2020.120275. [DOI] [PubMed] [Google Scholar]
- 124.Wang Y., Shahi P.K., Xie R., Zhang H., Abdeen A.A., Yodsanit N. A pH-responsive silica-metal-organic framework hybrid nanoparticle for the delivery of hydrophilic drugs, nucleic acids, and CRISPR-Cas9 genome-editing machineries. J Control Release. 2020;324:194–203. doi: 10.1016/j.jconrel.2020.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Noureddine A., Maestas-Olguin A., Saada E.A., LaBauve A.E., Agola J.O., Baty K.E. Engineering of monosized lipid-coated mesoporous silica nanoparticles for CRISPR delivery. Acta Biomater. 2020;114:358–368. doi: 10.1016/j.actbio.2020.07.027. [DOI] [PubMed] [Google Scholar]
- 126.Maxwell K.N., Breslow J.L. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci U S A. 2004;101:7100–7105. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Abifadel M., Varret M., Rabes J.P., Allard D., Ouguerram K., Devillers M. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 128.Blau N., van Spronsen F.J., Levy H.L. Phenylketonuria. Lancet. 2010;376:1417–1427. doi: 10.1016/S0140-6736(10)60961-0. [DOI] [PubMed] [Google Scholar]
- 129.Tuchman M. The clinical, biochemical, and molecular spectrum of ornithine transcarbamylase deficiency. J Lab Clin Med. 1992;120:836–850. [PubMed] [Google Scholar]
- 130.Tuchman M., Morizono H., Rajagopal B.S., Plante R.J., Allewell N.M. The biochemical and molecular spectrum of ornithine transcarbamylase deficiency. J Inherit Metab Dis. 1998;21(Suppl 1):40–58. doi: 10.1023/a:1005353407220. [DOI] [PubMed] [Google Scholar]
- 131.Deng S., Li X., Liu S., Chen J., Li M., Chew S.Y. Codelivery of CRISPR-Cas9 and chlorin e6 for spatially controlled tumor-specific gene editing with synergistic drug effects. Sci Adv. 2020;6 doi: 10.1126/sciadv.abb4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Dorand R.D., Nthale J., Myers J.T., Barkauskas D.S., Avril S., Chirieleison S.M. Cdk 5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science. 2016;353:399–403. doi: 10.1126/science.aae0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Finck A., Gill S.I., June C.H. Cancer immunotherapy comes of age and looks for maturity. Nat Commun. 2020;11:3325. doi: 10.1038/s41467-020-17140-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Riley R.S., June C.H., Langer R., Mitchell M.J. Delivery technologies for cancer immunotherapy. Nat Rev Drug Discov. 2019;18:175–196. doi: 10.1038/s41573-018-0006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gao Q.Q., McNally E.M. The dystrophin complex: structure, function, and implications for therapy. Comp Physiol. 2015;5:1223–1239. doi: 10.1002/cphy.c140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gee P., Lung M.S.Y., Okuzaki Y., Sasakawa N., Iguchi T., Makita Y. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat Commun. 2020;11:1334. doi: 10.1038/s41467-020-14957-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wehrens X.H., Lehnart S.E., Marks A.R. Intracellular calcium release and cardiac disease. Annu Rev Physiol. 2005;67:69–98. doi: 10.1146/annurev.physiol.67.040403.114521. [DOI] [PubMed] [Google Scholar]
- 138.Pan X., Philippen L., Lahiri S.K., Lee C., Park S.H., Word T.A. In vivo Ryr2 editing corrects catecholaminergic polymorphic ventricular tachycardia. Circ Res. 2018;123:953–963. doi: 10.1161/CIRCRESAHA.118.313369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang J., Gu B.J., Masters C.L., Wang Y.J. A systemic view of Alzheimer disease—insights from amyloid-beta metabolism beyond the brain. Nat Rev Neurol. 2017;13:612–623. doi: 10.1038/nrneurol.2017.111. [DOI] [PubMed] [Google Scholar]
- 140.Greenberg S.M., Bacskai B.J., Hernandez-Guillamon M., Pruzin J., Sperling R., van Veluw S.J. Cerebral amyloid angiopathy and Alzheimer disease—one peptide, two pathways. Nat Rev Neurol. 2020;16:30–42. doi: 10.1038/s41582-019-0281-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yan R., Vassar R. Targeting the beta secretase BACE1 for Alzheimer's disease therapy. Lancet Neurol. 2014;13:319–329. doi: 10.1016/S1474-4422(13)70276-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vassar R. BACE1: the beta-secretase enzyme in Alzheimer's disease. J Mol Neurosci. 2004;23:105–114. doi: 10.1385/JMN:23:1-2:105. [DOI] [PubMed] [Google Scholar]
- 143.Bates G.P., Dorsey R., Gusella J.F., Hayden M.R., Kay C., Leavitt B.R. Huntington disease. Nat Rev Dis Prim. 2015;1:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 144.Ekman F.K., Ojala D.S., Adil M.M., Lopez P.A., Schaffer D.V., Gaj T. CRISPR-Cas9-mediated genome editing increases lifespan and improves motor deficits in a Huntington's disease mouse model. Mol Ther Nucleic Acids. 2019;17:829–839. doi: 10.1016/j.omtn.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hardiman O., Al-Chalabi A., Chio A., Corr E.M., Logroscino G., Robberecht W. Amyotrophic lateral sclerosis. Nat Rev Dis Prim. 2017;3:17085. doi: 10.1038/nrdp.2017.85. [DOI] [PubMed] [Google Scholar]
- 146.Brown R.H., Jr., Al-Chalabi A. Amyotrophic lateral sclerosis. N Engl J Med. 2017;377:1602. doi: 10.1056/NEJMc1710379. [DOI] [PubMed] [Google Scholar]
- 147.Lim C.K.W., Gapinske M., Brooks A.K., Woods W.S., Powell J.E., Zeballos C.M. Treatment of a mouse model of ALS by in vivo base editing. Mol Ther. 2020;28:1177–1189. doi: 10.1016/j.ymthe.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Molday R.S., Kellner U., Weber B.H. X-linked juvenile retinoschisis: clinical diagnosis, genetic analysis, and molecular mechanisms. Prog Retin Eye Res. 2012;31:195–212. doi: 10.1016/j.preteyeres.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Strupaite R., Ambrozaityte L., Cimbalistiene L., Asoklis R., Utkus A. X-linked juvenile retinoschisis: phenotypic and genetic characterization. Int J Ophthalmol. 2018;11:1875–1878. doi: 10.18240/ijo.2018.11.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chou S.J., Yang P., Ban Q., Yang Y.P., Wang M.L., Chien C.S. Dual supramolecular nanoparticle vectors enable CRISPR/Cas9-mediated knockin of retinoschisin 1 gene—a potential nonviral therapeutic solution for X-Linked juvenile retinoschisis. 2020;7:1903432. doi: 10.1002/advs.201903432. Adv Sci (Weinh) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Nishiguchi K.M., Fujita K., Miya F., Katayama S., Nakazawa T. Single AAV-mediated mutation replacement genome editing in limited number of photoreceptors restores vision in mice. Nat Commun. 2020;11:482. doi: 10.1038/s41467-019-14181-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kent T., Chandramouly G., McDevitt S.M., Ozdemir A.Y., Pomerantz R.T. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat Struct Mol Biol. 2015;22:230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Truong L.N., Li Y., Shi L.Z., Hwang P.Y., He J., Wang H. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc Natl Acad Sci U S A. 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lamb T.D. Evolution of phototransduction, vertebrate photoreceptors and retina. Prog Retin Eye Res. 2013;36:52–119. doi: 10.1016/j.preteyeres.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 155.Gao X., Tao Y., Lamas V., Huang M., Yeh W.H., Pan B. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217–221. doi: 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.György B., Nist-Lund C., Pan B., Asai Y., Karavitaki K.D., Kleinstiver B.P. Allele-specific gene editing prevents deafness in a model of dominant progressive hearing loss. Nat Med. 2019;25:1123–1130. doi: 10.1038/s41591-019-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dash P.K., Kaminski R., Bella R., Su H., Mathews S., Ahooyi T.M. Sequential LASER ART and CRISPR treatments eliminate HIV-1 in a subset of infected humanized mice. Nat Commun. 2019;10:2753. doi: 10.1038/s41467-019-10366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mancuso P., Chen C., Kaminski R., Gordon J., Liao S., Robinson J.A. CRISPR based editing of SIV proviral DNA in ART treated non-human primates. Nat Commun. 2020;11:6065. doi: 10.1038/s41467-020-19821-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Knott G.J., Doudna J.A. CRISPR-Cas guides the future of genetic engineering. Science. 2018;361:866–869. doi: 10.1126/science.aat5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Komor A.C., Badran A.H., Liu D.R. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Caplan A.L., Parent B., Shen M., Plunkett C. No time to waste—the ethical challenges created by CRISPR: CRISPR/Cas, being an efficient, simple, and cheap technology to edit the genome of any organism, raises many ethical and regulatory issues beyond the use to manipulate human germ line cells. EMBO Rep. 2015;16:1421–1426. doi: 10.15252/embr.201541337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Brokowski C., Adli M. CRISPR ethics: moral considerations for applications of a powerful tool. J Mol Biol. 2019;431:88–101. doi: 10.1016/j.jmb.2018.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wilson R.C., Carroll D. The daunting economics of therapeutic genome editing. CRISPR J. 2019;2:280–284. doi: 10.1089/crispr.2019.0052. [DOI] [PubMed] [Google Scholar]
- 164.He W., Turkeshi A., Li X., Zhang H. Progress in systemic co-delivery of microRNAs and chemotherapeutics for cancer treatment by using lipid-based nanoparticles. Ther Deliv. 2020;11:591–603. doi: 10.4155/tde-2020-0052. [DOI] [PubMed] [Google Scholar]
- 165.Klinman D.M., Yamshchikov G., Ishigatsubo Y. Contribution of CpG motifs to the immunogenicity of DNA vaccines. J Immunol. 1997;158:3635–3639. [PubMed] [Google Scholar]
- 166.de Wolf H.K., Johansson N., Thong A.T., Snel C.J., Mastrobattista E., Hennink W.E. Plasmid CpG depletion improves degree and duration of tumor gene expression after intravenous administration of polyplexes. Pharm Res. 2008;25:1654–1662. doi: 10.1007/s11095-008-9558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yin H., Song C.Q., Suresh S., Wu Q., Walsh S., Rhym L.H. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat Biotechnol. 2017;35:1179–1187. doi: 10.1038/nbt.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Harris J.M., Chess R.B. Effect of pegylation on pharmaceuticals. Nat Rev Drug Discov. 2003;2:214–221. doi: 10.1038/nrd1033. [DOI] [PubMed] [Google Scholar]
- 170.Hoffman A.S., Lai J.J. Three significant highlights of controlled drug delivery over the past 55 years: PEGylation, ADCs, and EPR. Adv Drug Deliv Rev. 2020;158:2–3. doi: 10.1016/j.addr.2020.05.013. [DOI] [PubMed] [Google Scholar]
- 171.Suk J.S., Xu Q., Kim N., Hanes J., Ensign L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv Drug Deliv Rev. 2016;99:28–51. doi: 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ishida T., Harada M., Wang X.Y., Ichihara M., Irimura K., Kiwada H. Accelerated blood clearance of PEGylated liposomes following preceding liposome injection: effects of lipid dose and PEG surface-density and chain length of the first-dose liposomes. J Control Release. 2005;105:305–317. doi: 10.1016/j.jconrel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 173.Wang X.Y., Ishida T., Ichihara M., Kiwada H. Influence of the physicochemical properties of liposomes on the accelerated blood clearance phenomenon in rats. J Control Release. 2005;104:91–102. doi: 10.1016/j.jconrel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 174.Zhang L., Yang X., Lv Y., Xin X., Qin C., Han X. Cytosolic co-delivery of miRNA-34a and docetaxel with core-shell nanocarriers via caveolae-mediated pathway for the treatment of metastatic breast cancer. Sci Rep. 2017;7:46186. doi: 10.1038/srep46186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhao Z., Ukidve A., Kim J., Mitragotri S. Targeting strategies for tissue-specific drug delivery. Cell. 2020;181:151–167. doi: 10.1016/j.cell.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 176.Mitchell M.J., Billingsley M.M., Haley R.M., Wechsler M.E., Peppas N.A., Langer R. Engineering precision nanoparticles for drug delivery. Nat Rev Drug Discov. 2021;20:101–124. doi: 10.1038/s41573-020-0090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xu Y., Szoka F.C., Jr. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 178.Behr J.P. The Proton sponge: a trick to enter cells the viruses did not exploit. Chimica. 1997;51:34–36. [Google Scholar]
- 179.Wang Y., Huang L. A window onto siRNA delivery. Nat Biotechnol. 2013;31:611–612. doi: 10.1038/nbt.2634. [DOI] [PubMed] [Google Scholar]
- 180.Zhang H., Gerson T., Varney M.L., Singh R.K., Vinogradov S.V. Multifunctional peptide-PEG intercalating conjugates: programmatic of gene delivery to the blood–brain barrier. Pharm Res. 2010;27:2528–2543. doi: 10.1007/s11095-010-0256-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li W., Nicol F., Szoka F.C., Jr. GALA: a designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery. Adv Drug Deliv Rev. 2004;56:967–985. doi: 10.1016/j.addr.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 182.Zhang H., Vinogradov S.V. Short biodegradable polyamines for gene delivery and transfection of brain capillary endothelial cells. J Control Release. 2010;143:359–366. doi: 10.1016/j.jconrel.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Slone J., Huang T. The special considerations of gene therapy for mitochondrial diseases. NPJ Genom Med. 2020;5:7. doi: 10.1038/s41525-020-0116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhang X.H., Tee L.Y., Wang X.G., Huang Q.S., Yang S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang X., Wang Y., Wu X., Wang J., Wang Y., Qiu Z. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat Biotechnol. 2015;33:175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- 186.Wienert B., Wyman S.K., Richardson C.D., Yeh C.D., Akcakaya P., Porritt M.J. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science. 2019;364:286–289. doi: 10.1126/science.aav9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wienert B., Wyman S.K., Yeh C.D., Conklin B.R., Corn J.E. CRISPR off-target detection with DISCOVER-seq. Nat Protoc. 2020;15:1775–1799. doi: 10.1038/s41596-020-0309-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lazzarotto C.R., Nguyen N.T., Tang X., Malagon-Lopez J., Guo J.A., Aryee M.J. Defining CRISPR-Cas9 genome-wide nuclease activities with CIRCLE-seq. Nat Protoc. 2018;13:2615–2642. doi: 10.1038/s41596-018-0055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kim D., Kim J.S. Profiling Genome-wide specificity of CRISPR-Cas9 using digenome-seq. Methods Mol Biol. 2021;2162:233–242. doi: 10.1007/978-1-0716-0687-2_13. [DOI] [PubMed] [Google Scholar]
- 190.Frock R.L., Hu J., Meyers R.M., Ho Y.J., Kii E., Alt F.W. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Akcakaya P., Bobbin M.L., Guo J.A., Malagon-Lopez J., Clement K., Garcia S.P. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature. 2018;561:416–419. doi: 10.1038/s41586-018-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Guilinger J.P., Thompson D.B., Liu D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hu J.H., Miller S.M., Geurts M.H., Tang W., Chen L., Sun N. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Aschenbrenner S., Kallenberger S.M., Hoffmann M.D., Huck A., Eils R., Niopek D. Coupling Cas9 to artificial inhibitory domains enhances CRISPR-Cas9 target specificity. Sci Adv. 2020;6 doi: 10.1126/sciadv.aay0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cai W., Luo T., Mao L., Wang M. Spatiotemporal delivery of CRISPR/Cas9 genome editing machinery using stimuli-responsive vehicles. Angew Chem Int Ed Engl. 2021;60:8596. doi: 10.1002/anie.202005644. [DOI] [PubMed] [Google Scholar]
- 203.Chen X., Chen Y., Xin H., Wan T., Ping Y. Near-infrared optogenetic engineering of photothermal nanoCRISPR for programmable genome editing. Proc Natl Acad Sci U S A. 2020;117:2395–2405. doi: 10.1073/pnas.1912220117. [DOI] [PMC free article] [PubMed] [Google Scholar]