Abstract
Mice have provided critical mechanistic understandings of clinical traits underlying metabolic syndrome (MetSyn) and susceptibility to MetSyn in mice is known to vary among inbred strains. We investigated the diet- and strain-dependent effects on metabolic traits in the eight Collaborative Cross (CC) founder strains (A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ, NZO/HILtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ). Liver transcriptomics analysis showed that both atherogenic diet and host genetics have profound effects on the liver transcriptome, which may be related to differences in metabolic traits observed between strains. We found strain differences in circulating trimethylamine N-oxide (TMAO) concentration and liver triglyceride content, both of which are traits associated with metabolic diseases. Using a network approach, we identified a module of transcripts associated with TMAO and liver triglyceride content, which was enriched in functional pathways. Interrogation of the module related to metabolic traits identified NADPH oxidase 4 (Nox4), a gene for a key enzyme in the production of reactive oxygen species, which showed a strong association with plasma TMAO and liver triglyceride. Interestingly, Nox4 was identified as the highest expressed in the C57BL/6J and NZO/HILtJ strains and the lowest expressed in the CAST/EiJ strain. Based on these results, we suggest that there may be genetic variation in the contribution of Nox4 to the regulation of plasma TMAO and liver triglyceride content. In summary, we show that liver transcriptomic analysis identified diet- or strain-specific pathways for metabolic traits in the Collaborative Cross (CC) founder strains.
Keywords: Collaborative Cross, diet, genetic backgrounds, metabolic syndrome, transcriptomics
INTRODUCTION
Metabolic syndrome (MetSyn) is a cluster of clinical traits [including elevated blood lipids and glucose concentrations, increased blood pressure, and central obesity (1)] that is highly associated with the risk of diabetes and cardiovascular disease. In particular, the liver plays a central role in regulating these clinical traits and thus metabolic imbalance in the liver can affect susceptibility to MetSyn. For example, alterations in hepatic metabolism can induce dyslipidemia (2, 3) and production of novel metabolites associated with increased risk of MetSyn such as trimethylamine N-oxide (TMAO) (4–9). Specifically, TMAO is converted from trimethylamine (TMA) produced in the intestine by the activity of liver flavin monooxygenase 3 (FMO3) (10). Plasma levels of TMAO are determined by genetic variation (11, 12) and diet (13), and FMO3 may promote dyslipidemia by regulating several genes involved in hepatic gluconeogenesis and lipogenesis (14, 15). There are a number of factors that affect susceptibility to MetSyn including genetics and environmental queues such as diet. In particular, genetics has been shown to be an important factor significantly affecting the susceptibility of MetSyn in humans (16–18). Identifying the genetic architecture and biological pathways that modulate the risk of MetSyn is essential to discovering more effective therapeutic approaches.
The mouse has been indispensable for the study of MetSyn (19, 20) as both genetic and environmental factors can be well controlled in mouse models. The phenotypic spectrum present in various mouse strains provides an opportunity to discover genetic functions related to metabolic traits. However, most studies have been conducted with a small number of mouse strains with limited genetic variation. Almost exclusively mouse gene knockout studies are conducted in C57BL/6J mice, with a smaller number performed in FVB and 129/Sv. Studies of mice with targeted overexpression or inactivation of a gene often report specific phenotypic changes, but these effects are highly influenced by the background strain of the mice harboring the genetic mutation(s) (21). Thus, understanding the underlying genetic architecture remains important to further our understanding of MetSyn.
An alternative approach is to use forward genetic studies utilizing a wide variety of mice to investigate how natural variants affect MetSyn. Classically, these have been done in F2 crosses but more recently multiparent advanced generation intercross populations have been developed, such as the Collaborative Cross (CC). The CC mouse population is derived from five classic inbred mouse strains A/J, C57BL/6J (B6), 129S1/SvImJ (129), NOD/ShiLtJ (NOD), NZO/HILtJ (NZO), and three wild-derived strains CAST/EiJ (CAST), PWK/PhJ (PWK), and WSB/EiJ (WSB) (22). These eight CC founder strains are highly genetically diverse as they contain ∼40 million single-nucleotide polymorphisms (SNPs) and numerous insertions and deletions. The genetic diversity across the CC founder strains provides an opportunity to assess the effect of host genetics on metabolic traits. The genetic and phenotypic diversity of the combined eight founder strains is similar to the interindividual diversity of the human population. The utilization of the genomic sequence of the eight CC founder strains provides an unprecedented unique resource for genetic mapping and correlation studies (23–34)
Here, we evaluate the variability of metabolic traits and perform global liver transcriptomics from the eight CC founder strains fed a standard purified diet or an atherogenic diet. Our studies focus on female mice as a number of studies have reported that female mice are generally more susceptible to atherosclerosis than male mice (30, 32, 35–38). Our study shows that both diet and genetic background have a profound influence on metabolic traits and the associated transcriptional network. For example, analyses of differentially expressed genes (DEGs) and gene networks suggest that diet- or strain-specific DEGs and gene clusters are enriched for biological pathways known to affect metabolic traits. We identify a novel co-expression module associated with plasma TMAO and describe a candidate gene, NADPH oxidase 4 (Nox4), which is a hydrogen peroxide NADPH oxidase isoform. We found that the Nox4 gene showed a strong association with plasma TMAO and liver triacylglycerol (TG) in the liver transcriptome. Our results demonstrate the utility of leveraging the CC to understand dietary influences on the liver transcriptome and disease-associated traits.
METHODS
Ethics Statement
We followed all NIH animal welfare guidelines and animal care. The study protocols were approved by the North Carolina Research Campus (NCRC) Animal Care and Use Committee.
Study Design
Eight female mice from each of the eight different CC founder strains (A/J, C57BL/6J, 129S1/SvImJ, NOD/ShiLtJ, NZO/HILtJ, CAST/EiJ, PWK/PhJ, and WSB/EiJ) were purchased from Jackson Laboratories (Bar Harbor, ME) at 4 wk of age. The study design was reported previously (30). Briefly, mice were housed under standard conditions (12 h light:dark, temperature- and humidity-controlled conditions) with free access to water and a nutritionally purified AIN-93M diet (No. D10012M; Research Diets Inc., New Brunswick, NJ). Four weeks after AIN-93M administration, mice were assigned to either the AIN-93M diet or high-fat cholic acid (HFCA) diet (No. D12109C; Research Diets Inc.) for an additional 16 wk (n = 4 per diet per strain) (Supplemental Table S1; all Supplemental material is available at https://doi.org/10.6084/m9.figshare.13107797). After feeding this diet for 16 wk, mice were euthanized for tissue collection. Euthanasia of all mice was performed by cervical dislocation after anesthesia with isoflurane. For eight CC founder strains of mice, liver, gonadal fat, spleen, and heart were collected and weighed upon euthanasia.
Body Composition
Body composition (fat mass and lean mass) was assessed using EchoMRI-100H (Echo MRI LLC, Houston, TX) at 8 wk and 24 wk, respectively. Body fat and lean mass percentages were calculated by dividing fat mass by body weight and lean mass by body weight, respectively.
Plasma Clinical Metabolic Markers
Mice at 8 wk or 24 wk of age fasted for 4 h before blood collection via retro-orbital bleeding. Blood was collected into EDTA-containing tubes and plasma was separated by centrifugation at 10,000 g for 10 min. Plasma alanine aminotransferase (ALT), aspartate aminotransferase (AST), TG, total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), glucose, and urea were measured by the Biolis 24i Analyzer (Carolina Liquid Chemistries, Winston-Salem, NC). Very low-density lipoprotein cholesterol/low-density lipoprotein cholesterol (VLDL-C/LDL-C) levels were determined by subtracting high-density lipoprotein cholesterol (HDL-C) from total cholesterol. Insulin was assessed using the Alpco Mouse Ultrasensitive Insulin ELISA assay (Alpco, Salem, NH) and measured at 450 nm using a microplate reader (Bio-Tek, Winooski, VT).
Plasma Metabolite Analysis Using LC/MS/MS
TMAO analytes were measured by the Metabolomics Core Facility in the NCRC (24, 30). Briefly, plasma was extracted with internal standards TMAO-d9 (Cambridge Isotope Laboratories, Tewksbury, MA), creatinine-d3 (CDN Isotopes Inc., Quebec, Canada), choline-d9 (Cambridge Isotope Laboratories), and betaine-d9 (Sigma-Aldrich, St. Louis, MO), incubated on ice for 10 min, and centrifuged at 15,000 g for 2 min. The concentrations of TMAO, creatinine, choline, and betaine were quantified by using liquid chromatography-stable isotope dilution-multiple reaction monitoring mass spectrometry (LC-SID-MRM/MS). Chromatographic separations were conducted on an Atlantis Silica HILIC 3 μm 4.6 × 150 mm column (Waters Corp, Milford, MA) using a Waters ACQUITY UPLC system. The metabolites and their corresponding isotopes were monitored on a Waters TQ detector using characteristic precursor-product ion transitions: 76→58 for TMAO, 85→66 for TMAO-d9, 114→86 for creatinine, 117→89 for creatinine-d9, 104→45 for choline, 113→45 for choline-d9, 118→59 for betaine, and 127→68 for betaine-d9. Concentrations of each metabolite in samples were determined by calculating the peak area ratio of the metabolite versus its isotope.
Hepatic Triacylglycerol
Hepatic TG levels were quantified via Folch extraction. Mouse liver was collected, frozen, and stored at −80°C before analysis. Frozen mouse liver tissue was thoroughly homogenized for 5 min in 500 μL of a 2:1 vol/vol chloroform/methanol mix and then equilibrated for 15 min at room temperature. After adding 100 μL of 0.9% wt/vol NaCl to each sample, the samples were vortexed for 1 min and centrifuged at 2,000 g for 15 min at 4°C. The lower organic phase was separated and evaporated in Eppendorf tubes under a stream of nitrogen for 1 h. After evaporation, each tube was resuspended with 500 μL of a 0.5% Triton X-100/PBS solution, sonicated for 5 min using Bioruptor, and placed in a drying bath at 55°C for 5 min. Hepatic TG was measured using a colorimetric assay (Infinity, Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions as follows: 2 μL of the standards, samples, and blanks were pipetted into a 96-well plate in duplicate and 200 μL of the Infinity reagent was added to the 96-well plate. Absorbance (500/660 nm) was measured on a 96-well plate reader.
Metabolic Rate and Activity
Mice were placed into individual indirect calorimetry cages (Phenomaster, TSE SYSTEMS, Chesterfield, MO) at the week after 16 wk of the experimental diet challenge to obtain O2 consumption (V̇o2), respiratory exchange ratio (RER), and feed consumption measurements. After a 24-h acclimation period, data were collected during the following 24-h period. Basal activity was measured in three dimensions (x, y, and z) as breaks in the two infrared light beam frames that surrounded each cage. Rearing was detected by beam breaks in the z-axis and total physical activity was defined as the sum of beam breaks in all three axes in counts. Feed was available ad libitum and consumption was measured by weighing sensors that held containers for feed and water, respectively, and recorded the amount of feed or water consumed.
RNA-Seq Library Preparation and Sequencing
Total RNA was extracted from frozen liver samples with the Maxwell 16 LEV simplyRNA Tissue Kit (Promega, Madison, WI) according to the manufacturer’s protocol. The quality and amount of liver RNA were evaluated using the Qubit RNA HS assay kit (Thermo Fisher Scientific, Waltham, MA) and the Biorad Bioanalyzer Chip (Hercules, CA). RNA samples from 24 mice fed the AIN-93M diet and 24 mice fed the HFCA diet were submitted to the David H. Murdock Research Institute (Kannapolis, NC). The RNA-seq libraries were constructed from total RNA following the Illumina TruSeq RNA library construction protocol. The size of the adapted fragments in the libraries was determined by running an Agilent DNA 1000 Chip. In parallel, the DNA concentration in the libraries was quantified using Real-Time PCR (Kapa Biosystems, Wilmington, MA). The pooled libraries were sequenced on the Illumina HiSeq 2500 sequencing to achieve 100 bp paired-end reads in a total of 11 lanes (Illumina Inc., San Diego, CA). Seven pools were created from 44 samples, with 6–7 samples per pool in equimolar concentrations. Two B6 and two CAST samples from the control diet were deep sequenced with one sample per lane because B6 strain is the most commonly used laboratory mice and CAST strain is a wild-derived mouse strain that is genetically distinct from the B6 strain. Raw data were deposited at National Center for Biotechnology Information’s Gene Expression Omnibus (GEO accession GSE159992).
RNA-Seq Mapping and Quantification
Raw read data were filtered using HTStream (version 1.1.0, https://github.com/ibest/HTStream), which included screening for contaminants (such as rRNA and the sequencing control PHiX), PCR deduplication readout, quality-based trimming, adapter trimming, and overlapping paired-end reads. We randomly took 50% of the reads from the two B6 and two CAST deep sequenced samples. STAR (version 2.7.0f) (39) was used to align the processed data to custom reference mouse genomes constructed by incorporating genetic variants of eight founder strains into reference mouse genome GRCm38 using g2gtools (https://github.com/churchill-lab/g2gtools). Custom R code was then used for sequence read and alignment quality assessment as well as collating counts into a single table for downstream analysis. We obtained median 32 million pass-filter reads, 20 million uniquely mapped reads, and 17 million reads mapped to genes per library (Supplemental Table S2). We filtered in 12,502 transcripts with median counts per million (CPM) greater than 1 in 48 liver samples (Supplemental Table S3).
Differential Gene Expression and Enrichment Analysis
Diet or strain-specific differential expression genes (DEGs) analysis was performed using the R package “limma” version 3.11 (40) from TMM (trimmed mean of M values) normalized log2 transformed count per million (CPM) values.
Enrichment analyses for DEGs or modules were performed using enrichR (41) to generate enrichment terms and pathways from the Gene Ontology (GO) Biological Process 2018, Kyoto Encyclopedia of Genes and Genomes (KEGG) 2019 Mouse, and Jenson Diseases (42). This analysis identifies differential enrichment terms and pathways for the functional categories of DEGs or transcripts in the module. The GO Biological process 2018 contains 5,103 terms and 14,433 genes. Although it is clear that individual GO terms can be found in related classes of ontology, GO terms do not occupy strictly fixed levels in a hierarchy. Each of the GO terms identified is associated with a unique GO annotation number that relates to a specific function. Both the Gene Ontology website (http://geneontology.org/docs/faq/) and specific tool (enrichR) do not utilize a specific hierarchy, thus all available terms are used in the analysis.
Weighted Gene Co-Expression Network Analyses
Co-expression gene modules were calculated using Weighted Gene Co-expression Network Analysis (WGCNA) version 1.13 (100), which performs network construction by module detection. For the WGCNA analysis, log2 transformed 12,502 CPM measured in 48 liver samples from CC founder strains were included. We used a soft thresholding power of 9 by the scale-free topology criterion in the WGCNA package. We chose the “unsigned” network type to maintain the relationship of the negatively correlated gene and the “signed” topological overlap matrix (TOM) to exclude the connections influenced by noise (43, 44). We set 20 as the minimum number of genes to form a module. The network connectivity of each gene was calculated as the sum of the intensity of connectivity with all genes in the other network.
For each module, the first principal component (PC1) and the module eigengene (ME) were calculated and used for the correlation with metabolic traits. The average number of transcripts per module was 595, ranging from 44 (royalblue module) to 4,020 (turquoise module).
Assessing Genetic Variation at FMO3 and NOX4 Loci
To better understand the SNPs of the Fmo3 and Nox4 genes discovered as candidate genes, we compared 658 SNPs (Chromosome 1: 162,954,207–162,984,416) in Fmo3 and 2,641 SNPs (Chromosome 7: 87,246,136–87,398,699) in Nox4 in eight CC founder mouse genomes available from the Sanger Institute’s mouse database (www.sanger.ac.uk) and calculated SNP similarity between the reference genome, B6 strain, and the other seven strains. For example, in 2,641 Nox4 SNPs, the number of SNPs difference between A/J and B6 strain is 663. Therefore, dissimilarity between A/J and B6 for Nox4 SNPs is 663 divided by 2,641, which is 0.251 and the similarity (%) is 74.9% (100%–25.1%). The effects of SNP mutations on the protein function for the discovered candidate genes, Fmo3 and Nox4, were determined by a web-based tool, Protein Variation Effect Analyzer-PROVEAN (45) and Sorting Intolerant From Tolerant—SIFT (46).
Other Statistical Analysis
All statistical analyses were performed in R (v.3.5.3) (R Core Team) (47). Diet or strain effects were assessed using two-group Mann–Whitney U (Wilcoxon rank) or Kruskal–Wallis statistical test, respectively. Diet by strain interaction effect was assessed using a two-way ANOVA test. Tukey’s multiple comparison test was performed to compare groups with different diets and strains for plasma TMAO and liver TG traits. Spearman’s correlation was used to correlate the clinical traits and liver transcripts. The P values were adjusted using the Benjamini–Hochberg (BH) false discovery rate (FDR) procedure (48), and correlation coefficients and adjusted P value were visualized using the “pheatmap” package (49). Significance was determined with a P value <0.05.
RESULTS
Hepatic Transcriptomics Reveals Diet-Specific Differences in the CC Progenitors
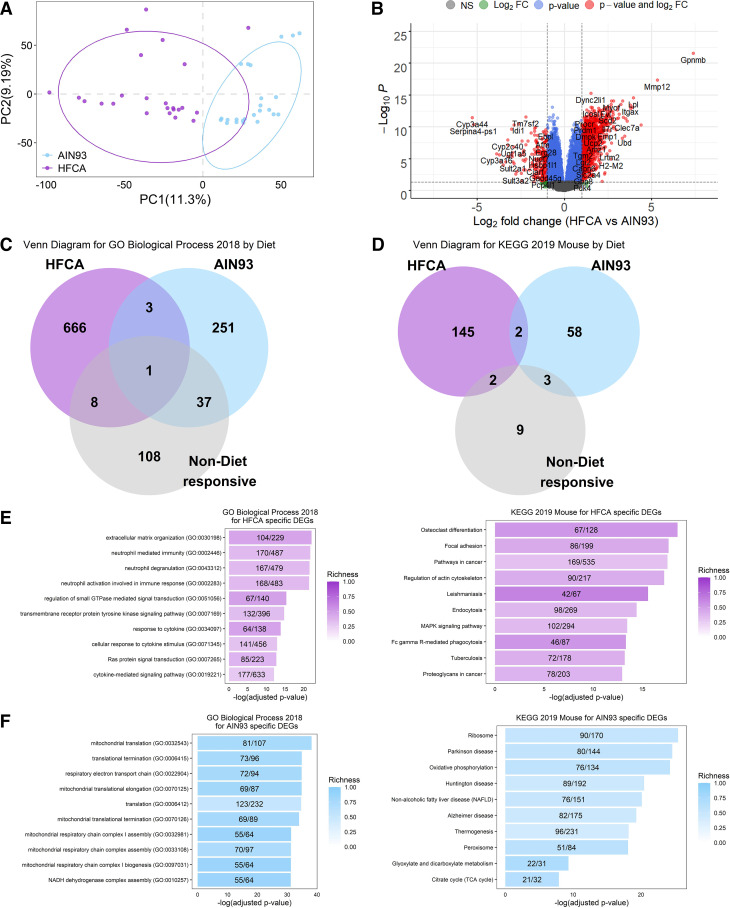
A number of studies have reported that thousands of genes are differentially expressed by a high-fat diet as compared to a control diet (50–53). We profiled global gene expression by RNA-Seq in three female mice fed the AIN-93M diet and three female mice fed a HFCA diet from each CC founder strains (n = 48 in total). We found that 12,502 transcripts were expressed in the liver and performed in differential gene expression analysis to identify transcriptional responses to diet perturbation regardless of strain by combining all eight founder strains. Principal component analysis revealed distinct differences in global gene expression between diets (Fig. 1A). A total of 6,411 genes showed significant differential expression (FDR adjusted P < 0.05; Supplemental Table S4) between mice fed the HFCA diet (3,157 genes were upregulated) compared with those fed the AIN-93M diet (3,254 genes were upregulated). These 6,411 genes we define as our “Core Diet DEGs.” A volcano plot showed that genes such as Gpnmb (Glycoprotein Nmb), Mmp12 (Matrix metalloproteinase-12), Lpl (Lipoprotein lipase), and Col1a1 (Collagen type I alpha 1 chain) previously reported as fatty liver-related genes (54–56) were also identified as HFCA diet-specific differentially expressed genes (DEGs) in this study (Fig. 1B).
Figure 1.
Effect of diet on liver gene expression in female eight Collaborative Cross (CC) founder strains mice. We identified the effect of an atherogenic diet on global liver gene expression in eight CC founder strains (n = 48). Principal component (PC) analysis (A) and Volcano plot (B) between high-fat and cholic acid (HFCA) diet and AIN-93M diet in liver gene expression in eight CC founder strains. B: horizontal dotted lines indicate adj. P < 0.05, vertical dotted gray lines indicate a twofold difference. Venn diagram to identify overlapping gene ontology (GO) Biological Process 2018 terms (C) and Kyoto Encyclopedia of Genes and Genomes (KEGG) 2019 Mouse pathways (D) between upregulated genes in HFCA diet, upregulated genes in AIN-93M diet, or nondiet responsive genes in enrichment analysis. Top 10 GO terms and KEGG pathways of upregulated genes in HFCA diet (E) and AIN-93M diet (F) identified in enrichment analysis. Pathways were ordered from top to bottom by significance (highest to lowest) and colored by gene richness.
We also determined which potential biological aspects of liver metabolism were reflected by the DEGs between the diets. Enrichment analysis revealed upregulated genes in a specific diet enriched in a number of GO Biological Processes (Fig. 1C) and KEGG pathways (Fig. 1D). In genes that were not identified as DEGs between the two diets (nondiet responsive genes: gray color denoted), a relatively small number of GO terms and KEGG pathways were identified compared with DEGs upregulated in a specific diet (Fig. 1, C and D). In particular, there was an upregulation of genes involved in immune response in the HFCA diet-fed mice whereas genes involved in mitochondrial function were upregulated in the AIN-93M diet-fed mice. GO Biological Processes and KEGG pathways that were highly enriched in HFCA diet-fed mice included “Extracellular matrix organization (GO: 00030196)” and “Neutrophil mediated immunity (GO: 0002446)” (−logP > 21) and “Osteoclast differentiation” (KEGG pathway) (−logP > 18). GO Biological Processes and KEGG pathways that were enriched in AIN-93M diet-fed mice included “Mitochondrial translation (GO: 0032543)” and “Respiratory electron transport chain (GO: 0022904)” (−logP > 34) and “Thermogenesis, Oxidative phosphorylation, and Non-alcoholic fatty liver disease” (KEGG pathway) (−logP > 19) (Fig. 1, E and F and Supplemental Tables S5 and S6).
DEGs between Two Diets by Eight Founder Strains Vary in Biological Pathways
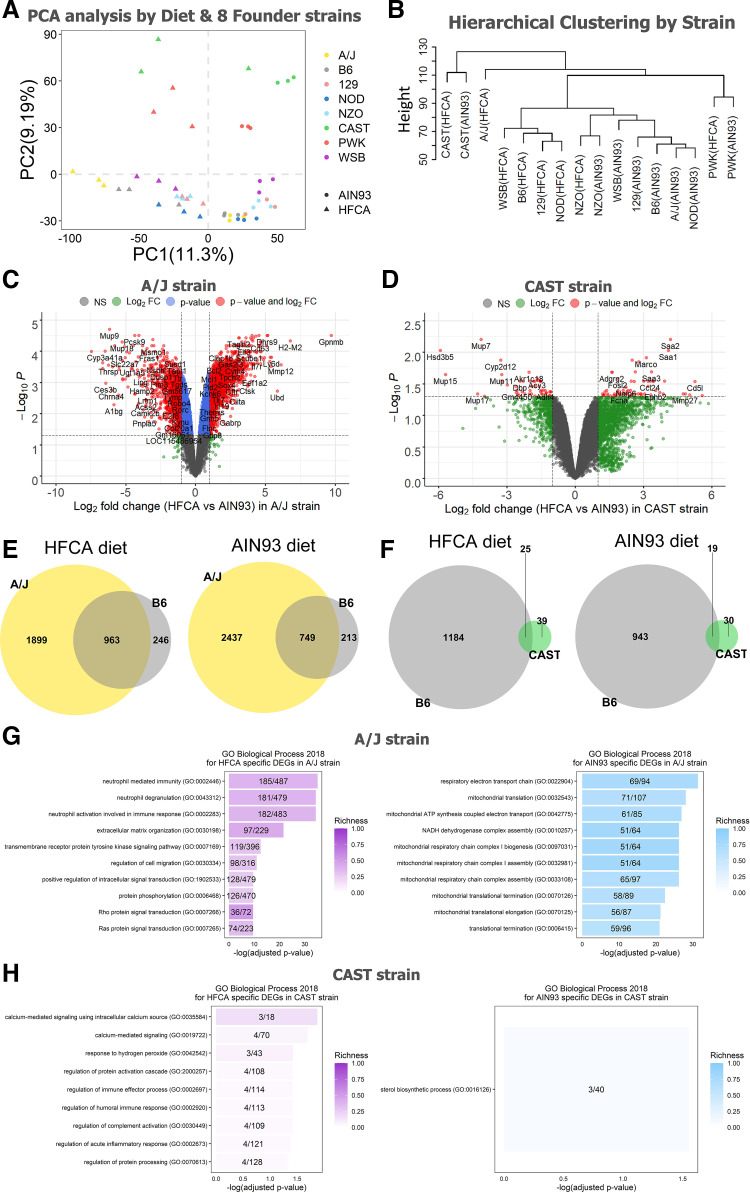
To investigate the effect of genetic background on the global hepatic gene expression, we next performed transcriptomic analysis of strain differences among progenitor strains of the CC. Hierarchical clustering and principal component analysis of the liver transcriptome demonstrate that transcript abundance was highly variable across the eight founder strains. This analysis reveals several interesting findings. First, the wild-derived founder strains, CAST and PWK, are distinct in their response to the other CC progenitor strains. Moreover, this difference is apparent for both diets (Fig. 2, A and B). Second, the B6, 129, NOD, and WSB strains showed distinct liver transcriptome patterns by diet and the NZO, CAST, and PWK strains were less affected by diet than other strains based on the hierarchical clustering (Fig. 2B). Finally, the AJ strain showed the most extreme diet response (Fig. 2B). Based on these results, we performed a DEG analysis between the HFCA diet-fed mice and the AIN-93M diet-fed mice for each strain to identify which strain was most or less responsive to the diet. As shown in Table 1, among the eight strains, the A/J strain showed the most significant difference in hepatic gene expression between the two diets (2,862 genes in the HFCA diet and 3,186 genes in the AIN-93M diet), and CAST was found to have a subdued diet response (64 genes in HFCA diet and 49 genes in AIN-93M diet), although we acknowledge a variable response to the HFCA in the CAST replicates. Furthermore, among diet-specific upregulated genes for each strain, the ratio of uniquely upregulated genes only in that strain was the highest in A/J strain (18.6% in HFCA diet and 27.7% in AIN-93M diet).
Figure 2.
Effect of genetic background on hepatic gene expression in the eight Collaborative Cross (CC) founder strains in female mice. Principal component analysis from AIN-93M diet or HFCA diet (A) and hierarchical clustering (B) determined that the major source of variation in gene expression was due to genetic variation among the eight strains. Venn diagrams to identify overlapping upregulated genes between A/J strain and B6 strain (C) and between CAST strain and B6 strain (D) in each diet. Volcano plots between high-fat and cholic acid (HFCA) diet and AIN-93M diet in liver gene expression in A/J strain (E) and CAST strain (F). Horizontal dotted lines indicate adj. P < 0.05, vertical dotted gray lines indicate a twofold difference. Top 10 GO terms of upregulated genes in HFCA diet and AIN-93M diet identified in enrichment analysis in A/J strain (G) and CAST strain (H). Pathways were ordered from top to bottom by significance (highest to lowest) and colored by gene richness. n = 6 mice for each founder, three for AIN-93M diet and three for HFCA diet-fed mice. A/J (yellow), B6 (gray), C57BL/6J; 129 (pink), 129S1/SvlmJ; NOD (blue), NOD/ShiLtJ; NZO (light blue), NZO/HILtJ; CAST (green), CAST/EiJ; PWJ (red), PWK/PhJ; WSB (purple), WSB/EiJ.
Table 1.
The number of DEGs between high-fat cholic acid diet and AIN-93M diet for each strain in liver gene expression in eight CC founder strains
| Strain | HFCA Diet-Upregulated Genes | HFCA Diet-Upregulated Genes that Are Unique To the Strain | AIN-93M Diet-Upregulated Genes | AIN-93M Diet-Upregulated Genes that Are Unique To the Strain |
|---|---|---|---|---|
| A/J | 2,862 | 532 (18.6%) | 3,186 | 881 (27.7%) |
| B6 | 1,209 | 86 (7.1%) | 962 | 72 (7.5%) |
| 129 | 1,352 | 77 (5.7%) | 772 | 85 (11.0%) |
| NOD | 1,141 | 103 (9.0%) | 750 | 103 (13.7%) |
| NZO | 2,438 | 452 (18.5%) | 2,225 | 362 (16.3%) |
| CAST | 64 | 6 (9.4%) | 49 | 9 (18.4%) |
| PWK | 414 | 63 (15.2%) | 490 | 88 (18.0%) |
| WSB | 2,482 | 322 (13.0%) | 2,006 | 380 (18.9%) |
| At least one strain | 4,503 | 4,725 | ||
| Overlap between all-strain and individual strain | 2,958 | 2,885 |
n = 48 (6 mice per eight strains and 24 mice per two diets). CC, Collaborative Cross; DEG, differential expression gene; HFCA, high-fat and cholic acid.
Based on the result of strain-specific DEGs between HFCA diet and AIN-93M diet, we also compared the differences in GO Biological Process 2018 terms and KEGG 2019 Mouse pathways between the two diets for each strain. We detected the highest number of 616 GO terms and 154 KEGG pathways in A/J while the number of GO (9 terms) and KEGG (9 pathways) in CAST was the least (Table 2). Therefore, we targeted the A/J and CAST strains, which showed the most extreme differences in DEG analysis. The top upregulated genes in each diet differed significantly between the two strains. For example, only four genes were overlapped in the top 100 upregulated genes between the two strains in both diets (Fig. 2, C and D). In the comparison of the A/J strain (most diet responsive) and the B6 strain (most utilized resource), we also found that 66.3% of the upregulated genes (1,899 out of 2,862 genes) in the HFCA diet-fed A/J strain and 76.5% of the upregulated genes (2,437 out of 3,186 genes) in the AIN-93M diet-fed A/J strain were uniquely identified only in the A/J strain (Fig. 2E). Likewise, 60.9% of upregulated genes (39 out of 64 genes) in HFCA diet-fed CAST strain and 61.2% of upregulated genes (30 out of 49 genes) in AIN-93M diet-fed CAST strain were uniquely identified only in the least diet responsive CAST strain (Fig. 2F).
Table 2.
The number of GO Biological Process 2018 terms and KEGG 2019 Mouse pathways between upregulated genes in HFCA diet, upregulated genes in AIN-93M diet, or non-diet responsive genes in enrichment analysis for each strain
| Strain | HFCA Diet-Upregulated Genes |
AIN-93M Diet-Upregulated Genes |
||
|---|---|---|---|---|
| No. GO | No. KEGG | No. GO | No. KEGG | |
| A/J | 616 | 154 | 230 | 64 |
| B6 | 155 | 86 | 69 | 28 |
| 129 | 309 | 108 | 48 | 28 |
| NOD | 216 | 89 | 21 | 13 |
| NZO | 479 | 119 | 191 | 53 |
| CAST | 9 | 9 | 1 | 4 |
| PWK | 82 | 35 | 59 | 20 |
| WSB | 477 | 124 | 163 | 55 |
n = 48 (6 mice per eight strains and 24 mice per two diets). GO, gene ontology; HFCA, high-fat and cholic acid; KEGG, Kyoto Encyclopedia of Genes and Genomes.
In terms of the GO Biological Process between the two strains, we found the immune response-related GO terms in both strains (Supplemental Table S7) and the A/J strain had a high richness in each GO term and the neutrophil-related immune response GO terms were mainly enriched (Fig. 2G) as shown in Fig. 1E. On the other hand, the CAST strain had a low richness in GO term and mainly enriched in GO terms such as calcium signaling, hydrogen peroxide, and acute inflammatory response (Fig. 2H). These results show that the hepatic gene expression pattern responding to the diet contains both Core Diet DEGs and DEGs unique to each strain.
Hepatic Transcriptional Network of CC Progenitors Enriched in Specific Functional Pathways
In addition to identifying transcripts whose abundance is affected by diet or genetic background, we were also interested in understanding the hepatic transcriptional network in the CC progenitors and its relationship to disease-related traits. For this reason, we performed weighted gene co-expression network analysis (WGCNA) to identify modules of highly co-expressed genes. Using WGCNA, we identified 20 co-expression gene modules in the liver transcriptome data as indicated by color names excluding a gray module containing unclustered genes in the network (Supplemental Fig. S1, A and B). The modules contain varying numbers of transcripts ranging from 44 to 4,020. A cluster dendrogram marks the modules as downward branches (Supplemental Fig. S1A). We calculated module eigengenes (ME) of each module to assess the transcript abundance pattern among the eight CC founder strains and two diets. Using GO and KEGG enrichment analysis, we identified 10 modules enriched for specific biological pathways (Table 3 and Supplemental Tables S8 and S9). The functional annotations of these ten modules include: “Fatty acid catabolic process (GO: 0009062)” for the brown module, “Steroid biosynthesis degradation” (KEGG pathway) for the green module, “Type 1 interferon signaling pathway (GO: 0060337)” for the magenta module, “mRNA processing (GO: 0006397)” for the red module, and “Neutrophil activation involved in immune response (GO:0002283)” for the turquoise module.
Table 3.
Gene set enrichment determined that each of the clusters was specifically enriched for GO biological process, KEGG pathway, and Jensen Disease
| Module (No. Genes) | GO Biological Process KEGG Pathway Jensen Disease |
Counts | Adjusted P Value |
|---|---|---|---|
| Turquoise (4,020) | Neutrophil activation involved in immune response (GO:0002283) Non-alcoholic fatty liver disease (NAFLD) Arthritis |
187 81 63 |
3.1 × 10−18 1.8 × 10−17 1.3 × 10−2 |
| Brown (1,151) | Fatty acid catabolic process (GO:0009062) Valine, leucine and isoleucine degradation 3-Methylcrotonyl-CoA carboxylase deficiency |
21 21 6 |
2.1 × 10−7 4.5 × 10−10 1.1 × 10−2 |
| Green (843) | Regulation of alcohol biosynthetic process (GO:1902930) Steroid biosynthesis |
20 19 |
1.0 × 10−15 1.2 × 10−8 |
| Red (700) | mRNA processing (GO:0006397) FoxO signaling pathway |
29 14 |
1.3 × 10−3 2.2 × 10−2 |
| Pink (476) | Cell communication by electrical coupling involved in cardiac conduction (GO:0086064)Cortisol synthesis and secretion Congenital adrenal hyperplasia |
6 20 9 |
7.6 × 10−4 2.7 × 10−14 1.9 × 10−5 |
| Magenta (441) | Type I interferon signaling pathway (GO:0060337) DNA replication Aicardi-Goutieres syndrome |
18 10 4 |
5.3 × 10−12 8.3 × 10−7 1.4 × 10−2 |
| Purple (259) | Muscle contraction (GO:0006936) Dilated cardiomyopathy (DCM) Cardiomyopathy |
50 14 9 |
4.4 × 10−56 3.0 × 10−9 9.2 × 10−12 |
| Cyan (136) | Platelet degranulation (GO:0002576) Complement and coagulation cascades Congenital afibrinogenemia |
13 13 8 |
1.4 × 10−8 8.7 × 10−12 1.4 × 10−6 |
| Midnight blue (127) | Positive regulation of vasculature development (GO:1904018) Cell adhesion molecules (CAMs) Ovarian hyperstimulation syndrome |
10 7 4 |
3.0 × 10−6 1.6 × 10−2 3.6 × 10−3 |
| Light cyan (125) | Signal recognition particle SSRP-dependent cotranslational protein targeting to membrane (GO:0006614) Ribosome Diamond-Blackfan anemia |
38 40 4 |
2.8 × 10−58 1.5 × 10−50 3.4 × 10−3 |
GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Effect of Diet and Strain on Hepatic Transcriptional Network of CC Progenitors
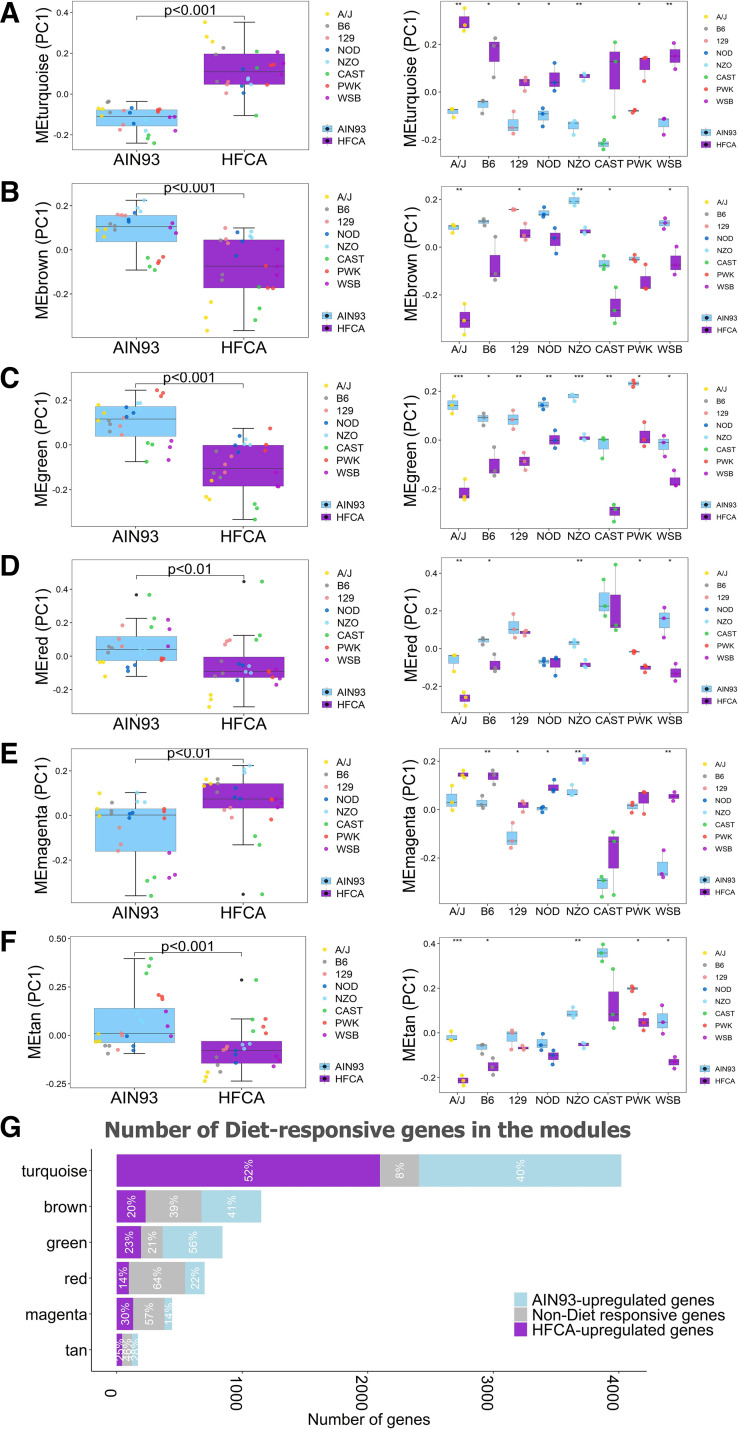
We next sought to delineate the effects of diet on these modules of highly connected genes. The modules can be characterized by their module eigengenes (MEs), which reflect the first principal component of the module. Nonparametric statistical tests of the effect of diet on ME levels were performed to identify diet-influenced modules. Among 20 co-expressed gene modules detected in WGCNA, 11 modules showed significant differences in MEs between diets (Fig. 3, A–F and Supplemental Table S10). Next, to investigate the connection between diet-specific DEGs and gene networks, we explored the proportion of “Core Diet DEGs” in each module (Supplemental Fig. S2). Of these 6,411 genes, 5,473 (85.3%) were contained in one of five modules within the network. Only 32 out of the 6,411 Core Diet DEGs were not associated with any module. The top four modules with the highest number of genes upregulated by the HFCA diet were turquoise (52.2%; 2,097 out of 4,020 genes), brown (20%; 230 out of 1,151 genes), green (23.0%; 194 out of 843 genes), and magenta (29.7%; 131 out of 441 genes). The top four modules with the highest number of genes upregulated by the AIN-93M diet were turquoise (40.1%; 1,612 out of 4,020 genes), green (56.5%; 476 out of 843 genes), brown (41%; 475 out of 1,151 genes), and red (22%; 154 out of 700 genes) (Fig. 3G and Supplemental Tables S11 and S12).
Figure 3.
Effect of diet and genetic background on hepatic co-expression gene modules in the eight Collaborative Cross (CC) founder strains in female mice. Differences of principal component (PC)1 of module eigengenes (ME) including turquoise (A), brown (B), green (C), red (D), magenta (E), and tan (F) module ME by diet and genetic background were plotted. G: number and proportion of diet or nondiet responsive genes in six modules. The x-axis is the number of genes and the y-axis is the color of each module. Length of the bar corresponding to each module is the number of module genes, and portions corresponding to purple, gray, and light blue colors in each bar are the number of genes upregulated by the high-fat and cholic acid (HFCA) diet (purple), nondiet responsive genes (gray), and genes upregulated by the AIN-93M diet (light blue). Proportion of each color is written in white letters in the bar. ***P < 0.001, **P < 0.01, *P < 0.05. n = 6 mice for each founder, three for AIN-93M diet (light blue color) and three for HFCA diet (purple color)-fed mice. A/J (yellow), B6 (gray), C57BL/6J; 129 (pink), 129S1/SvlmJ; NOD (blue), NOD/ShiLtJ; NZO (light blue), NZO/HILtJ; CAST (green), CAST/EiJ; PWJ (red), PWK/PhJ; WSB (purple), WSB/EiJ.
We next sought to identify strain-specific responses within the modules by examining both strain and strain by diet effects (Supplemental Table S10). Among the 20 modules, 18 were moduled by strain. One of the two modules without significant effects of strain was the turquoise module which contains the most diet-specific DEGs (P = 0.48; Fig. 3A and Supplemental Table S10). As shown in Table 1, the number of diet-specific DEGs identified in the A/J, NZO, and WSB strains was the highest, and the number of diet-specific DEGs identified in the CAST strain was the lowest across all modules (Supplemental Tables S11 and S12). Specifically, we highlighted six modules in which significant diet effects were identified in at least five strains that may indicate a critical strain by diet interaction (Fig. 3, A–F). We next described the brown and green modules with both strain and strain by diet effects as examples.
The brown module, which contains 1,151 transcripts, is highly enriched for genes involved in fatty acid catabolism (Supplemental Fig. S3B and Table 3). A previous report identified that hepatic fatty acid oxidation-related genes are downregulated by an atherogenic diet challenge in mice (57) and reflective of this, the brown module ME was higher in AIN-93M diet-fed mice than in HFCA diet-fed mice independent of strain (P < 0.001; Fig. 3B and Supplemental Table S10), and this trend was also observed in five individual strain analyses (P < 0.01 in A/J strain, P < 0.05 in 129 strain, P < 0.01 in NZO strain, P < 0.05 in CAST strain, and P < 0.05 in WSB strain). Furthermore, the number of genes upregulated by the AIN-93M diet (475 genes) in the brown module was 2.1 times higher than the number of genes upregulated by the HFCA diet (230 genes) (Fig. 3G and Supplemental Tables S11 and S12). In terms of strain-dependent effects, transcripts in this module were upregulated in NZO mice and downregulated in CAST mice, which may reflect differences in fatty acid catabolic process in the liver from these strains (Fig. 3B and Supplemental Table S10). Genes involved in fatty acid oxidation have increased expression in patients with fatty liver, which is considered to reduce reactive oxygen species produced by fatty acid oxidation (58). Thus, the NZO strain has the highest expression of fatty acid oxidation-related genes and has the highest adiposity and liver TG content, whereas the CAST strain has the lowest expression of the genes and is generally resistant to MetSyn.
As an additional example of strain and diet effects, we highlight the green module which contains 843 transcripts. A number of these are important for lipid processing such as low-density lipoprotein receptor (Ldlr), proprotein convertase subtilisin/kexin type 9 (Pcsk9), and many other steroid synthesis enzymes (Supplemental Fig. S3C and Supplemental Tables S8 and S9). The ME in the green module was higher in AIN-93M diet-fed mice than in HFCA diet-fed mice independent of strains (P < 0.001) and in all individual strains (Figs. 3C and Supplemental Table S10). In addition, the number of genes upregulated by the AIN-93M diet (476 genes) in the green module was 2.5 times higher than the number of genes upregulated by the HFCA diet (194 genes) (Fig. 3G and Supplemental Tables S11 and S12). A high-fat or an atherogenic diet is known to reduce the rate of synthesis and conversion of primary bile salts in humans and B6 mice (59, 60). In our study, HFCA-fed mice exhibited downregulation of the cholesterol/steroid synthesis pathway potentially due to bile acid accumulation in the liver. In terms of strain-dependent effects, transcripts in the green module were downregulated in the CAST strain and upregulated in the PWK strain (Fig. 3C and Supplemental Table S10). The CAST mice showed the highest plasma cholesterol concentration among all strains which may reflect a decreased expression of genes related to the cholesterol metabolism, including Ldlr which removes cholesterol from plasma LDL into the liver.
In addition to strain and diet effects, we also sought to identify a number of strains by diet interactions of the co-expression modules. Significant diet-by-strain interactions, assessed by a two-way ANOVA test, were observed in 12 modules. These include the above six modules, and the grey60, yellow, black, light yellow, green yellow, and midnight blue modules (Supplemental Table S10).
For example, the black and midnight blue modules showed opposite diet effects by genetic background. The ME of the midnight blue module, which showed KEGG pathway for cell adhesion molecules, was significantly higher in the AIN-93M diet-fed mice than in the HFCA diet-fed mice in the AJ and B6 strains, yet in the NZO and CAST strains the ME of the midnight blue module was higher in HFCA fed mice than in AIN-93 M fed mice (Supplemental Table S10). These results show that genetic background has a significant influence on hepatic gene networks in response to a control diet or an atherogenic diet.
Liver Transcriptome Co-Expression Modules Correlate with Metabolic Traits
Before assessing the relationship between gene modules and clinical traits, we determined the effect of diet on the 29 metabolic traits measured. Liver weight (P < 0.05), liver TG (P < 0.05), plasma ALT (P < 0.01), AST (P < 0.01), total cholesterol (P < 0.001), betaine (P < 0.05), and oxygen consumption per weight (P < 0.05) showed significant diet-dependent differences, assessed by the Wilcoxon test, regardless of genetic backgrounds during the 16-wk HFCA diet challenge period (Fig. 4 and Supplemental Table S13). Plasma TMAO concentration also increased 1.35-fold in HFCA diet-fed mice. This difference was not significant, but we noticed broad strain differences and speculated that the relationship between plasma TMAO and diet depends on the genetic background. We thus tested the clinical traits for strain-by-diet interactions and identified significant diet-by-strain interaction effects for food intake, plasma TG, glucose, and TMAO concentrations, as well as glucose/insulin ratio and oxygen consumption (Supplemental Table S13). Furthermore, food intake (P = 0.026), plasma TMAO level (P = 0.016), and oxygen consumption (P = 0.019) were significantly different between the two diets only when considering the diet-by-strain interaction effect by two-way ANOVA (Supplemental Table S13).
Figure 4.
Diet-dependent differences in key metabolic traits in eight Collaborative Cross (CC) founder strains. Liver weight and triglyceride (TG) and plasma aspartate aminotransferase (AST), alanine aminotransferase (ALT), total cholesterol, and betaine were higher in high-fat and cholic acid (HFCA) diet-fed mice (purple color), while oxygen consumption per body weight was higher in AIN-93M diet-fed mice (light blue color). Plasma trimethylamine N-oxide (TMAO) was higher in HFCA diet-fed mice, but not significant. A/J (yellow), B6 (gray), C57BL/6J; 129 (pink), 129S1/SvlmJ; NOD (blue), NOD/ShiLtJ; NZO (light blue), NZO/HILtJ; CAST (green), CAST/EiJ; PWJ (red), PWK/PhJ; WSB (purple), WSB/EiJ. ****P <0.001, **P < 0.01, *P < 0.05. Data were means ± S.E., n ≥ 4 mice/diet/strain.
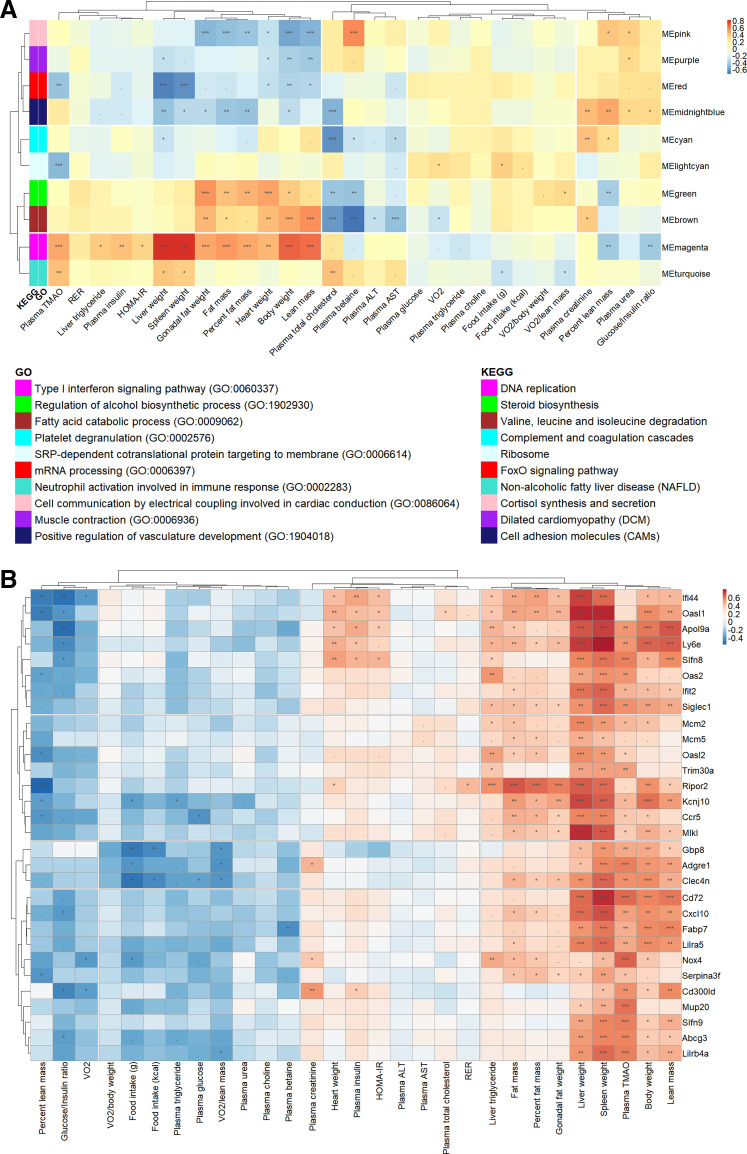
These data reveal that genetic background has a profound effect on metabolic traits in response to a control diet or an atherogenic diet. To assess the physiological significance of the gene networks in liver tissue, we investigated whether the modules were correlated with metabolic traits (Fig. 5A). For example, transcripts in the red module were enriched for the GO terms “mRNA processing (GO: 0006397) (GO term: −logP > 3) and “FoxO signaling pathway” (KEGG pathway: −logP > 2) and included many NADH:ubiquinone oxidoreductase and forkhead box O-related proteins (Supplemental Fig. S3D and Table 3). The ME in the red module was higher in AIN-93M diet-fed mice than in HFCA diet-fed mice independent of strain (P < 0.01; Fig. 3D and Supplemental Table S10), and this trend was also observed in five individual strain analyses (P < 0.01 in A/J strain, P < 0.05 in B6 strain, P < 0.01 in NZO strain, P < 0.05 in PWK strain, and P < 0.05 in WSB strain). The number of genes upregulated by the AIN-93M diet (154 genes) in the red module was 1.6 times higher than the number of genes upregulated by the HFCA diet (97 genes) (Fig. 3G and Supplemental Tables S11 and S12). In terms of strain-dependent effect, the ME in the red module was upregulated in the CAST strain and downregulated in the A/J strain (Figs. 3D and Supplemental Table S10). With regard to hepatic lipid metabolism, the FoxO signaling pathway has been shown to inhibit lipogenesis (61) and promote lipolysis (62). Therefore, our results suggest that HFCA diet-fed mice with increased hepatic lipogenesis have relatively downregulated FoxO signaling pathway, and CAST mice with the lowest liver TG have activated FoxO signaling pathway, resulting in decreased hepatic lipogenesis and increased lipolysis.
Figure 5.
Association of hepatic co-expression gene modules with metabolic traits in the eight Collaborative Cross (CC) founder strains in female mice. A: Spearman correlation between liver gene modules and metabolic traits in all mice. Module names were shown along the right axis, and top-enriched gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) terms in the legend. B: Spearman correlation between the top 30 high-fat and cholic acid (HFCA)-specific differential expression genes (DEGs) identified in the magenta module and metabolic traits in all mice. The P values were adjusted using the Benjamini–Hochberg (BH) false discovery rate (FDR) procedure. ***P < 0.001, **P < 0.01, *P < 0.05, .P < 0.10.
The magenta module is associated with a number of physiological traits including plasma TMAO concentration, plasma insulin, body composition, tissue weights such as liver, spleen, gonadal fat, and heart, and liver TG (Fig. 5A). Notably, the magenta module is enriched for “Type 1 interferon signaling pathway (GO: 0060337)” (GO term: −logP > 11) (Supplemental Fig. S3E and Table 3). Transcripts in the magenta module included various interferon regulatory factors, interleukin receptors, and chemokine (C-C motif) receptors and were upregulated in the liver of the B6 and NZO strains but downregulated in the liver from CAST (Figs. 3E and Supplemental Table S10). A recent study revealed that activation of type I interferon in CD8+ T cells is associated with insulin resistance in fatty liver patients and obese mice (63). Increased type I IFN signaling stimulates macrophage recruitment to lesions, promoting atherosclerosis in mice (64). In our study, various genes related to type I IFN signaling were upregulated in HFCA diet-fed mice, especially in B6 and NZO mice which are susceptible to MetSyn (Fig. 3E and Supplemental Table S10). This suggests that the increased hepatic lipid content and TMAO concentrations in B6 and NZO may be due to an upregulation or downregulation of transcripts involved in the type 1 interferon signaling pathway.
We further investigated the relationship between strain and diet in the magenta module. The ME in the magenta module was higher in HFCA diet-fed mice than in AIN-93M diet-fed mice independent of strain (P < 0.01; Fig. 3E and Supplemental Table S10), and this trend was also observed in five individual strain analyses (P < 0.01 in B6 strain, P < 0.05 for 129 strain, P < 0.05 for NOD strain, P < 0.01 in NZO strain, and P < 0.01 in WSB strain). Furthermore, the magenta module had a higher proportion of genes upregulated by HFCA-diet (29.7%) than that of genes upregulated by AIN-93M diet (13.6%) (Fig. 3G and Supplemental Tables S11 and S12). The ME (PC1) of the magenta module was the highest in NZO mice and the lowest in CAST mice (Fig. 3E and Supplemental Table S10), similar to the metabolic traits pattern by genetic backgrounds (Supplemental Table S13). In this regard, we speculated that the genes clustered in the magenta module may increase their expression by the HFCA diet challenge and regulate metabolic traits.
Among the 441 genes included in the magenta module, we first identified 30 genes with >twofold increased expression in HFCA versus AIN-93M groups by DEG analysis (Supplemental Table S14). Most of these 30 genes showed higher transcript abundance in laboratory-inbred strains (A/J, B6, 129, NOD, NZO, and WSB) than wild-derived strains (CAST and PWK). For example, the 30 genes that generally have the highest transcript abundance in NZO, A/J, and B6 strains also have the lowest transcript abundance in the CAST strain. Following a similar pattern, a number of clinically relevant MetSyn traits such as body composition, tissue weights, liver TG, and insulin resistance were highest in B6 and NZO mice and lowest in CAST strain (Supplemental Tables S13 and S14). In terms of diet-by-strain interaction for the 30 genes, laboratory-inbred strains (A/J, B6, 129, NOD, NZO, and WSB) generally showed diet-sensitive abundance changes, whereas wild-derived strains (CAST and PWK) showed a diet-resistant response (Supplemental Table S14).
To investigate the association of the top 30 HFCA-specific DEGs identified in the magenta module with metabolic traits, we performed Spearman correlation and hierarchical clustering between the genes and traits (Fig. 5B). In general, most of the genes showed positive correlations with body composition, liver weight, spleen weight, liver TG, insulin resistance, and plasma TMAO. Among them, genes showing significant correlations with body weight, plasma TMAO, and liver TG were Apol9a (apolipoprotein L 9a), Mcm2 (minichromosome maintenance complex component 2), Siglec1 (sialic acid binding Ig-like lectin 1), Ly6e (lymphocyte antigen 6 complex, locus E), Ripor2 (RHO family interacting cell polarization regulator 2), Slfn8 (schlafen 8), and Nox4 (NADPH oxidase 4). Among these genes, Apol9a and Slfn8 showed a significant correlation with the clinical indexes of insulin resistance, such as plasma insulin, homeostatic model assessment for insulin resistance (HOMA-IR), and glucose/insulin ratio (65). Finally, Nox4 had a number of clinically relevant correlations which we describe in discussion section.
Association of Hepatic Gene Modules with Metabolic Traits Point to Nox4-Associated Plasma TMAO and Liver TG Production
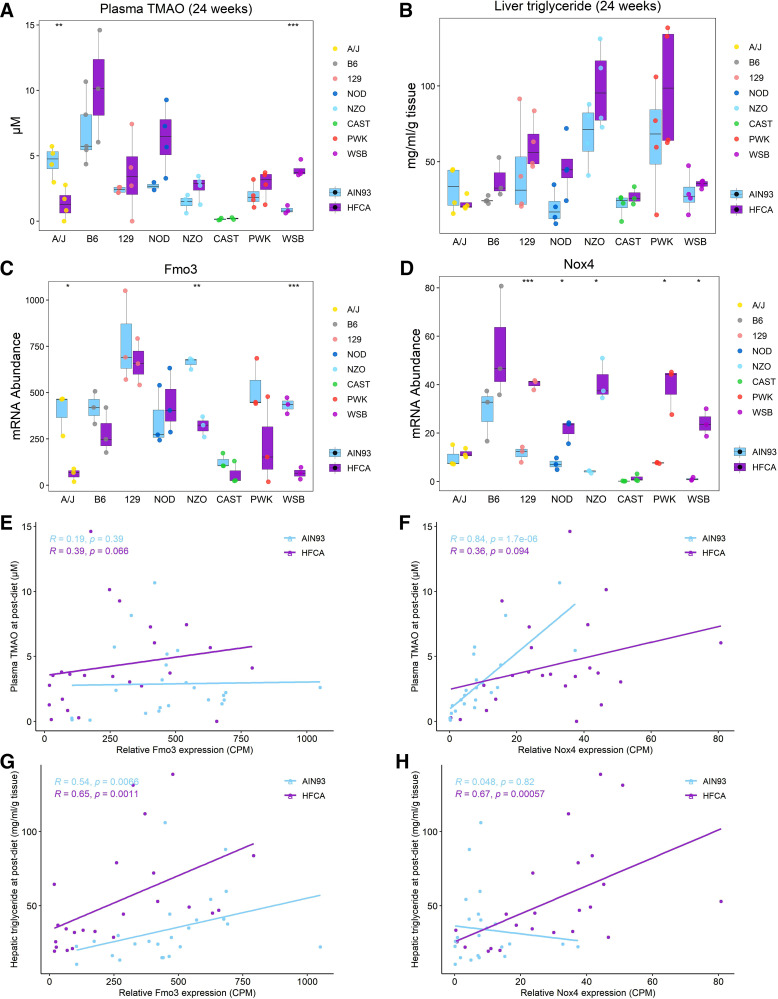
By referring to the literature and assessing the correlation with metabolic traits, we searched for putative genes involved in the regulation of MetSyn among the top 30 HFCA-specific DEGs identified in the magenta module. Interestingly, consistent with the WGCNA, DEG analysis, and strain effect analysis for magenta module genes, Nox4 was found to be a critical member of the magenta module. Among the genes identified in the magenta module (Supplemental Table S14), Nox4, a hydrogen peroxide NADPH oxidase isoform and the primary source of inflammation-induced oxidative stress, was a significant DEG by diet [log2fold change (HFCA/AIN93) = 1.76, adjusted P value = 4 × 10−4] and strain {log2fold change [6 strains (A/J, B6, 129, NOD, NZO, and WSB)/2 strains (CAST and PWK)] = 1.5, adjusted P value = 0.02}. In our study, the Nox4 gene showed the highest associations with plasma TMAO (adjusted P value = 6 × 10−5; Fig. 5B) and with liver TG (adjusted P value = 0.006; Fig. 5B). Nox4 transcript levels were highest in B6 mice and lowest in CAST mice. Another key gene regulating TMAO levels is flavin-containing monooxygenase 3 (Fmo3) which converts TMA produced from dietary precursors such as choline and carnitine into TMAO by NADPH-dependent oxygenation in the liver (66). Considering that oxidative stress is caused by a deficiency in detoxification mechanisms, we observed the effects of genetic backgrounds on plasma TMAO production and gene abundance of Fmo3 and Nox4. In terms of strain-dependent difference, hepatic Nox4 abundance and plasma TMAO were highest in the B6 strain (Fig. 6, A and D, and Supplemental Table S15), and hepatic Fmo3 and Nox4 abundance and plasma TMAO were lowest in the CAST strains (Fig. 6, A, C, and D, and Supplemental Table S15). In addition, plasma TMAO and Nox4 abundance showed an increased pattern in HFCA diet-fed mice except for A/J and CAST strains (Fig. 6, A and D, and Supplemental Table S15), and the only significant diet-dependent difference was observed in the WSB strain. To investigate the relationship between plasma TMAO and Fmo3 or Nox4, we performed the Spearman correlation between these transcripts and plasma TMAO concentration. Surprisingly, Fmo3 transcript abundance was not significantly correlated with plasma TMAO in either diets (Fig. 6E, AIN-93M diet: r = 0.19 and P = 0.39; HFCA diet: r = 0.39 and P = 0.066). Conversely, Nox4 transcript abundance was highly correlated with plasma TMAO (r = 0.84 and P = 1.7 × 10−6) in AIN-93M diet-fed mice and was the strongest correlation between TMAO and any of 12,502 expressed transcripts (Fig. 6F).
Figure 6.
Association of hepatic gene modules with metabolic traits points to Nox4-associated plasma trimethylamine N-oxide (TMAO) and creatinine production. Effect of diet or genetic background on plasma TMAO (A) and liver triacylglycerol (TG) (B), and hepatic expression of Fmo3 (C) and Nox4 (D). Spearman correlation between plasma TMAO and Fmo3 (E) or Nox4 (F) abundance. Spearman correlation between liver TG and Fmo3 (G) or Nox4 (H) abundance. ***P < 0.001, **P < 0.01, *P < 0.05, n = 6 mice for each founder, three for AIN-93M diet (light blue color) and three for high-fat and cholic acid (HFCA) diet-fed mice (purple color). A/J (yellow), B6 (gray), C57BL/6J; 129 (pink), 129S1/SvlmJ; NOD (blue), NOD/ShiLtJ; NZO (light blue), NZO/HILtJ; CAST (green), CAST/EiJ; PWJ (red), PWK/PhJ; WSB (purple), WSB/EiJ.
We next assessed how the two transcripts levels of these two genes were associated with hepatic lipid content, another MetSyn trait that was highly associated with the magenta module. In previous studies, FMO3 overexpression increased hepatic and plasma lipids in atherosclerosis susceptible mice (14) and NOX4-deficient mouse model was shown to have reduced fibrosis due to inhibition of TGF-β-induced apoptosis in epithelial cells (67). In our study, the liver TG and MEs for the brown module containing the Fmo3 gene and for the magenta module containing Nox4 gene were similarly high in NZO mice and low in CAST mice (Supplemental Tables S8, S13, and S16). Liver TG and Nox4 abundance showed an increased pattern in HFCA diet-fed mice except for A/J and CAST strains (Fig. 6, B and D, and Supplemental Table S16). Furthermore, Fmo3 expression was significantly correlated with liver TG in both diets (Fig. 6G, AIN-93M diet: r = 0.54 and P = 0.0066; HFCA diet: r = 0.65 and P = 0.0011), and Nox4 expression was highly correlated with liver TG in HFCA diet-fed mice (r = 0.67 and P = 0.00057; Fig. 6H).
Lastly, we investigated genome variation and haplotypes for the two genes to identify if there are unique haplotypes in the founder strains that could explain the strain variation in plasma TMAO and liver TG. We specifically assessed if the missense variants of the two genes could be functional variants that might explain the expression differences. We identified the total number of SNPs in the Fmo3 and Nox4 genes, calculated the similarity in the SNPs of these genes between the reference genome B6 strain and other strains, and obtained the number of missense variants and functional variants that may affect amino acid substitution using PROVEAN and SIFT in silico analysis. For example, we compared the similarity of each strain to the B6 strain in 658 SNPs in Fmo3 and 2,641 SNPs in Nox4. As expected, the wild-derived strains CAST and PWK contained the most genetic diversity at these loci and have considerable genetic variation as compared to the B6 strain (Supplemental Table S17). In particular, Nox4 showed the greatest diversity (37.37% similarity) in SNPs between B6 and CAST strains, and both liver Nox4 transcript abundance and plasma TMAO concentration had the greatest difference between B6 and CAST strains suggesting that the candidate gene Nox4 has shared haplotypes in the strains with shared phenotypes (Fig. 6, A and D, Supplemental Table S15). We also identified nine SNP missense variants from Fmo3 SNPs and six missense SNP variants from Nox4 SNPs, of which two variants from Fmo3 (rs37325482 at 162,967,784 bp and rs50797400 at 162,968,776 bp) and two variants from Nox4 (rs250243260 at 87,246,820 bp and rs217947741 at 87,246,834 bp) were predicted to have a known deleterious structural consequence by in silico analysis (Supplemental Table S18). Furthermore, we confirmed both Nox4 functional variants were caused by SNPs in the CAST strain (Supplemental Table S18).
DISCUSSION
In this study, we utilized the eight genetically diverse CC founder mouse strains fed AIN-93M or HFCA diet to assess the effect of diet or genetic backgrounds on MetSyn. We found that host genetics had a strong influence on the metabolic traits, including body composition, tissue weights, lipid panels, TMAO analytes, markers related to liver and kidney function, glucose metabolism, and energy expenditure in response to a range of metabolic stimuli. Consistent with previous studies, we found that of the eight strains, CAST had the lowest body adiposity, liver TG, and plasma TMAO levels, and the NZO mice were the most obese and insulin-resistant (68, 69). These results highlight the effects of genetic background on the liver transcriptome and phenotypic responses to diet, and also demonstrate the effectiveness of the CC founder strains in nutrigenomics studies.
The CC founder strains have been individually studied in several tissues, and Chick et al. (23) conducted liver proteome profiling across the eight CC founder strains. Our study represents an in-depth analysis of the effects of diet on the hepatic transcriptional network. To identify liver transcripts that underlie the diet- and strain-dependent differences in metabolic traits, we conducted transcriptomic analysis on liver collected from each strain-fed two different diets and identified and quantified 12,502 transcripts. Hierarchical clustering of the liver transcriptome indicated that mouse liver transcripts were clustered according to diet and strain. In particular, the wild-derived strains PWK and CAST, have altered transcript levels compared with other CC progenitor strains. This suggests that genetic variation has a profound impact on the liver transcriptome, which may be associated with differences in metabolic traits. Gene modules were determined using WGCNA, which clustered co-expressed transcripts, and these modules were enriched for specific functional terms and correlated with metabolic traits.
In this study, we also sought to find core modules with diet-responsive genes by elucidating the link between diet-specific DEGs and gene networks. We first identified 6,411 genes that were differentially regulated by diet independent of strain, and 9,228 genes responded by diet perturbation in at least one strain. Next, we identified 20 modules in gene network analysis and found that the top five modules (turquoise, brown, green, red, and magenta) contained 85.4% of 6,411 DEGs independent of strain and 67.5% of 9,228 DEGs that responded in at least one strain. These modules were enriched for functional annotations, associated with traits related to MetSyn, showed significant diet effects in at least five strains. These results show that diet responsive genes identified by all-strain or individual strain DEG analysis are segregated into specific gene networks that are enriched for functions associated with MetSyn phenotypes.
Correlations can be a method to build hypotheses to test causality. For example, the magenta module is enriched for type I interferon signaling pathway and positively correlated with body composition, tissue weights, insulin resistance (plasma insulin and HOMA-IR), liver TG, and plasma TMAO. In addition, the ME and the HFCA diet-specific DEG abundance in the magenta module were highest in the B6 and NZO strains that are susceptible to obesity and metabolic dysregulation associated with MetSyn. Conversely, the ME was the lowest in the CAST strain, which is resistant to MetSyn (30, 69, 70). The type I interferon signaling is involved in metabolic regulation, tissue inflammation, and MetSyn that are related to atherosclerosis, obesity, fatty liver, and type 1 diabetes (63, 71–75). Therefore, the role of type I interferon is critical in adipose tissue, hepatocytes, immune cells, and endothelial cells. Transcripts in the magenta module include interferon-induced protein 44 (Ifi44), interferon-induced protein with tetratricopeptide repeats 1 (Ifit1), C-X-C motif chemokine ligand 10 (Cxcl10), NADPH oxidase 4 (Nox4), and 2′-5′-oligoadenylate synthase-like protein 1 (Oasl1), which are known to be upregulated in the liver under high-fat diet feeding conditions or stimulation of type I interferon (63, 76–78). Recently IRF7, a master regulator of type I interferon response, showed increased expression in liver and adipose tissue from obese mice compared with controls, indicating increased type I interferon signaling (79). IRF7 knockout mice were resistant to diet-induced obesity, insulin resistance, and inflammation. The IRF7, together with our results, is associated with type I interferon signaling and modulates diet-induced obesity, insulin resistance, and their metabolic consequences. Other novel transcripts in the magenta module may also be important for type I interferon signaling and/or MetSyn.
Driven by our preliminary finding that plasma TMAO production can be modulated by FMO3 enzyme activity in the liver (36), we asked whether Fmo3 gene abundance was differentially abundant across the eight CC founder strains. Liver transcriptomic analysis revealed that hepatic Fmo3 expression is higher in B6, 129, and NZO mice and the lowest in CAST mice, consistent with the FMO3 protein abundance previously reported (23). FMO3 is a catalytic enzyme that converts TMA, produced from dietary precursors such as choline and carnitine by gut microbiota, into TMAO by host-dependent hepatic N-oxygenation in the liver (66). TMAO is mechanistically linked with MetSyn and other diseases, including atherosclerosis, obesity, fatty liver, type 2 diabetes mellitus, and chronic kidney disease (4–10), suggesting that the TMAO pathway may also be linked to the pathogenesis of MetSyn.
The fact that there is a positive correlation between Fmo3 expression and plasma TMAO production among mouse strains susceptible to atherosclerosis highlights the importance of FMO3 in the development of cardiovascular disease (80). However, in our study using mice with highly divergent genetic backgrounds, no significant correlation was observed between Fmo3 expression and plasma TMAO in either diet. For this reason, we searched for genes that are functionally similar to FMO enzymes and have an association with TMAO and focused on Nox4, which showed the highest correlation with plasma TMAO and was identified in the magenta module.
NOX4 is a hydrogen peroxide NADPH oxidase isoform and major producers of reactive oxidative species by transferring electrons from NADPH to molecular oxygen. NOX4 is found in various cardiovascular cells and tissues, which is involved in conditions related to MetSyn such as atherosclerosis (81, 82), hypertension (83), fatty liver (84, 85), insulin resistance (86), obesity (87), and kidney injury (88). Similar to NOX4, FMOs catalyze the NADPH-dependent oxidative metabolism of a variety of foreign chemicals, including dietary compounds, drugs, and environmental pollutants (89). Genetic analysis of patients with trimethylaminuria shows that the lack of FMO3 enzymatic activity often occurs in mutations that affect the binding of necessary cofactors FAD (90) or NADPH (91) highlighting the fundamental importance of these cofactors in the function of FMOs.
Our studies identified Nox4, as a potentially important gene related to TMAO concentrations. We note that this relationship is based on a transcriptional network and does not necessarily indicate a specific direct (i.e., enzymatic) action of Nox4 leading to TMAO production. Rather, our association highlights the complex transcriptional network relating TMAO concentrations. For example, Toll-like receptor 4 (TLR4), which is activated by the type I interferon signaling pathway, regulates the expression of Fmo3 (92), and TLR4 and NOX4 show direct interaction in several cell lines including hepatocytes (93–95), thereby affecting the production of inflammatory mediators that are encoded by genes identified in the magenta module. In addition, ablation of Nox4 lowers plasma homocysteine and betaine levels in mice, and NOX4 protects against acetaminophen-induced hepatotoxicity (96). FMO3 has also been shown to protect the liver from acetaminophen-induced hepatotoxicity (97), and TMAO precursors choline and betaine affect homocysteine levels as methyl donors in one-carbon metabolism (98). Our liver transcriptomic analysis revealed that Nox4, the only NADPH oxidase identified in the magenta module highly associated with MetSyn, has the highest correlation with plasma TMAO, as well as the highest expression in B6 and the lowest in CAST strain.
Our studies utilize eight strains that are genetically diverse and thus we assessed the genetic variation at the Nox4 locus. The Nox4 allele from the CAST strain contained a number of SNPs divergent from B6, and this difference is consistent with strain effects identified in liver Nox4 transcript and TMAO. More interestingly, Nox4 was revealed to be one of the genes that showed the highest correlation with liver TG among the genes identified in the magenta module, which is consistent with previous studies that demonstrated the association between Nox4 and fatty liver in the mouse and humans (67, 84, 85, 99). Both NOX4 and TMAO induce protein kinase R (PKR)/PKR-like endoplasmic reticulum kinase (PERK) activation (99, 100), leading to propagation of endoplasmic reticulum (ER) stress, which may contribute to the pathogenesis of fatty liver and MetSyn. These studies are further supported by associations between SNPs in NOX4 and metabolic syndrome in humans (101, 102). Our in silico analysis of the mouse variants suggests that functional variants of the Nox4 gene may cause deleterious consequence in the protein structure and affect metabolic traits. These observations point to a genetic underpinning of NOX4 in MetSyn, which is similar to the findings of the current study.
Here, we show that among the eight CC founder strains, B6 mice express a high level of Nox4 and have high production of TMAO. Why would B6 liver synthesize the most TMAO, which contributes to the development of MetSyn, and CAST liver synthesize the least? One possible explanation is that the B6 strain has an impaired mechanism to regulate TMAO production or renal excretion. B6 is one of the strains with the highest plasma TMAO production among other inbred strains (103), and it was reported to be more sensitive to damage caused by renal ischemia-reperfusion injury than the 129/Sv inbred strain (104). In addition, the CAST strain has the lowest hepatic expression of Fmo3 of the CC founder strains which could explain reduced FMO3 enzyme activity and TMAO production. Furthermore, studies utilizing the CC or the related Diversity Outbred population may shed further light on these strain differences.
In conclusion, our study provides a strong indication that host genetics affects the liver transcriptome under the intake of control or atherogenic diet. We also demonstrate that diet-by-strain interaction effects on the liver transcriptome are related to metabolic traits, suggesting that liver gene networks may underlie diet- or strain-dependent differences in MetSyn. The phenotype differences between mouse strains motivate us to find comparable phenotypic variations across the human population. Changes in plasma TMAO and liver TG similar to those seen in human MetSyn can be induced in B6 and NZO strains, but it should be taken into account that not all mouse strains developed MetSyn in this study. Understanding the changes in the liver transcriptome in response to diet and genetic background will be important to highlight the potential of precision nutrition and to understand interpersonal variability in disease risk. In this study, we saw dramatic strain variation in Nox4 expression in the mouse liver that could determine the ability to generate plasma TMAO and liver TG. Our results suggest that human genetic variations and variations in plasma TMAO and liver TG may contribute to the regulation of MetSyn.
GRANTS
This research was supported in part by R01HL128572 (to B. J. Bennett), a UNC Nutrition Obesity Research Consortium NORC pilot award from NIH P30DK056350 (to B. J. Bennett), and by USDA/ARS/Western Human Nutrition Research Center project funds 2032-51000-022-00D (to B. J. Bennett).
DISCLAIMERS
The USDA is an equal opportunity employer.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
B.J.B. conceived and designed research; A.O., J.A., and B.J.B. performed experiments; M.K., M.N.H., and B.D-J. analyzed data; M.K., M.N.H., and B.J.B. interpreted results of experiments; M.K. prepared figures; M.K. drafted manuscript; M.K., M.N.H., and B.J.B. edited and revised manuscript; M.K. and B.J.B. approved final version of manuscript.
ACKNOWLEDGMENTS
We acknowledge Nikhil Joshi (University of California, Davis Bioinformatics Core) for assistance with RNA-Seq mapping analysis.
REFERENCES
- 1.Huang PL. A comprehensive definition for metabolic syndrome. Dis Model Mech 2: 231–237, 2009. doi: 10.1242/dmm.001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamaguchi M, Kojima T, Takeda N, Nakagawa T, Taniguchi H, Fujii K, Omatsu T, Nakajima T, Sarui H, Shimazaki M, Kato T, Okuda J, Ida K. The metabolic syndrome as a predictor of nonalcoholic fatty liver disease. Ann Intern Med 143: 722–728, 2005. doi: 10.7326/0003-4819-143-10-200511150-00009. [DOI] [PubMed] [Google Scholar]
- 3.Marchesini G, Bugianesi E, Forlani G, Cerrelli F, Lenzi M, Manini R, Natale S, Vanni E, Villanova N, Melchionda N, Rizzetto M. Nonalcoholic fatty liver, steatohepatitis, and the metabolic syndrome. Hepatology 37: 917–923, 2003. doi: 10.1053/jhep.2003.50161. [DOI] [PubMed] [Google Scholar]
- 4.Barrea L, Annunziata G, Muscogiuri G, Di Somma C, Laudisio D, Maisto M, de Alteriis G, Tenore GC, Colao A, Savastano S. Trimethylamine-N-oxide (TMAO) as novel potential biomarker of early predictors of metabolic syndrome. Nutrients 10: 1971, 2018. doi: 10.3390/nu10121971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen YM, Liu Y, Zhou RF, Chen XL, Wang C, Tan XY, Wang LJ, Zheng RD, Zhang HW, Ling WH, Zhu HL. Associations of gut-flora-dependent metabolite trimethylamine-N-oxide, betaine and choline with non-alcoholic fatty liver disease in adults. Sci Rep 6: 19076, 2016. doi: 10.1038/srep19076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WHW, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19: 576–585, 2013. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lent-Schochet D, Silva R, McLaughlin M, Huet B, Jialal I. Changes to trimethylamine-N-oxide and its precursors in nascent metabolic syndrome. Horm Mol Biol Clin Investig 35, 2018. doi: 10.1515/hmbci-2018-0015. [DOI] [PubMed] [Google Scholar]
- 8.Schugar RC, Shih DM, Warrier M, Helsley RN, Burrows A, FergusonD , et al. The TMAO-producing enzyme flavin-containing monooxygenase 3 regulates obesity and the beiging of white adipose tissue. Cell Rep 19: 2451–2461, 2017[Erratum inCell Rep20: 279, 2017]. doi: 10.1016/j.celrep.2017.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stubbs JR, House JA, Ocque AJ, Zhang S, Johnson C, Kimber C, Schmidt K, Gupta A, Wetmore JB, Nolin TD, Spertus JA, Yu AS. Serum trimethylamine-N-oxide is elevated in CKD and correlates with coronary atherosclerosis burden. J Am Soc Nephrol 27: 305–313, 2016. doi: 10.1681/ASN.2014111063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WH, DiDonato JA, Lusis AJ, Hazen SL. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472: 57–63, 2011. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartiala J, Bennett BJ, Tang WH, Wang Z, Stewart AF, Roberts R, McPherson R, Lusis AJ, Hazen SL, Allayee H, Consortium CA; CARDIoGRAM Consortium. Comparative genome-wide association studies in mice and humans for trimethylamine N-oxide, a proatherogenic metabolite of choline and l-carnitine. Arterioscler Thromb Vasc Biol 34: 1307–1313, 2014. doi: 10.1161/ATVBAHA.114.303252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambert DM, Mamer OA, Akerman BR, Choiniere L, Gaudet D, Hamet P, Treacy EP. In vivo variability of TMA oxidation is partially mediated by polymorphisms of the FMO3 gene. Mol Genet Metab 73: 224–229, 2001. doi: 10.1006/mgme.2001.3189. [DOI] [PubMed] [Google Scholar]
- 13.Cho CE, Taesuwan S, Malysheva OV, Bender E, Tulchinsky NF, Yan J, Sutter JL, Caudill MA. Trimethylamine-N-oxide (TMAO) response to animal source foods varies among healthy young men and is influenced by their gut microbiota composition: a randomized controlled trial. Mol Nutr Food Res 61: 1600324, 2017. doi: 10.1002/mnfr.201600324. [DOI] [PubMed] [Google Scholar]
- 14.Shih DM, Wang Z, Lee R, Meng Y, Che N, Charugundla S, Qi H, Wu J, Pan C, Brown JM, Vallim T, Bennett BJ, Graham M, Hazen SL, Lusis AJ. Flavin containing monooxygenase 3 exerts broad effects on glucose and lipid metabolism and atherosclerosis. J Lipid Res 56: 22–37, 2015. doi: 10.1194/jlr.M051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warrier M, Shih DM, Burrows AC, Ferguson D, Gromovsky AD, Brown AL, Marshall S, McDaniel A, Schugar RC, Wang Z, Sacks J, Rong X, Vallim TA, Chou J, Ivanova PT, Myers DS, Brown HA, Lee RG, Crooke RM, Graham MJ, Liu X, Parini P, Tontonoz P, Lusis AJ, Hazen SL, Temel RE, Brown JM. The TMAO-generating enzyme flavin monooxygenase 3 is a central regulator of cholesterol balance. Cell Rep 10: 326–338, 2015. doi: 10.1016/j.celrep.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, Nemesh J, Lane CR, Schaffner SF, Bolk S, Brewer C, Tuomi T, Gaudet D, Hudson TJ, Daly M, Groop L, Lander ES. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26: 76–80, 2000. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 17.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38: 320–323, 2006. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 18.Hani EH, Boutin P, Durand E, Inoue H, Permutt MA, Velho G, Froguel P. Missense mutations in the pancreatic islet beta cell inwardly rectifying K+ channel gene (KIR6.2/BIR): a meta-analysis suggests a role in the polygenic basis of Type II diabetes mellitus in Caucasians. Diabetologia 41: 1511–1515, 1998. doi: 10.1007/s001250051098. [DOI] [PubMed] [Google Scholar]
- 19.Attie AD, Churchill GA, Nadeau JH. How mice are indispensable for understanding obesity and diabetes genetics. Curr Opin Endocrinol Diabetes Obes 24: 83–91, 2017. doi: 10.1097/MED.0000000000000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Getz GS, Reardon CA. Diet and murine atherosclerosis. Arterioscler Thromb Vasc Biol 26: 242–249, 2006. doi: 10.1161/01.ATV.0000201071.49029.17. [DOI] [PubMed] [Google Scholar]
- 21.Meng H, Vera I, Che N, Wang X, Wang SS, Ingram-Drake L, Schadt EE, Drake TA, Lusis AJ. Identification of Abcc6 as the major causal gene for dystrophic cardiac calcification in mice through integrative genomics. Proc Natl Acad Sci USA 104: 4530–4535, 2007. doi: 10.1073/pnas.0607620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Churchill GA, Gatti DM, Munger SC, Svenson KL. The diversity outbred mouse population. Mamm Genome 23: 713–718, 2012. doi: 10.1007/s00335-012-9414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chick JM, Munger SC, Simecek P, Huttlin EL, Choi K, Gatti DM, Raghupathy N, Svenson KL, Churchill GA, Gygi SP. Defining the consequences of genetic variation on a proteome-wide scale. Nature 534: 500–505, 2016. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coffey AR, Kanke M, Smallwood TL, Albright J, Pitman W, Gharaibeh RZ, Hua K, Gertz E, Biddinger SB, Temel RE, Pomp D, Sethupathy P, Bennett BJ. microRNA-146a-5p association with the cardiometabolic disease risk factor TMAO. Physiol Genomics 51: 59–71, 2019. doi: 10.1152/physiolgenomics.00079.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coffey AR, Smallwood TL, Albright J, Hua K, Kanke M, Pomp D, Bennett BJ, Sethupathy P. Systems genetics identifies a co-regulated module of liver microRNAs associated with plasma LDL cholesterol in murine diet-induced dyslipidemia. Physiol Genomics 49: 618–629, 2017. doi: 10.1152/physiolgenomics.00050.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huda MN, VerHague M, Albright J, Smallwood T, Bell TA, Que E, Miller DR, Roshanravan B, Allayee H, Pardo-Manuel de Villena F, Bennett BJ. Dissecting the genetic architecture of Cystatin C in Diversity Outbred mice. G3 (Bethesda) 10: 2529–2541, 2020. doi: 10.1534/g3.120.401275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller MP, Gatti DM, Schueler KL, Rabaglia ME, Stapleton DS, Simecek P, Vincent M, Allen S, Broman AT, Bacher R, Kendziorski C, Broman KW, Yandell BS, Churchill GA, Attie AD. Genetic drivers of pancreatic islet function. Genetics 209: 335–356, 2018. doi: 10.1534/genetics.118.300864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller MP, Rabaglia ME, Schueler KL, Stapleton DS, Gatti DM, Vincent M, Mitok KA, Wang Z, Ishimura T, Simonett SP, Emfinger CH, Das R, Beck T, Kendziorski C, Broman KW, Yandell BS, Churchill GA, Attie AD. Gene loci associated with insulin secretion in islets from nondiabetic mice. J Clin Invest 129: 4419–4432, 2019. doi: 10.1172/JCI129143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kemis JH, Linke V, Barrett KL, Boehm FJ, Traeger LL, Keller MP, Rabaglia ME, Schueler KL, Stapleton DS, Gatti DM, Churchill GA, Amador-Noguez D, Russell JD, Yandell BS, Broman KW, Coon JJ, Attie AD, Rey FE. Genetic determinants of gut microbiota composition and bile acid profiles in mice. PLoS Genet 15: e1008073, 2019. doi: 10.1371/journal.pgen.1008073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Connor A, Quizon PM, Albright JE, Lin FT, Bennett BJ. Responsiveness of cardiometabolic-related microbiota to diet is influenced by host genetics. Mamm Genome 25: 583–599, 2014. doi: 10.1007/s00335-014-9540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Que E, James KL, Coffey AR, Smallwood TL, Albright J, Huda MN, Pomp D, Sethupathy P, Bennett BJ. Genetic architecture modulates diet-induced hepatic mRNA and miRNA expression profiles in diversity outbred mice. Genetics 216: 241–259, 2020. doi: 10.1534/genetics.120.303481. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Smallwood TL, Gatti DM, Quizon P, Weinstock GM, Jung KC, Zhao L, Hua K, Pomp D, Bennett BJ. High-resolution genetic mapping in the diversity outbred mouse population identifies Apobec1 as a candidate gene for atherosclerosis. G3 (Bethesda) 4: 2353–2363, 2014. doi: 10.1534/g3.114.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyler AL, Ji B, Gatti DM, Munger SC, Churchill GA, Svenson KL, Carter GW. Epistatic networks jointly influence phenotypes related to metabolic disease and gene expression in Diversity Outbred mice. Genetics 206: 621–639, 2017. doi: 10.1534/genetics.116.198051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter JM, Gildea DE, Andreas JP, Gatti DM, Williams KA, Lee M, Hu Y, Zhang S, Program NCS, Mullikin JC, Wolfsberg TG, McDonnell SK, Fogarty ZC, Larson MC, French AJ, Schaid DJ, Thibodeau SN, Churchill GA, Crawford NP; NISC Comparative Sequencing Program. Mapping complex traits in a Diversity Outbred F1 mouse population identifies germline modifiers of metastasis in human prostate cancer. Cell Syst 4: 31–45.e36, 2017. doi: 10.1016/j.cels.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.AlSiraj Y, Chen X, Thatcher SE, Temel RE, Cai L, Blalock E, Katz W, Ali HM, Petriello M, Deng P, Morris AJ, Wang X, Lusis AJ, Arnold AP, Reue K, Thompson K, Tso P, Cassis LA. XX sex chromosome complement promotes atherosclerosis in mice. Nat Commun 10: 2631, 2019. doi: 10.1038/s41467-019-10462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bennett BJ, Davis RC, Civelek M, Orozco L, Wu J, Qi H, Pan C, Packard RRS, Eskin E, Yan M, Kirchgessner T, Wang Z, Li X, Gregory JC, Hazen SL, Gargalovic PS, Lusis AJ. Genetic architecture of atherosclerosis in mice: a systems genetics analysis of common inbred strains. PLoS Genet 11: e1005711, 2015. doi: 10.1371/journal.pgen.1005711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daugherty A. Mouse models of atherosclerosis. Am J Med Sci 323: 3–10, 2002. doi: 10.1097/00000441-200201000-00002. [DOI] [PubMed] [Google Scholar]
- 38.Hsu J, Smith JD. Genetic-genomic replication to identify candidate mouse atherosclerosis modifier genes. J Am Heart Assoc 2: e005421, 2013. doi: 10.1161/JAHA.112.005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21, 2013. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43: e47, 2015. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. Enrichr—interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128, 2013. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pletscher-Frankild S, Palleja A, Tsafou K, Binder JX, Jensen LJ. DISEASES: text mining and data integration of disease-gene associations. Methods 74: 83–89, 2015. doi: 10.1016/j.ymeth.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 43.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9: 559, 2008.doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4: Article17, 2005. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 45.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31: 2745–2747, 2015. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat Protoc 11: 1–9, 2016. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 47.R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing, 2016. [Google Scholar]
- 48.Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Statst Soc B 57: 289–300, 1995. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 49.Kolde R, Franzosa EA, Rahnavard G, Hall AB, Vlamakis H, Stevens C, Daly MJ, Xavier RJ, Huttenhower C. Host genetic variation and its microbiome interactions within the human microbiome project. Genome Med 10: 6, 2018. doi: 10.1186/s13073-018-0515-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cheng AA, Li W, Hernandez LL. Effect of high-fat diet feeding and associated transcriptome changes in the peak lactation mammary gland in C57BL/6 dams. Physiol Genomics 50: 1059–1070, 2018. doi: 10.1152/physiolgenomics.00052.2018. [DOI] [PubMed] [Google Scholar]
- 51.Lan X, Li D, Zhong B, Ren J, Wang X, Sun Q, Li Y, Liu L, Liu L, Lu S. Identification of differentially expressed genes related to metabolic syndrome induced with high-fat diet in E3 rats. Exp Biol Med (Maywood) 240: 235–241, 2015. doi: 10.1177/1535370214554531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon G, Cho KA, Song J, Kim YK. Transcriptomic analysis of high fat diet fed mouse brain cortex. Front Genet 10: 83, 2019.doi: 10.3389/fgene.2019.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Z, Zhao X, Chen L, Li Y, Chen Z, Wang Y, Zhou Z, Chu X. Integrated analysis of differentially expressed long noncoding RNAs and mRNAs associated with high-fat diet-induced hepatic insulin resistance in mice. Nutr Metab (Lond) 17: 45, 2020. doi: 10.1186/s12986-020-00467-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cazanave S, Podtelezhnikov A, Jensen K, Seneshaw M, Kumar DP, Min HK, Santhekadur PK, Banini B, Mauro AG, A MO, Vincent R, Tanis KQ, Webber AL, Wang L, Bedossa P, Mirshahi F, Sanyal AJ. The transcriptomic signature of disease development and progression of nonalcoholic fatty liver disease. Sci Rep 7: 17193, 2017. doi: 10.1038/s41598-017-17370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGettigan B, McMahan R, Orlicky D, Burchill M, Danhorn T, Francis P, Cheng LL, Golden-Mason L, Jakubzick CV, Rosen HR. Dietary lipids differentially shape nonalcoholic steatohepatitis progression and the transcriptome of kupffer cells and infiltrating macrophages. Hepatology 70: 67–83, 2019. doi: 10.1002/hep.30401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Remmerie A, Martens L, Scott CL. Macrophage subsets in obesity, aligning the liver and adipose tissue. Front Endocrinol (Lausanne) 11: 259, 2020. doi: 10.3389/fendo.2020.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuzawa N, Takamura T, Kurita S, Misu H, Ota T, Ando H, Yokoyama M, Honda M, Zen Y, Nakanuma Y, Miyamoto K, Kaneko S. Lipid-induced oxidative stress causes steatohepatitis in mice fed an atherogenic diet. Hepatology 46: 1392–1403, 2007. doi: 10.1002/hep.21874. [DOI] [PubMed] [Google Scholar]
- 58.Kohjima M, Enjoji M, Higuchi N, Kato M, Kotoh K, Yoshimoto T, Fujino T, Yada M, Yada R, Harada N, Takayanagi R, Nakamuta M. Re-evaluation of fatty acid metabolism-related gene expression in nonalcoholic fatty liver disease. Int J Mol Med 20: 351–358, 2007. [PubMed] [Google Scholar]
- 59.Bisschop PH, Bandsma RH, Stellaard F, ter Harmsel A, Meijer AJ, Sauerwein HP, Kuipers F, Romijn JA. Low-fat, high-carbohydrate and high-fat, low-carbohydrate diets decrease primary bile acid synthesis in humans. Am J Clin Nutr 79: 570–576, 2004. doi: 10.1093/ajcn/79.4.570. [DOI] [PubMed] [Google Scholar]
- 60.Renaud HJ, Cui JY, Lu H, Klaassen CD. Effect of diet on expression of genes involved in lipid metabolism, oxidative stress, and inflammation in mouse liver-insights into mechanisms of hepatic steatosis. PLoS One 9: e88584, 2014. doi: 10.1371/journal.pone.0088584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cook JR, Matsumoto M, Banks AS, Kitamura T, Tsuchiya K, Accili D. A mutant allele encoding DNA binding-deficient FoxO1 differentially regulates hepatic glucose and lipid metabolism. Diabetes 64: 1951–1965, 2015. doi: 10.2337/db14-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang W, Bu SY, Mashek MT, I OS, Sibai Z, Khan SA, Ilkayeva O, Newgard CB, Mashek DG, Unterman TG. Integrated regulation of hepatic lipid and glucose metabolism by adipose triacylglycerol lipase and FoxO proteins. Cell Rep 15: 349–359, 2016. doi: 10.1016/j.celrep.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghazarian M, Revelo XS, Nøhr MK, Luck H, Zeng K, Lei H, Tsai S, Schroer SA, Park YJ, Chng MHY, Shen L, D’Angelo JA, Horton P, Chapman WC, Brockmeier D, Woo M, Engleman EG, Adeyi O, Hirano N, Jin T, Gehring AJ, Winer S, Winer DA. Type I interferon responses drive intrahepatic T cells to promote metabolic syndrome. Sci Immunol 2: 7616, 2017. doi: 10.1126/sciimmunol.aai7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goossens P, Gijbels MJ, Zernecke A, Eijgelaar W, Vergouwe MN, van der Made I, Vanderlocht J, Beckers L, Buurman WA, Daemen MJ, Kalinke U, Weber C, Lutgens E, de Winther MP. Myeloid type I interferon signaling promotes atherosclerosis by stimulating macrophage recruitment to lesions. Cell Metab 12: 142–153, 2010. doi: 10.1016/j.cmet.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 65.Wallace TM, Levy JC, Matthews DR. Use and abuse of HOMA modeling. Diabetes Care 27: 1487–1495, 2004. doi: 10.2337/diacare.27.6.1487. [DOI] [PubMed] [Google Scholar]
- 66.Lang DH, Yeung CK, Peter RM, Ibarra C, Gasser R, Itagaki K, Philpot RM, Rettie AE. Isoform specificity of trimethylamine N-oxygenation by human flavin-containing monooxygenase (FMO) and P450 enzymes: selective catalysis by FMO3. Biochem Pharmacol 56: 1005–1012, 1998. doi: 10.1016/s0006-2952(98)00218-4. [DOI] [PubMed] [Google Scholar]
- 67.Carnesecchi S, Deffert C, Donati Y, Basset O, Hinz B, Preynat-Seauve O, Guichard C, Arbiser JL, Banfi B, Pache JC, Barazzone-Argiroffo C, Krause KH. A key role for NOX4 in epithelial cell death during development of lung fibrosis. Antioxid Redox Signal 15: 607–619, 2011. doi: 10.1089/ars.2010.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kreznar JH, Keller MP, Traeger LL, Rabaglia ME, Schueler KL, Stapleton DS, Zhao W, Vivas EI, Yandell BS, Broman AT, Hagenbuch B, Attie AD, Rey FE. Host genotype and gut microbiome modulate insulin secretion and diet-induced metabolic phenotypes. Cell Rep 18: 1739–1750, 2017. doi: 10.1016/j.celrep.2017.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mitok KA, Freiberger EC, Schueler KL, Rabaglia ME, Stapleton DS, Kwiecien NW, Malec PA, Hebert AS, Broman AT, Kennedy RT, Keller MP, Coon JJ, Attie AD. Islet proteomics reveals genetic variation in dopamine production resulting in altered insulin secretion. J Biol Chem 293: 5860–5877, 2018. doi: 10.1074/jbc.RA117.001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joost HG, Schurmann A. The genetic basis of obesity-associated type 2 diabetes (diabesity) in polygenic mouse models. Mamm Genome 25: 401–412, 2014. doi: 10.1007/s00335-014-9514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alsaggar M, Mills M, Liu D. Interferon beta overexpression attenuates adipose tissue inflammation and high-fat diet-induced obesity and maintains glucose homeostasis. Gene Ther 24: 60–66, 2017. doi: 10.1038/gt.2016.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen HJ, Tas SW, de Winther MPJ. Type-I interferons in atherosclerosis. J Exp Med 217: e20190459, 2020. doi: 10.1084/jem.20190459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marro BS, Legrain S, Ware BC, Oldstone MB. Macrophage IFN-I signaling promotes autoreactive T cell infiltration into islets in type 1 diabetes model. JCI Insight 4: e125067, 2019. doi: 10.1172/jci.insight.125067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Somers EC, Zhao W, Lewis EE, Wang L, Wing JJ, Sundaram B, Kazerooni EA, McCune WJ, Kaplan MJ. Type I interferons are associated with subclinical markers of cardiovascular disease in a cohort of systemic lupus erythematosus patients. PLoS One 7: e37000, 2012. doi: 10.1371/journal.pone.0037000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wieser V, Adolph TE, Grander C, Grabherr F, Enrich B, Moser P, Moschen AR, Kaser S, Tilg H. Adipose type I interferon signalling protects against metabolic dysfunction. Gut 67: 157–165, 2018. doi: 10.1136/gutjnl-2016-313155. [DOI] [PubMed] [Google Scholar]
- 76.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol 14: 36–49, 2014. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Paik YH, Kim J, Aoyama T, De Minicis S, Bataller R, Brenner DA. Role of NADPH oxidases in liver fibrosis. Antioxid Redox Signal 20: 2854–2872, 2014. doi: 10.1089/ars.2013.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhai Y, Qiao B, Gao F, Shen X, Vardanian A, Busuttil RW, Kupiec-Weglinski JW. Type I, but not type II, interferon is critical in liver injury induced after ischemia and reperfusion. Hepatology 47: 199–206, 2008. doi: 10.1002/hep.21970. [DOI] [PubMed] [Google Scholar]
- 79.Wang XA, Zhang R, Zhang S, Deng S, Jiang D, Zhong J, Yang L, Wang T, Hong S, Guo S, She ZG, Zhang XD, Li H. Interferon regulatory factor 7 deficiency prevents diet-induced obesity and insulin resistance. Am J Physiol Endocrinol Metab 305: E485–E495, 2013. doi: 10.1152/ajpendo.00505.2012. [DOI] [PubMed] [Google Scholar]
- 80.Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, Edwards PA, Hazen SL, Lusis AJ. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab 17: 49–60, 2013. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lozhkin A, Vendrov AE, Pan H, Wickline SA, Madamanchi NR, Runge MS. NADPH oxidase 4 regulates vascular inflammation in aging and atherosclerosis. J Mol Cell Cardiol 102: 10–21, 2017. doi: 10.1016/j.yjmcc.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schurmann C, Rezende F, Kruse C, Yasar Y, Lowe O, Fork C, van de Sluis B, Bremer R, Weissmann N, Shah AM, Jo H, Brandes RP, Schroder K. The NADPH oxidase Nox4 has anti-atherosclerotic functions. Eur Heart J 36: 3447–3456, 2015. doi: 10.1093/eurheartj/ehv460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bouabout G, Ayme-Dietrich E, Jacob H, Champy MF, Birling MC, Pavlovic G, Madeira L, Fertak LE, Petit-Demouliere B, Sorg T, Herault Y, Mudgett J, Monassier L. Nox4 genetic inhibition in experimental hypertension and metabolic syndrome. Arch Cardiovasc Dis 111: 41–52, 2018. doi: 10.1016/j.acvd.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 84.León-Mimila P, Villamil-Ramírez H, Li XS, Shih DM, Hui ST, Ocampo-Medina E, López-Contreras B, Morán-Ramos S, Olivares-Arevalo M, Grandini-Rosales P, Macías-Kauffer L, González-González I, Hernández-Pando R, Gómez-Pérez F, Campos-Pérez F, Aguilar-Salinas C, Larrieta-Carrasco E, Villarreal-Molina T, Wang Z, Lusis AJ, Hazen SL, Huertas-Vazquez A, Canizales-Quinteros S. Trimethylamine N-oxide levels are associated with NASH in obese subjects with type 2 diabetes. Diabetes Metab 47: 101183, 2021. doi: 10.1016/j.diabet.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rabelo F, Stefano JT, Cavaleiro AM, Lima RVC, de Campos Mazo DF, Carrilho FJ, Correa-Giannella ML, Oliveira CP. Association between the CYBA and NOX4 genes of NADPH oxidase and its relationship with metabolic syndrome in non-alcoholic fatty liver disease in Brazilian population. Hepatobiliary Pancreat Dis Int 17: 330–335, 2018. doi: 10.1016/j.hbpd.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 86.Den Hartigh LJ, Omer M, Goodspeed L, Wang S, Wietecha T, O’Brien KD, Han CY. Adipocyte-specific deficiency of NADPH oxidase 4 delays the onset of insulin resistance and attenuates adipose tissue inflammation in obesity. Arterioscler Thromb Vasc Biol 37: 466–475, 2017. doi: 10.1161/ATVBAHA.116.308749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang F, Lim HK, Morris MJ, Prior L, Velkoska E, Wu X, Dusting GJ. Systemic upregulation of NADPH oxidase in diet-induced obesity in rats. Redox Rep 16: 223–229, 2011. doi: 10.1179/174329211X13049558293713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jeong BY, Lee HY, Park CG, Kang J, Yu SL, Choi DR, Han SY, Park MH, Cho S, Lee SY, Hwang WM, Yun SR, Ryu HM, Oh EJ, Park SH, Kim YL, Yoon SH. Oxidative stress caused by activation of NADPH oxidase 4 promotes contrast-induced acute kidney injury. PLoS One 13: e0191034, 2018. doi: 10.1371/journal.pone.0191034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krueger SK, Williams DE. Mammalian flavin-containing monooxygenases: structure/function, genetic polymorphisms and role in drug metabolism. Pharmacol Ther 106: 357–387, 2005. doi: 10.1016/j.pharmthera.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang J, Tran Q, Lattard V, Cashman JR. Deleterious mutations in the flavin-containing monooxygenase 3 (FMO3) gene causing trimethylaminuria. Pharmacogenetics 13: 495–500, 2003. doi: 10.1097/00008571-200308000-00007. [DOI] [PubMed] [Google Scholar]
- 91.Fujieda M, Yamazaki H, Togashi M, Saito T, Kamataki T. Two novel single nucleotide polymorphisms (SNPs) of the FMO3 gene in Japanese. Drug Metab Pharmacokinet 18: 333–335, 2003. doi: 10.2133/dmpk.18.333. [DOI] [PubMed] [Google Scholar]
- 92.Zhang J, Chaluvadi MR, Reddy R, Motika MS, Richardson TA, Cashman JR, Morgan ET. Hepatic flavin-containing monooxygenase gene regulation in different mouse inflammation models. Drug Metab Dispos 37: 462–468, 2009. doi: 10.1124/dmd.108.025338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park HS, Jung HY, Park EY, Kim J, Lee WJ, Bae YS. Cutting edge: direct interaction of TLR4 with NAD(P)H oxidase 4 isozyme is essential for lipopolysaccharide-induced production of reactive oxygen species and activation of NF-kappa B. J Immunol 173: 3589–3593, 2004. doi: 10.4049/jimmunol.173.6.3589. [DOI] [PubMed] [Google Scholar]
- 94.Patel DN, Bailey SR, Gresham JK, Schuchman DB, Shelhamer JH, Goldstein BJ, Foxwell BM, Stemerman MB, Maranchie JK, Valente AJ, Mummidi S, Chandrasekar B. TLR4-NOX4-AP-1 signaling mediates lipopolysaccharide-induced CXCR6 expression in human aortic smooth muscle cells. Biochem Biophys Res Commun 347: 1113–1120, 2006. doi: 10.1016/j.bbrc.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 95.Singh A, Koduru B, Carlisle C, Akhter H, Liu RM, Schroder K, Brandes RP, Ojcius DM. NADPH oxidase 4 modulates hepatic responses to lipopolysaccharide mediated by Toll-like receptor-4. Sci Rep 7: 14346, 2017. doi: 10.1038/s41598-017-14574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Murray TV, Dong X, Sawyer GJ, Caldwell A, Halket J, Sherwood R, Quaglia A, Dew T, Anilkumar N, Burr S, Mistry RK, Martin D, Schroder K, Brandes RP, Hughes RD, Shah AM, Brewer AC. NADPH oxidase 4 regulates homocysteine metabolism and protects against acetaminophen-induced liver damage in mice. Free Radic Biol Med 89: 918–930, 2015. doi: 10.1016/j.freeradbiomed.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rudraiah S, Rohrer PR, Gurevich I, Goedken MJ, Rasmussen T, Hines RN, Manautou JE. Tolerance to acetaminophen hepatotoxicity in the mouse model of autoprotection is associated with induction of flavin-containing monooxygenase-3 (FMO3) in hepatocytes. Toxicol Sci 141: 263–277, 2014. doi: 10.1093/toxsci/kfu124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Craciunescu CN, Johnson AR, Zeisel SH. Dietary choline reverses some, but not all, effects of folate deficiency on neurogenesis and apoptosis in fetal mouse brain. J Nutr 140: 1162–1166, 2010. doi: 10.3945/jn.110.122044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bettaieb A, Jiang JX, Sasaki Y, Chao TI, Kiss Z, Chen X, Tian J, Katsuyama M, Yabe-Nishimura C, Xi Y, Szyndralewiez C, Schroder K, Shah A, Brandes RP, Haj FG, Torok NJ. Hepatocyte nicotinamide adenine dinucleotide phosphate reduced oxidase 4 regulates stress signaling, fibrosis, and insulin sensitivity during development of steatohepatitis in mice. Gastroenterology 149: 468–480.e410, 2015. doi: 10.1053/j.gastro.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen S, Henderson A, Petriello MC, Romano KA, Gearing M, Miao J, Schell M, Sandoval-Espinola WJ, Tao J, Sha B, Graham M, Crooke R, Kleinridders A, Balskus EP, Rey FE, Morris AJ, Biddinger SB. Trimethylamine N-oxide binds and activates PERK to promote metabolic dysfunction. Cell Metab 30: 1141–1151.e1145, 2019. doi: 10.1016/j.cmet.2019.08.021. [DOI] [PubMed] [Google Scholar]
- 101.He W, Wang Q, Gu L, Zhong L, Liu D. NOX4 rs11018628 polymorphism associates with a decreased risk and better short-term recovery of ischemic stroke. Exp Ther Med 16: 5258–5264, 2018. doi: 10.3892/etm.2018.6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Siqueira ER, Pereira LB, Stefano JT, Patente T, Cavaleiro AM, Silva Vasconcelos LR, Carmo RF, Moreira Beltrao Pereira LM, Carrilho FJ, Correa-Giannella ML, Oliveira CP. Association of a variant in the regulatory region of NADPH oxidase 4 gene and metabolic syndrome in patients with chronic hepatitis C. Eur J Med Res 20: 45, 2015. doi: 10.1186/s40001-015-0136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, Wagner MA, Bennett BJ, Li L, DiDonato JA, Lusis AJ, Hazen SL. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem 290: 5647–5660, 2015. doi: 10.1074/jbc.M114.618249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lu X, Li N, Shushakova N, Schmitt R, Menne J, Susnik N, Meier M, Leitges M, Haller H, Gueler F, Rong S. C57BL/6 and 129/Sv mice: genetic difference to renal ischemia-reperfusion. J Nephrol 25: 738–743, 2012. doi: 10.5301/jn.5000053. [DOI] [PubMed] [Google Scholar]