Abstract
Objective
To construct a polygenic risk score (PRS) for stroke and evaluate its utility in risk stratification and primary prevention for stroke.
Methods
Using a meta-analytic approach and large genome-wide association results for stroke and stroke-related traits in East Asians, we generated a combined PRS (metaPRS) by incorporating 534 genetic variants in a training set of 2,872 patients with stroke and 2,494 controls. We then validated its association with incident stroke using Cox regression models in large Chinese population-based prospective cohorts comprising 41,006 individuals.
Results
During a total of 367,750 person-years (mean follow-up 9.0 years), 1,227 participants developed stroke before age 80 years. Individuals with high polygenic risk had an about 2-fold higher risk of incident stroke compared with those with low polygenic risk (hazard ratio [HR] 1.99, 95% confidence interval [CI] 1.66–2.38), with the lifetime risk of stroke being 25.2% (95% CI 22.5%–27.7%) and 13.6% (95% CI 11.6%–15.5%), respectively. Individuals with both high polygenic risk and family history displayed lifetime risk as high as 41.1% (95% CI 31.4%–49.5%). Individuals with high polygenic risk achieved greater benefits in terms of absolute risk reductions from adherence to ideal fasting blood glucose and total cholesterol than those with low polygenic risk. Maintaining favorable cardiovascular health (CVH) profile could substantially mitigate the increased risk conferred by high polygenic risk to the level of low polygenic risk (from 34.6% to 13.2%).
Conclusions
Our metaPRS has great potential for risk stratification of stroke and identification of individuals who may benefit more from maintaining ideal CVH.
Classification of Evidence
This study provides Class I evidence that metaPRS is predictive of stroke risk.
Deaths due to stroke are expanding threats to global health. In China, stroke is the leading cause of death, accounting for 2.07 million deaths in 2017.1 Thus, early identification of high-risk individuals and advocation of healthy lifestyles or drug interventions are essential for primary prevention of stroke in China and worldwide.
Stroke is a complex disorder caused by both genetic and environmental factors. Genome-wide association studies (GWAS) have identified 42 genetic loci associated with stroke2,3 and hundreds of genetic loci associated with a range of stroke-related traits including blood pressure (BP), type 2 diabetes (T2D), lipids, body mass index (BMI), and atrial fibrillation (AF).4 The identification of these genetic variants provides an opportunity to evaluate whether genetic risk could lead to meaningful improvement in future risk prediction and stratification. Recently, polygenic risk scores (PRS) for stroke have been developed and evaluated in clinical utility in European-descent populations.5-10 However, their applicability to the Chinese population is questionable. Epidemiologic features of stroke vary substantially across countries, with much higher stroke incidence and proportion of hemorrhagic stroke in China than in Western populations.11 Marked differences in clinical risk factors and genetic background across populations may also affect the variability of stroke risk. Thus, it is of utmost importance to generate PRS for stroke in the Chinese population.
We constructed a combined PRS (metaPRS) for stroke by incorporating large-scale GWAS summary statistics from East Asian ancestry and then validated its utility in risk stratification and primary prevention for stroke in large prospective population-based cohorts.
Methods
The overall study design and workflow are given in figure 1.
Figure 1. Study Design and Workflow.
AF = atrial fibrillation; AUC = area under the receiver operating characteristic curve; BMI = body mass index; CAD = coronary artery disease; DBP = diastolic blood pressure; GWAS = genome-wide association study; HDL-C = high-density lipoprotein cholesterol; LDL-C = low-density lipoprotein cholesterol; MAP = mean arterial pressure; OR = odds ratio; PP = pulse pressure; PRS = polygenic risk score; SBP = systolic blood pressure; SNP = single nucleotide polymorphism; T2D = type 2 diabetes; TC = total cholesterol; TG = triglycerides; WC = waist circumference.
Study Population
We used a training set of case–control design to develop the metaPRS and externally validated its association with incident stroke in a large prospective cohort, the China-PAR project (Prediction for Atherosclerotic Cardiovascular Disease Risk in China). The training set consisted of 2,872 patients with stroke (2,548 ischemic and 324 hemorrhagic stroke) and 2,494 controls. Inpatients who were admitted with first-time acute stroke were recruited from local hospitals in 3 provinces (Jiangsu, Henan, and Shandong) in China between 2012 and 2017. All patients had a validated diagnosis of stroke and disease status was confirmed by a neurologist according to medical records of CT or MRI.12 Controls were randomly selected from individuals participating in a community-based survey of cardiovascular risk factors between 2013 and 2017 who were judged to be free of stroke by history, clinical examination, and self-report questionnaires.
Validation populations were from 3 cohorts of the China-PAR project: China Multicenter Collaborative Study of Cardiovascular Epidemiology 1998 (China MUCA 1998), International Collaborative Study of Cardiovascular Disease in Asia (InterASIA), and Community Intervention of Metabolic Syndrome in China & Chinese Family Health Study (CIMIC). Establishment and maintenance of these cohorts have been described in detail elsewhere.13 Briefly, the China MUCA 1998, InterASIA, and CIMIC were established in 1998, 2000–2001, and 2007–2008, respectively. According to unified questionnaires and laboratory protocols, both InterASIA and China MUCA 1998 cohorts were first followed up during 2007–2008, and all 3 cohorts were followed up in 2012–2015. Among a total of 43,881 participants from 3 cohorts with available blood samples and follow-up information, we further excluded 561 participants because of high genotype missing rate (>5.0%) or low mean sequencing depth (<30×), 1,352 participants aged <30 years or >75 years at baseline, and 962 participants with prevalent cardiovascular disease (stroke or myocardial infarction), leaving 41,006 participants for the current analysis.
Assessment of Major Risk Factors and Cardiovascular Health Metrics
At the baseline survey, participants were interviewed and underwent physical examinations and laboratory tests. A series of lifestyle risk factors and cardiometabolic conditions were collected by well-trained investigators according to standard protocols. We defined major stroke risk factors at baseline, including hypertension, dyslipidemia, diabetes, obesity (BMI ≥28 kg/m2), and family history of stroke. Hypertension was defined as systolic BP (SBP) ≥140 mm Hg or diastolic BP (DBP) ≥90 mm Hg or use of antihypertensive medication within the past 2 weeks. Dyslipidemia was defined as total cholesterol (TC) ≥240 mg/dL or high-density lipoprotein cholesterol (HDL-C) <40 mg/dL or triglycerides ≥200 mg/dL or low-density lipoprotein cholesterol (LDL-C) ≥160 mg/dL or on lipid-lowering medication. Diabetes was defined as fasting blood glucose (FBG) ≥126 mg/dL or the use of insulin or oral hypoglycemic agents. Family history of stroke was defined as any first-degree relative (father, mother, or sibling) with stroke according to the baseline questionnaire.
Seven cardiovascular health (CVH) metrics at baseline were also defined based on the American Heart Association (AHA) guidelines comprising 4 ideal cardiometabolic measurements (untreated TC <200 mg/dL, untreated SBP <120 mm Hg and DBP <80 mm Hg, untreated FBG <100 mg/dL, and BMI <24 kg/m2) and 3 ideal health behaviors (no current smoking, healthy diet, and moderate physical activity) (eTable 1 available from Dryad at doi.org/10.5061/dryad.zs7h44j7q).14 A modified healthy diet score for Chinese people was defined in accordance with AHA's recommended diet goals and the recent Chinese Guideline on Healthy Lifestyle to Prevent Cardiometabolic Diseases.15,16
Ascertainment of Incident Stroke Events
We collected information on incident stroke by interviewing participants or their proxies during the follow-up period and obtained medical records and death certificates for verification. For survival participants, we enquired information on the symptoms of diseases and medical examinations and treatments. For fatal cases, information on symptoms and medical examinations and treatments before death was also obtained. Copies of death certificates were acquired from the respondents, hospitals, or local administrations. Local investigators initially recorded fatal and nonfatal stroke events. All medical and death records were reviewed by the central adjudication committee at Fuwai Hospital (Beijing, China) to make the final diagnosis. Two committee members checked events independently, and their differences were discussed with additional committee members to conclude the final diagnosis. Causes of death were coded according to ICD-10. We defined incident stroke as a confirmed diagnosis of first-ever fatal or nonfatal stroke event during follow-up (I60–I69). Subtypes of stroke were classified as ischemic stroke (I63), hemorrhagic stroke (I60–I62), and unspecified stroke (I64–I69). Only first-ever stroke events were considered in the time to event analyses.
SNPs Selection and Genotyping
We selected 588 single-nucleotide polymorphisms (SNPs) that were of genome-wide significance in association with stroke (n = 42) and a range of stroke-related traits defined as main stroke risk factors including BP (n = 46), T2D (n = 89), lipids (n = 126), obesity (n = 79), and AF (n = 16) based on previous GWASs (eTable 2 and eTable 3). SNPs associated with coronary artery disease (CAD) (n = 199) were also included due to the shared etiologic pathways between stroke and CAD. We selected all the SNPs associated with stroke and CAD reported by either East Asian or European populations due to the limited loci identified in the East Asian population. For other traits, we mainly focused on the reported SNPs in East Asian ancestry.
Participants from both the training and validation set were genotyped using multiplex PCR targeted amplicon sequencing technology. We designed multiplexed primers targeting each SNP and amplified the target regions for high-throughput sequencing with Illumina Hiseq X10 sequencer. After excluding 10 SNPs with genotype call rate of <95%, 578 autosomal SNPs remained for subsequent analysis with call rate of 99.9% and median sequencing depth of 979× (eFigure 1 available from Dryad at doi.org/10.5061/dryad.zs7h44j7q). To evaluate genotyping reproducibility, 1,648 duplicate samples were genotyped, and the concordance rate was determined to be >99.4%.
Construction of the metaPRS
We generated specific PRS for each of 14 traits (stroke, CAD, T2D, AF, SBP, DBP, mean arterial pressure, pulse pressure, BMI, waist circumference, TC, LDL-C, triglycerides, and HDL-C) by adding the number of corresponding risk alleles (0/1/2) weighted by the effect size of each SNP on each trait. The effect sizes were extracted from large-scale GWASs for each trait conducted among East Asian populations and those containing Chinese samples were selected in priority if more than one GWAS was reported. Details of utilized GWAS studies are available in eTable 4. For example, the effect sizes for stroke were extracted from summary statistics of the largest GWAS conducted by Biobank Japan.17 For each trait, we generated 16 candidate PRS using different linkage disequilibrium (LD) metric r2 (0.2, 0.4, 0.6, 0.8) and significance thresholds (p value = 0.5, 0.05, 5 × 10−4, 5 × 10−6) based on trait-specific summary statistics. We estimated the r2 between SNPs based on the genotype data of our population-based cohorts using LD clumping procedure in PLINK version 1.9. The score with the largest odds ratio (OR) (per SD increase of PRS) for stroke estimated from a logistic regression model in the training set was selected as the optimal trait-specific PRS (eFigure 2).
Each optimal PRS was standardized to zero mean and unit SD. We employed the elastic-net logistic regression with 10-fold cross-validation using the R package “glmnet” to model the associations between the 14 optimal PRS and stroke and further to generate the metaPRS.18 This method has been used to construct PRS for stroke with consideration of correlation between distinct PRS.9 The best model with the highest cross-validated area under the receiver operating characteristic curve was selected as the final model, from which the adjusted coefficients ( ) for each PRS were obtained as weights. The final adjusted ORs for each PRS in the elastic-net logistic regression are shown in eFigure 3, in comparison with the univariate estimates (based on 1 PRS at a time). The metaPRS can be calculated as a weighted sum by converting PRS-level weights to SNP-level weights:
) for each PRS were obtained as weights. The final adjusted ORs for each PRS in the elastic-net logistic regression are shown in eFigure 3, in comparison with the univariate estimates (based on 1 PRS at a time). The metaPRS can be calculated as a weighted sum by converting PRS-level weights to SNP-level weights:
 |
where  ,…
,… are the SDs of each PRS in the training set,
are the SDs of each PRS in the training set,  are the SNP effect sizes (obtained from GWAS summary statistics) for the
are the SNP effect sizes (obtained from GWAS summary statistics) for the  th SNP in each PRS, and
th SNP in each PRS, and  is the genotype for the
is the genotype for the  th individual's
th individual's  th SNP. An SNP's effect size
th SNP. An SNP's effect size  was set to zero for the
was set to zero for the 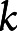 th score if the SNP was not included in that score. These procedures resulted in 534 SNPs finally included in the metaPRS. The information and weights of eligible SNPs are available in eTable 3.
th score if the SNP was not included in that score. These procedures resulted in 534 SNPs finally included in the metaPRS. The information and weights of eligible SNPs are available in eTable 3.
Statistical Analysis
Baseline characteristics of participants were described as means and SDs for continuous variables or percentages for categorical variables. The polygenic risk was categorized into low (lowest quintile of metaPRS), intermediate (2–4 quintiles of metaPRS), and high (highest quintile of metaPRS). Clinical risk score was evaluated with the China-PAR 10-year stroke risk equation comprising age, treated or untreated SBP, TC, HDL-C, current smoking, diabetes, waist circumference, geographic region, urbanization, and family history of stroke, as well as available interaction terms for age with risk factors that met predefined statistical criteria.19 CVH profiles were classified as favorable (6–7 ideal CVH metrics), intermediate (4–5 ideal CVH metrics), and unfavorable (0–3 ideal CVH metrics).20
Age-as-time-scale and stratified Cox proportional hazards regression models with strata defined as cohorts were used to estimate the hazard ratios (HRs) and 95% confidence intervals (CIs) for incident stroke associated with polygenic risk, major risk factors, and CVH metrics after adjustment of sex.21,22 We also evaluated the lifetime risk of incident stroke by age 80 years across polygenic risk, major risk factors, and CVH metrics by plotting sex-adjusted cumulative incidence curves using “survfit.coxph” (R package “survival”). A sensitive analysis using proportional subdistribution hazards regression models accounting for competing risk (3 states: stroke, nonstroke death, censored) was conducted to estimate the HRs and cumulative incidence of stroke associated with polygenic risk. C-index, net reclassification improvement (NRI), and integrated discrimination improvement (IDI) were used to evaluate risk predictions from models containing clinical risk score and plus metaPRS.23,24 We defined the absolute risk reductions (ARR) as the difference of the lifetime risk values between nonideal and ideal CVH groups. We used weighted least-squares models to assess the increasing trend in ARR across polygenic risk categories.25 We further applied relative excess risk due to interaction (RERI) to assess additive interaction between CVH metrics and polygenic risk.26 Bonferroni correction was applied to adjust for multiple testing. We considered 2-sided p values less than 0.007 (p value of less than 0.05 divided by the number of tests, i.e., 0.05/7) statistically significant when evaluating the 7 CVH metrics separately. All analyses were performed using R version 3.6.0 (R Foundation for Statistical Computing) or SAS statistical package, version 9.4 (SAS Institute Inc.).
Primary Research Question/Classification of Evidence
Could a PRS for stroke predict stroke incidence and guide primary prevention for stroke in a Chinese population? This study provides Class I evidence that a metaPRS is predictive of stroke risk.
Standard Protocol Approvals, Registrations, and Patient Consents
These studies were all approved by the institutional review board at Fuwai Hospital in Beijing. Written informed consent was obtained from each participant prior to data collection.
Data Availability
All data generated or analyzed during this study are included in this article and supplemental data.
Results
Study Population and the metaPRS
The training set consisted of 2,872 patients with stroke and 2,494 controls (eTable 5). The mean (SD) age of patients with stroke was 66.6 (9.8) years, which was similar to that of controls of 66.1 (10.3) years. As expected, patients with stroke had higher prevalence of hypertension, diabetes, and dyslipidemia than that of controls. Baseline characteristics for overall 41,006 participants in the validation prospective cohorts are presented in table 1. The mean (SD) age of participants at baseline was 51.9 (10.6) years, and 43.1% were male. Participants with high polygenic risk tended to have higher prevalence of cardiometabolic risk factors (hypertension, diabetes, dyslipidemia) and larger proportion of unfavorable CVH profiles. During a total of 367,750 person-years of follow-up (mean follow-up 9.0 years), 1,227 participants developed stroke before age 80 years, including 769 ischemic stroke, 355 hemorrhagic stroke, 21 ischemic stroke complicated with hemorrhagic stroke, and 124 unspecified subtype.
Table 1.
Baseline Characteristics of the Prospective Cohorts
We generated the stroke PRS using summary statistics derived from different ancestries and further compared their association magnitude with stroke in the training set. We observed that the ORs decayed markedly when using those from European population2 compared with those from East Asian population17 (eFigure 4). Also using effect sizes from East Asian population, we constructed another 13 optimal trait-specific PRS, which were correlated with each other to different degrees (eFigure 5). Finally, the metaPRS of stroke was generated by integrating 14 trait-specific PRS. In the prospective cohorts, the metaPRS had the most pronounced association with stroke than any other trait-specific PRS, and the HRs (95% CIs) for all stroke, ischemic stroke, and hemorrhagic stroke were 1.28 (1.21–1.36), 1.29 (1.20–1.39), and 1.30 (1.17–1.45) per SD increment, respectively (eFigure 6). The HRs of the metaPRS with incident stroke were only modestly attenuated after adjustment of major risk factors, including family history of stroke (eTable 6).
Lifetime Risk of Stroke Stratified by Polygenic Risk and Major Risk Factors
The metaPRS provided a significant gradient of stroke risk stratification across quintiles of metaPRS (p trend <0.001) (eFigure 7). Compared with participants with low polygenic risk (lowest quintile of metaPRS), those with high polygenic risk (highest quintile of metaPRS) had an about 2-fold higher risk of incident stroke (HR 1.99, 95% CI 1.66–2.38, p = 1.11 × 10−13) (figure 2). Individuals with high polygenic risk also had nearly 2-fold higher lifetime risk of stroke than those having low polygenic risk (25.2%, 95% CI 22.5%–27.7% vs 13.6%, 95% CI 11.6%–15.5%). The lifetime risk of stroke for 3 polygenic risk categories was only modestly attenuated after accounting for competing risk (eFigure 8). All the 5 major risk factors were significantly associated with incident stroke, with HRs (95% CIs) of 1.87 (1.56–2.23), 2.58 (2.29–2.91), 1.83 (1.53–2.19), 1.73 (1.50–2.01), and 1.62 (1.44–1.81), for family history of stroke, hypertension, diabetes, obesity, and dyslipidemia, respectively (eTable 7). There was marked variability in the lifetime risk of stroke across both polygenic risk categories and major risk factors. For example, the lifetime risk of stroke was 13.2% (95% CI 11.1%–15.1%) among individuals with low polygenic risk and without family history, whereas having either of the 2 risk factors presented almost the same lifetime risk (23.9%, 95% CI 21.1%–26.5% and 23.7%, 95% CI 13.4%–32.8%, respectively), and the presence of both could obtain the lifetime risk of stroke as high as 41.1% (95% CI 31.4%–49.5%) (figure 3). Substantial gradients of stroke risk were also observed within strata of polygenic and other 4 major risk factors (eFigure 9).
Figure 2. Lifetime Risk of Stroke Stratified by Polygenic Risk.

Age-as-time-scale and stratified Cox proportional hazards models with strata defined as cohorts were used to estimate the hazard ratios (HRs) (95% confidence intervals) and cumulative incidence curves of stroke by age 80 years adjusting for sex. PRS = polygenic risk score.
Figure 3. Lifetime Risk of Stroke According to Polygenic Risk and Family History of Stroke.

Age-as-time-scale and stratified Cox proportional hazards models with strata defined as cohorts were used to estimate the hazard ratios (HRs) (95% confidence intervals) and cumulative incidence curves of stroke by age of 80 years adjusting for sex. PRS = polygenic risk score.
We also evaluated improvements in clinical risk stratification when adding the metaPRS to clinical risk score, namely the China-PAR equation.19 The addition of the metaPRS to the China-PAR equation increased the C-index from 0.781 to 0.783 (difference, 0.2%; p = 0.004). Adding metaPRS to China-PAR equation improved reclassification with NRI of 2.34% (95% CI 0.56%–4.11%) and IDI of 0.10% (0.04%–0.16%) (eTable 8).
The Benefit of Ideal Cardiovascular Health Across Polygenic Risk Categories
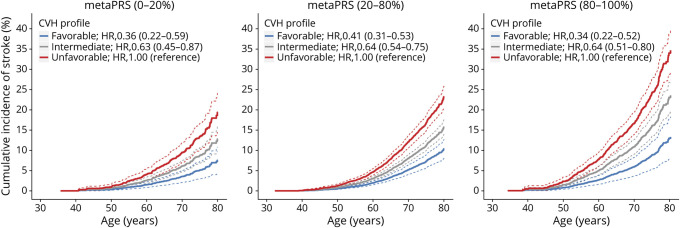
We then investigated to what extent the increased risk caused by high polygenic risk could be offset by maintaining ideal CVH individually or jointly. Among the 7 CVH metrics, adherence to ideal BP and FBG displayed the strongest protective effects, with HRs (95% CIs) of 0.39 (0.33–0.46) and 0.67 (0.59–0.75), respectively. Moreover, there were significant interactions between polygenic risk and ideal FBG or TC (p for interaction = 0.003 and 0.002, respectively) (eFigure 10). In terms of the ARRs, we noted significant gradients of benefits from 2 ideal cardiometabolic measurements (FBG and TC) across the low, intermediate, and high polygenic risk categories (both p for trend <0.007) (figure 4). When assessing the benefits of ideal CVH jointly, participants with favorable CVH (6–7 ideal metrics) in comparison with unfavorable CVH profiles showed 64%, 59%, and 66% lower risk of stroke in low, intermediate, and high polygenic risk categories, respectively (figure 5). In terms of ARRs of the lifetime risk, individuals with high polygenic risk (from 34.6% to 13.2%, ARR = 21.4%) received a 1.81-fold greater absolute benefit from favorable CVH than those with low polygenic risk (from 19.4% to 7.6%, ARR = 11.8%), substantially mitigating the increased risk in the high polygenic risk category to the level of the low polygenic risk category (eFigure 11).
Figure 4. Absolute Risk Reductions of Stroke From 4 Ideal Cardiometabolic Measurements Across Polygenic Risk Categories.
(A–D) Age-as-time-scale and stratified Cox proportional hazards models with strata defined as cohorts were used to estimate cumulative incidence of stroke by age of 80 years adjusting for sex. *p < 0.007 after Bonferroni correction. ARR = absolute risk reduction.
Figure 5. Relative and Absolute Risk of Stroke According to Polygenic Risk and Cardiovascular Health (CVH) Profile.
Age-as-time-scale and stratified Cox proportional hazards models with strata defined as cohorts were used to estimate the hazard ratios (HRs) (95% confidence intervals) and cumulative incidence curves of stroke by age of 80 years adjusting for sex. Favorable CVH = 6–7 ideal metrics; intermediate CVH = 4–5 ideal metrics; unfavorable CVH = 0–3 ideal metrics. PRS = polygenic risk score.
We repeated the analyses to test the associations of the metaPRS, major risk factors, and CVH profiles with incident stroke while restricting only to ischemic stroke or hemorrhagic stroke. The association patterns were similar for both stroke subtypes (eFigure 12 to eFigure 14).
Discussion
We developed and evaluated a metaPRS for stroke risk prediction in combination with the major clinical risk factors and CVH profile in Chinese population based on large prospective cohorts. The metaPRS alone or in combination with major clinical risk factors could substantially stratify individuals into different lifetime risk of stroke. Moreover, individuals with high polygenic risk derived greater benefit from ideal CVH metrics. Maintaining a favorable CVH profile could almost offset the high polygenic risk.
There is a large difference in the incidence rate of stroke between the Chinese and European populations (i.e., 333.7 per 100,000 person-years in our cohorts vs 97.1 per 100,000 person-years in UK Biobank).8 It is urgent and effective to optimize individualized stroke risk assessment by incorporating genetic information in the Chinese population. By integrating all genetic information from 14 stroke and stroke-related traits, the metaPRS we constructed was comparable to the genomic risk score derived from 3.2 million variants in the UK Biobank population,9 showing associations with ischemic stroke with HRs (95% CIs) of 1.29 (1.20–1.39) vs 1.26 (1.22–1.31) per SD increment of PRS, respectively. Our study also demonstrated that the prediction performance of stroke PRS highly depended on ancestry-matched summary statistics and highlighted the importance of constructing ancestry-specific PRS.27,28 More recently, a cohort study (3,038 individuals with 91 stroke events) from Japan also constructed a PRS for stroke and suggested that PRS may be useful for risk stratification.29 In comparison with this report, the current study demonstrated a more stable and substantial gradient of the risk for stroke, including both ischemic stroke and hemorrhagic stroke, across quintiles of polygenic risk. Our metaPRS seems to work equally well for hemorrhagic stroke as for ischemic stroke, probably due to integrating the overlapping clinical risk factors and the potential shared genetic pathways.3 Considering the distinct etiologic mechanism of stroke subtypes, it is necessary to develop PRS specific for hemorrhagic stroke in future studies when East Asian ancestry-specific genomic data are available.
Our large-scale prospective cohorts also allowed us to comprehensively estimate the absolute risk for incident stroke across polygenic risk categories, as well as in combination with major clinical risk factors. Individuals with high polygenic risk had nearly 2-fold higher risk of stroke than those with low polygenic risk, displaying similar lifetime risk to individuals with family history of stroke. That is, the polygenic risk could individually identify 20% of the general population early in life with equivalent stroke risk contributed by family history of stroke. On the other hand, high polygenic risk in combination with family history could identify individuals who had lifetime risk of more than 40.0%. The incorporation of the genetic risk and family history may be particularly useful to guide early life risk stratification for stroke. The addition of the metaPRS for stroke to clinical risk score was associated with a statistically significant, yet modest, improvement in the predictive accuracy and risk stratification for incident stroke. These results were consistent with previous studies related to the PRS of CAD developed in the European population.30,31 The potential advantage of the PRS, compared with clinical risk factors, is that it can be obtained by a one-off assessment from birth. Further studies are warranted to evaluate the feasibility and cost-effectiveness of implementing the PRS in clinical practice to guide earlier and more targeted prevention.
It has been found that the reduction of stroke risk by adhering to a healthy lifestyle was similar across all genetic risk strata in UK Biobank.8,10 However, the authors did not assess the benefits of adhering to the 3 ideal cardiometabolic measurements (BP, TC, and FBG). Our results demonstrated that maintaining the ideal cardiometabolic measurements could result in higher compensatory effects on stroke risk than the lifestyle factors. This means that congenital genetic high risk would be mitigated to some degree by maintaining ideal CVH, with a preference for the cardiometabolic measurements. Achieving ideal BP level rather than nonhypertension is the most noteworthy preventive measure for entire populations, whereas those with high polygenic risk derived greater benefit from achieving ideal FBG or TC level than those with low polygenic risk. It is encouraging that maintaining 6–7 ideal CVH metrics could almost offset the increased lifetime risk conferred by high polygenic risk. Given low proportions of ideal cardiovascular health globally32-34 and low control rate of major stroke risk factors (hypertension, T2D, dyslipidemia) in China,35 awareness of the high genetic risk may represent a future opportunity to efficiently improve risk factor management and primary prevention of stroke. Our results also emphasize that genetic risk assessment could be used to identify individuals at elevated risk who could benefit most from lifestyle changes or preventive treatment, highlighting great clinical implications for primary prevention.
Our study has several strengths, including the large sample size of prospective cohorts with well-defined phenotypes and high-quality outcomes, which enabled the rigorous assessment of genetic risk in combination with clinical risk factors and CVH profiles. Furthermore, we developed a PRS proved useful in Chinese population by integrating current large GWAS of stoke and stroke-related traits of East Asian ancestry. Nevertheless, several limitations should be noted. First, our metaPRS did not include all the variants that underlie stroke risk, which might underestimate the true effect. Further studies that construct PRS for stroke by capturing a full spectrum of genomic variants would provide additional gains in risk prediction and stratification. Second, lifestyle factors were self-reported and changes in health behavioral factors over time were not considered. However, possible measurement and classification errors are likely biased toward the null. Third, stratifying participants using both genetic and clinical risk factors resulted in relatively small samples in each stratum with wide CIs around point estimates. Independent validation of our analysis in other studies is required. Finally, this sample was restricted to the Chinese population. Therefore, further research is warranted to investigate to what degree these findings generalize to other East Asian populations.
We developed a metaPRS for stroke with good performance in stroke prediction and risk stratification, which has potential to identify individuals who would benefit most from lifestyle changes or preventive treatment. The practical applications of polygenic risk information for guiding primary prevention in the clinical setting remain to be defined in further studies.
Acknowledgment
The authors thank the Biobank Japan Project and Asian Genetic Epidemiology Network consortium for access to summary statistics data.
Glossary
- AF
atrial fibrillation
- AHA
American Heart Association
- ARR
absolute risk reduction
- BMI
body mass index
- BP
blood pressure
- CAD
coronary artery disease
- China MUCA 1998
China Multicenter Collaborative Study of Cardiovascular Epidemiology 1998
- China-PAR project
Prediction for Atherosclerotic Cardiovascular Disease Risk in China
- CI
confidence interval
- CIMIC
Community Intervention of Metabolic Syndrome in China & Chinese Family Health Study
- CVH
cardiovascular health
- DBP
diastolic blood pressure
- FBG
fasting blood glucose
- GWAS
genome-wide association studies
- HDL-C
high-density lipoprotein cholesterol
- HR
hazard ratio
- ICD-10
International Classification of Diseases, 10th revision
- IDI
integrated discrimination improvement
- InterASIA
International Collaborative Study of Cardiovascular Disease in Asia
- LD
linkage disequilibrium
- LDL-C
low-density lipoprotein cholesterol
- NRI
net reclassification improvement
- OR
odds ratio
- PRS
polygenic risk score
- SBP
systolic blood pressure
- SNP
single-nucleotide polymorphism
- T2D
type 2 diabetes
- TC
total cholesterol
Appendix. Authors

Footnotes
Class of Evidence: NPub.org/coe
Study Funding
This work was supported by National Natural Science Foundation of China (91857118, 82030102, 81773537), Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2017-I2M-1–004, 2019-I2M-2–003, 2016-I2M-1–009), and the National Key Research and Development Program of China (2017YFC0211700, 2018YFE0115300, 2017YFC0908401). The sources of funding had no role in study design, data collection, analyses, interpretation, or decision to submit the article for publication.
Disclosure
The authors report no disclosures relevant to the manuscript. Go to Neurology.org/N for full disclosures.
References
- 1.Zhou M, Wang H, Zeng X, et al. Mortality, morbidity, and risk factors in China and its provinces, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2019;394(10204):1145-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malik R, Chauhan G, Traylor M, et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montaner J, Ramiro L, Simats A, et al. Multilevel omics for the discovery of biomarkers and therapeutic targets for stroke. Nat Rev Neurol. 2020;16(5):247-264. [DOI] [PubMed] [Google Scholar]
- 4.Buniello A, MacArthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005-D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ibrahim-Verbaas CA, Fornage M, Bis JC, et al. Predicting stroke through genetic risk functions: the CHARGE Risk Score Project. Stroke. 2014;45(2):403-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malik R, Bevan S, Nalls MA, et al. Multilocus genetic risk score associates with ischemic stroke in case-control and prospective cohort studies. Stroke. 2014;45(2):394-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tada H, Shiffman D, Smith JG, et al. Twelve-single nucleotide polymorphism genetic risk score identifies individuals at increased risk for future atrial fibrillation and stroke. Stroke. 2014;45(10):2856-2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rutten-Jacobs LC, Larsson SC, Malik R, et al. Genetic risk, incident stroke, and the benefits of adhering to a healthy lifestyle: cohort study of 306 473 UK Biobank participants. BMJ. 2018;363:k4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abraham G, Malik R, Yonova-Doing E, et al. Genomic risk score offers predictive performance comparable to clinical risk factors for ischaemic stroke. Nat Commun. 2019;10(1):5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Said MA, Verweij N, van der Harst P. Associations of combined genetic and lifestyle risks with incident cardiovascular disease and diabetes in the UK Biobank study. JAMA Cardiol. 2018;3(8):693-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsai CF, Thomas B, Sudlow CL. Epidemiology of stroke and its subtypes in Chinese vs white populations: a systematic review. Neurology. 2013;81(3):264-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao Y, Zhu H, Zhu L, et al. A comprehensive contribution of genetic variations of the insulin-like growth factor 1 signalling pathway to stroke susceptibility. Atherosclerosis. 2020;296:59-65. [DOI] [PubMed] [Google Scholar]
- 13.Yang X, Li J, Hu D, et al. Predicting the 10-year risks of atherosclerotic cardiovascular disease in Chinese population: the China-PAR project (prediction for ASCVD risk in China). Circulation. 2016;134(19):1430-1440. [DOI] [PubMed] [Google Scholar]
- 14.Han C, Liu F, Yang X, et al. Ideal cardiovascular health and incidence of atherosclerotic cardiovascular disease among Chinese adults: the China-PAR project. Sci China Life Sci. 2018;61(5):504-514. [DOI] [PubMed] [Google Scholar]
- 15.Chinese Preventive Medicine Association. Chinese guideline on healthy lifestyle to prevent cardiometabolic diseases [in Chinese]. Zhonghua Yu Fang Yi Xue Za Zhi. 2020;54(3):256-277. [DOI] [PubMed] [Google Scholar]
- 16.Lloyd-Jones DM, Hong Y, Labarthe D, et al. Defining and setting national goals for cardiovascular health promotion and disease reduction: the American Heart Association's strategic Impact Goal through 2020 and beyond. Circulation. 2010;121(4):586-613. [DOI] [PubMed] [Google Scholar]
- 17.Ishigaki K, Akiyama M, Kanai M, et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat Genet. 2020;52(7):669-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1-22. [PMC free article] [PubMed] [Google Scholar]
- 19.Xing X, Yang X, Liu F, et al. Predicting 10-year and lifetime stroke risk in Chinese population. Stroke. 2019;50(9):2371-2378. [DOI] [PubMed] [Google Scholar]
- 20.Peloso GM, Beiser AS, Satizabal CL, et al. Cardiovascular health, genetic risk, and risk of dementia in the Framingham Heart Study. Neurology. 2020;95(10):e1341-e1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang K, Liang F, Yang X, et al. Long term exposure to ambient fine particulate matter and incidence of stroke: prospective cohort study from the China-PAR project. BMJ. 2019;367:l6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, Burnett RT, Kwong JC, et al. Spatial association between ambient fine particulate matter and incident hypertension. Circulation. 2014;129(5):562-569. [DOI] [PubMed] [Google Scholar]
- 23.Pencina MJ, D'Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23:2109-2123. [DOI] [PubMed] [Google Scholar]
- 24.Pencina MJ, D'Agostino RB Sr, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med. 2011;30(1):11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berg DD, Wiviott SD, Scirica BM, et al. Heart failure risk stratification and efficacy of sodium-glucose cotransporter-2 inhibitors in patients with type 2 diabetes mellitus. Circulation. 2019;140(19):1569-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R, Chambless L. Test for additive interaction in proportional hazards models. Ann Epidemiol. 2007;17(3):227-236. [DOI] [PubMed] [Google Scholar]
- 27.Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019;51(4):584-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duncan L, Shen H, Gelaye B, et al. Analysis of polygenic risk score usage and performance in diverse human populations. Nat Commun. 2019;10(1):3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hachiya T, Hata J, Hirakawa Y, et al. Genome-wide polygenic score and the risk of ischemic stroke in a prospective cohort: the hisayama study. Stroke. 2020;51(3):759-765. [DOI] [PubMed] [Google Scholar]
- 30.Elliott J, Bodinier B, Bond TA, et al. Predictive accuracy of a polygenic risk score-enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA. 2020;323(7):636-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosley JD, Gupta DK, Tan J, et al. Predictive accuracy of a polygenic risk score compared with a clinical risk score for incident coronary heart disease. JAMA. 2020;323(7):627-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bi Y, Jiang Y, He J, et al. Status of cardiovascular health in Chinese adults. J Am Coll Cardiol. 2015;65(10):1013-1025. [DOI] [PubMed] [Google Scholar]
- 33.Folsom AR, Yatsuya H, Nettleton JA, Lutsey PL, Cushman M, Rosamond WD. Community prevalence of ideal cardiovascular health, by the American Heart Association definition, and relationship with cardiovascular disease incidence. J Am Coll Cardiol. 2011;57(16):1690-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong C, Rundek T, Wright CB, Anwar Z, Elkind MS, Sacco RL. Ideal cardiovascular health predicts lower risks of myocardial infarction, stroke, and vascular death across whites, blacks, and Hispanics: the Northern Manhattan Study. Circulation. 2012;125(24):2975-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao D, Liu J, Wang M, Zhang X, Zhou M. Epidemiology of cardiovascular disease in China: current features and implications. Nat Rev Cardiol. 2019;16(4):203-212. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article and supplemental data.