Abstract
During mammalian spermatogenesis, meiosis is followed by a brief period of high transcriptional activity. At this time a large amount of mRNA is stored as messenger ribonucleoprotein (mRNP) particles. All subsequent processes of sperm maturation occur in the complete absence of transcription, primarily using proteins which are newly synthesized from these stored mRNAs. By expressing transgene mRNAs in the early haploid spermatids of mice, we have investigated the sequence requirements for determining whether specific mRNAs in these cells will be stored as mRNP particles or be assembled into polysomes. The results suggest that mRNAs which are transcribed in spermatids are assembled into mRNP particles by a mechanism that acts independently of mRNA sequence. Our findings reveal a fundamental similarity between the mechanisms of translational control used in spermatogenesis and oogenesis.
The haploid stages of spermatogenesis, termed spermiogenesis, and the prehaploid stages of oogenesis are two occasions in vertebrate development wherein large-scale assembly of mRNAs into messenger ribonucleoprotein (mRNP) particles occurs. In both cases, the sequestration of mRNA as mRNP particles temporarily represses translation and thereby serves as a mechanism that uncouples transcription and translation. Thus, since the late stages of spermiogenesis occur in transcriptionally inactive cells (29), mRNA sequestration provides a mechanism by which mRNAs can be synthesized prior to transcriptional arrest but not translated until their protein product is required (reviewed in references 13 and 21). In oogenesis, mRNA sequestration provides a mechanism by which the mRNAs required for meiotic maturation and the mRNAs required for the early stages of embryonic development can be preformed by the primary oocytes. As such, the oocyte is capable of completing meiosis without transcription, and the egg is poised for large-scale protein synthesis immediately after fertilization (34, 43, 44).
The translational regulatory system in oocytes must accommodate at least three classes of mRNAs. One class must be translated immediately, another class of mRNAs are translationally delayed until later maturation of the oocyte, and yet others are translationally delayed until after fertilization. Microinjection experiments using Xenopus oocytes have demonstrated that even though most mRNAs injected into oocytes are translationally active, nearly all mRNAs that are transcribed in the oocyte nucleus are translationally repressed in the cytoplasm by assembly into mRNP particles (2, 28, 51). Importantly, these studies indicate that translational repression in oocytes does not require specific sequence elements (2, 28). Rather, at least some proteins that are synthesized by oocytes, for example, linker histone B4 (6) or transcription factor TFIIIA (28, 50), are encoded by genes which contain specific signals that allow their mRNAs to be translated immediately. Interestingly, these signals are not encoded in the mRNAs per se (2). Rather, regulation is dependent on signals in the gene that determine the pathway of nuclear processing, termed the nuclear history, of the mRNA (28). A recent study indicates that the pathway chosen can be determined by the intron/exon organization of the gene (28).
Although translational repression in oocytes does not depend on specific sequences, at least some oocyte mRNAs do contain specific translational regulatory sequences (45). For example, in mouse oocytes, mRNA encoding the tissue-type plasminogen activator (tPA) contains regulatory elements in the 3′ untranslated region (3′-UTR) that play a role in delaying the translation of this mRNA until meiotic maturation of the oocyte (16). When the tPA 3′ regulatory sequences are cleaved from cytoplasmic mRNAs in situ, translational repression is unaffected; however, the oocyte subsequently fails to activate translation of the mRNA during meiotic maturation (47). The tPA 3′-UTR sequences can also direct masking of exogenous mRNAs injected into mouse oocytes (48). However, since translational repression of mRNAs transcribed in vivo is mRNA sequence independent (2, 28), the tPA 3′-UTR is more likely an activator responsible for inducing translation during meiotic maturation (47) than a repressor necessary for the dormant state of tPA mRNA in primary oocytes (48). A plausible model for translational regulation in oocytes is that the default mode for in vivo-transcribed mRNAs is to be translationally repressed. mRNAs required immediately are targeted to the polysomes by gene-specific regulation of mRNA nuclear history; mRNAs required for prezygotic maturation have specific sequences to allow activation at the correct time; mRNAs lacking activation signals remain repressed until fertilization.
In spermiogenesis, by contrast, a distinct mechanism of translational repression has been hypothesized (reviewed in references 3, 21, and 44). A classic study by Braun et al. (4) demonstrated that specific sequences on the mRNA encoding protamine 1 (Prm1), an mRNA which is translationally delayed in spermiogenesis (18), caused delayed translation of a reporter mRNA. Because the onset of protein accumulation from transgenes lacking these sequences was not delayed, it has been presumed that specific sequences from the prm1 gene are responsible for masking Prm1 mRNA by targeting it to mRNP particles (3, 8). A corollary of this hypothesis is that mRNAs which lack specific targeting sequences should be excluded from mRNP pools and instead translated immediately. Thus, although the assembly of mRNAs into mRNP particles is mRNA sequence independent in oocytes, it has been thought that mRNP assembly in spermatids requires specific mRNA sequences. Examples of mRNA sequence-specific translational repression have been documented for individual mRNAs in somatic cells, such as the ferritin subunit mRNAs (26) and the human immunodeficiency virus transcript (42). However, in adult testis, more than 70% of all mRNA is mRNP particle associated at any given time (17, 19, 51). Interestingly, although some mRNP particle-associated spermatid mRNAs share conserved sequences (12, 22), many other mRNAs that are sequestered in spermatids exhibit no obvious sequence similarities (21). These observations suggest that if assembly into mRNP particles were sequence dependent, a rather large number of distinct mRNA sequence elements would have to be specifically recognized by the targeting machinery.
Efforts to purify and identify proteins responsible for targeting Prm1 mRNA and related mRNAs to mRNP particles have resulted in the cloning and identification of several spermatid mRNP-associated proteins, including the poly(A)-binding proteins (11, 20), the Prm1 RNA-binding protein (25), the spermatid perinuclear RNA-binding protein (40), and the testis nuclear RNA-binding protein (41) (see references 14 and 21 for reviews). Whereas several of these proteins show greater affinity for Prm1 mRNA sequences than for arbitrary sequences, none have been shown to be required for assembling protamine mRNA into mRNP particles. Importantly, in considering the contrasting mechanisms of mRNP particle assembly proposed for spermatids and oocytes, the Y-box-containing FRGY and MSY proteins, which are major components of mRNPs in oocytes (2, 33), are also associated with spermatid mRNPs in vivo (23, 51). This observation suggests that the mechanisms of mRNP assembly are at least partially conserved between spermatids and oocytes.
In this study, we have examined the mRNA sequence requirements for sequestration of mRNAs into spermatid mRNP particles. We find that in spermatids, like in oocytes, assembly into mRNP particles occurs independently of the sequence of the mRNA. Our results suggest an unexpected and fundamental similarity between the processes of translational control in oocytes and spermatids.
MATERIALS AND METHODS
Transgene construction and production of transgenic mice.
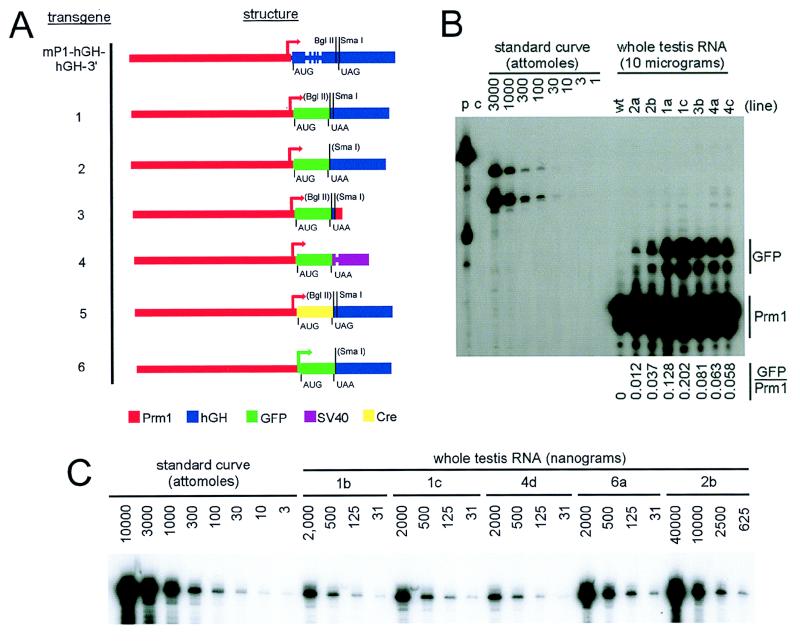
All transgenes were based on vector mP1-hGH (human growth hormone)-hGH-3′ (4, 31) and contained prm1 promoter sequences extending from the upstream HindIII site (−4100 from the cap site). In transgenes 1 to 5, the reporter genes were inserted at the synthetic BamHI linker at prm1 sequence position +91 from mP1-hGH-hGH-3′. Transgene 6 contained prm1 promoter sequences from −4100 to −1 from the cap site and initiated transcription at the first base of the synthetic linker upstream of the reporter gene. Transgenes 1 and 5 had hgh 3′ sequences extending from the BglII site in exon 5 (163 bp upstream of TAG) to the EcoRI site 800 bp downstream of BglII [633 bp downstream of the poly(A) site]). Transgenes 2 and 6 contain hgh 3′ sequences from the SmaI site 3 bp downstream of TAG to the EcoRI site. Transgene 3 contained the BglII/EcoRI hgh/prm1 terminator from plasmid mP1-hGH-mP1-3′ (4). Transgene 4 contained the simian virus 40 (SV40) small intron/poly(A) region isolated by using NotI/AflII from plasmid pEGFP-1 (Clontech Laboratories).
Three different structural genes were used: the parental genomic hgh reporter gene from the BamHI site 60 bp upstream of the translation initiation codon to the SmaI site 5 bp downstream of TAG (4); the modified green fluorescent protein (GFP) cDNA isolated from plasmid pEGFP-1; and the bacteriophage P1 Cre recombinase (46). All transgenes were linearized at the HindIII site 4,100 bp upstream of the cap site and were injected into the pronuclei of fertilized mouse eggs by using standard procedures (15). Mouse lines were named by a binary system wherein the transgene designation is followed by a unique letter for each founder bearing that transgene. Assays were performed on 6- to 12-week-old heterozygous males from crosses between heterozygous and wild-type parents. To estimate gene copy, serial dilutions of transgenic mouse tail DNA were prepared in wild-type mouse DNA. These samples were compared by quantitative PCR to a standard curve of pEGFP plasmid DNA diluted in wild-type mouse DNA.
As controls, the GFP-hGH region (without the Prm1 leader) or the Prm1-GFP-hGH region (including the 91-bp Prm1 leader) of transgene 1 was excised with BamHI/EcoRI or SpeI/EcoRI, respectively, and inserted under the control of the cytomegalovirus (CMV) promoter in vector pSCT-GAL-X556 (35). The mouse hepatoma cell line Hepa-1 c4c7 was cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Cells (106 cells/10-cm dish) were plated 24 h before transfection with 10 μg of supercoiled plasmid DNA, using 45 μl of Superfect reagent (Qiagen) as specified by the manufacturer. After 3 h, the cells were rinsed with 5 ml of medium and replenished with 10 ml of medium. After an additional 5 h (8 h posttransfection), dishes were rinsed with 1× phosphate-buffered saline (PBS) and quick-frozen at −80°C. Cytoplasmic lysates were prepared and used for velocity sedimentation within 48 h as described previously (36).
Fluorescence microscopy sample preparation.
Whole seminiferous tubules were gently teased out of decapsulated testes into 1× PBS. Tubules were placed on slides in 1× PBS with pieces of a broken coverslip as shims, covered, and observed by confocal fluorescence microscopy. For thick sections, whole decapsulated testes were fixed for 1 h at room temperature in 1× PBS containing 4% paraformaldehyde. These were embedded in 15% gelatin–1× PBS blocks, which were then fixed overnight at 4°C in 1× PBS containing 4% paraformaldehyde and washed in 1× PBS; sections roughly 100 μm thick were cut on a vibratome. To obtain thinner sections, the fixed decapsulated testes were embedded overnight at 4°C in 1× PBS containing 15% sucrose and then cast into blocks of 15% sucrose–7.5% gelatin. Blocks were frozen at −20°C, and 10-μm sections were cut on a cryostat. Vibratome and cryostat sections were collected on slides, flooded with 1× PBS containing 50% glycerol, covered with coverslips, and viewed immediately. In most lines (e.g., line 1a), the spermatid-specific GFP fluorescence was roughly 3 orders of magnitude above background for nonfixed tissues (Fig. 1A) and roughly 2 orders of magnitude above background for formaldehyde-fixed samples (not shown).
FIG. 1.
Transgene design and mRNA expression. (A) Transgene design. Colors refer to sequences derived from the sources indicated. Positions of translation initiation and termination codons are indicated. Fusion sites for the hgh 3′-UTR are indicated above the transgenes. Transcription initiation sites are denoted by bent arrows. Introns are shown as constrictions in the colored boxes in transgene mP1-hGH-hGH-3′ and transgene 4. (B) Relative GFP mRNA and Prm1 mRNA expression. At the top are indicated the samples in each lane. Mouse lines are designated by the transgene number followed by a unique letter designation for each founder carrying that transgene; “wt” denotes wild-type mice. RNase protection assays were performed with the indicated samples and the Prm1-GFP probe. Lane p contains a roughly 1:300 dilution of nondigested probe; lane c is a control lane containing probe hybridized to yeast RNA. Comparison to wild-type testis confirmed the identity of the transgene-specific signals. GFP-specific and Prm1-specific bands were excised from gels and were quantitated by liquid scintillation counting. Specific activities were corrected for differences in the radiolabeled UTP content of each protected fragment, and the ratios are presented below the autoradiogram. (C) GFP mRNA levels. The internal GFP probe, which does not hybridize to endogenous Prm1 mRNA and which gives an identical protected fragment with all of the GFP transgene mRNAs and with the synthetic control mRNA, was used to quantitate transgene mRNA levels. Assays were performed on RNA samples from each GFP transgenic mouse line and were quantitated by liquid scintillation counting of excised gel bands; data are presented in Table 1.
RNA preparation, velocity sedimentation, and RNase protection.
Total RNA was prepared from whole testes by sedimentation through CsCl cushions as described previously (37). For total RNA from cells sorted by fluorescence-activated cell sorting (FACS), samples in 1× PBS (sheath fluid) were collected directly into 1/10 the final volume of 10× TES (100 mM Tris [pH 7.5], 50 mM EDTA, 10% sodium dodecyl sulfate) containing 20 μg of proteinase K per ml. Samples (5 ml) were incubated at 55°C for 10 min. Each sample received 250 μl of 20% sarcosyl and 500 μl of 2.2 M ammonium acetate (pH 5), and each was extracted twice with 2 ml of phenol-chloroform. Nucleic acids were precipitated with isopropanol (5 ml) and resuspended in water, and high-molecular-weight RNA was precipitated on ice with a final concentration of 3 M LiCl. For measuring levels of nascent transcripts, nuclear RNA was purified as described previously (39). The RNA/DNA ratio in the nuclei preparations was 0.11.
Preparation and velocity sedimentation of testis and Hepa cell cytoplasms on exponential sucrose gradients, equivolume fractionation, and RNA extraction were performed as described previously (36, 38). A portion of the RNA from each fraction was denatured for 5 min at 75°C in 66% formamide, separated on 0.8% agarose gels, and stained with ethidium bromide to evaluate RNA integrity. Equal proportions of RNA from each fraction were used in each assay.
RNase protection experiments were performed as described previously (37). The following RNase protection probes were used. Endogenous Prm1 mRNA and mRNAs from transgenes having the GFP cassette fused at +91 (transgenes 1 through 4) were detected with a probe that spanned prm1 genomic sequences from the SpeI site at −40 to a synthetic BamHI linker fused at +91 (subcloned from plasmid mP1-hGH-hGH-3′ [31]) followed by the 21 bases of sequence between BamHI and NcoI from the polylinker of plasmid pEGFP-1. This probe gives a 91-base protected fragment for the endogenous Prm1 mRNA and an approximately 116-base protected fragment for correctly initiated transgene mRNAs. A second smaller band is also observed for all RNA species. Because a smaller protected fragment is observed with synthetic control mRNA in the standard curve, we suspect that the smaller bands represent an unstable region of the RNA-RNA duplex formed with this probe rather than mRNA heterogeneity. Comparisons to signals obtained with wild-type mice confirmed the identity of the GFP-specific signal (Fig. 1B). For quantitation of GFP mRNA levels and for detecting mRNA from transgene 6, an internal probe spanning sequences between 235 and 521 of plasmid pEGFP-1 was used. To generate synthetic GFP control mRNA, the 1,774-bp AvrII/EcoRI prm1–GFP–hgh–3′-UTR region from transgene 1 was inserted into EcoRI/XbaI-cut pBS+ (Stratagene), which was subsequently linearized with EcoRI and transcribed with T3 RNA polymerase to generate a 1,804-base transcript. Synthetic mRNA concentration was determined spectrophotometrically, and dilutions in yeast RNA carrier were used to generate a standard curve. For detecting transgenic Cre mRNA, an internal probe spanning 193 bases of sequence between BamHI and EcoRV of the Cre protein-coding sequences was used. To detect glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA, the previously described GAPDH probe (a 161-bp AccI fragment spanning sequences from +197 to +357 of the rat GAPDH cDNA) and conditions for stabilizing the rat-mouse hybrid were used (38). For GAPD-s mRNA, the probe used contained 13 bases of genomic sequences upstream of the cap site followed by cDNA sequences from the cap site to a synthetic XhoI linker inserted 30 bp downstream of the BamHI site in exon 2 (53) and thereby maps both the cap site and the exon 1/exon 2 junction.
Testis cell explants and FACS.
For each sort, both testes from a 6- to 9-week-old mouse were harvested and minced into 10 ml of ice-cold 1× PBS containing colcemid (20 μg/ml; Sigma) and 5 mM EGTA and were incubated on ice 15 min. Seminiferous tubules were transferred into fresh tubes containing 200 μl of hyaluronidase (10 mg/ml; Sigma) in 1× PBS and incubated at 35°C for 5 min. Samples received 10 μl of 1 M CaCl2 and 200 μl of crude collagenase (10 mg/ml; Sigma) and were incubated for an additional 5 min at 35°C. Samples then received 2 ml of 1× trypsin solution (Gibco/BRL) and 2 ml of Dispase solution (10 mg/ml; Gibco/BRL) in 1× PBS and were incubated on a slowly rotating 35°C incubator until dissociated (about 45 min). Proteases were quenched by addition of 2 ml of ice-cold fetal calf serum, and cells were passed twice through 35-μm nylon mesh filters. Cell suspensions were kept on ice or at 4°C during sorting. An aliquot of each explant was stained with trypan blue and assayed for cell viability.
Cell suspensions were sorted on either a Coulter FACS or a Beckman FACS apparatus, using 1× PBS for sheath fluid; gated cell samples were collected directly into lysis buffer for RNA analysis. Before and after preparative sorts, aliquots of 2 × 104 GFP-expressing (GFP+) and 2 × 104 non-GFP-expressing (GFP−) cells were collected and resorted to obtain quantitative estimates of cell purity.
RESULTS
Transgene design and expression.
The pioneering study by Braun et al. (4) on translational control of transgene expression in mouse spermatids used the hgh reporter gene under control of the mouse prm1 gene promoter, a promoter that is active only in the early spermatids of transgenic mice (31, 56). In that study, the effects of fusing the reporter gene to different 3′-UTRs were investigated. When the normal hgh 3′-UTR was used, no difference could be discerned between the time of transgene mRNA appearance and the onset of hGH protein accumulation. In contrast, when the hgh reporter gene was fused to the prm1 3′-UTR, a translational delay was observed. Both developmental and histochemical analyses showed that although hGH mRNA accumulated in the round spermatids, hGH protein was not detected until several days later, in the elongating spermatids (4). The presence or absence of 5′-UTR sequences was shown to be in consequential for this delay (3). It was concluded that the prm1 3′-UTR is sufficient to confer a translational delay on the hgh reporter gene, and it was inferred that these same sequences are responsible for causing the translational delay of Prm1 protein from the endogenous Prm1 mRNA. Because the translational delay of Prm1 protein synthesis is known to involve sequestration of the Prm1 mRNA as mRNP particles and their subsequent release into polysomes (10, 17, 19, 21), it has been posited that the 3′-UTR of Prm1 mRNA targets this mRNA to assemble into mRNP particles (3, 8).
If the above hypothesis is correct, then mRNAs lacking spermatid-specific mRNP targeting sequences should be excluded from mRNP particle pools and should instead assemble into polysomes and be translated immediately. To test this, the transgenes shown in Fig. 1A were expressed in mouse spermatids. To achieve expression specifically in spermatids, all of the transgenes are based on the prm1 expression system (4, 31). Three different reporter genes were used: (i) the genomic hGH cassette (transgene mP1-hGH-hGH-3′ [4]); (ii) the GFP cDNA (transgenes 1 through 4 and 6); and (iii) the bacteriophage P1 Cre recombinase gene (transgene 5 [46]). None of these reporter mRNAs exist normally in spermatids, and thus all were expected to lack any putative spermatid-specific mRNP targeting sequences. Because the study of Braun et al. (4) suggested that differences in 3′-UTR sequences might influence the subcellular targeting of the mRNA, we used three different terminators: the hgh and prm1 3′-UTRs used by Braun et al. (4) and the SV40 small intron/3′-UTR (Fig. 1A) (49).
The transgenes did not appear to perturb normal spermatogenic processes. In most cases, transgene mRNA levels were substantially lower than endogenous protamine mRNA levels (Fig. 1B and C; Table 1), making it unlikely that transgene mRNA could be saturating the translational control mechanisms. Moreover, all of the GFP lines exhibited normal male fertility, indicating that the mechanisms of spermatogenesis remained functional.
TABLE 1.
GFP transgene steady-state mRNA production
| Line | Haploid gene copy no. | GFP mRNA expression (molecules/ spermatida) | GFP mRNA molecules/geneb |
|---|---|---|---|
| 1a | 7 | 3,000 | 430 |
| 1b | 7 | 3,600 | 514 |
| 1c | 7 | 5,640 | 806 |
| Avg/gene | 583 | ||
| 2a | 6 | 180 | 30 |
| 2b | 23 | 1,200 | 52 |
| 2c | 8 | 660 | 82 |
| Avg/gene | 55 | ||
| 3a | 2 | 1,800 | 900 |
| 3b | 1 | 1,500 | 1,500 |
| 3c | 6 | 1,800 | 300 |
| Avg/gene | 900 | ||
| 4a | 14 | 1,800 | 130 |
| 4b | 9 | 2,280 | 253 |
| 4c | 10 | 2,400 | 240 |
| 4d | 4 | 2,520 | 630 |
| Avg/gene | 313 | ||
| 6a | 4 | 15,840 | 3,960 |
The number of spermatids represented in each assay was calculated based on the adult testis RNA/DNA ratio of 1.09 (37) an estimate of 0.33 pg of DNA per spermatid, and an estimate of 67% of all testis cells being spermatids (all stages included; only perhaps 20% of these will be the transcriptionally active round spermatids). Each value represents data from a single animal.
Since spermatids are haploid, the number of GFP mRNA molecules per transgene is the number of GFP mRNA molecules per spermatid (third column) divided by the haploid gene copy (second column).
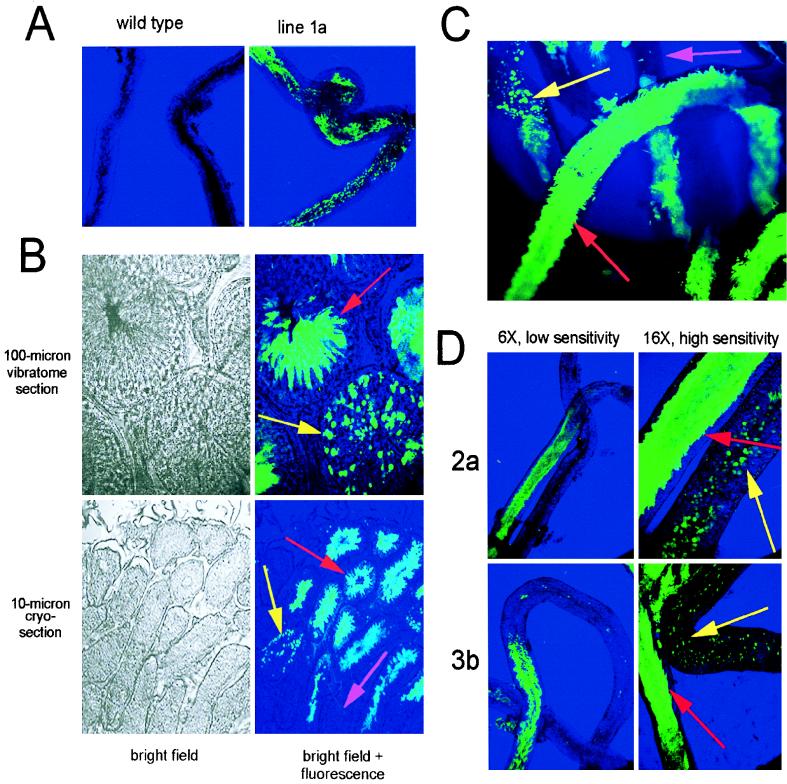
Confocal fluorescence microscopy was used to monitor GFP fluorescence in whole live seminiferous tubules (Fig. 2A and C), on 100-μm formaldehyde-fixed vibratome sections, and on 10-μm formaldehyde-fixed frozen testis sections (Fig. 1B) from heterozygous males of transgenic mouse line 1b containing the long hgh 3′-UTR. Strong GFP-specific fluorescence was detected in postmeiotic germ cells (Fig. 2B and C, red and yellow arrows) but not in regions where the prm1 promoter is expected to be inactive, such as domains of the seminiferous tubules containing only prehaploid germ cells (pink arrows) or in somatic cells. Fluorescence patterns were indistinguishable between all mouse lines bearing transgene 1 or 4 (not shown). Importantly, in mouse lines containing a transgene with either the long hgh 3′-UTR or the SV40 3′-UTR (transgene 1 or 4, respectively), strong GFP-specific fluorescence was observed not only in the late elongating spermatids (red arrows) but also in the early round spermatids (Fig. 2B and C, yellow arrows). This observation indicates that a large amount of GFP was being translated at this early stage. Thus, similar to the study by Braun et al. (4), we had generated mice that did not delay the onset of translation of the reporter gene mRNA.
FIG. 2.
Expression of GFP protein. (A) Confocal fluorescent microscopy with bright-field back-lighting on whole seminiferous tubules from wild-type or line 1a mice. (B) Confocal fluorescent and bright-field microscopy on 100-μm vibratome sections and on 10-μm cryosections of mouse line 1a testes. Yellow arrows indicate tubules with round spermatids; red arrows indicate tubules containing elongating spermatids; the pink arrow denotes a tubule with only prehaploid germ cells. (C) Whole live seminiferous tubules from mouse line 1a observed by confocal fluorescence microscopy showing regions with germ cells in the round spermatid stage (yellow arrow), the elongating spermatid stage (red arrow), and regions with only prehaploid germ cells (pink arrow). (D) Confocal fluorescent microscopy of seminiferous tubules from mouse lines 2a and 3b. Left, panels were photographed with a 6× objective and conditions used for panels A to C; right, panels photographed with a 16× objective and 30-fold-higher laser excitation energy. Arrows are as in other panels.
Under the conditions used to analyze mice bearing transgenes with either the long hgh 3′-UTR or the SV40 3′-UTR (transgene 1 or 4), mice bearing transgenes with the short hgh 3′-UTR or the prm1 3′-UTR (transgene 2 or 3) exhibited somewhat weaker GFP fluorescence. Although the fluorescence was predominantly in the elongating spermatids (Fig. 2D, left panels), a small amount of GFP could also be detected in round spermatids (Fig. 2D, right panels, yellow arrows). Whereas the low accumulation of transgene 2 mRNA may account for the reduced fluorescence in this line (see Discussion), differences in transgene mRNA levels (Table 1) could not account for the large differences in relative fluorescence intensity in round spermatids between line 1 or 4 and line 3 (Fig. 2). Rather, transgene 3 mRNA was less efficiently translated in round spermatids. This observation corroborates reports that the prm1 3′-UTR can delay translation of an mRNA (3, 4).
Distribution of transgene mRNAs in mRNP particles and polysomes.
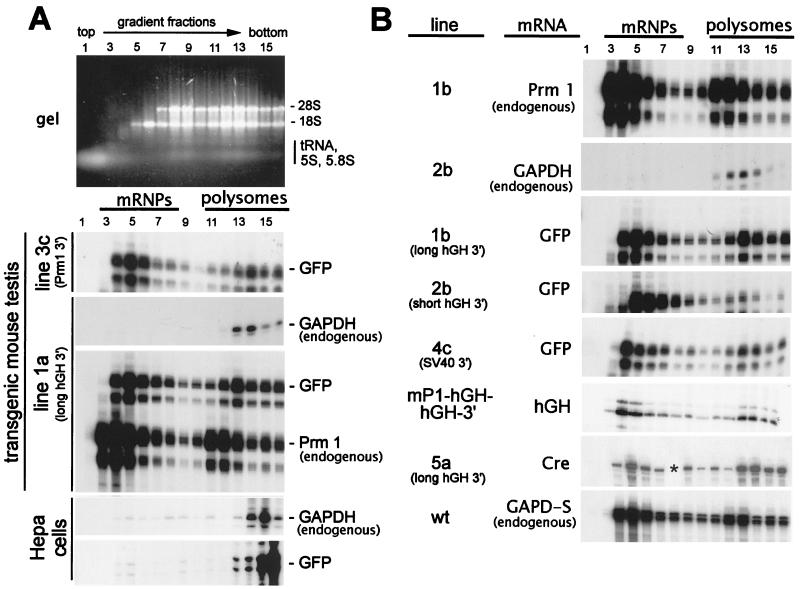
Although the results above confirm that the prm1 3′-UTR can delay translation of mRNAs during spermiogenesis, they do not indicate the mechanism. One possibility was that the prm1 3′-UTR specifically targets mRNAs to assemble into mRNP particles (3, 8). Alternatively, the prm1 3′-UTR might determine the timing of translational derepression of the mRNA. In the former case, mRNAs lacking specific targeting sequences would be excluded from assembling into mRNP particles; in the latter case mRNAs would assemble into mRNP particles nonspecifically and their release would be regulated. To distinguish these possibilities, we measured the proportion of transgene mRNA which was either mRNP particle or polysome associated in testes from each mouse line. During velocity sedimentation, particles migrate as a function of size; since polysomes are larger than mRNP particles, they sediment at a higher velocity (9). Figure 3 shows the results of representative RNase protection assays performed on the RNA fractions from adult testes. The endogenous Prm1 and GAPDH mRNAs served as controls for species that are either largely mRNP particle associated or predominantly polysome associated, respectively (Fig. 3) (19, 38). Transgene 3 contains the prm1 3′-UTR sequences which have been shown to impart a translational delay on mRNAs (reference 4; also see above). As expected, in testes from mice bearing this transgene, GFP mRNA was largely mRNP associated (Fig. 3A, line 3c). Importantly, however, in testes from mouse line 1a, which expresses the GFP gene fused to the long hGH 3′-UTR, a large proportion of the GFP mRNA was also assembled into mRNP particles (Fig. 3A, line 1a). Indeed, the distribution of GFP mRNA in line 1a was nearly indistinguishable from that of the endogenous Prm1 mRNA. Subsequent analyses on testes from mouse lines bearing the other GFP transgenes (transgenes 2 and 4, containing a shortened version of the hGH 3′-UTR or the SV40 3′-UTR, respectively) indicated that these mRNAs were also largely mRNP particle associated in adult testes (Fig. 3B). Because endogenous GAPDH mRNA in the samples was almost exclusively in the rapidly sedimenting polysomal fractions (Fig. 3), we could exclude the possibility that the slowly sedimenting mRNAs resulted from partial RNA degradation during sample preparation. Our results indicated that the prm1 3′-UTR was not required for assembly into mRNP particles.
FIG. 3.
Velocity sedimentation analysis of the distribution of mRNAs between mRNP particles and polysomes. Mouse testis or mouse Hepa cell cytoplasms were sedimented through exponential 10 to 85% sucrose gradients, and total RNA was purified from each fraction. At the top of panel A is shown an ethidium bromide-stained agarose gel of RNAs from a typical gradient (mouse line 1b). Gradient fractions (numbered from the top of the tube) are indicated. Below are representative autoradiograms of RNase protection assays on gradient fractions. The mouse line represented in each assay is listed at the left; adjacent to this is indicated the mRNA species being assayed. The positions of fractions containing mRNP particles and polysomes are indicated at the top. The Prm1 mRNA is much shorter than the other mRNAs and therefore assembles into smaller mRNP particles and smaller polysomes. For this reason, all Prm1 signals are shifted one to two fractions toward the top of the gradient (toward the left on the autoradiogram). As controls, GFP-hGH mRNA, either with (A, bottom) or without (not shown; both mRNAs gave similar results) the 91-base prm1 leader sequence, was expressed in mouse Hepa cells from the CMV promoter. The asterisk in lane 8 of the Cre sample in panel B (line 5a) denotes a gradient fraction for which the RNA pellet was lost during purification.
To ensure that assembly of the GFP mRNA into mRNP particles was spermatid specific and not due to a general translational defect in the mRNA, the Prm1 promoter from transgene 1 was replaced with the CMV promoter and the GFP-hGH transgene mRNA was expressed in mouse Hepa cells. Analyses of cytoplasmic preparations from transfected cells showed that GFP mRNA was predominantly polysomal (Fig. 3A, bottom), which confirmed that the transgene mRNA could be efficiently translated in somatic cells.
To ensure that the GFP cistron did not have a cryptic signal which targeted these mRNAs to assemble into mRNP particles, we expressed other nonspermatid mRNAs in mouse spermatids. mRNAs from either the parental transgene, mP1-hGH-hGH-3′ (4), or transgene 5, which contains the Cre cistron, were predominantly mRNP particle-associated (Fig. 3B). Thus, mRNP assembly was independent of sequences in either the reporter gene or the 3′-UTR.
All of the transgenes tested thus far were based on the constructs of Braun et al. (4), which utilized the prm1 cap site and included 91 bases of prm1-derived 5′-UTR. Previous work suggested that these sequences were neither necessary nor sufficient for imparting a translational delay on reporter gene mRNAs (3); however, it has been proposed that prm1 5′ sequences might interact with prm1 3′ sequences to target Prm1 mRNA to mRNP particles (44). To ensure that the prm1 leader sequences were not targeting the mRNAs to mRNP particles, transgene 6 was constructed (Fig. 1A). With this transgene, no prm1-derived sequences are transcribed or present in the mRNA. Confocal microscopy analysis of whole live seminiferous tubules from line 6a (Fig. 4A) showed GFP-specific fluorescence restricted to postmeiotic germ cells and present both in round (yellow arrow) and elongating (red arrow) spermatids. Velocity sedimentation analyses on adult testis from mouse line 6a showed that transgene 6 mRNA, like the other transgene mRNAs, was able to assemble into mRNP particles (Fig. 4C). We conclude that no specific mRNA sequences are required to direct the assembly of spermatid mRNAs into mRNP particles. Therefore, sequence-specific translational regulation via the prm1 3′-UTR likely acts by modulating the timing of release of the mRNA from mRNP particles rather than by targeting the mRNA to assemble into mRNP particles.
FIG. 4.
Expression and mRNP association of transgene mRNA from mouse line 6a. (A) Confocal fluorescent and bright-field microscopy of whole live seminiferous tubules. Due to the high expression of GFP protein in this line, very low excitation energy was used (about 3% of that used for line 1a in Fig. 2). Arrows are as in Fig. 2. (B) Relative levels of nascent GFP transcripts in nuclei from lines 1c and 6a. RNase protection assays were performed as described above on the indicated amounts of RNA isolated from whole testis (total) or purified nuclei (nuclear), using the internal GFP probe (upper two autoradiograms) or the GAPD-s probe (below). At the left are given the mouse line used and the identity of each protected fragment. The schematic at the bottom shows that GAPD-s pre-mRNA retaining intron 1 hybridizes to a 147-base region of the probe (dark hatched line below the RNAs); mRNA with exon 1 spliced onto exon 2 hybridizes to a 177-base region of the probe. To compare relative levels of spliced and unspliced transcripts, the radioactivity of each band was determined by liquid scintillation counting and was corrected for differences in the number of radiolabeled UTP residues in each protected fragment (31 and 49 for nonspliced and spliced, respectively). By assuming equal hybridization efficiency, we calculate that 43% of the GAPD-s transcripts in testis nuclei have not yet removed intron 1. (C) Velocity sedimentation analysis of GFP transgene mRNA and endogenous Prm1 mRNA. Assays were as in Fig. 3 except that for detecting GFP mRNA, the internal probe was used.
Transgene mRNA stability.
Mouse line 6a, bearing the GFP transgene that lacked prm1 5′-UTR sequences, was the strongest GFP-expressing mouse, producing so much GFP protein that the whole testes appeared lightly chartreuse under standard room illumination (wild-type testes and those from the other transgenic lines appear cream colored). Although most of the GFP was sloughed off in the polar bodies during late spermiogenesis, enough residual GFP remained to make the mature spermatozoon strongly fluorescent (not shown). Despite this high level of expression, these mice exhibited normal male fertility and a 50% progeny sex ratio.
Although line 6a contained relatively few copies of the transgene (four copies per haploid genome), expression of GFP protein in line 6a testis was reflected by high accumulation of GFP mRNA (Fig. 1C; Table 1). To distinguish whether the higher mRNA accumulation resulted from increased transcription of this transgene or increased stability of the mRNA, we measured levels of nascent transcripts in testis nuclei. Purified nuclei contain almost no fully processed mRNA (55), so transcripts measured in pure nuclear preparations represent nascent pre-mRNA levels (37, 39, 55). A probe for the spermatid-specific GAPD-s mRNA was used to evaluate the nuclear mRNA preparations because it will differentiate transcripts which retain intron 1 from those in which the intron has been removed (Fig. 4B). The results showed that although nonspliced exon 1 mRNA was too rare to detect in total RNA samples, it represented 43% of the GAPD-s transcripts in nuclei. The high enrichment of unspliced mRNA in the nuclear preparations is consistent with these samples containing only nascent pre-mRNAs. Despite containing nearly threefold more total GFP mRNA than line 1c, line 6a contained only 80% as much nuclear GFP mRNA (Table 1; Fig. 4B). This indicates that cytoplasmic mRNA from transgene 6 is roughly fourfold more stable than that from transgene 1.
Spermatid-specific expression of gapdh gene family members.
GAPDH mRNA is a valuable control because in whole testis lysates it is almost entirely polysomal (reference 38 and Fig. 3). However, in light of the findings above, we became curious about how GAPDH mRNA could be excluded from mRNP particles. It was possible that the gapdh gene had specific signals that prevented its mRNA from assembling into mRNP particles, as might be expected from the precedent set by mRNAs encoding TFIIIA or histone B4 in frog oocytes (6, 28, 50). Alternatively, since all assays to date had been performed with whole testis extracts, it was possible that, like other mRNAs which have been shown to be exclusively polysomal in whole testis lysates (21), GAPDH mRNA might be expressed only in a subset of cell types in the testis, specifically excluding the postmeiotic germ cells in which mRNP particles are assembled.
Spermatids rely heavily on glycolysis for energy production (30). As a part of their glycolytic machinery, spermatids express a gapdh gene family member named gapd-s (52, 53). Velocity sedimentation analyses showed that GAPD-s mRNA, unlike that encoding GAPDH, was predominantly associated with mRNP particles (Fig. 3B, bottom). Thus, although it has not previously been tested whether spermatids also express GAPDH, the one gapdh gene family member which is known to be expressed in spermatids does assemble into mRNP particles. Because GAPDH and GAPD-s appear to be enzymatically equivalent (53), it seemed unlikely that they would be differentially regulated in a single cell. Rather, we suspected that the gapdh gene was not expressed in spermatids.
Whereas cell type expression of testis mRNAs is commonly addressed by in situ hybridization, the sequence similarity between GAPDH and GAPD-s (52) complicated this approach. Therefore, to test whether GAPDH was expressed in spermatids, we wished to purify postmeiotic testis cells. Previous methods for isolating testicular cell types from juvenile testes based on unit-gravity sedimentation (1, 24) were deemed unsatisfactory for sorting cells from adult testis containing motile spermatozoa. Therefore, we took advantage of the GFP-expressing transgenic mouse lines to develop a method of separating postmeiotic germ cells from other adult testis cell types by FACS (Fig. 5).
FIG. 5.
Transgene mRNA, GAPDH mRNA, and Prm1 mRNA levels in explanted mouse line 4b testis cell populations purified by FACS. (A) Bright-field and fluorescent microscopy of trypan blue-stained cell explants. The yellow arrow denotes a trypan blue-stained (dead) cell. Red arrows indicate large multinucleate cells. Testis cells show extreme size variation; however, phase-contrast microscopy (not shown) revealed that most of the large cells in the micrographs are multinuclear (see Materials and Methods). (B) Preparative FACS on explanted cells. Left, distribution of cell types seen using forward and side scatter criteria; center, distribution of fluorescence intensities observed; right, gating parameters used in this study. Cells gated as GFP+ are green, cells gated as GFP− are red, and other cells (which were discarded) are gray. The colors in the left panel correspond to those in the right panel and thus give an indication of differences in the forward and side scatter properties of the various GFP+ and GFP− cell subpopulations. (C) RNase protection analyses of GFP mRNA from the transgene and the endogenous Prm1 and GAPDH mRNAs in the sorted GFP+ and GFP− cell populations and in the nonsorted explant. GFP and Prm1 mRNAs were detected with the Prm1-GFP probe, which maps the cap sites of both transcripts. Each lane contained 2 μg of total RNA from the indicated cell sample. The GFP+ sample contained 25% as much GAPDH mRNA as the GFP− sample, whereas FACS reanalysis of the GFP+ cell sample indicated that it contained 17.2% contamination with GFP− cells. By assuming that these were expressing the same amount of GAPDH mRNA per cell as cells in the pure GFP− population, we estimate that 69% (17.2% ÷ 25%) of the total GAPDH signal in the GFP+ sample arose from the contaminating GFP− cells (see text).
Many of the cell divisions of testicular germ cells are incomplete, leaving cytoplasmic bridges between sister cells (7, 32). Previous methods for explanting cells from testis (1, 24) disrupted these bridges to produce discrete cells. In our experience, assays for specific mRNAs indicated mRNA was depleted in samples isolated from cells prepared by this method (discussed in reference 38), possibly due to partial mRNA degradation or leakage after rupturing these bridges. The conditions developed here start by treatment with colcemid to depolymerize cytoskeletal microtubules, such that upon tissue dissociation, the cells fuse through their cytoplasmic bridges to form multinuclear cells (e.g., Fig. 5A, red arrows). Cell populations explanted by this method contained less than 0.1% dead cells as measured by trypan blue exclusion (Fig. 5A, yellow arrow). Moreover, levels of specific mRNAs measured in cells explanted by this method were indistinguishable from those in whole testis (not shown). Populations of GFP+ and GFP− cells were separated by FACS (Fig. 5B). Fluorescence microscopy and FACS reanalysis of sorted cell populations indicated that the GFP− cell samples contained no detectable GFP+ cells (<0.1%); the GFP+ cell populations contained 10 to 20% GFP− cells in each sort. The reason for the lower purity of the GFP+ samples is likely that single droplets containing one GFP+ and one GFP− cell will be sorted as GFP+. For our purposes, 10 to 20% carryover of GFP− cells was acceptable; results were mathematically corrected for the contribution of GFP− cells.
RNase protection analyses showed that GFP and Prm1 mRNAs were more than 100-fold enriched in the GFP+ cell population (Fig. 5C). In contrast, GAPDH mRNA was fourfold more abundant in the GFP− cell population. In the experiment shown, the GFP+ cell population contained 17.2% GFP− cells (see above), which accounts for 69% of the GAPDH mRNA in the GFP+ sample. Thus, levels of GAPDH mRNA were roughly 13-fold lower in the GFP+ postmeiotic cells than in the GFP− cell population. Due to its low level in these cells, we suspect that this GAPDH mRNA may be persisting in the earliest GFP-expressing stages from GAPDH mRNA which was transcribed during the prehaploid cell stages. Indeed, we are not aware of any example of an mRNA transcribed in spermatids which is excluded from assembly into mRNP particles. We conclude that mRNAs which are transcribed in round spermatids are assembled into mRNP particles by an mRNA sequence-independent mechanism.
DISCUSSION
Similarities between translational control in spermatogenesis and oogenesis.
In most if not all vertebrates, gametogenesis requires large-scale temporal uncoupling of the processes of transcription and translation, such that proteins can be synthesized in cells which are transcriptionally silent (44). In spermatogenesis, mRNP formation occurs in the early postmeiotic stages (13, 21), during which time the male gamete is specialized to serve as an efficient vector for fertilization. In oocytes, mRNA storage occurs in prehaploid stages and is required both for meiotic maturation (5, 47, 48) and to prepare the egg for the biosynthetic demands of early embryonic development (43, 45). The present study using spermatids, in combination with previous studies on translational repression in oocytes, reveals a fundamental similarity between the processes of uncoupling transcription and translation in spermatids and oocytes. Specifically, in each case, mRNAs synthesized by the last transcriptionally active stages of gametogenesis are assembled into mRNP particles by a mechanism that acts independently of the sequence of that mRNA.
At the molecular level, the mRNP particles in oocytes and spermatids are related. The Y-box proteins, which bind RNA with little or no sequence specificity (27, 54), are major components of mRNP particles from either source (2, 23, 51). Because the assembly of mRNP particles in both systems is sequence independent (references 2 and 28 and this study), it is plausible that non-sequence-specific RNA-binding proteins, like the Y-box proteins, are sufficient for mRNP particle assembly.
In frog oocytes, some genes encoding proteins that are required at high levels during oocyte maturation, such as the transcription factor TFIIIA, have signals to allow their mRNAs to assemble into polysomes and be translated immediately (50). Recently, Matsumoto et al. (28) showed that the cotranscriptional processing of an intron at the 5′ end of TFIIIA pre-mRNA promoted efficient translation, whereas insertion of an intron at the 3′ end of the gene led to translational silencing. Interestingly, the sequences of the mature mRNAs were identical. This suggests that the mechanism directing TFIIIA mRNA to be translated in oocytes is sensitive to the nuclear history of the mRNA (28). It is unknown whether certain mRNAs in spermatids might escape translational repression by a similar mechanism. Further studies will be required to distinguish whether mRNAs that are translated in the round spermatids are channeled into a translationally active state during nuclear maturation.
mRNA sequence-specific translational regulation in spermatids.
Our results show that the assembly of mRNAs into mRNP particles in spermatids is not sequence dependent; however, the timing of translation of individual mRNAs is. Braun et al. (4) have shown that the prm1 3′-UTR can impart a translational delay on the hgh reporter gene, and the present study extends this finding to show that these sequences can likewise delay translation of a GFP reporter gene. The results presented here suggest that the prm1 3′-UTR does not function to target mRNAs to assemble into mRNP particles but rather is a part of the timing mechanism that determines when various mRNAs within mRNP particles will be translated. This interpretation has implications for understanding the mechanisms of translational control in spermatids. For example, several RNA-binding proteins that are associated with spermatid mRNPs have been cloned in recent years (reviewed in reference 21). Based on the present study, one might expect those proteins with little or no sequence specificity for RNA binding to be likely candidates for mediating mRNP particle assembly, whereas proteins that exhibit specific binding would more likely be involved in timing the release of individual mRNAs for translation.
In mouse oocytes it has been shown that the timing of translation of tPA mRNA is determined by specific sequences in the 3′-UTR (47). These sequences can also repress translation of synthetic mRNAs microinjected into oocytes (48); however, it is uncertain whether translational repression of endogenous tPA mRNA requires these sequences. Indeed, it has previously been shown that translation of injected mRNAs in oocytes is likely an artifact of incorrect nuclear history; in vivo-transcribed mRNAs are generally not translationally active (2, 28). A clearer understanding of the role of the tPA 3′-UTR in translational repression will require studies similar to the present work to determine whether this region is necessary for assembly of in vivo-transcribed tPA mRNA into mRNP particles in the oocytes of transgenic mice.
Transgene mRNA stability and mRNP-polysome distribution.
Our mouse lines showed a 130-fold range in the number of mRNA molecules that accumulated per transgene (Table 1). Levels of mRNA accumulation per gene between independent mouse lines carrying the same transgene showed far less variation than was observed between lines carrying different transgenes (Table 1). This finding suggests that differences in mRNA levels were due, at least in part, to differences in relative mRNA stabilities. We tested this experimentally by comparing nuclear levels of nascent GFP pre-mRNA between lines 1c and 6a (Fig. 4B). The results confirmed that differences in steady-state GFP mRNA levels in these two lines were posttranscriptionally determined. Interestingly, transgene 6 mRNA not only is more stable than transgene 1 mRNA but also exhibits a smaller fraction of mRNA assembled into mRNP particles (compare Fig. 3 and 4C). Transgene 2 mRNA, which had the lowest mRNA accumulation (Table 1), showed the highest proportion of mRNA in mRNPs (Fig. 3B). One important observation from this study is that different distributions of mRNAs between mRNPs and polysomes in testis can be caused by processes other than differential translational regulation of the mRNAs. Further studies will be required to fully understand how differences in mRNA stability affect this distribution.
Future uses of GFP-expressing mice for studying spermiogenesis.
The technology developed here for purifying postmeiotic male germ cells from the spermatid-specific GFP+ transgenic mice by FACS may prove valuable for future studies on spermiogenesis. First, although we have not yet attempted culturing the explanted cells, it is possible that the protocol for fusing cells through their cytoplasmic bridges and isolating them as syncytia rather than rupturing the bridges may favor longer culture of these cells for investigating spermiogenic processes in vitro. Indeed, since the cytoplasmic bridges between mammalian spermatids are quite large, perhaps exceeding 0.3 nuclear diameters (32), the syncytial state may more closely approximate physiological conditions than do individual spermatids. Second, populations of postmeiotic cells purified by FACS may be valuable for numerous molecular studies. Here we compared specific mRNA levels between spermatids and all other testis cell types. For this work, high-yield recovery was required; therefore, we used conditions that gave some carryover of GFP− cells into GFP+ cell populations. Since the FACS scores each droplet rather than each cell, it is likely that the GFP− cells were carried through the sort by being in droplets of media with GFP+ cells. Nearly pure populations of GFP+ cells (<1% GFP− cells [not shown]) can be isolated by sorting the cells from more dilute samples. Although impractical where large preparative samples of cells are needed, such homogeneous populations may prove valuable for generating libraries for differential screening protocols. Moreover, additional criteria can be used to limit cell type diversity in the sorts. For example, in Fig. 5B, it is apparent that the cells from whole testis exhibit subpopulations that differ in their relative fluorescence, their forward light scatter (an indication of cell size), and their side light scatter (an indication of cellular substructure complexity). By restricting these parameters to sort for cells with specific properties, it may be possible to obtain pure samples of nearly all cell types from adult testis.
We have presented evidence that mRNAs transcribed in early spermatids are assembled into mRNP particles by a mechanism that acts independently of mRNA sequence. Methods developed here for isolating spermatids from mature testis will facilitate more detailed studies on the roles that mRNA nuclear history, sequence-specific mRNA-binding proteins, mRNA stability, and other regulatory processes play in determining and coordinating the timing of accumulation of specific proteins in spermatids.
ACKNOWLEDGMENTS
We thank D. Taylor and S. Barnett for technical assistance and training; A. Gaglio (Ventana Genetics) and J. Pierce and W. Green (University of Utah Flow Sorting Core Facility) for cell sorting; P. Flodby for suggesting the FACS separations; R. Palmiter and R. Braun for providing plasmids mP1-hGH-hGH-3′ and mP1-hGH-mP1-3′; J. Schmidt, A. Godwin, E. Leibold, C. Thummel, and K. Thomas for critically reading the manuscript; and our colleagues for thoughtful discussions.
This project was supported by the Howard Hughes Medical Research Foundation and the National Institutes of Health. E.E.S. was supported in part by the Mathers Charitable Foundation and is currently a Howard Hughes Fellow of the Life Sciences Research Foundation; E.S.H. was supported by training grant T32 DK07115 from the National Institutes of Health.
REFERENCES
- 1.Bellvé A R, Cavicchia J C, Millette C F, O’Brien D A, Bhatnagar Y M, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouvet P, Wolffe A P. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell. 1994;77:931–941. doi: 10.1016/0092-8674(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 3.Braun R E. Temporal translational regulation of the protamine 1 gene during mouse spermatogenesis. Enzyme. 1990;44:120–128. doi: 10.1159/000468752. [DOI] [PubMed] [Google Scholar]
- 4.Braun R E, Peschon J J, Behringer R R, Brinster R L, Palmiter R D. Protamine 3′ untranslated sequences regulate temporal translational control and subcellular localization of growth hormone in spermatids in transgenic animals. Genes Dev. 1989;3:793–802. doi: 10.1101/gad.3.6.793. [DOI] [PubMed] [Google Scholar]
- 5.de Vantéry C, Stutz A, Vassalli J-D, Schorderet-Slatkine S. Acquisition of meiotic competence in growing mouse oocytes is controlled at both translational and posttranslational levels. Dev Biol. 1997;187:43–54. doi: 10.1006/dbio.1997.8599. [DOI] [PubMed] [Google Scholar]
- 6.Dimitrov S, Dasso M C, Wolffe A P. Remodeling sperm chromatin in Xenopus laevis egg extracts: the role of core histone phosphorylation and linker histone B4 in chromatin assembly. J Cell Biol. 1994;126:591–601. doi: 10.1083/jcb.126.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dym M, Fawcett D W. Further observations on the number of spermatogonia, spermatocytes, and spermatids connected by intercellular bridges in the mammalian testis. Biol Reprod. 1971;4:195–204. doi: 10.1093/biolreprod/4.2.195. [DOI] [PubMed] [Google Scholar]
- 8.Fajardo M A, Haugen H S, Clegg C H, Braun R E. Separate elements in the 3-untranslated region of the mouse protamine 1 mRNA regulate translational repression and activation during murine spermatogenesis. Dev Biol. 1997;191:42–52. doi: 10.1006/dbio.1997.8705. [DOI] [PubMed] [Google Scholar]
- 9.Fan H, Penman S. Regulation of protein synthesis in mammalian cells. II. Inhibition of protein synthesis at the level of translational initiation during mitosis. J Mol Biol. 1970;50:655–670. doi: 10.1016/0022-2836(70)90091-4. [DOI] [PubMed] [Google Scholar]
- 10.Gedamu L, Iatrou K, Dixon G H. Isolation and characterization of trout testis protamine mRNAs lacking poly(A) Cell. 1977;10:443–451. doi: 10.1016/0092-8674(77)90031-9. [DOI] [PubMed] [Google Scholar]
- 11.Gu W, Kwon Y, Oko R, Hermo L, Hecht N B. Poly (A) binding protein is bound to both stored and polysomal mRNAs in the mammalian testis. Mol Reprod Dev. 1995;40:273–285. doi: 10.1002/mrd.1080400303. [DOI] [PubMed] [Google Scholar]
- 12.Han J R, Yiu G K, Hecht N B. Testis/brain RNA-binding protein attaches translationally repressed and transported mRNAs to microtubules. Proc Natl Acad Sci USA. 1995;92:9550–9554. doi: 10.1073/pnas.92.21.9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hecht N B. The making of a spermatoozoon: a molecular perspective. Dev Genet. 1995;16:95–103. doi: 10.1002/dvg.1020160202. [DOI] [PubMed] [Google Scholar]
- 14.Hecht N B. Molecular mechanisms of male germ cell differentiation. Bioessays. 1998;20:555–561. doi: 10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 15.Hogan B, Costantini F, Lacy E. Manipulating the mouse embryo. A laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 16.Huarte J, Belin D, Vassalli J-D. Plasminogen activator in mouse and rat oocytes: induction during meiotic maturation. Cell. 1985;43:551–558. doi: 10.1016/0092-8674(85)90184-9. [DOI] [PubMed] [Google Scholar]
- 17.Iatrou K, Dixon G H. The distribution of poly(A)+ and poly(A)− protamine messenger RNA sequences in the developing trout testis. Cell. 1977;10:433–441. doi: 10.1016/0092-8674(77)90030-7. [DOI] [PubMed] [Google Scholar]
- 18.Kleene K C, Distel R J, Hecht N B. Translational regulation of a protamine mRNA during spermatogenesis in the mouse. Dev Biol. 1984;105:71–79. doi: 10.1016/0012-1606(84)90262-8. [DOI] [PubMed] [Google Scholar]
- 19.Kleene K C. Multiple controls over the efficiency of translation of the mRNAs encoding transition proteins, protamines, and the mitochondrial capsule selenoprotein in late spermatids in mice. Dev Biol. 1993;159:720–731. doi: 10.1006/dbio.1993.1277. [DOI] [PubMed] [Google Scholar]
- 20.Kleene K C, Wang M-Y, Cutler M, Hall C, Shih D. Developmental expression of poly(A) binding protein mRNAs during spermatogenesis in the mouse. Mol Reprod Dev. 1994;39:355–364. doi: 10.1002/mrd.1080390403. [DOI] [PubMed] [Google Scholar]
- 21.Kleene K C. Patterns of translational regulation in the mammalian testis. Mol Reprod Dev. 1996;43:268–281. doi: 10.1002/(SICI)1098-2795(199602)43:2<268::AID-MRD17>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 22.Kwon Y K, Hecht N B. Binding of a phosphoprotein to the 3′ untranslated region of the mouse protamine 2 mRNA temporally represses its translation. Mol Cell Biol. 1993;13:6547–6557. doi: 10.1128/mcb.13.10.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon Y K, Murray M T, Hecht N B. Proteins homologous to the Xenopus germ cell-specific RNA binding proteins p54/p56 are temporally expressed in mouse male germ cells. Dev Biol. 1993;158:90–100. doi: 10.1006/dbio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 24.Lam D M K, Furrer R, Bruce W R. The separation, physical characterization, and differentiation kinetics of spermatogonial cells of the mouse. Proc Natl Acad Sci USA. 1970;65:192–199. doi: 10.1073/pnas.65.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee K, Fajardo M A, Braun R E. A testis cytoplasmic RNA-binding protein that has the properties of a translational repressor. Mol Cell Biol. 1996;16:3023–3034. doi: 10.1128/mcb.16.6.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leibold E A, Munro H N. Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5′ untranslated region of ferritin heavy- and light-subunit mRNAs. Proc Natl Acad Sci USA. 1988;85:2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marello K, La Rovere J, Sommerville J. Binding of Xenopus oocyte masking proteins to mRNA sequences. Nucleic Acids Res. 1992;20:5593–5600. doi: 10.1093/nar/20.21.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto K, Wassarman K M, Wolffe A P. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998;17:2107–2121. doi: 10.1093/emboj/17.7.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monesi V. Ribonucleic acid synthesis during mitosis and meiosis in the mouse testis. J Cell Biol. 1965;22:521–532. doi: 10.1083/jcb.22.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura M, Okinaga S, Arai K. Studies of metabolism of round spermatids: glucose as an unfavorable substrate. Biol Reprod. 1986;35:927–935. doi: 10.1095/biolreprod35.4.927. [DOI] [PubMed] [Google Scholar]
- 31.Peschon J J, Behringer R R, Brinster R L, Palmiter R D. Spermatid-specific expression of protamine 1 in transgenic mice. Proc Natl Acad Sci USA. 1987;84:5316–5319. doi: 10.1073/pnas.84.15.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips D M. Spermiogenesis. New York, N.Y: Academic Press; 1974. [Google Scholar]
- 33.Ranjan M, Tafuri S R, Wolffe A P. Masking mRNA from translation in somatic cells. Genes Dev. 1993;7:1725–1736. doi: 10.1101/gad.7.9.1725. [DOI] [PubMed] [Google Scholar]
- 34.Richter J D. Translational control in development: a perspective. Dev Genet. 1993;14:407–411. doi: 10.1002/dvg.1020140602. [DOI] [PubMed] [Google Scholar]
- 35.Rusconi S, Severne Y, Georgiev O, Galli I, Wieland S. A novel expression assay to study transcriptional activation. Gene. 1990;89:211–221. doi: 10.1016/0378-1119(90)90008-f. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt E E, Merrill G F. Changes in dihydrofolate reductase (DHFR) mRNA levels can account fully for changes in DHFR synthesis rates during terminal differentiation in a highly amplified myogenic cell line. Mol Cell Biol. 1991;11:3726–3734. doi: 10.1128/mcb.11.7.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt E E, Schibler U. Cell size regulation, a mechanism that controls cellular RNA accumulation: consequences on regulation of the ubiquitous transcription factors Oct1 and NF-Y, and the liver enriched transcription factor DBP. J Cell Biol. 1995;128:467–483. doi: 10.1083/jcb.128.4.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt E E, Schibler U. Developmental testis-specific regulation of mRNA levels and mRNA translational efficiencies for TATA-binding protein mRNA isoforms. Dev Biol. 1997;184:138–149. doi: 10.1006/dbio.1997.8514. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt E E, Ohbayashi T, Makino Y, Tamura T-A, Schibler U. Spermatid-specific overexpression of the TATA-binding protein gene involves recruitment of two potent testis-specific promoters. J Biol Chem. 1997;272:5326–5334. doi: 10.1074/jbc.272.8.5326. [DOI] [PubMed] [Google Scholar]
- 40.Schumacher J M, Lee K, Edelhoff S, Braun R E. Spnr, a murine RNA binding protein that is localized to cytoplasmic microtubules. J Cell Biol. 1995a;129:1023–1032. doi: 10.1083/jcb.129.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schumacher J M, Lee K, Edelhoff S, Braun R E. Distribution of Tenr, an RNA-binding protein, in a lattice-like network within the spermatid nucleus in the mouse. Biol Reprod. 1995b;52:1274–1283. doi: 10.1095/biolreprod52.6.1274. [DOI] [PubMed] [Google Scholar]
- 42.SenGupta D N, Berkhout B, Gatignol A, Zhou A, Silverman R H. Direct evidence for translational regulation by leader RNA and Tat protein of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1990;87:7492–7496. doi: 10.1073/pnas.87.19.7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spirin A S. On “masked” forms of messenger RNA in early embryogenesis and in other differentiating systems. Curr Top Dev Biol. 1966;1:1–38. [PubMed] [Google Scholar]
- 44.Spirin A S. Storage of messenger RNA in eukaryotes: envelopment with protein, translational barrier at 5′ side, or conformational masking by 3′ side? Mol Reprod Dev. 1994;38:107–117. doi: 10.1002/mrd.1080380117. [DOI] [PubMed] [Google Scholar]
- 45.Stebbins-Boaz B, Richter J D. Translational control during early development. Crit Rev Eukaryotic Gene Expr. 1997;7:73–94. doi: 10.1615/critreveukargeneexpr.v7.i1-2.50. [DOI] [PubMed] [Google Scholar]
- 46.Sternberg N, Sauer B, Hoess R, Abremski K. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by DNA methylation. J Mol Biol. 1986;187:197–212. doi: 10.1016/0022-2836(86)90228-7. [DOI] [PubMed] [Google Scholar]
- 47.Strickland S, Huarte J, Belin D, Vassalli A, Rickles R J, Vassalli J-D. Antisense RNA directed against the 3′ noncoding region prevents dormant mRNA activation in mouse oocytes. Science. 1988;241:680–684. doi: 10.1126/science.2456615. [DOI] [PubMed] [Google Scholar]
- 48.Stutz A, Conne B, Huarte J, Gubler P, Völkel V, Flandin P, Vassalli J-D. Masking, unmasking, and regulated polyadenylation cooperate in the translational control of a dormant mRNA in mouse oocytes. Genes Dev. 1998;12:2535–2548. doi: 10.1101/gad.12.16.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramani S, Mulligan R, Berg P. Expression of the mouse dihydrofolate reductase complementary deoxyribonucleic acid in simian virus 40 vectors. Mol Cell Biol. 1981;1:854–864. doi: 10.1128/mcb.1.9.854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tafuri S R, Wolffe A P. Selective recruitment of masked maternal mRNA from messenger ribonucleoprotein particles containing FRGY2 (mRNP4) J Biol Chem. 1993;257:24255–24261. [PubMed] [Google Scholar]
- 51.Tafuri S R, Familari M, Wolffe A P. A mouse Y box protein, MSY1, is associated with paternal mRNA in spermatocytes. J Biol Chem. 1993;268:12213–12220. [PubMed] [Google Scholar]
- 52.Welch J E, Schatte E C, O’Brien D A, Eddy E M. Expression of a glyceraldehyde 3-phosphate dehydrogenase gene specific to mouse spermatogenic cells. Biol Reprod. 1992;46:869–878. doi: 10.1095/biolreprod46.5.869. [DOI] [PubMed] [Google Scholar]
- 53.Welch J E, Brown P R, O’Brien D A, Eddy E M. Genomic organization of a mouse glyceraldehyde 3-phosphate dehydrogenase gene (gapd-s) expressed in post-meiotic spermatogenic cells. Dev Genet. 1995;16:179–189. doi: 10.1002/dvg.1020160210. [DOI] [PubMed] [Google Scholar]
- 54.Wolffe A P. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. Bioessays. 1994;16:245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- 55.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence of cotranscriptional splicing. Mol Cell Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zambrowicz B P, Harendza C J, Zimmermann J W, Brinster R L, Palmiter R D. Analysis of the mouse protamine 1 promoter in transgenic mice. Proc Natl Acad Sci USA. 1993;90:5071–5075. doi: 10.1073/pnas.90.11.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]