Abstract
Air pollution is a serious threat to our health and has become one of the major causes of many diseases including cardiovascular disease, respiratory disease, and cancer. The association between air pollution and various diseases has long been a topic of research interest. However, it remains unclear how air pollution actually impacts health by modulating several important cellular functions. Recently, some evidence has emerged about air pollution-induced epigenetic changes, which are linked with the etiology of various human diseases. Among several epigenetic modifications, DNA methylation represents the most prominent epigenetic alteration underlying the air pollution-induced pathogenic mechanism. Several other types of epigenetic changes, such as histone modifications, miRNA, and non-coding RNA expression, have also been found to have been linked with air pollution. Hypersensitivity pneumonitis (HP), one of the most prevalent forms of interstitial lung diseases (ILDs), is triggered by the inhalation of certain organic and inorganic substances. HP is characterized by inflammation in the tissues around the lungs’ airways and may lead to irreversible lung scarring over time. This review, in addition to other diseases, attempts to understand whether certain pollutants influence HP development through such epigenetic modifications.
Keywords: Air pollution, Particulate matter, Epigenetic modifications, Hypersensitivity pneumonitis, Epigenetic biomarkers
Introduction
Air pollution has become a global threat to human health as it predisposes the risks of various diseases including cancers. In today’s world, a whole range of industries such as petrochemical firms, chemical factories, fertilizer units, and metallurgical setups contribute to air pollution. Apart from the industrial sector, the main emissions from certain types of power stations, cars, railways, airways, combustion engines also cause air pollution. Certain field-cultivation techniques release ammonia in the air from heavily fertilized fields and consequently pollute the environment. Even natural disasters like forest fire, volcanic erosion, and dust storms pollute the air. Other than these, biomass fuel uses, agricultural burning and utilization of adulterated fuels are some of the sources of air pollution (Manisalidis et al. 2020). The use of wood and charcoal for cooking and the operation of computers, printers, and photocopy machines, among others, contribute to indoor air pollution. Rapid urbanization also contributes to air pollution as it involves huge construction works. Moreover, some people are exposed to air pollution due to their professions—for instance, mining workers, traffic police and drivers are exposed to large amounts of air pollutants (Shukla et al. 2019).
Air pollution is linked with a range of diseases like asthma, chronic bronchitis, chronic obstructive pulmonary disease (COPD), cardiovascular disease, metabolic disorders, various reproductive conditions, dysregulation in immune response, neurodegenerative disorders, and cancer (Breton and Marutani 2014; Kurt et al. 2016). In 2013, World Health Organization (WHO) declared outdoor air pollution as a leading environmental cause of death associated with cancer (WHO 2013). A new study has indicated that air pollution may increase the risk of death due to COVID-19, an infectious disease that emerged in 2019 and is caused by novel coronavirus SARS-CoV-2. It has been suggested that 15% deaths across the world due to covid-19 could be linked with long-term exposure to air pollution (Pozzer et al. 2020). Outdoor air is contaminated with various chemicals such as engine exhausts, solvents, metals, dusts, and particulate matter (PM). Although air pollution is linked with numerous diseases, the impact of pollutants at both cellular and molecular levels has not been explored in detail. At the cellular level, exposure to different air pollutants or other toxic elements may lead to apoptosis by targeting mitochondria either directly or indirectly. Airborne particles have been found to induce apoptosis associated with ROS production, cellular stress, and DNA fragmentation (Andreau et al. 2012). Air pollution also induces inflammation in tissues by influencing specific cytokines and may also impact our endocrine system and metabolism processes (Prada et al. 2020). Moreover, recent studies indicate that epigenetic alterations could be an important pathway through which environmental factors exert their effects (Alfano et al. 2018). Epigenetic is the alteration at the gene expression level without changing the underlying DNA sequence such as DNA methylation, histone modification, miRNA, and non-coding RNA expression (Weinhold 2006; Jin et al. 2011). Cancers and many other diseases are often associated with epigenetic modifications (Weinhold 2006). Exposure to environmental stimuli may result in epigenetic changes, which, in turn, can impact gene expression and predisposition to disease risk (Breton and Marutani 2014). Little is known about how specific air pollutants generate specific epigenetic marks associated with a particular disease. Understanding epigenetic alterations due to specific pollutants may lead to the development of biomarkers to assess the disease risk due to air pollution.
This narrative review provides an overview of the association between air pollution-induced epigenetic modification and disease development. It also explores whether such modification plays a role in HP pathogenesis. We have searched the PubMed and MEDLINE databases using the keywords ‘air pollution’, ‘air pollutant’, ‘air pollution-associated diseases’, ‘epigenetics’, and ‘hypersensitivity pneumonitis’. We have included all the original research articles published on the topic in the last 20 years. Overall, this review paper summarizes the findings of 235 articles, including 145 original research papers, 76 review articles, 6 systematic review and meta-analyses, 6 clinical trials, and 2 comparative studies. We have only included the studies where disease development is associated with air pollutants as well as with epigenetic changes. We have not included studies in which air pollution induces mutations or other alterations at the genetic level or causes other physiological changes associated with disease pathogenesis.
The objective of this review is to discuss the association between certain air pollutants, epigenetic modifications, and diseases linked with those pollutants. These epigenetic alterations could be the mechanisms underlying the diseases associated with air pollution. Such knowledge may guide the prevention and treatment of the diseases. We also discuss HP pathogenesis because certain pollutants could influence HP development, and epigenetic modifications could be a mechanism involved in this process. Moreover, this review will help researchers to develop epigenetic biomarkers for early detection and monitoring prognosis of diseases associated with air pollution. Other mechanisms involved in air pollution-associated disease development have not been discussed in this review article.
Air pollution: an overview
Ambient air pollution is a mixture of many different components comprising airborne PM, ozone, nitrogen dioxide (NO2), and volatile organic compounds, including benzene, carbon monoxide (CO), and sulfur dioxide (SO2). Primary pollutants are emitted directly into the air, while secondary pollutants are formed in the atmosphere from other components. Industrial sources, traffic, and residential heating are the major sources of air pollution (Breton and Marutani 2014; WHO 2013). In 2013, the International Agency for Research on Cancer (IARC) declared outdoor air pollution and the associated PM as a potential risk factor for lung cancer. Globally, the mutagenic potential of outdoor air exceeds the normal range over six orders of magnitude (revertants/m3) (Loomis et al. 2014; Cohen 2000). There is Increasing evidence to suggest that both acute and chronic exposure to air pollution is detrimental to human health and pregnant women, infants, and individuals with existing diseases, as they are more susceptible to the adverse effects of ambient air pollution (Kloog et al. 2012; Vieira 2015).
Particulate matters (PM)
PM is the sum of hazardous solid and liquid particles suspended in the air. They comprise both organic and inorganic particles such as dust, pollen, and liquid droplets. PM can be classified as coarse (diameter 10 μm; PM10), fine (diameter 2.5 μm; PM2.5), and ultrafine (0.1 μm; PM0.1) particles on the basis of their sizes (Ji and Khurana Hershey 2012). Coarse PM is generally produced from dust on roads, dust generated from construction works and industrial emissions, while combustion processes form fine particles composed of metals, organic molecules, sulfate, nitrate etc. These fine particles are capable of travelling long distances, at times more than 100 km (WHO 2006). However, PM concentration varies by location and is generally more toxic during working hours due to heavy traffic. PM-induced diseases are listed in detail in Table 1. Other than inducing diverse diseases, PM2.5 also alters epigenetic age, which is a marker of mortality and disease risk (White et al. 2019). Another work has shown that PM2.5 exposure promotes circadian rhythm disruption and metabolic dysfunction by downregulating histone deacetylases 2, 3, and 4, thereby altering the chromatin dynamics (Palanivel et al. 2020). Air pollution-induced changes in the transcriptomic profile has also been reported. PM2.5 exposure at birth is linked with differential gene expressions in early stages of life, which is relevant for complex diseases (Merid et al. 2021). It was observed that depending on the concentration and duration of PM2.5 exposure, a distinct transcriptomic and DNA methylomic profile is generated in bronchial epithelial cells (Huang et al. 2021).
Table 1.
Diverse disease risks due to air pollution-mediated epigenetic changes
| Air pollutants | Epigenetic mark | Diseases/Symptoms | References | |
|---|---|---|---|---|
| 1 | PM2.5 | Promoter methylation of the tumor suppressor p16 gene | Lung cancer | Soberanes et al. 2012 |
| 2 | PM2.5 | SOX2 promoter hypermethylation | Lung Cancer | Tantoh et al. 2019 |
| 3 | PM2.5 | miRNA mediated deregulation in gene expression . | Lung Cancer | Ning et al. 2019 |
| 4 | PM2.5 | H3K27ac (histone 3 lysine 27 acetylation) | Inflammatory responses | Liu et al. 2015 |
| 5 | PM2.5 | Alterations in LINE-1 methylation, methylation at CpG loci of TNF-α | Inflammation | Wang et al. 2020a |
| 6 | PM2.5 | Altered global DNA methylation, P16 gene promoter methylation, and DNMT activity, altered post-translational histone modification. | COPD | Leclercq et al. 2017 |
| 7 | PM2.5 | Mitochondrial DNA methylation | Heart rate variability | Byun et al. 2016 |
| 8 | PM2.5 | miRNA | Cardiovascular diseases | Chen et al. 2018 |
| 9 | PM 2.5 | Methylation at candidate CpGs in monocytes | Atherosclerosis | Chi et al. 2016 |
| 10 | PM 2.5 | Genome-wide DNA methylation | Cardiometabolic diseases | Li et al. 2018a |
| 11 | PM 2.5 | ICAM-1 promoter methylation | Glucose dysregulation | Peng et al. 2016 |
| 12 | PM2.5 | Regulation of extracellular vesicle-enriched miRNA (evmiRNAs) expression by DNA methylation | Altered systolic blood pressure | Rodosthenous et al. 2018 |
| 13 | PM2.5 | Altered methylation in clock genes (CRY1, CRY2, NPAS2 ) | Ischemic stroke | Cantone et al. 2020 |
| 14 | Prenatal exposure to PM2.5 | Cord blood DNA methylation of IGF2/H19 gene | Disease risk in later life | Wang et al. 2020b |
| 15 | Prenatal exposure to PM2.5 | Lower expression levels of the miR-17/92 cluster in cord blood | Cancer risk, disease predisposition in later life | Tsamou et al. 2020 |
| 16 | PM2.5, metals from traffic sources (Cd, Fe, Mn, and Ni) | DNA methylation in imprinted control regions(L3MBTL1, NNAT, PEG10, GNAS Ex1A, MCTS2, SNURF/SNRPN, IGF2R, and RB1) | Risk associated with non-small cell lung cancer, glioma, and bladder cancer | Liang et al. 2021 |
| 17 | PM2.5 and PM10 | Hypomethylation of tandem repeats SATα | Inflammation | Guo et al. 2014 |
| 18 | Ambient air pollutant: PM2.5, PM10, PAH, O3 | Hypermethylation of Foxp3 locus | Asthma | Janssen et al. 2013 |
| 19 | PM2.5, NOx | CpG-methylation, DMRs in circulating monocytes | Atherosclerosis pathogenesis | C Chi et al. 2021 |
| 20 | PM2.5, NO2 | DNA methylation on cytosine-phosphate-guanine (CpG) sites and differentially methylated regions (DMRs). | Inflammation and immune responses | Eze et al. 2020 |
| 21 | Traffic-related air pollution (TRAP) – PM2.5, PM10, NO2 and SO2 | DNA methylation and histone H3K9 acetylation | Respiratory disorders | Ding et al. 2017 |
| 22 | Exposure to PM2.5, PM10 during pregnancy | DNA methylation on CpG sites and on differentially methylated regions (DMRs) | Respiratory health | Gruzieva et al. 2019 |
| 23 | Ambient air pollutants: PM2.5, PM10, SO2, NO2, CO. | Genome-wide analysis of differential DNA methylation | Pulmonary disorders and cancers | Wang et al. 2020c |
| 24 | Traffic-related pollutants: PM2.5, black carbon, NO2, CO | LINE-1, Alu, and gene-specific DNA methylation tissue factor (F3), Toll-Like Receptor 2 (TLR-2), and ICAM-1) | Cardiovascular disease | Lisanti et al. 2013 |
| 25 | Prenatal exposures to PM2.5, PM10 | Altered DNA methylation in specific gene promoters | Childhood cardio-respiratory health | Breton et al. 2016 |
| 26 | Prenatal exposures to PM2.5, PM10 | Placental global DNA methylation | Placental adaptation at the level of global DNA methylation | Maghbooli et al. 2018 |
| 27 | PM10 | microRNA-101 | Blood pressure | Motta et al. 2016 |
| 28 | PM10 (most abundant elements: Al, Fe, water soluble organic fraction) | Alterations in DNA methylation in macrophages | Inflammation | Miousse et al. 2014 |
| 29 | Iron-rich combustion- and friction-derived nanoparticles (CFDNPs) that are abundantly present in airborne particulate matter | Lower H3K9me2/me3 and higher γ-H2A.X | Alzheimer's disease (AD) | Calderón-Garcidueñas et al. 2020 |
| 30 | Airborne PM and PM metallic components | Extracellular vesicle (EV) miRNAs | Inflammation and coagulation | Pavanello et al. 2016 |
| 31 | Ambient ultrafine particles (UFP, diameter Dp < 0.18 μm) | Noncoding RNAs | Neurodevelopmental and neurodegenerative disorders | Solaimani et al. 2017 |
| 32 | Ozone (O3) | miRNAs, namely miR-132, miR-143, miR-145, miR-199a*, miR-199b-5p, miR-222, miR-223, miR-25, miR-424, and miR-582-5p | Inflammatory diseases | Fry et al. 2014 |
| 33 | Ozone (O3) | DNA methylation of the apelin gene | Pulmonary edema | Miller et al. 2018 |
| 34 | Traffic-related air pollution (TRAP) | Nasal DNA methylation | Childhood asthma | Zhang et al. 2018b |
| 35 | Traffic-related air pollution | Differentially methylated CpG positions (DMPs) and differentially methylated regions (DMRs) | Asthma severity | Zhu et al. 2021 |
| 36 | Diesel exhaust particles (DEP) from traffic and house dust mite (HDM) | Changes in 5-methyl-cytosine (5mC) and 5-hydroxy-methylcytosine (5hmC) in the regulatory regions associated with transcription factors in airway epithelial cells | Airway diseases, asthma | Zhang et al. 2018a, 2018b, 2018c (July) |
| 37 | Diesel Exhaust (DE) | CpG site methylation | Allergic disease | Clifford et al. 2017 |
| 38 | Diesel Exhaust (DE) | CysLT-related gene methylation | Asthma, poor lung function | Rabinovitch et al. 2021 |
| 39 | Diesel Exhaust (DE) | CpG site methylation, DMRs in bronchial epithelial cells | Pulmonary pathologies | Cardenas et al. 2021 |
| 40 | Gestational exposure to Diesel Exhaust (DE) | DNA methylation in global regulatory regions | Cardiac hypertrophy, fibrosis and susceptibility to heart failure | Goodson et al. 2019 |
| 41 | Polycyclic aromatic hydrocarbons (PAHs) | Methylation in the FOXP3 locus | Asthma, allergic rhinitis | Hew et al. 2015 |
| 42 | PAH | Higher expression of let-7a, miR-146a-5p, and miR-155-5p and lower expression of IL-6, CXCL8, and TLR2 mRNAs | Inflammation | Li et al. 2020 |
| 43 | PAH, Nitro-PAH, PM2.5 | Promoter methylation of CDKN2A, APC, and MLH1 genes and hypomethylation of the LINE-1 | Cancer risk | Silva et al. 2019 |
| 44 | Phenanthrene (Phe) | CpG island methylation of FOXP3 gene | Atopic conditions | Liu et al. 2013 |
| 45 | Coke oven emissions (COE), PAHs | Hypomethylation of LINE-1 and AhRR gene | Lung cancer | Yang et al. 2018 |
| 46 | Air pollutants | Histone modifications (H3K9ac, H3K9me3, H3K27me3, and H3K36me3) | Blood pressure | Kresovich et al. 2017 |
| 47 | Air pollution | Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism | Alzheimer’s disease (AD) | Wu et al. 2017b |
| 48 | Environmental tobacco smoke | Alteration in gene-specific and global DNA methylation | Allergic asthma | Christensen et al. 2017 |
| 49 | Tobacco smoke exposure | CD14 methylation | Allergic diseases | Munthe-Kaas et al. 2012 |
| 50 | Environmental tobacco smoke | Alterations in methylation pattern of IFN-γ and Thy-1 promoters | Respiratory diseases | Cole et al. 2017 |
| 51 | Prenatal exposure to tobacco smoke | Alterations in DNA methylation of LINE1 and AluYb8. Changes in the methylation on CpGs in the promoter region of AXL and PTPRO genes. | Disease risk in later life | Breton et al. 2009 |
| 52 | Cigarette smoke | Hypomethylation on the glutamate-cysteine ligase catalytic subunit (GCLC) gene | COPD | Cheng et al. 2016 |
| 53 | Cigarette smoke | CpG methylation Of gene AIRE, PENK and SLC6A3 | Non-small cell lung cancer | Ma et al. 2019 |
| 54 | Cigarette smoke | DNA methylation in white blood cells | Cardiovascular diseases | Chatziioannou et al. 2017 |
| 55 | Cigarette smoke | DNA methylation | Schizophrenia | Ma et al. 2020 |
| 56 | Indoor air pollution caused by smoky coals with high benzo(a)pyrene (BaP) and quartz levels |
CDKN2A, DLEC1, CDH1, DAPK, RUNX3, APC, and WIF1 DNA methylation in the promoter region |
Primary non-small cell lung cancer (NSCLC) | Huang et al. 2018a |
| 57 | Biomass smoke | miR-126 and miR-155 | Cardiovascular diseases | Ruiz-Vera et al. 2019 |
| 58 | Second-hand smoke | Altered methylation of major repetitive DNA elements including LINE L1, IAP LTR and SINE B1 | Lung cancer | Tommasi et al. 2012 |
| 59 | Second-hand smoke | DNA methylation of several CpG loci | Bladder carcinoma | Wilhelm-Benartzi et al. 2011 |
| 60 | Second-hand smoke | CpG methylation within FOXP3 and IFNγ loci | Asthma | Runyon et al. 2012 |
| 61 | Smoke and second-hand smoke | Altered methylation of GSTP1, FHIT, and CDKN2A, SCGB3A1 and BRCA1genes | Breast cancer | Callahan et al. 2019 |
Persistent organic pollutants (POPs) and polycyclic aromatic hydrocarbons (PAHs)
In early 1900s, the entire biosphere started to become contaminated with toxic man-made substances called persistent organic pollutants (POPs) (Jones and de Voogt 1999). Only recently, there has been much concern about POPs as many of these chemicals cause perturbations in the endocrine and reproductive health of mammals (Gregoraszczuk and Ptak 2013). Once produced, these chemicals remain in our ecosystem for several years and continue to exert their harmful effect on human health in the form of diseases like cancer, birth defects, and neurological and immunological disorders (El-Shahawi et al. 2010). Certain POPs are associated with immunodeficiency in infants, while a few others are substantial risk factors for breast cancer (Toms et al. 2016; Mouly and Toms 2016). Brain development in infants may get affected due to exposure to POPs, as it has been shown that children exposed to POPs during infancy scored comparatively lower in intelligence and concentration measurements (Tran and Miyake 2017). POPs are also associated with gynecological disorders such as endometriosis, pre-term birth, and other diseases like learning disabilities and an increased incidence of diabetes (Gregoraszczuk and Ptak 2013; Ngwa et al. 2015).
The US Environmental Protection Agency (US EPA) suggests that among the POPs, dioxin-related cancer risk is greater than 1 in 1000 for current background levels of exposure. However, the quantitative cancer risk assessment information for dioxin is of varying ranges (Institute of Medicine (US) Committee on the Implications of Dioxin in the Food Supply 2003). Integrated Risk Information System (IRIS) has been providing quantitative cancer risk assessments for several other POPs including chlordane, DDE, DDT, dieldrin, hexachlorobenzene, alpha-hexachlorocyclohexane, and toxaphene (Gehlhaus et al. 2011). In the US, a number of pesticides have been banned or restricted in terms of usage due to their carcinogenic potential. Some of them are DDT, dimethylhydrazine, hexachlorobenzene, hexamethylphosphoramide, chlordecone, lindane, mirex, and nitrofen (Gunier et al. 2001). People with high exposure to pesticides have been identified with many different types of cancers including lung, brain, prostate, skin as well as blood and lymphatic system cancers (Blair and Freeman 2009).
Polycyclic aromatic hydrocarbons (PAHs) in the air are produced mainly by the burning of carbon-containing compounds like wood and fuel. They are also present in gasoline and diesel exhaust, cigarette smoke, and charcoal-broiled food. A number of studies have shown that exposure to PAHs is also linked with an increased incidence of lung, skin, and urinary tract cancers (Abdel-Shafy and Mansour 2016). A recent study has shown that PAH contributes to the biological age by changing the leukocyte telomere length and the mitochondrial DNA copy number, as well as by forming anti-benzo[a]pyrene diolepoxide adducts (Pavanello et al. 2020).
Effect of air pollution on human health
The most detrimental effect of air pollution is associated with lung cancer (Cohen and Pope 3rd. 1995). It was found that the incidence of lung cancer is correlated with urbanization variables other than smoking in a national Swedish cohort (Boffetta et al. 2001). Smoking was responsible for 85% and 20–40% of lung cancer incidences in men and women, respectively (Tomatis 1993). A study had shown that 4.3% of the lung cancer incidents in men and 10.5% in women in the Cracow region were due to the air pollution (Hemminki and Pershagen 1994). The nitroarenes from diesel exhaust which can be converted to aromatic amines are a threat to bladder cancer (Latifovic et al. 2015). Air pollution is also associated other type of cancers such childhood leukemia and breast cancer (Lien et al. 2017; Callahan et al. 2019). WHO has estimated that the air quality for 91% of the global population exceeds the pollution limit (WHO 2018). Ambient air pollution was responsible for 4.2 million deaths worldwide, contributing to 7.6% of all deaths in 2016. In 2018, ambient air pollution was responsible for 29% of all deaths and diseases due to lung cancer. It is well realized that air pollution remains one of the major causes for many diseases. In a study in 1994, more than 40% of COPD incidents was attributed to air pollution, while it was responsible for nearly 30% of ischemic heart disease, stroke, and other respiratory infections (Hemminki and Pershagen 1994). A recent study has indicated that air pollution can also bring out interstitial lung diseases (ILDs) (Goobie et al. 2020).
Air pollution and epigenetic modifications
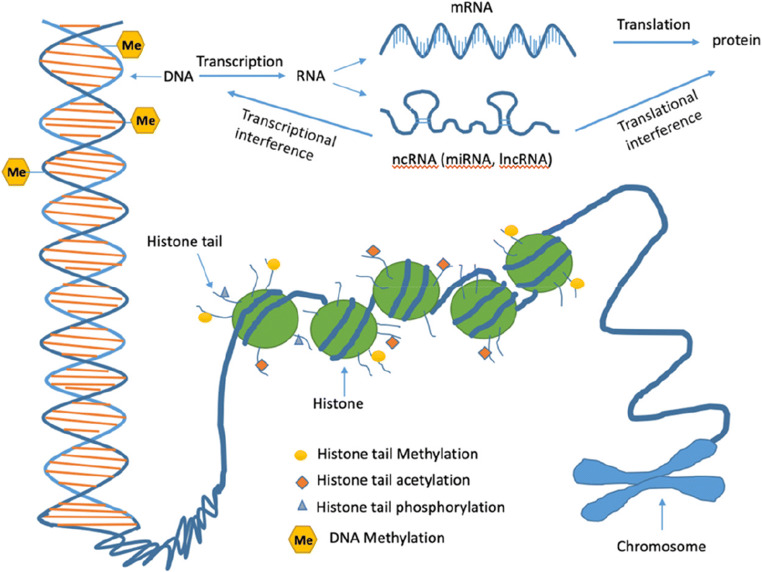
Epigenetics is the process where changes occur in gene functions without changing their underline DNA sequences. Such changes, which may get reflected in cellular and physiological traits, can be generated by environmental factors, besides being a part of the normal developmental phenomena (Sharma et al. 2010; Feil and Fraga 2012). DNA methylation and histone modifications are two key processes which alter gene functions without changing the DNA sequences (Fuks 2005). DNA methylation is a reversible process and are mediated by DNA methyltransferase (DNMT) and demethylase (Jin et al. 2011). Several studies indicated that a DNA methylation pattern can be greatly influenced by environmental factors like ambient air pollution, and these epigenetic changes are linked with diverse diseases (Plusquin et al. 2017; Ding et al. 2016; Xu et al. 2018). Histones can also be modified by several ways, such as acetylation, methylation, phosphorylation, and ubiquitylation. Micro-RNAs and ncRNAs play critical role in gene expression and contribute to epigenetic control in the process (Chuang and Jones 2007).
MicroRNAs are capable of regulating post-transcriptional gene expression including the expression of epigenetic factors like DNA methyltransferases, histone deacetylases, and polycomb group genes (Macfarlane and Murphy 2010; Filipowicz et al. 2008). Expression of miRNAs are associated with several developmental processes and may contribute to the onset of many diseases like cardiovascular diseases, cancer, metabolic diseases, and developmental disorders (Ha 2011). A diverse class of ncRNAs, such as snoRNAs, miRNAs, siRNAs, exRNAs, piRNAs, and long ncRNAs, can influence transcription and translation processes and play significant roles in health and diseases. They generally do not code proteins but are capable of regulating gene expression through the regulation of the chromatin structure and DNA methylation (Hombach and Kretz 2016). Moreover, mitrochondrial DNA (mtDNA) is also regulated at the epigenetic level in the form of mtDNA methylation (van der Wijst et al. 2017). Mitochondrial epigenetics are capable of influencing diverse cellular activities such as transcription regulation, cell division, cell cycle regulation, physiological homeostasis, bioenergetics, and disease pathologies (Stimpfel et al. 2018) (Fig. 1).
Fig. 1.
Epigenetic alterations in the mammalian system. (a) DNA methylation at the DNA bases, (b) different types of modifications at the histone tails, and (c) interference of the non-coding RNAs (ncRNAs) with the gene expression process
Chronic exposure to air pollution causes cytogenetic damage, DNA-strand breakage, epigenetic changes, and altered gene expression—all these changes are associated with higher risk factors for many diseases, especially for cancers (Ghorani-Azam et al. 2016). Studies have shown that organic extracts of outdoor PM induce changes in the DNA morphology in mammalian cell culture. Several cytotoxic effects, such as oxidative DNA damage, breakage in the DNA strand, altered telomere length, and mutation formation have been observed in human peripheral monocytes when exposed to such organic extracts (Cimino et al. 2014). DNA methylation pattern in the genome also gets altered upon exposure to certain pollutants (Martin and Fry 2018). Asbestos is not an essentially mutagenic substance; it may, however, impose the risk of cancer through an epigenetic mechanism (Kettunen et al. 2017). In addition, it is to be noted that an altered miRNA expression profile has been reported as an effect of PM exposure (Jardim 2011).
Epigenetic mechanisms are also involved in regulating many genes that are involved in inflammation and immune responses (Suárez-Álvarez et al. 2013). Therefore, aberrant environmental signals that may induce epigenetic changes could generate an imbalance in immunity. It was found that exposure to PM2.5, CO, and O3 altered the methylation pattern in many CpG sites for the immunoregulatory gene Foxp3, IL-4, IL-10, and IFN-g, thereby altering the immune response (Prunicki et al. 2021). Air pollution has also been found to have been associated with autoimmune diseases. Such pollutants may trigger pulmonary inflammation by altering the lung microenvironment and inducing epigenetic changes (Zhao et al. 2019). Besides abnormal immune responses, air pollution-induced epigenetic modifications could directly contribute to disease pathogenesis and such epigenetic changes could be inherited across generations.
Air pollution and DNA methylation
Emerging evidences suggest that air pollutants modulate epigenetic states, ranging from DNA methylation to miRNAs expression (Breton and Marutani 2014; Alfano et al. 2018). Epithelial cells from the aerodigestive tract of heavy smokers exhibit aberrant methylation of multiple genes associated with the pathogenesis of lung cancer (Zö Zöchbauer-Müller et al. 2003). Several genes that are related with cancer progression have also been found to be hypermethylated in the bronchial epithelium and peripheral lymphocytes of smokers (Sridhar et al. 2008). Exposures to metals induce DNA methylation as well as histone modifications (Martinez-Zamudio and Ha 2011). Metal-rich air particles generate H3K4me2 and H3K9ac patterns in specific promoters of transcriptionally active genes (Cantone et al. 2011). Mitochondrial DNA also gets methylated upon metal exposure (Byun et al. 2013). It is to be noted that fine PM is strongly pro-oxidant in nature, and mitochondrial DNA is more susceptible to oxidative damage due to lack of introns and histone proteins (Bhargava et al. 2018b). DNA from the sperm of mice exposed to steel plant air has been shown in a persistent hypermethylated state long after the exposure ended (Yauk et al. 2008). Epidemiological studies reveal that PM2.5 and PM10 exposures induce the hypomethylation of Alu and/or LINE1 (Long Interspersed Nuclear Element-1) elements in leukocytes and buccal cells (Terry et al. 2011; De Nys et al. 2018). PM exposure also alters DNA methylation in the NOS2A gene involved in nitric oxide production, which is important for cardiovascular and respiratory health (Breton et al. 2012; Breton et al. 2014). During pregnancy, especially in the first trimester, PM2.5 exposure is linked with global DNA hypomethylation in the placental tissue (Janssen et al. 2013). High levels of PM and ozone exposures are associated with hypermethylation of the FOXP3 gene in regulatory T cells (Nadeau et al. 2010). Direct exposure to PM2.5 also induces methylation of the p16 promoter (a tumor-suppressor gene) in alveolar epithelial cells as well as in the lungs of mice. The expression level of DNA methyltransferase 1 (DNMT1) is reported to be elevated in PM-exposed patients—this could be a mechanism to induce methylation in the p16 promoter region (Soberanes et al. 2012). In an attempt to establish a link between childhood asthma and air pollution, studies have demonstrated that exposure to PM2.5 induces DNA methylation in several CpG loci of inducible nitric oxide synthase (iNOS) genes, the major enzyme for nitric oxide production in the airways (Breton et al. 2012). Another study has shown that short-term exposure to air pollution alters DNA methylation at the promoter region of the iNOS gene (Salam et al. 2012). All these studies indicate distinct pathways through which air pollution exerts its phenotypic expression.
Expressions of many inflammatory and immune response genes are found to be altered due to DNA methylation linked with air pollution (Vawda et al. 2014). Air pollution-induced DNA methylation is often found to be associated with poor respiratory health outcomes (Rider and Carlsten 2019). Both short- and long-term exposures to NO2, CO, and PM2.5 lead to changes in the differentially methylated region (DMR) of the Foxp3 promoter region and the IL10 intronic region. These changes are long-lasting and significantly associated with asthma pathogenesis (Prunicki et al. 2018). The hypermethylation of interferon-gamma (IFN-γ) in effector T-cells and Foxp3 in regulatory T-cells has been observed as an effect of ambient air pollution (Kohli et al. 2012). In another trial, 15 healthy adult participants were exposed to concentrated ambient particles (CAPs). Exposure to fine CAPs reduced Alu methylation, while coarse CAPs reduced Toll-like receptor-4 (TLR4) methylation—a gene involved in asthma pathogenesis (Bellavia et al. 2013). Among other air pollutants, SO4 has been associated with altered methylation patterns in genes associated with asthma (Sofer et al. 2013). In addition to DNA repetitive elements, PM exposures alter the DNA methylation pattern for specific genes, including p53, p15, p16, APC, RASSF1A, HIC1, iNOS, hTERT, and IL-6, and such alterations influence the development of diverse diseases. However, each different gene and subfamily of repetitive DNA sequence responds independently to different exposure conditions (Pacchierotti and Spanò 2015).
PAH is also associated with DNA methylation. Neonatal exposure to PAH causes hypermethylated ACSL3 in the DNA of the white blood cells of the umbilical cord (Perera et al. 2009). PAHs can also cause a hypermethylation state in the LINE1 repetitive element (Lee et al. 2017). Airborne benzene exposure in humans also generates hypomethylation of the repetitive elements and specific genes (Bollati et al. 2007). The measurement of Alu and LINE-1 methylation gives an estimation of the global genomic DNA methylation content. Global hypomethylation and hypomethylation of transposable repetitive sequences are associated with reduced chromosomal stability and altered gene functions (Lisanti et al. 2013). It is possible that DNA methylation makes individuals susceptible to pollutant exposure. A study has shown that exposure to various pollutants alters ICAM-1 and VCAM-1 levels in blood and these effects are dependent on Alu, LINE-1, or TLR-2 methylation status (Bind et al. 2012).
Exposure to POPs such as DDT, DDE, HCH, chlordane, mirex, and PCBs also induce altered methylation in Alu and LINE1 sequences (Rusiecki et al. 2008). In a similar study, a healthy Korean cohort shows that the blood concentrations of different organochlorine pesticides are inversely associated with the Alu methylation level (Kim et al. 2010). Perfluoroalkyl substances (PFASs), a new class of POPs, are industrial wastes and have been found to be epigenetically active. A study has revealed that an increased level of serum perfluorooctanesulfonate (PFOS) is associated with LINE-1 methylation, while prenatal PFOS exposure could be associated with the hypomethylation in Alu repeats in cord blood (Watkins et al. 2014; Liu et al. 2018). Moreover, black carbon exposure was found to generate hypomethylation in the Alu region (Madrigano et al. 2011). Only a few studies have revealed the effect of PM10 or PM1 on epigenetic modifications. A combination of PM10, PM1, zinc, and iron exposures has been negatively associated with NOS3 methylation, while only zinc exposure has reduced endothelin-1 (EDN1) methylation. The reduced NOS3 and EDN1 methylation has led to an increase in the endogenous thrombin potential (ETP), a measurement of global coagulation, thereby providing a mechanism for environmental exposures on coagulation through epigenetic modifications (Tarantini et al. 2013). Overall, exposure to diesel exhaust particles (DEP) and other air pollutants may result in epigenetic changes at repeat elements and specific candidate genes, which in turn is likely to influence gene expression levels and contribute to disease pathogenesis. All these observations suggest that changes in the methylation pattern could be diverse and depend on several factors like the duration and length of exposure, co-exposure, targets, route of exposure, and the genetic makeup of hosts. Further investigations are required to establish a direct correlation between methylation patterns and specific pollutants.
Air pollution and histones
The effect of air pollution on histone modification is well established. However, measurement of histone modifications due to air pollution in large population studies remains unexplored. One study shows that inhaled nickel, arsenic, and iron are associated with increased histone3-lysine4 dimethylation (H3K4Me2) and histone3-lysine9 acetylation (H3K9Ac) on histones from blood leukocytes (Martinez-Zamudio and Ha 2011). PAH exposure also induces histone modifications in mice—for instance, exposure to benzo[a]pyrene (BaP), a type of PAH, decreases the acetylation of H3K14 in the steroidogenic acute regulatory protein (StAR) gene promoter, which is important for testosterone production (Liang et al. 2012). BaP exposure to HeLa cells generates H3K4Me3 and H3K9Ac modifications along with a reduced association of DNMT1 with the LINE-1 promoter. Cytosine methylation within the CpG island of the LINE-1 promoter is also reported to be reduced (Teneng et al. 2011). A knockout mice study shows that Sirt1, a member of class III histone deacetylase, can control the adverse effect of PM exposure on cardiopulmonary functions through the regulation of inflammation/coagulation responses in lungs following the exposure (Wu et al. 2012). Cigarette smoke exposure is also associated with an increased expression level of DNMT1 and a decreased level of DNMT3B (Xiao et al. 2015; Liu et al. 2007). Another study reveals that cigarette smoke exposure decreases histone deacetylase (HADC) activity and reduces HDAC1, HDAC2, and HDAC3 expressions in macrophages, thereby leading to an inflammatory response generation (Yang et al. 2006). Exposures to PM10 and DEP may cause an imbalance between histone acetyletransferase (HAT) and HDAC expression levels, as well as increase histone H4 acetylation in the promoter region of the specific genes, thereby generating inflammatory responses in human bronchial epithelial (HBE) cells (Gilmour et al. 2003; Cao et al. 2007).
Air pollution and miRNA
It is controversial whether to include miRNAs as an epigenetic mark. Almost two-thirds of the human genes are regulated by miRNAs and are thought to play a critical role in mediating biological responses to air pollution (Jardim 2011; Tomankova et al. 2011). In one study, human airway epithelial cells had been incubated with DEP and the miRNA expression profile was determined using miRNA microarray technique. In total, 197 miRNAs were found to be down or upregulated in response to the exposure, and a few among them involved in inflammatory and tumorigenic processes (Jardim et al. 2009). Exposure to PM also alters miRNA expression levels in the microvesicles derived from A549 alveolar cells and from the plasma of healthy steel plant workers (Bollati et al. 2015). These PM-induced changes often impose the risk of cardiovascular diseases. Literatures showing an association between ambient air pollution and miRNA is emerging. A small change in a single miRNA has the potential to influence a large number of genes working on its downstream mechanism (Catalanotto et al. 2016).
It is to be noted that miRNAs are involved in important cellular functions such as cell growth, differentiation, development, apoptosis, and metabolism. Dysregulations in miRNA expression are associated with diverse diseases including cancers (Mahmoud et al. 2021). A recent study has shown that a total of 25 miRNAs are deregulated in lung cancer as well as associated with air pollution exposure. It was suggested that miR-222, miR-21, miR-126-3p, miR-155, and miR-425 may serve as biomarkers for the assessment of risk factors for lung cancer in individuals who have been exposed to air pollution (Sima et al. 2021). Besides various types of cancers, miRNAs are also deregulated in different diseases such as stroke, Alzheimer’s disease, Parkinson’s disease, and multiple sclerosis (Mahmoud et al. 2021). All these diseases are also associated with air pollution (Kasdagli et al. 2019; Esmaeil Mousavi et al. 2017; Wu et al. 2017b). The presence of miRNAs in body fluids such as urine and serum as well as in tissues makes them suitable candidates for developing disease biomarkers.
Air pollution and non-coding RNA
Only a small number of known lncRNAs have been functionally characterized so far. lncRNAs are already emerging as potential regulators of air pollution-associated diseases (Karlsson and Baccarelli 2016). A recent work has shown that cigarette smoke extracts and PM2.5 treatment alter the expression level of a particular lncRNA, lung cancer progression-association transcript 1 (LCPAT1), in lung cancer cells and knockdown of LCPAT1 impairs the effect of treatment (Lin et al. 2018). An experiment reveals that lncRNA LOC101927514 is linked with the PM2.5-induced inflammation in bronchial 16HBE cells that are used as a respiratory inflammation model. RNAi inhibition of lncRNA LOC101927514 reduces inflammation induced by PM2.5. Moreover, lncRNA LOC101927514 binds with STAT3, a protein which is known to influence the inflammatory state of the cells. This indicates the possibility that lncRNA LOC101927514 could be a potential target for the treatment of PM2.5-induced respiratory inflammation (Tan et al. 2020). In another study, more than 500 lncRNAs have shown altered expression levels in rat embryos exposed to air pollution. Gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analyses of the deregulated lncRNAs indicate their involvement in neurological processes and congenital defects (Li et al. 2019). Smoking is another common risk factor for many diseases like COPD, asthma, lung cancer, and cardiovascular diseases; it influences the expression of lncRNAs in lung tissues (Wang et al. 2017). GO and KEGG pathway analysis show that these lncRNAs are linked with the pathways involved in the pathogenic processes of COPD (Zhang et al. 2018a). Another lncRNA, MALAT1 is involved in cigarette smoke extract-induced malignant changes in HBE cells (Ji et al. 2003).
lncRNA can influence miRNA expressions. In many cases, the crosstalk between lncRNAs and miRNAs are important regulators of gene expression. Sometimes intragenic miRNA sequences are embedded within lncRNA sequences, thus suggesting a post-transcriptional regulatory role of lncRNAs in miRNA regulations (Dykes and Emanueli 2017). These findings suggest that lncRNAs may function as novel biomarkers of environmental exposure—they are involved in interactions with various environmental factors and can regulate many cellular processes, thereby influencing disease pathogenesis associated with air pollution.
Air pollution and transgenerational epigenetic inheritance
Epigenetic changes may persist through several cell cycles or may get transmitted to subsequent generations without impacting the underlying DNA sequences (Henikoff and Greally 2016). Transgenerational epigenetic inheritance deals with epigenetic marks—i.e. DNA methylation, histone modifications, and non-coding RNAs—that are acquired in one generation and get stably transferred to the next generation through the gametes (Trerotola et al. 2015; Blake and Watson 2016). Transgenerational epigenetic inheritance is not a very common phenomenon. However, epigenetic changes can be inherited when they occur in gametes and are not completely erased due to reprogramming (Wei et al. 2015). Tumor-suppressor mismatch repair genes MLH1 and MSH2 are examples where epimutations are inherited through the germ lines (Hitchins et al. 2007; Chan et al. 2006). Imprinted genes provide another example of epigenetic inheritance. Generally, there are two functional copies (alleles) for each gene—one inherited from the mother and another from the father. One copy of the alleles which is heavily methylated is called the imprinted gene and it remains transcriptionally silenced (Barlow and Bartolomei 2014). miRNA is a regulatory molecule for gene expression in response to environmental stimuli, and such changes in miRNA expression may alter the gene expression so as to influence the gene expression even in the future generations.
It has been found that 5-methylcytosine (5mC) is the predominant DNA methylation in mammals. Methylation marks could be inherited if they are mitotically and meiotically stable and are capable to avoid epigenetic reprogramming in primordial germ cells as well as postfertilization embryos (Stenz et al. 2018). Recently, it has been thought that histone modifications and their regulatory enzymes can transfer the epigenetic signatures across generations. In C. elegans, histone3-lysine27 trimethylation (H3K27me3) marks can transfer the memory of repressing X-chromosome transgenerationally (Gaydos et al. 2014). The role of non-coding RNAs in transgenerational epigenetic inheritance is currently being researched (Larriba and del Mazo 2016).
In Table 1, we have summarised the connection between various air-pollutants, subsequent epigenetic modifications, and different disease risks. A few pollutants are associated with immune modulation through epigenetic changes and thus could play as a risk factor for HP development. It would be interesting to study whether there is direct correlation between HP development and exposure to those pollutants.
Air pollution and epigenetic modifications in hypersensitivity pneumonitis (HP)
Airborne pollutants have adverse effects in the lungs. Recent studies find an association between air pollution exposure and increased incidence of poor lung function and mortality in interstitial lung diseases (ILDs) (Johannson et al. 2015). In idiopathic pulmonary fibrosis (IPF), air pollution is associated with increased incidence and harmful effects on health (Harari et al. 2020). ILDs comprise an array of heterogeneous parenchymal lung diseases that occur due to fibrosis and inflammation of lung parenchyma. Considerable heterogeneity exists among ILDs, depending on a variety of causative agents, while in some cases the cause is unknown and therefore the disease is classified as idiopathic. Prevalence of ILDs depends largely on geographical locations and incumbent environmental conditions. IPF, sarcoidosis, and HP are the most regularly observed disorders amongst ILDs (Kalchiem-Dekel et al. 2018). According to data from Indian registry, HP accounts for 47.3% of all ILD cases and is the most frequent type of ILD in India. HP, also known as extrinsic allergic alveolitis (EAA), is a heterogeneous and immunologically-mediated lung disease. It results from the inhalation of certain organic substances of animal or plant origin and inorganic substances like low molecular weight chemical isocyanates. The most prevalent forms of HP are farmer’s lung disease, developed from bacterial and fungal exposure, and bird fancier’s lung that occur due to the inhalation of avian antigen found in bird droppings. Interestingly, the inhaled offending antigen does not induce the disease in every exposed individual. It is reported that nearly 5–10% of the exposed individuals will develop HP. Prevalence of chemical worker's lung is only 1–4.7% of the isocyanate-exposed workers, while farmer's lung is reported to have developed in 1–19% of the mouldy hay-exposed individual. It is also estimated that only about 6–20% of the avian antigen-exposed people develop bird fancier's lung. Such variations in disease incidence can be traced to the presence of predisposed genetic and epigenetic factors. Individuals who are genetically susceptible (first hit) develop HP when exposed to an inciting antigen (second hit). The second hit—i.e. the exposure to causative antigen—alone cannot induce HP development (Spagnolo et al. 2015). A recent study has indicated an association between air pollution level and HP occurrence in India (Singh et al. 2019). It is apparent that air pollution could be a risk factor for HP, but how air pollution could influence HP pathogenesis is largely unknown. It has been observed that exposure to specific air pollutants leads to inflammation and altered immune responses. Such exposure is also associated with epigenetic modifications. Therefore, we hypothesize that air pollution-induced epigenetic alterations influence immune responses, thereby making exposed individuals susceptible to the development of HP. The following section outlines the pathophysiology of HP in connection with epigenetic modifications induced by air pollution.
Immunopathogenesis of HP
HP is an inflammatory lung disease caused by the inhalation of organic or inorganic antigens (summarized in Table 2) less than 5 μm in diameter. According to the ‘two-hit’ hypothesis, the coexistence of inducing factors, such as antigen and promoting factors like genetic abnormalities or additional environmental exposures, may lead to an exaggerated immune reaction in the lungs and thereby causes inflammation. HP is characterized by alveolitis, which eventually results in granuloma formation and the development of fibrosis. The development of fibrosis leads to respiratory insufficiency, and finally, to the death of the patient (Selman et al. 2012).
Table 2.
Common HP-inducing antigens
| Name of the Disease | Origin | Antigen | Source of Antigen |
|---|---|---|---|
| 1. Farmer’s lung | Bacterial |
Micropolyspora faeni, Thermoactinomyces vulgaris |
Moldy hay |
| 2. Summer-type HP | Fungal | Penicillium frequentans | Contaminated old houses |
| 3. Hot tub lung | Mycobacterial | Mycobacterium avium complex (MAC), composed of M. avium and M. intracellularss | Contaminated water |
| 4. Bird fancier’s lung | Animal fur protein | Avian proteins | Feather bloom, avian serum(pigeons, chicken, turkey, goose, avian proteins),and excrement (mainly due to pigeon intestinal mucin) |
| 5. Chemical worker’s lung | Low-molecular weight chemicals | Diisocyanates, trimellitic anhydride | Polyurethane foams, spray paints, dyes, glues |
The pathogenesis of HP is complex and involves both Type III (immune complex-mediated) and Type IV (T cell-mediated) hypersensitivity reaction . Several studies have suggested that the alveolitis, (inflammation in alveoli branches) initiates with almost an exclusive predominance of neutrophils, eventually involving lymphocytes and becoming solely lymphocytic at later stages. The human lung contains approximately 1011 alveolar macrophages which play a major role in granuloma and fibrosis formation in HP. In the susceptible individual with repeated exposures, the disease-causing antigen forms an immune complex with the pre-existing antigen-specific antibody (IgG). This event triggers the activation of the complement cascade and results in the recruitment of macrophages, neutrophils, and lymphocytes at the site infection to induce the formation of granuloma (Patel et al. 2001). These immune cells also secrete several molecular mediators that induce the migration and stimulation of fibroblast to induce fibrosis. It is reported that chronic forms of HP are regulated by the T cells. Analyses of the bronchoalveolar lavage fluid (BALF) of chronic HP patients have suggested an involvement of Th2 T cells with a higher CD4+ / CD8+ ratio and a concomitant reduction in the number of γδT cells in disease development (Caillaud et al. 2012; Simonian et al. 2009). It is likely that the CD4 to CD8 ratio varies according to the type of the inhaled antigen. Activation of alveolar macrophages in the acute and sub-acute phases also promotes CD8+ T cells accumulation, granuloma formation, and subsequent development of fibrosis. Natural killer cells are also reported to be involved in HP pathogenesis (Korosec et al. 2007). Various reports have demonstrated that Th1 cytokine plays an essential role in disease pathogenesis (Barrera et al. 2008). The induction and perpetuation of the inflammatory responses in HP, including the recruitment of the different inflammatory cells, are mediated by a myriad of cytokines and chemokines. The pathophysiology of HP is not dependent on any one component of cellular immunity, but it is developed due to a multimodal interaction between these components, each of which may play a crucial role in disease progression.
Neutrophils are the body’s first line of defence against foreign bodies and plays a crucial role in inflammatory diseases like HP. After inhalation of the causative antigen, neutrophils accumulate in the alveoli and small airways of the lungs. Upon stimulation, neutrophils secrete various cytokines that are implicated in the pathogenesis of HP (Pardo et al. 2000). The secretion of IFN β, IL-10, and IL-6 by neutrophil is regulated by epigenetic modifications. The binding of two transcription factors, PU.1 and C/EBPβ, favours activation of the transcription of these three cytokines (Ostuni et al. 2016). Additionally, the matrix metalloproteinases (MMPs), collagenase-2 (MMP 8), and gelatinase B (MMP 2) produced by neutrophils are also reported to play a key role in the disease progression (Pardo et al. 2000). An epigenetic study on MMP2 reveals that its expression is dominated by DNA and histone modifications (Santiago-Ruiz et al. 2019). Patients with chronic HP have an increase of neutrophils loaded with MMP8 and MMP9, and many studies show that the expressions of MMPs are regulated by histone and DNA methylation (Campos et al. 2016).
Macrophages are predominantly involved in granuloma formation in HP (Suga et al. 1997). The lncRNA MEG3-4 has been identified as a tissue-specific regulator of inflammatory responses in alveolar macrophages during bacterial infection through the transcriptional regulation of immune response genes. It has been confirmed that the lncRNA MEG3-4 binds to the microRNA miR-138 in a competitive manner, with mRNA encoding the proinflammatory cytokine IL-1β, thereby increasing the abundance of IL-1β and enhancing the inflammatory response to bacterial infection in alveolar macrophages (Li et al. 2018b). A recent study on epigenome analysis has revealed that DNA methylation is involved during macrophage differentiation by changing the binding sites of transcription factors (Dekkers et al. 2019). T cells are also considered to be important regulators of HP development (Simonian et al. 2009). Epigenome maps combined with mechanistic studies, have demonstrated that T cells undergo extensive epigenome remodelling in response to signals, which has a strong effect on phenotypic stability and function of the lymphocytes (Schmidl et al. 2018). Moreover, it was shown that the second-hand smoke and ambient air pollution, which deteriorate respiratory health, were associated with DNA hypermethylation and a decreased expression of IFN-γ and Foxp3 in T cells population (Runyon et al. 2012).
External influence on HP development
Recently, Singh et al. have reported that ambient air pollution is a promoting factor in the development of HP. The study demonstrates that the odds of developing HP is 7% greater for every 10 μg/m3 increase in air pollution (PM2.5 level). The authors hypothesize that fine-particulate air pollutants lead to airway inflammation, reduce mucociliary clearance in the alveoli, and lead to an immune-mediated response that ultimately leads to the development of HP (Singh et al. 2019). It is apparent that more rigorous studies are necessary to establish this hypothesis. It is evident from the ‘Table 1’ that exposures to PM2.5, PM10, NO2, SO2, CO, and O3 were found to be associated with epigenetic changes like alteration in LINE-1 methylation, methylation on CpG sites of many immunoregulatory genes, histone modifications, and miRNA regulations. These epigenetic changes were again associated with inflammation and poor respiratory health. Therefore, exposure to those pollutants may alter immune responses, which could influence HP development. Moreover, HP pathogenesis involves many components of our immune systems that are under epigenetic regulations (Fernández-Morera et al. 2010). Wu et al. have reported methylation of EBF3 gene in peripheral blood samples of a Turkic ethnic group (Uygurs) with bird fancier’s lung. The authors hypothesized that EBF3 hypomethylation might be associated with IL 10 dysregulation in HP subjects (Wu et al. 2018). In another study on Chinese Uygur population by the same group, methylation of the gene Smad3 is reported. Smad 3 protein is one of the key players in the formation of fibrosis; it is likely that Smad3 gene hypomethylation promotes pulmonary fibrosis by increasing Smad3 mRNA expression (Wu et al. 2017a).
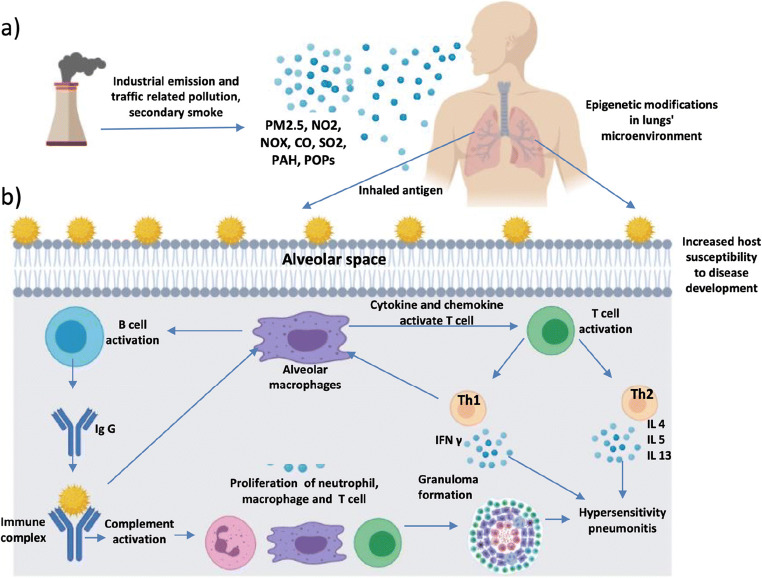
Diagnosis and effective management of HP often pose to be a challenge to the pulmonologists. HP is curable if diagnosed correctly in its early stages, i.e. before occurrence of irreversible lung damage. The prognosis of HP is not dependent only on the dose and duration of exposure to the causative agents; the genetic and environmental factors also play a major role in disease pathogenesis. Recent studies have shown that HP development is associated with ambient air pollution, which is known to be linked with inflammation and other lung diseases. An understanding of epigenetic alterations associated with air pollution has emerged in recent times and in many cases, such alterations are associated with diverse diseases including ILDs (Fig. 2).
Fig. 2.
Mechanism showing HP development by increasing host susceptibility through air pollution exposure. a) Emission of industrial and traffic-related air pollution. Pollutants reach the alveolar spaces in the lungs and induce epigenetic changes in the lungs’ microenvironment, thereby influencing host susceptibility to disease development. b) The interaction between the causative antigen deposited in alveolar spaces and the circulating IgG antibody forms precipitating antigen-antibody immune complexes in interstitial spaces. Immune complexes trigger activation of both complement and pulmonary macrophages. Activated macrophages secrete the cytokines IL-1 and TNF-a that promote adhesion of leukocytes to activated endothelium. Complement activation enhances vascular permeability and attracts inflammatory cells. As a consequence of recruitment and activation, inflammatory cells release toxic mediators that promote acute lung injury, which eventually leads to HP development
Future perspective: development of risk assessment biomarkers
Epigenetic signatures reflect changes in the cellular environment and can also be found in the human circulatory system in case of several diseases (Ladd-Acosta 2015; Bhargava et al. 2018a). These signatures seem promising and can be explored to identify robust biomarkers to monitor clinical outcomes as well as early detection of diseases. An ideal biomarker should show a high degree of specificity and sensitivity and ease of measurement; it should also be capable of detecting a disease in its early stage (Byrnes and Weigl 2018). Although the plasticity of the human epigenome makes it difficult to establish true epigenetic marks for a particular condition, diverse diseases show alterations in their epigenetic signatures. A recent study reports that epigenetic alterations take place even on seasonality and weather influence (Ricceri et al. 2014; Xu et al. 2020). A significant number of studies have been conducted to understand the epigenome during cancer. The cancer-specific hypermethylation of CpG islands shows clinical importance (Jin et al. 2011; Wilhelm-Benartzi et al. 2011). Epigenetic marks, which are established as an effect of air pollution, can also be used as biomarkers for risk assessments for diseases associated with air pollution.
Methylation in DNA sequences has several advantages. DNA is a relatively stable molecule. It can be collected from a large number of sources and stored for a long time after being collected from patients. Moreover, DNA methylation is a widespread change throughout the cancer genome and hence many points in the genome can be used for the assay to generate highly predictive models (Huang et al. 2018a, 2018b; Tommasi et al. 2012). Sophisticated techniques such as methylation-specific PCR (MSP), mass spectrometry-based methylation detection (EpiTYPER), MethyLight, and pyrosequencingetc, allow determination of the methylation state of a specific DNA location even from a minimum sample volume (Olkhov-Mitsel and Bapat 2012). This makes it feasible to evaluate the diagnostic and prognostic potential of the methylation state of a large number of DNA sequences for many cancer types. The Cancer Genome Atlas project (TCGA) provides a platform for information regarding DNA methylation changes in cancers (Lee 2016). A case control study in 2008 utilized a series of methylated genes to predict disease recurrence after surgery in Stage I lung cancer patients. The promoter hypermethylation of just four genes—CDH13, RASSF1A, APC, and CDKN2A (p16) was reported to predict tumor recurrence, thus establishing a link between gene methylation and tumor recurrence in lung cancer patients (Brock et al. 2008). This study established that the epigenetic status of tumors plays a critical role in clinical outcomes as well as in determining the aggressiveness of tumor development. MGMT (O6-methylguanine-DNA methyltransferase)-promoter methylation has been used to determine the treatment response among patients with glioblastoma (Butler et al. 2020). MGMT is a DNA-repair enzyme, and the efficiency of DNA repair in cancer cells plays an important role in therapeutic resistance. Thus, MGMT inactivation can sensitize such cells to therapeutic drugs (Verbeek et al. 2008). Many other epigenetic changes have started to emerge as predictive markers for the response to chemotherapy. BRCA1 hypermethylation is common in breast and ovarian cancers (Esteller et al. 2000). It has been found that BRCA1 methylation status can predict chemosensitivity and treatment response to cisplatin (Stefansson et al. 2012). The methylation of GSTP1 and MLH1 also predicts therapeutic outcomes in many cancer types (Shivapurkar and Gazdar 2010). Another oncogene, KRAS is itself capable of mediating epigenetic alterations, which are essential for cell transformation. KRAS has been found to promote DNA methylation by facilitating the recruitment of DNMT1 in the promoter regions of the specific genes (Serra et al. 2014). In H-Ras-transformed rat fibroblasts, MMP2 and clusterin gene expressions are suppressed by promoter DNA hypermethylation (Lund et al. 2006).
Aberrant histone modifications can also be utilized as predictive biomarkers to evaluate the progression and treatment outcomes in cancers. A significant number of proteins that regulate histone modifications are themselves mutated in tumors. The polycomb group protein Enhancer of Zeste 2 (EZH2) acts as a critical regulator in prostate cancer development. EZH2 is involved in chromatin silencing by methylating histone3 on lysine27 (H3K27me3). EZH2 has been found to be overexpressed in metastatic prostate cancer, and tumors that express a higher level of EZH2 are associated with poorer prognosis (Gan et al. 2018; Liu et al. 2019). In addition, EZH2 overexpression has shown poor clinical outcomes in other cancers such as esophageal cancer, breast cancer, head and neck cancer, and glioblastoma (He et al. 2010). Lysine-specific demethylase 1 (LSD1) is another histone-modifying protein, which is linked with cancer development. LSD-1 removes the mono- and di-methyl groups from H3K4 and functions as a transcriptional repressor (Majello et al. 2019). The role of LSD1 in breast cancer has not been clearly established. A study has shown that inactivation of LSD1 by both genetic and pharmacological approaches leads to aggressiveness in luminal breast cancer, while another study shows that LDS1 mediates chemoresistance in the disease (Verigos et al. 2019; Hu et al. 2019).
Bioinformatics, as a new emerging discipline, combines mathematics, information science, and biology and helps answer biological questions. There are several bioinformatics tools for DNA methylation analyses, but only a few platforms can correlate DNA methylation and gene expression for customized analyses. Several bioinformatics tools like COHCAP (Warden et al. 2013), PiiL tool (Moghadam et al. 2017), ViewBS (Huang et al. 2018a, 2018b), correlate DNA methylation with gene expression. MethHC databases can also be utilized to visualize DNA methylation and gene expression on a web platform (Huang et al. 2015). MethGET is another web-based bioinformatics platform that correlates genome-wide DNA methylation and gene expression (Teng et al. 2020). The Gene-Ontology (GO) database provides a useful tool to analyse the functions of all dysregulated genes (Harris et al. 2004). Database for Annotation, Visualization, and Integrated Discovery (DAVID) is another online bioinformatics tool that provide biological meaning behind gene profile (Dennis Jr et al. 2003). In addition to that, Reactome also provides intuitive bioinformatics tools for visualisation, interpretation and analysis of pathways associated with genes (Fabregat et al. 2017).
We anticipate that this review will provoke further investigation on the potential association between HP and air pollution, considering epigenetic modifications as a potential threat by which air pollution exerts its effect on disease development. It would be useful to identify a set of predictive biomarkers to assess the risk factors for the development and progression of the diseases associated with air pollution. However, in this review article we did not explore whether air pollution directly affects our genome by generating novel mutation, deletion, DNA breakage or chromosomal aberrations. We also did not discuss the genetic and immunological basis of HP development, although it could be crucial to understand the impact of air pollution in disease pathogenesis and may lead to the development of novel biomarkers to assess the risk of HP development due to air pollution.
Conclusion
The adverse effect of air pollution on our health is a matter of increasing concern. PM and other chemicals associated with air pollution pose various disease risks through both genetic and epigenetic alterations. Although the link between environmental exposure and disease risk due to epigenetic modifications is clearly established, little is known about the signaling pathways that connect the pollutants to the epigenome. Here we summarize the air pollutants that are associated with epigenetic modifications linked with increasing risk factors for several diseases. This discussion prompts the question whether these epigenetic changes could be utilized as risk assessment biomarkers for specific diseases associated with air pollution. Further research may identify more robust epigenetics marks that are associated a particular disease induced by specific pollutants. Moreover, such modifications may pass through several generations and predispose an individual to disease risks even without any direct exposure. These epigenetic alterations may serve as biomarkers for disease susceptibility for the next generation.
Recently, it has been indicated that HP is associated with ambient air pollution. Pollution-mediated epigenetic changes in HP is yet to be explored. Such changes may sensitize the immune system, thereby making individuals susceptible to developing HP when exposed to the causative agents. Future research may use the findings of this paper to more comprehensively understand HP pathogenesis as well as to develop biomarkers for early detection and disease prognosis.
Acknowledgements
This article is supported by the IMPRINT proposal number 4352. We are thankful to the Ministry of Human Resource Development, Government of India, New Delhi for the financial support.
Author contribution
Suranjana Mukherjee: conceptualization and preparation of the original manuscript.
Sanjukta Dasgupta: preparation of the original manuscript.
Pradyumna K. Mishra: conceptualization and vital inputs during manuscript preparation. Koel Chaudhury: conceptualization and critical review of the manuscript.
Funding
This article is supported by the IMPRINT proposal number 4352.
Data availability
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abdel-Shafy HI, Mansour MSM. A review on polycyclic aromatic hydrocarbons: Source, environmental impact, effect on human health and remediation. Egypt J Pet. 2016;25(1):107–123. doi: 10.1016/j.ejpe.2015.03.011. [DOI] [Google Scholar]
- Alfano R, Herceg Z, Nawrot TS, Chadeau-Hyam M, Ghantous A, Plusquin M. The impact of air pollution on our epigenome: how far is the evidence? (A Systematic Review) Curr Environ Health Rep. 2018;5(4):544–578. doi: 10.1007/s40572-018-0218-8. [DOI] [PubMed] [Google Scholar]
- Andreau K, Leroux M, Bouharrour A. Health and cellular impacts of air pollutants: from cytoprotection to cytotoxicity. Biochem Res Int. 2012;2012:493894. doi: 10.1155/2012/493894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow DP, Bartolomei MS. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol. 2014;6(2):a018382. doi: 10.1101/cshperspect.a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera L, Mendoza F, Zuñiga J, Estrada A, Zamora AC, Melendro EI, Ramírez R, Pardo A, Selman M. Functional diversity of T-cell subpopulations in subacute and chronic hypersensitivity pneumonitis. Am J Respir Crit Care Med. 2008;177(1):44–55. doi: 10.1164/rccm.200701-093OC. [DOI] [PubMed] [Google Scholar]
- Bellavia A, Urch B, Speck M et al (2013) DNA hypomethylation, ambient particulate matter, and increased blood pressure: findings from controlled human exposure experiments. J Am Heart Assoc. 2(3). e000212. 10.1161/JAHA.113.000212. Erratum in: J Am Heart Assoc. 2015;4(10):e001981. 10.1161/JAHA.115.001981\ [DOI] [PMC free article] [PubMed]
- Bhargava A, Bunkar N, Aglawe A, et al. Epigenetic biomarkers for risk assessment of particulate matter associated lung cancer. Curr Drug Targets. 2018;19(10):1127–1147. doi: 10.2174/1389450118666170911114342. [DOI] [PubMed] [Google Scholar]
- Bhargava A, Tamrakar S, Aglawe A, et al. Ultrafine particulate matter impairs mitochondrial redox homeostasis and activates phosphatidylinositol 3-kinase mediated DNA damage responses in lymphocytes. Environ Pollut. 2018;234:406–419. doi: 10.1016/j.envpol.2017.11.093. [DOI] [PubMed] [Google Scholar]
- Bind MA, Baccarelli A, Zanobetti A, et al. Air pollution and markers of coagulation, inflammation, and endothelial function: associations and epigene-environment interactions in an elderly cohort. Epidemiology. 2012;23(2):332–340. doi: 10.1097/EDE.0b013e31824523f0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair A, Freeman LB. Epidemiologic studies in agricultural populations: observations and future directions. J Agromed. 2009;14(2):125–131. doi: 10.1080/10599240902779436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake GE, Watson ED. Unravelling the complex mechanisms of transgenerational epigenetic inheritance. Curr Opin Chem Biol. 2016;33:101–107. doi: 10.1016/j.cbpa.2016.06.008. [DOI] [PubMed] [Google Scholar]
- Boffetta P, Järvholm B, Brennan P, Nyrén O. Incidence of lung cancer in a large cohort of non-smoking men from Sweden. Int J Cancer. 2001;94(4):591–593. doi: 10.1002/ijc.1507. [DOI] [PubMed] [Google Scholar]
- Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67(3):876–880. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- Bollati V, Angelici L, Rizzo G, et al. Microvesicle-associated microRNA expression is altered upon particulate matter exposure in healthy workers and in A549 cells. J Appl Toxicol. 2015;35(1):59–67. doi: 10.1002/jat.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton CV, Marutani AN. Air pollution and epigenetics: recent findings. Curr Envir Health Rpt. 2014;1:35–45. doi: 10.1007/s40572-013-0001-9. [DOI] [Google Scholar]
- Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180(5):462–467. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton CV, Salam MT, Wang X, Byun HM, Siegmund KD, Gilliland FD. Particulate matter, DNA methylation in nitric oxide synthase, and childhood respiratory disease. Environ Health Perspect. 2012;120(9):1320–1326. doi: 10.1289/ehp.1104439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton CV, Park C, Siegmund K, et al. NOS1 methylation and carotid artery intima-media thickness in children. Circ Cardiovasc Genet. 2014;7(2):116–122. doi: 10.1161/CIRCGENETICS.113.000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton CV, Gao L, Yao J, Siegmund KD, Lurmann F, Gilliland F. Particulate matter, the newborn methylome, and cardio-respiratory health outcomes in childhood. Environ Epigenet. 2016;2(2):dvw005. doi: 10.1093/eep/dvw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358(11):1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- Butler M, Pongor L, Su YT, et al. MGMT status as a clinical biomarker in glioblastoma. Trends Cancer. 2020;6(5):380–391. doi: 10.1016/j.trecan.2020.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes SA, Weigl BH. Selecting analytical biomarkers for diagnostic applications: a first principles approach. Expert Rev Mol Diagn. 2018;18(1):19–26. doi: 10.1080/14737159.2018.1412258. [DOI] [PubMed] [Google Scholar]
- Byun HM, Panni T, Motta V, et al. Effects of airborne pollutants on mitochondrial DNA methylation. Part Fibre Toxicol. 2013;10:18. doi: 10.1186/1743-8977-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun HM, Colicino E, Trevisi L, Fan T, Christiani DC, Baccarelli AA. Effects of Air Pollution and Blood Mitochondrial DNA Methylation on Markers of Heart Rate Variability. J Am Heart Assoc. 2016;5(4):e003218. doi: 10.1161/JAHA.116.003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud DM, Vergnon JM, Madroszyk A, Melloni BM, Murris M, Dalphin JC, French Group of Environmental Immunoallergic Bronchopulmonary Diseases Bronchoalveolar lavage in hypersensitivity pneumonitis: a series of 139 patients. Inflamm Allergy Drug Targets. 2012;11(1):15–19. doi: 10.2174/187152812798889330. [DOI] [PubMed] [Google Scholar]
- Calderón-Garcidueñas L, Herrera-Soto A, Jury N, et al. Reduced repressive epigenetic marks, increased DNA damage and Alzheimer's disease hallmarks in the brain of humans and mice exposed to particulate urban air pollution. Environ Res. 2020;183:109226. doi: 10.1016/j.envres.2020.109226. [DOI] [PubMed] [Google Scholar]
- Callahan CL, Bonner MR, Nie J, Wang Y, Tao MH, Shields PG, Marian C, Eng KH, Trevisan M, Freudenheim JL. Active and secondhand smoke exposure throughout life and DNA methylation in breast tumors. Cancer Causes Control. 2019;30(1):53–62. doi: 10.1007/s10552-018-1102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos K, Gomes CC, Farias LC, Silva RM, Letra A, Gomez RS. DNA methylation of MMP9 is associated with high levels of MMP-9 messenger RNA in periapical inflammatory lesions. J Endod. 2016;42(1):127–130. doi: 10.1016/j.joen.2015.10.002. [DOI] [PubMed] [Google Scholar]
- Cantone L, Nordio F, Hou L, et al. Inhalable metal-rich air particles and histone H3K4 dimethylation and H3K9 acetylation in a cross-sectional study of steel workers. Environ Health Perspect. 2011;119(7):964–969. doi: 10.1289/ehp.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantone L, Tobaldini E, Favero C, Albetti B, Sacco RM, Torgano G, Ferrari L, Montano N, Bollati V. Particulate air pollution, clock gene methylation, and stroke: effects on stroke severity and disability. Int J Mol Sci. 2020;21(9):3090. doi: 10.3390/ijms21093090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Bromberg PA, Samet JM. COX-2 expression induced by diesel particles involves chromatin modification and degradation of HDAC1. Am J Respir Cell Mol Biol. 2007;37(2):232–239. doi: 10.1165/rcmb.2006-0449OC. [DOI] [PubMed] [Google Scholar]
- Cardenas A, Fadadu RP, Van Der Laan L, Ward-Caviness C, Granger L, Diaz-Sanchez D, Devlin RB, Bind MA. Controlled human exposures to diesel exhaust: a human epigenome-wide experiment of target bronchial epithelial cells. Environ Epigenet. 2021;7(1):dvab003. doi: 10.1093/eep/dvab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. 2016;17(10):1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TL, Yuen ST, Kong CK, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet. 2006;38(10):1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- Chatziioannou A, Georgiadis P, Hebels DG, Liampa I, Valavanis I, Bergdahl IA, Johansson A, Palli D, Chadeau-Hyam M, Siskos AP, Keun H, Botsivali M, de Kok TM, Pérez AE, Kleinjans JC, Vineis P, Kyrtopoulos SA, EnviroGenomarkers project consortium Blood-based omic profiling supports female susceptibility to tobacco smoke-induced cardiovascular diseases. Sci Rep. 2017;7:42870. doi: 10.1038/srep42870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Li H, Cai J, et al. Fine particulate air pollution and the expression of micrornas and circulating cytokines relevant to inflammation, coagulation, and vasoconstriction. Environ Health Perspect. 2018;126(1):017007. doi: 10.1289/EHP1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Liu J, Li B, Liu S, Li X, Tu H. Cigarette Smoke-Induced Hypermethylation of the GCLC Gene Is Associated With COPD. Chest. 2016;149(2):474–482. doi: 10.1378/chest.14-2309. [DOI] [PubMed] [Google Scholar]
- Chi GC, Liu Y, MacDonald JW, et al. Long-term outdoor air pollution and DNA methylation in circulating monocytes: results from the Multi-Ethnic Study of Atherosclerosis (MESA) Environ Health. 2016;15(1):119. doi: 10.1186/s12940-016-0202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi CG, Liu Y, MacDonald JW, Reynolds LM, Enquobahrie DA, Fitzpatrick LA, Kerr KF, Budoff JM, Lee SI, Siscovick D, Kaufman DJ. Epigenome-wide analysis of long-term air pollution exposure and DNA methylation in monocytes: results from the Multi-Ethnic Study of Atherosclerosis. Epigenetics. 2021;5:1–17. doi: 10.1080/15592294.2021.1900028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S, Jaffar Z, Cole E, et al. Prenatal environmental tobacco smoke exposure increases allergic asthma risk with methylation changes in mice. Environ Mol Mutagen. 2017;58(6):423–433. doi: 10.1002/em.22097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61(5 Pt 2):24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- Cimino F, Speciale A, Siracusa L, et al. Cytotoxic effects induced in vitro by organic extracts from urban air particulate matter in human leukocytes. Drug Chem Toxicol. 2014;37(1):32–39. doi: 10.3109/01480545.2013.806529. [DOI] [PubMed] [Google Scholar]
- Clifford RL, Jones MJ, MacIsaac JL, et al. Inhalation of diesel exhaust and allergen alters human bronchial epithelium DNA methylation. J Allergy Clin Immunol. 2017;139(1):112–121. doi: 10.1016/j.jaci.2016.03.046. [DOI] [PubMed] [Google Scholar]
- Cohen AJ. Outdoor air pollution and lung cancer. Environ Health Perspect. 2000;108(Suppl 4):743–750. doi: 10.1289/ehp.00108s4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AJ, Pope CA., 3rd Lung cancer and air pollution. Environ Health Perspect. 1995;103(Suppl 8):219–224. doi: 10.1289/ehp.95103s8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole E, Brown TA, Pinkerton KE, et al. Perinatal exposure to environmental tobacco smoke is associated with changes in DNA methylation that precede the adult onset of lung disease in a mouse model. Inhal Toxicol. 2017;29(10):435–442. doi: 10.1080/08958378.2017.1392655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nys S, Duca RC, Nawrot T, et al. Temporal variability of global DNA methylation and hydroxymethylation in buccal cells of healthy adults: Association with air pollution. Environ Int. 2018;111:301–308. doi: 10.1016/j.envint.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Dekkers KF, Neele AE, Jukema JW, Heijmans BT, de Winther MPJ. Human monocyte-to-macrophage differentiation involves highly localized gain and loss of DNA methylation at transcription factor binding sites. Epigenetics Chromatin. 2019;12(1):34. doi: 10.1186/s13072-019-0279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Ding R, Jin Y, Liu X, et al. Characteristics of DNA methylation changes induced by traffic-related air pollution. Mutat Res Genet Toxicol Environ Mutagen. 2016;796:46–53. doi: 10.1016/j.mrgentox.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Ding R, Jin Y, Liu X, et al. Dose- and time- effect responses of DNA methylation and histone H3K9 acetylation changes induced by traffic-related air pollution. Sci Rep. 2017;7:43737. doi: 10.1038/srep43737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017;15(3):177–186. doi: 10.1016/j.gpb.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shahawi MS, Hamza A, Bashammakh AS, Al-Saggaf WT. An overview on the accumulation, distribution, transformations, toxicity and analytical methods for the monitoring of persistent organic pollutants. Talanta. 2010;80(5):1587–1597. doi: 10.1016/j.talanta.2009.09.055. [DOI] [PubMed] [Google Scholar]
- Esmaeil Mousavi S, Heydarpour P, Reis J, Amiri M, Sahraian MA. Multiple sclerosis and air pollution exposure: mechanisms toward brain autoimmunity. Med Hypotheses. 2017;100:23–30. doi: 10.1016/j.mehy.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst. 2000;92(7):564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- Eze IC, Jeong A, Schaffner E, et al. Genome-wide DNA methylation in peripheral blood and long-term exposure to source-specific transportation noise and air pollution: the SAPALDIA Study. Environ Health Perspect. 2020;128(6):67003. doi: 10.1289/EHP6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Sidiropoulos K, Viteri G, Forner O, Marin-Garcia P, Arnau V, D'Eustachio P, Stein L, Hermjakob H. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics. 2017;18(1):142. doi: 10.1186/s12859-017-1559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13(2):97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- Fernández-Morera JL, Calvanese V, Rodríguez-Rodero S, Menéndez-Torre E, Fraga MF. Epigenetic regulation of the immune system in health and disease. Tissue Antigens. 2010;76(6):431–439. doi: 10.1111/j.1399-0039.2010.01587.x. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Fry RC, Rager JE, Bauer R, et al. Air toxics and epigenetic effects: ozone altered microRNAs in the sputum of human subjects. Am J Physiol Lung Cell Mol Physiol. 2014;306(12):L1129–L1137. doi: 10.1152/ajplung.00348.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15(5):490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Gan L, Yang Y, Li Q, Feng Y, Liu T, Guo W. Epigenetic regulation of cancer progression by EZH2: from biological insights to therapeutic potential. Biomark Res. 2018;6:10. doi: 10.1186/s40364-018-0122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydos LJ, Wang W, Strome S. Gene repression. H3K27me and PRC2 transmit a memory of repression across generations and during development. Science. 2014;345(6203):1515–1518. doi: 10.1126/science.1255023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehlhaus MW, Gift JS, Hogan KA, Kopylev L, Schlosser PM, Kadry AR. Approaches to cancer assessment in EPA’s integrated risk information system. Toxicol Appl Pharmacol. 2011;254:170–180. doi: 10.1016/j.taap.2010.10.019. [DOI] [PubMed] [Google Scholar]
- Ghorani-Azam A, Riahi-Zanjani B, Balali-Mood M (2016) Effects of air pollution on human health and practical measures for prevention in Iran. J Res Med Sci 21:65. Published 2016 Sep 1. 10.4103/1735-1995.189646 [DOI] [PMC free article] [PubMed]
- Gilmour PS, Rahman I, Donaldson K, MacNee W. Histone acetylation regulates epithelial IL-8 release mediated by oxidative stress from environmental particles. Am J Physiol Lung Cell Mol Physiol. 2003;284(3):L533–L540. doi: 10.1152/ajplung.00277.2002. [DOI] [PubMed] [Google Scholar]
- Goobie GC, Nouraie M, Zhang Y, Kass DJ, Ryerson CJ, Carlsten C, Johannson KA. Air Pollution and Interstitial Lung Diseases: Defining Epigenomic Effects. Am J Respir Crit Care Med. 2020;202(9):1217–1224. doi: 10.1164/rccm.202003-0836PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson JM, MacDonald JW, Bammler TK, Chien WM, Chin MT. In utero exposure to diesel exhaust is associated with alterations in neonatal cardiomyocyte transcription, DNA methylation and metabolic perturbation. Part Fibre Toxicol. 2019;16(1):17. doi: 10.1186/s12989-019-0301-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoraszczuk EL, Ptak A. Endocrine-disrupting chemicals: some actions of POPs on female reproduction. Int J Endocrinol. 2013;2013:828532. doi: 10.1155/2013/828532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruzieva O, Xu CJ, Yousefi P, et al. Prenatal particulate air pollution and DNA methylation in newborns: an epigenome-wide meta-analysis. Environ Health Perspect. 2019;127(5):57012. doi: 10.1289/EHP4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunier RB, Harnly ME, Reynolds P, Hertz A, Von Behren J. Agricultural pesticide use in California: pesticide prioritization, use densities, and population distributions for a childhood cancer study. Environ Health Perspect. 2001;109(10):1071–1078. doi: 10.1289/ehp.011091071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Byun HM, Zhong J, Motta V, Barupal J, Zheng Y, Dou C, Zhang F, McCracken JP, Diaz A, Marco SG, Colicino S, Schwartz J, Wang S, Hou L, Baccarelli AA. Effects of short-term exposure to inhalable particulate matter on DNA methylation of tandem repeats. Environ Mol Mutagen. 2014;55(4):322–335. doi: 10.1002/em.21838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha TY. MicroRNAs in human diseases: from cancer to cardiovascular disease. Immune Netw. 2011;11(3):135–154. doi: 10.4110/in.2011.11.3.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari S, Raghu G, Caminati A, Cruciani M, Franchini M, Mannucci P. Fibrotic interstitial lung diseases and air pollution: a systematic literature review. Eur Respir Rev. 2020;29(157):200093. doi: 10.1183/16000617.0093-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L et al (2004) The Gene Ontology (GO) database and informatics resource, 32(Database issue):D258-61. Nucleic Acids Res. 10.1093/nar/gkh036 [DOI] [PMC free article] [PubMed]
- He LR, Liu MZ, Li BK, et al. High expression of EZH2 is associated with tumor aggressiveness and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Int J Cancer. 2010;127(1):138–147. doi: 10.1002/ijc.25031. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Pershagen G. Cancer risk of air pollution: epidemiological evidence. Environ Health Perspect. 1994;102(Suppl 4):187–192. doi: 10.1289/ehp.94102s4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Greally JM. Epigenetics, cellular memory and gene regulation. Curr Biol. 2016;26(14):R644–R648. doi: 10.1016/j.cub.2016.06.011. [DOI] [PubMed] [Google Scholar]
- Hew KM, Walker AI, Kohli A, et al. Childhood exposure to ambient polycyclic aromatic hydrocarbons is linked to epigenetic modifications and impaired systemic immunity in T cells. Clin Exp Allergy. 2015;45(1):238–248. doi: 10.1111/cea.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchins MP, Wong JJ, Suthers G, et al. Inheritance of a cancer-associated MLH1 germ-line epimutation. N Engl J Med. 2007;356(7):697–705. doi: 10.1056/NEJMoa064522. [DOI] [PubMed] [Google Scholar]
- Hombach S, Kretz M. Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol. 2016;937:3–17. doi: 10.1007/978-3-319-42059-2_1. [DOI] [PubMed] [Google Scholar]
- Hu X, Xiang D, Xie Y, et al. LSD1 suppresses invasion, migration and metastasis of luminal breast cancer cells via activation of GATA3 and repression of TRIM37 expression. Oncogene. 2019;38(44):7017–7034. doi: 10.1038/s41388-019-0923-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WY, Hsu SD, Huang HY, Sun YM, Chou CH, Weng SL, Huang HD. MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2015;43(Database issue):D856–D861. doi: 10.1093/nar/gku1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wu C, Fu Y, Guo L, Kong X, Cai H. Methylation analysis for multiple gene promoters in non-small cell lung cancers in high indoor air pollution region in China. Bull Cancer. 2018;105(9):746–754. doi: 10.1016/j.bulcan.2018.05.004. [DOI] [PubMed] [Google Scholar]
- Huang X, Zhang S, Li K, Thimmapuram J, Xie S, Wren J. ViewBS: a powerful toolkit for visualization of high-throughput bisulfite sequencing data. Bioinformatics. 2018;34(4):708–709. doi: 10.1093/bioinformatics/btx633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SK, Tripathi P, Koneva LA, Cavalcante RG, Craig N, Scruggs AM, Sartor MA, Deng F, Chen Y. Effect of concentration and duration of particulate matter exposure on the transcriptome and DNA methylome of bronchial epithelial cells. Environ Epigenet. 2021;7(1):dvaa022. doi: 10.1093/eep/dvaa022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Medicine (US) Committee on the Implications of Dioxin in the Food Supply . Dioxins and Dioxin-like Compounds in the Food Supply: Strategies to Decrease Exposure. Washington (DC): National Academies Press (US); 2003. [PubMed] [Google Scholar]
- Janssen BG, Godderis L, Pieters N, et al. Placental DNA hypomethylation in association with particulate air pollution in early life. Part Fibre Toxicol. 2013;10:22. doi: 10.1186/1743-8977-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardim MJ. microRNAs: implications for air pollution research. Mutat Res. 2011;717(1-2):38–45. doi: 10.1016/j.mrfmmm.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Jardim MJ, Fry RC, Jaspers I, Dailey L, Diaz-Sanchez D. Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environ Health Perspect. 2009;117(11):1745–1751. doi: 10.1289/ehp.0900756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Khurana Hershey GK. Genetic and epigenetic influence on the response to environmental particulate matter. J Allergy Clin Immunol. 2012;129(1):33–41. doi: 10.1016/j.jaci.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer. 2011;2(6):607–617. doi: 10.1177/1947601910393957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannson KA, Balmes JR, Collard HR. Air pollution exposure: a novel environmental risk factor for interstitial lung disease? Chest. 2015;147(4):1161–1167. doi: 10.1378/chest.14-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KC, de Voogt P. Persistent organic pollutants (POPs): state of the science. Environ Pollut. 1999;100(1-3):209–221. doi: 10.1016/S0269-7491(99)00098-6. [DOI] [PubMed] [Google Scholar]
- Kalchiem-Dekel O, Galvin JR, Burke AP, Atamas SP, Todd NW. Interstitial lung disease and pulmonary fibrosis: a practical approach for general medicine physicians with focus on the medical history. J Clin Med. 2018;7(12):476. doi: 10.3390/jcm7120476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson O, Baccarelli AA. Environmental health and long non-coding RNAs. Curr Environ Health Rep. 2016;3(3):178–187. doi: 10.1007/s40572-016-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasdagli MI, Katsouyanni K, Dimakopoulou K, Samoli E. Air pollution and Parkinson's disease: a systematic review and meta-analysis up to 2018. Int J Hyg Environ Health. 2019;222(3):402–409. doi: 10.1016/j.ijheh.2018.12.006. [DOI] [PubMed] [Google Scholar]
- Kettunen E, Hernandez-Vargas H, Cros MP, et al. Asbestos-associated genome-wide DNA methylation changes in lung cancer. Int J Cancer. 2017;141(10):2014–2029. doi: 10.1002/ijc.30897. [DOI] [PubMed] [Google Scholar]
- Kim KY, Kim DS, Lee SK, et al. Association of low-dose exposure to persistent organic pollutants with global DNA hypomethylation in healthy Koreans. Environ Health Perspect. 2010;118(3):370–374. doi: 10.1289/ehp.0901131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloog I, Coull BA, Zanobetti A, Koutrakis P, Schwartz JD. Acute and chronic effects of particles on hospital admissions in New-England. PLoS One. 2012;7(4):e34664. doi: 10.1371/journal.pone.0034664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli A, Garcia MA, Miller RL, et al. Secondhand smoke in combination with ambient air pollution exposure is associated with increasedx CpG methylation and decreased expression of IFN-γ in T effector cells and Foxp3 in T regulatory cells in children. Clin Epigenetics. 2012;4(1):17. doi: 10.1186/1868-7083-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korosec P, Osolnik K, Kern I, Silar M, Mohorcic K, Kosnik M. Expansion of pulmonary CD8+CD56+ natural killer T-cells in hypersensitivity pneumonitis. Chest. 2007;132(4):1291–1297. doi: 10.1378/chest.07-0128. [DOI] [PubMed] [Google Scholar]
- Kresovich JK, Zhang Z, Fang F, et al. Histone 3 modifications and blood pressure in the Beijing Truck Driver Air Pollution Study. Biomarkers. 2017;22(6):584–593. doi: 10.1080/1354750X.2017.1347961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt OK, Zhang J, Pinkerton KE. Pulmonary health effects of air pollution. Curr Opin Pulm Med. 2016;22(2):138–143. doi: 10.1097/MCP.0000000000000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd-Acosta C. Epigenetic signatures as biomarkers of exposure. Curr Environ Health Rep. 2015;2(2):117–125. doi: 10.1007/s40572-015-0051-2. [DOI] [PubMed] [Google Scholar]
- Larriba E, del Mazo J. Role of non-coding RNAs in the transgenerational epigenetic transmission of the effects of reprotoxicants. Int J Mol Sci. 2016;17(4):452. doi: 10.3390/ijms17040452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latifovic L, Villeneuve PJ, Parent MÉ, et al. Bladder cancer and occupational exposure to diesel and gasoline engine emissions among Canadian men. Cancer Med. 2015;4(12):1948–1962. doi: 10.1002/cam4.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq B, Platel A, Antherieu S, et al. Genetic and epigenetic alterations in normal and sensitive COPD-diseased human bronchial epithelial cells repeatedly exposed to air pollution-derived PM2.5. Environ Pollut. 2017;230:163–177. doi: 10.1016/j.envpol.2017.06.028. [DOI] [PubMed] [Google Scholar]
- Lee JS. Exploring cancer genomic data from the cancer genome atlas project. BMB Rep. 2016;49(11):607–611. doi: 10.5483/bmbrep.2016.49.11.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kalia V, Perera F, et al. Prenatal airborne polycyclic aromatic hydrocarbon exposure, LINE1 methylation and child development in a Chinese cohort. Environ Int. 2017;99:315–320. doi: 10.1016/j.envint.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chen R, Cai J, Cui X, Huang N, Kan H. Short-term exposure to fine particulate air pollution and genome-wide DNA methylation: a randomized, double-blind, crossover trial. Environ Int. 2018;120:130–136. doi: 10.1016/j.envint.2018.07.041. [DOI] [PubMed] [Google Scholar]
- Li R, Fang L, Pu Q, Bu H, Zhu P, Chen Z, Yu M, Li X, Weiland T, Bansal A, Ye SQ, Wei Y, Jiang J, Wu M. MEG3-4 is a miRNA decoy that regulates IL-1β abundance to initiate and then limit inflammation to prevent sepsis during lung infection. Sci Signal. 2018;11(536):eaao2387. doi: 10.1126/scisignal.aao2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Ma J, Li X, et al. Aberrantly expressed long non-coding RNAs in air pollution-induced congenital defects. J Cell Mol Med. 2019;23(11):7717–7725. doi: 10.1111/jcmm.14645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang T, Wang Y, Xu M, Zhang L, Li X, Liu Z, Gao S, Jia Q, Fan Y, Wang Z, Wu N, Zhang X, Dai Y, Kong F, Wang W, Duan H. Particulate matter air pollution and the expression of microRNAs and pro-inflammatory genes: Association and mediation among children in Jinan, China. J Hazard Mater. 2020;389:121843. doi: 10.1016/j.jhazmat.2019.121843. [DOI] [PubMed] [Google Scholar]
- Liang J, Zhu H, Li C, Ding Y, Zhou Z, Wu Q. Neonatal exposure to benzo[a]pyrene decreases the levels of serum testosterone and histone H3K14 acetylation of the StAR promoter in the testes of SD rats. Toxicology. 2012;302(2-3):285–291. doi: 10.1016/j.tox.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Liang Y, Hu L, Li J, Liu F, Jones KC, Li D, Liu J, Chen D, You J, Yu Z, Zhang G, Dong G, Ma H. Short-term personal PM2.5 exposure and change in DNA methylation of imprinted genes: panel study of healthy young adults in Guangzhou city, China. Environ Pollut. 2021;275:116601. doi: 10.1016/j.envpol.2021.116601. [DOI] [PubMed] [Google Scholar]
- Lien SA, Young L, Gau BS, KSP S. Meta-prediction of MTHFR gene polymorphism-mutations, air pollution, and risks of leukemia among world populations. Oncotarget. 2017;8(3):4387–4398. doi: 10.18632/oncotarget.13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Zhang X, Feng N, et al. LncRNA LCPAT1 Mediates Smoking/ Particulate Matter 2.5-Induced Cell Autophagy and Epithelial-Mesenchymal Transition in Lung Cancer Cells via RCC2. Cell Physiol Biochem. 2018;47(3):1244–1258. doi: 10.1159/000490220. [DOI] [PubMed] [Google Scholar]
- Lisanti S, Omar WA, Tomaszewski B, et al. Comparison of methods for quantification of global DNA methylation in human cells and tissues. PLoS One. 2013;8(11):e79044. doi: 10.1371/journal.pone.0079044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhou Y, Boggs SE, Belinsky SA, Liu J. Cigarette smoke induces demethylation of prometastatic oncogene synuclein-gamma in lung cancer cells by downregulation of DNMT3B. Oncogene. 2007;26(40):5900–5910. doi: 10.1038/sj.onc.1210400. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang L, Winterroth LC, et al. Epigenetically mediated pathogenic effects of phenanthrene on regulatory T cells. J Toxicol. 2013;2013:967029. doi: 10.1155/2013/967029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Xu J, Chen Y, et al. Characterization of genome-wide H3K27ac profiles reveals a distinct PM2.5-associated histone modification signature. Environ Health. 2015;14:65. doi: 10.1186/s12940-015-0052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Chen PC, Lien PC, Liao YP. Prenatal perfluorooctyl sulfonate exposure and Alu DNA hypomethylation in cord blood. Int J Environ Res Public Health. 2018;15(6):1066. doi: 10.3390/ijerph15061066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang G, Li Q, et al. Polycomb group proteins EZH2 and EED directly regulate androgen receptor in advanced prostate cancer. Int J Cancer. 2019;145(2):415–426. doi: 10.1002/ijc.32118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis D, Huang W, Chen G. The International Agency for Research on Cancer (IARC) evaluation of the carcinogenicity of outdoor air pollution: focus on China. Chin. J Cancer. 2014;33(4):189–196. doi: 10.5732/cjc.014.10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P, Weisshaupt K, Mikeska T, et al. Oncogenic HRAS suppresses clusterin expression through promoter hypermethylation. Oncogene. 2006;25(35):4890–4903. doi: 10.1038/sj.onc.1209502. [DOI] [PubMed] [Google Scholar]
- Ma B, Huang Z, Wang Q, Zhang J, Zhou B, Wu J. Integrative analysis of genetic and epigenetic profiling of lung squamous cell carcinoma (LSCC) patients to identify smoking level relevant biomarkers. BioData Min. 2019;12:18. doi: 10.1186/s13040-019-0207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Li J, Xu Y, Wang Y, Yao Y, Liu Q, Wang M, Zhao X, Fan R, Chen J, Zhang B, Cai Z, Han H, Yang Z, Yuan W, Zhong Y, Chen X, Ma JZ, Payne TJ, Xu Y, Ning Y, Cui W, Li MD. Identification of 34 genes conferring genetic and pharmacological risk for the comorbidity of schizophrenia and smoking behaviors. Aging (Albany NY) 2020;12(3):2169–2225. doi: 10.18632/aging.102735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane LA, Murphy PR. MicroRNA: biogenesis, function and role in cancer. Curr Genomics. 2010;11(7):537–561. doi: 10.2174/138920210793175895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigano J, Baccarelli A, Mittleman MA, et al. Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ Health Perspect. 2011;119(7):977–982. doi: 10.1289/ehp.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maghbooli Z, Hossein-Nezhad A, Adabi E, et al. Air pollution during pregnancy and placental adaptation in the levels of global DNA methylation. PLoS ONE. 2018;13(7):e0199772. doi: 10.1371/journal.pone.0199772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud MM, Sanad EF, Hamdy NM. MicroRNAs' role in the environment-related non-communicable diseases and link to multidrug resistance, regulation, or alteration. Environ Sci Pollut Res Int. 2021;28(28):36984–37000. doi: 10.1007/s11356-021-14550-w. [DOI] [PubMed] [Google Scholar]
- Majello B, Gorini F, Saccà CD, Amente S. Expanding the role of the histone lysine-specific demethylase LSD1 in cancer. Cancers (Basel) 2019;11(3):324. doi: 10.3390/cancers11030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manisalidis I, Stavropoulou E, Stavropoulos A, Bezirtzoglou E. Environmental and health impacts of air pollution: a review. Front Public Health. 2020;8:14. doi: 10.3389/fpubh.2020.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EM, Fry RC. Environmental influences on the epigenome: exposure- associated DNA methylation in human populations. Annu Rev Public Health. 2018;39:309–333. doi: 10.1146/annurev-publhealth-040617-014629. [DOI] [PubMed] [Google Scholar]
- Martinez-Zamudio R, Ha HC. Environmental epigenetics in metal exposure. Epigenetics. 2011;6(7):820–827. doi: 10.4161/epi.6.7.16250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merid SK, Bustamante M, Standl M, Sunyer J, Heinrich J, Lemonnier N, Aguilar D, Antó JM, Bousquet J, Santa-Marina L, Lertxundi A, Bergström A, Kull I, Wheelock ÅM, Koppelman GH, Melén E, Gruzieva O. Integration of gene expression and DNA methylation identifies epigenetically controlled modules related to PM2.5 exposure. Environ Int. 2021;146:106248. doi: 10.1016/j.envint.2020.106248. [DOI] [PubMed] [Google Scholar]
- Miller CN, Dye JA, Schladweiler MC, et al. Acute inhalation of ozone induces DNA methylation of apelin in lungs of Long-Evans rats. Inhal Toxicol. 2018;30(4-5):178–186. doi: 10.1080/08958378.2018.1483984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miousse IR, Chalbot MC, Aykin-Burns N, et al. Epigenetic alterations induced by ambient particulate matter in mouse macrophages. Environ Mol Mutagen. 2014;55(5):428–435. doi: 10.1002/em.21855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghadam BT, Zamani N, Komorowski J, Grabherr M. PiiL: visualization of DNA methylation and gene expression data in gene pathways. BMC Genomics. 2017;18(1):571. doi: 10.1186/s12864-017-3950-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta V, Favero C, Dioni L, et al. MicroRNAs are associated with blood-pressure effects of exposure to particulate matter: Results from a mediated moderation analysis. Environ Res. 2016;146:274–281. doi: 10.1016/j.envres.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouly TA, Toms LL. Breast cancer and persistent organic pollutants (excluding DDT): a systematic literature review. Environ Sci Pollut Res Int. 2016;23(22):22385–22407. doi: 10.1007/s11356-016-7577-1. [DOI] [PubMed] [Google Scholar]
- Munthe-Kaas MC, Bertelsen RJ, Torjussen TM, et al. Pet keeping and tobacco exposure influence CD14 methylation in childhood. Pediatr Allergy Immunol. 2012;23(8):747–754. doi: 10.1111/pai.12021. [DOI] [PubMed] [Google Scholar]
- Nadeau K, McDonald-Hyman C, Noth EM, et al. Ambient air pollution impairs regulatory T-cell function in asthma. J Allergy Clin Immunol. 2010;126(4):845–852.e10. doi: 10.1016/j.jaci.2010.08.008. [DOI] [PubMed] [Google Scholar]
- Ngwa EN, Kengne AP, Tiedeu-Atogho B, Mofo-Mato EP, Sobngwi E (2015) Persistent organic pollutants as risk factors for type 2 diabetes. Diabetol Metab Syndr 7:–41. Published 2015 Apr 30. 10.1186/s13098-015-0031-6 [DOI] [PMC free article] [PubMed]
- Ning J, Li P, Zhang B, Han B, Su X, Wang Q, Wang X, Li B, Kang H, Zhou L, Chu C, Zhang N, Pang Y, Niu Y, Zhang R. miRNAs deregulation in serum of mice is associated with lung cancer related pathway deregulation induced by PM2.5. Environ Pollut. 2019;254(Pt A):112875. doi: 10.1016/j.envpol.2019.07.043. [DOI] [PubMed] [Google Scholar]
- Olkhov-Mitsel E, Bapat B. Strategies for discovery and validation of methylated and hydroxymethylated DNA biomarkers. Cancer Med. 2012;1(2):237–260. doi: 10.1002/cam4.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostuni R, Natoli G, Cassatella MA, Tamassia N. Epigenetic regulation of neutrophil development and function. Semin Immunol. 2016;28(2):83–93. doi: 10.1016/j.smim.2016.04.002. [DOI] [PubMed] [Google Scholar]
- Pacchierotti F, Spanò M. Environmental impact on DNA methylation in the germline: state of the art and gaps of knowledge. Biomed Res Int. 2015;2015:123484. doi: 10.1155/2015/123484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanivel R, Vinayachandran V, Biswal S, Deiuliis JA, Padmanabhan R, Park B, Gangwar RS, Durieux JC, Ebreo Cara EA, Das L, Bevan G, Fayad ZA, Tawakol A, Jain MK, Rao S, Rajagopalan S. Exposure to air pollution disrupts circadian rhythm through alterations in chromatin dynamics. iScience. 2020;23(11):101728. doi: 10.1016/j.isci.2020.101728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo A, Barrios R, Gaxiola M, Segura-Valdez L, Carrillo G, Estrada A, Mejía M, Selman M. Increase of lung neutrophils in hypersensitivity pneumonitis is associated with lung fibrosis. Am J Respir Crit Care Med. 2000;161(5):1698–1704. doi: 10.1164/ajrccm.161.5.9907065. [DOI] [PubMed] [Google Scholar]
- Patel AM, Ryu JH, Reed CE. Hypersensitivity pneumonitis: current concepts and future questions. J Allergy Clin Immunol. 2001;108(5):661–670. doi: 10.1067/mai.2001.119570. [DOI] [PubMed] [Google Scholar]
- Pavanello S, Bonzini M, Angelici L, et al. Extracellular vesicle-driven information mediates the long-term effects of particulate matter exposure on coagulation and inflammation pathways. Toxicol Lett. 2016;259:143–150. doi: 10.1016/j.toxlet.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavanello S, Campisi M, Mastrangelo G, Hoxha M, Bollati V. The effects of everyday-life exposure to polycyclic aromatic hydrocarbons on biological age indicators. Environ Health. 2020;19(1):128. doi: 10.1186/s12940-020-00669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Bind MC, Colicino E, et al. Particulate Air Pollution and Fasting Blood Glucose in Nondiabetic Individuals: Associations and Epigenetic Mediation in the Normative Aging Study, 2000-2011. Environ Health Perspect. 2016;124(11):1715–1721. doi: 10.1289/EHP183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera F, Tang WY, Herbstman J, et al. Relation of DNA methylation of 5'-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS ONE. 2009;4(2):e4488. doi: 10.1371/journal.pone.0004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusquin M, Guida F, Polidoro S, et al. DNA methylation and exposure to ambient air pollution in two prospective cohorts. Environ Int. 2017;108:127–136. doi: 10.1016/j.envint.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozzer A, Dominici F, Haines A, Witt C, Münzel T, Lelieveld J. Regional and global contributions of air pollution to risk of death from COVID-19. Cardiovasc Res. 2020;116(14):2247–2253. doi: 10.1093/cvr/cvaa288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada D, López G, Solleiro-Villavicencio H, Garcia-Cuellar C, Baccarelli AA. Molecular and cellular mechanisms linking air pollution and bone damage. Environ Res. 2020;185:109465. doi: 10.1016/j.envres.2020.109465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunicki M, Stell L, Dinakarpandian D, et al. Exposure to NO2, CO, and PM2.5 is linked to regional DNA methylation differences in asthma. Clin Epigenetics. 2018;10:2. doi: 10.1186/s13148-017-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunicki M, Cauwenberghs N, Lee J, Zhou X, Movassagh H, Noth E, Lurmann F, Hammond SK, Balmes JR, Desai M, Wu JC, Nadeau KC. Air pollution exposure is linked with methylation of immunoregulatory genes, altered immune cell profiles, and increased blood pressure in children. Sci Rep. 2021;11(1):4067. doi: 10.1038/s41598-021-83577-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitch N, Jones MJ, Gladish N, Faino AV, Strand M, Morin AM, MacIsaac J, Lin DTS, Reynolds PR, Singh A, Gelfand EW, Kobor MS, Carlsten C. Methylation of cysteinyl leukotriene receptor 1 genes associates with lung function in asthmatics exposed to traffic-related air pollution. Epigenetics. 2021;16(2):177–185. doi: 10.1080/15592294.2020.1790802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricceri F, Trevisan M, Fiano V, et al. Seasonality modifies methylation profiles in healthy people. PLoS ONE. 2014;9(9):e106846. doi: 10.1371/journal.pone.0106846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider CF, Carlsten C. Air pollution and DNA methylation: effects of exposure in humans. Clin Epigenetics. 2019;11(1):131. doi: 10.1186/s13148-019-0713-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodosthenous RS, Kloog I, Colicino E, Zhong J, Herrera LA, Vokonas P, Schwartz J, Baccarelli AA, Prada D. Extracellular vesicle-enriched microRNAs interact in the association between long-term particulate matter and blood pressure in elderly men. Environ Res. 2018;167:640–649. doi: 10.1016/j.envres.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Vera T, Ochoa-Martínez ÁC, Pruneda-Álvarez LG, Zarazúa S, Pérez-Maldonado IN. Exposure to biomass smoke is associated with an increased expression of circulating miRNA-126 and miRNA-155 in Mexican women: a pilot study. Drug Chem Toxicol. 2019;42(3):335–342. doi: 10.1080/01480545.2018.1526181. [DOI] [PubMed] [Google Scholar]
- Runyon RS, Cachola LM, Rajeshuni N, et al. Asthma discordance in twins is linked to epigenetic modifications of T cells. PLoS One. 2012;7(11):e48796. doi: 10.1371/journal.pone.0048796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116(11):1547–1552. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salam MT, Byun HM, Lurmann F, Breton CV, Wang X, Eckel SP, Gilliland FD. Genetic and epigenetic variations in inducible nitric oxide synthase promoter, particulate pollution, and exhaled nitric oxide levels in children. J Allergy Clin Immunol. 2012;129(1):232–239.e7. doi: 10.1016/j.jaci.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Ruiz L, Buendía-Roldán I, Pérez-Rubio G, Ambrocio-Ortiz E, Mejía M, Montaño M, Falfán-Valencia R. MMP2 polymorphism affects plasma matrix metalloproteinase (MMP)-2 levels, and correlates with the decline in lung function in hypersensitivity pneumonitis positive to autoantibodies patients. Biomolecules. 2019;9(10):574. doi: 10.3390/biom9100574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidl C, Delacher M, Huehn J, Feuerer M. Epigenetic mechanisms regulating T-cell responses. J Allergy Clin Immunol. 2018;142(3):728–743. doi: 10.1016/j.jaci.2018.07.014. [DOI] [PubMed] [Google Scholar]
- Selman M, Pardo A, King TE., Jr Hypersensitivity pneumonitis: insights in diagnosis and pathobiology. Am J Respir Crit Care Med. 2012;186(4):314–324. doi: 10.1164/rccm.201203-0513CI. [DOI] [PubMed] [Google Scholar]
- Serra RW, Fang M, Park SM, Hutchinson L, Green MR. A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. Elife. 2014;3:e02313. doi: 10.7554/eLife.02313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivapurkar N, Gazdar AF. DNA methylation based biomarkers in non-invasive cancer screening. Curr Mol Med. 2010;10(2):123–132. doi: 10.2174/156652410790963303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A, Bunkar N, Kumar R, Bhargava A, Tiwari R, Chaudhury K, Goryacheva IY, Mishra PK. Air pollution associated epigenetic modifications: Transgenerational inheritance and underlying molecular mechanisms. Sci Total Environ. 2019;656:760–777. doi: 10.1016/j.scitotenv.2018.11.381. [DOI] [PubMed] [Google Scholar]
- Silva IR, Ramos MCAS, Arantes LMRB, Lengert AVH, Oliveira MA, Cury FP, Martins Pereira G, Santos AG, Barbosa F, Jr, Vasconcellos PC, Cuenin C, Herceg Z, Silveira HCS. Evaluation of DNA methylation changes and micronuclei in workers exposed to a construction environment. Int J Environ Res Public Health. 2019;16(6):902. doi: 10.3390/ijerph16060902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sima M, Rossnerova A, Simova Z, Rossner P., Jr The impact of air pollution exposure on the MicroRNA machinery and lung cancer development. J Pers Med. 2021;11(1):60. doi: 10.3390/jpm11010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonian PL, Roark CL, Born WK, O'Brien RL, Fontenot AP. Gammadelta T cells and Th17 cytokines in hypersensitivity pneumonitis and lung fibrosis. Transl Res. 2009;154(5):222–227. doi: 10.1016/j.trsl.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Collins BF, Bairwa M, Joshi JM, Talwar D, Singh N, Samaria JK, Mangal DK, Singh V, Raghu G. Hypersensitivity pneumonitis and its correlation with ambient air pollution in urban India. Eur Respir J. 2019;53(2):1801563. doi: 10.1183/13993003.01563-2018. [DOI] [PubMed] [Google Scholar]
- Soberanes S, Gonzalez A, Urich D, et al. Particulate matter Air Pollution induces hypermethylation of the p16 promoter Via a mitochondrial ROS-JNK-DNMT1 pathway. Sci Rep. 2012;2:275. doi: 10.1038/srep00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofer T, Baccarelli A, Cantone L, et al. Exposure to airborne particulate matter is associated with methylation pattern in the asthma pathway. Epigenomics. 2013;5(2):147–154. doi: 10.2217/epi.13.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solaimani P, Saffari A, Sioutas C, Bondy SC, Campbell A. Exposure to ambient ultrafine particulate matter alters the expression of genes in primary human neurons. Neurotoxicology. 2017;58:50–57. doi: 10.1016/j.neuro.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Spagnolo P, Rossi G, Cavazza A, Bonifazi M, Paladini I, Bonella F, Sverzellati N, Costabel U. Hypersensitivity pneumonitis: a comprehensive review. J Investig Allergol Clin Immunol. 2015;25(4):237–250. [PubMed] [Google Scholar]
- Sridhar S, Schembri F, Zeskind J, et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genomics. 2008;9:259. doi: 10.1186/1471-2164-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson OA, Villanueva A, Vidal A, Martí L, Esteller M. BRCA1 epigenetic inactivation predicts sensitivity to platinum-based chemotherapy in breast and ovarian cancer. Epigenetics. 2012;7(11):1225–1229. doi: 10.4161/epi.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenz L, Schechter DS, Serpa SR, Paoloni-Giacobino A. Intergenerational transmission of DNA methylation signatures associated with early life stress. Curr Genomics. 2018;19(8):665–675. doi: 10.2174/1389202919666171229145656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpfel M, Jancar N, Virant-Klun I. New challenge: mitochondrial epigenetics? Stem Cell Rev Rep. 2018;14(1):13–26. doi: 10.1007/s12015-017-9771-z. [DOI] [PubMed] [Google Scholar]
- Suárez-Álvarez B, Baragaño Raneros A, Ortega F, López-Larrea C. Epigenetic modulation of the immune function: a potential target for tolerance. Epigenetics. 2013;8(7):694–702. doi: 10.4161/epi.25201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga M, Yamasaki H, Nakagawa K, Kohrogi H, Ando M. Mechanisms accounting for granulomatous responses in hypersensitivity pneumonitis. Sarcoidosis Vasc Diffuse Lung Dis. 1997;14(2):131–138. [PubMed] [Google Scholar]
- Tan Y, Wang Y, Zou Y, et al. LncRNA LOC101927514 regulates PM2.5-driven inflammation in human bronchial epithelial cells through binding p-STAT3 protein. Toxicol Lett. 2020;319:119–128. doi: 10.1016/j.toxlet.2019.10.009. [DOI] [PubMed] [Google Scholar]
- Tantoh DM, Wu MF, Ho CC, Lung CC, Lee KJ, Nfor ON, Liaw YC, Hsu SY, Chen PH, Lin C, Chu HW, Liaw YC, Liaw YP. SOX2 promoter hypermethylation in non-smoking Taiwanese adults residing in air pollution areas. Clin Epigenetics. 2019;11(1):46. doi: 10.1186/s13148-019-0647-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantini L, Bonzini M, Tripodi A, et al. Blood hypomethylation of inflammatory genes mediates the effects of metal-rich airborne pollutants on blood coagulation. Occup Environ Med. 2013;70(6):418–425. doi: 10.1136/oemed-2012-101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teneng I, Montoya-Durango DE, Quertermous JL, Lacy ME, Ramos KS. Reactivation of L1 retrotransposon by benzo(a)pyrene involves complex genetic and epigenetic regulation. Epigenetics. 2011;6(3):355–367. doi: 10.4161/epi.6.3.14282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng CS, Wu BH, Yen MR, Chen PY. MethGET: web-based bioinformatics software for correlating genome-wide DNA methylation and gene expression. BMC Genomics. 2020;21(1):375. doi: 10.1186/s12864-020-6722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6(7):828–837. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomankova T, Petrek M, Gallo J, Kriegova E (2011) MicroRNAs: emerging regulators of immune-mediated diseases. Scand J Immunol. 10.1111/j.1365-3083.2011.02650.x [DOI] [PubMed]
- Tomatis L, editor. Indoor and outdoor air pollution and human cancer. Berlin Heidelberg: Springer; 1993. [Google Scholar]
- Tommasi S, Zheng A, Yoon JI, Li AX, Wu X, Besaratinia A. Whole DNA methylome profiling in mice exposed to second hand smoke. Epigenetics. 2012;7(11):1302–1314. doi: 10.4161/epi.22453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toms LM, Hearn L, Mueller JF, Harden FA. Assessing infant exposure to persistent organic pollutants via dietary intake in Australia. Food Chem Toxicol. 2016;87:166–171. doi: 10.1016/j.fct.2015.12.018. [DOI] [PubMed] [Google Scholar]
- Tran NQV, Miyake K. Neurodevelopmental Disorders and Environmental Toxicants: Epigenetics as an Underlying Mechanism. Int J Genomics. 2017;2017:7526592. doi: 10.1155/2017/7526592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trerotola M, Relli V, Simeone P, Alberti S. Epigenetic inheritance and the missing heritability. Hum Genomics. 2015;9(1):17. doi: 10.1186/s40246-015-0041-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsamou M, Nawrot TS, Carollo RM, Trippas AJ, Lefebvre W, Vanpoucke C, Vrijens K. Prenatal particulate air pollution exposure and expression of the miR-17/92 cluster in cord blood: Findings from the ENVIRONAGE birth cohort. Environ Int. 2020;142:105860. doi: 10.1016/j.envint.2020.105860. [DOI] [PubMed] [Google Scholar]
- van der Wijst MG, van Tilburg AY, Ruiters MH, Rots MG. Experimental mitochondria-targeted DNA methylation identifies GpC methylation, not CpG methylation, as potential regulator of mitochondrial gene expression. Sci Rep. 2017;7(1):177. doi: 10.1038/s41598-017-00263-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vawda S, Mansour R, Takeda A, et al. Associations between inflammatory and immune response genes and adverse respiratory outcomes following exposure to outdoor air pollution: a HuGE systematic review. Am J Epidemiol. 2014;179(4):432–442. doi: 10.1093/aje/kwt269. [DOI] [PubMed] [Google Scholar]
- Verbeek B, Southgate TD, Gilham DE, Margison GP. O6-Methylguanine-DNA methyltransferase inactivation and chemotherapy. Br Med Bull. 2008;85:17–33. doi: 10.1093/bmb/ldm036. [DOI] [PubMed] [Google Scholar]
- Verigos J, Karakaidos P, Kordias D, et al. The histone demethylase LSD1/ΚDM1A mediates chemoresistance in breast cancer via regulation of a stem cell program. Cancers (Basel) 2019;11(10):1585. doi: 10.3390/cancers11101585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira SE. The health burden of pollution: the impact of prenatal exposure to air pollutants. Int J Chron Obstruct Pulmon Dis. 2015;10:1111–1121. doi: 10.2147/COPD.S40214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chen L, Li D, et al. Microarray analysis of lung long non-coding RNAs in cigarette smoke-exposed mouse model. Oncotarget. 2017;8(70):115647–115656. doi: 10.18632/oncotarget.23362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, O'Brien KM, Xu Z, Sandler DP, Taylor JA, Weinberg CR. Long-term ambient fine particulate matter and DNA methylation in inflammation pathways: results from the Sister Study. Epigenetics. 2020;15(5):524–535. doi: 10.1080/15592294.2019.1699894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Plusquin M, Ghantous A, Herceg Z, Alfano R, Cox B, Nawrot TS. DNA methylation of insulin-like growth factor 2 and H19 cluster in cord blood and prenatal air pollution exposure to fine particulate matter. Environ Health. 2020;19(1):129. doi: 10.1186/s12940-020-00677-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zhao J, Wang Y, et al. Genome-wide DNA methylation analysis reveals significant impact of long-term ambient air pollution exposure on biological functions related to mitochondria and immune response. Environ Pollut. 2020;264:114707. doi: 10.1016/j.envpol.2020.114707. [DOI] [PubMed] [Google Scholar]
- Warden CD, Lee H, Tompkins JD, Li X, Wang C, Riggs AD, Yu H, Jove R, Yuan YC. COHCAP: an integrative genomic pipeline for single-nucleotide resolution DNA methylation analysis. Nucleic Acids Res. 2013;41(11):e117. doi: 10.1093/nar/gkt242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins DJ, Wellenius GA, Butler RA, Bartell SM, Fletcher T, Kelsey KT. Associations between serum perfluoroalkyl acids and LINE-1 DNA methylation. Environ Int. 2014;63:71–76. doi: 10.1016/j.envint.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Schatten H, Sun QY. Environmental epigenetic inheritance through gametes and implications for human reproduction. Hum Reprod Update. 2015;21(2):194–208. doi: 10.1093/humupd/dmu061. [DOI] [PubMed] [Google Scholar]
- Weinhold B. Epigenetics: the science of change. Environ Health Perspect. 2006;114(3):A160–A167. doi: 10.1289/ehp.114-a160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AJ, Kresovich JK, Keller JP, Xu Z, Kaufman JD, Weinberg CR, Taylor JA, Sandler DP. Air pollution, particulate matter composition and methylation-based biologic age. Environ Int. 2019;132:105071. doi: 10.1016/j.envint.2019.105071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2006) Regional Office for Europe & Joint WHO/Convention Task Force on the Health Aspects of Air Pollution. (2006). Health risks of particulate matter from long-range transboundary air pollution. https://apps.who.int/iris/handle/10665/107691
- WHO (2013) Air Pollution and Cancer. IARC Scientific Publication No. 161. Edited by Straif K, Cohen A, Samet J.https://publications.iarc.fr/Book-And-Report-Series/Iarc-Scientific-Publications/Air-Pollution-And-Cancer-2013
- WHO (2018) Ambient (outdoor) air pollution. 2018. https://www.who.int/airpollution/ambient/en/
- Wilhelm-Benartzi CS, Christensen BC, Koestler DC, et al. Association of secondhand smoke exposures with DNA methylation in bladder carcinomas. Cancer Causes Control. 2011;22(8):1205–1213. doi: 10.1007/s10552-011-9788-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Liu MC, Liang M, Fu J. Sirt1 protects against thrombomodulin down-regulation and lung coagulation after particulate matter exposure. Blood. 2012;119(10):2422–2429. doi: 10.1182/blood-2011-04-350413. [DOI] [PubMed] [Google Scholar]
- Wu C, Ding W, Li Q, Wang W, Deng M, Jin R, Pang B, Yang X. The correlation between pulmonary fibrosis and methylation of peripheral Smad3 in cases of pigeon breeder's lung in a Chinese Uygur population. Oncotarget. 2017;8(26):43104–43113. doi: 10.18632/oncotarget.17763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SM, Chen ZF, Young L, Shiao SP. Meta-prediction of the effect of methylenetetrahydrofolate reductase polymorphisms and air pollution on Alzheimer's disease risk. Int J Environ Res Public Health. 2017;14(1):63. doi: 10.3390/ijerph14010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Luo Z, Pang B, Wang W, Deng M, Jin R, Muhataer X, Li Y, Li Q, Yang X. Associations of Pulmonary Fibrosis with Peripheral Blood Th1/Th2 Cell Imbalance and EBF3 Gene Methylation in Uygur Pigeon Breeder's Lung Patients. Cell Physiol Biochem. 2018;47(3):1141–1151. doi: 10.1159/000490208. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Word B, Lyn-Cook L, Jr, Lyn-Cook B, Hammons G. Cigarette smoke condensate and individual constituents modulate DNA methyltransferase expression in human liver cells. SAGE Open Med. 2015;3:2050312115578317. doi: 10.1177/2050312115578317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CJ, Söderhäll C, Bustamante M, et al. DNA methylation in childhood asthma: an epigenome-wide meta-analysis. Lancet Respir Med. 2018;6(5):379–388. doi: 10.1016/S2213-2600(18)30052-3. [DOI] [PubMed] [Google Scholar]
- Xu R, Li S, Guo S, et al. Environmental temperature and human epigenetic modifications: a systematic review. Environ Pollut. 2020;259:113840. doi: 10.1016/j.envpol.2019.113840. [DOI] [PubMed] [Google Scholar]
- Yang SR, Chida AS, Bauter MR, et al. Cigarette smoke induces proinflammatory cytokine release by activation of NF-kappaB and posttranslational modifications of histone deacetylase in macrophages. Am J Physiol Lung Cell Mol Physiol. 2006;291(1):L46–L57. doi: 10.1152/ajplung.00241.2005. [DOI] [PubMed] [Google Scholar]
- Yang J, Liu Y, Zhang H, Zhang H, Wang W, Fan Y. Urinary 1-hydroxypyrene and smoking are determinants of LINE-1 and AhRR promoter methylation in coke oven workers. Mutat Res Genet Toxicol Environ Mutagen. 2018;826:33–40. doi: 10.1016/j.mrgentox.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Yauk C, Polyzos A, Rowan-Carroll A, et al. Germ-line mutations, DNA damage, and global hypermethylation in mice exposed to particulate air pollution in an urban/industrial location. Proc Natl Acad Sci U S A. 2008;105(2):605–610. doi: 10.1073/pnas.0705896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Sun D, Li D, et al (2018a) Long non-coding RNA expression patterns in lung tissues of chronic cigarette smoke induced COPD mouse model [published correction appears in Sci Rep. 2019 May 9;9(1):7398]. Sci Rep. 2018;8(1):7609. Published 2018 May 15. 10.1038/s41598-018-25702-3 [DOI] [PMC free article] [PubMed]
- Zhang X, Biagini Myers JM, Burleson JD, et al. Nasal DNA methylation is associated with childhood asthma. Epigenomics. 2018;10(5):629–641. doi: 10.2217/epi-2017-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen X, Weirauch MT, et al. Diesel exhaust and house dust mite allergen lead to common changes in the airway methylome and hydroxymethylome. Environ Epigenet. 2018;4(3):dvy020. doi: 10.1093/eep/dvy020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao CN, Xu Z, Wu GC, Mao YM, Liu LN, Qian-Wu DYL, Tao SS, Zhang Q, Sam NB, Fan YG, Zou YF, Ye DQ, Pan HF. Emerging role of air pollution in autoimmune diseases. Autoimmun Rev. 2019;18(6):607–614. doi: 10.1016/j.autrev.2018.12.010. [DOI] [PubMed] [Google Scholar]
- Zhu T, Zhang X, Chen X, Brown AP, Weirauch MT, Guilbert TW, Khurana Hershey GK, Biagini JM, Ji H. Nasal DNA methylation differentiates severe from non-severe asthma in African-American children. Allergy. 2021;76(6):1836–1845. doi: 10.1111/all.14655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zö Zöchbauer-Müller S, Lam S, Toyooka S, et al. Aberrant methylation of multiple genes in the upper aerodigestive tract epithelium of heavy smokers. Int J Cancer. 2003;107(4):612–616. doi: 10.1002/ijc.11458. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.