Abstract
Transcriptional activation by the estrogen receptor is mediated through its interaction with coactivator proteins upon ligand binding. By systematic mutagenesis, we have identified a group of conserved hydrophobic residues in the ligand binding domain that are required for binding the p160 family of coactivators. Together with helix 12 and lysine 366 at the C-terminal end of helix 3, they form a hydrophobic groove that accommodates an LXXLL motif, which is essential for mediating coactivator binding to the receptor. Furthermore, we demonstrated that the high-affinity binding of motif 2, conserved in the p160 family, is due to the presence of three basic residues N terminal to the core LXXLL motif. The recruitment of p160 coactivators to the estrogen receptor is therefore likely to depend not only on the LXXLL motif making hydrophobic interactions with the docking surface on the receptor, but also on adjacent basic residues, which may be involved in the recognition of charged residues on the receptor to allow the initial docking of the motif.
Estrogens regulate the transcription of target genes by binding to estrogen receptors (ERs) that function as ligand-dependent transcription factors. Transcriptional activation is mediated by two activation domains, AF1 at the N terminus and AF2, probably conserved in all members of the nuclear receptor superfamily, in the ligand binding domain (LBD) (8, 21, 30). Numerous proteins have been reported to interact with AF2, some of which have the properties of transcriptional coactivators (2, 13). One such target is the p160 (also known as RIP160) (6, 14) family of coactivators, characterized by three distinct genes coding for the proteins SRC1 (27), TIF2 (also known as GRIP1) (17, 33), and pCIP (also known as ACTR, AIB1, or RAC3) (1, 7, 19, 22); these proteins have molecular masses of approximately 160 kDa and similar overall domain structures. They appear to bind to most, if not all, nuclear receptors (NRs) in a ligand-dependent manner and potentiate transcription of target genes.
Recruitment of the p160 proteins to the ER is dependent on the integrity of a C-terminal helix, referred to as helix 12, and a lysine residue in helix 3 (8, 16). An additional interaction between p160 proteins and AF1 has been reported (35) that may be involved in the functional interaction between AF1 and AF2 (24). Structural analysis of the LBDs of a number of receptors suggests that the binding of a hormonal ligand results in the realignment of helix 12 (3, 28, 34, 36). Its importance in the ER is indicated by the observation that it is misaligned in the presence of an antiestrogen, raloxifene, which blocks AF2 activity (4). Thus, the p160 proteins are proposed to interact with residues in the vicinity of helices 3 and 12 which form a surface induced by the ligand. The interaction of the p160 proteins with receptors is mediated by LXXLL motifs, three of which are conserved in both sequence and spacing in all family members (15, 20, 31). Their relative affinity for the AF2 surface seems to vary according to the NR, with motif 2 being preferentially used for binding to the ER (10, 18, 32).
In this paper, we have mapped in detail residues in both the surface of the LBD and SRC1 required for estrogen-dependent interaction. We focused on residues in helices 3, 5, and 12 of the receptor, which together generate a hydrophobic patch that we predicted might interact with the LXXLL motif. In addition, we demonstrate that the LXXLL motif is sufficient for mediating receptor-coactivator interaction but that selectivity is determined by N-terminal residues adjacent to the motif.
MATERIALS AND METHODS
Plasmids.
Point mutations in the LBD (amino acids [aa] 313 to 599) of the mouse ER α (mERα) were introduced by recombinant PCR using Elongase (Gibco BRL). PCR fragments containing the desired mutation were inserted into the NdeI and BglII sites in the vector pSP6MORK (8). Mutations in helix 12 were generated by oligonucleotide-directed mutagenesis in pSP6MORK. All full-length mERα mutants were transferred into pSG5 as an EcoRI fragment for transient transfection. The mammalian expression vector pSG-Gal was created by amplifying the yeast Gal4 DNA binding domain (aa 1 to 147) by PCR using pSG424 as the template. The PCR product was digested with EcoRV and BglII and cloned into pSG5 digested with EcoRI (end filled with Klenow fragment) and BglII. The polylinker of pSG-Gal contains the following unique restriction sites: 5′-EcoRI-SmaI-BamHI-SacII-KpnI-BglII-3′. The LBD of wild-type and mutant receptors was fused to the Gal4 DNA binding domain by cloning PCR fragments encompassing Ser313 and Ile599 of mERα into pSG-Gal digested with EcoRI and BglII. The construct Gal-SRC1 (aa 570 to 780) was made by inserting an EcoRI-BamHI PCR fragment of the corresponding region of SRC1 into EcoRI-BglII-digested pSG-Gal. The LBD of wild-type and mutant receptors was fused to the acidic activation domain of VP16 (aa 410 to 490) by cloning an EcoRI-BamHI PCR fragment into EcoRI-BglII-digested pSGVP16 (5). Cloning and mutagenesis were verified by automated DNA sequencing.
The reporter pERE-tk-GL3 was constructed by transferring an HincII-BglII fragment from pEREBLCAT into SmaI-BglII-digested pGL3 Basic vector (Promega). For p5Gal-E1B-GL3, a PvuII-BamHI fragment from G5E1BCAT (8) was transferred into SmaI-BglII-digested pGL3 Basic vector.
Cell culture and transient transfection experiments.
COS-1 cells routinely were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS; Gibco). For transient transfection assays, COS-1 cells were plated in 24-well microtiter plates (Falcon) in phenol red-free DMEM containing 5% charcoal-dextran-stripped fetal bovine serum (DCFBS). Cells were transfected by calcium phosphate coprecipitation as described earlier (8). The transfected DNA included a pJ7-lacZ control plasmid (25) (45 ng), pERE-tk-GL3 or p5Gal-E1B-GL3 reporters (0.8 μg), and pSG5-based expression plasmids encoding either a full-length version or Gal4 fusion of mERα (20 ng) plus or minus pSG5-SRC1e (18) (100 ng). In mammalian two-hybrid experiments, expression plasmids for the Gal4 fusion of SRC1 (20 ng) and VP16 fusion of mERα LBD (20 ng) were used. A constant amount of DNA was maintained in each well with an appropriate amount of pSG5 expression vector. After 16 h, the cells were washed and then maintained in medium containing 5% DCFBS and phenol red-free DMEM in the presence or absence of 10−8 M 17β-estradiol for 24 h. Subsequently, cells were harvested and extracts were assayed for luciferase activity with the LucLite luciferase reporter gene assay kit (Packard) and for β-galactosidase activity with a Galato-Light chemiluminescent assay (Tropix). β-Galactosidase activity was used to correct for differences in transfection efficiency.
Gel retardation and ligand-binding assays.
For gel retardation and ligand-binding assays, the wild-type and mutant receptors were overexpressed in COS-1 cells by electroporation at 450 V and 250 μF in the presence of 18 μg of plasmid DNA. Cells were then plated out in DMEM containing 10% FBS and grown for 48 h. Whole-cell extracts were prepared in buffer containing 0.4 M KCl, 20 mM HEPES (pH 7.4), 1 mM dithiothreitol (DTT), 20% glycerol, and protease inhibitors. The protein content of cell extracts was determined by a colorimetric method (Bio-Rad). The DNA-binding activity of the mutant receptors was examined as described in reference 8. Cell extracts from transfected COS-1 cells were incubated with 32P-labelled double-stranded oligonucleotide probe containing a consensus estrogen response element from the vitellogenin A2 gene promoter. Binding reactions were performed in the presence of either preimmune serum or ERα-specific antibody MP16 (11). Receptor-DNA complexes were resolved from unbound DNA in 6% nondenaturing polyacrylamide gels in 0.5× Tris-borate-EDTA buffer and visualized by autoradiography.
Ligand-binding analysis of the wild-type and mutant receptors was performed essentially as described in reference 11 with 17β-[3H]estradiol (Amersham International), with the exception of the composition of the ligand binding buffer (20 mM HEPES [pH 7.7], 1.5 mM EDTA, 1 mM DTT, 0.1% bovine serum albumin, 10% glycerol). Scatchard analysis was performed over the range of 0.125 to 8 nM labelled steroid in the absence or presence of 500-fold excess of unlabelled 17β-estradiol.
GST pull-down assays.
Recombinant cDNAs in the pSP65 or pSG5 vector were transcribed and translated in vitro in reticulocyte lysate (Promega) in the presence of [35S]methionine (Amersham International) according to the manufacturer’s instructions. Glutathione S-transferase (GST) fusion proteins were induced and purified as described earlier (6). 35S-labelled proteins were incubated with GST fusion proteins in NETN buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 0.5% Nonidet P-40) containing 100 mM NaCl, unless otherwise stated, in the absence or presence of 17β-estradiol (10−6 M). Samples were subsequently washed and separated on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels. The bound proteins were visualized by fluorography. In peptide inhibition assays, individual peptide was dissolved in water and added to GST binding reaction mixtures immediately before the ligand.
RESULTS
A hydrophobic surface of the mERα LBD is required for coactivator binding.
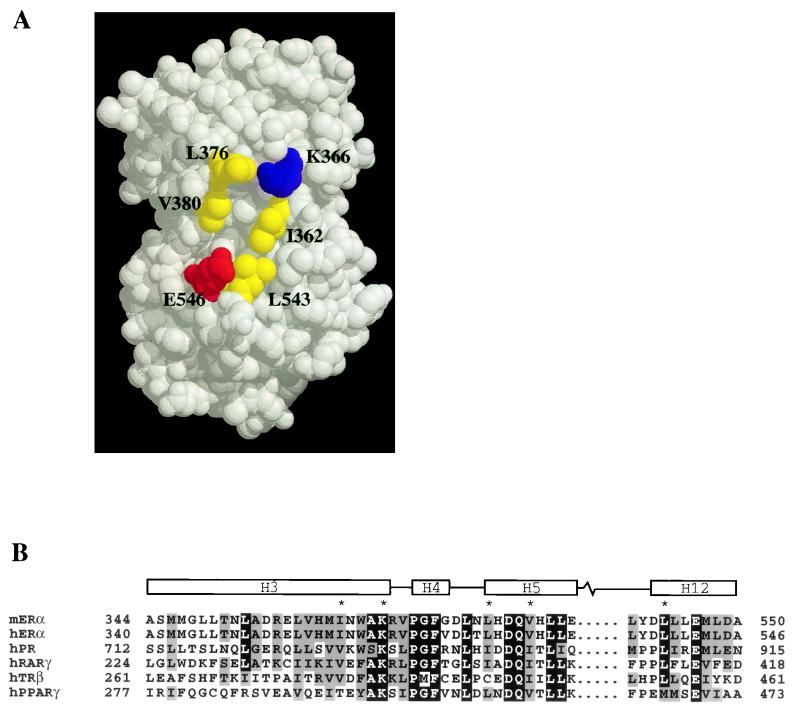
Inspection of the crystal structure of the human ERα (hERα) LBD in the presence of 17β-estradiol indicated that helix 12 and lysine 366 at the C-terminal end of helix 3, which are crucial for the ligand-dependent activation function of the receptor (8, 16), are located on the surface of the LBD (4). The observation that these two elements are not in close proximity to each other prompted us to speculate that they are only part of the surface responsible for the docking of p160 coactivator proteins. Because of the hydrophobic nature of the LXXLL motif, which is essential for mediating coactivator binding to the ER (15, 20, 31), we focused on a hydrophobic patch on the surface of the mERα LBD whose boundary seemed to be defined by helix 12 and lysine 366 (Fig. 1A). This hydrophobic patch seems to be composed mainly of three residues from helices 3 and 5, namely, I362, L376, and V380. Sequence analysis of the nuclear receptor superfamily revealed that the corresponding residues in other receptors are almost always hydrophobic (Fig. 1B) (12, 36).
FIG. 1.
Structure of the hERα LBD in the presence of 17β-estradiol. (A) Residues implicated in this study for participation in p160 coactivator binding are highlighted yellow (hydrophobic), red (acidic), and blue (basic). The residues are numbered as in mERα. The space-filled model was generated by RasMol and is based on the coordinates under the Protein Data Bank entry code 1ERE. (B) Sequence alignment of mERα LBD helices 3, 4, 5, and 12 with corresponding regions of members of the nuclear receptor superfamily whose agonist-bound crystal structures are solved. Note the absolute conservation of residues (marked with asterisks) in mERα and hERα, which are involved in coactivator binding. The boundaries for helices 3, 4, 5, and 12 are assigned according to the hERα LBD structure. The alignment was generated by Pileup (GCG) and formatted with MacBoxshade.
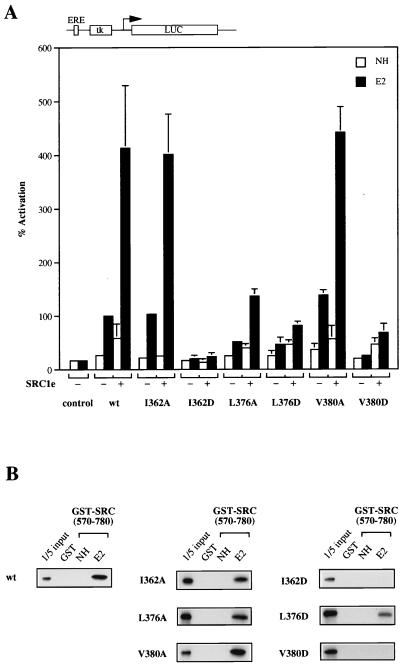
To assess the contribution of these residues to the transcriptional activity of mERα and its binding to coactivators, each of them was replaced by alanine. Full-length wild-type or mutant receptors were transiently transfected into COS-1 cells and tested for their ability to activate an ERE-tk-luciferase reporter gene. The transcriptional activity of the L376A mutant receptor was impaired, but nevertheless was stimulated by overexpressed SRC1e, a member of the p160 coactivator family (Fig. 2A). Mutant receptors bearing the mutation I362A or V380A activated the reporter gene to the same extent as the wild-type receptor. Their transcriptional activity could also be further enhanced by cotransfecting SRC1e. Consistent with the transient transfection assays, binding of in vitro-translated mutant receptors to GST-SRC1 (aa 570 to 780), which encompasses its receptor-interacting region, is comparable to that of wild-type mERα (Fig. 2B).
FIG. 2.
Surface hydrophobic residues in helices 3 and 5 of mERα are involved in coactivator binding. (A) COS-1 cells were transiently transfected with expression vector for wild-type (wt) or mutant receptors and pERE-tk-GL3 reporter in the absence (−) or presence (+) of 100 ng of full-length SRC1e. A cytomegalovirus promoter-driven pJ7-lacZ plasmid was cotransfected as the internal control. After transfection, cells were treated with ethanol vehicle alone (NH) or 17β-estradiol (E2) at 10−8 M for 24 h. Subsequently, cells were assayed for luciferase (LUC) and β-galactosidase activity. Normalized values are expressed as percentage of activity compared with that of wild-type mERα alone in the presence of β-estradiol (100%). The results shown represent the average of at least two independent experiments assayed in duplicate + standard errors. ERE, estrogen response element. (B) Binding activity of wild-type or mutant receptors to SRC1 in GST pull-down assay. In vitro-translated, [35S]methionine-labelled receptors were incubated with GST-SRC1 (aa 570 to 780) coupled to Sepharose beads in either the absence (NH) or presence (E2) of 10−6 M 17β-estradiol. Bound proteins were eluted and separated on SDS–10% polyacrylamide gels. Labelled proteins were detected by fluorography. The input lane represents 20% of the total volume of the lysate used in each reaction.
We next replaced I362, L376, and V380 with aspartic acid, a charged residue which might actively interfere with packing of hydrophobic side chains. All three mutant receptors had dramatically reduced transcriptional activity (the phenotype of the I362D mutation was also partially due to reduced protein expression [see Fig. 5]). There was no detectable binding of the I362D and V380D mutants to GST-SRC1 (aa 570 to 780), and reduced binding was observed for the L376D mutant. Our results imply that I362, L376, and V380 of mERα are in close proximity to the bound p160 coactivator. However, since removal of individual hydrophobic side chain from any of the three positions was insufficient to abolish coactivator binding by the receptor, these residues might be redundant in the formation of the coactivator interaction surface.
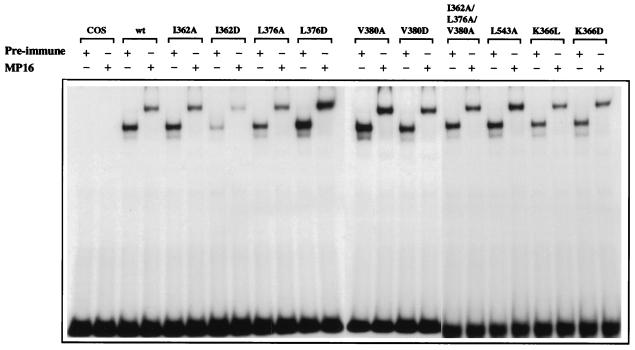
FIG. 5.
Mutant receptors bind to DNA with affinity similar to that of the wild-type receptor. Full-length wild-type (wt) or mutant receptors were transiently expressed in COS-1 cells. Equal amounts of receptor were analyzed for DNA binding in an electrophoretic mobility shift assay using a 32P-labelled oligonucleotide containing a single consensus estrogen response element from the vitellogenin A2 gene promoter. Binding reactions were performed either in the presence of ERα-specific antibody MP16 or preimmune serum. Protein-DNA complexes were separated on 6% native polyacrylamide gels and detected by autoradiography.
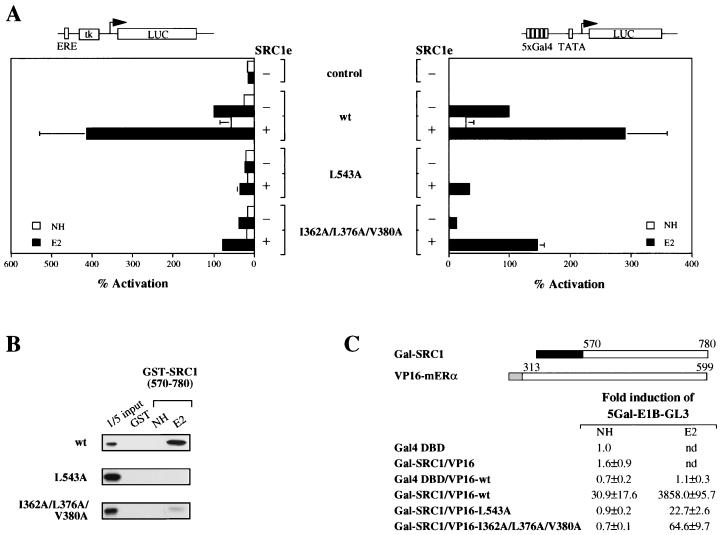
Of the four highly conserved hydrophobic residues in helix 12 of the mERα LBD, only L543 is exposed on the surface in the crystal structure. We have previously shown that alanine substitution of both L543 and L544 abrogated transcriptional activity of the mutant receptor (8). In light of the crystal structure, we tested whether L543 alone is required for coactivator binding and AF2 activity. The L543A mutant displayed negligible transcriptional activity when transiently transfected into COS-1 cells as a full-length receptor (Fig. 3A), in contrast to the phenotype observed for the single alanine substitution of I362, L376, or V380. To verify that the mutation is affecting AF2 alone and not interfering with possible cooperation between AF1 and AF2, a chimeric receptor consisting of the LBD with the L543A mutation fused to Gal4 DNA-binding domain was made. The chimeric receptor was unable to activate a Gal4 reporter gene in COS-1 cells, and weak activity was observed when SRC1e was overexpressed concomitantly (Fig. 3A). In GST pull-down experiments, no detectable binding was observed between the L543A mutant and GST-SRC1 (aa 570 to 780) (Fig. 3B). Hence, L543 seems to be essential for AF2 activity, at least in part due to its participation in coactivator binding.
FIG. 3.
Functional analysis of L543A and I362A-L376A-V380A mutant receptors. (A) Wild-type (wt) or mutant full-length (left) or chimeric receptors consisting of the LBD of mERα fused to the DNA binding domain of Gal4 (right) were transiently transfected into COS-1 cells. Luciferase (LUC) reporter genes as indicated were cotransfected in the presence (+) or absence (−) of 100 ng of full-length SRC1e, and pJ7-lacZ was used as an internal control. Data are presented as described for Fig. 2A. ERE, estrogen response element. (B) Binding of mutant receptors to GST-SRC1 (aa 570 to 780) in vitro was examined under the same conditions as described for Fig. 2B. (C) In vivo interaction of mutant mERα LBDs with SRC1 (aa 570 to 780). The expression vectors used are schematically represented with the numbers indicating the amino acid position in the full-length protein. The darkly shaded box represents the Gal4 DNA binding domain (aa 1 to 147), and the lightly shaded box represents the activation domain of VP16 (aa 410 to 490). HeLa cells were transiently transfected with the indicated expression vectors, together with a p5Gal-E1B-GL3 reporter gene and the pJ7-lacZ internal control plasmid. Following transfection, cells were treated with ethanol vehicle alone (NH) or 10−8 M 17β-estradiol (E2). After 24 h, cell extracts were prepared and assayed for luciferase and β-galactosidase activities. Normalized values are expressed as fold induction compared with that of the Gal4 DNA binding domain alone (set as 1). The results shown represent the average of at least two independent experiments assayed in duplicate ± standard error. nd, not determined.
Differential contribution of hydrophobic residues in AF2 activity.
It is apparent from the phenotypes of the mutant receptors that hydrophobic residues which form the putative coactivator interaction surface might not contribute equally to the AF2 activity of mERα. To extend this observation, we generated mutants with double and triple point mutations in which I362, L376, and V380 were replaced by alanine in all possible combinations. Alanine substitution for any two of the three residues failed to reduce the transcriptional activity of the receptor (data not shown). We observed a dramatic decrease in transcriptional activity only when all three residues were replaced with alanine, both as the full-length receptor or as the Gal4-chimeric receptor in COS-1 cells (Fig. 3A). Nevertheless, the triple mutant could be partially rescued by overexpressed SRC1e and was more active than the L543A mutant (Fig. 3A).
We tested whether the difference in transcriptional activity was correlated with the ability of these mutants to bind coactivator. In GST pull-down experiments, weak binding between the triple I362A-L376A-V380A mutant and GST-SRC1 (aa 570 to 780) was observed. There was no detectable binding between the L543A mutant and the same SRC1 construct (Fig. 3B). To obtain a quantitative comparison in vivo, mammalian two-hybrid interaction assays were conducted. We fused SRC1 (aa 570 to 780) to the Gal4 DNA binding domain and the LBD of the wild-type or mutant receptors to the VP16 acidic activation domain. Upon transient transfection into HeLa cells, interaction between receptor and SRC1 leads to activation of a Gal4 reporter gene. In these assays, the levels of interaction of the triple I362A-L376A-V380A and single L543A mutants with SRC1 were 60- and 168-fold lower, respectively, than that of the wild-type receptor (Fig. 3C). Together with the observation that L358A, F371A, and L383A mutants retain wild-type transcriptional activity (data not shown), our results suggest a hierarchy of conserved hydrophobic residues which form the coactivator-interacting surface by virtue of their differential contribution to the AF2 activity of the receptor.
Dual property of lysine 366 in mediating AF2 activity of mERα.
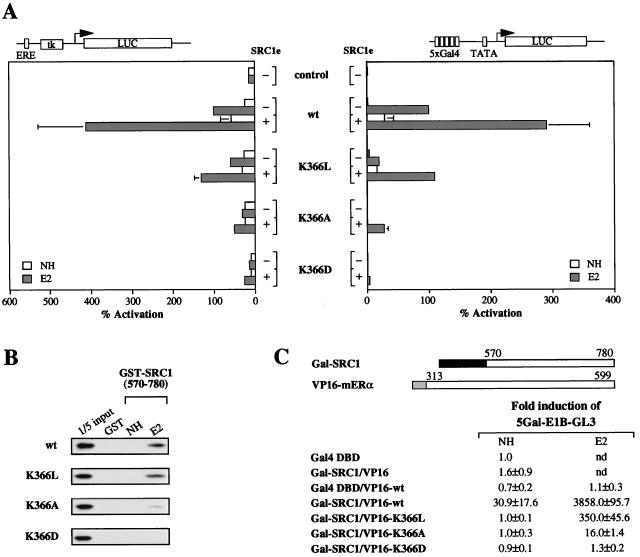
Lysine 366 is the only positively charged residue in the predominantly hydrophobic coactivator-interacting surface of mERα. It was shown previously that the K366A mutant exhibited negligible transcriptional activity and minimal binding to the coactivator SRC1 in vitro (16). However, it was unclear whether this effect was due to the lack of charge or the long aliphatic stem of the lysine side chain, since alanine is lacking both. To address this question, we generated a mutant receptor in which K366 was replaced by leucine, whose side chain mimics the aliphatic stem of lysine but is devoid of the terminal positive charge. In transiently transfected COS-1 cells, the transcriptional activity of the K366L mutant was reduced relative to that of the wild-type receptor but exceeded that of the K366A mutant when tested as full-length or Gal4-chimeric receptors (Fig. 4A). This intermediate activity was paralleled by the interaction of the K366L mutant with SRC1, which was reduced by 10-fold compared with wild-type mERα but was 20-fold greater than that of the K366A mutant in mammalian two-hybrid interaction assays (Fig. 4C). This suggests that the terminal charge of K366 is required for optimal transcriptional activity and coactivator binding, but the aliphatic stem of its side chain is sufficient for the partial activity observed.
FIG. 4.
Mutation of K366 reveals its dual property in AF2 activity. (A) COS-1 cells were transiently transfected with expression vector for wild-type (wt) or mutant receptors (left) or Gal4-chimeric receptors (right), the pJ7-lacZ internal control plasmid, and the luciferase (LUC) reporter plasmid as indicated. Data are presented as described for Fig. 2A. (B) Binding of mutant receptors to GST-SRC1 (aa 570 to 780) was examined under the same conditions as described for Fig. 2B. (C) In vivo interaction of mutant mERα LBDs with SRC1 (aa 570 to 780) in transiently transfected HeLa cells. Data are presented as described for Fig. 3C. nd, not determined.
Next, we substituted aspartic acid and arginine for K366. The K366D mutant had negligible transcriptional activity (Fig. 4A) and displayed no binding to SRC1 both in vitro (Fig. 4B) and in vivo (Fig. 4C), a phenotype more severe than that of the K366A mutant. However, the K366R mutation had no effect on the transcriptional activity of the receptor (data not shown). The first result implies that there is an absolute requirement for a positive charge, while the latter suggests that the exact positioning of the positive charge is not crucial. Taken together, we conclude that both the terminal positive charge and the aliphatic stem of the K366 side chain are involved in mediating the AF2 activity of the receptor.
Mutation of residues which constitute the coactivator interaction surface does not affect ligand binding or DNA binding.
Expression of the wild-type and mutant receptors was verified by Western blotting (data not shown). To ensure that mutations at the coactivator interaction surface had no effect on the integrity of the LBD structure, ligand-binding assays were performed. All receptor proteins bound 17β-estradiol with similar affinities (Table 1). In addition, they bound to DNA as dimers in a gel retardation assay (Fig. 5). The identity of the receptor-DNA complex was confirmed by the supershift observed in the presence of the ERα-specific antibody MP16. Therefore, alterations in the transcriptional activity of the mutant receptors were attributed to defects in coactivator interactions rather than ligand or DNA binding.
TABLE 1.
Estrogen binding activity of wild-type and mutant receptorsa
| Receptor | Kd (nM) |
|---|---|
| Wild-type mERα | 0.33 |
| I362A-L376A-V380A | 0.28 |
| I362D | 0.49 |
| L376D | 0.86 |
| V380D | 0.90 |
| K366L | 0.31 |
| K366D | 0.33 |
Extracts prepared from transiently transfected COS-1 cells were analyzed for ligand binding activity. The dissociation constant (Kd) for ligand binding for each mutant was determined by Scatchard analysis.
Specificity of LXXLL motifs to the mERα coactivator interaction surface.
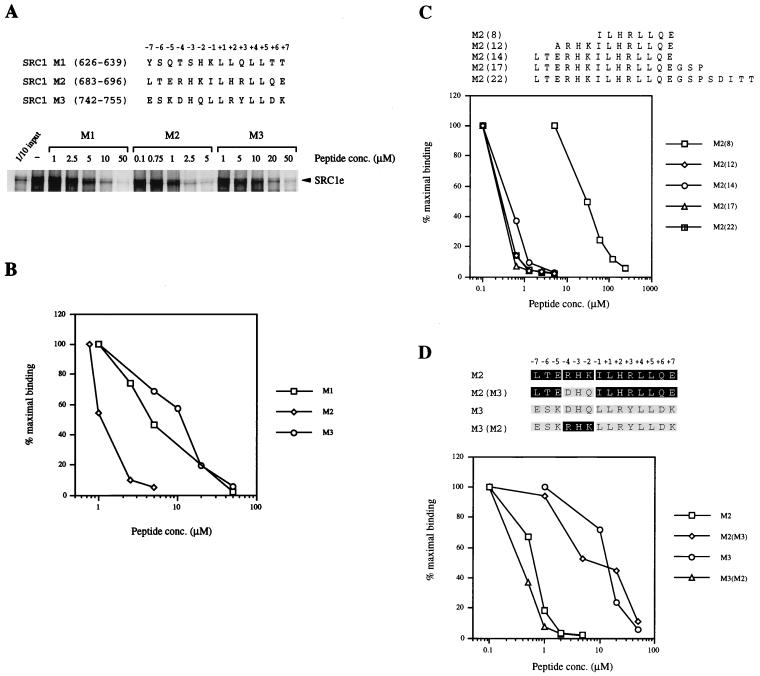
Having probed the coactivator interaction surface of the mERα LBD, we attempted to identify potential determinants for high-affinity binding of p160 coactivators. There are three LXXLL motifs in the receptor interaction domain of each of the p160 coactivator family members. Previous work indicated that motif 2 in SRC1 is preferentially used in mediating interaction between mERα and SRC1 in vitro (18). To test the affinity of different SRC1 motifs toward the docking site in greater detail, increasing concentrations of 14-mer peptides M1, M2, and M3, encompassing SRC1 motifs 1, 2, and 3, respectively, were used to compete for the in vitro binding of GST-mERα LBD with SRC1e. Inhibition of SRC1 binding by the M2 peptide was approximately eightfold better than that by the M1 and M3 peptides (Fig. 6A and B), implying that SRC1 motif 2 has a higher affinity to the mERα LBD. Next, a panel of M2 peptides ranging from 8- to 22-mers [designated M2(8) to M2(22)] were used to investigate whether the length of the peptide would affect its ability to inhibit receptor-coactivator interaction. Inhibition by the M2(12) peptide was about 100-fold stronger than that by the M2(8) peptide; however, further extension of the peptide at the N or C termini did not increase the degree of inhibition (Fig. 6C). This suggests that the determinants of SRC1 motif 2 for its high-affinity binding to the mERα docking site are N terminal to the minimal LXXLL motif. In particular, we noted a cluster of three basic residues at positions −2 to −4 of motif 2, which are conserved across all p160 coactivator family members. To determine whether the three basic residues are sufficient to confer specificity, we synthesized an M3 peptide with residues at positions −2 to −4 substituted for by the corresponding basic residues of SRC1 motif 2. The resultant peptide, M3(M2), had an inhibition profile similar to that of the native M2 peptide (Fig. 6D). Conversely, when we replaced the three basic residues in M2 with the corresponding residues from motif 3, M2(M3) behaved in a manner similar to that of the native M3 peptide (Fig. 6D). Finally, we tested the effect of replacing the individual basic residues with alanine, but found that all three mutant peptides were capable of inhibiting the binding of SRC1 to a similar extent as the wild-type peptide (unpublished data). Thus, our results suggest that the preference for motif 2 when the ER and SRC1 interact is conferred, at least in part, by basic residues N terminal to the minimal LXXLL motif. Moreover, these three residues seem to be sufficient for transforming a low-affinity motif into a high-affinity motif for the docking site on the ER.
FIG. 6.
Differential inhibition of mERα-SRC1 interaction in vitro by an LXXLL motif containing peptides. (A) Comparison of peptides encompassing either SRC1 LXXLL motif 1, 2, or 3. A GST fusion protein of mERα LBD which had been coupled to Sepharose beads was incubated with in vitro-translated [35S]methionine-labelled SRC1e protein and increasing amounts of LXXLL motif-containing peptide, in the presence of 10−6 M 17β-estradiol. Bound labelled proteins were eluted, separated on SDS–10% polyacrylamide gels, and detected by fluorography. The input lane represents 10% of the total volume of the lysate used in each reaction. (B) Graphical representation of results from Fig. 6A. The amount of bound SRC1e protein was quantified with a PhosphorImager and is expressed as a percentage of maximal binding relative to the amount of bound proteins in the absence of any LXXLL motif-containing peptide (100%). At least two independent experiments were performed, and the data shown are from one representative experiment. (C) Effect of flanking residues on inhibition of mERα-SRC1 interaction by motif 2-containing peptide. Increasing amounts of M2 peptide, the length of which varied from 8 to 22 residues, were used to inhibit interaction between GST-mERα LBD and [35S]methionine-labelled SRC1e protein in an assay described for panel A. Data are presented as described for panel B and are from one representative experiment. At least two independent experiments were performed. (D) Three basic residues N terminal to LXXLL core motif 2 confer high-affinity binding to mERα. Residues −4DHQ−2 of SRC1 motif 3 were replaced by residues −4RHK−2 from the corresponding positions of motif 2 in a 14-mer peptide and vice versa. Increasing amounts of wild-type or mutant peptides were incubated with GST-mERα LBD and [35S]methionine-labelled SRC1e protein in an assay described for panel A. Data from one representative experiment are presented as described for panel B. At least two independent experiments were performed.
DISCUSSION
Transcriptional activation by the ER is achieved through its interaction with coactivator proteins upon ligand binding. It has been shown that the recruitment of the p160 family of coactivators is dependent upon the integrity of a short hydrophobic motif, LXXLL, three of which are conserved in individual family members (15, 20, 31). Here we identified a cluster of residues in the LBD of mERα which comprise an interaction surface to allow docking of the motif. In addition, we demonstrate that the preferential binding of SRC1 motif 2 can be accounted for by the presence of three basic residues N-terminal to the core LXXLL motif which enhance its binding affinity to the receptor in vitro.
The coactivator interaction surface of mERα LBD which is composed mainly of hydrophobic residues from helices 3, 5, and 12 closely resembles a similar surface described for human thyroid hormone receptor β (hTRβ) (9, 12). More importantly, side chains of residues characterized here, namely, I362, L376, V380, and L543 (Fig. 1A), were shown to make van der Waals contacts with side chains of the three LXXLL motif leucines and of the isoleucine immediately N terminal to the motif in the crystal structure of the agonist- bound hERα LBD complexed with GRIP1 NR box II peptide (29). While the aspartic acid substitutions allowed us to show that these residues are in close contact with the motif in functional assays, the alanine substitutions led to the notion that they could be divided into two classes. One class, including L358, I362, F371, L376, V380, and L383, are likely to contribute to the optimal binding of coactivators, but are dispensable, since removal of one or two side chains of these residues had little effect on receptor function. In contrast, L543 is essential for ligand-dependent coactivator binding and AF2 activity of mERα. This residue was shown to make intramolecular van der Waals contacts with residues in helix 3 in the crystal structure of agonist-bound hERα (4). Therefore, we postulate that L543 plays a pivotal role not only in coactivator binding per se, but also in the completion and stabilization of the coactivator interaction surface upon ligand binding. Hence, the L543A mutation might destabilize the position of helix 12 in addition to obliterating an essential contact with the LXXLL motif.
In the structure of the agonist-bound hERα LBD complexed with NR box II peptide, K366 and E546 were shown to form hydrogen bonds with the main chain of the peptide (29), similar to the observation made in the holo-PPARγ-SRC1 cocrystal structure (26). This led to the suggestion that these oppositely charged residues, which are situated at opposite ends of the coactivator interaction surface, might serve as a “charge clamp” and stabilize the helical structure of the peptide. Although the phenotypes of the K366L (Fig. 4) and E546A (reference 8 and unpublished data) mutants implied that the charges of these residues might be involved in p160 coactivator binding by mERα, our assays did not allow us to distinguish between their roles in recognition or equilibrium binding. However, SRC1 binding to mERα (18) and the ability of a peptide containing SRC1 motif 2 to inhibit such binding occurs at a high salt concentration in vitro (unpublished data), suggesting that electrostatic interaction between mERα and SRC1 is dispensable for equilibrium binding. It is conceivable that the initial recognition of the peptide by the docking surface of the receptor results from the complementarity between the surface of the LBD and the LXXLL motif. However, given that the GRIP1 NR box II peptide used in crystalization studies is not structured on its own (9), recognition may be achieved in other ways. For example, the polarity of the surface, imposed by K366 and E546, may favor the formation of helical structure of the peptide in one orientation. On the other hand, K366 and E546 could be recognized directly by flanking residues of the LXXLL motif which do not appear to participate in equilibrium binding. Since the SRC1 moiety in the holo-PPARγ-SRC1 complex appeared to be largely unstructured except for a short helix containing the LXXLL motif (26), we speculate that the mechanism of recognition postulated for the NR box II peptide might also be applicable for native p160 coactivator proteins.
The ER has been demonstrated to interact preferentially with motif 2 in SRC1 (18) or TIF2 (32). This motif was also shown to bind with highest affinity to the TRβ (9), while alternative motifs are preferentially utilized by other receptors (10, 23). Sequence alignment indicates that the degree of conservation of a particular LXXLL motif in different p160 coactivators is greater than that between different motifs within any one p160 protein. Since such conservation sometimes extends beyond the minimal LXXLL sequence, it is conceivable that residues flanking the LXXLL motif may confer preferential binding of particular motifs to different receptors. Using a peptide inhibition assay, we confirmed our previous observation that SRC1 motif 2 has the highest affinity for binding to the LBD of ER and identified three basic residues, N terminal to the core LXXLL motif, as determinants for such high-affinity binding. Residues adjacent to motif 2 were also shown to modulate its affinity with the TRβ (9), although the relative contributions of the N- and C-terminal residues were not assessed. The basic residues seem to function as a unit independently of other residues flanking the motif, since replacement of −4DHQ−2 from motif 3 with −4RHK−2 from motif 2 was sufficient to confer high-affinity binding to the ER.
The specificity determinants N terminal to the LXXLL motif are disordered in the structure of the agonist-bound hERα complexed with NR box II peptide (29). Therefore, they are unlikely to form stable interactions with residues of ER in equilibrium binding. It was proposed that these basic residues might be accommodated by a shallow groove between H5 and H12 in TRβ (9). Alternatively, we envisage that the three basic residues may be involved in long-range recognition of surface features of ER which are not necessarily in the proximity of the coactivator docking site. Although not detected in our peptide inhibition assays, microinjection studies demonstrated that residues C terminal to SRC1 motif 2 could also serve as specificity determinants for ER binding (23). It is tempting to speculate that the same principle of long-range recognition might be applicable. When these findings were taken together, a stepwise model of p160 coactivator binding to ER emerged. The first step involves the flanking residues of the LXXLL motif, whose primary function is to direct the core motif to a broad area of the receptor which encompasses the coactivator interaction surface. Once the LXXLL motif is in the vicinity of the surface, polarity imposed by K366 and E546 directs the formation and docking of the helix, which contains the motif in one orientation due to the dipole intrinsic to helical structure. Finally, specific hydrophobic and electrostatic interactions between the motif and the receptor ensue, resulting in stable interaction of the coactivator with the receptor.
ACKNOWLEDGMENTS
We thank I. Goldsmith and staff for oligonucleotides, G. Clark and staff for DNA sequencing, and Nicola O’Reilly for peptides. We are grateful to Geoff Greene and coworkers for communicating results prior to publication. We also thank Paul Freemont, Peter Verrijzer, Roger White, Shaun Cowley, Paul Bates, and members of the Molecular Endocrinology Laboratory for discussions and comments on the manuscript.
This work was supported by the Imperial Cancer Research Fund.
REFERENCES
- 1.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X-Y, Sauter G, Kallioniemi O-P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 2.Beato M, Herrlich P, Schutz G. Steroid-hormone receptors—many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 3.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Crystal-structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature. 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 4.Brzozowski A M, Pike A C W, Dauter Z, Hubbard R E, Bonn T, Engstrom O, Ohman L, Greene G L, Gustafsson J-A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 5.Butler A J, Parker M G. Coup-TF-II homodimers are formed in preference to heterodimers with RXR-alpha or TR-beta in intact-cells. Nucleic Acids Res. 1995;23:4143–4150. doi: 10.1093/nar/23.20.4143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cavaillès V, Dauvois S, Danielian P S, Parker M G. Interaction of proteins with transcriptionally active estrogen receptors. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;190:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 8.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding X F, Anderson C M, Ma H, Hong H, Uht R M, Kushner P J, Stallcup M R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 11.Fawell S E, Lees J A, White R, Parker M G. Characterization and colocalization of steroid binding and dimerization activities in the mouse estrogen receptor. Cell. 1990;60:953–962. doi: 10.1016/0092-8674(90)90343-d. [DOI] [PubMed] [Google Scholar]
- 12.Feng W, Ribeiro R C J, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 13.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 14.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins—possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 15.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 16.Henttu P M A, Kalkhoven E, Parker M G. AF-2 activity and recruitment of steroid receptor coactivator 1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol Cell Biol. 1997;17:1832–1839. doi: 10.1128/mcb.17.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong H, Kohli K, Garabedian M J, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor coactivator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 20.Le Douarin B, Nielsen A L, Garnier J-M, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 21.Lees J A, Fawell S E, Parker M G. Identification of two transactivation domains in the mouse oestrogen receptor. Nucleic Acids Res. 1989;17:5477–5488. doi: 10.1093/nar/17.14.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McInerney E M, Tsai M J, Omalley B W, Katzenellenbogen B S. Analysis of estrogen-receptor transcriptional enhancement by a nuclear hormone-receptor coactivator. Proc Natl Acad Sci USA. 1996;93:10069–10073. doi: 10.1073/pnas.93.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murakami A, Grinberg D, Thurlow J, Dickson C. Identification of positive and negative regulatory elements involved in the retinoic acid/cAMP induction of Fgf-3 transcription in F9 cells. Nucleic Acids Res. 1993;21:5351–5359. doi: 10.1093/nar/21.23.5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 27.Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 28.Renaud J-P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 29.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 30.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 31.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 32.Voegel J J, Heine M J, Tini M, Vivat V, Chambon P, Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:101–108. [PMC free article] [PubMed] [Google Scholar]
- 34.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 35.Webb P, Nguyen P, Shinsako J, Anderson C, Feng W, Nguyen M P, Chen D, Huang S M, Subramanian S, McKinerney E, Katzenellenbogen B S, Stallcup M R, Kushner P J. Estrogen receptor activation function 1 works by binding p160 coactivator proteins. Mol Endocrinol. 1998;12:1605–1618. doi: 10.1210/mend.12.10.0185. [DOI] [PubMed] [Google Scholar]
- 36.Wurtz J-M, Bourguet W, Renaud J-P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]