Abstract
Multiple adenovirus (Ad) early proteins have been shown to inhibit transcription activation by p53 and thereby to alter its normal biological functioning. Since these Ad proteins affect the activity of p53 via different mechanisms, we examined whether this inhibition is target gene specific. In addition, we analyzed whether the same Ad early proteins have a comparable effect on transcription activation by the recently identified p53 homologue p73. Our results show that the large E1B proteins very efficiently inhibited the activity of p53 on the Bax, p21Waf1, cyclin G, and MDM2 reporter constructs but had no effect on the activation of the same reporter constructs by p73, with the exception of some inhibition of the Bax promoter by Ad12 E1B. The repressive effect of the E1A proteins on p53 activity is less than that seen with the large E1B proteins, but the E1A proteins inhibit the activity of both p53 and p73. We could not detect significant inhibition of p53 functions by E4orf6, but a clear repression of the transcription activation by p73 by this Ad early protein was observed. In addition, we found that stable expression of the Ad5 E1A and that of the E1B protein both caused increased p73 protein expression. The large E1B and the E4orf6 proteins together do not target the p73 protein for rapid degradation after adenoviral infection, as has previously been found for the p53 protein, probably because the large E1B protein does not interact with p73. Our results suggest that the p53 and p73 proteins are both inactivated after Ad infection and transformation but via distinct mechanisms.
By regulating the expression of different target genes, p53 can affect important cellular processes like cell cycle progression and apoptosis. Multiple adenovirus early (Ad E) proteins have been shown to inhibit the transcription activation potential of p53 via different mechanisms and thereby to impair its normal biological functioning. The first Ad E protein identified as inhibiting p53 activity was the large E1B protein (38). Yew and colleagues have shown that in the presence of the large E1B protein p53 can still bind to its consensus sequence but that the transcription-repressive regions present in the large E1B protein inhibit the transcription activation potential of p53 (39). In addition, we and others have shown that the E1A proteins also can inhibit transcription activation by p53 (27, 30). The E1A proteins can directly interact with the p300 protein, which not only serves as a cofactor for p53 transactivation but also activates its sequence-specific DNA binding by acetylation of the C terminus of p53 (6, 7, 12). The third Ad E protein which has been reported to inhibit transcription activation by p53 is the E4orf6 protein. Dobner and coworkers reported that E4orf6 inhibits the activity of p53 by a direct interaction with the C terminus of p53 which inhibits binding of TAF32 to the transcription activation domain of p53 (2). It is currently unclear why Ads produce such a number of different proteins all causing repression of the transcriptional activity of p53. It suggests that inactivation of p53 is very important for the Ad replicative cycle. However, since all these Ad E proteins inhibit p53 via different mechanisms we hypothesized that gene-specific effects might exist.
Recently, Kaghad and colleagues have identified the p73 gene, which has significant homology with p53 (10). Lack of expression of the gene was observed in neuroblastoma cells, indicating that dysregulation of this p53 homologue might contribute to tumorigenesis. In addition, it was found that p73 can activate p53-responsive genes (9, 10). In this study, we examined whether the transcriptional activity of p73 was inhibited by the Ad E proteins in a way similar to that in which they affect p53.
The Ad E proteins not only inhibit the activity of p53 but also affect the half-life of the tumor suppressor protein. We and others have shown that, after Ad infection, the complex of the large E1B and the E4orf6 protein causes enhanced degradation of the p53 protein (22, 23, 28). In contrast, when cells are transformed by the Ad E1 region and no E4orf6 protein is coexpressed, both the E1A (14) and the E1B (34) proteins cause stabilization of p53. After transformation of rat cells with the oncogenic adenovirus type 12 (Ad12), highly stabilized p53 was found in the nucleus (41), while after transformation by the nononcogenic virus Ad5 or Ad2 the p53 protein is detected mainly in the cytoplasm, and the appearance of specific cytoplasmic bodies containing the large E1B protein together with the p53 protein has been reported (41).
In this study, we examined the effects of the Ad E proteins on the activation of different responsive genes by p53 in transient transfections. We found a very strong repressive effect by the large E1B proteins, while intermediate effects were found for the E1A proteins. In contrast, the large E1B proteins had no effect on the transcription activation by p73, with the exception of some inhibition of the Bax promoter by Ad12 E1B. The effects of E1A on p73 were comparable with the repression of p53. Furthermore, we found that the E4orf6 proteins could significantly inhibit the activity of p73, while no effect on p53 was detectable. In addition, we analyzed the effects of these Ad E proteins on endogenous p73 expression levels and found that the Ad5 E1A as well as the large E1B protein causes increased levels of p73 protein. However, no interaction between the large Ad5 E1B protein and p73 could be detected, which might explain why no down-regulation of p73 was detectable after Ad5 infection.
MATERIALS AND METHODS
Tissue culture and cell lines.
Hep3B (21) and G401 (36) cells were grown in Dulbecco’s modified Eagle’s medium plus 8% fetal bovine serum (FBS). The stable G401 transfectants 21K-C1 and 21K-C8 (21C1 and 21C8, respectively); 21K/E1A-C3 and 21K/E1A-C24 (5/21C3 and 5/21C24, respectively); 5E1B-C2, 5E1B-C3, and 5E1B-C5 (G55C1, G55C3, and G55C5, respectively); 12E1B-C1, 12E1B-C3, and 12E1B-C7 (G54C1, G54C3, and G54C7, respectively); and E4-C3 and E4-C7 (GE4C3 and GE4C7, respectively) have been described previously (28–30) and were grown on the same medium with addition of G418 (300 μg/ml). Primary HER (human embryonic retina), Ad5-transformed HER XC1 and HER XC2, and Ad12-transformed HER RC4 and HER RC6 cell lines have been described previously (33) and were cultured on Eagle’s minimal essential medium supplemented with vitamins, nonessential and essential amino acids, glucose, and 10% FBS.
Plasmids, transient transfections, and luciferase assays.
Hep3B cells were transiently transfected by the calcium phosphate precipitation method (35) on six-well plates. From the pGL2-NA(mdm2)-luc (MDM2Luc) (4), pGL3-CyclinG-Luc (CyclinGLuc) (41), pGL3-Bax-luc (BaxLuc) (16), IGF-BP3-BoxB-luc (IGF-BP3Luc) (1), and pGL3-p21-luc (p21Waf1Luc) (3) reporter constructs, 2.5 μg was used. The cotransfection experiments as presented in Fig. 1 were performed with 10 ng of pCMV-p53 or 10 ng of pCMVneo together with 1.0 μg of cytomegalovirus (CMV)-E1B21K (31), CMV-12E1B (34), pCMV-5E1B (34), pRSV-E1A (18), pCMV-E4orf6 (2), or CMVneo. The same cotransfections were performed for the experiments whose results are shown in Fig. 3 but now with 100 ng of pCMV-p73 (hemagglutinin [HA] tagged) or 100 ng of pCMVneo in addition to 1.0 μg of pRSV-12SE1A, pRSV-12SE1A(111–123), or pRSV-12SE1A(4–25) mutant. The titration experiments whose results are shown in Fig. 2 were performed with 2.5 μg of the reporter constructs. Increasing concentrations of either pCMV-p53 or pCMV-p73 were used as described in the figure legends. With the addition of pCMVneo, the total amount of CMV-containing plasmid was adjusted to 1.0 μg in all precipitates. In all transfections, the total amount of DNA was adjusted to 10 μg with salmon sperm DNA, and all precipitates were made in triplicate. Exponentially growing cells were transfected overnight. The next morning, cells were washed twice with phosphate-buffered saline (PBS), and 24 to 28 h later, lysates were made in luciferase lysis buffer (0.1 M Tris-H2PO4 [pH 7.8], 1% Triton, 15% glycerol, 2 mM dithiothreitol, 8 mM MgCl2). After a 15-min incubation at room temperature, cell debris was removed by 1 min of centrifugation. The luciferase activity was measured with a Lumac/3M luminometer (Biocounter) in 10 or 20 μl of the lysate after addition of 100 μl of luciferin (Boehringer Mannheim). The protein concentrations of the lysates were determined by the Bradford (Bio-Rad) assay, and the luciferase activity in 1 μg of protein lysate was calculated. A number of the transfections were also performed with a pCMVLacZ internal control. The data obtained were similar, but since both E1A and high concentrations of p53 affected the activity of the internal control plasmid, corrections of the luciferase activity were made based on the protein concentrations of the lysates. To check the expression of the transfected constructs by Western immunoblotting, transfections were performed exactly the same way as described above. At 24 h posttransfection, lysates were made in E1A buffer (50 mM Tris-HCl [pH 7.4], 250 mM NaCl, 0.1% Triton, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM orthovanadate, 1 mM leupeptin, 0.5 mM trypsin inhibitor, and 0.1 mM aprotinin). Lysates were cleared by centrifugation at 14,000 rpm (Eppendorf 5417R) for 10 min. Protein concentrations of the lysates were measured by the Bradford assay (Bio-Rad).
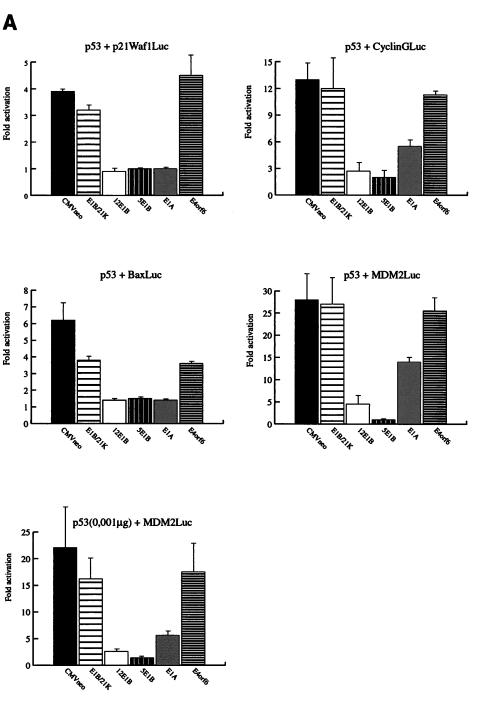
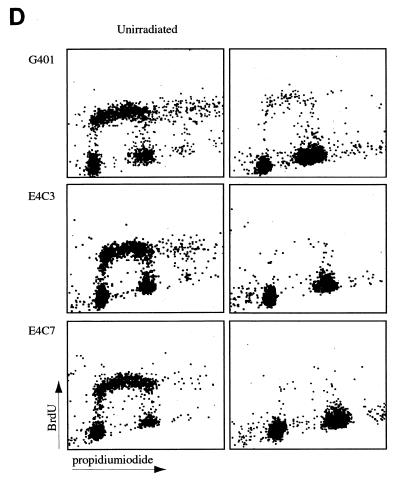
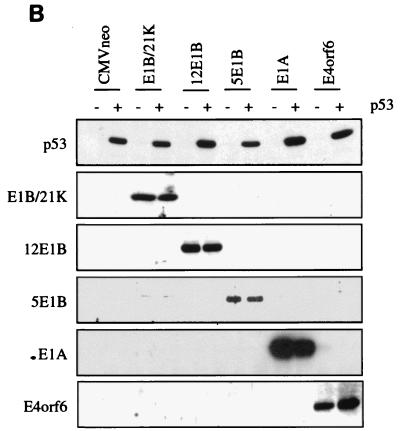
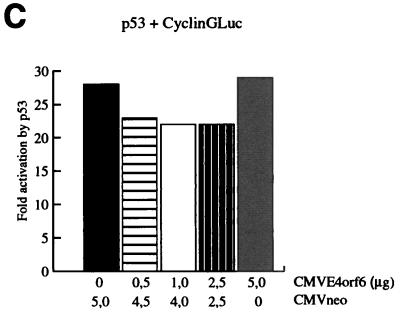
FIG. 1.
Distinct effects of the Ad E1A, E1B, and E4orf6 proteins on the regulation of different responsive genes by p53. (A) Transient transfections were performed in Hep3B cells. Twenty-four hours posttransfection, lysates were made and the luciferase activity was measured. Subsequently, the activation of the reporter constructs by CMV-p53 (0.01 μg) in the presence or absence of the different Ad E proteins (1.0 μg) was calculated. The cotransfection of the MDM2Luc construct was performed with 0.01 μg and with a 10-fold-lower amount of CMV-p53 (0.001 μg). The total amount of DNA in the precipitates was adjusted to 10 μg with salmon sperm DNA. All transfections were performed in triplicate at least twice, and the error bars in the figure represent the variations between the independent experiments. (B) Expression of the transfected expression plasmids was verified by Western immunoblotting. (C) Cotransfection of increasing concentrations of CMV-E4orf6 did not cause inhibition of the activation of the CyclinGLuc reporter construct by 0.01 μg of CMV-p53. (D) Parental G401 cells and stable transfectants expressing the E4orf6 protein were either mock irradiated or irradiated with X rays (5 Gy). Eighteen hours postirradiation, cells were labelled with BrdU and analyzed by FACScan. A significantly decreased number of cells in S phase was detected for the G401 cells as well as for the E4orf6 stable transfectants.
FIG. 3.
(A) The effects of E1A, E1B, and E4orf6 on regulation of different p53-responsive constructs by p73. The p21Waf1Luc, BaxLuc, CyclinGLuc, and MDM2Luc reporter constructs were transiently transfected in Hep3B cells. Activation of the reporter constructs by CMV-p73 (0.1 μg) was measured after cotransfection of the control CMVneo vector (1.0 μg) or after cotransfection of the indicated Ad expression plasmids (1.0 μg). In the case of the MDM2Luc reporter construct, only 0.01 μg of CMV-p73 was used. Twenty-four hours posttransfection, lysates were made and the luciferase activity was measured. All transfections were performed at least two times in triplicate. The given data represent the mean values of these experiments, and the error bars indicate the variations between the independent experiments. (B) Expression of p73 in the cotransfections was verified by Western immunoblotting. (C) The effects of 12SE1A and E1A mutants [12S(Δ4–25) lacking p300 binding domain and 12S(Δ111–123) lacking pocket protein binding domain] on regulation of the BaxLuc reporter construct by p73. (D) A side-by-side comparison of the effects of the Ad proteins on the transcription activation by p53 and p73 on the various reporter constructs.
FIG. 2.
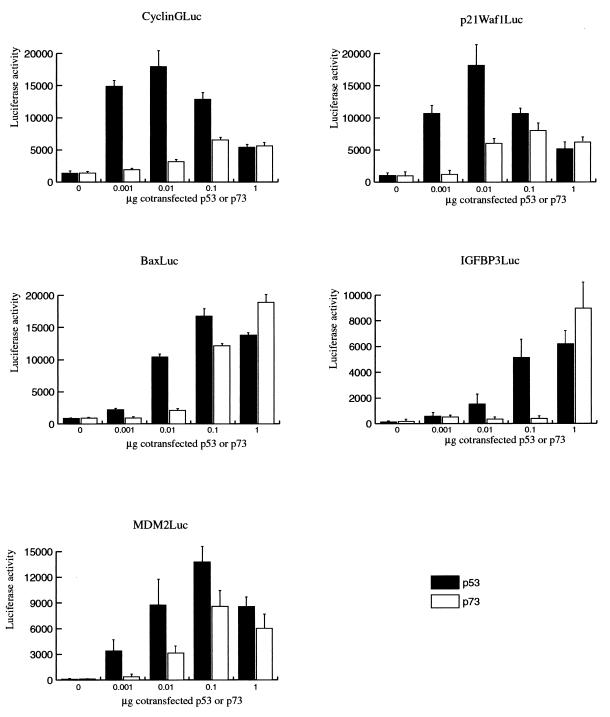
Comparison of the effects of p53 and p73 on different p53-responsive reporter constructs. Increasing concentrations of the CMV-p53 and CMV-p73 expression plasmids were cotransfected with the CyclinGLuc, p21Waf1Luc, BaxLuc, IGF-BP3Luc, and MDM2Luc reporter constructs. At 24 h posttransfection, lysates were made and the luciferase activity was measured. All precipitates were made in triplicate, and the error bars represent the variations among three independent precipitates.
Virus techniques.
The wild-type Ad5 was grown and purified as previously described (28). The concentration of the virus was determined by plaque assay in 911 cells as described previously (28). For the infection experiments, the cells were split 1 day prior to infection and seeded out on 5-cm-diameter petri dishes. The next day, cells were infected with 100 PFU/cell diluted in a volume of 500 μl of PBS–2% FBS. Afterwards, the virus was removed and normal tissue culture medium was added to the cells. Twenty-four hours postinfection, the cells were washed twice with ice-cold PBS and lysates were made in E1A buffer as described in “Plasmids, transient transfections, and luciferase assays” above.
Western immunoblotting.
For the analysis of the p73 and p53 expression in primary and Ad-transformed HER cells, lysates were made in radioimmunoprecipitation assay buffer (20 mM triethanolamine [pH 7.4], 140 mM NaCl, 0.1% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 0.1% Triton, 1 mM PMSF, 1 mM orthovanadate, 1 mM leupeptin, 0.5 mM trypsin inhibitor, and 0.1 mM aprotinin). Protein concentrations of the lysates were measured by the Bradford assay (Bio-Rad), and 40 μg of the lysates was examined by Western immunoblotting. For the analysis of the G401 cells and the stable transfectants, lysates were made in E1A buffer as described in “Plasmids, transient transfections, and luciferase assays,” and 25 μg of these lysates was examined by Western immunoblotting. The indicated amounts of lysate were boiled for 5 min in Laemmli sample buffer and subsequently separated on SDS–10% polyacrylamide gels. Subsequently, the proteins were blotted onto Protran nitrocellulose membranes (Schleicher and Schuell). The membranes were incubated with the antibody DO-1 (SanverTECH, p53 specific), RSA3 (E4orf6 specific), A1C6 (5E1B specific), T8A9 (12E1B specific), M73 (E1A specific), 1G11 (21K-E1B specific), or anti-p73. After incubation with horseradish peroxidase-conjugated mouse, rabbit, or rat secondary antibodies, immune complexes were detected by enhanced chemiluminescence (Amersham).
Cell cycle analysis.
Exponentially growing G401, E4-C3, and E4-C7 cells were mock irradiated or irradiated with X rays (5 Gy). Eighteen hours after treatment, unirradiated and irradiated cells were labelled for 30 min with 20 μM bromodeoxyuridine (BrdU). The cells were washed twice with PBS and harvested by trypsinization. The cells were washed again with PBS and fixed in 70% ethanol overnight. Subsequently, the cells were centrifuged and the pellet was resuspended in 2 N HCl. Cells were incubated for 12 min at 37°C and, after washing, incubated with anti-BrdU (Becton Dickinson). Afterwards, cells were incubated with propidium iodide and RNase, and 5,000 cells were analyzed on a FACScan flow cytometer (Becton Dickinson) for DNA content and BrdU incorporation. The fluorescence-activated cell sorting was used to determine the effects of p73 expression on the cell cycle. For the analysis of the cell cycle profile, an expression vector for green fluorescent protein (GFP)-tropomyosin fusion was added to each transfection. Thirty-six hours after transfection, cells were washed once in PBS and incubated with propidium iodide (2 μg of propidium iodide per ml, 0.6% Nonidet P-40, and 100 μg of RNase per ml). Five thousand transfected GFP-positive cells were analyzed on a FACScan flow cytometer.
Immunofluorescence.
Primary HER, HER RC6, and HER XC1 cells were grown on coverslips. The cells were rinsed twice with PBS, fixed with 80% acetone for 5 min at room temperature, and incubated for 1 h with T8A9 (anti-12E1B), 9C10 (anti-5E1B), rabbit polyclonal anti-p53 (SanverTECH), or a mixture of anti-p53 monoclonal antibodies (Ab 122 and Ab 421) or anti-p73. Subsequently, the cells were incubated for 1 h with anti-rabbit-rhodamine, anti-mouse-fluorescein isothiocyanate, or anti-rat-fluorescein isothiocyanate antibodies.
Metabolic labelling and immunoprecipitations.
Exponentially growing cells were rinsed twice with PBS and maintained for 0.5 h in methionine-cysteine-free minimal essential medium supplemented with 2% FBS. [35S]methionine (100 μCi per dish) was then added directly to the medium, and cells were incubated at 37°C for an additional 3 h. After labelling, the cells were washed twice with ice-cold PBS and cell lysates were prepared in E1A buffer as modified by Giordano (50 mM Tris HCl [pH 7.4], 0.25 M NaCl, 0.1% Triton X-100, 5 mM EDTA), supplemented with protease inhibitor cocktail (Complete; Boehringer Mannheim) and 0.5 M PMSF. Equal amounts of radioactivity were precleared with protein A beads. Precleared lysates were precipitated with either 12CA5 (anti-HA) or A1C6 (Ad5 E1B/55K specific). Immunoprecipitations were performed as described previously (29). Immunoprecipitated proteins were separated on SDS–10% polyacrylamide gels, prepared for fluorography with Amplify (Amersham Science), dried, and exposed to Kodak XAR-5 film at −80°C.
RESULTS
Distinct effects of E1B, E1A, and E4orf6 on the activation of different p53-responsive genes.
It has been reported that the large E1B, E4orf6, and E1A proteins can inhibit transcription activation by p53 but all via different mechanisms. To test the hypothesis that this might lead to gene-specific regulation, transient transfections were performed in p53-deficient Hep3B cells. Cotransfection of 0.01 μg of CMV-p53 leads to significant activation of the p21Waf1, Bax, cyclin G, and MDM2 reporter constructs in the presence of the CMVneo control vector (Fig. 1A). On all four reporter genes, we found strong repression of the p53-mediated transcription activation after cotransfection of the oncogenic Ad12 and the nononcogenic Ad5 large E1B proteins. Previous reports have shown that the small E1B protein could not inhibit transcription activation by p53 (24, 26, 31). Indeed, no effect of the E1B/21-kDa protein was found on the activity of p53 on the p21Waf1, cyclin G, and MDM2 reporter constructs, but a slight inhibition was measured on the activity of p53 on the Bax reporter construct. Interestingly, gene-specific effects were found after cotransfection of E1A. On the p21Waf1 and Bax reporter constructs, inhibition of p53 activity by E1A was as strong as the effects of the large E1B proteins. However, the activity of p53 on the cyclin G and MDM2 reporter constructs was only partially repressed by E1A. Expression of p53 and the cotransfected Ad E proteins was checked by Western immunoblotting (Fig. 1B). The cyclin G and MDM2 reporter constructs showed the strongest activation by p53. It might be that, due to this strong activation by p53, only a partial effect of E1A could be detected. When the amount of p53 expression plasmid in the cotransfections was reduced from 0.01 to 0.001 μg, a slightly reduced activation of the MDM2Luc construct was observed, but now the repressive effect due to cotransfection of E1A was significantly stronger (Fig. 1A). Surprisingly, no significant inhibition of the transcription activation by p53 was observed by cotransfection of E4orf6, although we could clearly detect E4orf6 expression by Western immunoblotting (Fig. 1B). As can be seen in Fig. 1C, even after cotransfection of 5 μg of the CMV-E4orf6 plasmid, no repression of the activation of the CyclinGLuc construct by p53 was found. We also tested the effect of E4orf6 on Gal4-p53 fusion constructs in Saos-2 cells and found no repression of the transcriptional activity of p53 by E4orf6 (data not shown). Finally, we examined whether stable expression of the E4orf6 protein could inhibit the function of endogenous p53. Parental G401 cells and two independent G401-E4orf6 stable transfectants were mock irradiated or irradiated with X rays (5 Gy). We have previously shown that the E4orf6 protein is highly expressed in these transfectants and can cause enhanced degradation of the p53 protein together with the large E1B protein after infection with a ΔE4 Ad (28). Cells were labelled with BrdU at 18 h postirradiation and analyzed by fluorescence-activated cell sorting. As can be seen in Fig. 1D, in the presence of E4orf6, p53 can still perform its cell cycle regulatory function as in the parental G401 cells. This result indicates that the E4orf6 protein can inhibit neither cotransfected nor endogenous p53. It is not clear why our results are in contrast with the earlier reports (2, 17), but possibly cell-type- and/or reporter-specific effects play a role.
Comparison of the effects of p73 and p53 on different p53-responsive genes.
We were interested to investigate whether the Ad E proteins could also inhibit transcription activation by the p53 homologue p73. Therefore, we first performed titrations with CMV-p53 and CMV-p73 constructs to examine which amount of the p73 expression vector is required in the cotransfection experiments to be able to make a comparison with the p53 effects. Increasing amounts of the two expression vectors were cotransfected with the different reporter constructs. As can be seen in Fig. 2, p73 could activate the promoters of the cell-cycle-regulatory proteins p21Waf1 and cyclin G, but not up to the same level as p53. In contrast, our results show that p73 can activate the Bax and IGF-BP3 promoters up to the same level as p53, although higher concentrations of p73 appear to be required. When the same titrations were carried out with the MDM2Luc construct, we found that both proteins can very efficiently activate the p53-responsive element in the MDM2 gene, although the maximal activation by p73 remains a fraction lower than that by p53. All the titration experiments were repeated with independent batches of CMV-p53 and CMV-p73 expression constructs, which resulted in similar effects (data not shown).
On all five reporter constructs, we found that activation by p53 occurred at very low amounts of expression plasmids while higher amounts of the p73 expression plasmid appeared to be required. Since both genes are driven by a CMV promoter, it suggests that the p53 protein is already active at lower concentrations, but it cannot be excluded that in the transfection experiments the p53 protein is better expressed than the p73 protein. The results in Fig. 2 show strong activation of p21Waf1Luc and CyclinGLuc with low concentrations of CMV-p53, while cotransfection of high concentrations of the expression plasmid resulted in a decreased activation of the reporter constructs. We have previously reported this phenomenon for an artificial reporter construct (31), and this effect is probably caused by the inhibition of transcription by p53, which at a high concentration overrules its transcription-activating capacity. Further study is required to find out whether p73 also can function as a repressor of transcription.
Effects of the Ad E proteins on the activation of transcription by p73.
To analyze the effects of the independent Ad E proteins on p73, again transient transfections were performed in Hep3B cells and the results are presented in Fig. 3A. Transfection of 0.1 μg of p73 leads to a low but significant activation of the p21Waf1 and cyclin G reporter constructs and to very strong activation of the BaxLuc construct. To study the effects on the activation of MDM2Luc, we used a 10-fold-lower p73 concentration as we did for p53 in Fig. 1A. In contrast to the strong inhibition by the large E1B proteins on the activation of all four reporter constructs by p53, no effects were found for the Ad5 large E1B protein on the p73 activity on any of the four reporter constructs. Also, the large E1B protein of Ad12 is unable to repress the transactivation by p73 of the p21Waf1, cyclin G, and MDM2 reporter constructs. However, some repression of the transactivation of the BaxLuc construct (2.6-fold) by Ad12 E1B was found. The Ad12 E1B protein had no effect on the basal level of the BaxLuc construct, suggesting that the observed effect is, at least in part, via the inhibition of p73 activity. E1A, however, appear to inhibit the transactivation by p73 as efficiently as the transactivation by p53 on all these reporter constructs. To investigate whether the repression by E1A involves the binding of p300 to E1A as reported for p53 (7, 12), we used two E1A mutants. The E1A(Δ111–123) mutant, which is unable to bind pocket proteins, represses transactivation by p73 as efficiently as does E1A. In contrast, the E1A(Δ4–25) mutant, which cannot bind p300, is unable to inhibit transactivation by p73, suggesting a role for p300 in the regulation of transcription by p73 (Fig. 3C). Very interestingly, we observed a significant repression of the p73 activity by E4orf6, mainly on the BaxLuc and the MDM2Luc constructs, in contrast to the effect on p53. Expression of p73 (Fig. 3B) and all the cotransfected Ad E proteins (data not shown) has been checked by Western immunoblotting. Relative inhibition by Ad E proteins of transcription activation by p53 and p73 is summarized in Fig. 3D.
p73 expression and localization in Ad12- and Ad5-transformed human cells.
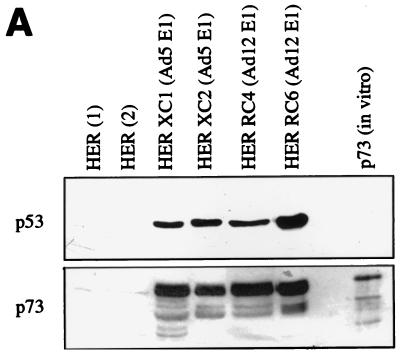
Not only have the Ad E proteins been found to regulate the activity of p53, but also the stability and cellular localization of the tumor suppressor protein are significantly altered by the same Ad proteins (14, 28, 34, 40). To examine whether Ad transformation also leads to stabilization of p73, lysates of untransformed and Ad5 E1- and Ad12 E1-transformed HER were analyzed by Western immunoblotting. As can be seen in Fig. 4A, in primary HER cells only very weak expression of the p73 protein is detectable. However, in both the Ad5-transformed (XC1 and XC2) and the Ad12-transformed (RC4 and RC6) HER lysates, strongly increased levels of p73 were found.
FIG. 4.
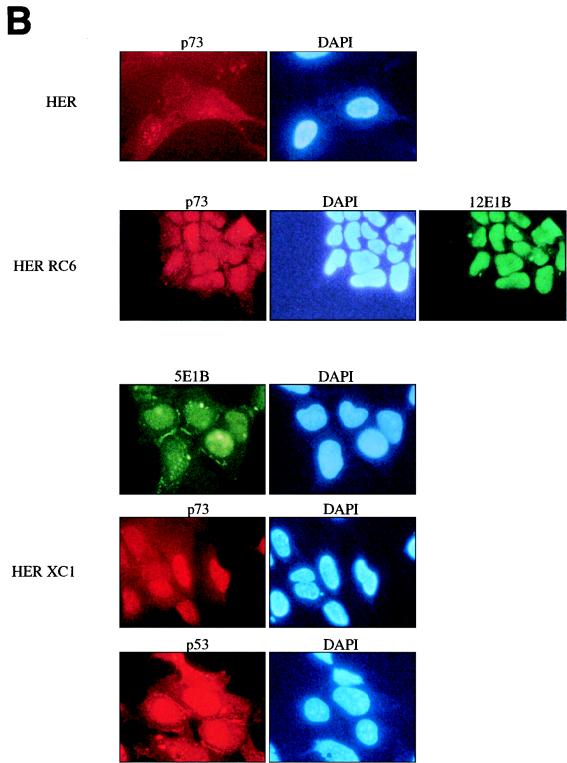
p73 expression and localization in Ad-transformed cells. (A) The p73 protein level in two independent lysates of primary HER cells, Ad5-transformed HER cells (HER XC1 and HER XC2) and Ad12-transformed HER cells (HER RC4 and HER RC6), was examined by Western immunoblotting (see Materials and Methods) and compared with the p53 expression in the same lysates. For all lysates, 40 μg of protein was loaded on an SDS–10% polyacrylamide gel. As a positive control, in vitro-translated p73 was loaded. (B) The localization of p73 in primary and Ad5- and Ad12-transformed HER cells was examined by immunofluorescence. Low p73 expression was observed in primary HER cells, while in the Ad12-transformed HER RC6 cells high p73 expression is detectable in the nucleus. In the Ad5-transformed HER XC1 cells, high p73 expression is detectable in the nucleus while both p53 and Ad5 E1B/55K are also found in the cytoplasm. DAPI, 4′,6-diamidino-2-phenylindole.
We subsequently examined the cellular localization of p73 in the primary and transformed HER cells by immunofluorescence (Fig. 4B). In primary HER cells, we found low expression of p73 in the nucleus. High expression of p73 was found in the Ad12-transformed cells, and as was previously reported for p53, in these cells p73 was localized in the nucleus. Also, expression of the Ad12 large E1B protein was mainly localized in the nucleus. In the Ad5-transformed HER cells, the large E1B protein was expressed in the nucleus and in the cytoplasm, localized in speckles and in the membranes. In Ad5-transformed human cells, the existence of the cytoplasmic bodies is not as clear as has previously been described for Ad5-transformed rat cells (40). In the Ad5 cells, the p73 protein is localized in the nucleus, while p53 protein shows a nuclear and cytoplasmic localization similar to that of the large Ad5 E1B protein.
Both E1A and large E1B proteins cause increased p73 protein expression.
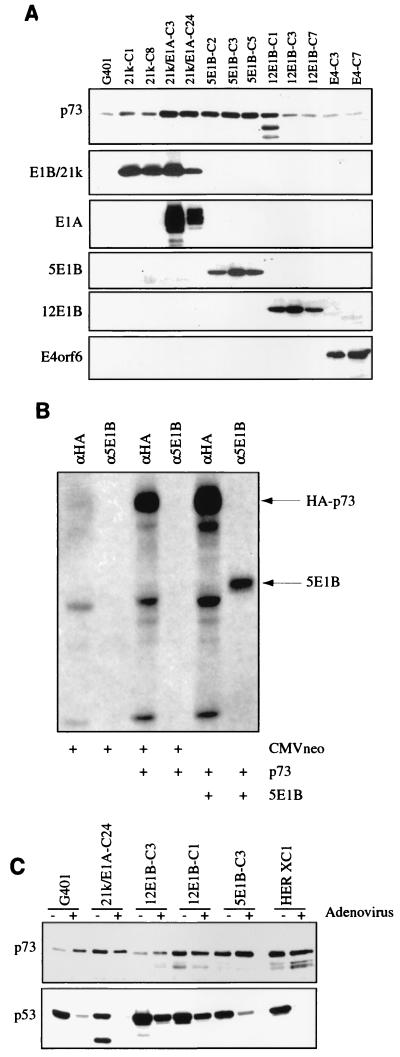
To analyze whether the enhanced p73 expression in the Ad-transformed cells was caused by the E1A or the E1B protein, a panel of independent stable transfectants derived from G401 cells, which are from a rhabdoid tumor of the kidney expressing wild-type p53, were examined by Western immunoblotting. As can be seen in Fig. 5A, expression of the small E1B or the E4orf6 protein does not significantly alter the expression level of p73. However, in the transfectants expressing the E1A or the Ad5 large E1B protein, increased levels of p73 were found, indicating that these Ad E proteins have an effect on the expression of p73 similar to that on p53. In two of the three stable transfectants expressing the Ad12 large E1B protein, no increase in the p73 expression could be detected. In the Ad12 E1B-expressing transfectant (12E1B-C1) showing elevated levels of the p73 protein, besides the full-length protein other bands were found. The nature of these alternative proteins is not clear, but they might represent products of alternative splice variants. Further study is required to determine the exact effect of the Ad12 large E1B protein on the stability of p73.
FIG. 5.
The Ad5 E1A and large E1B proteins cause increased p73 expression in stable transfectants. (A) Lysates were made of exponentially growing parental G401 cells and the stable transfectants expressing the indicated Ad E proteins. Twenty-five micrograms of the lysates was loaded on an SDS-polyacrylamide gel and examined by Western immunoblotting for expression of p73 and the expression of the Ad E proteins. (B) No association between Ad5 large E1B and p73. Hep3B cells, transiently transfected with CMVneo or CMV-p73 alone or in the presence of CMV-5E1B, were labelled with 35S-methionine, and immunoprecipitations on labelled lysates were performed with the indicated antibodies. The precipitates were separated by SDS-polyacrylamide gel electrophoresis, followed by autoradiography. (C) Expression of p53 and p73 in various G401 stable transfectants expressing the viral E proteins and HER XC1 cells after infection with wild-type Ad5. Twenty-four hours postinfection, lysates were made and p53 and p73 expression was analyzed by Western immunoblotting.
No association between p73 and the Ad5 large E1B protein.
To investigate whether p73 and the Ad5 large E1B protein could form a complex in vivo, we cotransfected the HA-tagged p73 expression vector and CMV-5E1B/55K into Hep3B cells. After metabolic labelling, lysates were immunoprecipitated with anti-HA and anti-E1B/55K antibodies (Materials and Methods). As can be seen in Fig. 5B, both the p73 and the E1B/55K protein are well expressed, but no coprecipitation of p73 with Ad5 E1B/55K or vice versa could be detected, indicating that under these conditions no association of p73 and E1B/55K takes place. Under the same conditions, the interaction between p53 and Ad5 E1B/55K can be easily detected, as we and others have shown repeatedly. The same conclusions can be drawn from immunoprecipitation-Western blot studies on stable transfectants of the G401 cells expressing the Ad5 E1B/55K protein (data not shown). The lack of association between p73 and E1B/55K has recently been shown by others as well (15, 23).
It has previously been shown that after Ad infection the large Ad5 E1B protein together with the E4orf6 protein can target p53 for active degradation. Since we did not detect an association between the Ad5 large E1B protein and p73, we expected that no down-regulation of p73 protein level would take place in cells after Ad infection. Indeed, as can be seen in Fig. 5C, neither for the parental G401 cells nor for any of the stable transfectants expressing either high or low levels of p73 nor for Ad5-transformed HER cells was a decrease in p73 levels observed after Ad infection.
Distinct effects of Ad E proteins on p73-induced G1 arrest.
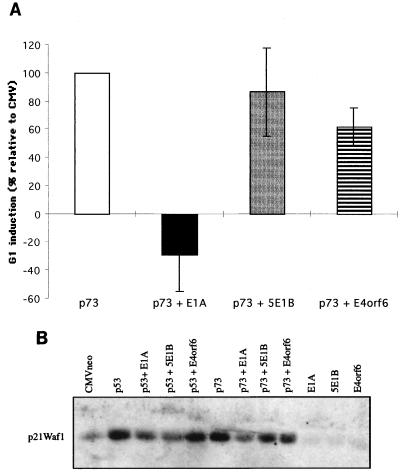
To investigate the effect of the Ad proteins on a G1 arrest imposed by p73, we transfected Saos-2 cells with p73 in the absence or presence of the Ad E proteins and analyzed the cell cycle profile by FACScan analysis. p73 expression was found to result in a moderate increase in G1-phase cells, with an increase in the percentage of G1-phase cells between 8 and 20%, depending on the experiment. To be able to compare the effects of the Ad E proteins between different experiments, in each experiment the increase in G1-phase cells caused by p73 compared to vector-only-transfected cells was set at 100%. It was found that in the presence of E1A the p73 effect is completely abolished. Even in the presence of p73, E1A was actually able to stimulate S-phase entry. Coexpression of the Ad5 large E1B protein showed no significant effect, which is in concordance with the results described above. In the presence of E4orf6, the G1 arrest by p73 was only partly rescued (40% reduction). This limited effect can be explained by the observation that E4orf6 inhibits the induction of the p21Waf1 promoter in transient expression assays only about twofold (Fig. 3A). The effect of p73 expression on the cell cycle corresponds quite well with the regulation of the endogenous levels of p21Waf1 protein after transfection of p73. Coexpression of Ad5 E1A almost completely abolishes the p73-mediated increase in p21Waf1 (Fig. 6B). The E4orf6 protein results in only a limited reduction of p21Waf1. Also, the large Ad5 E1B protein appears to have some effect on the p73-mediated induction of p21Waf1 expression, but this effect can mainly be attributed to an effect of 5E1B on the basal level of p21Waf1.
FIG. 6.
Effects of Ad E proteins on p73-induced G1 arrest and p73-induced endogenous p21Waf1 expression. (A) pCMV-p73 was transfected either alone or together with the indicated plasmids in the presence of GFP-tropomyosin into Saos-2 cells. Forty-eight hours after transfection, cells were collected and the cell cycle profile of GFP-positive cells was analyzed by FACScan. The effects of the adenoviral proteins on G1 increase by p73 are shown relative to the increase in G1 caused by the expression of p73 alone, which was set to 100%. The given data represent the mean values of four different experiments, and the error bars indicate the variations between the independent experiments. (B) Expression of endogenous p21Waf1 after transient transfection of Saos-2 cells by p73 alone or in the presence of the indicated plasmids. Forty-eight hours after transfection, lysates were made and the p21Waf1 expression was analyzed by Western blot analysis.
DISCUSSION
In the early phase of Ad infection, multiple proteins which abolish the activity of p53 are expressed. The E1A and E1B proteins both inhibit the transcription activation by p53, the small E1B protein inhibits p53-mediated apoptosis, and the large E1B and the E4orf6 proteins together target p53 for active degradation. The fact that such a number of Ad E proteins are involved in the inactivation of p53 suggests that inactivation of the p53 protein is very important for Ad infection. The recently identified p73 protein has significant homology with p53 (10). In addition, it has been shown that p73 can activate the p53-responsive p21Waf1 promoter, suggesting that p73 might control cell cycle progression like p53, and moreover, p73-induced apoptosis has also been observed previously (9, 10). Since the p53 and p73 proteins not only have sequence homology but also seem to perform the same functions, we were interested to investigate whether the Ad E proteins would have a similar effect on the features of p73 and p53. We first compared the effects of CMV-p53 and CMV-p73 on different reporter constructs and found that both p53 and p73 could very efficiently activate the Bax and IGF-BP3 reporter constructs, while the activation of the p21Waf1 and cyclin G reporter constructs by p73 was significantly lower than that by p53. These results suggest that both proteins efficiently activate the apoptotic pathway but that the expression of cell-cycle-regulatory proteins is more strongly induced by p53 than by p73.
We subsequently examined the effects of the Ad E proteins on the activation of the different reporter constructs by p73. Our results show that the large E1B proteins, the strongest inhibitors of p53, in general do not alter the activity of p73. Mutation analysis performed by Lin and coworkers has previously identified a number of amino acids in the N terminus of p53 which are crucial for large Ad5 E1B binding (13). Some of these amino acids are not conserved in p73, already suggesting that the large Ad5 E1B protein might not directly interact with p73. Indeed, we could not detect a direct association between the Ad5 large E1B protein and p73. The lack of interaction between p73 and E1B was recently reported by others as well (15, 23). Surprisingly, the induction of the Bax promoter was partly inhibited by the large Ad12 E1B protein. The mechanism by which this occurs is not clear yet. It is possible that the induction of the Bax promoter by p73 is via a synergism between p73 and another putative transcription factor also binding to the Bax promoter, which also might explain the strong induction of the Bax promoter by p73. The Ad12 E1B protein then somehow prevents this synergism, resulting in the activation of the Bax promoter by p73 itself.
We previously showed that, for the active degradation of p53 by the large E1B and E4orf6 proteins, direct interaction between the large E1B protein and p53 was essential. Based on these observations, we predicted that p73 would not be targeted for active degradation after wild-type Ad infection, which was what we found. Furthermore, p73 was found to be resistant to degradation after cotransfection of Ad5 E1B and E4orf6 expression plasmids (23). Interestingly, p73 is also resistant to human papillomavirus E6-mediated degradation, in contrast to the effect on p53 (20).
In contrast to the large E1B, the E1A proteins cause significant repression of the transcription activation by p73. Since an E1A mutant defective in p300 binding did not effectively inhibit the transcription activation by p73, it is proposed that the p300/CREB-binding proteins are also involved in the regulation of transcription by the p73 protein.
Unexpectedly, we did not observe inhibition of the p53-mediated transcription activation by the E4orf6 protein. This result, however, is in line with a recent publication which also described a lack of effect of E4orf6 on p53 activity (23). It has previously been reported that inhibition of the transcriptional activity of p53 by E4orf6 is mediated by inhibited binding of TAF32 to the transcription activation domain of p53 (2). Possibly, in our assay system TAF32 does not play an essential role, so that coexpression of E4orf6 does not result in inactivation of the transcriptional activity of p53. Interestingly, the E4orf6 protein could partly inhibit the transcriptional activity of p73 on the reporter constructs tested, with a strong effect seen only on the Bax promoter. The mechanism of this effect is unclear as yet. Further study is required to determine whether E4orf6 can directly associate with p73 or whether this is an indirect effect.
All in all, it can be hypothesized that, in the early phase of Ad infection, when those early proteins are expressed, distinct Ad E proteins are involved in inhibition of the transcription activation by both p53 and p73, although an effect on p73 activity during Ad infection has not been proven directly.
Increased p73 protein level could be detected in Ad-transformed cells, and in a more detailed analysis, we found that Ad5 E1A and the large E1B proteins might independently cause an increased p73 protein level. The mechanism by which the large E1B proteins can stabilize p53 has not yet been determined, but it can be hypothesized that in this process the MDM2 protein plays a crucial role. The MDM2 protein has been shown to be an important regulator of the half-life of p53 (8, 11), and stabilization of p53 in tumor cells might be due to the fact that mutant p53 cannot induce MDM2 expression. Not only might the large E1B protein inhibit the expression of the MDM2 protein by repressing the p53 transcriptional activity, but it has also been shown that the Ad5 large E1B protein binds to the same domain of p53 as the MDM2 protein and might prevent binding of the MDM2 protein to p53 (13). A similar mechanism does not explain the increased expression of the p73 protein, since first we did not observe inhibition of the transcriptional activity of p73 by the large E1B proteins and second no direct interaction between p73 and the Ad5 large E1B proteins could be detected. Further study is required to clarify the mechanisms by which the large E1B proteins increase the p73 expression levels.
It has previously been reported that the E1A proteins can stabilize the p53 protein (14). Since the E1A protein can also inhibit the p53-mediated expression of MDM2, the increased levels of p53 might be due to decreased MDM2 expression. If this is indeed the mechanism by which E1A can enhance the p53 levels, then p73 will probably be regulated in the same way since p73-induced MDM2 expression is also inhibited by the E1A proteins. Recently, it has also been shown that p300 might have a function in the MDM2-mediated degradation of p53 (5). Inhibition of p300 by E1A might thus also repress this degradation pathway, resulting in stabilized p53 and possibly also p73.
In summary, we can conclude that the E1A proteins seem to have a similar effect on p53 and on p73, but these proteins are differently affected by the large E1B and E4orf6 proteins. However, the final effect is that both the p53 and the p73 proteins are functionally inactivated as a result of both infection and transformation by Ad. Apart from the p73 gene, the p53 family contains at least one other member: the KET/p51/p40/p63 protein (19, 25, 32, 37). It will be interesting to investigate whether the different forms of this p53 homologue can be inactivated by the Ad E proteins as well.
ACKNOWLEDGMENTS
We thank D. Caput for the generous gift of the CMV-p73 construct and the anti-p73 rabbit polyclonal serum. We thank T. Shenk for the kind gift of the RSA3 antibody; M. Oren for the p21Waf1Luc, MDM2Luc, and CyclinGLuc constructs; J. C. Reed for the BaxLuc construct; and N. Kley for the IGF-BP3Luc construct. Furthermore, we thank R. Bernards for helpful discussion.
This project was supported by a grant from the Dutch Cancer Society.
W.T.S. and A.S. contributed equally to the work.
REFERENCES
- 1.Buckbinder L, Talbott R, Velasco-Miguel S, Takenaka I, Faha B, Seizinger B R, Kley N. Induction of the growth inhibitor IGF-binding protein 3 by p53. Nature. 1995;377:646–649. doi: 10.1038/377646a0. [DOI] [PubMed] [Google Scholar]
- 2.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–1473. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 3.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 4.Friedlander P, Haupt Y, Prives C, Oren M. A mutant p53 that discriminates between p53-responsive genes cannot induce apoptosis. Mol Cell Biol. 1996;16:4961–4971. doi: 10.1128/mcb.16.9.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z X, Kumar S, Howley P M, Livingston D M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 6.Gu W, Roeder R G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 7.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 8.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 9.Jost C A, Marin M C, Kaelin W G., Jr p73 is a human p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 10.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan J C, Valent A, Minty A, Chalon P, Lelias J M, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90:809–819. doi: 10.1016/s0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 11.Kubbutat M H, Jones S N, Vousden K H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 12.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 13.Lin J, Chen J, Elenbaas B, Levine A J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 14.Lowe S W, Ruley H E. Stabilization of the p53 tumor suppressor is induced by adenovirus 5 E1A and accompanies apoptosis. Genes Dev. 1993;7:535–545. doi: 10.1101/gad.7.4.535. [DOI] [PubMed] [Google Scholar]
- 15.Marin M C, Jost C A, Irwin M S, DeCaprio J A, Caput D, Kaelin W G., Jr Viral oncoproteins discriminate between p53 and the p53 homolog p73. Mol Cell Biol. 1998;18:6316–6324. doi: 10.1128/mcb.18.11.6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyashita T, Reed J C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 17.Nevels M, Rubenwolf S, Spruss T, Wolf H, Dobner T. The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA. 1997;94:1206–1211. doi: 10.1073/pnas.94.4.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Offringa R, Gebel S, van Dam H, Timmers M, Smits A, Zwart R, Stein B, Bos J L, van der Eb A, Herrlich P. A novel function of the transforming domain of E1a: repression of AP-1 activity. Cell. 1990;62:527–538. doi: 10.1016/0092-8674(90)90017-9. [DOI] [PubMed] [Google Scholar]
- 19.Osada M, Ohba M, Kawahara C, Ishioka C, Kanamaru R, Katoh I, Ikawa Y, Nimura Y, Nakagawara A, Obinata M, Ikawa S. Cloning and functional analysis of human p51, which structurally and functionally resembles p53. Nat Med. 1998;4:839–843. doi: 10.1038/nm0798-839. [DOI] [PubMed] [Google Scholar]
- 20.Prabhu N S, Somasundaram K, Satyamoorthy K, Herlyn M, El-Deiry W S. p73beta, unlike p53, suppresses growth and induces apoptosis of human papillomavirus E6-expressing cancer cells. Int J Oncol. 1998;13:5–9. doi: 10.3892/ijo.13.1.5. [DOI] [PubMed] [Google Scholar]
- 21.Puisieux A, Galvin K, Troalen F, Bressac B, Marcais C, Galun E, Ponchel F, Yakicier C, Ji J, Ozturk M. Retinoblastoma and p53 tumor suppressor genes in human hepatoma cell lines. FASEB J. 1993;7:1407–1413. doi: 10.1096/fasebj.7.14.8224613. [DOI] [PubMed] [Google Scholar]
- 22.Querido E, Marcellus R C, Lai A, Charbonneau R, Teodoro J G, Ketner G, Branton P E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71:3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roth J, Konig C, Wienzek S, Weigel S, Ristea S, Dobbelstein M. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J Virol. 1998;72:8510–8516. doi: 10.1128/jvi.72.11.8510-8516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sabbatini P, Chiou S K, Rao L, White E. Modulation of p53-mediated transcriptional repression and apoptosis by the adenovirus E1B 19K protein. Mol Cell Biol. 1995;15:1060–1070. doi: 10.1128/mcb.15.2.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15:1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- 26.Shen Y, Shenk T. Relief of p53-mediated transcriptional repression by the adenovirus E1B 19-kDa protein or the cellular Bcl-2 protein. Proc Natl Acad Sci USA. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Somasundaram K, El-Deiry W S. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene. 1997;14:1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- 28.Steegenga W T, Riteco N, Jochemsen A G, Fallaux F J, Bos J L. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene. 1998;16:349–357. doi: 10.1038/sj.onc.1201540. [DOI] [PubMed] [Google Scholar]
- 29.Steegenga W T, Shvarts A, van Laar T, van der Eb A J, Jochemsen A G. Altered phosphorylation and oligomerization of p53 in adenovirus type 12-transformed cells. Oncogene. 1995;11:49–57. [PubMed] [Google Scholar]
- 30.Steegenga W T, van Laar T, Riteco N, Mandarino A, Shvarts A, van der Eb A J, Jochemsen A G. Adenovirus E1A proteins inhibit activation of transcription by p53. Mol Cell Biol. 1996;16:2101–2109. doi: 10.1128/mcb.16.5.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steegenga W T, Van Laar T, Shvarts A, Terleth C, Van der Eb A J, Jochemsen A G. Distinct modulation of p53 activity in transcription and cell-cycle regulation by the large (54 kDa) and small (21 kDa) adenovirus E1B proteins. Virology. 1995;212:543–554. doi: 10.1006/viro.1995.1512. [DOI] [PubMed] [Google Scholar]
- 32.Trink B, Okami K, Wu L, Sriuranpong V, Jen J, Sidransky D. A new human p53 homologue. Nat Med. 1998;4:747–748. doi: 10.1038/nm0798-747. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 33.Vaessen R T, Houweling A, Israel A, Kourilsky P, van der Eb A J. Adenovirus E1A-mediated regulation of class I MHC expression. EMBO J. 1986;5:335–341. doi: 10.1002/j.1460-2075.1986.tb04217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van den Heuvel S J, van Laar T, The I, van der Eb A J. Large E1B proteins of adenovirus types 5 and 12 have different effects on p53 and distinct roles in cell transformation. J Virol. 1993;67:5226–5234. doi: 10.1128/jvi.67.9.5226-5234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van der Eb A J, Graham F L. Assay of transforming activity of tumor virus DNA. Methods Enzymol. 1980;65:826–839. doi: 10.1016/s0076-6879(80)65077-0. [DOI] [PubMed] [Google Scholar]
- 36.Weissman B E, Saxon P J, Pasquale S R, Jones G R, Geiser A G, Stanbridge E J. Introduction of a normal human chromosome 11 into a Wilms’ tumor cell line controls its tumorigenic expression. Science. 1987;236:175–180. doi: 10.1126/science.3031816. [DOI] [PubMed] [Google Scholar]
- 37.Yang A, Kaghad M, Wang Y, Gillett E, Fleming M D, Dotsch V, Andrews N C, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;3:305–316. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 38.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 39.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]
- 40.Zantema A, Fransen J A, Davis-Olivier A, Ramaekers F C, Vooijs G P, DeLeys B, Van der Eb A J. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology. 1985;142:44–58. doi: 10.1016/0042-6822(85)90421-0. [DOI] [PubMed] [Google Scholar]
- 41.Zantema A, Schrier P I, Davis-Olivier A, van Laar T, Vaessen R T, van der Eb A J. Adenovirus serotype determines association and localization of the large E1B tumor antigen with cellular tumor antigen p53 in transformed cells. Mol Cell Biol. 1985;5:3084–3091. doi: 10.1128/mcb.5.11.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zauberman A, Lupo A, Oren M. Identification of p53 target genes through immune selection of genomic DNA: the cyclin G gene contains two distinct p53 binding sites. Oncogene. 1995;10:2361–2366. [PubMed] [Google Scholar]