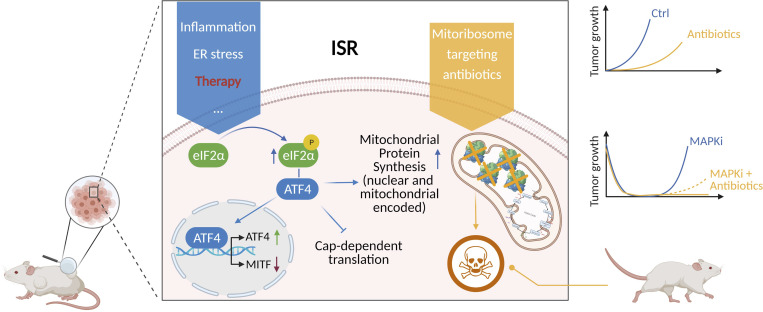
Vendramin et al. demonstrate that the integrated stress response in melanoma promotes cell plasticity and drives therapy resistance by boosting mitochondrial translation. Therefore, repurposing of mitoribosome-targeting antibiotics offers a salvage strategy for the treatment of patients with limited therapeutic options.
Abstract
The ability to adapt to environmental stress, including therapeutic insult, contributes to tumor evolution and drug resistance. In suboptimal conditions, the integrated stress response (ISR) promotes survival by dampening cytosolic translation. We show that ISR-dependent survival also relies on a concomitant up-regulation of mitochondrial protein synthesis, a vulnerability that can be exploited using mitoribosome-targeting antibiotics. Accordingly, such agents sensitized to MAPK inhibition, thus preventing the development of resistance in BRAFV600E melanoma models. Additionally, this treatment compromised the growth of melanomas that exhibited elevated ISR activity and resistance to both immunotherapy and targeted therapy. In keeping with this, pharmacological inactivation of ISR, or silencing of ATF4, rescued the antitumoral response to the tetracyclines. Moreover, a melanoma patient exposed to doxycycline experienced complete and long-lasting response of a treatment-resistant lesion. Our study indicates that the repurposing of mitoribosome-targeting antibiotics offers a rational salvage strategy for targeted therapy in BRAF mutant melanoma and a therapeutic option for NRAS-driven and immunotherapy-resistant tumors.
Graphical Abstract
Introduction
Overcoming resistance to cancer therapy remains a major clinical challenge. Despite recent breakthroughs in targeted therapy and immune checkpoint blockade (ICB; Sosman et al., 2012; Larkin et al., 2015), the clinical response to these therapies is often incomplete and/or transient. These findings indicate that combining various therapeutic agents and modalities will be needed to overcome treatment resistance (Holohan et al., 2013). Given that the development of new drugs is very slow (Pushpakom et al., 2019) and is affected by high attrition rates (Waring et al., 2015), the repurposing of existing de-risked compounds offers a fast track for the clinical implementation of novel and high-order drug combinations. However, a deeper understanding of the mechanisms underlying drug resistance is essential before novel combinations can be rationally designed.
The most commonly accepted explanation for the inexorable development of therapy resistance invokes specific genetic alterations that are acquired by chance before or during treatment (Holohan et al., 2013). However, recent findings indicate that a subset of cancer cells are capable of surviving therapeutic insult by engaging specific adaptation programs that confer them with drug-tolerant phenotypes (Sharma et al., 2010; Roesch et al., 2013; Ravindran Menon et al., 2015; Su et al., 2017; Shen et al., 2019). These drug-tolerant persister (DTP) cells provide a pool, commonly referred to as minimal residual disease (MRD), from which stable resistance is established. These findings indicate that eradicating MRD before stable resistance is acquired may offer new promising therapeutic avenues (Boumahdi and de Sauvage, 2020; Rambow et al., 2019). However, characterization of the cellular composition of MRD using single-cell approaches has recently highlighted that implementing such a strategy will come with its own challenges. Indeed, coemergence (within the same MRD lesion) of four very distinct drug-tolerant states was observed following exposure of proto-oncogene B-Raf (BRAF) mutant melanoma to MAPK-targeted therapy (Rambow et al., 2018). These included the starved melanoma cell (SMC) state sharing transcriptomic features of nutrient-deprived cells (Rambow et al., 2018), a neural crest stem-like cell (NCSC) state, an invasive or mesenchymal-like state that was recently renamed undifferentiated state (Tsoi et al., 2018; Rambow et al., 2018), and a hyperdifferentiated state. The two de-differentiated states, NCSC and undifferentiated/mesenchymal, which harbor cancer stem cell features, are considered particularly important drivers of tumor recurrence (Boshuizen et al., 2018; Rambow et al., 2018). Unfortunately, there are currently no clinically compatible approaches known to efficiently cotarget these two distinct cell populations. Whether these (and the other additional DTP subpopulations), despite harboring distinct gene expression signatures, exhibit common and actionable vulnerabilities has therefore become a key question.
Although the metabolic profile of cancer cells varies across patients, tumor types, and subclones within a tumor, there is emerging evidence that mitochondrial bioenergetics, biosynthesis, and signaling are required for tumorigenesis. Accordingly, several recent studies have demonstrated that mitochondrial biology is potentially a promising new area for cancer therapy (Weinberg and Chandel, 2015; Jagust et al., 2019). Critically, as our understanding of the biology of MRD across multiple tumor types increases, it is becoming clear that DTP cells exhibit a strong dependence on mitochondrial biology, even higher than that of their drug-naive counterparts (Jagust et al., 2019; Sharon et al., 2019; Davis et al., 2020). For instance, relapse-initiating cells in B-progenitor acute lymphoblastic leukemia are characterized by elevated levels of mitochondrial metabolism (Dobson et al., 2020). Similarly, a slow-cycling population of melanoma cells that emerge in cultures exposed to vemurafenib or cisplatin exhibits elevated oxidative phosphorylation, and targeting mitochondrial respiration blocks their emergence and delays drug resistance (Roesch et al., 2013). It remains unclear, however, whether metabolic reprogramming is the only underlying cause of this increased mitochondrial dependence and whether (all) DTP cells that coemerge following BRAF and MAPK kinase (MEK) coinhibition are equally sensitive to mitochondria-targeting agents.
DTP cells from multiple cancer types often exhibit elevated activation of the integrated stress response (ISR; Deng and Haynes, 2017) following drug exposure (Almanza et al., 2019; McConkey, 2017). As the activating transcription factor 4 (ATF4) is a key mediator of the ISR, its role in drug tolerance is increasingly recognized (Rzymski et al., 2009; Jewer et al., 2020; Ojha et al., 2019). ISR is an adaptive translation program, triggered by several intracellular and extracellular stressors, that converges on the phosphorylation and activation of the eukaryotic translation initiation factor 2 (eIF2) complex. Activation of this pathway eventually results in reduced global translation and entry into a quiescent state essential to survive the environmental stress (Min and Spencer, 2019). However, when the stress becomes overwhelming, the same pathway triggers apoptosis (Verheyden et al., 2019). Emerging evidence indicates that in mammals, the ISR is essential to convey mitochondrial disfunctions to the nucleus through a process known as retrograde signaling (Quirós et al., 2017). Conversely, the effect of ISR activation on mitochondrial activities is less clear. The observation that the rate of mitochondrial translation adapts to the influx of nuclear-encoded mitochondrial proteins (Richter-Dennerlein et al., 2016) raised the possibility that DTP cells may also reduce their mitochondrial translational rate. In contrast to this prediction, however, we show herein that activation of ISR in DTP cells promotes selective translation of a subset of mRNAs encoding for mitochondrial proteins, and thereby generates an anterograde signaling from the cytosol-boosting mitochondrial translation. This mechanism may explain the exquisite sensitivity of DTP cells to mitochondria-targeting agents such as uncouplers, and importantly identifies mitochondrial translation as one critical sensitive node.
Interestingly, specific antibiotics can be repurposed to inhibit mitochondrial protein synthesis. Like the bacteria from which they originate, the mitochondrial translational machinery of eukaryotic cells, in particular mitoribosomes, are inhibited by tetracyclines (Kalghatgi et al., 2013; Moullan et al., 2015). The antitumor efficacy of these antibiotics has been demonstrated in vitro (Ahler et al., 2013) and in vivo in hematologic malignancies (Škrtić et al., 2011; D’Andrea et al., 2016; Zhang et al., 2017; Ravà et al., 2018), with several clinical trials in acute myeloid leukemia and double-hit lymphomas currently ongoing (Reed et al., 2016). Critically, we show herein that targeting mitochondrial protein synthesis with antibiotics of the tetracycline family prevented emergence of most (three out of four) drug-tolerant subpopulations and delayed and even prevented the development of resistance to MAPK inhibition in BRAF mutant preclinical patient-derived xenograft (PDX) melanoma models (Patton et al., 2021) as well as in one melanoma patient. Finally, we also show efficacy of this treatment on models derived from patients with limited therapeutic options such as uveal melanomas (UMs), BRAF WT cutaneous melanomas, or melanomas with intrinsic or acquired resistance to targeted therapies and ICB.
Together, these findings indicate that targeting mitochondrial translation with antibiotics of the tetracycline family should be exploited to rationally design anticancer therapeutic regimens. Importantly, given the widespread clinical use of such agents, these approaches could be rapidly implemented into the clinic.
Results
Activation of the ISR increases mitochondrial translation
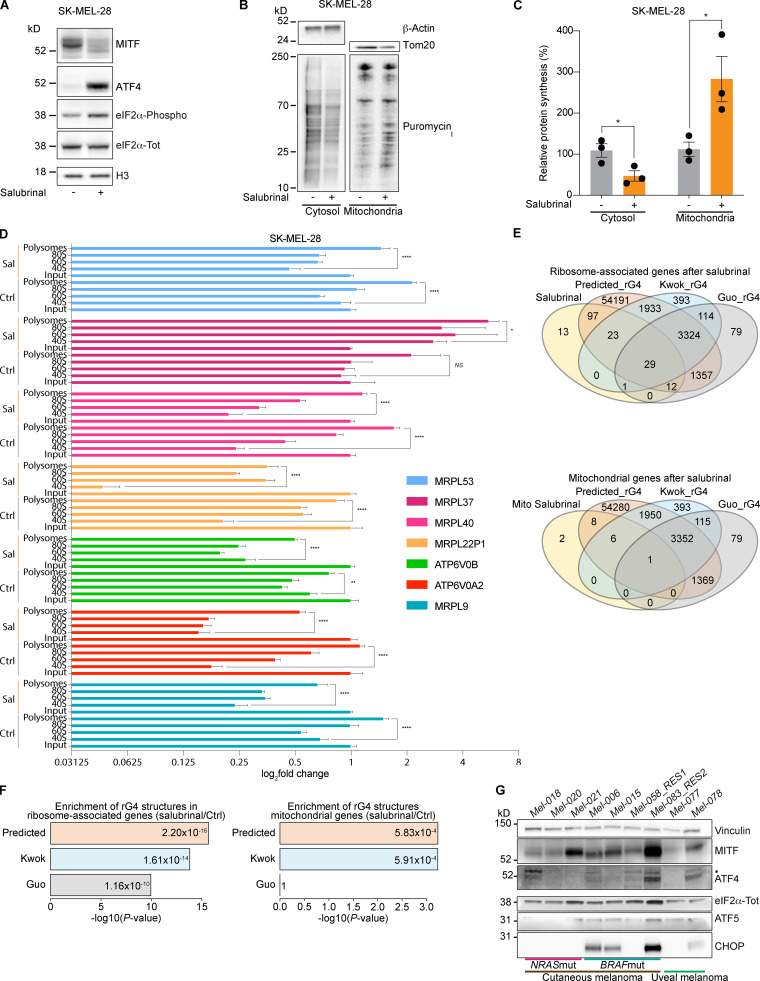
Phenotype switching into an undifferentiated drug-tolerant state can be induced in vitro by activating the ISR, leading to an ATF4-dependent down-regulation of the microphthalmia-associated transcription factor (MITF; Falletta et al., 2017). Accordingly, exposure of drug-naive melanoma cells to salubrinal, an ISR agonist (Boyce et al., 2005), increased levels of ATF4 and caused a concomitant down-regulation of MITF (Fig. 1 A).
Figure 1.
Activation of the ISR increases mitochondrial translation. (A) Western blotting of SK-MEL-28 cells 72 h after treatment with salubrinal (+, 20 µM) or DMSO (–). Representative images of three independent experiments. (B) Western blotting of cells described in A, after a 10-min pulse with puromycin (10 µM) and subsequent cytosol-mitochondria fractionation. Representative images of three independent experiments. (C) Quantification of protein synthesis (%), measured by calculating the intensity of the puromycin signal on western blot, in SK-MEL-28 cells as described in B. Data are mean ± SEM of three different biological replicates. *, P < 0.05 by Student’s t test. (D) RT-qPCR of cells described in A and Fig. S1 A for mitochondrial encoded genes. Ctrl, DMSO; Sal, salubrinal. Error bars represent mean ± SD of three independent experiments. NS, P > 0.05; *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by Dunnett’s test. (E) Intersection of total ribosome-associated mRNAs (top) or mitochondrial mRNAs (bottom) after treatment with salubrinal and genes containing RNA rG4 structures in their mRNA in the entire genome, as predicted by using the QGRS Mapper tool (Kikin et al., 2006) and the Kwok (Kwok et al., 2016) and Guo (Guo and Bartel, 2016) datasets of experimentally validated rG4s. (F) Enrichment in rG4 elements in the mitochondrial (right) or total (left) mRNA associated with ribosomes after treatment with salubrinal. The displayed enrichment was calculated by comparing the proportion of rG4s in these two sets of transcripts with the proportion of rG4s in the whole transcriptome as predicted by QGRSMapper (Predicted_rG4) or experimentally validated (Kwok_rG4 and Guo_rG4). P values were calculated by Fisher’s test (Benjamini–Hochberg corrected value). (G) Western blotting of a panel of drug-naive melanoma PDX models. Mut, mutant; RES1, resistant to BRAFi; RES2, resistant to BRAFi + MEKi and anti-PD-1 + anti-CTLA-4; •, ATF4 band.
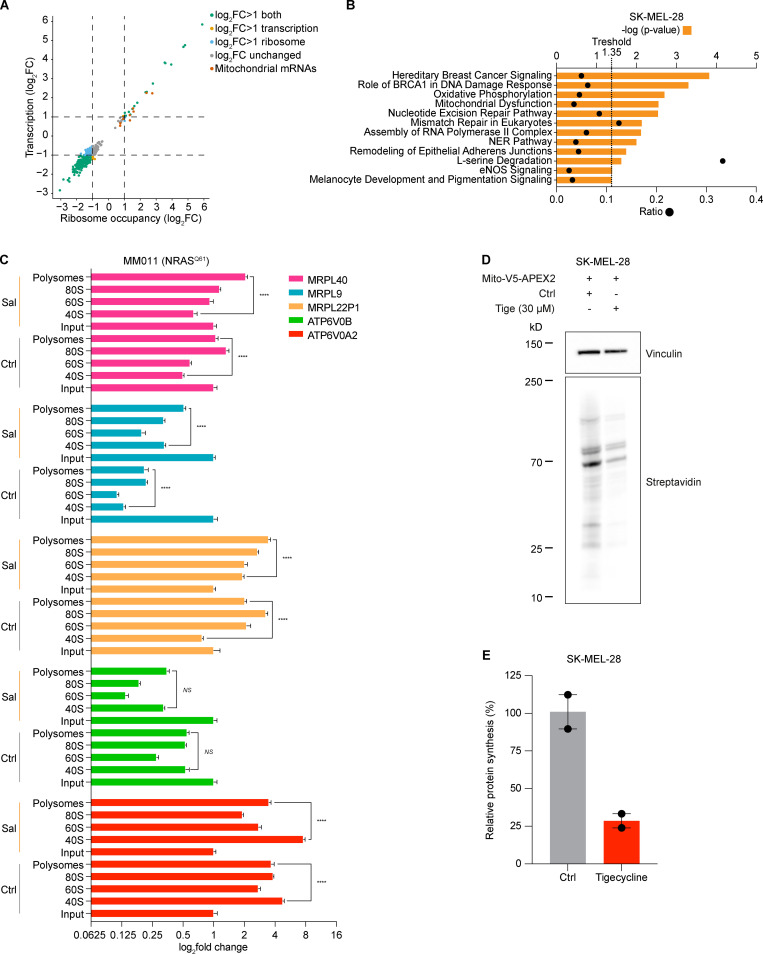
Considering that mitochondrial translation adapts to the influx of nuclear-encoded mitochondrial proteins, the observation that drug-tolerant cells down-regulate cytosolic protein synthesis suggests that these cells may also reduce the activity of their mitochondrial translation machinery. Surprisingly, however, puromycin incorporation assay followed by mitoplast isolation upon salubrinal treatment demonstrated that ISR activation caused instead a dramatic increase in mitochondrial translation, despite the expected overall decrease in cytosolic translation (Fig. 1, B and C). To further investigate the underlying mechanism, we identified translationally regulated mRNAs upon salubrinal treatment by performing polysome profiling analyses followed by RNA sequencing. We identified 382 transcripts whose association with ribosomes significantly (adjusted P value <0.05) changed in response to ISR activation and thus in response to phenotype switching and acquisition of therapy resistance. As expected, the vast majority of the transcripts (90%) were depleted from the ribosomal fractions upon ISR activation, while only 10% showed enrichment (Fig. S1 A). Among those, 2.3% were mitochondrial mRNAs and, accordingly, Ingenuity Pathway Analysis showed enrichment for mitochondria-related terms (Fig. S1 B). These findings were further validated in both BRAF mutant (Fig. 1 D) and neuroblastoma RAS (NRAS) mutant (Fig. S1 C) cell lines by performing polysome profiling followed by quantitative RT-PCR (RT-qPCR) upon salubrinal treatment. Since the influx of nuclear-encoded mitochondrial proteins from the cytosol directly regulates mitochondrial translation (Richter-Dennerlein et al., 2016), the latter observation provides a likely explanation for the observed increase in mitochondrial protein synthesis.
Figure S1.
Activation of ISR increases mitochondrial translation. (A) Ribosome occupancy (ribosome) and transcription of genome-wide polysome profile between salubrinal (20 µM, 72 h)-treated and control (DMSO) SK-MEL-28 cells. Color-coded genes have an adjusted P value <0.05 (DESeq) in the differential expression analysis of the ribosomal fraction. (B) Ingenuity Pathway Analysis of most differentially expressed genes in RNA obtained from polysome profiling of cells described in Fig. 1 A, with orange bars indicating −log(P value) and dots indicating ratios. (C) RT-qPCR of RNA obtained from polysome profiling of MM011 (NRASQ61) cells 72 h after treatment with salubrinal (Sal, 20 µM) or DMSO (Ctrl) for mitochondrial encoded genes. Error bars represent mean ± SD of three independent experiments. NS, P > 0.05; ****, P < 0.0001 by Student’s t test. (D) Western blotting of SK-MEL-28 cells 48 h after transient transfection with the mito-V5-APEX2 plasmid (constitutive expression) and 24 h after treatment with tigecycline (Tige) or PBS (Ctrl). Representative images of two independent experiments. (E) Quantification of protein synthesis (%), measured by calculating the intensity of the streptavidin signal on western blot, in SK-MEL-28 cells as described in D. Data are mean ± SEM of two independent experiments. NER, nucleotide excision repair; eNOS: endothelial NOS.
Strikingly, in silico analysis highlighted that 69.3% (122 of 176) of all ribosome-bound mRNAs in salubrinal-treated cells contain G-quadruplexes (rG4s; P = 2.4 × 1005 Benjamini–Hochberg corrected value; Fig. 1, E and F). In keeping with this, validated and/or predicted rG4s were identified in 88% (15 of 17) of the ribosome-associated mRNAs encoding mitochondrial proteins. rG4s, which are noncanonical four-stranded structures formed by guanine-rich sequences, have been implicated in all steps of RNA metabolism (Dumas et al., 2021). The above observations suggest that rG4s may contribute to the specific recruitment of selected transcripts to the translation machinery in cells undergoing ISR (Fig. 1, E and F).
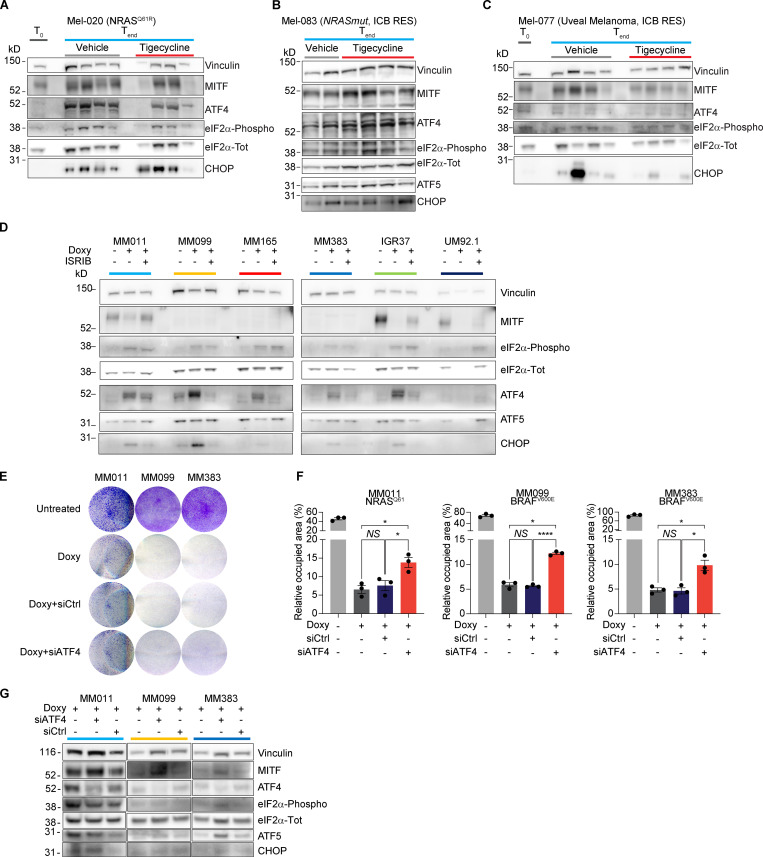
Overall, these data identify mitochondrial translation as a putative Achilles’ heel of cells engaging the ISR pathway, such as drug-tolerant cells. Of note, ISR signaling is activated not only in response to drug treatment: western blot analysis detected activation of the ISR, as demonstrated by the detection of P-eIF2α, ATF4, and C/EBP homologous protein (CHOP), in non–drug-exposed human melanoma lesions, including lesions that were resistant to both targeted and immune therapies (Fig. 1 G). These data therefore indicate that targeting mitochondrial translation may offer a promising therapeutic approach in a substantial number of patients/cases.
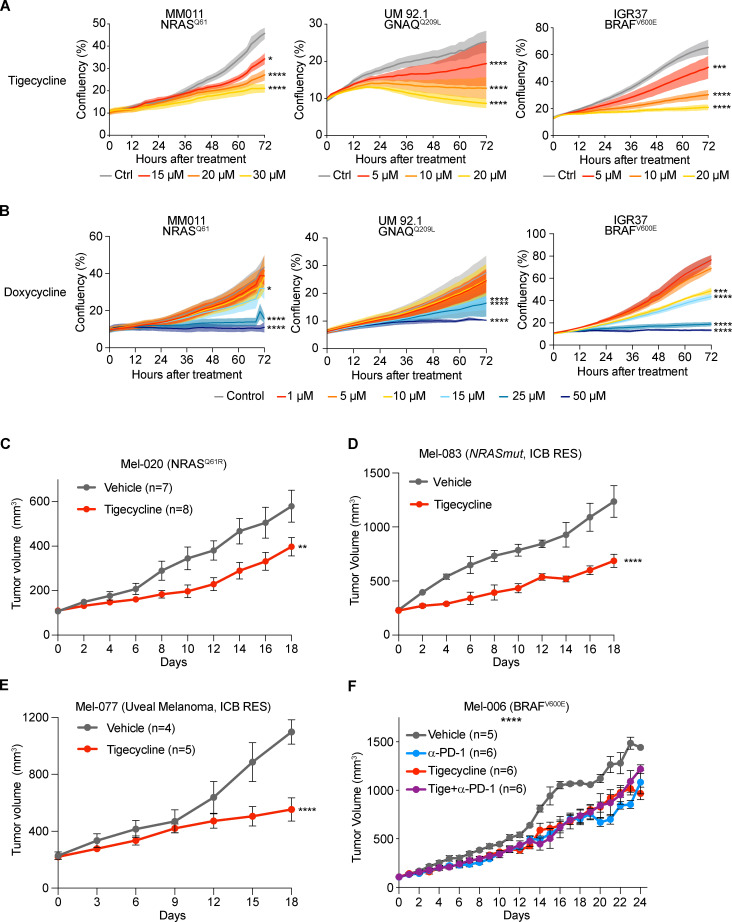
Tigecycline overcomes acquired resistance to MAPK inhibitors in vivo and significantly increases overall survival (OS) and progression-free survival (PFS)
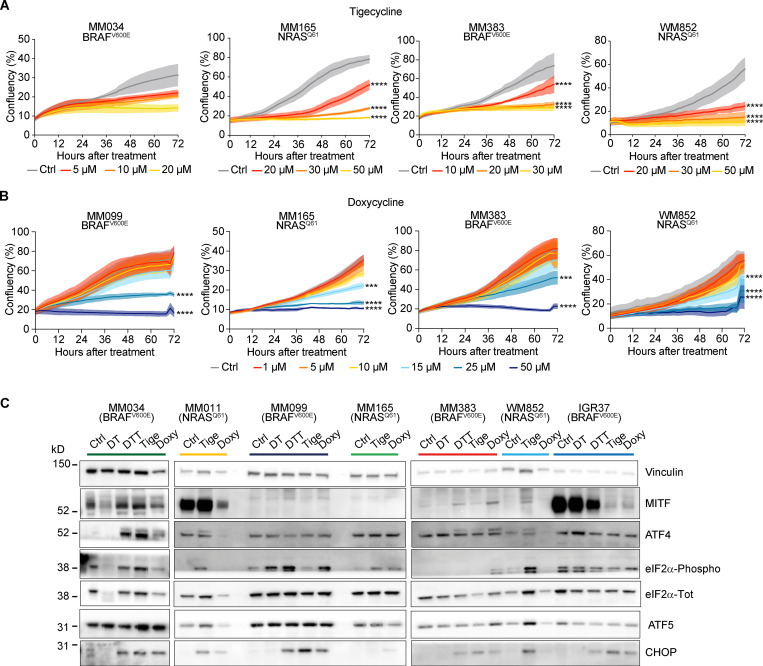
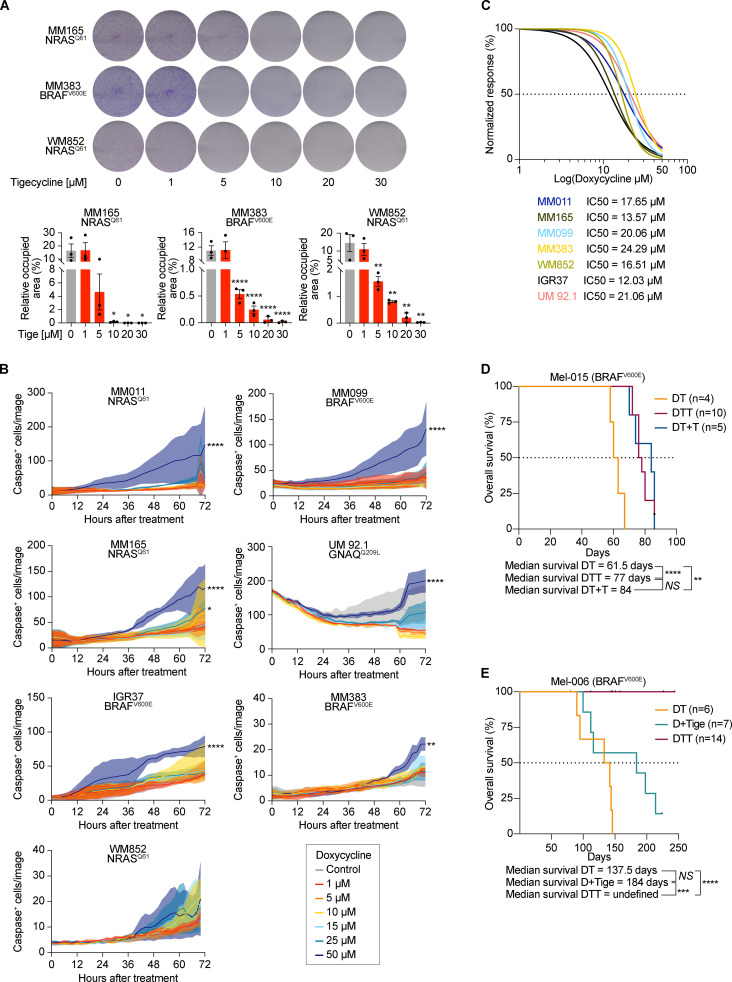
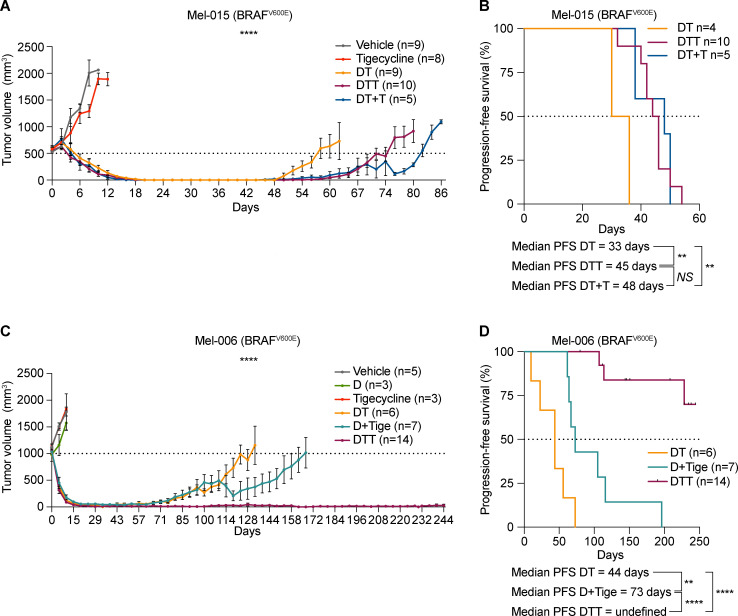
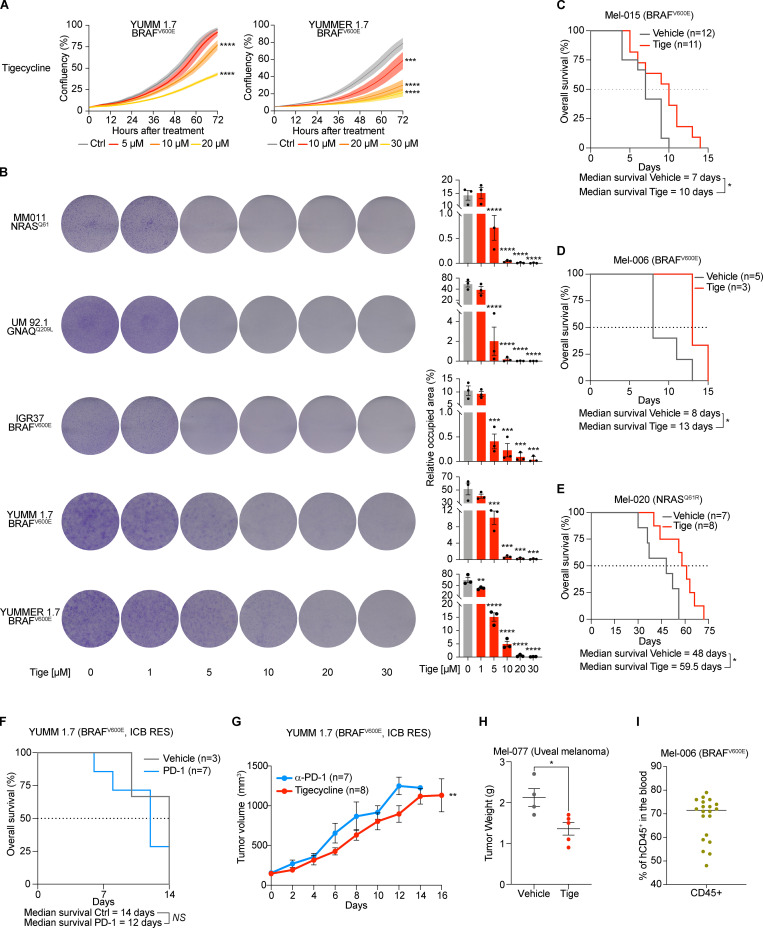
We reasoned that the increased recruitment of transcripts encoding mitochondrial proteins in cells harboring an activated ISR may sensitize them to antibiotics that target mitochondrial translation. To test this hypothesis, we exposed several human melanoma cell lines (Table 1) that harbor transcriptional profiles reminiscent of two critical drug-tolerant states, namely invasive/undifferentiated and NCSCs, to increasing concentrations of both tigecycline and doxycycline. Both antibiotics promoted a significant and dose-dependent decrease in cell growth in all cell lines tested (Fig. 2, A and B; and Fig. S2, A–C). Importantly, western blot analysis for ISR regulators confirmed that the ISR pathway is activated in these cell lines before treatment. Moreover, exposure to the antibiotics alone or in combination with MAPK inhibitors (MAPKi) further exacerbated proteotoxic stress, as illustrated by an increase in CHOP activation (Fig. 2 C). Conversely, treatment with antibiotics of MM034, which do not display expression of ATF4 (Fig. 2 C), did induce a certain degree of growth inhibition; however, the decrease did not reach statistical significance (Fig. 2 A). These observations indicated that tetracyclines may offer a therapeutic option for ablation of the residual tumor cells emerging following exposure to MAPKi. We therefore tested whether addition of tigecycline to dabrafenib-trametinib (DT), a standard-of-care treatment for patients with BRAF mutant melanoma, delayed or prevented the onset of resistance in two different BRAFV600E melanoma PDXs (Mel-006 and Mel-015). As expected, all mice from both cohorts initially responded to DT but eventually developed resistance (Fig. 3 and Fig. S2, D and E). In the Mel-015 cohort, the addition of antibiotics, whether added from the start of the treatment (dabrafenib-trametinib-tigecycline [DTT]) or after lesions reached MRD (DT + T [tigecycline]), significant delayed the development of resistance (Fig. 3 A) and increased PFS and OS (Figs. 3 B and S2 D). Strikingly, in the Mel-006 cohort, DTT treatment resulted in complete remission in 11 of 14 mice (Fig. 3, C and D; and Fig. S2 E). Importantly, the curative nature of this treatment was confirmed by interrupting the DTT treatment in another cohort of mice after tumors had regressed (Fig. 3, C and D; and Fig. S2 E). Notably, dabrafenib plus tigecycline (D + Tige) was as effective as DT treatment at inducing tumor regression in this particular PDX model. This treatment even significantly increased PFS and OS compared with DT (Fig. 3, C and D; and Fig. S3 B). Thus, addition of tigecycline successfully delays and/or prevents acquisition of resistance to targeted therapy in BRAF mutant skin melanoma preclinical models.
Table 1. Cell lines used in the study.
| Cell line | Origin | Mutation | Characteristics |
|---|---|---|---|
| SK-MEL-28 | Human | BRAFV600E; TP53 | Proliferative |
| MM011 | Human | NRASQ61 | Proliferative |
| MM034 | Human | BRAFV600E | Proliferative |
| MM099 | Human | BRAFV600E | Invasive |
| MM165 | Human | NRASQ61 | Invasive |
| MM383 | Human | BRAFV600E | NCSC |
| WM852 | Human | NRASQ61 | NCSC |
| IGR37 | Human | BRAFV600E | Immunotherapy resistant |
| YUMM 1.7 | Mouse | BRAFV600E; PTEN−/−; CDKN2−/− | Immunotherapy resistant |
| YUMMER 1.7 | Mouse | BRAFV600E; PTEN−/−; CDKN2−/− | Immunotherapy sensitive |
| UM 92.1 | Human | GNAQQ209L | Uveal melanoma |
Mutational and phenotypic status of cell lines used.
Figure 2.
Tetracyclines exacerbate the activation of the ISR and affect the viability of multiple drug–tolerant states. (A) Cell growth (measured as percentage of cell confluency) of MM034 (BRAF mutant, proliferative), MM165 (NRAS mutant, invasive), MM383 (BRAF mutant, NCSC-like), and WM852 (NRAS mutant, NCSC-like) cell lines upon exposure to increasing concentrations of tigecycline for 72 h. Data are mean ± SEM of three independent experiments. ****, P < 0.0001 by Dunnett’s test. (B) Cell growth (measured as percentage of cell confluency) of MM099 (BRAF-mutant, invasive), MM165 (NRAS mutant, invasive), MM383 (BRAF-mutant, NCSC-like), and WM852 (NRAS mutant, NCSC-like) cell lines upon exposure to increasing concentrations of doxycycline for 72 h. Data are mean ± SEM of three independent experiments. ***, P < 0.001; ****, P < 0.0001 by Dunnett’s test. (C) Western blotting of a panel of different melanoma cell lines treated with DMSO (Ctrl), DT (20 and 4 nM, respectively), DTT (20 nM, 4 nM, and 20 µM respectively), tigecycline (Tige, 20 µM), or doxycycline (Doxy, 20 µM) for 72 h. Representative image of three independent experiments.
Figure S2.
Tetracyclines affect the growth of multiple drug–tolerant states. (A) Top: Colony formation assays with cells described in Fig. 3 A. The violet color is due to crystal violet, a compound that binds intracellular DNA and protein, thus highlighting the cells attached to the plate. Representative image of three independent experiments. Bottom: Quantification of colony formation assays of cells described in top panel presented as the mean density (percentage of area occupancy) ± SEM of three independent experiments. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by Dunnett’s test. (B) Caspase activity (measured as average number of caspase+ cells per image) of cells described in Figs. 2 B and 5 B. Data are presented as mean ± SEM of three independent experiments. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by Dunnett’s test. (C) Growth inhibition curves and half-maximal inhibitory concentration (IC50) values of several cutaneous melanoma cell lines and one UM (92.1) cell line upon exposure to increasing concentrations of doxycycline. Data are mean ± SEM of three independent experiments. (D) Kaplan–Meier plot showing OS of mice described in Fig. 3 A. DT (n = 4), DTT (n = 10), dabrafenib + trametinib with the addition of tigecycline at MRD (DT + T, n = 5). NS, P > 0.05; **, P < 0.01; ****, P < 0.0001 by log-rank (Mantel–Cox) test. (E) Kaplan–Meier plot showing OS of mice described in Fig. 3 C. DT (n = 6), D + Tige, n = 7), DTT (n = 14). NS, P > 0.05; ***, P < 0.001; ****, P < 0.0001 by log-rank (Mantel–Cox) test.
Figure 3.
Tigecycline overcomes acquired resistance to MAPK inhibitors in vivo and significantly increases survival. (A) Tumor volume of cohorts of Mel-015 BRAFV600 PDX mice treated with vehicle (DMSO, n = 9), tigecycline (n = 8), DT (n = 9), DTT (n = 10), or DT with the addition of tigecycline at MRD (DT + T, n = 5). Data are mean ± SEM of different biological replicates. ****, P < 0.0001 by two-way ANOVA. One of the mice in the DT + T cohort had to be euthanized on day 76, thereby explaining the rapid drop in the growth curve. (B) Kaplan–Meier plot showing PFS of mice described in A. DT (n = 4), DTT (n = 10), DT with the addition of tigecycline at MRD (DT + T, n = 5). NS, P > 0.05; **, P < 0.01 by log-rank (Mantel–Cox) test. (C) Tumor volume of cohorts of Mel-006 BRAFV600E PDX mice treated with vehicle (DMSO, n = 5), dabrafenib (D, n = 3), tigecycline, DT (n = 6), dabrafenib + tigecycline (D + Tige, n = 7), or DTT (n = 14). Data are mean ± SEM of different biological replicates. ****, P < 0.0001 by two-way ANOVA. Two mice in the D + T cohort had to be euthanized between day 110 and 120, thereby explaining the rapid drop in the growth curve. (D) Kaplan–Meier plot showing PFS of mice described in C. DT (n = 6), dabrafenib + tigecycline (D + Tige, n = 7), DTT (n = 14). **, P < 0.01; ****, P < 0.0001 by log-rank (Mantel–Cox) test.
Figure S3.
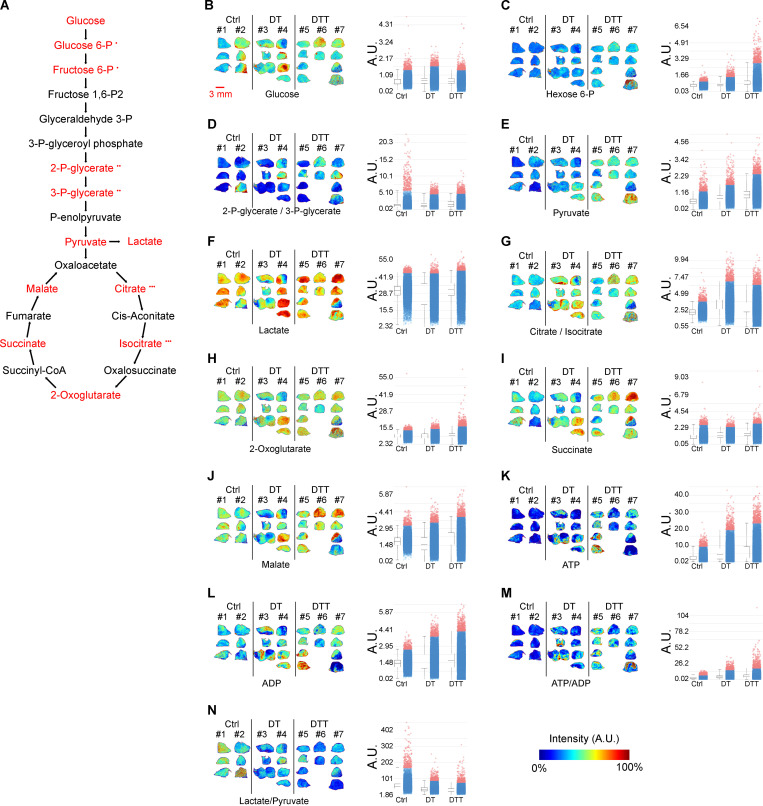
Tigecycline overcomes acquired resistance to MAPK inhibitors in vivo in the absence of significant metabolic rewiring. (A) Molecules participating in glycolysis, Krebs cycle, and energy production were measured in negative mode using a MALDI-TOF instrument, acquired at 50-µm resolution, and assigned according to matched theoretical value of measured ion adduct with mass accuracy 30 ppm. Matched compounds are highlighted in red. •, the same theoretical value for different compounds which cannot be distinguished. (B–N) MALDI-TOF performed on Mel006 BRAFV600E PDX tumors treated with vehicle (Ctrl, n = 2), DT (n = 2), and DTT (n = 3). Images (left) are displayed in color scheme with scaled intensities normalized by root mean square method to ADP intensities (left) and to pyruvate intensity (middle), respectively, with quantification (arbitrary units [A.U.], right). (B) Glucose [M + Cl]−; m/z 215; scale 0–305%. (C) Glucose 6-phosphate [M-H]− and fructose 6-phosphate [M-H]−; m/z 259; scale 0–703%. (D) 2-Phospho-glycerate [M-H]− and 3-phospho-glycerate [M-H]−; m/z 184; scale 0–590%. (E) Pyruvate [M-H]−; m/z 87; scale 0–218%. (F) Lactate [M-H]−; m/z 89; scale 0–134%. (G) Citrate [M-H]− and isocitrate [M-H]−; m/z 191; scale 0–204%. (H) 2-Oxoglutarate [M-H2O-H]−; m/z 153; scale 0–446%. (I) Succinate [M-H]−, m/z 117; scale 0–357%. (J) Malate [M-H]−, m/z 133; scale 0–175%. (K) ATP [M-H]−; m/z 506; scale 0–212%. (L) ADP [M-H]−; m/z 426; scale 0–164%. (M) ATP to ADP ratio; scale 0–914%. (N) Lactate to pyruvate ratio; scale 0–377%. Scale bar (displayed in B) = 3 mm.
Tigecycline suppresses known drug-resistant cell populations
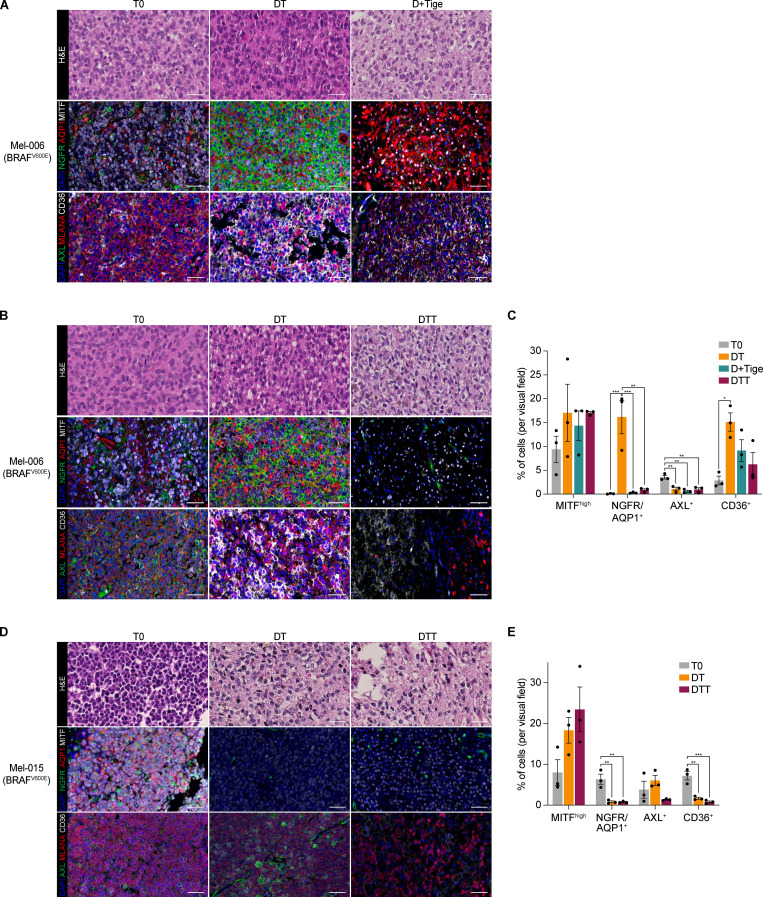
We have now provided evidence that tetracyclines affect the growth of both mesenchymal and NCSC de-differentiated drug-tolerant cells in vitro. To assess the sensitivity of these and other drug-tolerant subpopulations in vivo, we treated Mel-006 with either D + Tige or DT until reaching MRD. Compared with DT treatment, the D + Tige combination successfully eradicated most of the NCSCs (NGFR+AQP1+), undifferentiated/mesenchymal-like cells (AXL+), and SMCs (CD36+) drug-tolerant cells (Fig. 4, A–C). Similarly, in two different BRAF mutant PDX cohorts (Mel-006 and Mel-015) treated with the triple DTT combination, antibiotic treatment led to the eradication of NCSCs (NGFR+AQP1+), undifferentiated/mesenchymal-like cells (AXL+) and pseudo-starved cell population SMCs (CD36+). The hyperpigmented cell population (MITF+MLANA+) was the only drug-tolerant population to resist tigecycline combinatorial treatments (Fig. 4, B–E). Note that, consistent with tigecycline targeting mitochondrial translation and not metabolism, spatial metabolomics indicated that the antibiotic-containing treatment (DTT) did not induce any significant metabolic shift compared with DT alone (Fig. S3, A–N). These data indicate that a combination of antibiotics and MAPKis affects known drug-resistant populations by targeting mitochondrial translation in both de-differentiated (NCSCs) and undifferentiated/invasive states in the absence of significant metabolic rewiring.
Figure 4.
Tigecycline efficiently suppresses known drug-resistant cell populations. (A) Top: Representative H&E staining of Mel-006 BRAFV600E PDX tumors before treatment (T0, left), upon treatment with MAPKis at MRD (DT, middle), and upon treatment with dabrafenib and tigecycline (D + Tige, right) at MRD. Middle: Representative immunofluorescence staining against MITF (white), NGFR (green), and AQP1 (red) before treatment (T0) and at MRD of DT and D-Tige lesions. Bottom: Representative immunofluorescence staining against CD36 (white), AXL (green), and Melan-A (MLANA; red) before treatment (T0) and at MRD of DT and D-Tige lesions. Slides were counterstained with DAPI (blue). Scale bar = 50 µm. (B) Top: Representative H&E staining of Mel-006 BRAFV600E PDX tumors before treatment (T0, left), upon treatment at MRD with MAPKis (DT, middle), and upon treatment with DTT (right). Middle: Representative immunofluorescence staining against MITF (white), NGFR (green), and AQP1 (red) before treatment (T0) and at MRD of DT and DTT lesions. Bottom: Representative immunofluorescence staining against CD36 (white), AXL (green), and MLANA (red) before treatment (T0) and at MRD of DT and DTT lesions. Slides were counterstained with DAPI (blue). Scale bar = 50 µm. (C) Quantification of the immunostainings described in A and B presented as the mean percentage of positive cells per visual field per each antibody ± SEM of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA with Tukey’s multiple comparisons test. (D) Top: Representative H&E staining of Mel-015 BRAFV600E PDX tumors before treatment (T0, left), upon treatment at MRD with MAPK inhibitors (DT, middle), and upon treatment with DTT (right). Middle: Representative immunofluorescence staining against MITF (white), NGFR (green), and AQP1 (red) before treatment (T0) and at MRD of DT and DTT lesions. Bottom: Representative immunofluorescence staining against CD36 (white), AXL (green), and MLANA (red) before treatment (T0) and at MRD of DT and DTT lesions. Slides were counterstained with DAPI (blue). Scale bar = 50 µm. (E) Quantification of the immunostainings described in D presented as the mean percentage of positive cells per visual field per each antibody ± SEM of three independent experiments. **, P < 0.01; ***, P < 0.001 by one-way ANOVA with Tukey’s multiple comparisons test.
Tetracyclines restrain the growth of therapy-resistant melanoma
Data in Fig. 2 C indicate that cells intrinsically resistant to MAPK inhibition (see MM011) and/or immunotherapy (see IGR37) may also be sensitive to mitochondrial translation inhibition. Therefore, we exposed BRAF WT melanoma cell lines, including a UM line and the immunotherapy-resistant IGR37 and YUMM 1.7 cell lines (together with the sensitive counterpart YUMMER 1.7; see Table 1) to increasing concentrations of tigecycline or doxycycline (Fig. 5, A and B; and Fig. S4, A and B). Both antibiotics promoted a significant and dose-dependent decrease in cell growth in all cell lines. We next tested the efficacy of these antibiotics on the in vivo growth of several therapy-resistant PDX models (Fig. 5, C–E). Daily treatment with tigecycline significantly delayed the growth of NRAS mutant PDXs (Mel-020 and Mel-083), including one model derived from a patient that had progressed on immunotherapy (Mel-083; Fig. 5, C and D; and Fig. S4 E). Milder, yet significant, results were also obtained in the drug-naive BRAFV600E mutant cutaneous melanoma models (Mel-006 and Mel-015), which exhibit sensitivity to MAPKi (Fig. S4, C and D). Likewise, C57BL/6 mice engrafted with the immunotherapy-resistant YUMM 1.7 cells (Fig. S4 F) displayed sensitivity to daily monotreatment of tigecycline (Fig. S4 G). Furthermore, tigecycline treatment of the UM PDX model Mel-077 and NRASQ61R mutant cutaneous melanoma Mel-083, both derived from patients that had progressed on immune checkpoint inhibitors (ICI, pembrolizumab + temozolomide for Mel-077 and ipilimumab + nivolumab for Mel-083), was also sufficient to significantly delay tumor growth (Fig. 5, D and E; and Fig. S4 H). Lastly, to investigate whether the antibiotic treatment negatively impact the response to ICIs, the Mel-006 model was engrafted in hu-NOG-EXL mice humanized with CD34+ cells (Fig. S5 G). Mice were treated with anti–PD-1 alone or in combination with tigecycline (50 mg/kg) once tumors reached 100 mm3 (Fig. 5 F). Treatment with tigecycline did not overtly ameliorate, nor impair, the response to anti–PD-1 (Fig. 5 F). Similar results were obtained with YUMMER allografts (data not shown). Together, these data indicate that although tetracycline does not sensitize to ICI, monotreatment with this antibiotic is sufficient to significantly delay progression of melanoma lesions that are insensitive to targeted and immune therapies in vivo.
Figure 5.
Tetracyclines restrain the growth of therapy-resistant melanomas. (A) Cell growth (measured as a percentage of cell confluency) of MM011 (NRAS mutant), UM 92.1 (GNAQ mutant, UM), and IGR37 (resistant to immunotherapy) cell lines upon exposure to increasing concentrations of tigecycline for 72 h. Data are mean ± SEM of three independent experiments. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 by Dunnett’s test. (B) Cell growth (measured as percentage of cell confluency) of cells described in A upon exposure to increasing concentrations of doxycycline for 72 h. Data are mean ± SEM of three independent experiments. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 by Dunnett’s test. (C) Tumor volume of cohorts of Mel-020 NRASQ61R PDX mice treated with vehicle (DMSO, n = 7) or tigecycline (n = 8). Data are mean ± SEM of different biological replicates. **, P < 0.01 by two-way ANOVA. (D) Tumor volume of cohorts of Mel-083 (resistant to BRAFi + MEKi and anti–PD-1 + anti-CTLA-4) PDX mice treated with vehicle (DMSO, n = 2) or tigecycline (n = 4). Data are mean ± SEM of different biological replicates. ****, P < 0.0001 by two-way ANOVA. (E) Tumor volume of cohorts of Mel-077 UM PDX mice treated with vehicle (DMSO, n = 4) or tigecycline (n = 5). Data are mean ± SEM of different biological replicates. ****, P < 0.0001 by two-way ANOVA. (F) Tumor volume of cohorts of YUMMER 1.7 (BRAFV600E) mouse xenografts treated with α-PD-1 (n = 6), tigecycline (n = 6), a combination of the two (n = 6), or vehicle (n = 5). Data are mean ± SEM of different biological replicates. ****, P < 0.0001 by two-way ANOVA with Geisser–Greenhouse correction.
Figure S4.
Tigecycline restrains the growth of BRAF WT and ICB-resistant melanomas and increases OS. (A) Cell growth (measured as percentage of cell confluency) of YUMM 1.7 (resistant to immunotherapy) and YUMMER 1.7 (immunotherapy-sensitive) cell lines upon exposure to increasing concentrations of tigecycline for 72 h. Data are mean ± SEM of three independent experiments. ***, P < 0.001; ****, P < 0.0001 by Dunnett’s test. (B) Left: Colony formation assays with cells described in Fig. 2 A and A treated with increasing concentrations of tigecycline. The violet color is due to crystal violet, a compound that binds intracellular DNA and protein, thus highlighting the cells attached to the plate. Representative image of three independent experiments. Right: Quantification of colony formation assays of cells described in Fig. 2 A and A presented as the mean density (percentage of area occupancy) ± SEM of three independent experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Dunnett’s test. (C) Kaplan–Meier plot showing OS of Mel-015 BRAFV600E PDX mice treated with vehicle (DMSO, n = 12) or tigecycline (n = 11). *, P < 0.05 by log-rank (Mantel–Cox) test. (D) Kaplan–Meier plot showing OS of Mel-006 BRAFV600E PDX mice treated with vehicle (DMSO, n = 5) or tigecycline (n = 3). *, P < 0.05 by log-rank (Mantel–Cox) test. (E) Kaplan–Meier plot showing OS of mice described in Fig. 5 C. Vehicle (DMSO, n = 7) or tigecycline (n = 8). *, P < 0.05 by log-rank (Mantel–Cox) test. (F) Kaplan–Meier plot showing OS of YUMM 1.7 mouse xenografts treated with α-PD-1 (n = 7) or vehicle (n = 3). NS, P > 0.5 by log-rank (Mantel–Cox) test. ICB RES, ICB resistant. (G) Tumor volume of cohorts of YUMM 1.7 (BRAFV600E) mouse xenografts treated with α-PD-1 (n = 7) or tigecycline (n = 8). Data are mean ± SEM of different biological replicates. **, P < 0.01 by two-way ANOVA. (H) Tumor weight of Mel-077 cohorts of mice described in Fig. 5 E. Vehicle (DMSO, n = 4), tigecycline (n = 5). *, P < 0.05 by Student’s t test. (I) Percentage of human CD45 positive (hCD45+) in the blood of Mel-006 mice described in Fig. 5 F. n = 19.
Figure S5.
Activation of the ISR predicts durable responses to antibiotic treatment. (A) Western blotting of Mel-020 (NRASQ61R) PDX tumors described in Fig. 5 C collected right before the start of the treatment (T0) or at the end of the experiment (Tend) after receiving a daily dose of vehicle or tigecycline. (B) Western blotting of Mel-083 (NRAS mutant, ICB resistant) PDX tumors described in Fig. 5 D. (C) Western blotting of Mel-077 (UM) PDX tumors described in Fig. 5 E collected right before the start of the treatment (T0) or at the end of the experiment (Tend) after receiving a daily dose of vehicle or tigecycline. (D) Western blotting of cells described in Fig. 6 C. Representative image of three independent experiments. (E) Colony formation assays of MM011 (NRASQ61), MM099 (BRAFV600E), and MM383 (BRAFV600E) cells treated with doxycycline (Doxy, 37.5 µM) and transfected with either a control siRNA (siCtrl, 20 nM) or an siRNA targeting ATF4 (siATF4, 20 nM) or untreated for 72 h. The violet color is due to crystal violet, a compound that binds intracellular DNA and protein, thus highlighting the cells attached to the plate. Representative image of three independent experiments. (F) Quantification of colony formation assays of cells described in E, presented as the mean density (percentage of area occupancy) ± SEM of three independent experiments. NS, P > 0.05; *, P < 0.05; ****, P < 0.0001 by Dunnett’s test. (G) Western blotting of cells described in E. Representative image of three independent experiments.
Doxycycline sensitizes refractory lesions to MAPKi in preclinical settings and in one patient
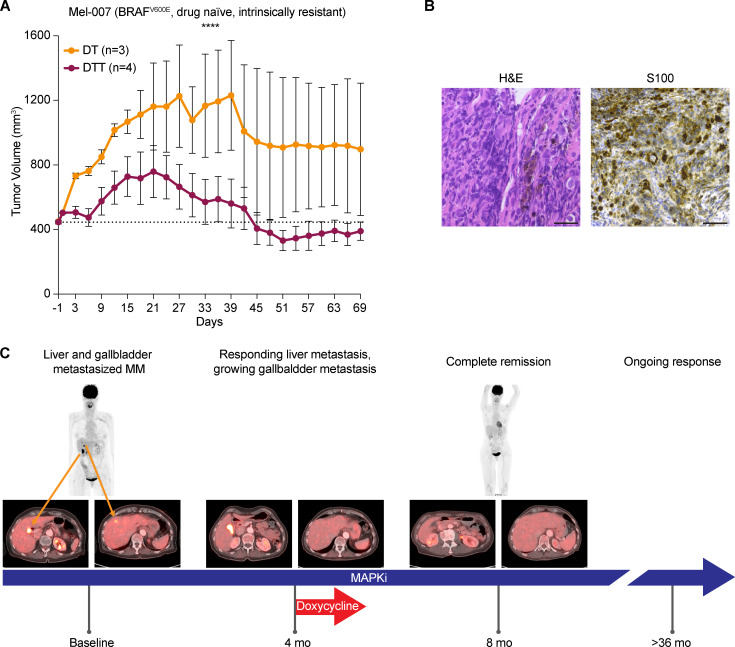
We also assessed whether tetracyclines can overcome intrinsic resistance to targeted therapy. To this end, we derived a cutaneous melanoma PDX model (Mel-007) from a lesion that exhibited intrinsic resistance to a combination of BRAF-MEK inhibitors. Exposure of a cohort of these mice to DT versus DTT showed that the addition of tigecycline promoted a more robust antitumor effect and even promoted tumor regression, in contrast to DT treatment (Fig. 6 A).
Figure 6.
Doxycycline sensitizes to MAPKi in preclinical models and in one melanoma patient. (A) Tumor volume of cohorts of Mel-007 BRAFV600 PDX mice (resistant to MAPKi) treated with DT (n = 3) or DTT (n = 4). Data are mean ± SEM of different biological replicates. ****, P < 0.0001 by two-way ANOVA. (B) Histology and immunohistochemistry of the bile duct biopsy confirming the presence of melanoma metastasis in one melanoma patient. Scale bar = 25 µm. (C) Case presentation of a patient with MAPK inhibitory therapy and sporadic exposure to doxycycline. Baseline PET-CT revealed liver (lower left, right-hand image) and gallbladder metastasis (lower left, left-hand image). First response assessment showed disappearance of the liver metastasis but a persistent gallbladder lesion (4 mo of treatment). Adding doxycycline to MAPK inhibitory therapy showed shrinkage after two more cycles of treatment (CT scan not depicted), which was subsequently confirmed by PET-CT after 8 mo on treatment, revealing a complete metabolic response. After >36 mo, the patient is still responding to the treatment.
Doxycycline is a broad-spectrum antibiotic that also belongs to the tetracycline family. It exerts its antibacterial action by binding to the 30S ribosomal subunit and thereby blocking ribosome biogenesis. It can be administered orally for extended periods of time with only minor adverse effects in patients (Tan et al., 2011).
To assess the relevance of our in vitro and preclinical findings, we followed the clinical course of one 73-yr-old female patient at our clinic. The patient was diagnosed with stage III melanoma in 2011 and with a relapse of her disease in 2016 after an episode of jaundice, which was related to a tumor mass in her gallbladder and bile duct. Histology and immunohistochemistry of the bile duct biopsy were performed (Fig. 6 B). H&E (left) shows a highly anaplastic tumor with large epithelioid cells and pleomorphic nuclei; the presence of brown pigment associated with these cells was highly suggestive for a melanoma metastasis. The immunostaining for S100 (right), one of the most sensitive markers to identify melanoma, was diffusely positive at nuclear and cytoplasmic levels, therefore confirming the diagnosis of melanoma metastasis. Baseline positron-emission tomography/computed tomography (PET-CT) showed metastatic tumor at the aforementioned locations as well as in the liver and in the gallbladder (Fig. 6 C). Subsequently BRAF-MEK inhibitor therapy was initiated. Response assessment after four cycles showed that while the liver metastasis had responded to the treatment, the gallbladder did not. Due to a skin infection (an acne-like rash) the patient received doxycycline during her fifth BRAF-MEK inhibitor cycle for a total of 12 d. Strikingly, the gallbladder metastasis started to regress after six cycles, as measured by CT scan, and the PET-CT scan after eight cycles showed a complete response. The patient was under BRAF-MEK inhibitor therapy and had a persistent response for >36 mo (Fig. 6 C). This particular case study highlights the potential clinical benefit of using tetracyclines to sensitize intrinsically resistant lesions to MAPK inhibitors.
Activation of the ISR predicts durable responses to an antibiotic treatment
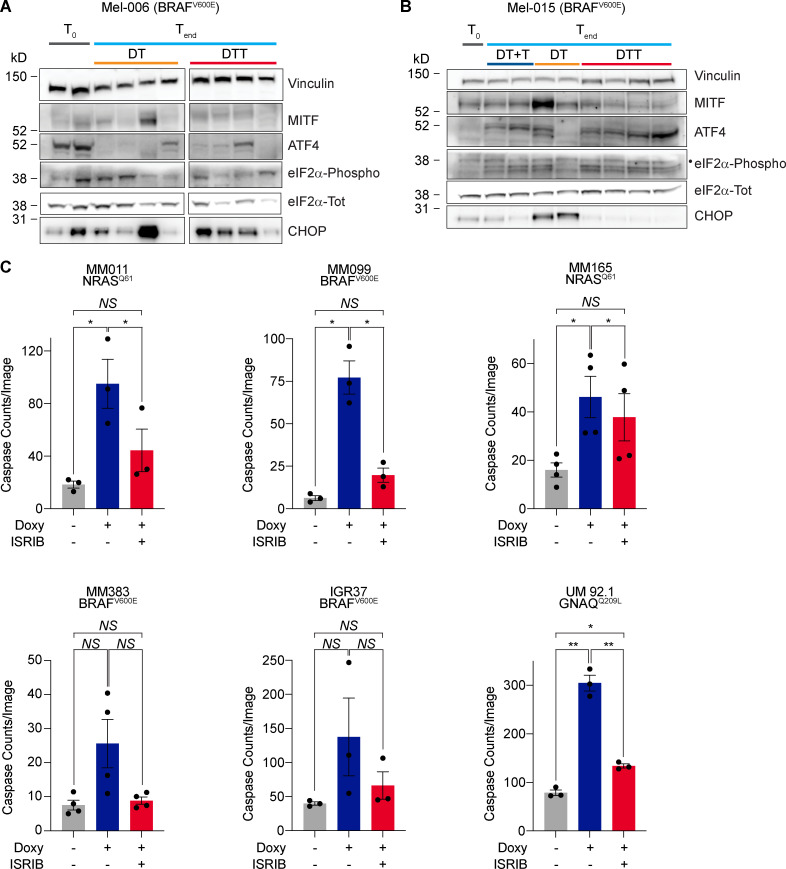
We postulated that the differential efficacy of the antibiotic combinatorial treatment in preventing the development of resistance between Mel-006 and Mel-015 BRAFV600E mutant melanoma PDXs may result from a difference in the kinetic and/or extent at which the ISR pathway is activated before applying the therapy. Consistently, ATF4 activation was readily detectable by western blotting, before the start of the therapy (T0), in the Mel-006 PDX model, an observation that may explain the high sensitivity to the antibiotic treatment and the long-lasting response (Figs. 1 G and 7 A). In contrast, ATF4 activation could only be detected in lesions from Mel-015 following therapy exposure (Figs. 1 G and 7 B). Importantly, basal ATF4 levels were also detectable in drug-naive lesions from the tigecycline-sensitive models Mel-077 UM model and the Mel-020 and Mel-083 NRASQ61R mutant models (Fig. S5, A–C). This observation may explain why the OS and PFS were comparable in cohorts exposed to the antibiotics at MRD or from the beginning of the treatment. These data suggest that the extent of the response to mitoribosome-targeting antibiotics is dependent on the level of chronic ISR activation. In keeping with this, cotreatment with the ISR inhibitor ISRIB, which blocks signaling downstream of the eIF2α kinase and thereby dampens ATF4 activation and ISR engagement (Fig. S5 D), reduced CHOP induction (Fig. S5 D) and abolished the apoptotic response observed in a series of melanoma cells lines exposed to doxycycline (Fig. 7 C). Consistently, silencing of ATF4 was also sufficient to overcome doxycycline-induced growth inhibition (Fig. S5, E and F). Altogether, these data indicate that dependency on mitochondrial translation is a consequence of ATF4-mediated ISR activation. Importantly, these findings also identify ATF4 as a predictive biomarker for the antitumor efficacy of the antibiotic treatment.
Figure 7.
Sensitivity to tetracyclines is dictated by ISR. (A) Western blotting of Mel-006 (BRAFV600E) PDX tumors collected right before the start of the treatment (T0) or at the end of the experiment (Tend) after receiving a daily dose of DT or DTT. (B) Western blotting of Mel-015 (BRAFV600E) PDX tumors collected right before the start of the treatment (T0) or at the end of the experiment (Tend) after receiving a daily dose of DT, DTT, or dabrafenib + trametinib with the addition of tigecycline at MRD (DT + T). •, eIF2α-Phospho band. (C) Caspase counts per image in a panel of cell lines treated with doxycycline (Doxy, 50 µM) ± ISRIB (200 nM) for 72 h. Data are mean ± SEM of three or more different biological replicates. NS, P > 0.05; *, P < 0.05; **, P < 0.01 by Student’s t test.
Discussion
Cancer cells have the ability to survive a wide range of insults—including therapy exposure—through activation of an ATF4-ISR–dependent translation remodeling. We show herein that this pathway, in addition to promoting an overall decrease in cytosolic translation, enhances mitochondrial protein synthesis and selective translation of mRNAs encoding mitochondrial proteins in the cytosol and thereby establishes an unexpected vulnerability to inhibitors of mitoribosome assembly or function. Remarkably, this vulnerability can be targeted using the FDA-approved and widely used mitoribosome-targeting antibiotics. Accordingly, we show that tigecycline and doxycycline can decrease the growth and viability of a wide range of melanoma cells, exhibiting elevated ISR activity, including de-differentiated DTP cells or cells that have acquired resistance to targeted and immune therapies. We show that ATF4 inhibition or pharmacological inactivation of ISR rescues the antitumor effects of such antibiotics, thus confirming that this dependence relies on elevated ISR activity.
It has been proposed that internal ribosome entry site–dependent translation is one mechanism allowing translation of transcripts encoding survival proteins in cells exposed to stress and engaging an ISR. In fact, the translation of ATF4 itself depends on the internal ribosome entry site (Chan et al., 2013). Here we show that transcripts that are recruited to the ribosomes are enriched in rG4 sequences, raising the possibility that these elements also contribute to the selective translation of a series of transcripts, including those encoding mitochondrial proteins, in stressed cells.
An emerging theme in cancer biology and drug resistance is the reliance of different lesions and DTP cells, across multiple cancer types, on mitochondrial integrity and functions (Chen, 2012; Haq et al., 2013; Jagust et al., 2019; Faubert et al., 2020; Davis et al., 2020). There is even evidence that primary resistance to immunotherapy may also be driven by the up-regulation of mitochondrial translation (Jerby-Arnon et al., 2018; Poźniak et al., 2019). For instance, it has been shown that DTP cells exhibit elevated levels of oxidative phosphorylation, and that targeting mitochondrial respiration can delay resistance to targeted therapy (Roesch et al., 2013). Unfortunately, the use of mitochondrial respiration uncouplers to treat obesity and other metabolic diseases led to serious systemic side effects in patients (Childress et al., 2018). These observations have dampened the excitement toward these molecules and delayed their exploitation in the clinic. Importantly, we show herein that metabolic reprogramming is not the only underlying cause of the increased mitochondrial dependency of DTP cells. Our data therefore provide an alternative approach for the targeting of mitochondrial biology, through the repurposing of mitoribosome-targeting antibiotics. We argue that this approach may even be less susceptible to the development of resistance through metabolic adaptation. Accordingly, we show that tetracyclines addition to the standard-of-care targeted therapy (i.e., DT) extended PFS and often prevented relapse in a BRAF mutant preclinical PDX model (Mel-006). Mechanistically, we showed that antibiotic treatment compromises the emergence and/or survival of multiple drug–tolerant subpopulations, including NCSCs, mesenchymal/undifferentiated, and pseudo-starved subpopulations at MRD. This is an exciting observation given that these de-differentiated melanoma cells were shown to contribute to the development of resistance to various anti-melanoma therapies, including T cell transfer therapy (Landsberg et al., 2012; Lee et al., 2020; Mehta et al., 2018; Rambow et al., 2018; Boshuizen et al., 2020).
Importantly, although providing a significant increase in survival, the triple DTT combination did not prevent relapse in another BRAF mutant preclinical PDX model (Mel-015). The relatively lower benefit from the tigecycline treatment in this model correlated with undetectable levels of ISR activation (as measured by lack of ATF4 expression) before treatment. ATF4 levels also predicted responses of therapy-resistant melanomas, such as UM and BRAF WT cutaneous melanomas, to the tetracycline treatment. These observations offer a new avenue for the treatment of these diseases with no, or only limited, therapeutic options and indicate that ATF4 levels are predictive of responses to tetracycline exposure, either as a single agent or in combination with standard of care.
Recent reports highlighted that brain metastasis across different cancers (Chen et al., 2007; Chen, 2012; Cheng et al., 2019), including melanoma (Fischer et al., 2019; Sundstrøm et al., 2019), display increased oxygen consumption and are therefore more dependent on OXPHOS. Considering that tetracycline family antibiotics can cross the blood–brain barrier (Nau et al., 2010), this approach may also be a viable option for patients with this life-threatening and frequently untreatable condition.
Note that, in addition to their ability to inhibit mitochondrial translation, it remains possible that the antitumor effect of tetracyclines is multifactorial. Recent findings highlighted that mitochondrial translation deficiencies impair lysosome acidification and, thereby, autophagy (Yagi et al., 2021). Given the importance of autophagy in the biology of drug tolerance (Kinsey et al., 2019), this observation may also partly explain the efficacy of tetracyclines in delaying/preventing the development of resistance to MAPKi. Moreover, both dabrafenib and trametinib are partially metabolized by CYP3A4 (Lawrence et al., 2014), which is inhibited by tetracycline. Its coadministration may therefore raise the active concentrations of dabrafenib and trametinib and thereby enhance the antitumor efficacy of DT (Bassi et al., 2004). We also cannot rule out that, in addition to cancer cell intrinsic effects, the dramatic responses observed in the preclinical and clinical settings are a consequence of tetracycline-mediated remodeling of the tumor microenvironment, such as the (intratumor) microbiome. This may be important, as the efficacy of anti–PD-1 therapy has been shown to be affected by the microbiome (Matson et al., 2018; McQuade et al., 2019). However, most common symbiotic bacteria are resistant to doxycycline, which is a broad-spectrum antibiotic that can be administered orally for long periods of time with no or minor toxic effects (Tan et al., 2011). Its administration is therefore unlikely to affect significantly the patients’ microbiome.
In conclusion, our data indicate that the repurposing of mitoribosome-targeting antibiotics offers a new promising therapeutic avenue for the treatment of a large spectrum of melanoma patients, including those with limited therapeutic options. Moreover, we provide evidence that patient stratification should be guided by ATF4 levels, which could be used as an accompanying biomarker to predict efficacy. Importantly, this drug-repositioning strategy can be easily implemented in the clinic.
Materials and methods
Study design
The objectives of this study were (a) to identify novel metastatic melanoma vulnerabilities, (b) to prevent or delay acquisition of therapy resistance, (c) to sensitize therapy-resistant lesions to therapy, and (d) to exploit this knowledge for therapeutic benefit in a preclinical setting. To test this objective, we made use of several clinically relevant mouse models of metastatic cutaneous and UM with different mutational backgrounds and therapy sensitivities (i.e., with intrinsic or acquired therapy resistance profiles). Written informed consent was obtained from patients, and all procedures involving human samples were approved by the Universitair Ziekenhuis (UZ) Leuven/Katholieke Universiteit (KU) Leuven Medical Ethical Committee (S63799) and performed in accordance with the principles of the Declaration of Helsinki and with General Data Protection Regulations. The mouse experiments were approved by the KU Leuven animal ethical committee under Ethical Committee for Animal Testing (Ethische Commissie Dierproven, ECD) P038-2015 and P164-2019 and performed in accordance with the internal, national, and European guidelines of animal care and use. Mice were evaluated for OS, PFS, and tumor growth daily. Cell lines were monitored for cell growth and cell death. The melanoma patient was monitored for tumor load and tumor response to the treatments. Each in vitro experiment was repeated at least three times (as specified in the figure legend) to ensure reproducibility. For in vivo experiments, sample size was calculated using the software G power v3.1.9.2 with a power of 80–95% and an α-error probability of 0.05%, assuming an effect size of 0.25 (based on previous experiments and pilot studies). This number was not altered during the experiment. This study included several cell cultures, two animal models, and one melanoma patient. Investigators were not blinded.
Cell lines
MM383 and WM852 cell lines (a kind gift from Göran Jönsson), SK-MEL-28 cells, and the IGR37 cell line (a kind gift from Irwin Davidson) were grown in RPMI 1640 (Gibco BRL Invitrogen) supplemented with 10% FBS (Gibco BRL Invitrogen). The patient-derived low-passage MM cell lines (a gift from G.-E. Ghanem) were grown in F-10 (Gibco BRL Invitrogen), supplemented with 10% FBS (Gibco BRL Invitrogen). The YUMM 1.7 and YUMMER 1.7 cell lines (a kind gift from Marcus Bosenberg; Meeth et al., 2016; Wang et al., 2017) were grown in DMEM/F12 (50:50; Gibco BRL Invitrogen) medium supplemented with 10% FBS (Gibco BRL Invitrogen). The UM92.1 cell line (a kind gift from Aart Jochemsen) were grown in a 1:1 mixture of RPMI 1640 (Gibco BRL Invitrogen) and DMEM/F12 (Gibco BRL Invitrogen) medium supplemented with 10% FBS (Gibco BRL Invitrogen).
All cell lines used are of human origin, save for the YUMM 1.7 and YUMMER 1.7 cell lines, derived from male C57BL/6J (Mus musculus). Gender of the patients from whom the cell cultures were derived is as follows (female: F; male: M): SK-MEL-28, M; MM034 and MM011, F; MM099, M; MM165, M; IGR37, M; YUMM 1.7, M; UM92.1, F. All cell lines were confirmed negative for mycoplasma before the study using the MycoAlert Mycoplasma Detection Kit (Lonza) according to the manufacturer’s specifications.
Cell viability assays
For colony formation assays, cells were plated in 6-well plates at the appropriate density (1.5 × 104/well in the case of the MM011, MM034, MM099, MM165, MM383, WM852, IGR37, YUMM 1.7, and YUMMER 1.7 cells and 8 × 104/well in the case of the UM92.1 cells). Cells were then treated with increasing amounts of tigecycline and cultured for 3 d (YUMM 1.7, YUMMER 1.7, and UM 92.1), 5 d (WM852 and IGR37), or 7 d (MM011, MM034, MM165, and MM383). Cells were then washed twice with PBS, fixed, and stained for 15 min with 1% crystal violet in 35% methanol solution. Surface occupancy was measured using ImageJ.
For siRNA-mediated knockdown, cells were plated in duplicate in 6-well plates at the confluency described above and transfected with Lipofectamine 2000 (Thermo Fisher Scientific) according to manufacturer instructions with 25 nM of nontargeting siRNA (control, SMART-pool; Dharmacon) or with 25 nM siATF4 (SMART-pool; Dharmacon). 72 h after transfection, cells were collected (for protein extraction) or stained with crystal violet and quantified as described above.
For IncuCyte proliferation assays, cells were plated in 96-well plates (TPP) at the appropriate density (MM011, MM034, MM165, MM099, IGR37, and MM383, 1.5 × 104/well; WM852, 5 × 103/well; YUMM 1.7, YUMMER 1.7, and UM 92.1, 2.5 × 103/well). Cells were treated with increasing amounts of tigecycline or doxycycline and cultured for 72 h. Apoptotic cells were labeled with IncuCyte Caspase 3/7 Green Apoptosis Assay Reagent (Essen BioScience). Four images per well were taken at 2-h intervals using an IncuCyte ZOOM system (Essen BioScience). The percentage of cell confluency and fluorescent green counts indicating apoptotic cells were measured and analyzed with IncuCyte ZOOM software.
Polysome profiling
SK-MEL-28 cells (5 × 15-cm dishes per each condition) were plated to have 70% confluency after 72 h. The following day, cells were treated with 20 µM salubrinal (Sigma-Aldrich). 72 h after the start of the treatment, cells were treated with 100 µg/ml of cycloheximide (Sigma-Aldrich) for 12 min at 37°C, collected, and resuspended in lysis buffer (30 mM Tris-HCl, 150 mM KCl, 10 mM MgCl2 supplemented with 1 mM DTT [Sigma-Aldrich], 100 µg/ml cycloheximide, 20 U/µl SUPERase-IN RNase Inhibitor [Invitrogen, Thermo Fisher Scientific], and 1× Halt Protease and Phosphatase Inhibitor Single-Use Cocktail [Life Technologies] before the start of the experiment). Lysates were then incubated agitating at 4°C for 35 min, and then centrifuged at 17,000 rcf for 15 min at 4°C. Lysates were loaded on a sucrose gradient (the linear sucrose gradient 5–20% was generated from two different solutions, sucrose 5 and 20%, made with buffer G (20 mM Tris-HCl, 100 mM KCl, 10 mM MgCl2 supplemented with 1 mM DTT and 100 µg/ml cycloheximide before the start of the experiment). Samples were then centrifugated in an SW41Ti rotor (Beckman Coulter) at 37,000 rpm for 170 min at 4°C. The fractions were obtained with a Biological LP System (Bio-Rad). 14 fractions were collected from each sample, with each fraction having a final volume of 600 µl. From the initial 14 fractions, 4 final samples were obtained (by pulling together some of them): 40S, 60S, 80S, and polysomes.
RNA extraction
For RNA extraction from the fractions, each fraction was digested at 37°C for 90 min in a digestion mix of proteinase K (final concentration 100 µg/ml; Sigma-Aldrich) and 1% SDS. Phenol acid chloroform (5:1; Sigma-Aldrich) and 10 mM NaCl were then added. Samples were centrifuged at 16,000 rcf for 5 min at 4°C. The upper aqueous phase was transferred in a new tube, and 1 ml of isopropanol was added. Samples were stored at −80°C overnight to precipitate the RNA. The following day, the samples were centrifuged at 16,000 rcf for 40 min at 4°C. The supernatant was discarded, and the pellet was washed with 500 µl of 70% EtOH, then centrifuged again at 16,000 rcf for 5 min at 4°C. Pellet were air dried and resuspended in diethylpyrocarbonate water. No DNase treatment was performed (not needed). Input RNAs were extracted with TRIzol (Invitrogen) according to the manufacturer’s instructions.
RT-qPCR
RNA was reverse transcribed using the High-Capacity complementary DNA Reverse Transcription Kit (Thermo Fisher Scientific) on a Veriti 96-well thermal cycler (Thermo Fisher Scientific). Gene expression was measured by qPCR on a QuantStudio 5 (Thermo Fisher Scientific) and normalized in qbase + 3.0 (Biogazelle) using 28S and 18S as reference genes (for polysome profiling experiments) or the average of HPRT, TBP, and UBC. Sequences of the primers are indicated in Table S1.
RNA sequencing and data analysis
Samples were prepared for sequencing with TruSeq Stranded Total RNA kit (Illumina) according to the manufacturer’s instructions. Libraries were sequenced on an Illumina NextSeq according to the manufacturer’s instructions, generating PE high output configuration cycles: (R1: 100) − (I1: 6) − (I2: 0) − (R2: 100). Quality control of raw reads was performed with FastQC v0.11.5. Adapters were filtered with eautils v1.2.2.18. Splice-aware alignment was performed with STAR against the human hg19 human genome assembly. The number of allowed mismatches was 2. Reads that mapped to more than one site of the reference genome were discarded. The minimal score of alignment quality to be included in count analysis was 10. Resulting Sequence Alignment/Map (SAM) and binary SAM alignment files were handled with Samtools v0.1.19.24. Quantification of reads per gene was performed with HT-Seq count v0.5.3p3. Count-based differential expression analysis was done with R-based (R Foundation for Statistical Computing) Bioconductor package DESeq. Reported P values were adjusted for multiple testing with the Benjamini–Hochberg procedure, which controls false discovery rate. Differentially expressed genes were selected at false discovery rate 0.05. For the comparison of transcriptional and ribosome association changes in Fig. S1 A, the R package ggplot2 was used. The data generated in this study are accessible through GEO with accession number GSE177075.
rG4 element analysis
rG4 elements were predicted within the mRNA of the selected gene lists by using the QGRS Mapper tool (https://bioinformatics.ramapo.edu/QGRS/index.php; Kikin et al., 2006) with default parameters, and transcript sequences from the GENCODE V32 release, hg38 human genome assembly (https://www.gencodegenes.org). Being the rG4 score distribution bimodal with two well-separated peaks, the high-confidence score threshold was selected to be 21, i.e., the value corresponding to the point at which the first peak fell to 0 predicted rG4s before the start of the second, higher-scoring peak. Experimentally determined in vitro rG4s were obtained from the Kwok et al. (2016) and Guo and Bartel (2016) datasets. Enrichment of rG4s in the selected gene lists was computed by Fisher’s exact test, using the whole transcriptome as a background. Finally, the enrichment of regulatory factor binding sites (RNA-binding proteins and microRNAs) and cis-elements sites in the 5′ and 3′ UTRs of rG4-containing mRNAs was computed by the Regulatory Enrichment function of the AURA2 database (Dassi et al., 2014), using a 0.05 threshold on the BH-adjusted enrichment P value.
Puromycin incorporation assay (SUNsET)
SUNsET was performed as described in Schmidt et al. (2009). Briefly, ≈80% confluent adherent cells were washed twice in 1× PBS and subsequently pulsed with puromycin-containing medium (InvivoGen, 10 µg ml−1) for 10 min. The cells were then supplemented with normal medium for 60 min before downstream applications (chase). Due to the fact that puromycin is a structural analogue of aminoacyl transfer RNAs, it gets incorporated into the nascent polypeptide chain and prevents elongation. Therefore, when used for reduced amounts of time, puromycin incorporation in neosynthesized proteins directly reflects the rate of mRNA translation in vitro. Puromycin incorporation was measured by western blotting using an antibody that recognizes puromycin.
Cellular fractionation and mitoplast isolation
Briefly, mitochondria were purified from 4–6 × 107 cells using a mitochondria isolation kit for cultured cells (Thermo Fisher Scientific) according to manufacturer instructions; all buffers were supplemented with 60 U ml−1 SUPERase-In (Ambion) and 1× Halt Protease and Phosphatase Inhibitor Single-Use Cocktail (Life Technologies). Mitoplasts were obtained by incubating purified mitochondria in RNase A–containing hypotonic buffer (Hepes, pH 7.2, supplemented with 1× Halt Protease and Phosphatase Inhibitor Single-Use Cocktail and 10 µg/ml RNase A [Roche]) for 20 min on ice and subsequently incubated for 10 additional min at room temperature in order to remove all possible cytosolic RNA contaminants. The purified mitoplasts were then washed three times with mitoplast isolation buffer (250 nM mannitol, 5 mM Hepes, pH 7.2, 0.5 mM EGTA, 1 mg/ml BSA supplemented with 60 U ml−1 Superase-In [Ambion] and 1× Halt Protease and Phosphatase Inhibitor Single-Use Cocktail).
Ascorbic acid peroxidase (APEX) assay
SK-MEL-28 cells were seeded at 70% confluency in 15-cm dishes (one per condition) and transiently transfected with 10 µg mito-V5-APEX2 plasmid (#72480; Addgene; Lam et al., 2015) using Lipofectamine 2000 (Thermo Fisher Scientific). 24 h after transfection, the medium was removed, and the cells were subsequently treated with either 1× PBS (control) or 30 µM tigecycline. 24 h after treatment, which corresponds to 48 h after transfection, cells were labeled by a 30-min incubation with 500 µM biotin-phenol at 5% CO2, 37°C (Iris Biotech), followed by a 1-min incubation with 1 mM H2O2 at room temperature. Medium was quickly removed, and the biotin-phenol reaction was quenched by three washes with quencher solution (10 mM sodium ascorbate, 5 mM Trolox, and 10 mM sodium azide diluted in PBS). The cells were then also washed in 1× PBS, scraped in 4 ml cold 1× PBS, and collected for lysis. Each sample was lysed in 50 µl polysome lysis buffer supplemented with 1× Halt Protease and Phosphatase Inhibitor Single-Use Cocktail.
Western blotting
Western blotting experiments were performed using the following primary antibodies: vinculin (V9131, clone hVIN-1; Sigma-Aldrich, 1:5,000), histone 3 (#4499, clone D1H2; Cell Signaling Technology, 1:1,000), MITF (ab12039; Abcam, 1:1,000), ATF4 (#11815, clone D4B8; Cell Signaling Technology, 1:1,000), ATF5 (SAB4500895; Sigma-Aldrich, 1:500), eIF2α-Tot (#5324, clone D7D3; Cell Signaling Technology, 1:1,000), eIF2α-Phospho-S51 (#3398, clone D9G8; Cell Signaling Technology, 1:1,000), CHOP (#2895, clone L63F7, 1:1000), puromycin (MABE343, clone 12D10; Merck-Millipore, 1:10,000), streptavidin (#N1000; ThermoFisher Scientific, 1:5,000), β-actin (#4970, clone 13E5; Cell Signaling Technology, 1:1,000), and Tom20 (sc-17764, clone F-10; Santa Cruz Biotechnology, 1:1,000). The following HRP-linked secondary antibodies were used: anti-mouse IgG (NA931-1ML; Sigma-Aldrich, 1:10,000) and anti-rabbit IgG (NA934-1ML; Sigma-Aldrich, 1:10,000). Relative protein levels were measured using ImageJ.
PDX experiments
The cutaneous melanoma PDX models are part of the Trace collection (https://www.uzleuven-kuleuven.be/lki/trace/trace-leuven-pdx-platform) and were established using metastatic melanoma lesions derived from patients undergoing surgery as part of standard treatment at UZ Leuven. The UM model derives from a lymph node metastasis and is a kind gift of M. Herlyn (The Wistar Institute, Philadelphia, PA). Written informed consent was obtained from patients and all procedures involving human samples were approved by the UZ Leuven/KU Leuven Medical Ethical Committee (S63799) and performed in accordance with the principles of the Declaration of Helsinki and with GDPR regulations. The experiments were approved by the KU Leuven animal ethical committee under ECDs P038-2015 and P164-2019 and performed in accordance with the internal, national, and European guidelines of animal care and use. Tumor pieces were implanted subcutaneously in the interscapular fat pad of female NMRI nude BomTac:NMRI-Foxn1nu, 4-wk-old females (Taconic Biosciences). Mice were maintained in a semi-specific pathogen–free facility under standard housing conditions with continuous access to food and water. The health and welfare of the animals was supervised by a designated veterinarian. The KU Leuven animal facilities comply with all appropriate standards (cages, space per animal, temperature [22°C], light, humidity, food, and water), and all cages are enriched with materials that allow the animals to exert their natural behavior. Mice used in the study were maintained on a diurnal 12-h light/dark cycle. PDX models Mel-006, Mel-015, and Mel-020 were derived from a female, male, and female drug-naive patients, respectively. The UM Mel-077 sample was derived from a male patient progressing on pembrolizumab and temozolomide. Mel-018, Mel-021, and Mel-078 were derived from male, female, and male patients, respectively.
Once tumors reached 250 mm3 for Mel-077 and Mel-020, 500 mm3 for Mel-015, or 1,000 mm3 for Mel-006, the mice were enrolled into treatment cohorts. Mice from distinct batches were randomly assigned to the different experimental groups. Mice were treated daily by oral gavage with a capped dose of 600 dabrafenib µg–6 trametinib µg (DT), in 250 µl total volume, and tigecycline (50 mg/kg) was administered daily by i.p. injection.
Humanized huNOG-EXL (NOD.Cg-Prkdcscid Il2rgtm1Sug Tg(SV40/HTLV-IL3,CSF2)10-7Jic/JicTac), engrafted at the age of 6 wk with CD34+ hematopoietic stem cells from two different donors, were purchased from Taconic Biosciences. The percentage of humanization was 20–80% as estimated by FACS for CD45+ cells 10 wk after engraftment. The mice were engrafted with the Mel-006 human melanoma model described before at the age of 23 wk. When the tumor reached the size of 100 mm3, treatment with tigecycline (50 mg/kg i.p.) and/or nivolumab (10 mg/kg twice a week i.p.) was initiated. This experiment was approved by the KU Leuven animal ethical committee under ECD P210/2018.
No specific randomization method was used. According to animal welfare guidelines, mice were sacrificed when tumors reached a volume of 2,500 mm3 or when body weight decreased >20% from the initial weight. Mice used in this study never reached or exceeded these limits.
Allografts
5 × 104 YUMMER 1.7 cells were injected subcutaneously in the interscapular fat pad of 4-wk-old C57BL/6 males (males were chosen since females frequently rejected the implantation and had sporadic tumor ulceration). Mice were maintained under standard housing conditions with continuous access to food and water. The health and welfare of the animals was supervised by a designated veterinarian. The KU Leuven animal facilities comply with all appropriate standards (cages, space per animal, temperature (22°C), light, humidity, food, water), and all cages are enriched with materials that allow the animals to exert their natural behavior. Mice used in the study were maintained on a diurnal 12 h light/dark cycle. The experiments were approved by the KU Leuven animal ethical committee under ECD P049-2019 and performed in accordance with the internal, national, and European guidelines of animal care and use. Once the tumors reached 50 mm3 (for YUMMER 1.7), mice were enrolled in the treatment cohort. Mice from distinct batches were randomly assigned to the different experimental groups. Mice were treated with tigecycline (Bio Connect) i.p., 50 mg/kg daily, with anti–PD-1 (Ultra-LEAF Purified anti-mouse CD279 antibody, clone RMP1-14) i.p., 10 mg/kg twice per week, for 3 wk, for a total of six injections.
No specific randomization method was used. According to animal welfare guidelines, mice were sacrificed when tumors reach a volume of 2,500 mm3 or when body weight decreased >20% from the initial weight. Mice used in this paper never reached nor exceeded these limits.
Immunofluorescence on PDX biopsies
Fluorescent staining was performed using Opal staining reagents, which use individual tyramide signal amplification–conjugated fluorophores to detect various targets within an immunofluorescence assay. In brief, samples were fixed with 4% paraformaldehyde and embedded in paraffin. Serially cut sections of 5 µm were stained with H&E for routine light microscopy and used for immunohistochemistry.
Depending on the antibody, antigen retrieval (AR) was performed in citrate buffer at pH 6.0 or EDTA buffer at pH 9. Deparaffinized sections were then incubated overnight with primary antibodies against AQP1 (#AB2219; Millipore, 1:3,000), NGFR (8238; Cell Signaling Technology, 1:1,000), MITF (#HPA003259; Sigma-Aldrich, 1:100), MLANA (#HPA048662; Sigma-Aldrich, 1:200), CD36 (#HPA002018; Sigma-Aldrich, 1:200), AXL (#AF154; R&D Systems, 1:50), and S100 (#Z0311; Dako, 1:100). Subsequently, the slides were washed in PBS, pH 7.2, and incubated for 10 min at room temperature with Opal Polymer HRP Mouse Plus Rabbit secondaries (PerkinElmer). After another wash in PBS, the slides were incubated at room temperature for 10 min with one of the following Alexa Fluorescent tyramides (PerkinElmer) included in the Opal 4 color kit (NEL810001KT; Akoya Biosciences) to detect antibody staining, prepared according to the manufacturer’s instructions: Opal 520, Opal 570, and Opal 690 (dilution 1:50). Stripping of primary and secondary antibodies was performed by placing the slides in a plastic container filled with AR; microwave technology was used to bring the liquid to 100°C (2 min), and the sections were then microwaved for an additional 15 min at 75°C. Slides were allowed to cool in the AR buffer for 15 min at room temperature and were then rinsed with deionized water and 1× Tris-buffered saline with Tween 20. After three additional washes in deionized water, the slides were counterstained with DAPI for 5 min and mounted with ProLong Gold Antifade Mountant (P10144; Thermo Fisher Scientific). Slides were scanned for image acquisition using Zeiss AxioScan Z.1 and ZEN2 software. Images were quantified using QuPath. In brief, three different regions per section were quantified. Every dot in the graph represents the percentage of each quantified population. For MITF/NGFR/AQP1 Opal staining, melanocytic/proliferative cells were quantified based on MITFhigh expression, and only NGFR+/AQP1+ cells were considered to be NCSCs. In AXL/CD36/MLANA Opal staining, mesenchymal cells were quantified based on AXL positivity, and to identify SMCs, we quantified the presence of CD36+ cells.
Matrix-assisted laser desorption/ionization (MALDI)
Frozen cryosections were removed from the freezer, quickly placed into cryomolds, covered with a 10% solution of gelatin, and frozen in a 2-methylbutane/liquid nitrogen bath. Samples were then stored at −80°C until further processing. For cryosectioning, samples were taken from −80°C and placed into a cryostat (Leica CM1950) chamber for 1 h before cutting. Sections of 10-µm thickness were thaw-mounted onto indium tin oxide (ITO) glass (Ossila), washed subsequently with hexane and 2-propanol, and coated with a solution composed of 0.05% poly-L-lysine (Sigma-Aldrich) and 0.1% NP-40 detergent substitute. Sections were dried in a desiccator for 15 min, sealed in foil inside a plastic container, evacuated, and stored at −80°C until further processing. Samples were then transferred to room temperature, left to temper for 15 min, and dried in a desiccator for 15 min. Samples were then sprayed with a matrix solution of 9-aminoacridine (5 mg/ml in 70% ethanol [vol/vol]). A TM-Sprayer 3 (HTX-Technologies) was used to cover the tissue with matrix solution with the following settings: nozzle temperature 95°C, 16 passes, flow 0.06 ml/min, nozzle velocity 1,200 mm/min, track spacing 3 mm, CC pattern, gas pressure 10 psi, gas flow rate 2 liter/min, drying time 5 s, nozzle height 40 mm, and propelled with 50% methanol (vol/vol). After spraying, samples were dried in a desiccator for an additional 15 min.
Spectral images were taken with a rapifleX MALDI–time of flight (TOF)/TOF spectrometer (Bruker) in negative mode using the 355-nm smartbeam 3D laser with a spatial resolution of 50 × 50 µm within the range of m/z 20–1,100, with constant laser fluence of 44% and laser frequency of 5 kHz. 200 shots were accumulated from every position. Samples were measured in random order. Calibration was done externally using red phosphorus (Sigma-Aldrich), achieving precision ≤5 ppm.
For histology, tissues on ITO glass with matrix were washed three times with 70% ethanol, each time for 2 min. After washing in water, samples were stained with H&E, dehydrated through an ethanol series, washed in xylene (Leica ST5020), and mounted in Pertex (Leica CV5030). Samples were then scanned with Axio Scan.Z1 (Zeiss) operated by Zen 2 software (blue edition; Zeiss). Scanned images were processed with white balance corrected to background and exported as JPG files using ZEN 2.5 lite software (Zeiss). For image processing, single raw data files were processed at once using SCiLS Lab software (v2020a, SCiLS; Bruker). Raw data were smoothed with a convolution algorithm (width 20), and all subsequent computations and image rendering were done with root mean square normalization, with the exception of ATP/ADP ratio and lactate/pyruvate ratio, for which ADP or pyruvate raw peak values, respectively, were used to calculate the ratio. Regions of interest were corrected with H&E scans. Spectra were exported as CSV files and processed in mMass software (v5.5.0, Martin Strohalm). Peak picking was performed, and matrix-related peaks were removed. The resulting peak list was imported back to SCiLS Lab and used for computations.
Receiver operating characteristic analysis was performed to find discriminative values for two selected regions. All tissues from the respective groups (control, DT, or DTT) were used for computations. The operation was performed with all individual spectra, with a random subset of spectra set up properly according to the number of spectra in respective regions. Peaks with an area under the curve value >0.9 or <0.1 were considered significant. Based on morphological assessment, five region types were selected and marked by a trained pathologist: tumor tissue, fibrotic tissue, necrosis, white adipose tissue, and muscle tissue. Classification Pipeline was performed in SCiLS software to assign tissue type automatically. Resulting areas of tumor tissue were selected, and receiver operating characteristic analysis was performed to find differences in metabolite expression. All tissues from the respective groups (control, DT, or DTT) were used for computations. The operation was performed with all individual spectra with a random subset of spectra set up properly accordingly to the number of spectra in respective regions. Peaks with an area under the curve value above 0.9 or less than 0.1, respectively, were considered as significant.
Identification was done using modified HMDB v4 database (using compounds from serum, cerebrospinal fluid, urine, sweat, and saliva, excluding lipids, drugs, and metabolites; https://hmdb.ca/) considering [M-H]−, [M-H-H2O]−, [M + Cl]−, [M + Na-2H]−, [M + Na-2H-H2O]−, [M + K-2H]−, and [M + K-2H-H2O]− adducts within a tolerance of 30 ppm.
Case report
The patient was treated with BRAF-MEK inhibitors for stage IV malignant melanoma according to standard of care. PET-CT scans were done at baseline and for confirmation of remission. Every 2 mo, the patient underwent radiographical assessment with either PET-CT or regular CT scan.
Statistical analyses
All data are graphed as mean ± SEM. In animal experiments, n indicates the number of animals used. When comparing two treatment cohorts over time, statistical significance was calculated by two-way ANOVA. When comparing three or more groups, statistical significance was calculated by one-way ANOVA with Dunnett’s multiple comparisons test. When comparing Kaplan–Meier plots, statistical significance was calculated by log-rank (Mantel–Cox) test. Statistical tests used are specified in the corresponding figure legends. Throughout all figures, statistical significance was considered at P < 0.05 (*, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001). All statistical analyses were performed with GraphPad Prism v8.4.2 (464), April 8, 2020, for Mac OS Catalina.
Online supplemental material
Fig. S1 shows the increase in mitochondrial translation upon activation of the ISR as seen through sequencing of a BRAF mutant cell line and confirmed by qPCR in a NRAS mutant cell line. Fig. S2 shows that tetracyclines affect multiple drug–tolerant states and thus increase OS in PDX models. In Fig. S3, a spatial metabolomic experiment shows that addition of tigecycline to targeted therapy does not significantly rewire melanoma metabolism in vivo. Fig. S4 shows that tigecycline significantly affects the growth of immune and targeted therapy–resistant cells and lesions. Fig. S5 shows that the effect of tetracyclines can be rescued by knocking down ATF4. Table S1 lists primers used in the study.
Supplementary Material
lists primers used in the study.
Acknowledgments
The authors wish to thank Ghanem E. Ghanem (Jules Bordet Institute, Brussels, Belgium), Marcus Bosenberg (Yale University, New Haven, CT), Irwin Davidson (Institute of Genetic and Molecular and Cellular Biology, Strasbourg, France), Aart Jochemsen (Leiden University, Leiden, Netherlands), and Göran Jönsson (Lund University, Lund, Sweden) for kindly making gifts of some of the melanoma cell lines used in this study. We also thank Amanda Lund for comments and suggestions. We are grateful to Petr Makovicky and Ashkan Zareie (Czech Centre for Phenogenomics) for their help with the MALDI-TOF analysis.
The present study was funded by Kom op tegen Kanker (Stand Up To Cancer), the Flemish Cancer Society, and by a KU Leuven C1 grant. E. Leucci is funded by the Melanoma Research Alliance Amanda and Jonathan Eilian young investigator award (2018). Trace staff are supported by Stichting Tegen Kanker grant 2016-054. V. Katopodi is a recipient of a Research Foundation – Flanders PhD fellowship (1S47519N). S. Cinque is a recipient of a Research Foundation – Flanders PhD fellowship (1SD1620N). The Czech Centre for Phenogenomics and Trace are members of the EuroPDX Consortium. L. Kucer and J. Rozman are supported by the Czech Academy of Sciences (RVO 68378050, LM2018126), the Ministry of Education, Youth and Sports (project CZ.02.1.01/0.0/0.0/16_013/0001789), and European Structural and Investment Funds.
Author contributions: R. Vendramin designed and performed in vitro and allograft experiments and supervised the work of A. Konnova and Z. Knezevic. A. Konnova performed cell treatments (tigecycline, doxycycline, and ISRIB) followed by colony assays and Incucyte measurements and collected the samples for western blots in Figs. 2 C and S5 D. Z. Knezevic performed the drug treatments in combination with ATF4 silencing and collected the samples for western blots in Fig. S5 F. R. Vendramin, A. Konnova, and Z. Knezevic performed allograft experiments. S. Cinque and V. Katopodi performed the in vitro experiments in Fig. 1 and ran western blots. S. Adnane performed the UM PDX experiment, contributed to the in vitro experiments, and extracted proteins from tumors. Y. Verheyden performed bioinformatic analysis. P. Karras and F.M. Bosisio performed and interpreted multiplex IHC. E. Demesmaeker and L. Rizzotto performed and designed cutaneous melanoma PDX experiments. I. Gladwyn-Ng assisted with the design of the humanized PDX experiments. L. Kucera and J. Rozman performed and analyzed spatial metabolomics. E. Dassi and S. Millevoi designed and analyzed rG4-related experiments. O. Bechter provided and analyzed the clinical case. E. Leucci and R. Vendramin analyzed and interpreted the data. E. Leucci acquired funding. J-C. Marine and E. Leucci designed the research study and wrote the manuscript with input from all the authors.
References
- Ahler, E., Sullivan W.J., Cass A., Braas D., York A.G., Bensinger S.J., Graeber T.G., and Christofk H.R.. 2013. Doxycycline alters metabolism and proliferation of human cell lines. PLoS One. 8:e64561. 10.1371/journal.pone.0064561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almanza, A., Carlesso A., Chintha C., Creedican S., Doultsinos D., Leuzzi B., Luís A., McCarthy N., Montibeller L., More S., et al. 2019. Endoplasmic reticulum stress signalling - from basic mechanisms to clinical applications. FEBS J. 286:241–278. 10.1111/febs.14608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassi, P.U., Onyeji C.O., and Ukponmwan O.E.. 2004. Effects of tetracycline on the pharmacokinetics of halofantrine in healthy volunteers. Br. J. Clin. Pharmacol. 58:52–55. 10.1111/j.1365-2125.2004.02087.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshuizen, J., Koopman L.A., Krijgsman O., Shahrabi A., van den Heuvel E.G., Ligtenberg M.A., Vredevoogd D.W., Kemper K., Kuilman T., Song J.Y., et al. 2018. Cooperative targeting of melanoma heterogeneity with an AXL antibody-drug conjugate and BRAF/MEK inhibitors. Nat. Med. 24:203–212. 10.1038/nm.4472 [DOI] [PubMed] [Google Scholar]
- Boshuizen, J., Vredevoogd D.W., Krijgsman O., Ligtenberg M.A., Blankenstein S., de Bruijn B., Frederick D.T., Kenski J.C.N., Parren M., Brüggemann M., et al. 2020. Reversal of pre-existing NGFR-driven tumor and immune therapy resistance. Nat. Commun. 11:3946. 10.1038/s41467-020-17739-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumahdi, S., and de Sauvage F.J.. 2020. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug Discov. 19:39–56. 10.1038/s41573-019-0044-1 [DOI] [PubMed] [Google Scholar]
- Boyce, M., Bryant K.F., Jousse C., Long K., Harding H.P., Scheuner D., Kaufman R.J., Ma D., Coen D.M., Ron D., and Yuan J.. 2005. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 307:935–939. 10.1126/science.1101902 [DOI] [PubMed] [Google Scholar]
- Chan, C.P., Kok K.H., Tang H.M.V., Wong C.M., and Jin D.Y.. 2013. Internal ribosome entry site-mediated translational regulation of ATF4 splice variant in mammalian unfolded protein response. Biochim. Biophys. Acta. 1833:2165–2175. 10.1016/j.bbamcr.2013.05.002 [DOI] [PubMed] [Google Scholar]
- Chen, E.I.2012. Mitochondrial dysfunction and cancer metastasis. J. Bioenerg. Biomembr. 44:619–622. 10.1007/s10863-012-9465-9 [DOI] [PubMed] [Google Scholar]
- Chen, E.I., Hewel J., Krueger J.S., Tiraby C., Weber M.R., Kralli A., Becker K., Yates J.R. III, and Felding-Habermann B.. 2007. Adaptation of energy metabolism in breast cancer brain metastases. Cancer Res. 67:1472–1486. 10.1158/0008-5472.CAN-06-3137 [DOI] [PubMed] [Google Scholar]
- Cheng, G., Zhang Q., Pan J., Lee Y., Ouari O., Hardy M., Zielonka M., Myers C.R., Zielonka J., Weh K., et al. 2019. Targeting lonidamine to mitochondria mitigates lung tumorigenesis and brain metastasis. Nat. Commun. 10:2205. 10.1038/s41467-019-10042-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childress, E.S., Alexopoulos S.J., Hoehn K.L., and Santos W.L.. 2018. Small Molecule Mitochondrial Uncouplers and Their Therapeutic Potential. J. Med. Chem. 61:4641–4655. 10.1021/acs.jmedchem.7b01182 [DOI] [PubMed] [Google Scholar]
- D’Andrea, A., Gritti I., Nicoli P., Giorgio M., Doni M., Conti A., Bianchi V., Casoli L., Sabò A., Mironov A., et al. 2016. The mitochondrial translation machinery as a therapeutic target in Myc-driven lymphomas. Oncotarget. 7:72415–72430. 10.18632/oncotarget.11719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassi, E., Re A., Leo S., Tebaldi T., Pasini L., Peroni D., and Quattrone A.. 2014. AURA 2: Empowering discovery of post-transcriptional networks. Translation (Austin). 2:e27738. 10.4161/trla.27738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, R.T., Blake K., Ma D., Gabra M.B.I., Hernandez G.A., Phung A.T., Yang Y., Maurer D., Lefebvre A.E.Y.T., Alshetaiwi H., et al. 2020. Transcriptional diversity and bioenergetic shift in human breast cancer metastasis revealed by single-cell RNA sequencing. Nat. Cell Biol. 22:310–320. 10.1038/s41556-020-0477-0 [DOI] [PubMed] [Google Scholar]
- Deng, P., and Haynes C.M.. 2017. Mitochondrial dysfunction in cancer: Potential roles of ATF5 and the mitochondrial UPR. Semin. Cancer Biol. 47:43–49. 10.1016/j.semcancer.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson, S.M., García-Prat L., Vanner R.J., Wintersinger J., Waanders E., Gu Z., McLeod J., Gan O.I., Grandal I., Payne-Turner D., et al. 2020. Relapse-Fated Latent Diagnosis Subclones in Acute B Lineage Leukemia Are Drug Tolerant and Possess Distinct Metabolic Programs. Cancer Discov. 10:568–587. 10.1158/2159-8290.CD-19-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas, L., Herviou P., Dassi E., Cammas A., and Millevoi S.. 2021. G-Quadruplexes in RNA Biology: Recent Advances and Future Directions. Trends Biochem. Sci. 46:270–283. 10.1016/j.tibs.2020.11.001 [DOI] [PubMed] [Google Scholar]
- Falletta, P., Sanchez-Del-Campo L., Chauhan J., Effern M., Kenyon A., Kershaw C.J., Siddaway R., Lisle R., Freter R., Daniels M.J., et al. 2017. Translation reprogramming is an evolutionarily conserved driver of phenotypic plasticity and therapeutic resistance in melanoma. Genes Dev. 31:18–33. 10.1101/gad.290940.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert, B., Solmonson A., and DeBerardinis R.J.. 2020. Metabolic reprogramming and cancer progression. Science. 368:eaaw5473. 10.1126/science.aaw5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, G.M., Jalali A., Kircher D.A., Lee W.C., McQuade J.L., Haydu L.E., Joon A.Y., Reuben A., de Macedo M.P., Carapeto F.C.L., et al. 2019. Molecular profiling reveals unique immune and metabolic features of melanoma brain metastases. Cancer Discov. 9:628–645. 10.1158/2159-8290.CD-18-1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, J.U., and Bartel D.P.. 2016. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 353:aaf5371. 10.1126/science.aaf5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haq, R., Shoag J., Andreu-Perez P., Yokoyama S., Edelman H., Rowe G.C., Frederick D.T., Hurley A.D., Nellore A., Kung A.L., et al. 2013. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 23:302–315. 10.1016/j.ccr.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holohan, C., Van Schaeybroeck S., Longley D.B., and Johnston P.G.. 2013. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 13:714–726. 10.1038/nrc3599 [DOI] [PubMed] [Google Scholar]
- Jagust, P., de Luxán-Delgado B., Parejo-Alonso B., and Sancho P.. 2019. Metabolism-Based Therapeutic Strategies Targeting Cancer Stem Cells. Front. Pharmacol. 10:203. 10.3389/fphar.2019.00203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby-Arnon, L., Shah P., Cuoco M.S., Rodman C., Su M.J., Melms J.C., Leeson R., Kanodia A., Mei S., Lin J.R., et al. 2018. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell. 175:984–997.e24. 10.1016/j.cell.2018.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewer, M., Lee L., Leibovitch M., Zhang G., Liu J., Findlay S.D., Vincent K.M., Tandoc K., Dieters-Castator D., Quail D.F., et al. 2020. Translational control of breast cancer plasticity. Nat. Commun. 11:2498. 10.1038/s41467-020-16352-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalghatgi, S., Spina C.S., Costello J.C., Liesa M., Morones-Ramirez J.R., Slomovic S., Molina A., Shirihai O.S., and Collins J.J.. 2013. Bactericidal antibiotics induce mitochondrial dysfunction and oxidative damage in Mammalian cells. Sci. Transl. Med. 5:192ra85. 10.1126/scitranslmed.3006055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikin, O., D’Antonio L., and Bagga P.S.. 2006. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 34(Web Server):W676–W682. 10.1093/nar/gkl253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey, C.G., Camolotto S.A., Boespflug A.M., Guillen K.P., Foth M., Truong A., Schuman S.S., Shea J.E., Seipp M.T., Yap J.T., et al. 2019. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 25:620–627. 10.1038/s41591-019-0367-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, C.K., Marsico G., Sahakyan A.B., Chambers V.S., and Balasubramanian S.. 2016. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods. 13:841–844. 10.1038/nmeth.3965 [DOI] [PubMed] [Google Scholar]
- Lam, S.S., Martell J.D., Kamer K.J., Deerinck T.J., Ellisman M.H., Mootha V.K., and Ting A.Y.. 2015. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods. 12:51–54. 10.1038/nmeth.3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg, J., Kohlmeyer J., Renn M., Bald T., Rogava M., Cron M., Fatho M., Lennerz V., Wölfel T., Hölzel M., and Tüting T.. 2012. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature. 490:412–416. 10.1038/nature11538 [DOI] [PubMed] [Google Scholar]
- Larkin, J., Lao C.D., Urba W.J., McDermott D.F., Horak C., Jiang J., and Wolchok J.D.. 2015. Efficacy and Safety of Nivolumab in Patients With BRAF V600 Mutant and BRAF Wild-Type Advanced Melanoma: A Pooled Analysis of 4 Clinical Trials. JAMA Oncol. 1:433–440. 10.1001/jamaoncol.2015.1184 [DOI] [PubMed] [Google Scholar]
- Lawrence, S.K., Nguyen D., Bowen C., Richards-Peterson L., and Skordos K.W.. 2014. The metabolic drug-drug interaction profile of Dabrafenib: in vitro investigations and quantitative extrapolation of the P450-mediated DDI risk. Drug Metab. Dispos. 42:1180–1190. 10.1124/dmd.114.057778 [DOI] [PubMed] [Google Scholar]
- Lee, J.H., Shklovskaya E., Lim S.Y., Carlino M.S., Menzies A.M., Stewart A., Pedersen B., Irvine M., Alavi S., Yang J.Y.H., et al. 2020. Transcriptional downregulation of MHC class I and melanoma de- differentiation in resistance to PD-1 inhibition. Nat. Commun. 11:1897. 10.1038/s41467-020-15726-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson, V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.-L., Luke J.J., and Gajewski T.F.. 2018. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 359:104–108. 10.1126/science.aao3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConkey, D.J.2017. The integrated stress response and proteotoxicity in cancer therapy. Biochem. Biophys. Res. Commun. 482:450–453. 10.1016/j.bbrc.2016.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade, J.L., Daniel C.R., Helmink B.A., and Wargo J.A.. 2019. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 20:e77–e91. 10.1016/S1470-2045(18)30952-5 [DOI] [PubMed] [Google Scholar]
- Meeth, K., Wang J.X., Micevic G., Damsky W., and Bosenberg M.W.. 2016. The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res. 29:590–597. 10.1111/pcmr.12498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, A., Kim Y.J., Robert L., Tsoi J., Comin-Anduix B., Berent-Maoz B., Cochran A.J., Economou J.S., Tumeh P.C., Puig-Saus C., and Ribas A.. 2018. Immunotherapy resistance by inflammation-induced dedifferentiation. Cancer Discov. 8:935–943. 10.1158/2159-8290.CD-17-1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min, M., and Spencer S.L.. 2019. Spontaneously slow-cycling subpopulations of human cells originate from activation of stress-response pathways. PLoS Biol. 17:e3000178. 10.1371/journal.pbio.3000178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moullan, N., Mouchiroud L., Wang X., Ryu D., Williams E.G., Mottis A., Jovaisaite V., Frochaux M.V., Quiros P.M., Deplancke B., et al. 2015. Tetracyclines Disturb Mitochondrial Function across Eukaryotic Models: A Call for Caution in Biomedical Research. Cell Rep. 10:1681–1691. 10.1016/j.celrep.2015.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau, R., Sörgel F., and Eiffert H.. 2010. Penetration of drugs through the blood-cerebrospinal fluid/blood-brain barrier for treatment of central nervous system infections. Clin. Microbiol. Rev. 23:858–883. 10.1128/CMR.00007-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha, R., Leli N.M., Onorati A., Piao S., Verginadis I.I., Tameire F., Rebecca V.W., Chude C.I., Murugan S., Fennelly C., et al. 2019. Er translocation of the mapk pathway drives therapy resistance in BRAF-mutant melanoma. Cancer Discov. 9:396–415. 10.1158/2159-8290.CD-18-0348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, E.E., Mueller K.L., Adams D.J., Anandasabapathy N., Aplin A.E., Bertolotto C., Bosenberg M., Ceol C.J., Burd C.E., Chi P., et al. 2021. Melanoma models for the next generation of therapies. Cancer Cell. 39:610–631. 10.1016/j.ccell.2021.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poźniak, J., Nsengimana J., Laye J.P., O’Shea S.J., Diaz J.M.S., Droop A.P., Filia A., Harland M., Davies J.R., Mell T., et al. 2019. Genetic and environmental determinants of immune response to cutaneous melanoma. Cancer Res. 79:2684–2696. 10.1158/0008-5472.CAN-18-2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushpakom, S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C., et al. 2019. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 18:41–58. 10.1038/nrd.2018.168 [DOI] [PubMed] [Google Scholar]
- Quirós, P.M., Prado M.A., Zamboni N., D’Amico D., Williams R.W., Finley D., Gygi S.P., and Auwerx J.. 2017. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216:2027–2045. 10.1083/jcb.201702058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambow, F., Rogiers A., Marin-Bejar O., Aibar S., Femel J., Dewaele M., Karras P., Brown D., Chang Y.H., Debiec-Rychter M., et al. 2018. Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell. 174:843–855.e19. 10.1016/j.cell.2018.06.025 [DOI] [PubMed] [Google Scholar]
- Rambow, F., Marine J.C., and Goding C.R.. 2019. Melanoma plasticity and phenotypic diversity: therapeutic barriers and opportunities. Genes Dev. 33:1295–1318. 10.1101/gad.329771.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravà, M., D’Andrea A., Nicoli P., Gritti I., Donati G., Doni M., Giorgio M., Olivero D., and Amati B.. 2018. Therapeutic synergy between tigecycline and venetoclax in a preclinical model of MYC/BCL2 double-hit B cell lymphoma. Sci. Transl. Med. 10:eaan8723. 10.1126/scitranslmed.aan8723 [DOI] [PubMed] [Google Scholar]
- Ravindran Menon, D., Das S., Krepler C., Vultur A., Rinner B., Schauer S., Kashofer K., Wagner K., Zhang G., Bonyadi Rad E., et al. 2015. A stress-induced early innate response causes multidrug tolerance in melanoma. Oncogene. 34:4448–4459. 10.1038/onc.2014.372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, G.A., Schiller G.J., Kambhampati S., Tallman M.S., Douer D., Minden M.D., Yee K.W., Gupta V., Brandwein J., Jitkova Y., et al. 2016. A Phase 1 study of intravenous infusions of tigecycline in patients with acute myeloid leukemia. Cancer Med. 5:3031–3040. 10.1002/cam4.845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter-Dennerlein, R., Oeljeklaus S., Lorenzi I., Ronsör C., Bareth B., Schendzielorz A.B., Wang C., Warscheid B., Rehling P., and Dennerlein S.. 2016. Mitochondrial Protein Synthesis Adapts to Influx of Nuclear-Encoded Protein. Cell. 167:471–483.e10. 10.1016/j.cell.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch, A., Vultur A., Bogeski I., Wang H., Zimmermann K.M., Speicher D., Körbel C., Laschke M.W., Gimotty P.A., Philipp S.E., et al. 2013. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 23:811–825. 10.1016/j.ccr.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzymski, T., Milani M., Singleton D.C., and Harris A.L.. 2009. Role of ATF4 in regulation of autophagy and resistance to drugs and hypoxia. Cell Cycle. 8:3838–3847. 10.4161/cc.8.23.10086 [DOI] [PubMed] [Google Scholar]
- Schmidt, E.K., Clavarino G., Ceppi M., and Pierre P.. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 6:275–277. 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- Sharma, S.V., Lee D.Y., Li B., Quinlan M.P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M.A., et al. 2010. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 141:69–80. 10.1016/j.cell.2010.02.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon, D., Cathelin S., Mirali S., Di Trani J.M., Yanofsky D.J., Keon K.A., Rubinstein J.L., Schimmer A.D., Ketela T., and Chan S.M.. 2019. Inhibition of mitochondrial translation overcomes venetoclax resistance in AML through activation of the integrated stress response. Sci. Transl. Med. 11:eaax2863. 10.1126/scitranslmed.aax2863 [DOI] [PubMed] [Google Scholar]
- Shen, S., Faouzi S., Bastide A., Martineau S., Malka-Mahieu H., Fu Y., Sun X., Mateus C., Routier E., Roy S., et al. 2019. An epitranscriptomic mechanism underlies selective mRNA translation remodelling in melanoma persister cells. Nat. Commun. 10:5713. 10.1038/s41467-019-13360-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Škrtić, M., Sriskanthadevan S., Jhas B., Gebbia M., Wang X., Wang Z., Hurren R., Jitkova Y., Gronda M., Maclean N., et al. 2011. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 20:674–688. 10.1016/j.ccr.2011.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosman, J.A., Kim K.B., Schuchter L., Gonzalez R., Pavlick A.C., Weber J.S., McArthur G.A., Hutson T.E., Moschos S.J., Flaherty K.T., et al. 2012. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N. Engl. J. Med. 366:707–714. 10.1056/NEJMoa1112302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y., Wei W., Robert L., Xue M., Tsoi J., Garcia-Diaz A., Homet Moreno B., Kim J., Ng R.H., Lee J.W., et al. 2017. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc. Natl. Acad. Sci. USA. 114:13679–13684. 10.1073/pnas.1712064115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundstrøm, T., Prestegarden L., Azuaje F., Aasen S.N., Røsland G.V., Varughese J.K., Bahador M., Bernatz S., Braun Y., Harter P.N., et al. 2019. Inhibition of mitochondrial respiration prevents BRAF-mutant melanoma brain metastasis. Acta Neuropathol. Commun. 7:55. 10.1186/s40478-019-0712-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, K.R., Magill A.J., Parise M.E., and Arguin P.M.. Centers for Disease Control and Prevention . 2011. Doxycycline for malaria chemoprophylaxis and treatment: report from the CDC expert meeting on malaria chemoprophylaxis. Am. J. Trop. Med. Hyg. 84:517–531. 10.4269/ajtmh.2011.10-0285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoi, J., Robert L., Paraiso K., Galvan C., Sheu K.M., Lay J., Wong D.J.L., Atefi M., Shirazi R., Wang X., et al. 2018. Multi-stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell. 33:890–904.e5. 10.1016/j.ccell.2018.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyden, Y., Goedert L., and Leucci E.. 2019. Control of nucleolar stress and translational reprogramming by lncRNAs. Cell Stress. 3:19–26. 10.15698/cst2019.01.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Perry C.J., Meeth K., Thakral D., Damsky W., Micevic G., Kaech S., Blenman K., and Bosenberg M.. 2017. UV-induced somatic mutations elicit a functional T cell response in the YUMMER1.7 mouse melanoma model. Pigment Cell Melanoma Res. 30:428–435. 10.1111/pcmr.12591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring, M.J., Arrowsmith J., Leach A.R., Leeson P.D., Mandrell S., Owen R.M., Pairaudeau G., Pennie W.D., Pickett S.D., Wang J., et al. 2015. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 14:475–486. 10.1038/nrd4609 [DOI] [PubMed] [Google Scholar]
- Weinberg, S.E., and Chandel N.S.. 2015. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 11:9–15. 10.1038/nchembio.1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi, M., Toshima T., Amamoto R., Do Y., Hirai H., Setoyama D., Kang D., and Uchiumi T.. 2021. Mitochondrial translation deficiency impairs NAD+ -mediated lysosomal acidification. EMBO J. 40:e105268. 10.15252/embj.2020105268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Xu L., Zhang F., and Vlashi E.. 2017. Doxycycline inhibits the cancer stem cell phenotype and epithelial-to-mesenchymal transition in breast cancer. Cell Cycle. 16:737–745. 10.1080/15384101.2016.1241929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists primers used in the study.