Abstract
The aim of this study was to develop a diagnostic strategy for esophageal squamous cell carcinoma (ESCC) that combines plasma metabolomics with machine learning algorithms. Plasma-based untargeted metabolomics analysis was performed with samples derived from 88 ESCC patients and 52 healthy controls. The dataset was split into a training set and a test set. After identification of differential metabolites in training set, single-metabolite-based receiver operating characteristic (ROC) curves and multiple-metabolite-based machine learning models were used to distinguish between ESCC patients and healthy controls. Kaplan-Meier survival analysis and Cox proportional hazards regression analysis were performed to investigate the prognostic significance of the plasma metabolites. Finally, twelve differential plasma metabolites (six up-regulated and six down-regulated) were annotated. The predictive performance of the six most prevalent diagnostic metabolites through the diagnostic models in the test set were as follows: arachidonic acid (accuracy: 0.887), sebacic acid (accuracy: 0.867), indoxyl sulfate (accuracy: 0.850), phosphatidylcholine (PC) (14:0/0:0) (accuracy: 0.825), deoxycholic acid (accuracy: 0.773), and trimethylamine N-oxide (accuracy: 0.653). The prediction accuracies of the machine learning models in the test set were partial least-square (accuracy: 0.947), random forest (accuracy: 0.947), gradient boosting machine (accuracy: 0.960), and support vector machine (accuracy: 0.980). Additionally, survival analysis demonstrated that acetoacetic acid was an unfavorable prognostic factor (hazard ratio (HR): 1.752), while PC (14:0/0:0) (HR: 0.577) was a favorable prognostic factor for ESCC. This study devised an innovative strategy for ESCC diagnosis by combining plasma metabolomics with machine learning algorithms and revealed its potential to become a novel screening test for ESCC.
Keywords: Diagnostic, Esophageal squamous cell carcinoma (ESCC), Metabolomics, Machine learning, Prognostic
Highlights
-
•
Six most prevalent diagnostic plasma metabolites were identified in ESCC.
-
•
Plasma-metabolite-based machine learning models (PLS, RF, GBM, and SVM) for ESCC diagnosis.
-
•
Acetoacetic acid was an unfavorable prognostic factor, while PC (14:0/0:0) was a favorable prognostic factor for ESCC.
1. Introduction
Esophageal squamous cell carcinoma (ESCC), a predominant histological subtype of esophageal cancer, occurs most frequently in developing countries such as China [1]. It is difficult to diagnose ESCC at an early stage due to a lack of typical symptoms as well as specific and sensitive biomarkers of this tumor [2]. Consequently, patients are often diagnosed at a relatively advanced stage, usually accompanied by lymph node metastasis and invasion. Unfortunately, no effective treatment has been reported available for such plight currently [3]. Despite significant improvements in diagnostic modalities and treatments including surgery, radiation, chemotherapy, and their combination, the prognosis for ESCC still remains unsatisfactory [4,5]. Furthermore, the 5-year overall survival rate is about 20% and only 1% for those with advanced stages [6]. Therefore, early detection is extremely important with an urgent need for a novel and accurate means by which to diagnose ESCC.
A hallmark of malignancy is metabolic changes [7] through which cancer cells reprogram normal metabolic pathways that support uncontrolled proliferation. Some of the most striking alterations include elevation of glycolysis [8], up-regulation of amino acid [9] and lipid metabolism [10,11], as well as macromolecule biosynthesis [12]. Thus, the investigation of metabolic perturbations in cancer may be a promising means to discover novel cancer biomarkers and therapeutic targets.
Metabolomics is a powerful and efficient tool for the discovery of metabolic biomarkers and targets within biological specimens. Previously, plasma- and tissue-based ESCC metabolomic studies have identified potential diagnostic and prognostic metabolites (e.g., tryptophan and kynurenine), suggesting metabolic reprogramming to be associated with the initiation and development of ESCC [2,[13], [14], [15], [16], [17]]. Also, liquid biopsy for cancer diagnosis has many advantages, such as relatively low invasiveness [18]. Herein, plasma/serum-based metabolomics is an attractive approach for the discovery of ESCC diagnostic biomarkers.
For diagnosis, metabolomics data analysis requires statistical and machine learning-based classification methods [19]. Machine learning is a type of computer algorithm focusing on prediction through pattern recognition [20]. Principal component analysis (PCA) and partial least-square discriminant analysis (PLS-DA) are the two most widely used multivariate analysis methods for metabolomic studies [21]. In addition to these two well-known methods, the application of other machine learning algorithms, such as random forest (RF), gradient boosting machine (GBM), and support vector machine (SVM), has emerged in recent years and shown a promising diagnostic potential when combined with metabolomics data [19,22]. However, when reviewing previously published ESCC metabolomics studies, most of them were found to solely focus on the discovery of differential metabolites, and use single-metabolite diagnostic models with a rare evaluation of multiple-metabolite-based machine learning models [15,16,23]. Therefore, further studies employing a combination of plasma metabolomics and machine learning algorithms are encouraged, which might have potential clinical usefulness.
Initially, an untargeted plasma metabolomics study was conducted in a cohort consisting of 88 ESCC patients and 52 healthy controls. Based on the differential metabolites annotated, both single-metabolite-based receiver operating characteristic (ROC) curves and multiple-metabolite-based machine learning models, including PLS, RF, GBM, and SVM, were established in the training set (n = 100). In the test set, the predictive performance of the machine learning models had an accuracy range of 0.947–0.980, which was higher than that of single-metabolite models (an accuracy range of 0.653–0.887). These findings indicate that plasma-metabolites-based machine learning models are an excellent diagnostic strategy for ESCC.
2. Materials and methods
2.1. Reagents
High-performance liquid chromatography (HPLC) grade acetonitrile and methanol were purchased from Tedia Co. Inc. (Fairfield, OH, USA). HPLC grade formic acid was purchased from Roe Scientific Inc. (Newark, DE, USA). Distilled water was from Wahaha Group Co., Ltd. (Hangzhou, China).
2.2. Plasma samples
ESCC plasma samples were collected from 88 patients recruited after histopathologic confirmation of ESCC and radical resection at Zhejiang Cancer Hospital, China, from May 2010 to December 2012. The clinical stages of ESCC patients were determined based on the American Joint Committee on Cancer 8th edition staging system [24]. Participants were followed up until December 2017, and the overall survival (OS) from their surgery to the date of death or the last follow-up visit was evaluated. Healthy controls, recruited from our health examination center, were matched with ESCC patients based on age and sex. Fasting blood samples were collected from preoperative patients and healthy controls at approximately 8 a.m., with plasma immediately separated from whole blood by centrifugation at 1000 g, 4 °C for 10 min. Di-potassium salt of ethylenediaminetetraacetic acid was used as the anticoagulant. All the plasma samples were stored at −80 °C until analysis. The basic characteristics of these samples are listed in Table 1.
Table 1.
Clinical information of the patients.
| Feature | ESCC patients | Healthy controls | Refs. |
|---|---|---|---|
| Sex | |||
| Male | 70 (79.5%) | 32 (60.4%) | |
| Female | 18 (20.5%) | 20 (39.6%) | |
| Age | |||
| ≥ 60 | 52 (59.1%) | 22 (42.3%) | |
| < 60 | 36 (40.9%) | 30 (57.7%) | |
| Smoking | |||
| Yes | 63 (71.6%) | - | |
| No | 25 (28.4%) | - | |
| Drinking | |||
| Yes | 55 (62.5%) | - | |
| No | 33 (37.5%) | - | |
| pTa stage | [24] | ||
| 2 | 12 (13.6%) | - | |
| 3 | 76 (86.4%) | - | |
| pNa stage | [24] | ||
| 0 | 43 (48.8%) | - | |
| 1 | 28 (31.8%) | - | |
| 2 | 15 (17.1%) | - | |
| 3 | 2 (2.3%) | - | |
| pTNMa stage | [24] | ||
| I | 6 (6.8%) | - | |
| II | 37 (42.0%) | - | |
| III | 43 (48.9%) | - | |
| IV | 2 (2.3%) | - | |
Pathological TNM classification was used based on the American Joint Committee on Cancer, 8th edition.
The study protocol was performed in accordance with the Declaration of Helsinki, approved by the Research Ethics Committee of Zhejiang Cancer Hospital, with written informed consent obtained from all individuals.
2.3. Sample preparation
Sample preparation was conducted according to Huang et al. [25]. Briefly, each plasma sample (50 μL) was thawed on ice and immediately mixed with 200 μL of ice-cold acetonitrile. After vortexing for 1 min, the mixture was centrifuged at 16,000 g, 4 °C for 15 min. The supernatant (150 μL) was transferred into a fresh tube and lyophilized till dry. Residues were dissolved by mixing for 1 min with 80 μL of a solution consisting of 25% acetonitrile and 75% water. After centrifuging for 15 min at 16,000 g and 4 °C, 60 μL of the supernatant was transferred into a sample vial. An aliquot of 5 μL supernatant was used for liquid chromatography–mass spectrometry (LC-MS) analysis.
2.4. LC-MS analysis
An Ultimate 3000 UPLC system (Dionex, Idstein, Germany) linked to a Q Exactive Orbitrap mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) was used for this un-targeted metabolomics study. Separation was performed on an ACQUITY UPLC HSS T3 column (2.1 mm × 100 mm, 1.8 μm, Waters, Milford, MA, USA) at 35 °C. The mobile phase was water containing 0.1% (V/V) formic acid (phase A) and acetonitrile (phase B), with a flow rate of 0.3 mL/min. The linear gradients of phase B were as follows: 2% for 0–1 min, 2%–100% for 1–10 min, 100%–2% for 10–13 min, and 2% for 13–16 min. Linear gradients of phase A changed accordingly complementary to that of phase B. The electrospray voltages were 3.5 kV in positive mode and 2.5 kV in negative mode. The probe heater temperatures were set at 320 °C and 350 °C in positive and negative modes, respectively. The sheath gas was set at 35 and 40 arb in positive and negative modes, respectively. For collecting MS/MS spectra, a data-dependent acquisition mode for top 10 ions was conducted with a mass resolution of 17,500 and stepped collision energies of 10, 20, and 40 eV.
Quality control (QC) samples were prepared by pooling re-dissolved samples in equal amounts (15 μL) and periodically analyzed throughout the entire analytical run to monitor instrument stability.
2.5. Metabolomics data analysis
Metabolomics data analysis was performed according to Yang et al. [26]. Briefly, R package XCMS (version 3.8.2) was utilized for processing mass raw data, including peak detection, retention time alignment, peak matching, and correction. R package MetaX package (version 1.4.16) was used for ion filtration based on the following exclusion criteria: (1) ions not detected in over 50% of all QC samples or over 80% of all non-QC samples; (2) ions with relative standard deviation > 30% in QC samples. QC-based robust LOESS signal correction was applied to reduce the influence of signal shift.
The cohort data (n = 140, ESCC case (C): 88, healthy control (N): 52) were randomly divided into a training set (n = 100, C: 63, N: 37) and a test set (n = 40, C: 25, N: 15). To discover differential metabolites, an unsupervised PCA was first conducted to investigate the trends for all samples in the training set. Then supervised PLS-DA was performed to identify the most discriminating ion features between ESCC plasma and non-cancerous counterparts based on VIP values. Finally, those with VIP > 1, false discovery rate (FDR) < 0.05, and |log2 (fold change)| > 0.584 were defined as differential ion features. Metabolite annotation was performed using Progenesis QI (Waters, Milford, MA, USA) software based on METLIN (http://metlin.scripps.edu) and HMDB (http://www.hmdb.ca/). Metabolism pathway analysis was conducted using the online tool, Metaboanalyst (https://www.metaboanalyst.ca/MetaboAnalyst/home.xhtml).
2.6. Development of diagnostic models using single-metabolite ROC curves and metabolite-based machine learning models
For single metabolites, ROC curves were first analyzed for each metabolite in the training set. Youden's index (sum of sensitivity and specificity minus one) was used as a criterion for selecting the optimum cut-off point for each metabolite. With cut-off points for each metabolite, predictive classes were calculated for each unknown sample in the test set. The predictive performance including accuracy, sensitivity, and specificity was then calculated for the test set.
For metabolite-based machine learning modeling, data in the training set were preprocessed with “scaling” and “centering”. The same preprocessing methods with the same parameters were applied to the test set. Algorithms including PLS, RF, GBM, and SVM were investigated for cancerous and non-cancerous classification. R package caret (version 6.0–85) was utilized to train and test PLS, RF, and GBM models, while R package e1071 (version 1.7–3) was used to train and test SVM model. Ten repeated and five-fold cross-validation was performed to train the models PLS, RF, and GBM, and optimization was conducted using R package caret, in which the number of components in PLS, “mtry” in RF, as well as “n.trees”, “interaction.depth”, “shrinkage”, and “n.minobsinnode” in GBM, were tuned. For the SVM model, linear kernel was used and value of “cost” was screened from 1 to 10. To reduce model complexity, models with different amounts of top features, which were ranked in each model, were tested. Predictive accuracy in the test set was used to evaluate the predictive performance of models.
2.7. Survival analysis for plasma metabolites in ESCC
Kaplan-Meier curves were used to identify the relationships between metabolite levels in ESCC patients and their OS through log-rank test with a median split. Proportional hazard regression was performed for each metabolite to calculate the hazard ratio (HR) value. Factors with P values < 0.05 were considered significantly prognostic.
2.8. Statistical analysis
Statistical analysis was performed using R software (version 3.6.2). Normality of the variables was tested by the Shapiro-Wilk normality test in R. Cox proportional hazard regression analysis was conducted using R package survival (version 3.1–8). ROC analysis was performed by R package pROC (version 1.15.3). Student's t-test was used to compare the means between two groups, whereas ANOVA test was used to compare the means among three or more groups. A two-tailed P value < 0.05 was considered to be statistically significant.
3. Results
3.1. Metabolic shift in plasma of ESCC patients
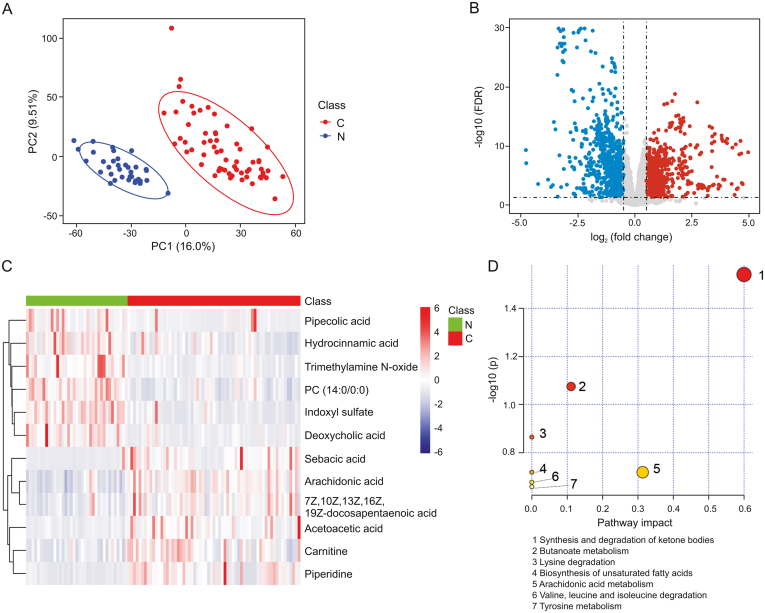
Un-targeted metabolomics was performed to investigate differential metabolites within the plasma of ESCC patients and healthy controls. A total of 3090 metabolic features in electrospray ionization positive (ESI+) mode and 3399 metabolic features in electrospray ionization negative (ESI-) mode were obtained from the metabolomics data. PCA analysis demonstrated a significant separation trend in plasma between ESCC patients and healthy controls, indicating a metabolic shift in ESCC plasma (Fig. 1). Furthermore, PLS-DA analysis demonstrated that ESCC patients were markedly separated from healthy controls, suggesting a global metabolic shift between the two groups (Fig. 2A). Volcano plots illustrating differential metabolomic features are shown in Fig. 2B. A total of 840 differential metabolic features were obtained based on the criteria VIP > 1, |log2 fold change| > 0.584, and FDR < 0.05. Among these, 12 features were annotated with specific metabolites (Table 2, Table S1). The heatmap demonstrated that 12 differential metabolites were able to distinguish ESCC patients from healthy controls (Fig. 2C). Pathway analysis of the 12 differential metabolites revealed that the top 3 significant metabolism pathways were synthesis and degradation of ketone bodies, butanoate metabolism, and lysine degradation (Fig. 2D).
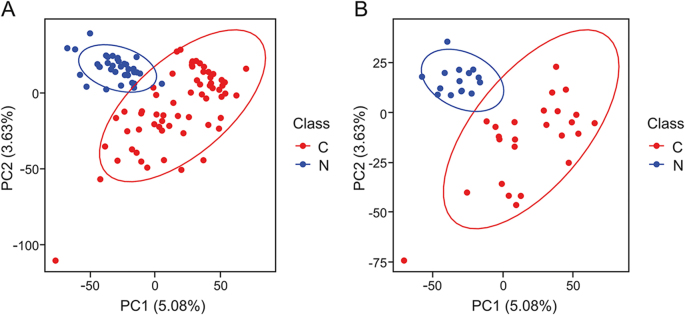
Fig. 1.
Principle component analysis (PCA) score plot of plasma metabolic profiles of ESCC patients and healthy controls. (A) Training set and (B) test set. C: ESCC patient; N: healthy control.
Fig. 2.
Metabolic shift in plasma of ESCC patients compared with healthy controls. (A) PLS-DA score plot derived from partial least-squares discriminant analysis in the training set; (B) differential ion features were defined as VIP > 1, |log2 FC| > 0.584, and an FDR < 0.05; (C) heatmap analysis of 12 plasma differential metabolites revealed a metabolic shift in ESCC patients compared with healthy controls; (D) pathway analysis of 12 differential metabolites. FC: fold change; FDR: Benjamini-Hochberg false discover rate; C: ESCC patient; N: healthy control.
Table 2.
Summary of the 12 differential metabolites found in plasma of ESCC patients and healthy controls a.
| Metabolite | Ion mode | m/z | RT (min) | FDR | log2 FC | VIP value | AUC |
|---|---|---|---|---|---|---|---|
| Indoxyl sulfate | Negative | 212.0018 | 5.2 | 5.4 × 10-12 | −1.68 | 1.75 | 0.937 |
| PC (14:0/0:0) | Positive | 468.3071 | 9.0 | 2.0 × 10-11 | −1.06 | 1.72 | 0.908 |
| Arachidonic acid | Negative | 303.2332 | 12.0 | 4.0 × 10-10 | 0.82 | 1.63 | 0.888 |
| Deoxycholic acid | Negative | 391.2859 | 9.6 | 1.3 × 10-9 | −2.00 | 1.61 | 0.885 |
| Piperidine | Positive | 86.0968 | 1.3 | 3.3 × 10-8 | 1.95 | 1.47 | 0.810 |
| Trimethylamine N-oxide | Positive | 76.0762 | 1.0 | 2.0 × 10-7 | −1.65 | 1.43 | 0.889 |
| Carnitine | Positive | 162.1119 | 1.3 | 8.8 × 10-6 | 0.84 | 1.33 | 0.758 |
| 7Z,10Z,13Z,16Z,19Z-docosapentaenoic acid | Negative | 329.2490 | 12.1 | 2.1 × 10-5 | 0.99 | 1.21 | 0.812 |
| Pipecolic acid | Positive | 130.0860 | 1.0 | 2.5 × 10-5 | −1.54 | 1.20 | 0.724 |
| Hydrocinnamic acid | Positive | 151.0749 | 7.6 | 1.3 × 10-4 | −0.78 | 1.11 | 0.779 |
| Sebacic acid | Negative | 201.1126 | 6.8 | 1.6 × 10-4 | 1.67 | 1.05 | 0.897 |
| Acetoacetic acid | Positive | 103.0392 | 0.9 | 3.4 × 10-4 | 1.16 | 1.06 | 0.811 |
Differential metabolites were discovered with the training set, in which a total of 100 samples including 63 from ESCC patients and 37 from healthy controls were evaluated. RT: retention time; FDR: Benjamini-Hochberg false discover rate; FC: fold change; AUC: as area under the ROC curve.
3.2. Predictive performance of single-metabolite models
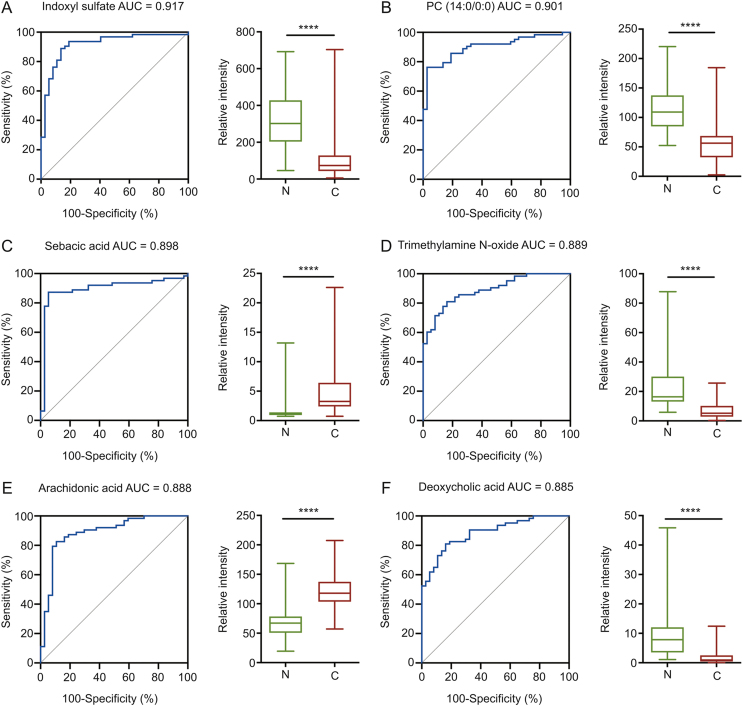
For single metabolite-based biomarker development, ROC curve analysis for metabolites in the training set showed that there were six metabolites with AUC values of over 0.85: indoxyl sulfate, phosphatidylcholine (PC) (14:0/0:0), sebacic acid, trimethylamine N-oxide, arachidonic acid, and deoxycholic acid (Fig. 3). These six metabolites were further used to develop single-metabolite-based diagnostic models for ESCC. After calculating the predictive classes for the unknown samples in the test set, confusion matrices were obtained, and the testing predictive performance of each metabolite is listed in Table 3. Arachidonic acid displayed the highest predictive accuracy (0.887, 95%CI: 0.732–0.958), followed by sebacic acid (0.867, 95%CI: 0.701–0.943), indoxyl sulfate (0.850, 95%CI: 0.701–0.942), PC (14:0/0:0) (0.825, 95%CI: 0.672–0.926), deoxycholic acid (0.773, 95%CI: 0.644–0.910), and trimethylamine N-oxide (0.653, 95%CI: 0.535–0.834).
Fig. 3.
Receiver operating characteristic (ROC) curves of single-metabolite models and boxplots of peak intensity distribution; (A) indoxyl sulfate, (B) PC (14:0/0:0), (C) sebacic acid, (D) trimethylamine N-oxide, (E) arachidonic acid, (F) deoxycholic acid. AUC: area under the curve. Two-tailed student's t-test was used with P value < 0.05 considered significant. ∗∗∗∗ P < 0.0001.
Table 3.
Predictive performance of different diagnostic models with the test seta.
| Model | Predictive performance |
|||
|---|---|---|---|---|
| Accuracy (95%CI) | Sensitivity | Specificity | ||
| Single metabolite model | ||||
| Arachidonic acid | 0.887 | (0.732, 0.958) | 0.933 | 0.887 |
| Sebacic acid | 0.867 | (0.701, 0.943) | 0.800 | 0.933 |
| Indoxyl sulfate | 0.850 | (0.701, 0.942) | 0.920 | 0.733 |
| PC (14:0/0:0) | 0.825 | (0.672, 0.926) | 0.760 | 0.933 |
| Deoxycholic acid | 0.773 | (0.644, 0.910) | 0.880 | 0.773 |
| Trimethylamine N-oxide | 0.653 | (0.535, 0.834) | 0.467 | 0.840 |
| Machine learning model | ||||
| PLS b | 0.947 | (0.830, 0.994) | 0.960 | 0.933 |
| RF c | 0.947 | (0.831, 0.994) | 0.960 | 0.933 |
| GBM d | 0.960 | (0.830, 0.994) | 0.830 | 0.994 |
| SVM e | 0.980 | (0.868, 0.999) | 0.960 | 1.000 |
A total of 40 samples for the test set, including 25 samples from ESCC patients and 15 samples from healthy controls.
Partial least-square.
Random forest.
Gradient boosting machine.
Support vector machine.
3.3. Predictive performance of multiple-metabolite-based machine learning models
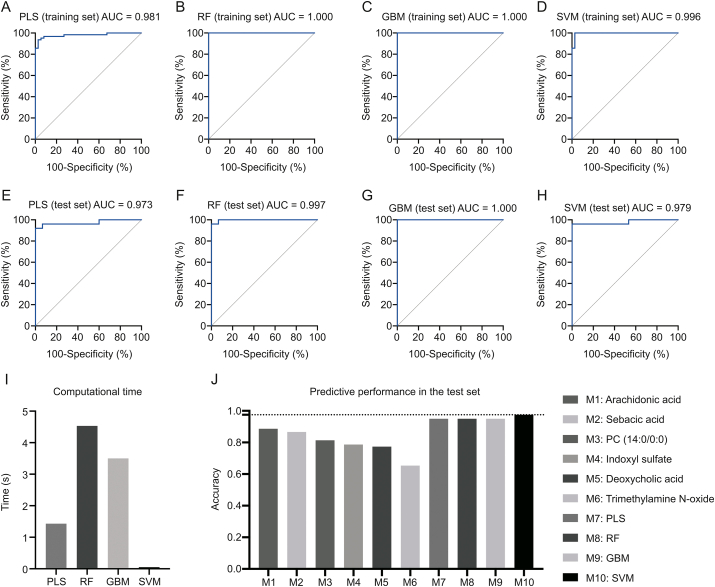
For the PLS model, the optimized number of components used in the model was 1, and the AUC of the ROC curve (AUCROC) was 0.981 (95%CI: 0.906–1.000) in the training set (Fig. 4A), and 0.973 (95%CI: 0.924–1.000) in test set (Fig. 4E). The predictive accuracies in the training set and the test set were 0.955 (95%CI: 0.887–0.984) and 0.947 (95%CI: 0.830–0.994), respectively (Table 3 and Table S2).
Fig. 4.
ROC curves of different machine learning models of training set: (A) PLS; (B) RF; (C) GBM; (D) SVM. ROC curves of different machine learning models of test set: (E) PLS; (F) RF; (G) GBM; (H) SVM. (I) Computational times of different machine learning models. (J) Predictive performance of six single-metabolite and four multiple-metabolite-based machine learning models in the test set. PLS: partial least-square; RF: random forest; GBM: gradient boosting machine; SVM: support vector machine.
For the RF model, the optimized value for entry was 2, and the AUCROC was 1.000 (95%CI: 0.906–1.000) in the training set (Fig. 4B), and 0.997 (95%CI: 0.989–1.000) in test set (Fig. 4F). The predictive accuracies in the training set and the test set were 1.000 (95%CI: 0.964–1.000) and 0.947 (95%CI: 0.831–0.994), respectively (Table 3 and Table S2).
For the GBM model, the final optimized model had the following parameters: n.trees value of 150, interaction depth value of 2, shrinkage value of 0.1, n.minobsinnode value of 10, with the AUCROC 1.000 (95%CI: 0.906–1.000) in the training set (Fig. 4C), and 1.000 (95%CI: 1.000–1.000) in test set (Fig. 4G). The predictive accuracies in the training set and the test set were 1.000 (95%CI: 0.964–1.000) and 0.960 (95%CI: 0.830–0.994), respectively (Table 3 and Table S2).
For the SVM model, linear SVM was finally selected with “C-classification” as the type, “cost” value of 3, and 16 support vectors, with the AUCROC of 0.996 (95%CI: 0.866–1.000) in the training set (Fig. 4D), 0.979 (95%CI: 0.935–1.000) in test set (Fig. 4H). The predictive accuracies in the training set and the test set were 0.987 (95%CI: 0.946–0.999) and 0.980 (95%CI: 0.868–0.999), respectively (Table 3 and Table S2).
In comparison to the single-metabolite models, the four metabolite-based machine learning models displayed higher predictive performance (Fig. 4J; Table 3), demonstrating the ascendancy of combined metabolomics data and machine learning approaches. Among the four machine learning models, SVM showed the highest accuracy of 0.980 in the test set among the four models. In terms of computational time (Fig. 4I), the fastest model was SVM (0.05 s), followed by PLS (1.43 s), GBM (3.5 s), and RF (4.53 s). Taken together, four machine learning models, especially SVM, were all considered as promising diagnostic models for ESCC.
3.4. Feature metabolite selection
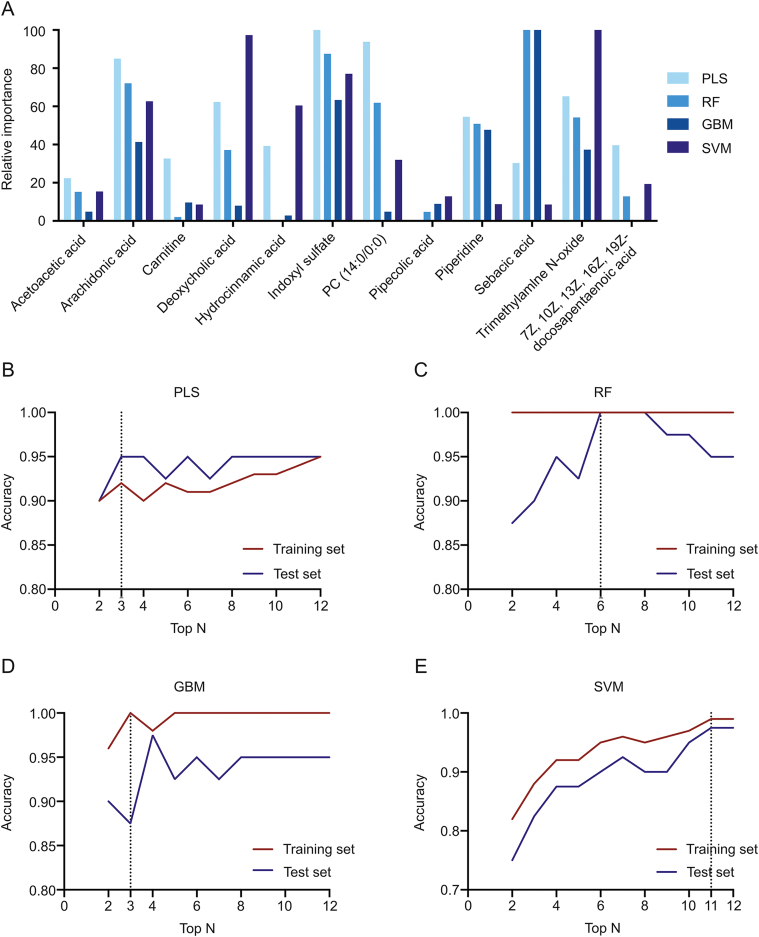
By ranking the feature importance of annotated metabolites via different machine learning models, it was reported from all models that several metabolites were of high importance, including indoxyl sulfate, arachidonic acid, and trimethylamine N-oxide. On the other hand, some were of low importance, including acetoacetic acid, pipecolic acid, and carnitine. Moreover, inter-model variations in feature importance of the same metabolite were noted. For instance, deoxycholic acid was reported of relatively high importance in PLS, RF, and SVM, while of relatively low importance in GBM (Fig. 5A).
Fig. 5.
(A) Feature importance of 12 metabolites in different machine learning models, and (B–E) machine learning models with different feature metabolites. The curves of predictive accuracy values increase as the number of feature metabolites grows in the (B) PLS model, (C) RF model, (D) GBM model, and (E) SVM model.
To reduce model complexity, optimization of machine learning models can be achieved through the use of fewer variables. Thus, models with different amounts of top features were investigated (from top 2 to top 12). The results showed that three models (i.e., PLS, RF, and GBM) reached accuracies of over 0.900 for the training set and over 0.850 for the test set, for the top 3 variables (Figs. 5B–D). A constant accuracy of 1.000 was reported in the training set of RFs, while in the corresponding test set a downward trend was observed when modeling with top 8–12 (Fig. 5C). The SVM model showed an escalating accuracy and achieved 0.987 for the training set and 0.980 for the test set (Fig. 5E).
3.5. Prognostic value of plasma metabolites for ESCC
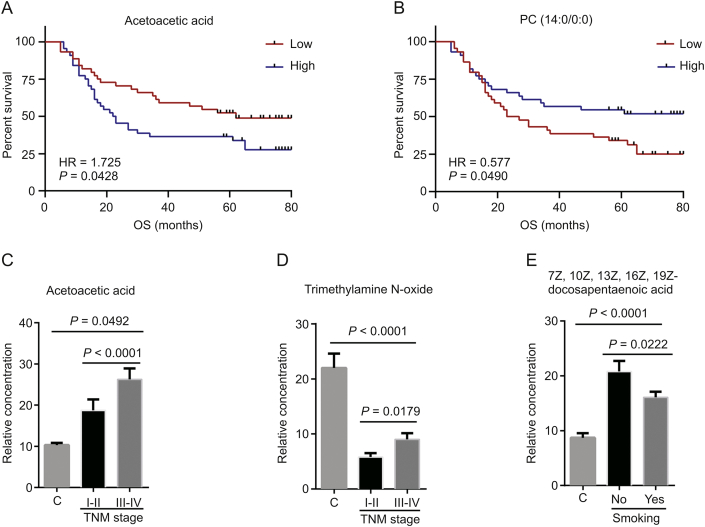
To assess the prognostic value of the 12 differential metabolites, survival analysis was performed, and the results demonstrated acetoacetic acid to be negatively associated with OS for ESCC, with an HR of 1.752 (95%CI: 1.012–3.033) (Fig. 6A). PC (14:0/0:0) was positively related to OS, with an HR of 0.577 (95%CI: 0.333–1.002) (Fig. 6B). Two metabolites, acetoacetic acid and trimethylamine N-oxide, were significantly increased in ESCC patients with advanced stages (TNM III–IV) compared with early stages (TNM I–II) (Figs. 6C and D). Compared with healthy controls, ESCC patients had an evident increase in acetoacetic acid levels, while a significant decrease in trimethylamine N-oxide level. With regard to smoking status, the levels of 7Z, 10Z, 13Z, 16Z, and 19Z-docosapentaenoic acid were lower in smoking ESCC patients than in non-smoking ones, while the level of this metabolite was higher in ESCC patients than in healthy controls independent of their smoking status (Fig. 6E). Based on these results, plasma acetoacetic acid was an unfavorable prognostic factor for ESCC and might be related to the progression of ESCC.
Fig. 6.
Prognostic significance of plasma metabolites. Kaplan–Meier survival curves for ESCC patients stratified by plasma metabolites with a median-split: (A) acetoacetic acid; (B) PC (14:0/0:0). Relative plasma concentrations of (C) acetoacetic acid, and (D) trimethylamine N-oxide among healthy controls, ESCC patients with early stages (I, II) and patients with advanced stages (III, IV). Relative plasma concentration of (E) 7Z, 10Z, 13Z, 16Z, 19Z-docosapentaenoic acid among healthy controls, smoking ESCC patients and non-smoking ones. Log-rank test was used with P value < 0.05 considered significant. Two-tailed student's t-test and ANOVA test were used with P value < 0.05 considered significant.
4. Discussion
ESCC patients survive longer when diagnosed at an early stage. Therefore, it is urgent to develop accurate and convenient diagnostic methods for early stage ESCC diagnosis. Previous metabolomic studies have demonstrated metabolic reprogramming to be a significant feature of ESCC, with the associated metabolites considered potential diagnostic biomarkers [2,[14], [15], [16], [17],23,27]. Plasma/serum are the most common clinical fluid biopsies. These specimens are an excellent and relatively non-invasive source of precise, rapid, and real-time diagnostic biomarkers [28]. Previously, several plasma/serum metabolomic studies [15,23,27] have found a group of metabolites differentially present in ESCC patients compared with healthy controls. For example, Cheng et al. [2] have found an increase in tryptophan metabolites including kynurenine, 5-hydroxytryptamine, 5-hydroxytryptophan, and 5-hydroxyindole-3-acetic acid in ESCC serum. Mir et al. [15] have revealed a dysregulation of serum PC in ESCC patients. Liu et al. [23] have demonstrated six plasma phospholipids, phosphatidylserine, phosphatidic acid, phosphocholine, phosphatidylinositol, phosphatidylethanolamine, and sphinganine 1-phosphate, to be significantly up-regulated in ESCC. However, these studies have limitations as follows: diagnostic significance of the metabolites was not clearly elucidated by proper validation design, such as splitting the data set into a training set and a test set; current multivariate analysis methods used for metabolomics data are PCA and PLS-DA (one form of PLS when Y is categorical), which can result in classifications that are over-optimistic or over-fitting. In order to have an in-depth understanding of the diagnostic significance of ESCC plasma metabolites and to enhance the clinical application of metabolomics, the present study developed and assessed metabolite-based machine learning models to discriminate between plasma samples of ESCC patients and healthy controls.
Four machine learning algorithms, PLS, RF, GBM, and SVM, are all widely used in the healthcare field, particularly in the area of medical diagnosis. However, with the exception of PLS, the other three machine learning algorithms have not been fully investigated for metabolomic data analysis yet. Based on our results, the metabolite-based machine learning models used in this study showed satisfactory predictive performance (accuracy range of 0.947–0.980), which was significantly higher than that of single-metabolite-based models (accuracy range of 0.653–0.887). The SVM exhibited highest predictive performance among the four models, both in the training set (0.987) and in the test set (0.980). Meanwhile, it had the lowest computational cost, altogether indicating SVM may be most suitable for analysis of large metabolomics data sets. Taken together, this study demonstrated machine learning methods other than PLS to be useful for clinical metabolomics studies, encouraging the use of combined metabolomics and machine learning approaches for the development of diagnostic cancer tools.
In the present study, a significant relationship was observed between TNM stage and acetoacetic acid, which was herein evidenced as the most prominent metabolite, possessing both diagnostic and prognostic value. Acetoacetic acid was originally considered as a ketone body, mainly produced in the liver during periods of nutrient deprivation, that served as high-energy fuel for extrahepatic tissues like the brain, heart, and skeletal muscle [29]. Consistent with our results, other studies have reported up-regulated ketone bodies in ESCC cancerous tissues [13]. These were characterized as an accumulation of ketone bodies (acetone and acetoacetic acid) as well as up-regulated ketone transporter-monocarboxylate transporter 1 in ESCC [13,30]. Taken together, these results suggested a potential functional role for acetoacetic acid in ESCC. However, contradicting results regarding ketones in cancer also exist. For example, Poff et al. [31] claimed cancer cells to be unable to efficiently utilize ketones, while ketones slow the proliferation of tumor cells. Martinez-Outschoorn et al. [32] illustrated an opposite effect of ketones which increased the stemness of cancer cells, resulting in recurrence, metastasis, and poor clinical outcomes in breast cancer. Therefore, it is essential to clarify whether ESCC cancer cells utilize ketones as an energy resource as well as to determine the biological role of acetoacetic acid in ESCC.
Lipids are essential to cancer cell structure, signal transduction, and cancer cell proliferation [[33], [34], [35]]. Our results showed a significantly decreased PC (14:0/0:0) level in ESCC, which is favorable to this cancer. Similarly, Mir et al. [15] also detected a group of decreased serum phosphatidylcholines, such as PC (18:2/0:0), PC (18:1/18:2), and PC (20:4/0:0) in ESCC. Meanwhile, Kamphorst et al. [34] proposed the capability of cancer cells in lipid uptake and utilization from the circulation through macro-pinocytosis. Collectively, these findings suggested that alterations in circulating lipids may be associated with enhanced lipid consumption by cancer cells. It is herein evident that lower circulating PC (14:0/0:0) levels, corresponding to higher consumption of PC (14:0/0:0) by cancer cells, are related to poorer survival of ESCC.
Our results exhibited an indicative decrease in indoxyl sulfate. The highest AUC value in ROC analysis was observed for this metabolite in the training set when performing single-metabolite-based diagnostic model analysis (AUC = 0.917). Its AUC value in test set, though not the greatest, was also very high. Moreover, great importance of indoxyl sulfate was observed in all multiple-metabolite-based diagnostic models combined with machine learning models. These results thus suggest that indoxyl sulfate might be a promising diagnostic biomarker for ESCC. In addition, a previous study revealed that indoxyl sulfate is related to microbial tryptophan catabolism [36]. In accordance with these results, Cheng et al. [2] reported disturbed tryptophan metabolism in ESCC. Altogether, tryptophan metabolism, especially microbial tryptophan catabolism, is potentially associated with ESCC initiation or progression.
Deoxycholic acid is a secondary bile acid, the metabolic by-product of intestinal bacteria. It is known to increase intracellular production of reactive oxygen as well as reactive nitrogen species, and higher levels are associated with increased frequencies of colon cancer [[37], [38], [39]]. Additionally, deoxycholic acid has multifunctional biological activities, such as disrupting the intestinal mucosal barrier [40] and enhancing Wnt signaling [41]. However, there have been no reports about deoxycholic acid in ESCC yet; thus it is worthy to further investigate its biological functions.
7Z,10Z,13Z,16Z,19Z-docosapentaenoic acid is an Omega-3 polyunsaturated fatty acid with 5 double bonds in 7-, 10-, 13-, 16-, 19- positions. The present study detected a significant increase in 7Z,10Z,13Z,16Z,19Z-docosapentaenoic acid levels in ESCC patients, which indicates a diagnostic potential for ESCC. Consistently, Liu et al. [42] have recently reported that people with high docosapentaenoic acid levels are vulnerable to lung cancer, indicating a biological role of docosapentaenoic acid in cancer initiation and development. Therefore, more work is needed to investigate the potential mechanisms of docosapentaenoic acid in ESCC.
This study also presented conflicting results: down-regulated trimethylamine N-oxide was found in ESCC patients at early stage compared with healthy controls, while an accretion was denoted at advanced stage in comparison to early stage, though the levels were still lower than those of healthy controls. A plausible explanation for such outcome is that levels of trimethylamine-N-oxide are determined by two factors: trimethylamine production from precursor molecules such as choline and l-carnitine by the metabolism of gut microbiota; and dietary intake of trimethylamine-N-oxide-rich foods such as high-choline or high-carnitine diet [43]. Accordingly, plasma levels of trimethylamine-N-oxide in ESCC patients might be altered by both changes in dietary compositions and intestinal bacteria. Meanwhile, controversial results are suggested in previous studies. Bae et al. [44] postulated a positive correlation between incidence of colorectal cancer (CRC) and plasma levels of trimethylamine N-oxide in US women, while Guertin et al. [45] indicated no correlation between this metabolite and risks of CRC. It remains opaque, thus requiring further researches to investigate, whether an increase in trimethylamine N-oxide level is a cause or a consequence of cancer.
Limitations of this study must be addressed. First, the sample size was relatively small for machine learning algorithms. A larger cohort is needed to validate model performance and finely optimize model parameters. Second, metabolite annotation efficiency was relatively low due to a lack of an in-house database and online MS2 spectral data, resulting in many diagnostic metabolite ions not being annotated. Third, the current prediction capacity of machine learning models is limited to two classes of plasma samples, so more diverse samples are needed to improve future performance of machine learning algorithms. Last but not least, the detailed function of the identified metabolites, such as acetoacetic acid, is required to be further clarified.
In conclusion, this study successfully established plasma metabolite-based machine learning models to distinguish ESCC cancer patients from healthy controls, demonstrating that the combination of metabolomics and machine learning is a novel and efficient diagnostic strategy for ESCC and possibly for other cancers. Although this study was a pilot in nature, due to relatively small sample size and limited diversity within the training set, the results could encourage future applications of machine learning algorithms to clinical metabolomics studies and accordingly aid medical diagnostic development. In addition to the discovery of diagnostic metabolites, this study explored progression-associated metabolites, which may provide potential prognostic biomarkers for ESCC. The findings of this study contribute to an understanding of the molecular pathogenesis of ESCC and provide useful information for individualized cancer therapy. In summary, further studies with larger cohorts should be conducted through a combined application of metabolomics and machine learning. This approach is promising in cancer diagnosis and will contribute significantly to cancer treatment.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81672315, 81802276, and 81302840), Key R&D Program Projects in Zhejiang Province (Grant No. 2018C04009), and 1022 Talent Training Program of Zhejiang Cancer Hospital.
Footnotes
Peer review under responsibility of Xi’an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2020.11.009.
Contributor Information
Su Zeng, Email: zengsu@zju.edu.cn.
Weimin Mao, Email: maowm@zjcc.org.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Liang H., Fan J.H., Qiao Y.L. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol. Med. 2017;14:33–41. doi: 10.20892/j.issn.2095-3941.2016.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng J., Jin H., Hou X. Disturbed tryptophan metabolism correlating to progression and metastasis of esophageal squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2017;486:781–787. doi: 10.1016/j.bbrc.2017.03.120. [DOI] [PubMed] [Google Scholar]
- 3.Tiasto V., Mikhailova V., Gulaia V. Esophageal cancer research today and tomorrow: lessons from algae and other perspectives. AIMS Genet. 2018;5:75–90. doi: 10.3934/genet.2018.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohashi S., Miyamoto S., Kikuchi O. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. 2015;149:1700–1715. doi: 10.1053/j.gastro.2015.08.054. [DOI] [PubMed] [Google Scholar]
- 5.Hulscher J.B., van Sandick J.W., de Boer A.G. Extended transthoracic resection compared with limited transhiatal resection for adenocarcinoma of the esophagus. N. Engl. J. Med. 2002;347:1662–1669. doi: 10.1056/NEJMoa022343. [DOI] [PubMed] [Google Scholar]
- 6.Testa U., Castelli G., Pelosi E. Esophageal cancer: genomic and molecular characterization, stem cell compartment and Clonal evolution. Medicines (Basel) 2017;4:67. doi: 10.3390/medicines4030067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pavlova N.N., Thompson C.B. The emerging hallmarks of cancer metabolism. Cell Metabol. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng J. Energy metabolism of cancer: glycolysis versus oxidative phosphorylation (Review) Oncol. Lett. 2012;4:1151–1157. doi: 10.3892/ol.2012.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vettore L., Westbrook R.L., Tennant D.A. New aspects of amino acid metabolism in cancer. Br. J. Canc. 2020;122:150–156. doi: 10.1038/s41416-019-0620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo X., Cheng C., Tan Z. Emerging roles of lipid metabolism in cancer metastasis. Mol. Canc. 2017;16:76. doi: 10.1186/s12943-017-0646-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbet C., Feron O. Emerging roles of lipid metabolism in cancer progression. Curr. Opin. Clin. Nutr. Metab. Care. 2017;20:254–260. doi: 10.1097/MCO.0000000000000381. [DOI] [PubMed] [Google Scholar]
- 12.Weinberg S.E., Chandel N.S. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L., Chen J., Chen L. 1H-NMR based metabonomic profiling of human esophageal cancer tissue. Mol. Canc. 2013;12:25. doi: 10.1186/1476-4598-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu J., Chen Y., Zhang R. Global metabolomics reveals potential urinary biomarkers of esophageal squamous cell carcinoma for diagnosis and staging. Sci. Rep. 2016;6:35010. doi: 10.1038/srep35010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mir S.A., Rajagopalan P., Jain A.P. LC-MS-based serum metabolomic analysis reveals dysregulation of phosphatidylcholines in esophageal squamous cell carcinoma. J. Proteomics. 2015;127:96–102. doi: 10.1016/j.jprot.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Jin H., Qiao F., Chen L. Serum metabolomic signatures of lymph node metastasis of esophageal squamous cell carcinoma. J. Proteome Res. 2014;13:4091–4103. doi: 10.1021/pr500483z. [DOI] [PubMed] [Google Scholar]
- 17.Sun C., Li T., Song X. Spatially resolved metabolomics to discover tumor-associated metabolic alterations. Proc. Natl. Acad. Sci. U. S. A. 2019;116:52–57. doi: 10.1073/pnas.1808950116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Rhun E., Seoane J., Salzet M. Liquid biopsies for diagnosing and monitoring primary tumors of the central nervous system. Canc. Lett. 2020;480:24–28. doi: 10.1016/j.canlet.2020.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Alakwaa F.M., Chaudhary K., Garmire L.X. Deep learning accurately predicts estrogen receptor status in breast cancer metabolomics data. J. Proteome Res. 2018;17:337–347. doi: 10.1021/acs.jproteome.7b00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orlenko A., Kofink D., Lyytikainen L.P. Model selection for metabolomics: predicting diagnosis of coronary artery disease using automated machine learning. Bioinformatics. 2020;36:1772–1778. doi: 10.1093/bioinformatics/btz796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long N.P., Lim D.K., Mo C. Development and assessment of a lysophospholipid-based deep learning model to discriminate geographical origins of white rice. Sci. Rep. 2017;7:8552. doi: 10.1038/s41598-017-08892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cuperlovic-Culf M. Machine learning methods for analysis of metabolic data and metabolic pathway modeling. Metabolites. 2018;8:4. doi: 10.3390/metabo8010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R., Peng Y., Li X. Identification of plasma metabolomic profiling for diagnosis of esophageal squamous-cell carcinoma using an UPLC/TOF/MS platform. Int. J. Mol. Sci. 2013;14:8899–8911. doi: 10.3390/ijms14058899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D., Zheng Y., Wang Z. Comparison of the 7th and proposed 8th editions of the AJCC/UICC TNM staging system for esophageal squamous cell carcinoma underwent radical surgery. Eur. J. Surg. Oncol. 2017;43:1949–1955. doi: 10.1016/j.ejso.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Huang Q., Tan Y., Yin P. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Canc. Res. 2013;73:4992–5002. doi: 10.1158/0008-5472.CAN-13-0308. [DOI] [PubMed] [Google Scholar]
- 26.Yang Z., Song Z., Chen Z. Metabolic and lipidomic characterization of malignant pleural effusion in human lung cancer. J. Pharmaceut. Biomed. Anal. 2020;180:113069. doi: 10.1016/j.jpba.2019.113069. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H., Wang L., Hou Z. Metabolomic profiling reveals potential biomarkers in esophageal cancer progression using liquid chromatography-mass spectrometry platform. Biochem. Biophys. Res. Commun. 2017;491:119–125. doi: 10.1016/j.bbrc.2017.07.060. [DOI] [PubMed] [Google Scholar]
- 28.Marrugo-Ramirez J., Mir M., Samitier J. Blood-based cancer biomarkers in liquid biopsy: a promising non-invasive alternative to tissue biopsy. Int. J. Mol. Sci. 2018;19:2877. doi: 10.3390/ijms19102877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Puchalska P., Crawford P.A. Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metabol. 2017;25:262–284. doi: 10.1016/j.cmet.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X., Chen X., Liu F. Monocarboxylate transporter 1 is an independent prognostic factor in esophageal squamous cell carcinoma. Oncol. Rep. 2019;41:2529–2539. doi: 10.3892/or.2019.6992. [DOI] [PubMed] [Google Scholar]
- 31.Poff A.M., Ari C., Arnold P. Ketone supplementation decreases tumor cell viability and prolongs survival of mice with metastatic cancer. Int. J. Canc. 2014;135:1711–1720. doi: 10.1002/ijc.28809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Outschoorn U.E., Prisco M., Ertel A. Ketones and lactate increase cancer cell "stemness," driving recurrence, metastasis and poor clinical outcome in breast cancer: achieving personalized medicine via Metabolo-Genomics. Cell Cycle. 2011;10:1271–1286. doi: 10.4161/cc.10.8.15330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menendez J.A., Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Canc. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 34.Kamphorst J.J., Cross J.R., Fan J. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bi J., Ichu T.-A., Zanca C. Oncogene amplification in growth factor signaling pathways renders cancers dependent on membrane lipid remodeling. Cell Metabol. 2019;30:525–538.e8. doi: 10.1016/j.cmet.2019.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hubbard T.D., Murray I.A., Perdew G.H. Indole and tryptophan metabolism: endogenous and dietary routes to ah receptor activation. Drug Metab. Dispos. 2015;43:1522–1535. doi: 10.1124/dmd.115.064246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernstein H., Bernstein C., Payne C.M. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J. Gastroenterol. 2009;15:3329–3340. doi: 10.3748/wjg.15.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ocvirk S., O'Keefe S.J. Influence of bile acids on colorectal cancer risk: potential mechanisms mediated by diet - gut microbiota interactions. Curr. Nutr. Rep. 2017;6:315–322. doi: 10.1007/s13668-017-0219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T., Apte U. Bile acid metabolism and signaling in cholestasis, inflammation, and cancer. Adv. Pharmacol. 2015;74:263–302. doi: 10.1016/bs.apha.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L., Dong W., Wang S. Deoxycholic acid disrupts the intestinal mucosal barrier and promotes intestinal tumorigenesis. Food Funct. 2018;9:5588–5597. doi: 10.1039/c8fo01143e. [DOI] [PubMed] [Google Scholar]
- 41.Cao H., Luo S., Xu M. The secondary bile acid, deoxycholate accelerates intestinal adenoma-adenocarcinoma sequence in Apc (min/+) mice through enhancing Wnt signaling. Fam. Cancer. 2014;13:563–571. doi: 10.1007/s10689-014-9742-3. [DOI] [PubMed] [Google Scholar]
- 42.Liu J., Zhou H., Zhang Y. Docosapentaenoic acid and lung cancer risk: a Mendelian randomization study. Cancer Med. 2019;8:1817–1825. doi: 10.1002/cam4.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan C.W.H., Law B.M.H., Waye M.M.Y. Trimethylamine-N-oxide as one hypothetical link for the relationship between intestinal microbiota and cancer - where we are and where shall we go? J. Canc. 2019;10:5874–5882. doi: 10.7150/jca.31737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bae S., Ulrich C.M., Neuhouser M.L. Plasma choline metabolites and colorectal cancer risk in the Women's Health Initiative Observational Study. Canc. Res. 2014;74:7442–7452. doi: 10.1158/0008-5472.CAN-14-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guertin K.A., Li X.S., Graubard B.I. Serum trimethylamine N-oxide, carnitine, choline, and betaine in relation to colorectal cancer risk in the alpha tocopherol, beta carotene cancer prevention study. Cancer Epidemiol. Biomark. Prev. 2017;26:945–952. doi: 10.1158/1055-9965.EPI-16-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.