Abstract
We provide evidence that the human DNA ligase III gene encodes a mitochondrial form of this enzyme. First, the DNA ligase III cDNA contains an in-frame ATG located upstream from the putative translation initiation start site. The DNA sequence between these two ATG sites encodes an amphipathic helix similar to previously identified mitochondrial targeting peptides. Second, recombinant green fluorescent protein harboring this sequence at its amino terminus was efficiently targeted to the mitochondria of Cos-1 monkey kidney cells. In contrast, native green fluorescent protein distributed to the cytosol. Third, a series of hemagglutinin-DNA ligase III minigene constructs were introduced into Cos-1 cells, and immunocytochemistry was used to determine subcellular localization of the epitope-tagged DNA ligase III protein. These experiments revealed that inactivation of the upstream ATG resulted in nuclear accumulation of the DNA ligase III protein, whereas inactivation of the downstream ATG abolished nuclear localization and led to accumulation within the mitochondrial compartment. Fourth, mitochondrial protein extracts prepared from human cells overexpressing antisense DNA ligase III mRNA possessed substantially less DNA ligase activity than did mitochondrial extracts prepared from control cells. DNA end-joining activity was also substantially reduced in extracts prepared from antisense mRNA-expressing cells. From these results, we conclude that the human DNA ligase III gene encodes both nuclear and mitochondrial enzymes. DNA ligase plays a central role in DNA replication, recombination, and DNA repair. Thus, identification of a mitochondrial form of this enzyme provides a tool with which to dissect mammalian mitochondrial genome dynamics.
For some time it was thought that the mammalian mitochondria lacked the capacity to repair damaged DNA (18). This hypothesis had its origin in the observation that pyrimidine dimers produced within the mitochondrial DNA (mtDNA) of cultured cells were not repaired, in marked contrast to similar lesions produced in the nuclear genomes of these same cells (4, 15, 21). However, it is now clear that this organelle is not entirely deficient in DNA repair activity. For example, it appears that base excision repair of oxidized DNA occurs in mtDNA of mammalian cells (8, 23, 34, 40). In addition, a number of studies have revealed that mammalian cultured cells can repair mtDNA damage caused by a number of chemical agents, including cisplatin (15), bleomycin (33), streptozotocin (22), and 4-nitroquinoline (36). More recently, it was shown that mammalian mitochondrial extracts possess DNA nonhomologous end-joining and homologous recombination activities (14, 41). Taken together, these findings indicate that mammalian mitochondria likely possess a number of DNA repair pathways. A series of recent observations suggest that mutations to the mitochondrial genome may be associated with a variety of human pathologies (5, 9, 10, 38, 49), thus highlighting the need for a greater understanding of the molecular biology of mtDNA repair.
A common feature of nearly all DNA repair pathways is the requirement for DNA ligase activity. While a great deal is known about mammalian nuclear DNA ligase activity (42, 43), very little is known about mammalian mitochondrial DNA ligase(s). Nearly two decades ago, a report describing ligase activity in the mitochondria was published (16). More recently, a DNA ligase activity was purified from Xenopus laevis mitochondrial extracts and shown to be immunologically related to the human nuclear DNA ligase III protein (23). Thus, identification and cloning of a mitochondrial DNA ligase gene would provide a valuable tool with which to dissect mtDNA repair pathways.
The results of Pinz and Bogenhagen (23) raised the possibility that the DNA ligase III gene could encode both mitochondrial and nuclear proteins. As these investigators pointed out, the human DNA ligase III cDNA contains an in-frame ATG upstream of the putative translation initiation ATG site (3, 48). Inspection of the cDNA sequence flanked by these two ATG codons (herein referred to as M1 and M2, for methionines 1 and 2) revealed that it encodes a positively charged amphipathic α helix similar to the leader sequence present on the amino terminus of mitochondrial matrix proteins (46, 47). Thus, if translation initiated at M1, a mitochondrial protein could be produced, whereas translation initiated at M2 would presumably produce the nonmitochondrial, nuclear form. Alternate translation initiation has been shown to result in the production of both mitochondrial and nonmitochondrial proteins from the same gene in both yeasts (1, 35, 45) and vertebrates (6, 39).
In this report, we demonstrate that the upstream 5′ end of the DNA ligase III cDNA functions as a mitochondrial targeting sequence. Translation from M1 leads to the mitochondrial protein, while translation from M2 results in the previously identified nuclear protein. Mitochondrial protein extracts from cells overexpressing antisense DNA ligase III mRNA show marked reduction in both DNA ligase and DNA end-joining activities. Based on these results, we conclude that the human DNA ligase III gene encodes both nuclear and mitochondrial proteins.
MATERIALS AND METHODS
Cloning of human DNA ligase III cDNA.
Reverse transcriptase-mediated PCR (RT-PCR) was used to amplify nucleotides 73 to 333 and 73 to 3102 of the human DNA ligase cDNA from HT1080 human fibrosarcoma-derived cells (24). The nucleotide positions given herein are based on the DNA ligase III cDNA sequence from GenBank accession no. X84740 (3, 48). The primers used for PCR amplification of nucleotides 73 to 333 of the DNA ligase III cDNA were LIG3(5′)S (AAGCTTATGTCTTTGGCTTTCAAG) and LIG3(5′)AS (AAGCTTCTCACAGGGGCCACT) (nucleotides in italics indicate HindIII linker sequence).
The full-length coding sequence of the DNA ligase III cDNA was cloned via RT-PCR using primers LIG3(FL)HA-S (GGATCCATGTCTTTGGCTTTCAAGATCTTCT) and LIG3(FL)HA-AS (GGATCCCTAGGCGTAGTCGGGGACGTCGTAGGGGTAGCAGGGAGCTACCAGTCTCC) (the stop codon is in boldface; the underlined sequence encodes a hemagglutinin [HA] tag), as a consequence of which the PCR product encodes a DNA ligase protein with an HA epitope at its carboxyl terminus. All oligonucleotide sequences were synthesized at the Microchemical Facility (University of Minnesota) and are depicted in the 5′-3′ orientation.
Plasmid constructs. (i) DNA ligase III-GFP fusion constructs.
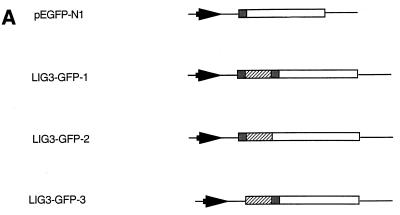
The 5′ portion of the DNA ligase III cDNA (from nucleotides 73 to 333) was introduced into the multiple cloning site of plasmid pEGFP-N1 (Clontech). Cloning was performed such that the putative mitochondrial targeting sequence of the DNA ligase III cDNA was in frame with the ATG codon of the GFP gene (thereby producing plasmid pLIG3-GFP-1). Similarly, the mitochondrial targeting sequence of the cytochrome c oxidase gene was cloned into the same region of pEGFP-N1. Others have shown that constructs of this nature produce GFP fusion proteins that are targeted to the mitochondria (26). This vector (pCOXVIII-GFP) was used as a positive control. Site-directed mutagenesis (see below) was performed on plasmid pLIG3-GFP-1 to change the DNA ligase III cDNA-encoded ATG (nucleotides 73 to 75) to ATC, thereby creating plasmid pLIG3-GFP-2. Similarly, site-directed mutagenesis was used to modify the GFP gene-encoded ATG codon from plasmid pLIG3-GFP-1 to ATC, thus creating plasmid pLIG3-GFP-3.
(ii) HA-tagged DNA ligase III.
Using RT-PCR, an HA tag was cloned in frame with the 3′ end of the human DNA ligase III cDNA. This PCR product was used to create the expression vector pLIG3-FL. By using site-directed mutagenesis (see below), the ATG at positions 73 to 75 of the human DNA ligase III cDNA was mutated to ATC to create plasmid pLIG3-FL1. Similarly, the ATG at positions 334 to 336 was changed to ATC, creating plasmid pLIG3-FL2.
(iii) DNA ligase III cDNA antisense.
An antisense expression plasmid was created by introducing a portion of the human DNA ligase III cDNA (nucleotides 73 to 333) into the HindIII site of pREP4 (Invitrogen), to create plasmid pREP4dnlAS.
Site-directed mutagenesis.
Single-stranded DNA was prepared from plasmid constructs, and point mutations were generated by the method of Kunkel et al. (13). Putative translation initiation sequences (ATG) were altered to ATC. The ATG-to-ATC changes were included with additional alterations to create a new ClaI restriction endonuclease recognition sequence. New plasmid constructs were screened by restriction digestion using ClaI and by DNA sequencing.
Transfection of mammalian cells. (i) Transient transfection by DEAE-dextran.
To study intracellular localization of the various GFP-DNA ligase III fusion proteins, the expression plasmids described above were transfected into African green monkey kidney-derived Cos-1 cells that had been grown on coverslips. Briefly, cells were incubated in serum-free medium containing DEAE-dextran and 10 μg of DNA at 37°C for 3 to 4 h (17). At the end of the incubation period, cells were treated with 10% dimethyl sulfoxide in HEPES-buffered saline for 60 s and washed with serum-free medium three times. Cells were allowed to grow for 24 to 48 h in growth medium containing 9% fetal bovine serum prior to analysis (see below).
(ii) Stable transfection by electroporation.
Human fibrosarcoma-derived HT1080 cells were transfected with plasmids pREP4 and pREP4dnlAS via electroporation (11). Stable transfectants were isolated based on their resistance to hygromycin.
Immunocytochemistry.
Transfected HT1080 cells were processed for immunocytochemistry by previously described procedures (2). At 48 h posttransfection, cells were fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) for 20 min. The cells were washed with PBS three times and incubated in 50 mM NH4Cl in PBS for 10 min. Cells were treated with 0.1% Triton X-100 in PBS and then washed for 30 min with 0.2% gelatin in PBS. The cells were incubated with the primary antibody for 1 h at 37°C and stained with either rhodamine- or fluorescein-conjugated antibody (Pierce). The cells were washed four times with PBS and then analyzed by confocal fluorescence microscopy.
Confocal microscopy.
Transfected mammalian cells on coverslips were transferred to a petri dish containing PBS and scanned with a Bio-Rad MRC-1024 confocal system. A 60× water immersion objective was used, and the samples were excited at 488 nm with the emission fixed at 605 nm. The lipophilic cationic probe 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolylcarbocyanine io-dide (JC-1) was used as a marker for mitochondrial localization (27). Cos-1 cells on coverslips were transferred to PBS and incubated with JC-1 at a final concentration of 3 μM for 25 min at 37°C. The PBS solution containing the dye was removed and replaced with fresh PBS. Cells were immediately visualized under the confocal microscope. Digitized fluorescent images were analyzed with Adobe Photoshop.
Preparation of mitochondrial protein extracts.
Mitochondria were isolated from transfected HT1080 cells as previously described (7). Briefly, cells were homogenized and centrifuged three times to remove unbroken cells, cell debris, and nuclei. Mitochondria were collected from the supernatant by centrifugation at 20,000 × g for 20 min. The isolated mitochondrial fraction was washed two to three times, and protein extracts prepared by hypo-osmotic shock. Purity was assessed by determining cytochrome c oxidase activity (37).
Adenylation reaction.
Adenylation reactions were carried out on the mitochondrial protein extracts as described previously (44). Reactions were carried out in a 20-μl volume containing 60 mM Tris-Cl (pH 8.0), 10 mM MgCl2, 5 mM dithiothreitol, 50 μg of bovine serum albumin per ml, [α-32P]ATP, and 5 μg of mitochondrial protein extract. Following a 15-min incubation at room temperature, reactions were stopped by the addition of sodium dodecyl sulfate loading buffer followed by boiling. The ligase-adenylate complex was separated on 10% denaturing polyacrylamide gel, fixed with 10% acetic acid, dried, and exposed to a phosphorimager screen. The amount of the adenylated protein product was quantified with the program IP Lab Gel.
DNA end-joining reaction.
DNA end-joining reactions were carried out by the method of North et al. (20). Circular pUC118 plasmid DNA was linearized with PstI. Linearized plasmid (1 μg) was incubated with 10 μg of mitochondrial protein extract in 70 mM Tris (pH 7.5) buffer in the presence of 10 mM MgCl2, 10 mM dithiothreitol, and 1 mM ATP in a 25-μl reaction volume. Reactions were carried out at 14°C for 2 h. Following treatment of samples with proteinase K for 30 min at 37°C, samples were separated on a 1% agarose gel. The gel was transferred to a nitrocellulose membrane and probed with radioactive pUC118 (specific activity of 5 × 106 cpm/μg of DNA).
RESULTS
Analysis of the DNA ligase III cDNA sequence.
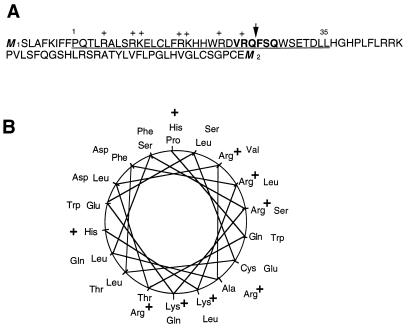
It has previously been proposed that the start site for protein translation of the human DNA ligase III cDNA is at the ATG encoded by nucleotides 334 to 336 (M2 [Fig. 1A]). However, the sequence upstream of this site contains a second in-frame ATG codon (nucleotides 73 to 75; M1 [Fig. 1A]). The conclusion that protein translation initiates at M2 was based on its sequence context, which was considered more favorable for translation initiation (3, 48) than that associated with M1. Closer examination of the upstream sequence indicates that translation from M1 would generate an amino-terminal peptide that is amphipathic in nature. Secondary structure analysis of this peptide was carried out with pHDsec, a network method (28, 29). The predicted helical region of the amino-terminal peptide (Fig. 1A, underlined sequence) strongly resembles a mitochondrial targeting signal (Fig. 1B) (30, 46, 47). This sequence was also analyzed using PSORT, an algorithm that recognizes mitochondrial targeting signals using discriminate analysis from values of partial amino acid composition. Processing of proteins imported into the mitochondria occurs due to the action of a processing peptidase that recognizes a consensus sequence specified as (V/A/S)-R-X-(F/L)-(S/A), where X denotes an arbitrary amino acid residue (31). As Fig. 1A indicates, the DNA ligase III cDNA sequence appears to encode a well-conserved cleavage site (VRQFSQ) for the mitochondrial processing peptidase (47). Based on this analysis, we concluded that the putative N-terminal region resulting from translation from the M1 resembles a mitochondrial targeting sequence.
FIG. 1.
The human DNA ligase III cDNA contains a putative mitochondrial targeting sequence. (A) Deduced amino acid sequence of the human DNA ligase III cDNA upstream sequence from the first in-frame ATG (M1) to the second ATG (M2). M2 has previously been designated the translation initiation site for the nuclear ligase III protein (3, 47). The underlined sequence represents the alpha-helical region as predicted by the program pHDsec (27, 28). Plus signs overlie positively charged residues; the amino acids in bold comprise the putative cleavage recognition sequence, and the arrow indicates the putative cleavage site. (B) Amino acid residues 1 through 35 (underlined in panel A) were plotted as specified by Schiffer and Edmundson (30). Positively charged residues are indicated.
Expression of DNA ligase III-GFP constructs in mammalian cells.
To test whether the peptide encoded by positions 73 to 333 of the human DNA ligase III cDNA could function as a mitochondrial targeting sequence, the relevant portion of the DNA ligase III cDNA upstream sequence was cloned in frame with GFP in plasmid pEGFP-N1 (Fig. 2A). This change was engineered such that the initiation ATG of the GFP is replaced M2 of the human DNA ligase III. The constructs were transiently transfected into Cos-1 cells, and the subcellular localization of GFP fusion proteins was monitored by confocal fluorescence microscopy 24 to 36 h after transfection.
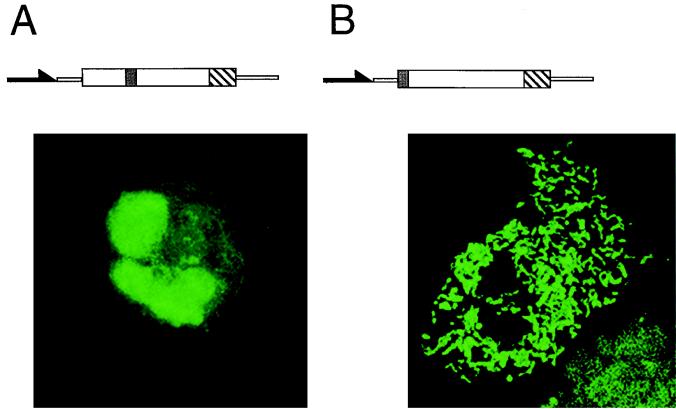
FIG. 2.
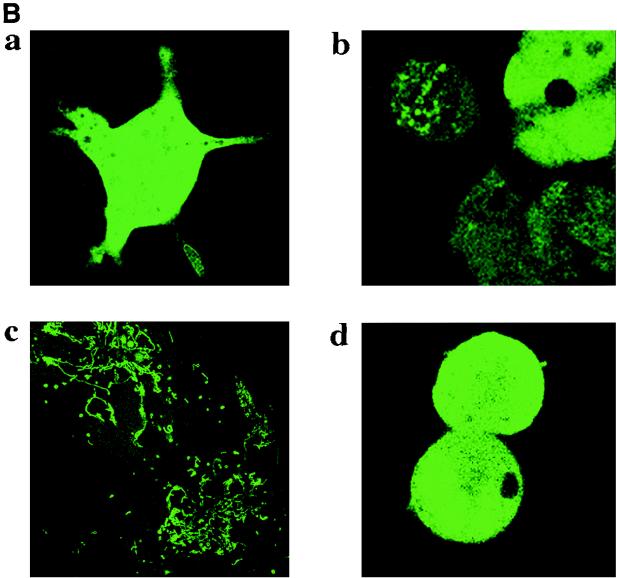
Subcellular localization of DNA ligase III-GFP fusion proteins. (A) DNA ligase III-GFP fusion constructs. White boxes, GFP coding region; hatched boxes, DNA ligase III cDNA nucleotides 73 to 333; shaded boxes, ATG codons; arrowheads; cytomegalovirus promoter; thin lines, vector sequences. Construction of vectors is described in Materials and Methods. (B) Subcellular localization of DNA ligase III-GFP fusion proteins. Cos-1 cells were transiently transfected with GFP plasmid constructs pEGFP-N1 (a), pLIG3-GFP-1 (b), pLIG3-GFP-2 (c), and pLIG3-GFP-3 (d) and observed under a Bio-Rad confocal microscope 24 h following transfection.
Figure 2B shows confocal images of Cos-1 cells transfected with these plasmid constructs. Cells transfected with plasmid pEGFP revealed pancellular distribution of the GFP protein (Fig. 2B, panel a). When plasmid pLIG3-GFP-1 was transfected into these cells, 10 to 20% of the cells showed punctate fluorescence, while the remaining cells exhibited fluorescence characteristic of cytoplasmic localization (Fig. 2B, panel b). We hypothesized that the cytosolic fluorescence seen in cells transfected with pLIG3-GFP-1 could result from translation initiating at the methionine codon of the GFP gene. The sequence surrounding this site is far more favorable for translation than that associated with the DNA ligase III M1 (3, 48). It thus seemed reasonable to propose that in this construct translation initiation from M1 would occur at a low level. To address this possibility, the ATG of the GFP gene was mutated to ATC by site-directed mutagenesis, thereby creating plasmid pLIG3-GFP-2. Confocal imaging of cells transfected with this construct (Fig. 2B, panel c) revealed a rod-shaped fluorescence pattern typical of mitochondria (26). To confirm that mitochondrial localization resulted from translation initiation at DNA ligase III M1, we constructed an additional mutant (pLIG3-GFP-3) in which this ATG was altered to ATC while the GFP-encoded ATG was left unaltered. Confocal imaging of cells transfected with pLIG3-GFP-3 is shown in Fig. 2B, panel d. The fluorescence observed in these cells was primarily cytosolic, and the thread-like mitochondrial signal was absent. An additional construct in which both ATG sites were altered to ATC did not produce any detectable intracellular fluorescence (data not shown).
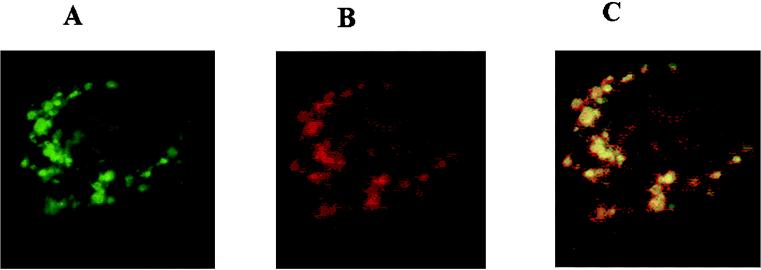
When cells expressing GFP were scanned at very low laser strength (0.3 to 3%), a very high fluorescence yield was obtained. Further, control cells which were not transfected with the plasmid did not show green fluorescence. However, we wished to exclude the possibility that the punctate pattern obtained with pLIG3-GFP-2 was due to low levels of autofluorescence produced by cellular components. Figure 3 shows the fluorescence pattern of cells transfected with plasmid pLIG3-GFP-2, incubated with anti-GFP monoclonal antibody, and stained with rhodamine-conjugated anti-mouse antibody. Figure 3A shows cells excited at 488 nm, displaying fluorescence from GFP; Fig. 3B shows cells excited at 568 nm, depicting the fluorescence of rhodamine. Both rhodamine and GFP fluorescence exhibited a punctate pattern of expression in cells transfected with the plasmid pLIG3-GFP-2. Figure 3C depicts the merged images of GFP and rhodamine fluorescence. As one would expect based on examination of the two separate panels, there was essentially complete overlap of the fluorescence images, indicating that the punctate fluorescence is due to the presence of the GFP protein and not due to low amounts of autofluorescence of cellular components.
FIG. 3.
Colocalization within the mitochondria of GFP autofluorescence and rhodamine-labeled anti-GFP antibody. Panels show immunofluorescence of Cos-1 cells transfected with plasmid pLIGIII-GFP-2. (A) GFP fluorescence; (B) cells incubated with anti-GFP monoclonal antibody and then stained with rhodamine labeled anti-mouse immunoglobulin G antibody; (C) merged images of GFP and rhodamine fluorescence from panels A and B.
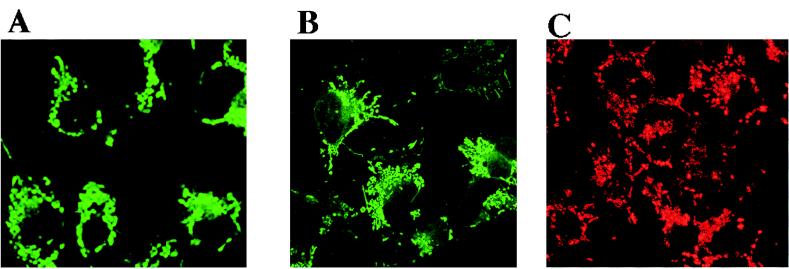
We wanted to confirm that the punctate fluorescence exhibited by cells transfected with pLIG3-GFP-2 was indeed indicative of mitochondrial localization of the fusion protein. Therefore, as a positive control, we produced a plasmid expression construct in which the mitochondrial targeting sequence of cytochrome c oxidase subunit VIII was cloned in frame with the GFP gene (pCOXVIII-GFP). Other investigators have shown that GFP fusion constructs essentially identical to this one localize to the mitochondrial compartment in a variety of cell types (2, 26). We found the intracellular distribution of fluorescence seen in confocal images of cells transfected with either pCOXVIII-GFP (Fig. 4A) or pLIG3-GFP-2 (Fig. 4B) to be quite similar. As a further control, we incubated untransfected Cos-1 cells with the lipophilic cationic probe JC-1. The membrane potential of mitochondria promotes a directional uptake of JC-1 into the matrix, with the subsequent formation of the J-aggregates that cause mitochondria to fluoresce red (25). As Fig. 4C indicates, the mitochondrion-specific fluorescence seen in Cos-1 cells stained with JC-1 is essentially identical to the pattern seen in Fig. 4A or B. Based on these results, we can conclude that the peptide encoded by residues 73 through 333 of the DNA ligase III cDNA is capable of functioning as a mitochondrial targeting sequence.
FIG. 4.
Residues 73 to 333 of the DNA ligase III cDNA encode a functional mitochondrial targeting sequence. Confocal images show Cos-1 cells transfected with plasmid pCOXVIII-GFP (A), Cos-1 cells transfected with pLIG3-GFP-2 (B), and untransfected Cos-1 cells (C) stained with 3 μM JC-1, a fluorescent dye that specifically stains mitochondria.
Expression of HA-tagged DNA ligase III in mammalian cells.
While these data indicate that this sequence is capable of functioning as a mitochondrial targeting sequence, they do not by themselves address the physiological relevance of this observation. We therefore created a series of expression constructs that would direct expression of HA-tagged DNA ligase III minigenes. First, we used RT-PCR to amplify nucleotides 73 to 3102 of the DNA ligase III cDNA. The primer used to clone the 3′ end of this cDNA encoded an HA epitope tag. Site-directed mutagenesis was then used to inactivate M1 or M2. Thus, we constructed two plasmids, pLIG3-FL-1 and pLIG3-FL-2 (Fig. 5). These plasmids were separately transfected into Cos-1 cells, and immunocytochemistry was performed using a fluorescein-labeled anti-HA monoclonal antibody. As Fig. 5 indicates, cells transfected with pLIG3-FL-1 display fluorescence localized to the nucleus, with some diffuse cytoplasmic signal observed, while cells transfected with pLIG3-FL-2 yielded a rod-like fluorescence pattern, consistent with mitochondrial localization. These results are consistent with the hypothesis that the DNA ligase III cDNA encodes two separate proteins. They suggest that translation initiation from the M1 led to the production of a ligase enzyme containing an amino-terminal mitochondrial targeting sequence. These data further suggest that translation from the M2 results in the production of a nucleus-specific ligase protein.
FIG. 5.
Mitochondrial or nuclear targeting of HA epitope-tagged DNA ligase III minigene products. Mammalian expression vectors harbored HA-tagged DNA ligase III minigenes in which either M1 (pLIG-FL-1) or M2 (pLIG-FL-2) was inactivated. Filled arrows, cytomegalovirus promoter from expression vector; white boxes, DNA ligase III cDNA; hatched boxes, simian virus 40 polyadenylation signal; shaded boxes, ATG codons M1 and M2 (the absence of these boxes indicates that the relevant ATG site has been inactivated). Expression constructs harboring HA-tagged DNA ligase III proteins were used to transiently transfect Cos-1 cells, and subcellular localization was visualized by immunocytochemistry using fluorescein-labeled anti-HA monoclonal antibody (confocal images of cells transfected with pLIG-FL-1 or pLIG-FL-2 are displayed beneath the expression constructs).
Effect of antisense DNA ligase III expression in mammalian cells.
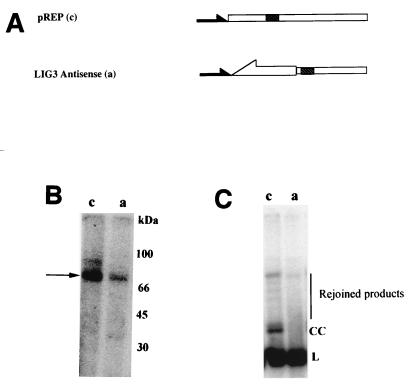
We wanted to provide independent evidence supporting our conclusion that the DNA ligase III gene encodes two separate proteins, one nuclear and the other mitochondrial. We hypothesized that if the DNA ligase III cDNA indeed encodes a mitochondrial form of this enzyme, overexpression of antisense DNA ligase III mRNA should interfere with translation of this protein. We therefore cloned nucleotides 73 to 333 of the DNA ligase III cDNA into the expression vector pREP4 in the antisense orientation, thereby creating plasmid pREP4dnlAS (Fig. 6). The rationale behind choosing this portion of the DNA ligase III cDNA (which encodes the mitochondrial targeting sequence identified above) was to inhibit the expression of ligase III specifically without altering the levels of other DNA ligases that share considerable sequence similarity (43). Plasmid pREP4dnlAS or the unmodified expression vector pREP4 was electroporated into human HT1080 cells, and pools of approximately 100 stable transfectants each were isolated. Mitochondrial protein extracts were prepared from the pooled transfectants, and the amount of DNA ligase activity present within them was determined by measuring protein adenylation following incubation with [α-32P]ATP (44). The same amount of protein (5 μg) was used in all assays. Figure 6B reveals mitochondrial extracts prepared from both control and antisense mRNA-expressing cells contain a major adenylated protein of approximately 80 kDa. However, cells expressing the antisense DNA ligase III mRNA (lane a) exhibit substantially reduced levels of this protein. To confirm that levels of DNA ligase activity were lower in antisense mRNA-expressing cells than in control cells, we incubated linearized plasmid DNA along with 10 μg of mitochondrial extract prepared from control and antisense mRNA-expressing cells and measured their ability to rejoin this plasmid by using Southern blot hybridization. Figure 6C shows a representative Southern blot pattern of PstI-linearized pUC118 treated with mitochondrial protein extracts from pREP4-transfected HT1080 cells as a control (lane c) and cells expressing the antisense DNA ligase III (lane a). The control lane shows formation of closed circular and other high-molecular-weight products.
FIG. 6.
Expression of DNA ligase III antisense mRNA reduces mitochondrial adenylation activity. (A) DNA ligase III antisense mRNA constructs. pREP4 plasmid (c; Invitrogen) has a Rous sarcoma virus promoter (filled arrow) and a simian virus 40 polyadenylation site (hatched box). Nucleotides 73 to 330 of the human DNA ligase III cDNA (open arrow) were cloned in the reverse orientation into pREP4 to create pREP4dnlAS (a). (B) Mitochondrial adenylation activity. Equal amounts of mitochondrial protein extracts (5 μg) prepared from pREP4-transfected cells (lane c) or cells transfected with pREP4dnlAS (lane a) were incubated with [α-32P]ATP, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and exposed to a phosphorimager screen. (C) Mitochondrial DNA end-joining activity. DNA end-joining reactions were carried out on 5 μg of mitochondrial extracts prepared from human cells transfected with pREP4 (lane c) or pREP4dnlAS (lane a) as described in Materials and Methods. L, linear substrate DNA; CC, covalently closed circular plasmid DNA; vertical line, high-molecular-weight end-joined DNA products.
To more quantitatively assess the effect of antisense mRNA expression of mitochondrial ligase activity, we performed a series of adenylation and end-joining reactions using the mitochondrial protein extracts described above. They reveal that both adenylation and end-joining activities are approximately 75% lower in mitochondrial extracts prepared from antisense mRNA-expressing cells than in similar extracts prepared from control cells. Since the amounts of cytochrome c oxidase activity present in both antisense mRNA- and control-transfected cells were indistinguishable (data not shown), we conclude that expression of the DNA ligase III antisense mRNA had no general effect on mitochondrial protein levels. We repeated the above analyses on pools of HT1080 clones obtained from independent transfections with pREP4 or pREP4dnlAS. In the latter experiments, we again observed that cells transfected with pREP4dnlAS possessed significantly lower levels of mitochondrial DNA ligase activity than control cells transfected with pREP4 control vector (data not shown).
DISCUSSION
We present evidence that the human DNA ligase III gene encodes a mitochondrial form of ligase. Sequence analysis of the DNA ligase III cDNA reveals the presence of two ATG codons that are in frame with the protein-coding region of this enzyme (23). The deduced amino acid sequence encoded by the region spanning these two ATG sites (M1 and M2 [Fig. 1A]) resembles a mitochondrial targeting peptide. It is hypothesized that translation from the upstream M1 site would produce a mitochondrial protein, whereas translation from the downstream M2 would produce a nuclear form. In this study, we show that the putative mitochondrial targeting peptide encoded by the DNA ligase III cDNA can function to localize a heterologous protein, in this case GFP, to the mitochondria of cultured mammalian cells. Using an HA-tagged DNA ligase III minigene, we show that mitochondrial and nuclear localization result from translation initiating from M1 and M2, respectively. Finally, we show that cells expressing antisense DNA ligase III had dramatically reduced levels of DNA ligase activity compared to control cells.
Our results allow us to conclude that the DNA ligase III gene encodes a mitochondrial protein. We have further concluded that this protein is targeted to this compartment by the mitochondrial targeting sequence encoded by residues 73 through 333 of the cDNA. We do not know whether one or two forms of mRNA are produced. In principle, one can imagine that there are two separate transcriptional initiation sites present in this gene. Alternatively, it is conceivable that a single mRNA molecule is produced and that there are two separate translational start sites. Based on its sequence context, M1 is likely to be a relatively weak start site, with only an R-3 (where R is a pyrimidine). In contrast, M2 is located within a far more attractive environment for translation initiation (YNNATGG, where Y is a purine). It has been proposed that mRNA species in which the upstream-most in-frame AUG codon is unfavorable initiate translation at both the first and second AUGs through a leaky scanning mechanism, thus producing two proteins from a single mRNA species (12). In the case of ligase III cDNA, translation initiation from the first M1 is likely to be very inefficient. In contrast, one would predict far more efficient translation from M2. This represents an attractive proposition, since there is roughly 100-fold more DNA in the mammalian nucleus than in the mitochondria of these cells (32), and this would provide an elegant mechanism to ensure proper relative levels of ligase protein expression in the two organelles.
There are a number of examples in which two different forms of a protein are encoded by the same gene in both yeast and animals. The Mod5 mRNA of Saccharomyces cerevisiae, which encodes a tRNA modification enzyme, has two in-frame AUGs. Protein translation from the first AUG yields a product that is imported into the mitochondria, while the protein product translated from the second AUG is localized in the cytoplasm (35). In the case of eukaryotic cells, the UNG gene, encoding uracil DNA glycosylase, yields two alternate forms of transcripts. One of these mRNAs encodes a protein containing a nuclear localization signal, while the second mRNA encodes a protein possessing a mitochondrial targeting signal (47). A single copy of the decarboxylase gene present in the goose genome encodes a mitochondrial protein found in extremely low levels in the liver and a cytosolic form present in higher amounts in uropygial glands (6). The mitochondrial and cytosolic forms of rat liver fumarase protein have identical amino acid sequences, suggesting that the two isozymes are encoded by a single gene and that their precursors may be encoded by a single species of mRNA (39).
We detected a dramatic reduction in DNA ligase protein activity in mitochondrial protein extracts prepared from cells that expressed antisense DNA ligase III mRNA. At present, given the limitations of antisense-based technology, it is not possible to conclude whether the absence of complete inhibition of ligase expression in the mitochondria indicates the presence of an additional mitochondrial ligase or instead is due to inefficiency of antisense-based gene inactivation. The major adenylated protein product that we detected in mitochondrial extracts treated with [α-32P]ATP migrated with an apparent molecular mass of approximately 80 kDa. This size is similar to that of DNA ligase II, which is thought to be derived from DNA ligase III as a result of proteolysis (3, 43, 48). If the 80-kDa mitochondrial adenylation product were derived from a larger precursor, this could explain the small amount of higher-molecular-weight product seen in Fig. 6B, lane a. Additional experimentation will be required to determine whether other minor forms of DNA ligases are present in the mitochondria.
In vitro reconstitution experiments indicate that DNA ligase III is likely to be involved in mitochondrial base excision repair (23). It has further been suggested that this enzyme plays a role in recombinational repair (3), suggesting that this molecule could play a role in the homologous recombination activity detected in mammalian mitochondrial extracts (41). Finally, the involvement of DNA ligase III in mitochondrial DNA end joining (this study) suggests a potential role for this protein in the repair of DNA double-strand breaks in this organelle. These observations suggest that the DNA ligase III protein plays a major role in mitochondrial DNA repair in mammals. Thus, identification of mitochondrial ligase offers a useful tool to dissect the various repair processes that occur within this organelle.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institute of Health (CA R29 CA61906), the American Heart Association (9601390), and the American Cancer Society (DHP-171). U.L. was supported by a fellowship from the American Heart Association.
Greg Coffey provided much-appreciated editorial advice.
REFERENCES
- 1.Beltzer J P, Morris S R, Kohlhaw G B. Yeast LEU4 encodes mitochondrial and non-mitochondrial forms of alpha-isopropylmalate synthase. J Biol Chem. 1988;263:368–374. [PubMed] [Google Scholar]
- 2.Brini M, Marsault R, Bastianutto C, Pozzan T, Rizzuto R. Nuclear targeting of aequorin: a new approach for measuring Ca2+ concentration in intact cells. Cell Calcium. 1994;16:259–268. doi: 10.1016/0143-4160(94)90089-2. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Tomkinson A E, Ramos W, Mackey Z B, Danehower S, Walter C A, Schultz R A, Besterman J M, Husain J. Mammalian DNA ligase III: molecular cloning, chromosomal localization and expression in spermatocytes undergoing meiotic recombination. Mol Cell Biol. 1995;15:5412–5422. doi: 10.1128/mcb.15.10.5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clayton D A, Doda J N, Friedberg E C. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc Natl Acad Sci USA. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corral-Debrinski M, Horton T, Lott M T, Shoffner M, McKeem A C, Beal M F, Graham B H, Wallace D C. Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics. 1994;23:471–476. doi: 10.1006/geno.1994.1525. [DOI] [PubMed] [Google Scholar]
- 6.Courchesne-Smith C, Jang S H, Shi Q, DeWille J, Sasaki G, Kolattukudy P E. Cytoplasmic accumulation of a normally mitochondrial malonyl-CoA decarboxylase by the use of an alternate transcription start site. Arch Biochem Biophys. 1992;298:576–86. doi: 10.1016/0003-9861(92)90452-3. [DOI] [PubMed] [Google Scholar]
- 7.Darley-Usmar V M, Rickwood D, Wilson M T. Mitochondria: a practical approach. New York, N.Y: IRL Press; 1987. [Google Scholar]
- 8.Driggers W J, LeDoux S P, Wilson G L. Repair of oxidative damage within the mitochondrial DNA of RINr 38 cells. J Biol Chem. 1993;268:22042–22045. [PubMed] [Google Scholar]
- 9.Hamblet N S, Castora F J. Elevated levels of the Kearns-Sayre syndrome mitochondrial DNA deletion in temporal cortex of Alzheimer’s patients. Mutat Res. 1997;379:253–262. doi: 10.1016/s0027-5107(97)00158-9. [DOI] [PubMed] [Google Scholar]
- 10.Horton T M, Graham B H, Corral-Debrinski M, Shoffner M, Kaufman A E, Beal M F, Wallace D C. Marked increase in mitochondrial DNA deletion levels in the cerebral cortex of Huntington’s disease patients. Neurology. 1995;45:1879–1883. doi: 10.1212/wnl.45.10.1879. [DOI] [PubMed] [Google Scholar]
- 11.Keown W A, Campbell C R, Kucherlapati R S. Methods for introducing DNA into mammalian cells. Methods Enzymol. 1990;185:527–357. doi: 10.1016/0076-6879(90)85043-n. [DOI] [PubMed] [Google Scholar]
- 12.Kozak M. Interpreting cDNA sequences: some insights from studies on translation. Mamm Genome. 1996;7:563–574. doi: 10.1007/s003359900171. [DOI] [PubMed] [Google Scholar]
- 13.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;155:166–178. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 14.Lakshmipathy U, Campbell C. Double strand break rejoining by mammalian mitochondrial extracts. Nucleic Acids Res. 1999;27:1198–1204. doi: 10.1093/nar/27.4.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LeDoux S P, Wilson G L, Beecham E J, Stevnsner T, Wassermann K, Bohr V A. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–1973. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 16.Levin C J, Zimmerman S B. A DNA ligase from mitochondria of rat liver Biochem. Biophys Res Commun. 1976;69:514–520. doi: 10.1016/0006-291x(76)90551-9. [DOI] [PubMed] [Google Scholar]
- 17.Lopata M, Cleveland D W, Sollner-Webb B. High level transient expression of chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984;12:5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miquel J. An update on the mitochondrial-DNA mutation hypothesis of cell aging. Mutat Res. 1992;275:209–216. doi: 10.1016/0921-8734(92)90024-j. [DOI] [PubMed] [Google Scholar]
- 19.Nagelhus T A, Slupphang G, Lindmo T, Krokan H E. Cell cycle regulation and subcellular localization of the major human uracil-DNA glycosylase. Exp Cell Res. 1995;220:292–297. doi: 10.1006/excr.1995.1318. [DOI] [PubMed] [Google Scholar]
- 20.North P, Ganesh A, Thacker J. The rejoining of double strand breaks in DNA by human cell extracts. Nucleic Acids Res. 1990;18:6205–6210. doi: 10.1093/nar/18.21.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pascucci B, Versteegh A, van Hoffen A, van Zeeland A A, Mullenders L H, Dogliotti E. DNA repair of UV photoproducts and mutagenesis in human mitochondrial DNA. J Mol Biol. 1997;273:417–427. doi: 10.1006/jmbi.1997.1268. [DOI] [PubMed] [Google Scholar]
- 22.Pettepher C C, LeDoux S P, Bohr V A, Wilson G L. Repair of alkali-labile sites within the mitochondrial DNA of RINr 38 cells after exposure to the nitrosourea streptozotocin. J Biol Chem. 1991;266:3113–3137. [PubMed] [Google Scholar]
- 23.Pinz K G, Bogenhagen D F. Efficient repair of abasic sites in DNA by mitochondrial enzymes. Mol Cell Biol. 1998;18:1257–1265. doi: 10.1128/mcb.18.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasheed S, Nelson-Rees W A, Toth E M, Arnstein P, Gardner M B. Characterization of a newly derived human sarcoma cell line (HT-1080) Cancer. 1974;33:1027–1033. doi: 10.1002/1097-0142(197404)33:4<1027::aid-cncr2820330419>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 25.Reers M, Smith T W, Chen L B. J-aggregate formation of a carbocyanine as a quantitative fluorescent indicator of membrane potential. Biochemistry. 1991;30:4480–4486. doi: 10.1021/bi00232a015. [DOI] [PubMed] [Google Scholar]
- 26.Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent proteins as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- 27.Robins P, Lindahl T. DNA ligase IV from HeLa cell nuclei. J Biol Chem. 1996;271:24257–24261. doi: 10.1074/jbc.271.39.24257. [DOI] [PubMed] [Google Scholar]
- 28.Rost B, Sanders C. Prediction of protein structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 29.Rost B, Sanders C, Schneider R. pHD—an automatic mail server for protein secondary structure prediction. CABIOS. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 30.Schiffer M, Edmundson A B. Use of helical wheels to represent the structure of proteins and to identify segments with helical potential. Biophys J. 1967;7:21–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider G, Sjoling S, Wallin E, Wrede P E, Glaser E, von Heijne G. Feature-extraction from endopeptidase cleavage sites in mitochondrial targeting peptides. Proteins Struct Funct Genet. 1998;30:19–60. [PubMed] [Google Scholar]
- 32.Shadel G S, Clayton D A. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 33.Shen C C, Wertelecki W, Driggers W J, LeDoux S P, Wilson G L. Repair of mitochondrial DNA damage induced by bleomycin in human cells. Mutat Res. 1995;337:19–23. doi: 10.1016/0921-8777(95)00008-8. [DOI] [PubMed] [Google Scholar]
- 34.Slupphaug G, Markussen F H, Olsen L C, Aasland R, Aarsaether N, Bakke O, Krokan H E, Helland D E. Nuclear and mitochondrial forms of human uracil-DNA glycosylase are encoded by the same gene. Nucleic Acids Res. 1993;21:2579–2584. doi: 10.1093/nar/21.11.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slusher L B, Gillman E C, Martin N C, Hopper A K. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc Natl Acad Sci USA. 1991;88:9789–9793. doi: 10.1073/pnas.88.21.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snyderwine E G, Bohr V A. Gene- and strand-specific damage and repair in Chinese hamster ovary cells treated with 4-nitroquinoline 1-oxide. Cancer Res. 1992;52:4183–4189. [PubMed] [Google Scholar]
- 37.Storrie B, Madden E A. Isolation of subcellular organelles. Methods Enzymol. 1990;182:203–225. doi: 10.1016/0076-6879(90)82018-w. [DOI] [PubMed] [Google Scholar]
- 38.Suomalainen A, Kaukonen J, Amati P, Timonen R, Haltia M, Weissenbach J, Zeviani M, Somer H, Peltonen L. An autosomal locus predisposing to deletions of mitochondrial DNA. Nat Genet. 1995;9:146–151. doi: 10.1038/ng0295-146. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki T, Sato M, Yoshida T, Tuboi S. Rat liver mitochondrial and cytosolic fumarases with identical amino acid sequences are encoded from a single gene. J Biol Chem. 1989;264:2581–2586. [PubMed] [Google Scholar]
- 40.Taffe B G, Larminat F, Laval J, Croteau D L, Anson R M, Bohr V A. Gene-specific nuclear and mitochondrial repair of formamidopyrimidine DNA glycosylase-sensitive sites in Chinese hamster ovary cells. Mutat Res. 1996;364:183–192. doi: 10.1016/s0921-8777(96)00031-6. [DOI] [PubMed] [Google Scholar]
- 41.Thyagarajan B, Padua R A, Campbell C. Mammalian mitochondrial extracts have homologous recombination activity. J Biol Chem. 1996;271:27536–27543. doi: 10.1074/jbc.271.44.27536. [DOI] [PubMed] [Google Scholar]
- 42.Tomkinson A E, Levin D S. Mammalian DNA ligases. Bioessays. 1997;19:893–901. doi: 10.1002/bies.950191009. [DOI] [PubMed] [Google Scholar]
- 43.Tomkinson A E, Mackey Z B. Structure and function of mammalian DNA ligases. Mutat Res. 1998;407:1–9. doi: 10.1016/s0921-8777(97)00050-5. [DOI] [PubMed] [Google Scholar]
- 44.Tomkinson A E, Roberts E, Daly G, Totty N F, Lindahl T. Three distinct DNA ligases in mammalian cells. J Biol Chem. 1991;286:21728–24261. [PubMed] [Google Scholar]
- 45.Ueda M, Kawachi H, Atomi H, Tanaka A. Peroxisomal and mitochondrial carnitine acetyltransferase isozymes of the n-alkane-assimilating yeast, Candida tropicalis, occurred by alternative initiation of translation from the transcripts of a single gene. Biochim Biophys Acta. 1998;1397:213–22. doi: 10.1016/s0167-4781(98)00009-8. [DOI] [PubMed] [Google Scholar]
- 46.von Heijne G. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 1986;5:1335–1342. doi: 10.1002/j.1460-2075.1986.tb04364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Heijne G, Steppuhn J, Herrmann R G. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- 48.Wei Y-F, Robins P, Carter K, Caldecott K, Pappin D J C, Yu G-L, Wang R-P, Shell B K, Nash R A, Schar P, Barnes D E, Haseltine W A, Lindahl T. Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination. Mol Cell Biol. 1995;15:3206–3216. doi: 10.1128/mcb.15.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeviani M, Tiranti V, Piantadosi C. Mitochondrial disorders. Medicine. 1998;77:59–72. doi: 10.1097/00005792-199801000-00006. [DOI] [PubMed] [Google Scholar]