Abstract
Introduction
Dipeptidyl peptidase-4 (DPP-4) inhibitors have various cellular effects that are associated with vascular protection. Here, we examined whether vildagliptin protected endothelial function in diabetic rats and explored the involved mechanism.
Material and methods
Experimental diabetic rats were obtained by feeding a high-fat diet and administering an intraperitoneal injection of streptozotocin. Rats were randomly divided into four groups: controls (CON), diabetes (DM), diabetes + low dose of vildagliptin (Lvil, 10 mg/kg/day), and diabetes + high dose of vildagliptin (Hvil, 20 mg/kg/day). The metabolic parameters, endothelial function, and whole miRNA expression were measured.
Results
After a 12-week treatment, vildagliptin-treated rats showed a significant reduction in blood glucose and blood lipid levels. Moreover, vildagliptin recovered aortic endothelial function in diabetic rats. We identified 31 miRNAs that were differentially expressed in the Hvil group compared with the diabetic group. Importantly, through miRNA target biological function and pathway analysis, we found that vildagliptin activated miR-190-5p to inhibit Ccl2 expression and inhibited miR-134-5p and miR-375-3p to increase Bdnf and Pdk1 expression in the aorta.
Conclusions
Our present study indicates that vildagliptin can recover endothelial function in diabetic rats. Anti-inflammatory and anti-apoptosis mechanisms and endothelial moderation may be the intervention targets of vildagliptin to protect the cardiovascular system through miRNA regulation.
Keywords: glucagon-like peptide-1, dipeptidyl peptidase-4 inhibitor, endothelial function, diabetes, miRNA
Introduction
An increased incidence of type 2 diabetes mellitus has become a big public health problem worldwide. Diabetes is one of the most important risk factors for cardiovascular disease. Increasing research has revealed the correlation between diabetes and the increased risk of cardiovascular disease.
MicroRNAs are a group of small non-coding RNAs that bind to target genes, leading to the inhibition of gene expression in both healthy and sick hosts. Recent studies have shown that the disorder of miRNA expression leads to various metabolic diseases [1]. In the cardiovascular system, miRNAs play an important role in endothelial cell (EC) and vascular smooth muscle cell (VSMC) biology [2, 3]. miR-126 negatively regulates the vascular endothelial growth factor (VEGF) pathway in mouse EC [4, 5]. miR-92a is abundant in human EC. Overexpression of miR-92a blocked new blood vessel growth both in human umbilical vein endothelial cells (HUVECs) and zebrafish [6]. When limb ischemia and myocardial infarction in mice were treated with miR-92a antagomir to inhibit miR-92a expression, blood vessel growth and function were recovered [6]. miR-21 is also highly expressed in the arteries of arteriosclerosis obliterans [7]. Depletion of miR-21 prevents VSMC proliferation and activates cellular apoptosis [8].
Glucagon-like peptide 1 (GLP-1) is one of the most important incretin hormones and promotes insulin secretion after nutrient ingestion. However, GLP-1 is quickly digested by dipeptidyl peptidase-4 (DPP-4) in the circulation.
Dipeptidyl peptidase-4 inhibitors maintain glucose homeostasis by inhibiting the enzymatic activity of DPP-4 and increasing the concentration of GLP-1 in the circulation [9–11]. Dipeptidyl peptidase-4 is highly expressed in EC [12, 13]. DPP-4 can moderate vascular function through GLP-1 independent pathways [14]. Increasing evidence has shown the cardiovascular protection function of DPP-4 inhibitors [15]. Vildagliptin, a DPP-4 inhibitor, was approved to treat diabetes by the European Agency in 2008. In addition to glucose control, DPP-4 inhibitors also protect cardiovascular function. Sitagliptin attenuates infarct size in myocardial ischemia in rodents [16]. Vildagliptin attenuates the depressed heart rate variability (HRV), cardiac dysfunction, and cardiac mitochondrial dysfunction in rats fed a high-fat diet [17, 18]. Vildagliptin combined with metformin provides better cardiovascular outcomes than single therapy [19]. Because DPP-4 is found on endothelial cells, DPP-4 inhibitors may also have direct effects on the cardiovascular system, independent of the incretin system. Stromal cell-derived factor (SDF)-1α is known as a substrate for DPP-4, and DPP-4 inhibitors could increase SDF-1 concentrations. SDF-1α participates in healing of the injured myocardium [20]. Another proposed mechanism may be the reduction in inflammation by DPP-4 [21]. However, the exact mechanism of DPP-4 cardiovascular protection is still not clear.
Several studies have shown that some key miRNAs contributed to pancreatic β cells in GLP-1 agonist-treated diabetic rodents and patients, such as miR-7 [22], miR-9 [22], miR-375 [22–24], miR-27a [25], miR-139-5p [26], miR-132 [27], miR-212 [27]. However, the effect of GLP-1 agonists or DPP-4 inhibitors on the diabetic cardiovascular system is still unknown.
The purpose of this study was to find candidate miRNAs involved in the mechanism of vildagliptin in cardiovascular protection. We applied whole miRNA expression arrays and bioinformatics to seek the key miRNA and pathways in vildagliptin-treated diabetic rats.
Material and methods
Animal experiment
All animal experiments were approved by the Animal Care Committee of the Peking Union Medical Hospital Animal Ethics Committee (Project XHDW-2015-0051, 15 Feb 2015), and all efforts were made to minimize suffering. Five-week-old male Sprague-Dawley (SD) rats (163.3 ±18.8 g) were purchased from the Institute of Laboratory Animal Science, Chinese Academy of Medical Sciences and Peking Union Medical College (Beijing, China, SCXK-2014-0013). All rats were fed in a 12 : 12 h light-dark cycle (lights on between 7 AM to 7 PM) under controlled temperature (21 ±1°C) and humidity (50–60%). After adaptation, rats were divided into four groups: normal control (CON, n = 6), diabetic (DM, n = 6), Lvil (low dosage of vildagliptin, n = 6), and Hvil (high dosage of vildagliptin, n = 6). Normal control rats were fed a standard rodent diet (kcal %: 10% fat, 20% protein, and 70% carbohydrate; 3.85 kcal/gm). Meanwhile, the other three groups received a high-fat (HF) diet (kcal %: 45% fat, 20% protein, and 35% carbohydrate; 4.73 kcal/gm, Research Diet, New Brunswick, NJ, USA). Four weeks later, the diabetic, Lvil and Hvil groups were intraperitoneally injected with a low dosage of streptozotocin (STZ, 30 mg/kg of body weight). Rats with a fasting blood glucose > 11.1 mmol/l were considered diabetic. Then, the Lvil and Hvil groups were orally given 10 mg or 20 mg of vildagliptin/kg of body weight/day, respectively. The control and diabetic groups were administered normal saline. After 12 weeks of treatment, rats had food withdrawn overnight, and then they were anesthetized (ketamine 100 mg/kg i.p., Pharmacia and Upjohn Ltd., Crawley, UK). Blood samples were collected from the intraorbital retrobulbar plexus. Then, the rats were sacrificed by decapitation. The thoracic aortas were immediately collected. Parts of the aortas were placed in cold Krebs solution (120 mmol/l of NaCl, 4.7 mmol/l of KCl, 1.18 mmol/l of KH2PO4, 2.25 mmol/l of CaCl2, 24.5 mmol/l of NaHCO3, 1.2 mmol/l of MgSO4·7 H2O, 11.1 mmol/l of glucose, and 0.03 mmol/l of EDTA) aerated with 95% O2 and 5% CO2 and then were cut into 3 mm long rings for the vascular reactivity assay. The other aortas were frozen in liquid nitrogen and stored at –80°C for microarray and real-time PCR assays.
Body weight and fasting blood glucose measurements
The rats were weighed every 4 weeks. Fasting blood glucose (FBG) levels were measured using a Bayer Contour TS glucometer (Hamburg, Germany).
Oral glucose tolerance test
Glucose tolerance was determined using an oral glucose tolerance test (OGTT) at the end of 12 weeks of treatment. After 10 h of deprivation of food, rats were orally given 2 g glucose/kg of body weight. Blood glucose levels were measured before and 30, 60, and 120 min after glucose administration. The area under the curve (AUC) was obtained using the linear trapezoid method [28].
Serum insulin and lipid panel measurements
Fasting insulin levels were measured with an enzyme-linked immunosorbent assay kit (Millipore, Billerica, MA, USA). The homeostasis model assessment of insulin resistance (HOMA-IR) was calculated by the following formula: FBG (mmol/l) x fasting insulin (μIU/ml)/22.5. Serum total cholesterol (TC), triglyceride (TG), high-density lipoprotein-cholesterol (HDL-C) and low-density lipoprotein-cholesterol (LDL-C) were measured with the enzyme end-point method (Roche Diagnostics, GmbH, Mannheim, Germany).
Vascular reactivity measurement
The measurement of vascular reactivity was performed as described previously [29]. The aortic rings were placed in a tissue chamber filled with Krebs solution bubbled with 95% O2 and 5% CO2. The isometric contractile force of the rings was continuously measured by the BL-410 biological function system (Chengdu Tai Meng Science and Technology Co., Ltd., Chengdu, China). The basal passive tension was kept at 1.0 g. The relaxant responses of the endothelium-intact aortic rings precontracted with phenylephrine (Phe, 10–7 mmol/l) to acetylcholine (Ach, 10–10–10–4 mmol/l) were measured.
RNA extraction and miRNA expression analysis
Total RNA from the aorta was obtained using the mirVana RNA Isolation Kit (Ambion, San Paulo, SP, Brazil) and was quantified using the NanoDrop ND-2000 (Thermo Scientific). cDNA was synthesized using a TaqMan microRNA Reverse Transcription kit (Life Technologies, Rockville, MA, USA). miRNA profiles were obtained using an Affymetrix Multispecies miRNA 4.0 Array (Affymetrix Technologies, Santa Clara, CA, USA) in the Hvil group and DM group. Briefly, double-stranded cDNAs were synthesized from cRNAs. The second-cycle cDNAs were then biotin-labeled and hybridized onto a microarray. After washing and staining, the microarrays were scanned by an Affymetrix Scanner 3000 7G (Santa Clara, CA, USA). Raw data were obtained from array images by using Command Console Software (version 4.0, Affymetrix Technologies, Santa Clara, CA, USA). Raw data were normalized using an RMA algorithm in Expression Console software (version 1.4.1, Affymetrix Technologies, Santa Clara, CA, USA). After the significance (one-way ANOVA) analysis, differentially expressed miRNA were selected according to certain criteria (p < 0.05 and fold change > 1.5). A heat map was generated using TIGR MeV (MultiExperiment Viewer) software (http://www.tm4.org/mev.html) [30]. The microarray raw data have been submitted to the Gene Expression Omnibus (GEO) repository (GSE102195, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=qhijqekubnupdmv&acc=GSE102195 for reviewers only). The validated gene targets for differentially expressed miRNAs were searched in MiRTarVase Database version 6.0 (http:// http://mirtarbase.mbc.nctu.edu.tw/, released Sep 2015) [31].
miRNA quantitative PCR analysis
Three differentially expressed miRNAs were validated by qPCR analysis using the TaqMan PCR kit (Applied Biosystems, Foster City, CA, USA) and ABI 7700 (Applied Biosystems, Foster City, CA, USA). The reactions were incubated in a 96-well plate at 95°C for 10 min, followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. A comparative Ct method [32] was used to compare each group and the normal group. The relative levels of miRNAs were normalized to U6.
Target gene, inflammation gene, and apoptosis gene quantitative PCR analysis
Quantitative PCR analysis was performed using the SYBR Green method in ABI 7700 (Applied Biosystems, Foster City, CA, USA). The primers are listed in Table I. The reaction conditions were as follows: 48°C for 30 min, followed by 95°C for 15 min, and then 40 cycles of 95°C for 15 s and 55°C for 1 min. All signals were normalized to CypA (cytochrome P450 A). The comparative Ct method was used to assess the difference between treatment groups and the control group.
Table I.
Oligonucleotide sequences for qPCR analysis
| Gene symbol | GenBank ID | Forward primer | Reverse primer | Product size |
|---|---|---|---|---|
| Bndf | NM_001270630 | CTGTCGCACGGTCCCCATT | CCCGGTCTCATCAAAGCCTG | 102 |
| Ccl2 | NM_031530 | TGATCCCAATGAGTCGGCTG | TGGACCCATTCCTTATTGGGG | 127 |
| Pdk1 | NM_031081 | CCACCAGCCAGCTGTATGAC | CACTAGGAATGCCAGGGGAC | 120 |
| IL6 | NM_012589 | AGCGATGATGCACTGTCAGA | GGAACTCCAGAAGACCAGAGC | 127 |
| Casp3 | NM_012922 | GGAGCTTGGAACGCGAAGAA | ACACAAGCCCATTTCAGGGT | 169 |
Bdnf – brain-derived neurotrophic factor, Ccl2 – C-C motif chemokine ligand 2, Pdk1 – 3-phosphoinositide dependent protein kinase-1, IL6 – interleukin 6, Casp3 – caspase 3.
Target gene function and pathway analysis
To understand the biological function and pathways, target genes of miRNAs underwent gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses using DAVID (Data-base for annotation, visualization and integrated discovery) software (http://david.abcc.ncifcrf.gov/) [33]. The interaction of miRNA and genes was analyzed by Cytoscape software (http://www.cytoscape.org) [34].
Statistical analysis
All results were described as the mean ± standard deviation (SD). Variables were compared using ANOVA followed by Student’s t test. A p-value of < 0.05 was considered significant. GraphPad Prism software (version 5.0, San Diego, CA, USA) was used to complete the analyses.
Results
Effect of vildagliptin treatment on metabolic parameters in diabetic rats
Compared with the vehicle group, vildagliptin treatment reduced fasting blood glucose and AUC in OGTT (p < 0.01; Table II). Vildagliptin also reduced serum fasting insulin levels and the HOMA-IR index (p < 0.01; Table II). In blood lipids, vildagliptin suppressed TC and LDL-C levels (p < 0.01; Table II).
Table II.
The effects of vildagliptin on metabolic parameters in DM rats after treatment for 12 weeks
| Parameter | Groups | |||
|---|---|---|---|---|
| CON | DM | Lvil | Hvil | |
| Body weight [g] | 554.96 ±20.36 | 479.04 ±10.24** | 508.97 ±14.61**## | 496.26 ±21.49** |
| Fasting blood glucose [mmol/l] | 5.14 ±0.36 | 21.04 ±2.15** | 14.27 ±1.36**## | 16.88 ±1.78**## |
| AUCgluose [mmol/l/h] | 15.92 ±1.78 | 53.25 ±2.22** | 39.96 ±3.50**## | 42.03 ±5.59**## |
| Insulin [mIU/l] | 38.89 ±1.97 | 54.7384 ±6.39** | 37.15 ±6.09## | 36.98 ±7.06## |
| HOMA index | 8.87 ±0.62 | 50.95 ±5.61** | 23.37 ±3.35**## | 27.88 ±6.47**## |
| TC [mmol/l] | 1.39 ±0.05 | 2.40 ±0.36** | 1.66 ±0.27*## | 1.55 ±0.21## |
| TG [mmol/l] | 0.60 ±0.14 | 1.44 ±0.30** | 1.48 ±0.32** | 1.31 ±0.28** |
| HDL-C [mmol/l] | 0.59 ±0.08 | 0.42 ±0.08** | 0.59 ±0.10## | 0.50 ±0.08 |
| LDL-C [mmol/l] | 0.2 ±0.07 | 0.53 ±0.10** | 0.25 ±0.05## | 0.22 ±0.05## |
CON – control, DM – diabetes mellitus, Lvil – low dose of vildagliptin, Hvil – high dose of vildagliptin.
P < 0.05
p < 0.01 vs. normal control group;
p < 0.01 vs. diabetes group.
Effect of vildagliptin treatment on vascular dysfunction in diabetic rats
Vascular function was impaired by HF/STZ treatment (p < 0.01; Table III) and normalized by vildagliptin therapy (p < 0.01; Table III).
Table III.
Vasorelaxation responses (%) to Ach of thoracic aortic rings after treatment
| Group | Ach [mol/l] | ||||||
|---|---|---|---|---|---|---|---|
| 1 × 10–10 | 1 × 10–9 | 1 × 10–8 | 1 × 10–7 | 1 × 10–6 | 1 × 10–5 | 1 × 10–4 | |
| CON | 94.3 ±5.1 | 90.7 ±5.7 | 70.4 ±6.8 | 46.3 ±5.1 | 30.2 ±3.1 | 21.4 ±1.9 | 9.3 ±1.0 |
| DM | 97.4 ±2.1 | 95.7 ±4.9 | 80.4 ±5.3 | 56.4 ±5.7 | 41.7 ±4.3** | 35.3 ±3.2** | 25.4 ±2.7** |
| Lvil | 95.8 ±4.3 | 91.8 ±4.9 | 72.1 ±5.3 | 48.3 ±5.1 | 32.4 ±3.7 | 24.5 ±2.8 | 11.3 ±1.0## |
| Hvil | 95.8 ±4.1 | 92.3 ±5.3 | 78.4 ±4.3 | 51.3 ±4.9 | 37.4 ±4.1 | 31.3 ±3.1 | 15.3 ±1.6## |
CON – control, DM – diabetes mellitus, Lvil – low dose of vildagliptin, Hvil – high dose of vildagliptin.
P < 0.01 vs. control group;
p < 0.01 vs. diabetes group.
miRNA array result
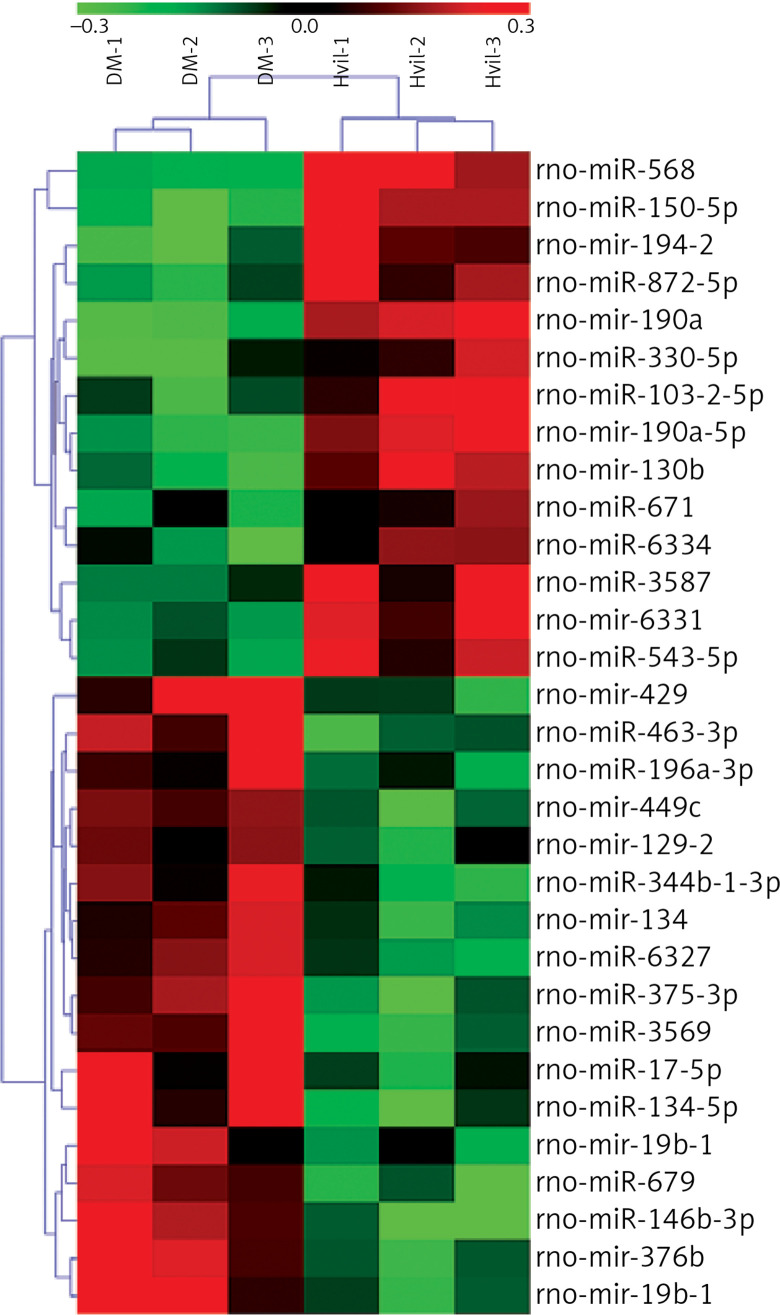
We identified 31 miRNAs that were differentially expressed in the Hvil group compared with the diabetic group (fold change > 1.5, p < 0.05; Table IV). In these 31 differentially expressed miRNAs, miR-568, miR-190a-5p, mir-190a, mir-194-2, miR-330-5p, miR-3587, miR-671, miR-103-2-5p, mir-130b, miR-872-5p, miR-6334, mir-6331, miR-150-5p and miR-543-5p were up-regulated; however, mir-449c, miR-463-3p, miR-344b-1-3p, miR-196a-3p, mir-129-2, mir-134, mir-376b, miR-375-3p, miR-17-5p, mir-19b-1, miR-6327, miR-679, mir-429, miR-3569, miR-134-5p, miR-146b-3p, and mir-19b-1 were down-regulated in the Hvil group compared with the diabetic group. The hierarchical clustering of these differentially expressed miRNAs showed that miRNA expression between the Hvil group and diabetic group was distinct (Figure 1).
Table IV.
Differential expression miRNAs identified in Hvil group (fold change < 1.5, p < 0.05)
| miRNA | Fold change | P-value | Mature sequence |
|---|---|---|---|
| rno-miR-568 | 2.616 | 0.0074 | AUGUAUAAAUGUAUACACAC |
| rno-miR-190a-5p | 2.234 | 0.0017 | UGAUAUGUUUGAUAUAUUAGGU |
| rno-mir-190a | 1.995 | 0.0009 | UGCAGGCCUCUGUGUGAUAUGUUUGAUAUAUUAGGUUGUUAUUUAAUCCAACUAUAUAUCAAGCAUAUUCCUACAGUGUCUUGCC |
| rno-mir-194-2 | 1.993 | 0.0162 | UGGCUCCCACCCCCUGUAACAGCAACUCCAUGUGGAAGUGCCCACUGAUUCCAGUGGGGCUGCUGUUAUCUGGGGUGGAGGCUGG |
| rno-miR-330-5p | 1.906 | 0.0329 | UCUCUGGGCCUGUGUCUUAGGC |
| rno-miR-3587 | 1.868 | 0.0482 | AACACUGAUUUCAAAUGGUGC |
| rno-miR-671 | 1.798 | 0.0480 | UCCGGUUCUCAGGGCUCCACC |
| rno-miR-103-2-5p | 1.757 | 0.0287 | AGCUUCUUUACAGUGCU |
| rno-mir-130b | 1.724 | 0.0043 | GGCUUGCUGGACACUCUUUCCCUGUUGCACUACUGUGGGCCUCUGGGAAGCAGUGCAAUGAUGAAAGGGCAUCCGUCAGGCC |
| rno-miR-872-5p | 1.650 | 0.0284 | AAGGUUACUUGUUAGUUCAGG |
| rno-miR-6334 | 1.648 | 0.0424 | CCAGGCUCUCCCAGCUGCCGGC |
| rno-mir-6331 | 1.611 | 0.0107 | CUCACACCGACAACUUUGAGGUGGCAACAGCUUUGGUGGCUUAGUUCUUUGUGCUUGGUUUGGGAGACACUGAGACUAACUCACCUGUCUGUUGUUGCUGUUAAAUCUUAAUCCAGAGGAACU |
| rno-miR-150-5p | 1.590 | 0.0411 | UCUCCCAACCCUUGUACCAGUG |
| rno-miR-543-5p | 1.554 | 0.0102 | AAGUUGCCCGCGUGUUUUUCG |
| rno-mir-449c | 0.666 | 0.0130 | AAGCGUGGGUGUGUCAGACAGGCAGUGCAUUGCUAGCUGGCUGUUAGAACUUCGUCCCAACAGUGGCUAGCUGCACUACCCUCUGCUGCACUUAGAAGC |
| rno-miR-463-3p | 0.666 | 0.0167 | UGAUAGACGCCAAUUUGGGUAG |
| rno-miR-344b-1-3p | 0.660 | 0.0224 | GAUAUAACCAAAGCCCGACUGU |
| rno-miR-196a-3p | 0.658 | 0.0448 | UCGGCAACAAGAAACUGCCUGA |
| rno-mir-129-2 | 0.649 | 0.0425 | AGACUGCCCUUCGCGAAUCUUUUUGCGGUCUGGGCUUGCUGUACAUAACUCAAUAGCCGGAAGCCCUUACCCCAAAAAGCAUUCGCGGAGGGCGCGC |
| rno-mir-134 | 0.627 | 0.0187 | CAGGGUGUGUGACUGGUUGACCAGAGGGGCGUGCACUUUGUUCACCCUGUGGGCCACCUAGUCACCAACCCUC |
| rno-mir-376b | 0.622 | 0.0187 | UUUGGUAUUUAAAAGGUGGAUAUUCCUUCUAUGGUUACGUGCUUCCUGGAUAAUCAUAGAGGAACAUCCACUUUUUCAGUAUCAAA |
| rno-miR-375-3p | 0.615 | 0.0151 | UUUGUUCGUUCGGCUCGCGUGA |
| rno-miR-17-5p | 0.580 | 0.0403 | CAAAGUGCUUACAGUGCAGGUAG |
| rno-mir-19b-1 | 0.539 | 0.0405 | CACUGGUCUAUGGUUAGUUUUGCAGGUUUGCAUCCAGCUGUAUAAUAUUCUGCUGUGCAAAUCCAUGCAAAACUGACUGUGGUGGUG |
| rno-miR-6327 | 0.523 | 0.0097 | CGGACUGUAGAUCCAUCUC |
| rno-miR-679 | 0.499 | 0.0114 | CAAGGUCUUCCUCACAGUAGC |
| rno-mir-429 | 0.495 | 0.0280 | UGCCUGCUGAUGGAUGUCUUACCAGACAUGGUUAGAUCUGGAUGUAUCUGUCUAAUACUGUCUGGUAAUGCCGUCCAUCCAUGGC |
| rno-miR-3569 | 0.478 | 0.0197 | GGAGGACAGCAGACUCAGGUC |
| rno-miR-134-5p | 0.453 | 0.0390 | UGUGACUGGUUGACCAGAGGGG |
| rno-miR-146b-3p | 0.418 | 0.0094 | CCUAGGGACUCAGUUCUGGUG |
| rno-mir-19b-1 | 0.390 | 0.0280 | CACUGGUCUAUGGUUAGUUUUGCAGGUUUGCAUCCAGCUGUAUAAUAUUCUGCUGUGCAAAUCCAUGCAAAACUGACUGUGGUGGUG |
Figure 1.
Heat map of differentially expressed miRNAs in Hvil and diabetic groups. Red represents up-regulation; green indicates down-regulation
DM – diabetes mellitus, Hvil – high dose of vildagliptin.
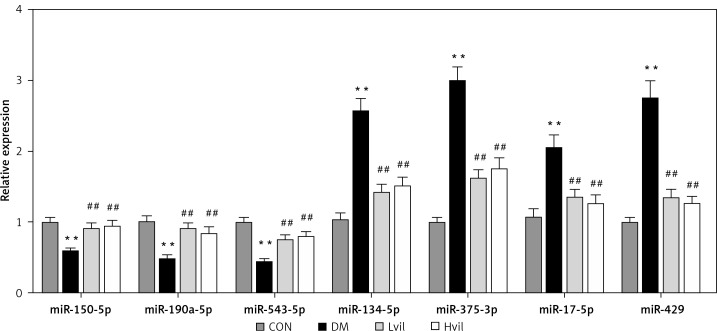
Validation of miRNAs by qPCR
For the validation of the miRNA array results, we chose 7 miRNAs to perform miRNA qPCR. These 7 miRNAs have experimental validated target genes in the miRTarBase Database. The results showed that seven differentially expressed miRNAs tested by miRNA array were changed significantly in the Hvil group compared with the diabetic group (Figure 2). The results of the qPCR assay agreed with the miRNA array.
Figure 2.
Validation of differentially expressed miRNAs by using qPCR. Data are presented as mean ± SD (n = 6)
**P < 0.01 vs. control group, ##p < 0.01 vs. diabetic group.
CON – control, DM – diabetes mellitus, Lvil – low dose of vildagliptin, Hvil – high dose of vildagliptin.
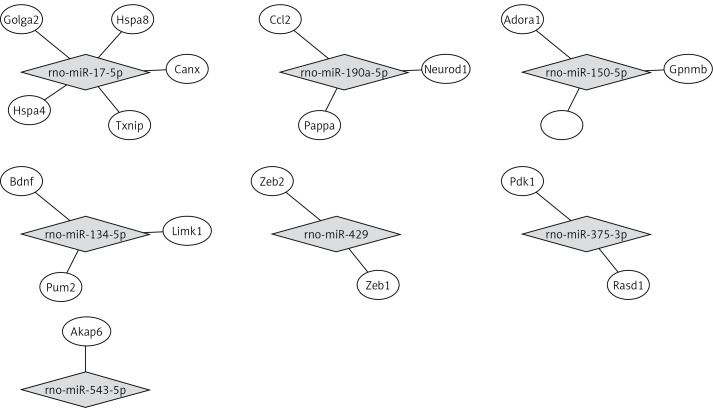
miRNA target biological function and pathway analysis
By using the miRTarBase Database, we found 19 validated target genes from 7 miRNAs (Table V). Figure 3 shows a miRNA-mRNA interaction network using Cytoscape. To understand the function of the target genes of differentially expressed miRNAs, we performed biological function and pathway analysis using DAVID software. We found that the sarcoplasmic reticulum, focal adhesion in the cellular component category and the antigen processing and presentation pathway were involved in the target gene function (Table VI).
Table V.
Validated miRNA target for differentially expressed miRNAs
| miRNA | Regulation | Target genes |
|---|---|---|
| rno-miR-150-5p | Up | Adora1, Gpnmb |
| rno-miR-190a-5p | Up | Ccl2, Neurod1, Pappa |
| rno-miR-543-5p | Up | Akap6 |
| rno-miR-134-5p | Down | Bdnf, Limk1, Pum2 |
| rno-miR-375-3p | Down | Rasd1, Pdk1 |
| rno-miR-17-5p | Down | Hspa8, Canx, Golga2, Txnip, Hspa4 |
| rno-miR-429 | Down | Zeb1, Zeb2 |
Figure 3.
The interaction between miRNA and target genes
Table VI.
The miRNA target gene-associated biological function category and pathway (p < 0.05)
| Term ID | Term name | P-value | Genes | Fold enrichment |
|---|---|---|---|---|
| GO analysis: | ||||
| 0016529 | Sarcoplasmic reticulum | 0.0217 | AKAP6, RASD1 | 85.894 |
| 0005925 | Focal adhesion | 0.0468 | LIMK1, VIM, HSPA8 | 8.103 |
| KEGG pathway: | ||||
| 0004612 | Antigen processing and presentation | 0.0059 | HSPA4, CANX, HSPA8 | 23.332 |
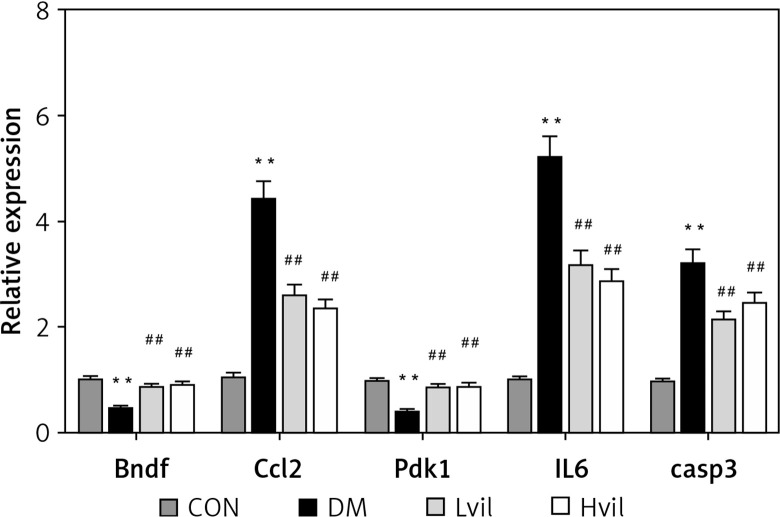
Target gene qPCR
Real-time PCR results showed that the expression of brain-derived neurotrophic factor (Bdnf) and 3-phosphoinositide-dependent protein kinase-1 (Pdk1) in the Hvil group was significantly higher than in the diabetic group; however, the expression of chemokine (C-C) motif ligand 2 (Ccl2) in the Hvil group was significantly lower than in the diabetic group (Figure 4).
Figure 4.
Target genes, inflammation gene, and apoptosis gene expression by using qPCR. Data are presented as mean ± SD (n = 6)
**P < 0.01 vs. control group, ##p < 0.01 vs. diabetic group. CON – control, DM – diabetes mellitus, Lvil – low dose of vildagliptin, Hvil – high dose of vildagliptin, Bdnf – brain-derived neurotrophic factor, Ccl2 – C-C motif chemokine ligand 2, Pdk1 – 3-phosphoinositide dependent protein kinase-1, IL6 – interleukin 6, Casp3 – caspase 3.
Inflammation gene and apoptosis gene expression
Diabetic rats had higher interleukin 6 (Il6) and caspase 3 (casp3) gene expression than those in the control group (p < 0.01; Figure 4). Vildagliptin treatment reduced Il6 and Casp3 gene expression in diabetic aorta (p < 0.01; Figure 4).
Discussion
In our study, we found that vildagliptin reduced blood glucose, TC and LDL-C, and moderated insulin sensitivity. Similar results were obtained for the effect of DPP-4 inhibitors on glucose and lipid metabolism. Saxagliptin treatment for 24 weeks improved total cholesterol in type-2 diabetic patients [35].
We also found that vildagliptin moderated endothelial function in diabetic rats. Increasing evidence has shown that DPP-4 inhibitors improve vascular function. Alogliptin leads to dose-dependent relaxation of aortic rings, even under GLP-1 receptor antagonist pretreatment [14]. In HUVECs, alogliptin induces phosphorylation of the Akt-eNOS pathway and nitric oxide (NO) release [14]. DPP-4 inhibitors improved NO release and reduced inflammation in endothelial cells [36].
We found that miR-134-5p was down-regulated in the vildagliptin-treated group. Bdnf is a target of miR-134-5p [37]. We also found that the expression of Bdnf was up-regulated in the vildagliptin-treated group. BDNF is a major member of the neurotrophin (NT) family. Patients suffering acute coronary syndrome (ACS) [38], acute myocardial infarction (MI) [39], metabolic syndrome [40] and type-2 diabetes [41, 42] have decreased circulating BDNF levels. Decreased circulating BDNF levels have been associated with endothelial dysfunction [43]. Endothelial dysfunction is associated with cardiovascular diseases and risk factors such as hypertension [44]. BDNF is considered a link between endothelial dysfunction and cardiovascular diseases. Recently, evidence has shown that BDNF expression is high in the rat heart and aorta. Cardiovascular BDNF is prominent in ECs [45]. Spontaneously hypertensive rats have reduced expression of BDNF in ECs. Physical training normalizes the endothelial BDNF levels in both normal rats and spontaneously hypertensive rats [45]. Thus, vildagliptin may inhibit the expression of miR-134-5p to stimulate the expression of Bdnf and moderate endothelial function.
Moreover, we found that miR-190a-5p expression was up-regulated in the vildagliptin group. Ccl2 is one of the target genes of miR-190a-5p [46]. CCL2, also known as MCP-1, was the first human chemokine to be described [47, 48]. CCL2 can attract monocytes, such as macrophages, and microglia, in vitro and in vivo [49, 50]. CCL2 is an important chemokine for atherogenesis. Pro-inflammatory mediators induce atherosclerotic inflammation in the aorta [51, 52]. We also found that vildagliptin reduced Il6 expression in diabetic aorta. Thus, vildagliptin could activate miR-190a-5p to inhibit CCL2 expression to inhibit inflammation in the aorta.
In our research, miR-375-3p was down-regulated in the vildagliptin-treated group. Pdk1 is one of the target genes of miR-375-3p [53]. PDK1 has an anti-apoptotic function in the development of atherosclerosis [54]. The activation of PDK1 could activate the PI3K/Akt pathway. The PI3K/Akt pathway plays a prominent role in cell survival, metabolism, growth and proliferation by directly regulating apoptotic proteins such as Bcl-2, caspase-3, caspase-9 and Bax [55, 56]. In our study, vildagliptin inhibited the expression of Casp3 in diabetic aorta. Thus, vildagliptin may inhibit Casp3 through reducing miR-375-3p in diabetic aorta.
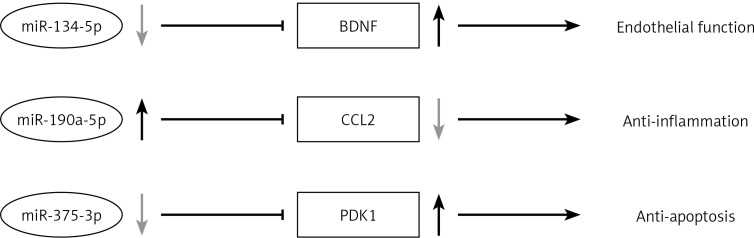
Our present study shows that vildagliptin elicits vascular protection in diabetic rats. This study provides promising evidence of DPP-4 inhibitors’ application in diabetic patients with cardiovascular disorders. Vildagliptin activates miR-190a-5p and inhibits miR-134-5p and miR-375-3p expression in the aorta to activate Bdnf and Pdk1. Vildagliptin inhibits Ccl2 to moderate endothelial function and inhibits inflammation and apoptosis (Figure 5). Involved miRNAs function still needs to be verified in vivo and in intro. Further study of the key miRNAs in the pathogenesis of the cardiovascular system will help us find the potential therapeutic targets.
Figure 5.
Vildagliptin inhibit miR-134-5p in aorta to activate Bdnf to moderate endothelial function; activate miR-190a-5p to inhibit Ccl2; and inhibit miR-375-3p to activate Pdk1 to inhibit apoptosis.
Bdnf – brain-derived neurotrophic factor, Ccl2 – C-C motif chemokine ligand 2, Pdk1 – 3-phosphoinositide dependent protein kinase-1.
Acknowledgments
This work was supported by the grants from National Key R&D Program of China (2017YFC1309603), National Key Research and Development Program of China (2016YFA0101002), National Natural Science Foundation of China (No. 81170736, 81570715, 81870579, 81870545), National Natural Science Foundation for Young Scholars of China (No. 81300649), China Scholarship Council foundation (201308110443), PUMC Youth Fund (33320140022) and Fundamental Research Funds for the Central Universities, and Scientific Activities Foundation for Selected Returned Overseas Professionals of Human Resources and Social Security Ministry. We are very grateful to Beijing Compass Biotechnology Company for excellent technical assistance with the microarray experiments.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Sliwinska A, Kasinska MA, Drzewoski J. MicroRNAs and metabolic disorders − where are we heading? Arch Med Sci. 2017;13:885–96. doi: 10.5114/aoms.2017.65229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quintavalle M, Condorelli G, Elia L. Arterial remodeling and atherosclerosis: miRNAs involvement. Vascul Pharmacol. 2011;55:106–10. doi: 10.1016/j.vph.2011.08.216. [DOI] [PubMed] [Google Scholar]
- 3.Elia L, Condorelli G. RNA (Epi)genetics in cardiovascular diseases. J Mol Cell Cardiol. 2015;89:11–6. doi: 10.1016/j.yjmcc.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Fish JE, Santoro MM, Morton SU, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–84. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261–71. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonauer A, Carmona G, Iwasaki M, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–3. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 7.Wang M, Li W, Chang GQ, et al. MicroRNA-21 regulates vascular smooth muscle cell function via targeting tropomyosin 1 in arteriosclerosis obliterans of lower extremities. Arterioscler Thromb Vasc Biol. 2011;31:2044–53. doi: 10.1161/ATVBAHA.111.229559. [DOI] [PubMed] [Google Scholar]
- 8.Ji R, Cheng Y, Yue J, et al. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of MicroRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–88. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- 9.Dicker D. DPP-4 inhibitors: impact on glycemic control and cardiovascular risk factors. Diabetes Care. 2011;34(Suppl 2):S276–8. doi: 10.2337/dc11-s229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chrysant SG, Chrysant GS. Clinical implications of cardiovascular preventing pleiotropic effects of dipeptidyl peptidase-4 inhibitors. Am J Cardiol. 2012;109:1681–5. doi: 10.1016/j.amjcard.2012.01.398. [DOI] [PubMed] [Google Scholar]
- 11.Kubota Y, Miyamoto M, Takagi G, et al. The dipeptidyl peptidase-4 inhibitor sitagliptin improves vascular endothelial function in type 2 diabetes. J Korean Med Sci. 2012;27:1364–70. doi: 10.3346/jkms.2012.27.11.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drucker DJ, Nauck MA. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet. 2006;368:1696–705. doi: 10.1016/S0140-6736(06)69705-5. [DOI] [PubMed] [Google Scholar]
- 13.Moritoh Y, Takeuchi K, Asakawa T, Kataoka O, Odaka H. Chronic administration of alogliptin, a novel, potent, and highly selective dipeptidyl peptidase-4 inhibitor, improves glycemic control and beta-cell function in obese diabetic ob/ob mice. Eur J Pharmacol. 2008;588:325–32. doi: 10.1016/j.ejphar.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 14.Shah Z, Pineda C, Kampfrath T, et al. Acute DPP-4 inhibition modulates vascular tone through GLP-1 independent pathways. Vascul Pharmacol. 2011;55:2–9. doi: 10.1016/j.vph.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomovic K, Lazarevic J, Kocic G, Deljanin-Ilic M, Anderluh A, Smelcerovic A. Mechanisms and pathways of anti-inflammatory activity of DPP-4 inhibitors in cardiovascular and renal protection. Med Res Rev. 2019;39:404–22. doi: 10.1002/med.21513. [DOI] [PubMed] [Google Scholar]
- 16.Ye Y, Keyes KT, Zhang C, Perez-Polo JR, Lin Y, Birnbaum Y. The myocardial infarct size-limiting effect of sitagliptin is PKA-dependent, whereas the protective effect of pioglitazone is partially dependent on PKA. Am J Physiol Heart Circ Physiol. 2010;298:H1454–65. doi: 10.1152/ajpheart.00867.2009. [DOI] [PubMed] [Google Scholar]
- 17.Apaijai N, Pintana H, Chattipakorn SC, Chattipakorn N. Cardioprotective effects of metformin and vildagliptin in adult rats with insulin resistance induced by a high-fat diet. Endocrinology. 2012;153:3878–85. doi: 10.1210/en.2012-1262. [DOI] [PubMed] [Google Scholar]
- 18.Apaijai N, Pintana H, Chattipakorn SC, Chattipakorn N. Effects of vildagliptin versus sitagliptin, on cardiac function, heart rate variability and mitochondrial function in obese insulin-resistant rats. Br J Pharmacol. 2013;169:1048–57. doi: 10.1111/bph.12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apaijai N, Chinda K, Palee S, Chattipakorn S, Chattipakorn N. Combined vildagliptin and metformin exert better cardioprotection than monotherapy against ischemia-reperfusion injury in obese-insulin resistant rats. PLoS One. 2014;9:e102374. doi: 10.1371/journal.pone.0102374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fadini GP, Avogaro A. Potential manipulation of endothelial progenitor cells in diabetes and its complications. Diabetes Obes Metab. 2010;12:570–83. doi: 10.1111/j.1463-1326.2010.01210.x. [DOI] [PubMed] [Google Scholar]
- 21.Dobrian AD, Ma Q, Lindsay JW, et al. Dipeptidyl peptidase IV inhibitor sitagliptin reduces local inflammation in adipose tissue and in pancreatic islets of obese mice. Am J Physiol Endocrinol Metab. 2011;300:E410–21. doi: 10.1152/ajpendo.00463.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo C, Sun YQ, Li Q, Zhang JC. MiR-7, miR-9 and miR-375 contribute to effect of Exendin-4 on pancreatic beta-cells in high-fat-diet-fed mice. Clin Invest Med. 2018;41:E16–E24. doi: 10.25011/cim.v41i1.29459. [DOI] [PubMed] [Google Scholar]
- 23.Lee IS, Park KC, Yang KJ, et al. Exenatide reverses dysregulated microRNAs in high-fat diet-induced obese mice. Obes Res Clin Pract. 2016;10:315–26. doi: 10.1016/j.orcp.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 24.Keller DM, Clark EA, Goodman RH. Regulation of micro-RNA-375 by cAMP in pancreatic beta-cells. Mol Endocrinol. 2012;26:989–99. doi: 10.1210/me.2011-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao Y, Xu Y, Wang W, Zhang J, Li Q. Glucagon-like peptide-1 improves beta-cell dysfunction by suppressing the miR-27a-induced downregulation of ATP-binding cassette transporter A1. Biomed Pharmacother. 2017;96:497–502. doi: 10.1016/j.biopha.2017.10.049. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Su L, Gong YY, et al. Downregulation of miR-139-5p contributes to the antiapoptotic effect of liraglutide on the diabetic rat pancreas and INS-1 cells by targeting IRS1. PloS One. 2017;12:e0173576. doi: 10.1371/journal.pone.0173576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang J, Li J, Keller MP, et al. Induction of miR-132 and miR-212 expression by glucagon-like peptide 1 (GLP-1) in rodent and human pancreatic beta-cells. Mol Endocrinol. 2015;29:1243–53. doi: 10.1210/me.2014-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Q, Sun X, Xiao X, et al. Maternal chromium restriction leads to glucose metabolism imbalance in mice offspring through insulin signaling and Wnt signaling pathways. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17101767.E1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumoto S, Shimabukuro M, Fukuda D, et al. Azilsartan, an angiotensin II type 1 receptor blocker, restores endothelial function by reducing vascular inflammation and by increasing the phosphorylation ratio Ser(1177)/Thr(497) of endothelial nitric oxide synthase in diabetic mice. Cardiovasc Diabetol. 2014;13:30. doi: 10.1186/1475-2840-13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saeed AI, Bhagabati NK, Braisted JC, et al. TM4 micro-array software suite. Methods Enzymol. 2006;411:134–93. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 31.Chou CH, Chang NW, Shrestha S, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44:D239–47. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 34.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenstock J, Aguilar-Salinas C, Klein E, Nepal S, List J, Chen R; Investigators CVS. Effect of saxagliptin monotherapy in treatment-naive patients with type 2 diabetes. Curr Med Res Opin. 2009;25:2401–11. doi: 10.1185/03007990903178735. [DOI] [PubMed] [Google Scholar]
- 36.Mason RP, Jacob RF, Kubant R, et al. Effect of enhanced glycemic control with saxagliptin on endothelial nitric oxide release and CD40 levels in obese rats. J Atheroscler Thromb. 2011;18:774–83. doi: 10.5551/jat.7666. [DOI] [PubMed] [Google Scholar]
- 37.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–66. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 38.Manni L, Nikolova V, Vyagova D, Chaldakov GN, Aloe L. Reduced plasma levels of NGF and BDNF in patients with acute coronary syndromes. Int J Cardiol. 2005;102:169–71. doi: 10.1016/j.ijcard.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 39.Lorgis L, Amoureux S, de Maistre E, et al. Serum brain-derived neurotrophic factor and platelet activation evaluated by soluble P-selectin and soluble CD-40-ligand in patients with acute myocardial infarction. Fundam Clin Pharmacol. 2010;24:525–30. doi: 10.1111/j.1472-8206.2009.00790.x. [DOI] [PubMed] [Google Scholar]
- 40.Chaldakov GN, Fiore M, Stankulov IS, et al. Neurotrophin presence in human coronary atherosclerosis and metabolic syndrome: a role for NGF and BDNF in cardiovascular disease? Prog Brain Res. 2004;146:279–89. doi: 10.1016/S0079-6123(03)46018-4. [DOI] [PubMed] [Google Scholar]
- 41.Krabbe KS, Nielsen AR, Krogh-Madsen R, et al. Brain-derived neurotrophic factor (BDNF) and type 2 diabetes. Diabetologia. 2007;50:431–8. doi: 10.1007/s00125-006-0537-4. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Zhang SW, Khandekar N, et al. Reduced serum levels of oestradiol and brain derived neurotrophic factor in both diabetic women and HFD-feeding female mice. Endocrine. 2017;56:65–72. doi: 10.1007/s12020-016-1197-x. [DOI] [PubMed] [Google Scholar]
- 43.Golden E, Emiliano A, Maudsley S, et al. Circulating brain-derived neurotrophic factor and indices of metabolic and cardiovascular health: data from the Baltimore Longitudinal Study of Aging. PLoS One. 2010;5:e10099. doi: 10.1371/journal.pone.0010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin BJ, Anderson TJ. Risk prediction in cardiovascular disease: the prognostic significance of endothelial dysfunction. Can J Cardiol. 2009;25(Suppl A):15A–20A. doi: 10.1016/s0828-282x(09)71049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prigent-Tessier A, Quirié A, Maguin-Gaté K, et al. Physical training and hypertension have opposite effects on endothelial brain-derived neurotrophic factor expression. Cardiovasc Res. 2013;100:374–82. doi: 10.1093/cvr/cvt219. [DOI] [PubMed] [Google Scholar]
- 46.Kuang H, Han D, Xie J, Yan Y, Li J, Ge P. Profiling of differentially expressed microRNAs in premature ovarian failure in an animal model. Gynecol Endocrinol. 2014;30:57–61. doi: 10.3109/09513590.2013.850659. [DOI] [PubMed] [Google Scholar]
- 47.Rollins BJ. Monocyte chemoattractant protein 1: a potential regulator of monocyte recruitment in inflammatory disease. Mol Med Today. 1996;2:198–204. doi: 10.1016/1357-4310(96)88772-7. [DOI] [PubMed] [Google Scholar]
- 48.Yoshimura T, Robinson EA, Tanaka S, Appella E, Kuratsu J, Leonard EJ. Purification and amino acid analysis of two human glioma-derived monocyte chemoattractants. J Exp Med. 1989;169:1449–59. doi: 10.1084/jem.169.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshimura T, Leonard EJ. Human monocyte chemoattractant protein-1: structure and function. Cytokines. 1992;4:131–52. [PubMed] [Google Scholar]
- 50.Rollins BJ. JE/MCP-1: an early-response gene encodes a monocyte-specific cytokine. Cancer Cells. 1991;3:517–24. [PubMed] [Google Scholar]
- 51.Ait-Oufella H, Taleb S, Mallat Z, Tedgui A. Recent advances on the role of cytokines in atherosclerosis. Arterioscler Thromb Vasc Biol. 2011;31:969–79. doi: 10.1161/ATVBAHA.110.207415. [DOI] [PubMed] [Google Scholar]
- 52.Verhamme P, Quarck R, Hao H, et al. Dietary cholesterol withdrawal reduces vascular inflammation and induces coronary plaque stabilization in miniature pigs. Cardiovasc Res. 2002;56:135–44. doi: 10.1016/s0008-6363(02)00515-1. [DOI] [PubMed] [Google Scholar]
- 53.El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E. miR-375 targets 3’-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes. 2008;57:2708–17. doi: 10.2337/db07-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Yang C, Zhang L, Yang P. MicroRNA-210 induces endothelial cell apoptosis by directly targeting PDK1 in the setting of atherosclerosis. Cell Mol Biol Lett. 2017;22:3. doi: 10.1186/s11658-017-0033-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao H, Wang H, Peng J. Hispidulin induces apoptosis through mitochondrial dysfunction and inhibition of P13k/Akt signalling pathway in HepG2 cancer cells. Cell Biochem Biophys. 2014;69:27–34. doi: 10.1007/s12013-013-9762-x. [DOI] [PubMed] [Google Scholar]
- 56.Pathania AS, Guru SK, Verma MK, et al. Disruption of the PI3K/AKT/mTOR signaling cascade and induction of apoptosis in HL-60 cells by an essential oil from Monarda citriodora. Food Chem Toxicol. 2013;62:246–54. doi: 10.1016/j.fct.2013.08.037. [DOI] [PubMed] [Google Scholar]