Abstract
Cortical surface registration is an essential step and prerequisite for surface-based neuroimaging analysis. It aligns cortical surfaces across individuals and time points to establish cross-sectional and longitudinal cortical correspondences to facilitate neuroimaging studies. Though achieving good performance, available methods are either time consuming or not flexible to extend to multiple or high dimensional features. Considering the explosive availability of large-scale and multimodal brain MRI data, fast surface registration methods that can flexibly handle multimodal features are desired. In this study, we develop a Superfast Spherical Surface Registration (S3Reg) framework for the cerebral cortex. Leveraging an end-to-end unsupervised learning strategy, S3Reg offers great flexibility in the choice of input feature sets and output similarity measures for registration, and meanwhile reduces the registration time significantly. Specifically, we exploit the powerful learning capability of spherical Convolutional Neural Network (CNN) to directly learn the deformation fields in spherical space and implement diffeomorphic design with “scaling and squaring” layers to guarantee topology-preserving deformations. To handle the polar-distortion issue, we construct a novel spherical CNN model using three orthogonal Spherical U-Nets. Experiments are performed on two different datasets to align both adult and infant multimodal cortical features. Results demonstrate that our S3Reg shows superior or comparable performance with state-of-the-art methods, while improving the registration time from 1 min to 10 sec.
Keywords: Surface registration, Spherical U-Net, unsupervised learning, diffeomorphism, convolutional neural networks
I. Introduction
CORTICAL surface registration is a fundamental task in population-based neuroimaging studies and has been an increasingly important research topic for decades. Accurate registration of the convoluted cerebral cortex is important for establishing cortical correspondences across individuals and time points, thus facilitating the subsequent analysis, e.g., group comparison or longitudinal studies.
Motivated by the inherent spherical topology of the cerebral cortex, many surface registration methods [1]-[5] model the cortical surface as a 2D closed manifold and map it onto a sphere, which can thus offer a simpler and more accurate geometry for aligning cortical structure and function than 3D volumetric registration approaches [6], [7]. After mapping to the spherical space, surface registration aims to estimate the spherical deformation field, following which the underlying spherical coordinate system is warped to establish inter-subject or intra-subject feature correspondences across surfaces.
Conventional spherical registration methods are typically designed to solve an optimization problem on the spherical space that aligns vertices with similar feature patterns, while enforcing smoothness constraints on the spherical deformations. For example, FreeSurfer [2] estimates a vertex-wise deformation field by minimizing the mean squared error between the source and target cortical features maps using a gradient descent optimization strategy. Using the same similarity measure, Spherical Demons [1] employs Demons algorithm [8] and Gauss-Newton method to significantly improve the computational efficiency (from the order of hours to minutes). Additionally, Spherical Demons extends one parameter subgroups of diffeomorphisms [9] from Euclidean space to spherical space to guarantee a diffeomorphic registration, which is theoretically invertible and thus can effectively avoid self-intersections and preserve topology of cortical surfaces. These classical methods [1], [2], [5] are originally developed for aligning cortical folding-based features, e.g., average convexity and mean curvature, which, however, are not always proper criterion for cortical functional alignment as revealed by recent studies [3], [6], [10]. Therefore, some works try to integrate high dimensional spectral embedding features [11], functional activation or connectivity features [12], [13] for cortical surface registration. However, these methods are still hardly extendable to include multiple or high dimensional features due to their specific mathematical modeling. Multimodal Surface Matching (MSM) [3] first proposes to deal with multimodal features, including cortical folding, function, and connectivity, by modeling registration as a discrete labelling problem, thus offering flexibility in the selection of features sets and similarity metrics. However, the discrete nature of MSM brings higher computational burden and makes it unsuitable for diffeomorphic implementation. Alternatively, MSM restricts the deformation size to avoid self-intersections, which may provide suboptimal solutions for large deformations.
Another common limitation in previous methods is that the optimization is performed between each pair of surfaces, which is computationally expensive. As far as we know, Spherical Demons provides the fastest solution, which still takes 1 minute for registration of a pair of surfaces. When handling large-scale neuroimaging studies, the increased computation time is more and more likely to be a barrier for subsequent analyses.
Consequently, a spherical surface registration method that can be flexibly extended to align multimodal features with faster and diffeomorphic implementation is highly needed. To this end, in this study, we develop an end-to-end Superfast Spherical Surface Registration (S3Reg) framework based on the following three considerations.
1). End-to-end Unsupervised Learning:
Recently, deep learning-based methods have greatly advanced and accelerated 3D volumetric image registration in medical imaging, especially using the Convolutional Neural Network (CNN) models with GPU implementations [14]-[16]. Most earlier approaches [17], [18] use supervised learning, where quasi-ground-truth deformations are derived via conventional registration tools or simulations, which introduces extra cumbersome preprocessing or possible bias for real data applications. To solve this issue, recent works [14]-[16], [19], [20], including VoxelMorph [15], employ end-to-end learning strategy to directly predict the deformation field given input image pairs in an unsupervised learning framework. We note that these methods not only promise a significant speedup for medical image registration with comparable performance to conventional registration tools, but also offer flexibility in the choice of input feature sets and output loss functions.
Inspired by this, we formulate S3Reg as a novel end-to-end unsupervised learning framework on the spherical space based on VoxelMorph framework [15] and our recently developed Spherical U-Net [21]. Benefiting from the end-to-end architecture, S3Reg allows multiple alternative similarity metrics and also enables different weightings for different features and areas. Meanwhile, S3Reg only needs to learn one global optimization function for aligning any pair of surfaces, thus yielding a superfast registration process, with less than 10 seconds for registration of a pair of surfaces.
2). Topology-preserving Deformations:
Another challenge in registration is to preserve the topology of cortical surfaces during deformation, as topology-incorrect deformations induces fatal error (e.g., self-intersections) when warping the source to target. In volumetric registration, some methods [15], [20] rely on diffusion regularizers to encourage spatially smooth deformations, which do not strictly guarantee topology-preserving deformations. Alternatively, diffeomorphism [22], especially a stationary velocity field model related to a diffeomorphism through the exponential mapping [9], is popular as it ensures the topology under the theory of flows of vector fields, and can be conveniently implemented by several differentiable “scaling and squaring” layers in CNNs [14], [23]. In spherical surface registration, Spherical Demons also adopts the extended diffeomorphism [24] parameterized by stationary velocity field on tangent spaces. In this work, we extend the “scaling and squaring” layers in Euclidean space to spherical space based on the underlying theories presented in Spherical Demons [1] to guarantee diffeomorphic deformations.
3). Choice of Spherical CNNs:
To apply deep learning on spherical surfaces, some earlier studies use the equirectangular parameterized sphere data [25]-[27], leading to different weights at different regions on the sphere, and introducing artificial discontinuities at poles and 0° longitude. In a concurrent work, Cheng et al. [28] used a sphereNet [27] for unsupervised cortical surface registration. They first projected the spherical surface data to a 2D image grid using the equirectangular projection method, and then directly applied the conventional image registration framework [14] in Euclidean space to predict the deformation field in the 2D projected images using a standard 2D U-Net [29]. After the similarity and smoothness losses were enforced on the 2D image grid to train the U-Net, the predicted deformation field and warped image were finally projected back to the sphere. Although they used different weights on different latitudes to account for the unbalanced sampling points introduced by the equirectangular projection, the discontinuities at poles are not well addressed. This is potentially catastrophic because the points near the poles cannot be moved across the poles in the 2D equirectangular projected images (where the poles are projected as two lines) and thus the displacement across the poles cannot be correctly predicted (see Fig. 2 for more interpretations). Directly performing convolution and pooling on icosahedron discretized spherical surface with uniformly sampled vertices thus become popular, both in computer vision [30]-[32] and cortical surface analysis [21], [33], [34]. Some of them perform convolution on the tangent space by weighting the kernel and resampled data points in the tangent plane [30], [33], [34], which introduces heavy computational burden by repeatedly re-interpolating the surface. Other works propose to directly use the 1-ring kernel on icosahedron discretized surfaces for 3D shape recognition [31] and omnidirectional image segmentation [32]. For cortical surfaces, Parvathaneni et al. [35] uses the global differential operators [32] as the convolution kernel for parcellation. Our Spherical U-Net [36] directly uses the 1-ring kernel and extends U-Net [29] to spherical space by replacing all operations (convolution, pooling, and transposed convolution) with their spherical operation counterparts. It thus possesses the same excellent ability with the standard U-Net in the image space to learn both contextual and localization information, which has been demonstrated with the state-of-the-art performance in various tasks [21], [37], [38]. However, the 1-ring kernel was originally designed to be azimuthally rotation equivariant/invariant. In this design, to establish a reference direction on sphere, the local coordinates are inversed across poles, thus introducing inherent discontinuity at poles. While SO(3) rotation equivariant spherical CNNs [39], [40] did exist, they aim to learn a global representation for classification or regression tasks, thus are not suitable for segmentation or registration, which needs to estimate vertex-wise local representations.
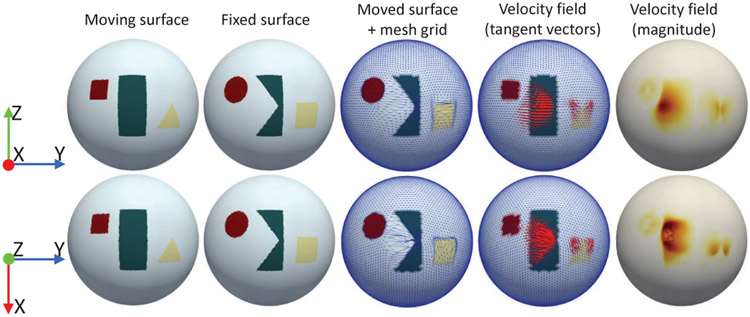
Fig. 2.
An example shows the polar distortion issue when registering the moving surface to the fixed surface using one single spherical U-Net. The first row and second row show the registration results near the equator and near the north pole, respectively. We can see that the predicted deformation field, represented by either the final warped mesh grid or velocity field, are dramatically distorted and topology-incorrect at polar regions. Specifically, the number of folded triangles [42] is 0 for the moved surface in the first row and 118 in the second row (total 20,480 faces), respectively. Especially, the last column shows smaller magnitude at the center near north pole, indicating that it is impossible to learn the particular displacement vectors that move vertices across the pole, due to the inherent discontinuity of the 1-ring kernel design at poles.
As a result, in this work, we adopt Spherical U-Net as our backbone spherical CNN in our S3Reg framework with modification to address the polar distortion issue. Specifically, during training stage, polar regions are masked and disregarded, and three complementary Spherical U-Nets are trained jointly for three orthogonal directions. In testing stage, the prediction results from three models are fused to obtain the final deformation fields.
Moreover, as an optional strategy, we propose to integrate the consistency of cortical parcellation maps in the loss function thanks to the flexibility of S3Reg framework, when cortical parcellation maps are available during training. To the best of our knowledge, this is also the first study that leverages parcellation maps for improving surface registration. To summarize, our main contributions in this work include:
We present a novel end-to-end unsupervised learning framework, S3Reg, for spherical cortical surface registration, thus offering significant flexibility in the choice of feature sets and similarity metrics.
We demonstrate effective extension of “scaling and squaring” layers to sphere and integration of it in spherical CNN yield diffeomorphic surface registration results.
We propose a novel spherical CNN model consisting of three complementary orthogonal Spherical U-Nets with a weighting strategy that effectively solves the intrinsic polar distortion issue in spherical registration.
We demonstrate comparable accuracy to the state-of-the-art methods in registering multimodal cortical features for both adult and infant cortical surfaces, while improving the computational time from 1 minute to 10 seconds.
II. Background
A. VoxelMorph Framework
VoxelMorph [15] is a constructive and popular deformable image registration framework that learns to predict voxel-wise non-linear correspondences between a fixed image and a moving image based on CNNs. In training stage, it aims to learn a parametric function that maps all voxels of one image to another by minimizing the loss function composed of similarity metric and smoothness constraint on all training image pairs. In testing stage, for any new pair of images, the deformation field can be quickly computed by evaluating the function using the learned parameters.
Let F be the fixed image and M be the moving image. VoxelMorph models the registration function fθ(F, M) = ϕ using a CNN similar to U-Net [29], where ϕ is the deformation field and θ are the learnable parameters of the function f. Then the objective is:
| (1) |
where M ∘ ϕ represents M warped by ϕ, measures feature similarity, imposes spatial smoothness on ϕ, and λs is a regularization parameter. The goal is thus to find optimal parameters that minimizes on a training set D:
| (2) |
As a result, this learning-based framework provides great flexibility in the choices of similarity metric and smoothness constraint, which can be optimized by stochastic gradient descent or Adam optimizer [41] in an end-to-end way without ground-truth deformation fields. Our S3Reg framework will build on this idea to avoid the need of ground-truth and profit from its flexibility, as well as its efficiency by learning a single parametric function for all surface pairs.
B. Diffeomorphic Registration
Although our method can incorporate multiple deformation representations, we choose to work with diffeomorphisms, in particular with a stationary velocity field representation [9]. A stationary velocity field is related to a diffeomorphism through the exponential mapping . In this case, it can be given at time 1 by integrating the stationary velocity field over t = [0, 1] in the following ordinary differential equation (ODE):
| (3) |
with the initial condition ϕ(0) = Id representing the identity transformation, i.e., .
However, to compute such exponential of velocity vectors is numerically difficult. Herein, we follow volumetric registration methods [14], [23] that integrate the stationary velocity field over time t using the scaling and squaring layers [9], and further extend them to spherical surface in S3Reg framework. Specifically, given an initial deformation field ϕ(1/2T), where T is empirically set to 6, ϕ(1) can be obtained using the recurrence ϕ(1/2t–1) = ϕ(1/2t) ∘ ϕ(1/2t). Note that in volumetric registration, the initial deformation is generally obtained by simply adding the velocity vectors:
| (4) |
where x denotes the spatial locations. While for surface registration, we follow the basic ideas in Spherical Demons [1] to obtain the initial deformation and will elaborate it in Sec. III-B.
III. S3Reg Framework
A. Overall Architecture
As shown in Fig. 1, the parametrization of fθ(F, M) for spherical surface registration is based on Spherical U-Net architecture [21]. We assume that F and M are spherical surface maps containing single-channel feature for simplicity, e.g., mean curvature, average convexity, myelin content or functional gradient density, defined on the sphere S2 parameterized by icosahedron subdivisions [43], though it is very straightforward to extend to high dimensional features. The input to the Spherical U-Net is the concatenation of F and M. It is then processed using repetitive spherical convolution and pooling layers through the encoder path. Then a decoder path composed of spherical transposed convolution and convolution layers with skip connections to the encoder generates the high-resolution feature maps. At the final layer, the vertex-wise filter weighting is used to map the 64-component feature maps to the desired 2 channels. The output from the Spherical U-Net is the velocity field represented by tangent vectors on the surface.
Fig. 1.
Overview of our S3Reg framework. We learn the velocity fields using three orthogonal Spherical U-Nets for each equator region at different orientations. The final velocity field is obtained by rotating and fusing the three velocity fields. This velocity field is then used to warp the moving surface using spherical transform layer. Optionally, auxiliary information such as parcellation maps can be leveraged during training.
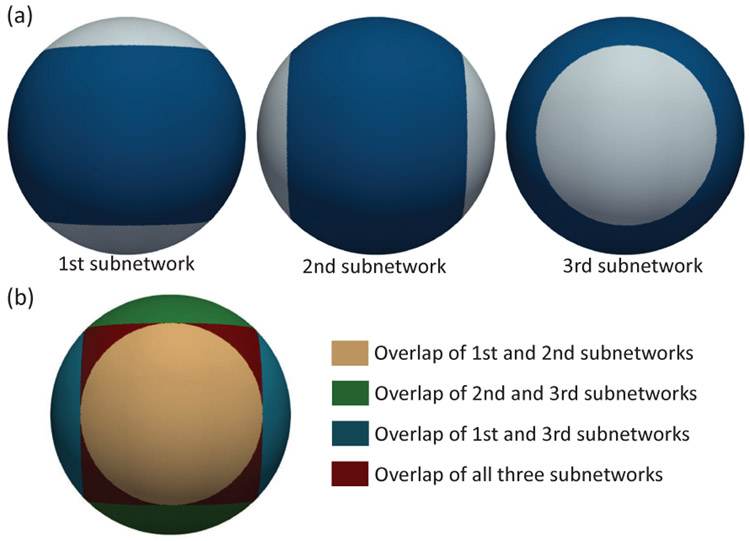
However, as mentioned earlier, the 1-ring kernel in Spherical U-Net was originally designed to be azimuthally rotation equivariant/invariant, which inevitably leads to distortions in the prediction results for the vertices near the poles, as illustrated in Fig. 2 using two synthetic surfaces. The fundamental reason for this distortion is that in the convolution operation, the 1-ring kernel is translated along the latitude from 0 to 360 but only along the longitude from 0 to 180, i.e., from the north pole to the south pole. This is required to establish a reference direction on the sphere, i.e., the z-axis in this design. Therefore, the 1-ring kernel is reversed when translated across the poles, which leads to the discontinuities at poles. Since predicting the deformation at polar regions would result in distortion and predicting the deformation at the equatorial regions would not, we proposed to rotate the sphere along y-axis by 90 and predict the deformation only around the equatorial regions. In this way, the original polar regions’ deformation can also be effectively predicted after rotation, because it is now near the equator. After the deformation field is predicted, we rotate it back to its original polar region, thus addressing the polar distortion issue. However, if we only rotate the sphere once, the deformation field is predicted twice in some regions but only one time in other regions. This will result in different weights (2:1) and biased results at different regions on sphere. To make the weights and results more unbiased, we proposed to rotate the sphere one more time along z-axis. Now the deformation field is predicted twice in the majority of regions and three times in the remaining small regions, as shown in Fig. 3(b). Therefore, it is easier for the network to train and learn to overcome the less biased weights. Theoretically, arbitrary rotations that can move the polar regions to be not overlapped with original polar regions are fine but obviously not an efficient way to predict the deformation field in the non-orthogonal regions due to 1) the complicated computations of the deformations’ rotation and fusion; 2) the difficulties in balancing different weights between different regions. Finally, we empirically found that 3 orthogonal sphere-networks along x, y, z axes is a good tradeoff between accuracy and computational complexity (with more subnetworks, the deformations are predicted more times and should be more accurate but slower).
Fig. 3.
(a) Mask maps of different subnetworks for addressing the polar distortion issue. Dark blue regions will be trained and predicted by each subnetwork while white regions will be ignored. (b) Overlap regions trained by different subnetworks.
Specifically, suppose for a spherical surface with N vertices , we obtain the second sphere by rotating 90° along y-axis and then the third sphere by rotating 90° along z-axis, i.e., , , where R is the rotation matrix, represents the vertices on i-th sphere. For convenience, we now also represent as . Accordingly, we construct three subnetworks independently for the spheres in 3 orientations, as shown in Fig. 1, named three orthogonal Spherical U-Nets architecture. Each subnetwork predicts a separate velocity field (, , in Fig. 1) for registering the corresponding rotated moving surface and the rotated atlas surface. Then in each subnetwork, a binary mask is designed to disregard the influences of vertices near poles: , where r is the radius of the sphere, v(z) is the z value of vertex v. Fig. 3(a) shows the binary masks for different subnetworks. Fig. 3(b) intuitively shows the overlap regions of the 3 orthogonal Spherical U-Nets, based on which the final velocity field can be obtained by fusion. We adopt early fusion to fuse velocity field instead of deformation field ϕ to reduce potential computational redundancy.
To fully exploit the rich information in the three orthogonal Spherical U-Nets, besides the regular similarity and smoothness term in loss function, we additionally enforce the consistency among three velocity fields, , , and :
| (5) |
where is a 3×2 orthonormal basis on the tangent space at vn, and maps the tangent vector from the tangent space to the 3D space, Oi,j represents the overlap vertices of i-th and j-th velocity field, e.g., O1,2 = { and }, and ∣Oi,j∣ is the number of vertices in Oi,j.
B. Spherical Transform Layer
Following the basics in Spherical Demons [1], we represent the deformation field . It maps a point vn ∈ S2 to another point ϕ(vn) ∈ S2. We also parameterize ϕ using a stationary velocity field . The stationary velocity field is predicted and fused from the three orthogonal Spherical U-Nets for each vertex with size N × 2. As introduced earlier, we now use the scaling and squaring layer to compute the spherical deformation ϕ using . Firstly, we compute the initial deformation field ϕ(1/2T):
| (6) |
where T is empirically set to 6, and maps the tangent vector from the tangent space to the 3D space. This means ϕ(1/2T) maps the vn on moving surface to ϕ(1/2T)(vn) on fixed surface. Note that both vn and ϕ(1/2T)(vn) are on the sphere. Then ϕ can be computed using the recurrence ϕ(1/2t–1) = ϕ(1/2t) ∘ ϕ(1/2t), which is the operation in each scaling and squaring layer. Numerically, for each warp operation, we use the barycentric interpolation to compute the deformation ϕ(1/2t) (ϕ(1/2t)(vn)), which means vn is first moved to ϕ(1/2t)(vn) and then moved to ϕ(1/2t)(ϕ(1/2t)(vn)). With this definition, we establish 1-to-1 correspondence between and , and thus also between and , when the angle α between vn and ϕ(1/2T)(vn) is less than π/2, which is a reasonable assumption in spherical surface registration. Hence, given a surface with N vertices , the spherical deformation , or equivalently the velocity field , together with a choice of an interpolation function define the deformation ϕ everywhere on S2. It is worth noting that the length of in our definition is equal to tan(α) rather than sin(α) in Spherical Demons or the geodesic distance α. Though these three definitions are approximately equal when α is very small, our definition with fewer computation steps is more convenient and faster in implementation.
Now for each deformed vertex ϕ(vn), we choose to directly warp the moving surface M so that the feature value at ϕ(vn) on the fixed surface F(ϕ(vn)) is computed using barycentric interpolation. Then we can directly compare M(vn) and F(ϕ(vn)) in the loss function. Conclusively, all the operations in the spherical transform layer are differentiable and thus can backpropagate error to train the parameters in three orthogonal Spherical U-Nets architecture.
C. Loss Functions
With all abovementioned definitions, we rewrite the objective as:
| (7) |
where λs and λcon is the weight for smooth and velocity field consistency terms.
For the similarity term, we choose both mean squared distance (MSD) used in Spherical Demons [1] and correlation coefficient [3] used in MSM:
| (8) |
where λcc is the weight for correlation term, cov(·, ·) is the covariance and σ is the standard deviation.
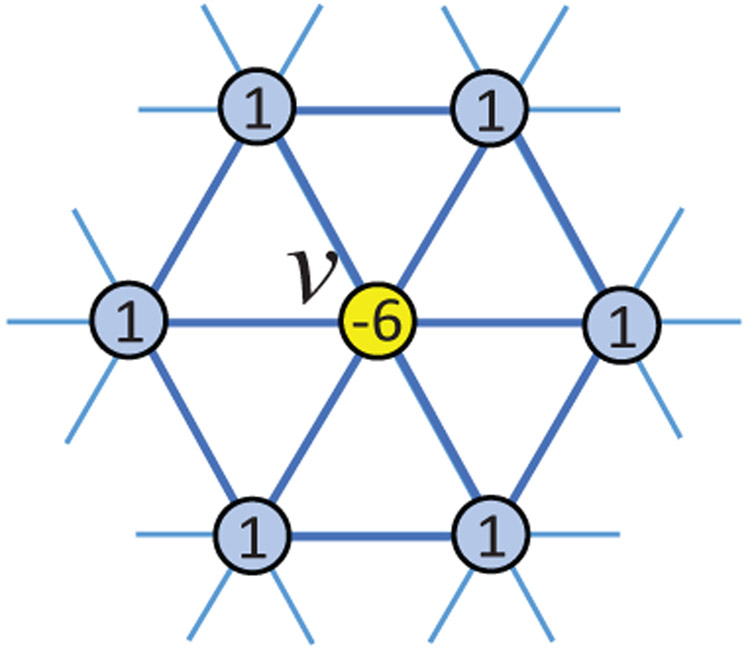
Although our method guarantees a diffeomorphic registration, it is still conditioned on the initial deformation field, which is required to be sufficiently smooth and small [1]. Otherwise, it is still possible to generate topology-incorrect deformations as shown in the second row in Fig. 2. Therefore, based on the tangent velocity field, we propose a new operator ∇s on 1-ring filter (Fig. 4) approximating the spherical gradients to explicitly enforce smoothness on the velocity field. Accordingly, penalizes local spatial variations in , where represents the local 1-ring velocity vectors of vertex vn. It can be efficiently computed by the 1-ring convolution in the Spherical U-Net architecture.
Fig. 4.
Smooth operator ∇s on spherical surface approximating spherical gradients.
D. Optional Auxiliary Parcellation Consistency Loss
A parcellation map divides the cortical surface into small parcels and assigns each vertex a cortical region of interest (ROI) label, thus providing rich information of cortical maps. Cortical parcellation maps based on anatomical or functional features are sometimes available during training, and can be annotated by human experts or automated algorithms [21]. Thanks to the flexibility of S3Reg, we can additionally integrate features like parcellation maps as auxiliary information during training.
Intuitively, if a deformation field ϕ represents accurate cortical correspondences, the ROIs in F and M ∘ ϕ should overlap well. Accordingly, let pF and pM be the parcellation maps for F and M, and pM ∘ ϕ be the warped parcellation maps, we compute the surface area overlap for each ROI using the Dice score:
| (9) |
where k denotes the vertices labeled as k, ∣ · ∣ represents the number of vertices in this ROI. Then we define the auxiliary parcellation consistency loss averaged over all K ROIs:
| (10) |
In summary, the full objective to train our S3Reg framework can be written as:
| (11) |
where λparc is the weight for the parcellation consistency term.
IV. Experiments
We performed a series of experiments that register multiple cortical features on both adult and infant cortical surfaces, and demonstrated comparable accuracy with significantly reduced registration time compared to the state-of-the-art methods: Spherical Demons [1], FreeSurfer [2] and MSM [3]. For illustration, we focus on atlas-based registration, a common task in population-based analysis, although our method is not limited to that. In all experiments, we register each individual surface to an atlas surface fairly using all methods.
A. Experimental Setup
1). Data and preprocessing:
For adult brain MRI data, we used the NAMIC cortical surface dataset1 and conducted the experiments on left hemisphere. This dataset consists of 39 cortical surfaces constructed from MR images using FreeSurfer [44]. Each surface was mapped onto the sphere and represented as a spherical image with 2 folding-based geometric features at each vertex, i.e., sulc (average convexity) and curv (mean curvature) [43]. Basically, the mean curvature measures the cortical folding in a fine view, and the average convexity measures the cortical folding in a coarse view. Each surface was manually parcellated by a neuroanatomist into 35 regions based on major sulci and gyri [45]. We used FreeSurfer atlas surface [43] as the fixed surface in this experiment. Therefore, with ground truth parcellation maps, the aim here is to validate the usefulness of our framework on parcellation of cortical surfaces and at the same time show the effectiveness comprehensively with some ablation studies.
We further validated our method on another dataset with 102 infants around 1 year of age. The cortical surfaces were reconstructed using an infant-specific pipeline [46]-[50], and further mapped onto the sphere. Each surface was represented with multimodal features, including 2 geometric features, i.e., sulc and curv, and 2 function-related features, i.e., myelin content and functional gradient density, and a parcellation map obtained using the method in [21] based on geometric features. Myelin content and functional gradient density are both reliable and meaningful functional features closely related to cortical functional areas [10]. The myelin maps were computed using the ratio of T1w/T2w images. The infant resting-state fMRI data (rs-fMRI) were acquired during natural sleeping. Besides the conventional HCP fMRI processing steps [51], we especially used the following strategies [52]: (1) Onetime resampling and denoising of functional signals completed in the native image space; (2) Deep learning-based noisy component removal for fast and robust fMRI denoising. The fMRI time series of each vertex on the middle cortical surface was extracted. The map of gradient density of functional connectivity was then computed on the cortical surface using the method in [53], [54]. As there are no ground truth parcellation maps of this dataset, we focus on extensive validation of the generalization ability of our method on multimodal features. The atlas we used for this dataset was constructed from other 83 subjects with similar ages by co-registration of them using Spherical Demons [1].
2). Baseline Methods:
We used the official codes of Spherical Demons2, MSM3 and FreeSurfer (7.1.0) for their experiments. Before these non-rigid methods start, we performed a rigid registration for initialization as in Spherical demons [1] and FreeSurfer [43], based on an exhaustive searching among a range of rotations along each axis. Specifically, we have 4 rounds of searching, and the range for searching at each round is [−64,64], [−32,32], [−16,16], and [−8,8], respectively. The criterion used to evaluate the similarity between the moved surface and the fixed surface is the mean square error of sulc. Then we run Spherical Demons and FreeSurfer with their default parameters: registering two surfaces at 4 levels of icosahedron subdivisions (4, 5, 6, 7, with 2,562, 10,242, 40,962, 163,842 vertices, respectively) to align sulc, sulc, sulc, curv, respectively. Based on the sulc-driven alignment, we registered functional features at 6th level using the same configuration as curv for infant cortical surfaces. For MSM, as the default parameters cannot get satisfactory results, we used a set of optimized parameters for each dataset. We made the regularization term larger and the input smoothing term smaller. It was still run by default for 2 geometric features (first sulc, then curv) at four levels (4, 4, 5, 6 for sulc, then 4, 5, 5, 6 for curv) using 3 iterations at each level. For functional features, we used the same configuration as curv, but only run at 6th level to be consistent with Spherical Demons. To have a fair comparison with our method, we run all the conventional methods on an Intel Core i7-8700 CPU.
3). Implementation of S3Reg:
We implemented our S3Reg framework in PyTorch [55] and used Adam optimizer [41] with a learning rate decreasing from 1e-3 to 1e-6 in 100 epochs to train the models. Feature values were firstly normalized by subtracting the mean and dividing by the standard deviation. Unlike one-shot volumetric registration, cortical surface registration prefers registering multiple features in multiple resolutions in a coarse-to-fine manner as performed in previous methods. Therefore, we also implement our default registration algorithm based on S3Reg in a coarse-to-fine multi-level style. We trained 4 models at 4 levels (3, 4, 5, 6) for aligning sulc, sulc, sulc, curv, respectively, as our default model for aligning geometric features, referred as S3Reg-multi-sulc-curv. All 4 models were trained separately. Specifically, we first trained a model for registering sulc feature at a low level (level 3). After the training of this network was done, we saved the model and inferred the deformation field at level 3 for registering sulc. The deformation field at level 3 was then upsampled to level 4 and used to warp cortical surfaces at level 4. We then trained the new model at level 4 for registering the corresponding feature on the warped surfaces. We repeated this process until level 6 for registering curv. For upsampling the deformation field from the current level to the next level, we used barycentric interpolation to generate the moved vertices and then mapped them onto the sphere. For downsampling operation on the spherical surface, we simply extract the vertices (and their attached features) on the low-level surfaces to obtain the downsampled spherical surfaces from a high-level surface. We did not apply any smoothing before this downsampling process. To show the superiority of multi-level registration, we trained a model to register sulc only on 6th level subdivision (S3Reg-single-sulc), then curv on 6th level (S3Reg-single-sulc-curv). To further show the improvement enabled by the optional auxiliary parcellation maps, we added the parcellation consistency loss at the last level of S3Reg-multi-sulc-curv, which is referred to as S3Reg-multi-sulc-curv+parc. For functional features, we used the same registration strategy as in Spherical Demons and MSM.
We used 60% surfaces for training, 10% surfaces for validation, and 30% surfaces for testing. λcc and λcon for training models at each level is consistently 1. λs is 8, 10, 12, 16 for sulc at each level, 40 for curv, myelin and functional gradient density at 6th level. λparc is 10 at 6th level. We run our method on a PC with an NVIDIA RTX2080 Ti GPU and an Intel Core i7-8700 CPU.
4). Evaluation Metrics:
To quantitatively evaluate the registration performance, we adopt the widely used Dice ratio of warped parcellation maps as one of the metrics. As better alignment of geometric features leads to higher agreement of anatomical boundaries, higher Dice ratio suggests better registration performance. Note that the Dice here are directly computed based on two warped parcellation maps, not between manual parcellation map and predicted one obtained using additional classifier based on registered features. Because we focus on a fair comparison of registration methods and do not want to involve any extra classifiers. In addition, we also compute mean absolute error (MAE) and Pearson correlation coefficient (PCC) of the registered cortical surface maps to directly evaluate the alignment of registered features. Therefore, a higher PCC and a lower MAE indicate a better alignment. We evaluate these metrics by comparing any two registered surfaces in the testing set, as after registration all surfaces should be in the same spherical space. Note that since we also used MSD and PCC in the loss functions to train the network, the MAE and CC here are only for additionally evaluating the within-group spatial normalization accuracy. As a more unbiased metric, Dice is a more compelling and reliable measurement for demonstrating the superiority of an approach [56].
B. Results on Adult Cortical Surfaces
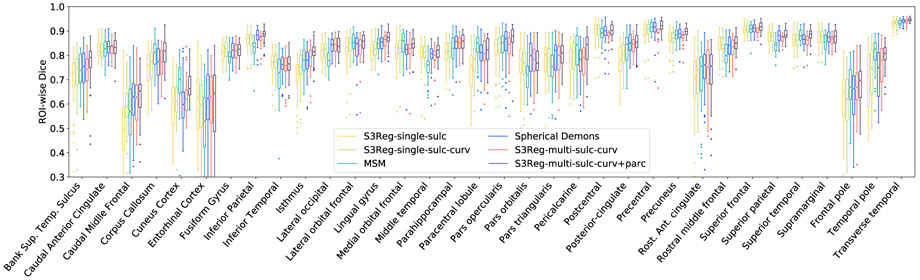
Table I presents the overall registration performance on NAMIC dataset using different methods. We can see that S3Reg-multi-sulc-curv+parc consistently achieves better results than other methods, indicating that auxiliary parcellation maps and coarse-to-fine multi-level registration did help improve the alignment. Of note, parcellation maps were not used during testing, but only in the training stage. The only one exception, MAE of curv, does not show significant difference to S3Reg-multi-sulc-curv and MSM, which may suggest S3Reg-multi-sulc-curv and MSM are already approximating the lower bound of the MAE of curv. Without auxiliary parcellation information, our default S3Reg model, i.e., S3Reg-multi-sulc-curv, achieves better alignment on sulc than MSM and Spherical Demons, which may profit from the strong and robust learning of particular MSD and correlation loss matching the evaluation metrics. On the other hand, MSM achieves slightly better alignment on curv. This is because only MSM aligns curv multiple times at different levels based on the sulc-driven registration. In terms of Dice, as Spherical Demons was originally developed and performed parcellation experiments on this dataset, the default parameters in Spherical Demons may contribute to a better Dice result, close to our default S3Reg model. Fig. 5 shows detailed ROI-wise Dice values for different registration methods. Our best model produced highest results in 25 regions out of 34 regions. In the remaining 9 sub-optimal regions, only 4 regions (cuneus cortex, paracentral lobule, pars orbitalis, temporal pole) shows significant difference. Nevertheless, as these methods all provide flexible regularization or smoothness parameters to trade off alignment accuracy against deformation field’s smoothness, the results are still tunable. However, in terms of run-time, it learns one global optimization function for aligning all surface pairs and therefore improves the computational efficiency significantly. Even it takes about 12 hours to train the models completely, it reduces the time from 1 minute to 13 seconds in testing fairly using the same CPU as other methods. Taking advantage of GPUs, our S3Reg further reduces the registration time to less than 10 seconds. Note that the speed improvement on GPU compared to CPU is not very big. This is because the most time-consuming process, i.e., the interpolation of features and deformation field, is still relatively slow on GPU because of the triangles searching process.
TABLE I.
Comparison of registration performance using different methods on NAMIC dataset
| PCC of sulc | PCC of curv | MAE of sulc | MAE of curv | Dice | CPU Time | GPU Time | |
|---|---|---|---|---|---|---|---|
| S3Reg-single-sulc | 0.70250.0675* | 0.18200.1383* | 3.05540.5613* | 0.17610.0291* | 0.76770.0432* | 3.5s | 1.1s |
| S3Reg-single-sulc-curv | 0.76620.0638* | 0.36580.1237* | 2.66330.5351* | 0.15290.0264* | 0.79560.0412* | 6.7s | 4.6s |
| MSM | 0.79720.0400* | 0.43930.1031 | 2.50340.4301* | 0.14090.0232 | 0.79630.0399* | 8min | - |
| Spherical Demons | 0.80050.0439* | 0.40260.1088* | 2.45230.4357* | 0.14670.0243* | 0.80840.0388 | 1min | - |
| FreeSurfer | 0.79910.0530* | 0.40910.1057* | 2.46200.4850* | 0.14610.0251* | 0.80100.0401* | 30min | - |
| S3Reg-multi-sulc-curv | 0.81960.0466 | 0.43380.1154 | 2.28590.4338 | 0.14080.0256 | 0.80740.0382 | 13.0s | 9.1s |
| S3Reg-multi-sulc-curv+parc | 0.83380.0437 * | 0.44870.1100 * | 2.15420.4160 * | 0.14000.0241 | 0.81080.0381 * | 13.0s | 9.1s |
indicates statistically significant difference compared to S3Reg-multi-sulc-curv (p<0.05). The bold and italic numbers indicate first and second best results, respectively.
Fig. 5.
Boxplots for ROI-wise Dice using different methods.
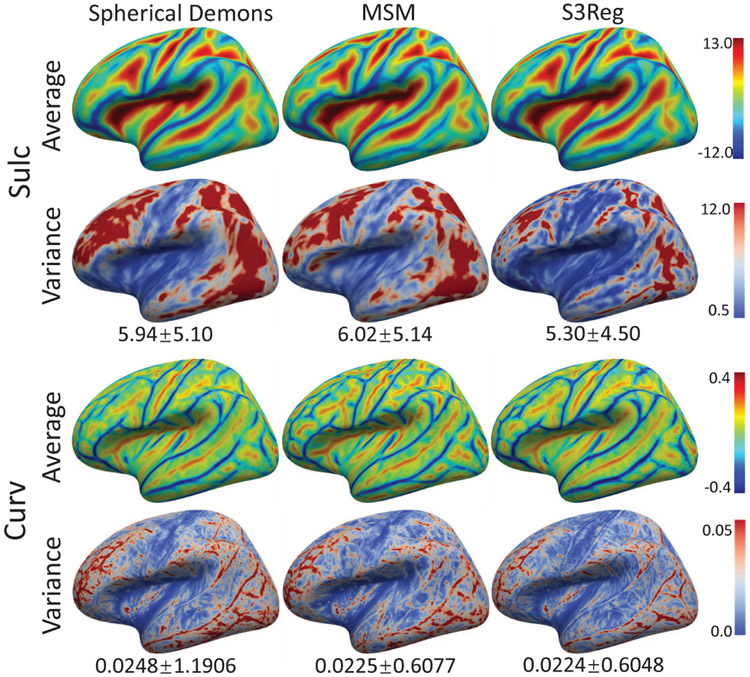
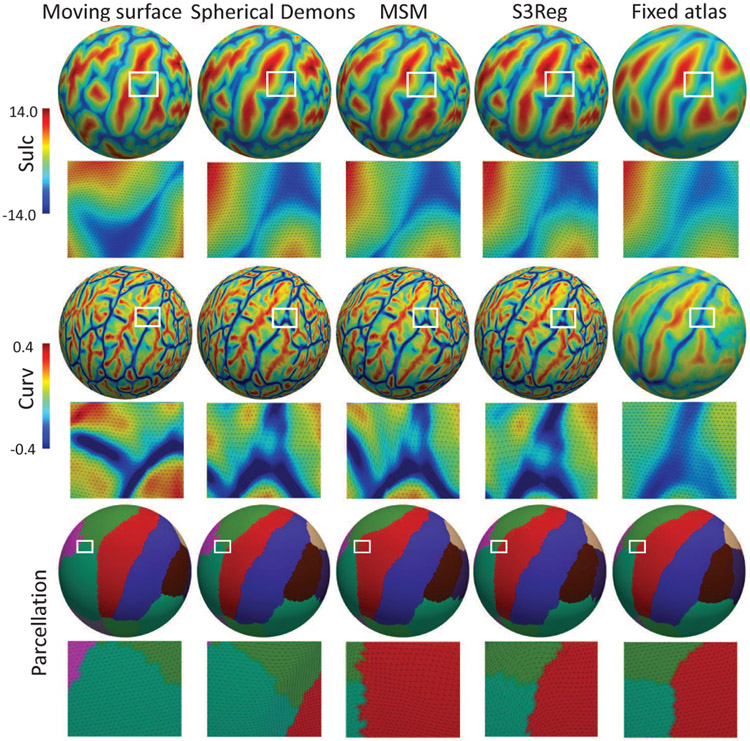
Fig. 6 shows the group-average and variance maps of the testing subjects in NAMIC dataset after registering to the FreeSurfer atlas using different methods. We can see our S3Reg presents shaper folding patterns in many regions and shows lower variance among all testing subjects, indicating better alignment of these two geometric features. Fig. 7 provides a representative example of registration using different methods. We can see that all methods achieve very similar results for sulc and curv alignment. While for the warped parcellation maps of this surface, our model leveraging the parcellation maps during training leads to better alignment with the atlas at the postcentral gyrus boundary.
Fig. 6.
Group-average and variance maps of NAMIC dataset after registration using different methods. We compute the mean variance and its standard deviation as in [5] and show them below each variance map.
Fig. 7.
An example of registering a moving surface to atlas using different methods. S3Reg represents S3Reg-multi-sulc-curv+parc here.
C. Validation on Infant Cortical Surfaces
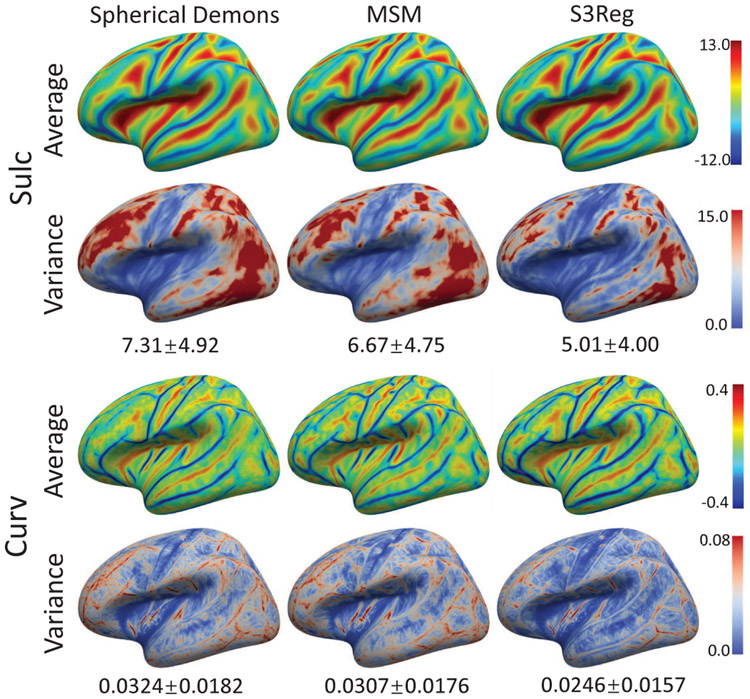
Table II and Fig. 8 shows the quantitative performance and qualitative group-average maps of the testing infant cortical surfaces after registration to the atlas using different methods based on geometric features, sulc and curv. The results are consistent with NAMIC dataset. Still, our S3Reg achieves better alignment in sulc maps, while MSM achieves better results in curv maps, and Spherical Demons and S3Reg obtain highest Dice. In many regions, e.g., the rostral middle frontal gyrus and temporal-occipital junction cortex, our method still presents sharper folding patterns, indicating a better alignment across subjects.
TABLE II.
Comparison of registration performance based on geometric features for infant cortical surfaces.
| Spherical Demons | MSM | S3Reg | |
|---|---|---|---|
| PCC of sulc | 0.69820.0521* | 0.71030.0431* | 0.73590.0526 |
| PCC of curv | 0.27490.0811* | 0.32680.0762 * | 0.29550.0810 |
| MAE of sulc | 2.76690.3528* | 2.66600.3168* | 2.37300.3362 |
| MAE of curv | 0.19330.0205* | 0.17900.0189 * | 0.18530.0197 |
| Dice | 0.78580.0583 | 0.76590.0552* | 0.79030.0566 |
indicates statistically significant difference compared to S3Reg (p<0.05).
Fig. 8.
Group average and variance maps of infant cortical surfaces after registration using different methods.
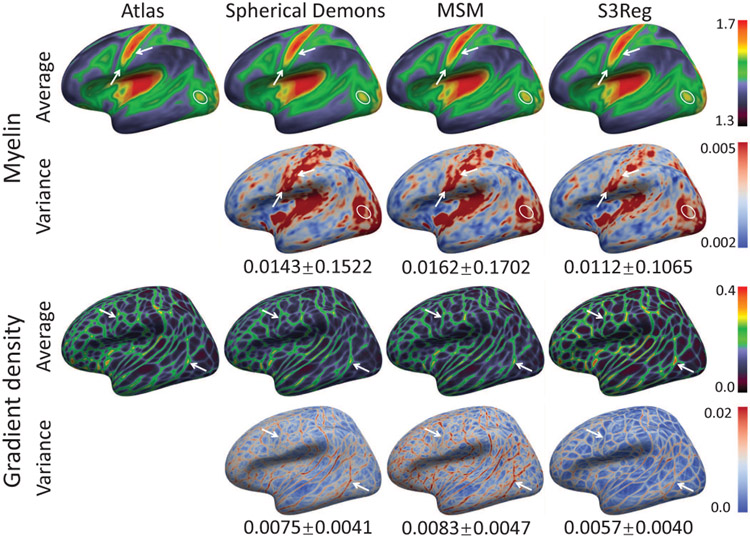
Further, to validate the generalization ability and scalability of our method to register functional features, based on the final sulc-driven alignment at 6th level icosahedron subdivision, we registered the myelin content and functional gradient density maps, respectively. We note that the initial alignment based on geometric features using different methods may affect the registration performance of functional features. To have a fair comparison between different methods, we choose the Spherical Demons results as the initial alignment for all methods, as it directly provides a higher resolution surface with aligned features at 7th level, while the results should be the same regardless of the choice of initialization methods. Table III summarizes the performance of functional features-driven registration on infant cortical surfaces. With the same parameters as when registering curv, Spherical Demons and MSM show lower generalization to novel functional features, while our S3Reg still achieves promising results. Fig. 9 shows the myelin and functional gradient density maps of the atlas, group average and variance of testing surface maps after registration using different methods. As can be seen, our S3Reg presents a shaper and clearer average map than the original atlas, Spherical Demons and MSM, indicating an effective individual-to-atlas registration in this group. Since the group average map could be regarded as a new population-representative atlas, the sharper new atlas by our S3Reg may also benefit the subsequent analysis significantly.
TABLE III.
Comparison of registration performance based on functional features for infant cortical surfaces.
| Spherical Demons | MSM | S3Reg | |
|---|---|---|---|
| PCC of myelin | 0.69290.1127 | 0.63620.1225 | 0.71960.1082 |
| MAE of myelin | 0.10760.0312 | 0.11270.0327 | 0.09700.0301 |
| PCC of FGD | 0.43400.0867 | 0.41150.1163 | 0.51540.0740 |
| MAE of FGD | 0.08740.0132 | 0.09140.0180 | 0.07410.0121 |
FGD represents functional gradient density. The results all show significant difference compared to each other.
Fig. 9.
The first and third row show myelin and functional gradient density maps of original atlas and the new atlas generated by averaging all registered surface maps using different methods. The second and fourth row show their corresponding variance maps.
D. Validation on Abnormal Cortical Surfaces
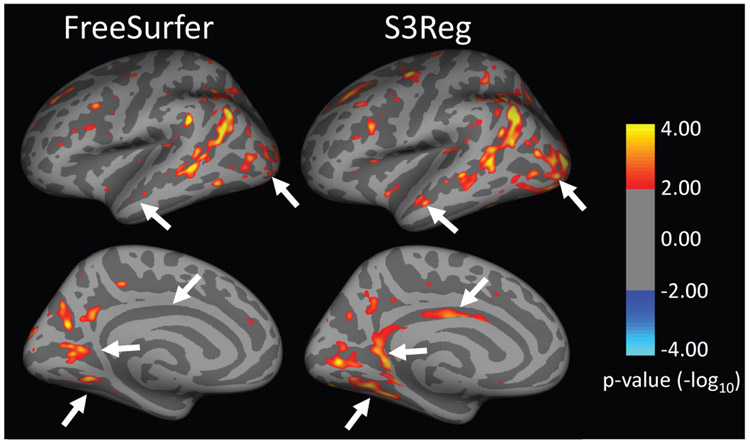
Since all the above results are based on healthy people, we additionally performed an experiment to register the cortical surfaces from Alzheimer’s Disease Neuroimaging Initiative (ADNI) database [57], to better validate our method on abnormal brains in a prospective study. We directly applied the model trained with the adult data to register the cortical surfaces from ADNI dataset to the FreeSurfer average template. Note that we did not use any ADNI data for training or fine-tuning the model. The dataset consists of a total of 428 participants from the ADNI-1 cohort, divided into 2 groups with matched demographics: healthy controls (HC) (n=229, mean age: 76.0±5.1 years, 48% female) and Alzheimer’s disease (AD) (n=199, mean age: 75.6±7.3 years, 48.5% female). A 1.5T scan with sufficient image quality from each subject’s baseline visit was selected. All cortical surfaces were reconstructed using FreeSurfer [43] and further mapped onto the sphere. Considering FreeSurfer’s popularity in the neuroimaging field, we compare our method with FreeSurfer’s registration tool (mris_register) in this experiment. Starting from the same spherical surfaces, we aligned them onto the fsaverage surface [43] using FreeSurfer and our S3Reg, respectively. Then, a vertex-wise comparison of cortical thickness between the two groups was performed using the general linear model (GLM) in FreeSurfer. Cortical surface maps showing statistically significant differences between AD and HC after correction for multiple comparisons were generated for FreeSurfer and our S3Reg, respectively, as shown in Fig. 10. The results of both registration methods show significant cortical thinning in AD in the lateral temporal cortex, inferior parietal cortex, dorsal superior frontal gyrus, lateral and medial occipital cortex. However, our S3Reg reveals more and larger clusters of significant thinning regions than FreeSurfer, especially in the superior temporal sulcus, lateral occipital cortex, dorsal superior frontal gyrus, fusiform gyrus, parahippocampal gyrus, isthmus and posterior cingulate cortex, as indicated by the arrows. These results suggest that our S3Reg aligns cortical surfaces across individuals more accurately, and thus is more effective and computationally efficient (with only 10s for S3Reg versus 30min for FreeSurfer for each surface) in detecting disease effects.
Fig. 10.
Statistical comparison of cortical thickness differences between Alzheimer’s disease (AD) and healthy control groups on the left hemisphere based on different registration methods. Results have been corrected for multiple comparisons using false discovery rating with q rate of 0.05. The color bar shows the logarithmic scale of p-values (−log10). Warmer colors (positive values) represent statistically significant cortical thinning in AD; cooler colors (negative values) represent statistically significant cortical thickening in AD.
V. Discussion
In this paper, we present the novel S3Reg framework for spherical cortical surface registration. Benefiting from the impressive advance of CNN-based deep learning techniques and the end-to-end unsupervised learning architecture, S3Reg offers superfast, diffeomorphic, accurate and flexible registrations for multimodal cortical features. As a versatile but preliminary method, there still remain several questions to be addressed.
First of all, since alignment of multimodal MRI data is a highly complex problem and there is no consensus over the best strategy for optimal cortical alignment [3], a flexible registration framework that can be easily extended to align multimodal features (such as structure, function, connectivity, or combination of them) is highly needed. We believe that our method will encourage investigations of optimal alignment strategy by automatically learning weightings for different feature combinations and regions using the powerful deep learning ability. Nevertheless, much of the focus of this paper is on serial alignment of multimodal features to mainly validate the accuracy and speed on preliminary experiments from a broad set of potential applications. Note that the choice of features and their corresponding order (sulc, sulc, sulc, curv in 3, 4, 5, 6 levels, respectively) in this paper to align cortical geometric features follows the popular configurations [1]-[3], [10], which are empirical and not proven to be the optimal solution. Further validation and exploration in finding more biologically-meaningful alignments using different alignment strategies and high dimensional features will be investigated in future works. Since our framework is very generic, cortical features derived from other imaging modalities, e.g., cortical diffusivity and fiber connectivity computed from diffusion MRI [58]-[60] and activation maps from task functional MRI, can also be conveniently used in our method. Moreover, besides cortical surfaces, our framework is also applicable to register surface features of any structures with a spherical topology, e.g., shape and connectivity features of subcortical structures.
Concerning parameters’ fine-tuning, it is intuitive that a larger weight for the smoothness term (λs) would result in a smoother deformation field but lower feature alignment accuracy, while a larger weight for the feature similarity term would lead to better feature alignment but may result in a less smoothed and even topologically-incorrect deformation field. In terms of the velocity field consistency loss (λcon), we found a weight 1.0 is sufficient to achieve consistency between different predictions from different subnetworks and further increasing it has no apparent influence on the results. However, a weight lower than 1.0 would result in inconsistency between predictions. The parcellation consistency loss (λparc) shares a similar pattern as the velocity field consistency loss. We found that the upper bound is 10.0 for the weight of the parcellation consistency loss to improve the Dice score. Regarding the correlation term and mean squared distance term, we found there is no significant difference between using which of them. This is because both similarity terms are positively correlated to the feature alignment, either of which can effectively detect the similarity between moved surface and fixed surface. Therefore we set correlation loss’s weight (λcc) as 1.0.
In terms of speed improvement, we want to clarify that the comparison is solely based on the registration process, i.e., on the same staring point and end point in the neuroimaging processing pipeline, which means it is also a solid speedup for the whole neuroimaging analysis pipelines. On the other hand, as compared methods don’t have GPU version codes, our method takes advantage of GPU to further speedup the registration process to less than 10 seconds. Since GPU is increasingly popular for computations in recent years, this is a valuable contribution to the community and we will realease our models to the public soon to greatly advance large-scale neuroimaging analyses.
It is also worth noting that the variance between the adult and infant cortical surfaces did exist and would decrease the registration accuracy if directly applying the model trained on one dataset to another dataset. The reason is that infant cortical surfaces have large differences with adult cortical surfaces [46], in terms of cortical size, shape, folding and functional features. Moreover, there also exists substantial site effects between these two datasets, caused by different scanners, imaging protocols, and image processing pipelines. Therefore, directly applying the model trained on the adult/infant dataset to the infant/adult dataset inevitably leads to a certain degree of performance decrease compared to re-training or fine-tuning the model on the target datasets, which is a general and important issue of CNN-based deep learning approaches [61]. Nevertheless, we have shown the performance of our method on an independent adult dataset, i.e., ADNI collected at multiple sites. The results have demonstrated a satisfying generalization ability of our method on different adult datasets, which will need to be further extensively validated in large-scale neuroimaging studies in the future. Finally, in order to improve the generalization ability of our registration model, we can incorporate some domain adaption techniques [62] or domain invariant features [63] to partially overcome the domain differences between adults and infants, or some harmonization techniques [38] to firstly remove the site effects and then train the network. In this way, a more powerful registration model with increased generalization ability that works well on different datasets could be obtained, which will be further investigated in our future work.
VI. Conclusion
In this work, we proposed a versatile deep learning-based framework, S3Reg, for cortical surface registration on the spherical space. We showed that the characteristics of end-to-end unsupervised learning, combined with 3 complementary orthogonal Spherical U-Nets can effectively solve the pole distortion issue and learn the deformation field on sphere without ground-truth deformation field. To guarantee a topology-preserving deformation, we integrate stationary velocity fields through novel spherical scaling and squaring differentiable layers. As shown in the paper, our S3Reg is flexible to incorporate multimodal features from structural and functional images for cortical surface registration. Importantly, compared to state-of-the-art non-learning-based methods, S3Reg runs superfast, while still achieves better or similar registration accuracy, making it preferable for large-scale neuroimaging data analyses. Furthermore, our method is independent of the choices of similarity metrics and smoothness constraints, thus providing many potential directions on learning-based spherical surface registration, and also for other genus-0 organs and computer graphics or vision applications.
Acknowledgments
This work was supported in part by National Institutes of Health (NIH) grants: MH116225 (to G. Li) and MH117943 (to L. Wang and G. Li).
Footnotes
We used the MSM algorithm in fsl/6.0.0.
Contributor Information
Fenqiang Zhao, Key Laboratory of Biomedical Engineering of Ministry of Education, Zhejiang University, Hangzhou, 310027, China; Department of Radiology and BRIC, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27514, USA..
Zhengwang Wu, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27514, USA.
Fan Wang, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27514, USA.
Weili Lin, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27514, USA.
Shunren Xia, Key Laboratory of Biomedical Engineering of Ministry of Education, Zhejiang University, Hangzhou, 310027, China.
Dinggang Shen, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27514, USA.
Li Wang, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27514, USA.
Gang Li, Department of Radiology and BRIC, University of North Carolina at Chapel Hill, Chapel Hill, NC, 27514, USA.
References
- [1].Yeo BT, Sabuncu MR, Vercauteren T, Ayache N, Fischl B, and Golland P, “Spherical demons: fast diffeomorphic landmark-free surface registration,” IEEE transactions on medical imaging, vol. 29, no. 3, pp. 650–668, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fischl B, Sereno MI, Tootell RB, and Dale AM, “High-resolution intersubject averaging and a coordinate system for the cortical surface,” Human brain mapping, vol. 8, no. 4, pp. 272–284, 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Robinson EC, Jbabdi S, Glasser MF, Andersson J, Burgess GC, Harms MP, Smith SM, Van Essen DC, and Jenkinson M, “Msm: a new flexible framework for multimodal surface matching,” Neuroimage, vol. 100, pp. 414–426, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Robinson EC, Garcia K, Glasser MF, Chen Z, Coalson TS, Makropoulos A, Bozek J, Wright R, Schuh A, Webster M et al. , “Multimodal surface matching with higher-order smoothness constraints,” Neuroimage, vol. 167, pp. 453–465, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lyu I, Kang H, Woodward ND, Styner MA, and Landman BA, “Hierarchical spherical deformation for cortical surface registration,” Medical image analysis, vol. 57, pp. 72–88, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Coalson TS, Van Essen DC, and Glasser MF, “The impact of traditional neuroimaging methods on the spatial localization of cortical areas,” Proceedings of the National Academy of Sciences, vol. 115, no. 27, pp. E6356–E6365, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li G, Wang L, Yap P-T, Wang F, Wu Z, Meng Y, Dong P, Kim J, Shi F, Rekik I et al. , “Computational neuroanatomy of baby brains: A review,” NeuroImage, vol. 185, pp. 906–925, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Vercauteren T, Pennec X, Perchant A, and Ayache N, “Diffeomorphic demons: Efficient non-parametric image registration,” NeuroImage, vol. 45, no. 1, pp. S61–S72, 2009. [DOI] [PubMed] [Google Scholar]
- [9].Arsigny V, Commowick O, Pennec X, and Ayache N, “A log-euclidean framework for statistics on diffeomorphisms,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2006, pp. 924–931. [DOI] [PubMed] [Google Scholar]
- [10].Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M et al. , “A multi-modal parcellation of human cerebral cortex,” Nature, vol. 536, no. 7615, pp. 171–178, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lombaert H, Sporring J, and Siddiqi K, “Diffeomorphic spectral matching of cortical surfaces,” in International Conference on Information Processing in Medical Imaging. Springer, 2013, pp. 376–389. [DOI] [PubMed] [Google Scholar]
- [12].Nenning K-H, Liu H, Ghosh SS, Sabuncu MR, Schwartz E, and Langs G, “Diffeomorphic functional brain surface alignment: Functional demons,” NeuroImage, vol. 156, pp. 456–465, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Conroy BR, Singer BD, Guntupalli JS, Ramadge PJ, and Haxby JV, “Inter-subject alignment of human cortical anatomy using functional connectivity,” NeuroImage, vol. 81, pp. 400–411, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dalca AV, Balakrishnan G, Guttag J, and Sabuncu MR, “Unsupervised learning for fast probabilistic diffeomorphic registration,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2018, pp. 729–738. [Google Scholar]
- [15].Balakrishnan G, Zhao A, Sabuncu MR, Guttag J, and Dalca AV, “An unsupervised learning model for deformable medical image registration,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2018, pp. 9252–9260. [Google Scholar]
- [16].Niethammer M, Kwitt R, and Vialard F-X, “Metric learning for image registration,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2019, pp. 8463–8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Krebs J, Mansi T, Delingette H, Zhang L, Ghesu FC, Miao S, Maier AK, Ayache N, Liao R, and Kamen A, “Robust non-rigid registration through agent-based action learning,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2017, pp. 344–352. [Google Scholar]
- [18].Rohé M-M, Datar M, Heimann T, Sermesant M, and Pennec X, “Svfnet: Learning deformable image registration using shape matching,” in International conference on medical image computing and computer-assisted intervention. Springer, 2017, pp. 266–274. [Google Scholar]
- [19].de Vos BD, Berendsen FF, Viergever MA, Sokooti H, Staring M, and Išgum I, “A deep learning framework for unsupervised affine and deformable image registration,” Medical image analysis, vol. 52, pp. 128–143, 2019. [DOI] [PubMed] [Google Scholar]
- [20].Zhou Y, Pang S, Cheng J, Sun Y, Wu Y, Zhao L, Liu Y, Lu Z, Yang W, and Feng Q, “Unsupervised deformable medical image registration via pyramidal residual deformation fields estimation,” arXiv preprint arXiv:2004.07624, 2020. [Google Scholar]
- [21].Zhao F, Xia S, Wu Z, Duan D, Wang L, Lin W, Gilmore JH, Shen D, and Li G, “Spherical u-net on cortical surfaces: methods and applications,” in International Conference on Information Processing in Medical Imaging. Springer, 2019, pp. 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Beg MF, Miller MI, Trouvé A, and Younes L, “Computing large deformation metric mappings via geodesic flows of diffeomorphisms,” International journal of computer vision, vol. 61, no. 2, pp. 139–157, 2005. [Google Scholar]
- [23].Krebs J, Delingette H, Mailhé B, Ayache N, and Mansi T, “Learning a probabilistic model for diffeomorphic registration,” IEEE transactions on medical imaging, vol. 38, no. 9, pp. 2165–2176, 2019. [DOI] [PubMed] [Google Scholar]
- [24].Olver PJ, Applications of Lie groups to differential equations. Springer Science & Business Media, 2000, vol. 107. [Google Scholar]
- [25].Tateno K, Navab N, and Tombari F, “Distortion-aware convolutional filters for dense prediction in panoramic images,” in Proceedings of the European Conference on Computer Vision (ECCV), 2018, pp. 707–722. [Google Scholar]
- [26].Hu H-N, Lin Y-C, Liu M-Y, Cheng H-T, Chang Y-J, and Sun M, “Deep 360 pilot: Learning a deep agent for piloting through 360 sports videos,” in 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). IEEE, 2017, pp. 1396–1405. [Google Scholar]
- [27].Coors B, Paul Condurache A, and Geiger A, “Spherenet: Learning spherical representations for detection and classification in omnidirectional images,” in Proceedings of the European Conference on Computer Vision (ECCV), 2018, pp. 518–533. [Google Scholar]
- [28].Cheng J, Dalca AV, Fischl B, and Zllei L, “Cortical surface registration using unsupervised learning,” NeuroImage, vol. 221, p. 117161, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention. Springer, 2015, pp. 234–241. [Google Scholar]
- [30].Zhao Q, Zhu C, Dai F, Ma Y, Jin G, and Zhang Y, “Distortion-aware cnns for spherical images.” in IJCAI, 2018, pp. 1198–1204. [Google Scholar]
- [31].Rao Y, Lu J, and Zhou J, “Spherical fractal convolutional neural networks for point cloud recognition,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2019, pp. 452–460. [Google Scholar]
- [32].Jiang C, Huang J, Kashinath K, Marcus P, Niessner M., et al. “Spherical cnns on unstructured grids,” arXiv preprint arXiv:1901.02039, 2019. [Google Scholar]
- [33].Wu Z, Li G, Wang L, Shi F, Lin W, Gilmore JH, and Shen D, “Registration-free infant cortical surface parcellation using deep convolutional neural networks,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2018, pp. 672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Seong S-B, Pae C, and Park H-J, “Geometric convolutional neural network for analyzing surface-based neuroimaging data,” Frontiers in Neuroinformatics, vol. 12, p. 42, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Parvathaneni P, Bao S, Nath V, Woodward ND, Claassen DO, Cascio CJ, Zald DH, Huo Y, Landman BA, and Lyu I, “Cortical surface parcellation using spherical convolutional neural networks,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2019, pp. 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhao F, Xia S, Wu Z, Wang L, Chen Z, Lin W, Gilmore JH, Shen D, and Li G, “Spherical u-net for infant cortical surface parcellation,” in 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). IEEE, 2019, pp. 1882–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhao F, Wu Z, Wang L, Lin W, Gilmore JH, Xia S, Shen D, and Li G, “Spherical deformable u-net: Application to cortical surface parcellation and development prediction.” IEEE Transactions on Medical Imaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhao F, Wu Z, Wang L, Lin W, Xia S, Shen D, Li G, Consortium UBCP et al. , “Harmonization of infant cortical thickness using surface-to-surface cycle-consistent adversarial networks,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2019, pp. 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Esteves C, Allen-Blanchette C, Makadia A, and Daniilidis K, “Learning so (3) equivariant representations with spherical cnns,” in Proceedings of the European Conference on Computer Vision (ECCV), 2018, pp. 52–68. [Google Scholar]
- [40].Cohen TS, Geiger M, Köhler J, and Welling M, “Spherical cnns,” arXiv preprint arXiv:1801.10130, 2018. [Google Scholar]
- [41].Kingma DP and Ba J, “Adam: A method for stochastic optimization,” arXiv preprint arXiv:1412.6980, 2014. [Google Scholar]
- [42].Möller T, “A fast triangle-triangle intersection test,” Journal of graphics tools, vol. 2, no. 2, pp. 25–30, 1997. [Google Scholar]
- [43].Fischl B, “Freesurfer,” Neuroimage, vol. 62, no. 2, pp. 774–781, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dale AM, Fischl B, and Sereno MI, “Cortical surface-based analysis: I. segmentation and surface reconstruction,” Neuroimage, vol. 9, no. 2, pp. 179–194, 1999. [DOI] [PubMed] [Google Scholar]
- [45].Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT et al. , “An automated labeling system for subdividing the human cerebral cortex on mri scans into gyral based regions of interest,” Neuroimage, vol. 31, no. 3, pp. 968–980, 2006. [DOI] [PubMed] [Google Scholar]
- [46].Li G, Wang L, Shi F, Gilmore JH, Lin W, and Shen D, “Construction of 4d high-definition cortical surface atlases of infants: Methods and applications,” Medical image analysis, vol. 25, no. 1, pp. 22–36, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Li G, Nie J, Wang L, Shi F, Gilmore JH, Lin W, and Shen D, “Measuring the dynamic longitudinal cortex development in infants by reconstruction of temporally consistent cortical surfaces,” Neuroimage, vol. 90, pp. 266–279, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wang L, Li G, Shi F, Cao X, Lian C, Nie D, Liu M, Zhang H, Li G, Wu Z et al. , “Volume-based analysis of 6-month-old infant brain mri for autism biomarker identification and early diagnosis,” in International conference on medical image computing and computer-assisted intervention. Springer, 2018, pp. 411–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Sun L, Zhang D, Lian C, Wang L, Wu Z, Shao W, Lin W, Shen D, Li G, Consortium UBCP et al. , “Topological correction of infant white matter surfaces using anatomically constrained convolutional neural network,” NeuroImage, vol. 198, pp. 114–124, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang F, Lian C, Wu Z, Zhang H, Li T, Meng Y, Wang L, Lin W, Shen D, and Li G, “Developmental topography of cortical thickness during infancy,” Proceedings of the National Academy of Sciences, vol. 116, no. 32, pp. 15 855–15 860, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR et al. , “The minimal preprocessing pipelines for the human connectome project,” Neuroimage, vol. 80, pp. 105–124, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hu D, Zhang H, Wu Z, Wang F, Wang L, Smith JK, Lin W, Li G, and Shen D, “Disentangled-multimodal adversarial autoencoder: Application to infant age prediction with incomplete multimodal neuroimages,” IEEE Transactions on Medical Imaging, vol. 39, no. 12, pp. 4137–4149, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Huang Y, Wang F, Wu Z, Chen Z, Zhang H, Wang L, Lin W, Shen D, Li G, U. B. C. P. Consortium et al. , “Construction of spatiotemporal infant cortical surface functional templates,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2020, pp. 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gordon EM, Laumann TO, Adeyemo B, Huckins JF, Kelley WM, and Petersen SE, “Generation and evaluation of a cortical area parcellation from resting-state correlations,” Cerebral cortex, vol. 26, no. 1, pp. 288–303, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Paszke A, Gross S, Chintala S, Chanan G, Yang E, DeVito Z, Lin Z, Desmaison A, Antiga L, and Lerer A, “Automatic differentiation in pytorch,” 2017. [Google Scholar]
- [56].Rohlfing T, “Image similarity and tissue overlaps as surrogates for image registration accuracy: widely used but unreliable,” IEEE transactions on medical imaging, vol. 31, no. 2, pp. 153–163, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jack CR Jr, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell JL, Ward C et al. , “The alzheimer’s disease neuroimaging initiative (adni): Mri methods,” Journal of Magnetic Resonance Imaging, vol. 27, no. 4, pp. 685–691, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li K, Guo L, Li G, Nie J, Faraco C, Zhao Q, Miller LS, and Liu T, “Cortical surface based identification of brain networks using high spatial resolution resting state fmri data,” in 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. IEEE, 2010, pp. 656–659. [Google Scholar]
- [59].Li G, Liu T, Ni D, Lin W, Gilmore JH, and Shen D, “Spatiotemporal patterns of cortical fiber density in developing infants, and their relationship with cortical thickness,” Human brain mapping, vol. 36, no. 12, pp. 5183–5195, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rekik I, Li G, Yap P-T, Chen G, Lin W, and Shen D, “Joint prediction of longitudinal development of cortical surfaces and white matter fibers from neonatal mri,” NeuroImage, vol. 152, pp. 411–424, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shen D, Wu G, and Suk H-I, “Deep learning in medical image analysis,” Annual review of biomedical engineering, vol. 19, pp. 221–248, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].He Y, Carass A, Zuo L, Dewey BE, and Prince JL, “Self domain adapted network,” in International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2020, pp. 437–446. [Google Scholar]
- [63].Zhong T, Zhao F, Pei Y, Ning Z, Liao L, Wu Z, Niu Y, Wang L, Shen D, Zhang Y et al. , “Dika-nets: Domain-invariant knowledge-guided attention networks for brain skull stripping of early developing macaques,” NeuroImage, p. 117649, 2020. [DOI] [PubMed] [Google Scholar]