Abstract
Circulating tumor DNA (ctDNA) molecular residual disease (MRD) following curative-intent treatment strongly predicts recurrence in multiple tumor types, but whether further treatment can improve outcomes in patients with MRD remains unclear. We applied CAPP-Seq ctDNA analysis to 218 samples from 65 patients receiving chemoradiation therapy (CRT) for locally advanced NSCLC, including 28 patients receiving consolidation immune checkpoint inhibition (CICI). Patients with undetectable ctDNA after CRT had excellent outcomes whether or not they received CICI. Among such patients, one died from CICI-related pneumonitis, highlighting the potential utility of only treating patients with MRD. In contrast, patients with MRD after CRT who received CICI had significantly better outcomes than patients who did not receive CICI. Furthermore, the ctDNA response pattern early during CICI identified patients responding to consolidation therapy. Our results suggest that CICI improves outcomes for NSCLC patients with MRD and that ctDNA analysis may facilitate personalization of consolidation therapy.
Introduction
Lung cancer is the leading cause of cancer deaths worldwide1. Patients with unresectable locoregionally advanced non-small cell lung cancer (NSCLC) are primarily managed with definitive chemoradiation (CRT)2, but long-term survival is poor and most patients develop progressive disease3. Consolidation immune checkpoint inhibition (CICI) following CRT was recently shown to improve progression-free and overall survival in patients with NSCLC4. However, a significant fraction of such patients are cured by CRT alone5, and tumors recur in the majority of patients despite CICI therapy4, suggesting opportunities for further personalization of treatment. Furthermore, since gross disease is either cleared by CRT or difficult to distinguish from normal tissue changes induced by radiation therapy, tracking response to CICI is usually not possible using current standard-of-care radiologic studies.
Tumors release circulating tumor DNA (ctDNA) into peripheral blood that can be detected and quantified in liquid biopsies to monitor treatment responses6. Detection of circulating tumor DNA molecular residual disease (MRD) following curative-intent treatment predicts the development of progressive disease across multiple tumor types7–11, and we previously showed that detection of MRD following CRT predicts the development of progressive disease in NSCLC with high sensitivity and specificity12. However, it remains unclear if CICI can improve outcomes in these patients, especially because CICI may only be able to improve outcomes in patients with residual disease levels too low to be detected by ctDNA analysis. Accordingly, we performed a retrospective study examining whether ctDNA could serve as a biomarker both to identify NSCLC patients treated with CRT who might benefit from CICI and to track treatment responses.
Results
Patient characteristics and pre-treatment ctDNA detection
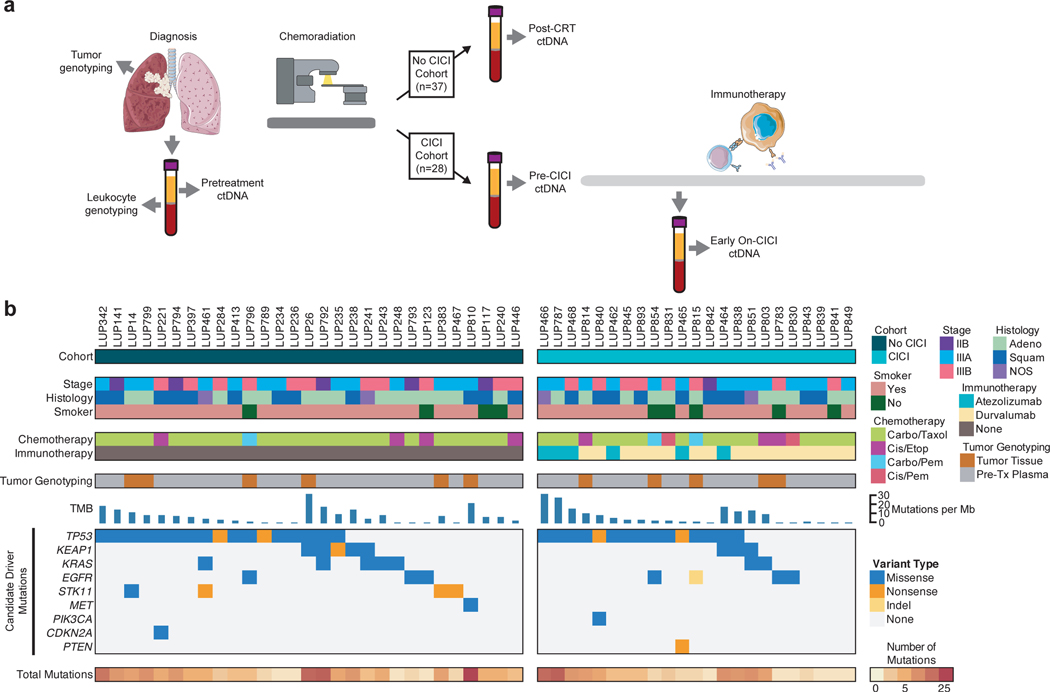
Using Cancer Personalized Profiling by deep Sequencing (CAPP-Seq), we analyzed 218 blood and tissue samples that had been prospectively collected for the purpose of ctDNA analysis from two cohorts of patients with localized NSCLC treated with definitive CRT with or without CICI (Fig. 1a). The first cohort of 37 patients received CRT without CICI as part of the therapeutic care standard prior to regulatory approval of durvalumab (No CICI cohort), with 17 patients having been previously reported in a prior study12. Plasma samples were collected pretreatment and within 4 months of completing all first line therapy for localized NSCLC (post-CRT). The second cohort consisted of 28 patients receiving CRT followed by CICI (CICI cohort). Plasma samples were collected pretreatment, a median of 1 week after completing CRT prior to starting CICI (pre-CICI), and a median of 11 weeks into CICI therapy (early on-CICI). The majority of patients in the CICI cohort received commercial durvalumab (79%) and the remainder received atezolizumab as part of the DETERRED trial (NCT02525757). All patients had unresectable stage IIB-IIIB NSCLC with most having locally advanced disease. The median radiation dose was 66 Gy delivered in 30 fractions, and carboplatin/paclitaxel was the most common concurrent chemotherapy regimen in both cohorts. Median follow-up was 19 months from the start of CRT. Baseline patient characteristics were not significantly different between the two cohorts (Supplementary Table S1).
Fig. 1: Study schema and pretreatment genotypes for patients with locoregionally advanced non-small cell lung cancer treated with chemoradiation and consolidation immunotherapy.
a, Schematic of treatment, genotyping, and ctDNA monitoring for the two patient cohorts. Tumor genotyping was performed using tumor tissue if available or pretreatment plasma in combination with peripheral blood leukocytes. Plasma samples were collected for ctDNA quantification pretreatment and within 4 months of completing all planned radiation and chemotherapy (post-CRT) in patients treated without consolidation immune checkpoint inhibition (No CICI cohort). In patients who received consolidation immune checkpoint inhibition (CICI cohort), plasma samples were collected pretreatment, a median of 1 week after completing chemoradiation before starting consolidation immune checkpoint inhibition (Pre-CICI), and a median of 11 weeks into consolidation immune checkpoint inhibition (Early On-CICI). b, Plot of patient characteristics and tumor variants for patients in both cohorts with variants identified from tumor tissue or pretreatment plasma. Each column represents pretreatment information for a single patient. The top heat maps depict key patient characteristics. The middle bar graph displays tumor mutational burden (TMB) extrapolated from the mutation rate measured by CAPP-Seq. The bottom heat maps show mutations in candidate driver genes along with total number of single nucleotide variants detected.
In order to enable tumor-informed ctDNA MRD detection which increases sensitivity compared to tumor-naive cfDNA analysis, tumor genotyping was performed before first line therapy, with 11 patients profiled using tumor tissue and the remaining 54 patients genotyped using pretreatment plasma (Fig. 1b). We observed the expected frequency of mutations in canonical lung cancer driver genes in both cohorts13,14, including in genes such as TP53, KEAP1, KRAS, EGFR, STK11, MET, PIK3CA, CDKN2A, and PTEN (Supplementary Table S2-5). The median tumor mutational burden was 6.2 non-synonymous mutations per Mb as inferred from CAPP-Seq12. Pretreatment ctDNA was detected in 78% of patients in the No CICI cohort with a median concentration of 25.1 haploid genome equivalents per ml (hGE/ml) and 75% of patients in the CICI cohort with a median concentration of 21.6 hGE/ml (P=0.86 for percent detected and P=0.99 for median concentration). Thirteen patients (20%) were excluded from the ctDNA monitoring analysis because tumor tissue was not available and no variants were detected in the pretreatment plasma sample using tumor-naïve calling. The pre-CICI sample for LUP464 was excluded from analysis due to insufficient cell-free DNA input for sequencing. A median of 8 variants were monitored in patients with tumor genotyping performed on tumor tissue, and a median of 4 variants were monitored in patients with tumor genotyping performed from pre-treatment plasma.
Monitoring ctDNA in patients with NSCLC treated with CRT with or without CICI
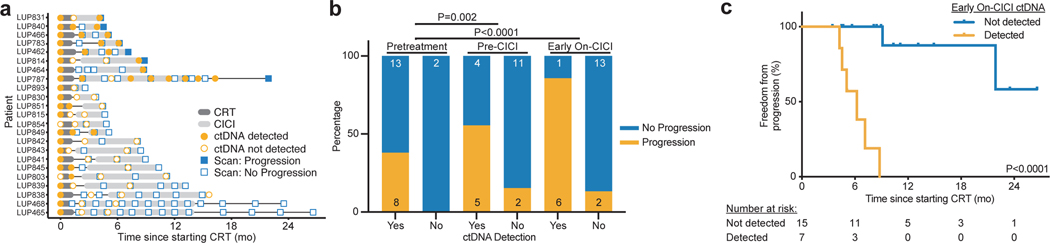
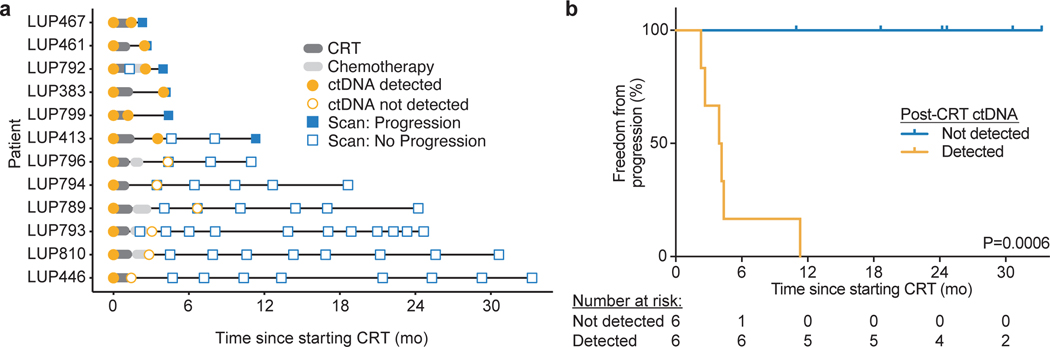
We first sought to validate our previous observation that MRD after CRT alone can predict the development of progressive disease12. Excluding the 17 patients analyzed in our previous study, 12 patients in the No CICI cohort had variants called from tumor tissue or pre-treatment plasma. Of these previously uncharacterized patients, ctDNA was detected in 6 patients at the post-CRT sample (50%, Extended Data 1a). Freedom from progression (FFP) 24 months after starting CRT was 0% in patients with detectable ctDNA as compared with 100% in patients with undetectable ctDNA (P=0.0006, Extended Data 1b). These results validate our prior observation that MRD is highly prognostic for risk of progressive disease after CRT12.
Next, we performed serial ctDNA monitoring in the CICI cohort before and early during CICI (Fig. 2a). Among the 8 patients who progressed, ctDNA was detected prior to or at the time of disease progression in all patients (100%), with mean lead time of 4.1 months between ctDNA detection and radiographic progression. Detection of ctDNA pre-CICI or early on-CICI was a strong predictor of risk of disease progression (Fig. 2b). Six of the 7 patients (86%) with ctDNA detected in the early on-CICI sample developed progressive disease compared with 2 of the 15 patients (13%) with ctDNA not detected. In patients with detectable ctDNA in the early on-CICI sample, we observed 0% FFP at 12 months after starting CRT as compared with 87.5% in patients with undetectable ctDNA (P<0.0001, HR 84.4, 95% CI 12.3–579.9, Fig. 2c). These results demonstrate that detectable ctDNA after starting CICI is highly prognostic.
Fig. 2: ctDNA changes during therapy are associated with outcomes in NSCLC patients treated with chemoradiation therapy (CRT) and consolidation immune checkpoint inhibition (CICI).
a, Event chart showing timing of therapy, progression based on RECIST 1.1 evaluation of imaging, and results of ctDNA testing for each patient in the CICI cohort. b, Proportion of patients with ctDNA detected or not detected pretreatment, pre-CICI, and early on-CICI in the CICI cohort who developed progressive disease during follow-up. The number of patients in each group is displayed on the graph. P values were calculated using two-sided Fisher’s exact tests. c, Kaplan-Meier analysis of freedom from progression in the CICI cohort stratified by ctDNA detection in the early on-CICI sample (n=15 not detected, n=7 detected). P value was calculated using a two-sided log-rank test.
Undetectable ctDNA after CRT
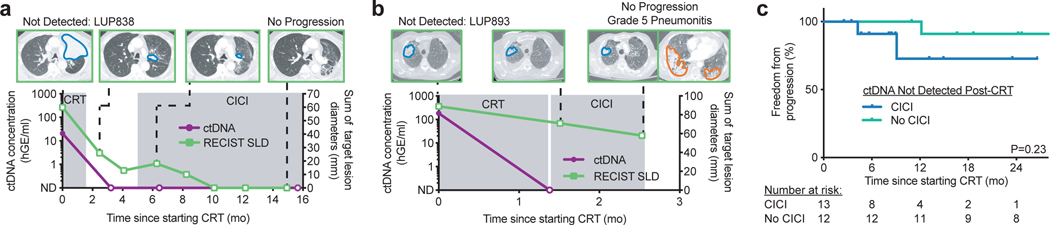
We next sought to determine if ctDNA detection after CRT could identify patients who are most likely to benefit from CICI. The majority of patients in our CICI cohort had undetectable ctDNA after CRT (n=13; 59%). Outcomes of these patients were excellent, with a one-year FFP of 80%. Among the patients in this group, two clinical vignettes are instructive (LUP838 and LUP893, Fig. 3a,b). Most patients had courses similar to LUP838, who had detectable ctDNA prior to CRT for stage IIIA lung squamous cell carcinoma (Fig. 3a). Specifically, 1.5 months after completing CRT and before starting CICI, ctDNA was undetectable. At the early on-CICI time point 1.5 months into consolidation immunotherapy and at last follow-up 15 months after starting CRT, ctDNA remained undetectable and there was no evidence of disease.
Fig. 3: Patients with ctDNA not detected after chemoradiation therapy (CRT) may not benefit from consolidation immune checkpoint inhibition (CICI).
a, Example of longitudinal CT imaging with sum of target lesion longest diameters measured according to RESIST 1.1 (RECIST SLD, right y-axis) and ctDNA concentrations (left y-axis) for a patient in the CICI cohort with ctDNA not detected after CRT. ctDNA remained not detected early on-CICI and at last follow-up with no evidence of disease. Tumors are outlined in blue on the CT images. A total of 10 patients in the CICI cohort had similar ctDNA testing and clinical outcomes. b, Example of longitudinal CT imaging and ctDNA concentrations for the one patient in the CICI cohort with ctDNA not detected after CRT who developed grade 5 pneumonitis during CICI. Pneumonitis is outlined in orange on the CT image. c, Kaplan-Meier analysis of freedom from progression in patients with ctDNA not detected pre-CICI treated with CICI (n=13) and patients with ctDNA not detected post-CRT treated without CICI (n=12). P value was calculated using a two-sided log-rank test.
In stark contrast, the experience of a second patient in this group (LUP893) highlights how the personalization of CICI based on MRD could have clinical utility. LUP893 received CRT with carboplatin and paclitaxel and 66 Gy in 30 fractions to the right upper lobe for stage IIIB lung squamous cell carcinoma, and ctDNA became undetectable (Fig. 3b). Shortly after starting CICI, he developed worsening cough and shortness of breath and died of respiratory failure despite aggressive management. Autopsy showed an acute and organizing pneumonia in the bilateral lungs outside of the irradiated region consistent with drug-induced pneumonitis most likely from immunotherapy15. There was no evidence of residual carcinoma.
To explore if the potential toxicity associated with CICI might be avoidable in patients such as LUP893, we compared outcomes of patients with undetectable ctDNA after CRT who received CICI (n=13) to patients with negative ctDNA after CRT but did who not receive CICI (n=12). Strikingly, FFP between these two groups was statistically indistinguishable (P=0.23, Fig. 3c). While our analysis was not powered to detect small differences in outcome, these results suggest that patients with negative ctDNA after CRT may not derive a large benefit from CICI.
Detectable ctDNA after CRT and ctDNA dynamics during CICI
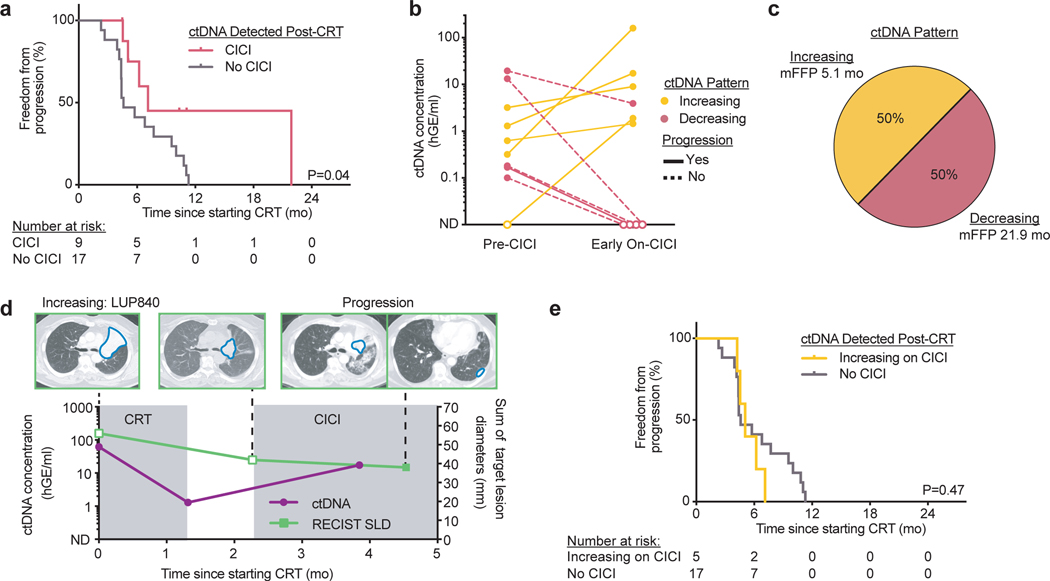
In contrast to patients with undetectable ctDNA after CRT, patients with detectable ctDNA after CRT in the CICI cohort (n=9, 41%) had significantly better FFP than patients with detectable ctDNA after CRT in the No CICI cohort (n=17, P=0.04, Fig. 4a). To determine if ctDNA kinetics during CICI could further clarify which patients benefit from CICI, we compared ctDNA concentrations at the pre-CICI and early on-CICI blood draws. In patients with ctDNA detected at either of these time points, we observed two ctDNA response patterns (Fig. 4b,c): 1. Rising ctDNA concentrations early during CICI treatment (“Increasing”, 50%); 2. Decreasing ctDNA concentrations early during CICI treatment (“Decreasing”, 50%).
Fig. 4: ctDNA dynamics predict benefit from consolidation immune checkpoint inhibition (CICI) after chemoradiation therapy (CRT).
a, Kaplan-Meier analysis of freedom from progression in patients with ctDNA detected pre-CICI treated with CICI (n=9) and patients with ctDNA detected post-CRT treated without CICI (n=17). P value was calculated using a two-sided log-rank test. b, ctDNA concentrations for the two ctDNA patterns observed in patients from the CICI cohort with ctDNA detected in the pre-CICI or early on-CICI samples. “Increasing”: ctDNA concentration increases early on-CICI. “Decreasing”: ctDNA concentration decreases early on-CICI. Only patients with evaluable pre-CICI and early on-CICI samples are included. Patients with disease progression in the follow-up period are denoted with a solid line, and patients without disease progression are denoted with a dashed line. c, Percentage of patients and median freedom from progression (mFFP) for each ctDNA pattern. d, Example of longitudinal CT imaging and ctDNA concentrations for a patient with the ctDNA “Increasing” pattern. ctDNA was detected pre-CICI and increased during CICI in a patient who developed pleural metastases, suggesting ctDNA can identify a lack of response to CICI. A total of 4 patients in the CICI cohort had similar ctDNA testing and clinical outcomes. e, Kaplan-Meier analysis of freedom from progression in patients with the ctDNA “Increasing” pattern during CICI (n=5) and patients with ctDNA detected post-CRT treated without CICI (n=17). P value was calculated using a two-sided log-rank test.
Rising ctDNA levels during CICI portended poor outcomes. All such patients with increasing ctDNA developed progressive disease within 4.5 months of starting CICI, indicating a lack of response to immunotherapy. In one such patient (LUP840), ctDNA concentration decreased but remained detectable after undergoing CRT with 72 Gy in 30 fractions for Stage IIIB lung squamous cell carcinoma (Fig. 4d). Only 1.5 months into CICI, the ctDNA concentration rose 13-fold and a chest CT 3 weeks later demonstrated pleural metastases. As a group, patients with an “Increasing” ctDNA pattern during CICI had statistically indistinguishable outcomes from patients with detectable ctDNA after CRT but who did not receive CICI (P=0.47, Fig. 4e). These results suggest that rising ctDNA concentrations early during CICI identify patients who have primary resistance to immunotherapy.
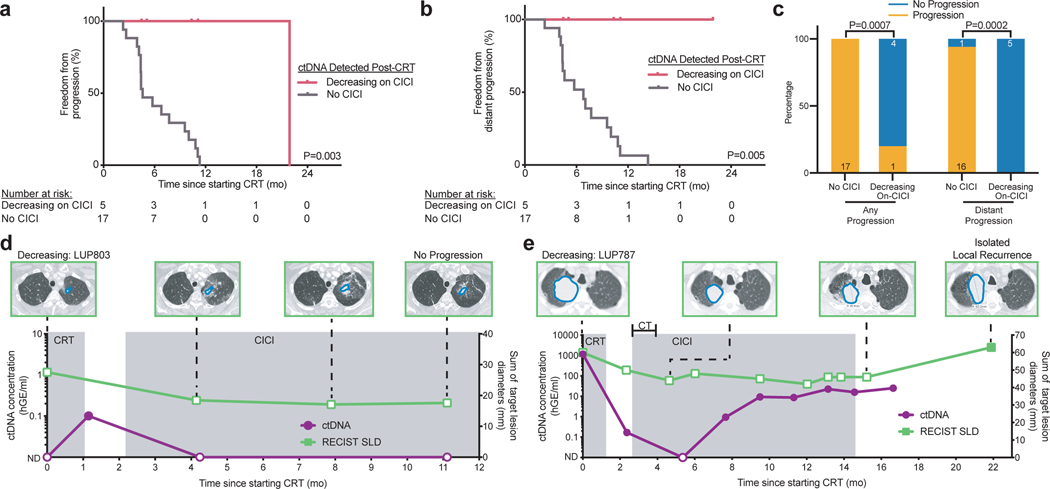
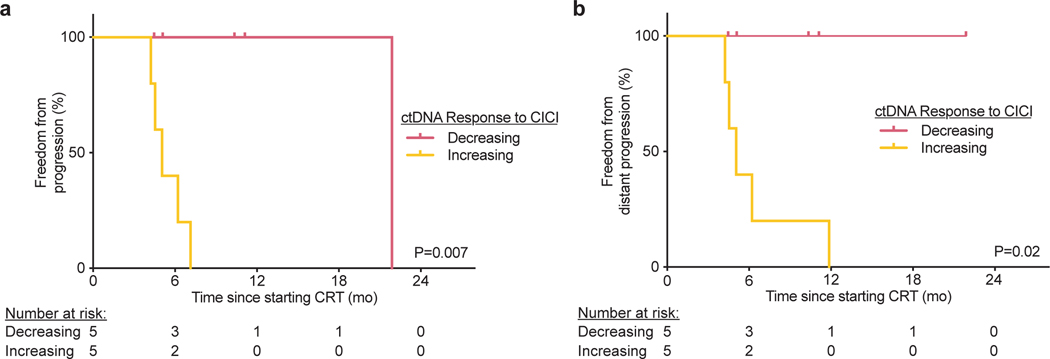
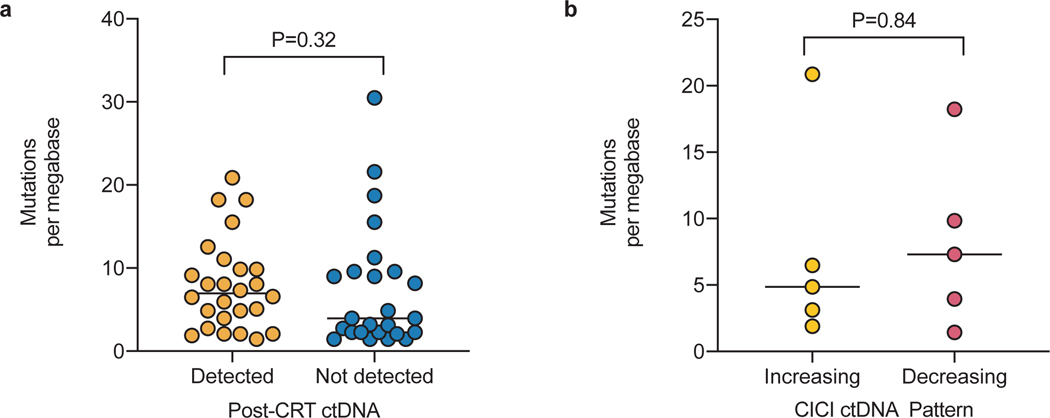
Lastly, outcomes in patients with decreasing ctDNA during CICI were excellent, with a 1-year FFP of 100%. Strikingly, FFP of patients with a “Decreasing” ctDNA response pattern was significantly longer than patients with detectable ctDNA after CRT who did not receive CICI (P=0.003, Fig. 5a). Furthermore, none of the CICI patients developed distant progression compared to 94% of patients with detectable ctDNA after CRT who did not receive CICI (P=0.005, Fig. 5b,c). Similarly, patients with a “Decreasing” ctDNA response pattern had significantly better FFP (P=0.007) and freedom from distant progression (P=0.02) than patients with an “Increasing” ctDNA response pattern (Extended Data 2). Pretreatment TMB level was not associated with likelihood of ctDNA detection after CRT or the ctDNA response pattern to CICI (Extended Data 3).
Fig. 5: Decreasing ctDNA concentration during consolidation immune checkpoint inhibition (CICI) identifies MRD-positive patients with improved outcomes.
a-b, Kaplan-Meier analysis of (a) freedom from progression and (b) freedom from distant progression in patients with ctDNA detected after chemoradiation therapy with the ctDNA “Decreasing” pattern during CICI (n=5) and patients with ctDNA detected after chemoradiation therapy not treated with CICI (n=17). P values were calculated using a two-sided log-rank test. c, Proportion of patients with ctDNA detected after chemoradiation therapy not treated with CICI or patients with the ctDNA “Decreasing” pattern during CICI who developed any disease progression and distant progression. The number of patients in each group is displayed on the graph. P values were calculated using two-sided Fisher’s exact tests. d-e, Longitudinal CT imaging, sum of target lesion longest diameters measured by RESIST 1.1 (RECIST SLD, right y-axis), and ctDNA concentrations (left y-axis) for two patients with the ctDNA “Decreasing” pattern during CICI. A total of 5 patients in the CICI cohort had similar ctDNA testing with 1 of 5 patients developing disease progression. d, ctDNA was detected following CRT and converted to not detected during CICI in a patient with no evidence of disease progression at last follow-up. e, ctDNA was detected following CRT and temporarily became not detected during CICI with two cycles of concurrent carboplatin and paclitaxel (CT) before increasing 14 months prior to progression in a patient with an isolated local failure 22 months after starting CRT. Tumors are outlined in blue on the CT images.
Two patients with a “Decreasing” ctDNA response pattern and the longest follow-up both had prolonged disease-free intervals. LUP803 had ctDNA detected post-CRT with 66 Gy in 24 fractions to a small left upper lobe stage IIIA lung adenocarcinoma, suggesting persistent disease (Fig. 5d). Two months into CICI, ctDNA was no longer detectable. Follow-up imaging demonstrated post-radiation changes with no evidence of progressive disease 11 months after starting CRT. In another case, LUP787 underwent CRT with 66 Gy in 30 fractions to a large right upper lobe Stage IIIA lung adenocarcinoma and had a significant drop in ctDNA after treatment (Fig. 5e). Three months into CICI with two cycles of concurrent carboplatin and paclitaxel, ctDNA became undetectable before rising again three months later. The patient developed a late isolated local recurrence 22 months after starting CRT. These results suggest that ctDNA monitoring after CRT and early during CICI could identify the subset of patients most likely to benefit from CICI.
Discussion
Multiple studies across a variety of solid tumor types have demonstrated an extremely high risk of recurrence in patients with detectable MRD after definitive local therapy7–12. In this context, a limited number of published cases have suggested that adjuvant chemotherapy can lower ctDNA levels after definitive therapy9,11,16,17. However, an open question in the field has been whether further systemic therapy can not only decrease MRD in a subset of patients but more importantly improve ultimate clinical outcomes. To the best of our knowledge, our results provide the first evidence that systemic therapy (in this case immunotherapy) can potentially improve the clinical outcomes of patients with detectable MRD. Furthermore, our findings suggest that ctDNA analysis could potentially guide the decision to administer CICI after CRT and distinguish responders and non-responders early during CICI.
In our study, patients with ctDNA detected after chemoradiotherapy who received CICI had superior FFP when compared to patients with ctDNA detected post-CRT who did not receive consolidation immunotherapy. ctDNA kinetics early during CICI further clarified which patients benefited from consolidation therapy, with patients whose ctDNA concentration decreased during CICI having superior outcomes compared to patients with a rising ctDNA concentration. Additional studies will be necessary to determine if ctDNA analysis can determine the optimal duration of immunotherapy in patients responding to treatment. Patients with a rising ctDNA concentration early on-CICI progressed at a similar rate to patients with ctDNA detected after CRT alone, suggesting that ctDNA analysis early during CICI can rapidly identify non-responders that might benefit from escalation of treatment or changing systemic therapy. In our study, the early on-CICI sample was collected a median of 11 weeks into immunotherapy, but it is possible that responders could be identified even earlier during CICI. For example, in patients with metastatic lung cancer, ctDNA changes within 8 weeks after starting anti-PD1 or anti-PD-L1 therapy are strongly associated with treatment response18–20.
Patients in whom ctDNA was no longer detected post-CRT had low rates of progression irrespective of whether or not they were treated with CICI. It should be noted that our study was not powered to detect a small benefit from CICI in these patients, and a much larger study would be necessary to definitively demonstrate non-inferiority for withholding consolidation immunotherapy in patients without MRD. However, our results suggest that personalization of CICI based on the presence of MRD after CRT or change in ctDNA concentrations during CICI could be a powerful approach for rational therapy selection that balances predicted risk versus benefit as well as costs associated with consolidation immunotherapy. While there is a high threshold for withholding a therapy associated with a survival benefit in a large randomized trial4, rare but severe toxicities can be seen in patients treated with immunotherapy21. Indeed, one patient in our cohort who had negative ctDNA after CRT but died from drug-induced pneumonitis is an example of this. Still, withholding CICI in patients with negative ctDNA after CRT would risk harming patients with false negative MRD measurements who would miss an opportunity to potentially benefit from CICI. That said, we observed that only 8.3% of patients in our No CICI cohort with undetectable MRD after CRT developed progressive disease. Based on the 10.7% overall survival benefit at 24 months for CICI on the PACIFIC trial, which led to FDA approval of this treatment strategy4, this suggests that 112 ctDNA MRD-negative patients would need to be treated with CICI for a single such patient to benefit.
Randomized studies will be required to definitively test the clinical utility of personalizing CICI using ctDNA MRD analysis before this approach could routinely be applied to patients. However, even in light of the large number of ctDNA MRD-negative patients needed to treat suggested by our results, such studies may be challenging to run given potential concerns about withholding CICI in patients with false negative ctDNA MRD results. One potential approach to address this concern would be to perform serial ctDNA testing in patients with undetectable ctDNA after CRT and to initiate CICI if ctDNA becomes detectable at a later time point. The fact that ctDNA was detected in all patients prior to or at the time of relapse in our study and that previous studies11,12,16,17,22 have shown that repeat ctDNA surveillance can improve sensitivity for predicting disease relapse support such an approach.
In order to maximize sensitivity for MRD detection, we restricted analysis to patients in whom tumor tissue was available or somatic mutations could be identified in pre-treatment plasma using tumor-naïve genotyping. As a result, 20% of patients were excluded from the monitoring analysis. Additionally, one pre-CICI sample was excluded from analysis due to cell-free DNA input less than the minimum requirement of 10 ng. Because the limit of detection of CAPP-Seq is dependent on the number of reporters monitored, and more variants were identified from tumor tissue than pretreatment plasma, the sensitivity of CAPP-Seq was higher in patients with available tumor tissue. Of note, there was a similar percentage of patients with tumor tissue available in both cohorts (16% in the No CICI cohort and 18% in the CICI cohort).
Additional biomarkers are needed for predicting which patients with detectable MRD will benefit from CICI. An exploratory post-hoc analysis of the PACIFIC trial suggested that PD-L1 expression may be associated with response to CICI4. Unfortunately, PD-L1 staining was not routinely performed for patients in our cohort. Separately, tumor mutation burden has been associated with response to immune checkpoint inhibitors in some metastatic lung cancer studies23. However, this has not been demonstrated in the consolidation setting, and we did not observe an association of TMB with response to CICI in our patients. Given the small proportion of patients who have MRD and responded to CICI, larger studies will be necessary to identify robust predictive factors.
Limitations of our study include the modest samples size and limited follow up time, which were due to CICI only being FDA approved ~1.5 years ago. Additionally, although plasma samples were collected prospectively for ctDNA profiling, our analyses were retrospective, and patients were not randomly assigned to treatment cohorts. As a result, unmeasured confounding factors could potentially have impacted clinical outcomes. Furthermore, chemotherapy, radiotherapy, and immunotherapy regimens were not standardized across patients, and while unlikely, small differences in treatment could potentially have affected patient outcomes. Finally, it is important to note that the high sensitivity afforded by our tumor-informed ctDNA MRD detection approach differentiates it from most ctDNA detection methods currently available clinically. Our results should therefore not be automatically extrapolated to other techniques for ctDNA detection.
Taken together, our results demonstrate that CICI appears to improve outcomes in a subset of patients with MRD following CRT for locoregionally advanced NSCLC. Patients without MRD after CRT appear to have excellent outcomes irrespective of CICI treatment, and rising ctDNA during CICI appears to strongly predict risk of future disease progression. Importantly, prospective trials will be required to test whether personalization of CICI based on MRD has clinical utility before such an approach could be routinely applied to patients.
Methods
Study design and patients
This study was approved by the Stanford Institutional Review Board under protocol 33868. The samples analyzed in this study were prospectively collected for ctDNA analysis as part of two studies focused on molecular analysis of lung cancers and other tumors (NCT00349830 and MDACC LAB09–0983) and a Phase II trial assessing the safety and feasibility of adding PD-L1 blockade to chemoradiation for NSCLC (NCT02525757). Consecutive patients from these protocols were included in the current study if they had American Joint Committee on Cancer 7th edition stage IIB-IIIB NSCLC and were treated with curative-intent concurrent chemoradiation. Patients from part II of NCT02525757 who received concurrent immunotherapy during chemoradiation were excluded.
Power considerations
We hypothesized that patients with ctDNA detected after chemoradiation whose ctDNA decreased during consolidation immunotherapy would have improved FFP defined by RECIST 1.1 criteria from the start of chemoradiation therapy compared to patients with ctDNA detected after chemoradiation who did not receive CICI. The study statistical plan used the assumption that patients with detectable ctDNA after CRT who did not receive CICI would have a median FFP of 4 months12, assumed a hazard ratio of 0.2 for patients with ctDNA detected after chemoradiation whose ctDNA decreased during consolidation immunotherapy, and expected a median follow up of 24 months. Furthermore, we expected 66% of patients to have detectable ctDNA after chemoradiation5 and 25% of patients treated with consolidation immunotherapy to display decreased ctDNA during immunotherapy based on the progression-free survival results of the PACIFIC trial4. Based on these assumptions, a total of 16 patients per group would achieve a power of 80% to detect a difference between the patients with ctDNA responses during CICI and patients with ctDNA detected after chemoradiation who did not receive CICI at an alpha of 0.0524. We aimed for at least 20 patients per group to account for an 80% probability of detecting ctDNA in pretreatment plasmas by tumor-naïve genotyping of cfDNA12.
As a secondary aim, we sought to validate our previous finding that detection of ctDNA following curative-intent therapy for lung cancer is associated with inferior FFP defined by RECIST 1.1 criteria from the start of chemoradiation therapy compared to patients with ctDNA not detected. Expecting a median FFP of 4 months in patients with ctDNA detected after chemoradiation therapy and a hazard ratio of 43.4 based on our previous data12, 9 patients with a median follow up of 24 months would be required to achieve a power of 80% to detect a difference between the two groups at an alpha of 0.05 if 66% of patients have ctDNA detected after chemoradiation. We aimed for 11 patients in our validation cohort treated with chemoradiation therapy alone to account for an 80% probability of detecting ctDNA by tumor-naïve genotyping12.
Clinical procedures and blood collection
Patients underwent pretreatment imaging with chest computed tomography (CT), whole body positron emission tomography (PET)-CT, and brain MRI. All patients received radiation therapy with concurrent chemotherapy. When administered, consolidation immunotherapy was started a median of 4 weeks after completion of chemoradiation with a planned duration of 12 months. Patients treated on NCT02525757 (n=6) received concurrent carboplatin and paclitaxel for 2 cycles during consolidation immunotherapy with atezolizumab. All other patients (n=22) treated with consolidation immunotherapy received durvalumab. Plasma prior to all treatment was collected from all patients. Genotyping was performed using tumor tissue when available or otherwise pretreatment plasma. Matched leukocytes were used in both cases to identify germline SNPs and to also eliminate clonal hematopoiesis somatic variants. For the 37 patients treated with chemoradiation therapy alone, a second blood sample was collected within 4 months of completing radiation and chemotherapy. Seventeen of the patients treated with chemoradiation therapy alone were reported in a previous study and were analyzed as previously described12. For the 28 patients who received consolidation immunotherapy, a second blood sample was collected a median of 1 week after completing chemoradiation therapy and prior to the start of consolidation immunotherapy, and a third blood sample was collected at least 2 weeks after the initiation of consolidation immunotherapy at a median of 11 weeks into treatment. LUP893 died prior to the collection of a blood sample during consolidation immunotherapy. Additional follow-up samples were analyzed for a subset of patients. All samples were collected with informed consent in accordance with the Declaration of Helsinki.
ctDNA library preparation and sequencing
Tumor, plasma, and leukocyte samples were analyzed by CAPP-Seq as previously described with minor modifications12,25,26. Briefly, venous blood collected in K2EDTA or CellSave tubes was centrifuged and plasma was stored at −80°C. The remaining plasma-depleted whole blood was also stored at −80°C for germline DNA isolation from leukocytes. Formalin-fixed, paraffin embedded (FFPE) tumor sections were available for a subset of patients for isolation of tumor DNA. Genomic DNA from tumor and leukocytes was fragmented prior to library preparation using a Covaris S2 sonicator. Cell-free DNA (cfDNA) was extracted from plasma using the QiaAmp Circulating Nucleic Acid Kit (Qiagen) according to the manufacturer’s instructions. A maximum of 55 ng of cfDNA (median of 32 ng) was input into sequencing library preparation using the KAPA LTP Library Prep Kit (Kapa Biosystems), with minor modifications to the manufacturer’s instructions. Samples with less than 10 ng of cfDNA available for library prep were excluded from the analysis. Hybridization-based enrichment of target sequences from multiplexed samples was performed using a custom designed selector pool of biotinylated oligonucleotides (Roche NimbleGen) prior to sequencing using 2 × 150 bp paired-end reads with eight-base indexing on an Illumina HiSeq 4000.
Analysis of sequencing data and somatic genotyping
Sequencing data were processed using a custom bioinformatics pipeline, and EGFR exon 19 deletions and single nucleotide variants were called as previously described25,26. Tumor-naïve variant calling in pretreatment plasma was performed using an approach that achieved 95% specificity in an independent cohort of 26 control cfDNA samples. Briefly, we removed single nucleotide polymorphisms identified in germline samples from any patient in the cohort, variants lying in repeat, intronic, intergenic, or pseudogene regions, variants without duplex-support, low depth positions (less than or equal to 50% of the median sample depth), variants with a population allele frequency of greater than or equal to 0.001 in the gnomAD database (Broad Institute), and variants present in greater than 5% of a cohort 150 germline samples run using the same selector. To enrich for tumor-derived mutations, we also required variants to have greater than 2 deduped reads in short DNA fragments less than 150 bp or 220–300 bp in length27. Variants with allele frequencies above the selector-wide background in the matched germline sample were considered to be due to clonal hematopoiesis and were removed. Finally, truncal mutations were enriched by removing variants with an allele fraction less than one-fifth the allele fraction of the most prevalent variant. Variants called from tumor tissue were filtered using a similar strategy except with a minimum allele frequency threshold of 5% and no fragment size requirement or filtering for truncal mutations. Patients without tumor tissue available for genotyping were excluded from the monitoring analysis if no variants were called in the pretreatment plasma sample.
Monitoring for ctDNA MRD
To query the presence of ctDNA at each time point a previously-described Monte Carlo-based ctDNA detection index was used to monitor for somatic single nucleotide variants identified pretreatment25. The ctDNA detection index threshold for each patient was set to achieve ≥90% specificity in 54 held-out control cfDNA samples analyzed using the same selector. ctDNA was classified as detected if the detection index was less than the detection threshold for a given time point. The ctDNA allele fraction for each sample was calculated by averaging the allele fractions of all variants used for monitoring, and the ctDNA concentration was calculated by multiplying the mean allele fraction by the plasma cfDNA concentration measured by Qubit (Thermo Fisher Scientific). Each haploid genomic equivalent was assumed to have a mass of 3.3 pg. Patient ctDNA dynamics were classified as having “Increasing” or “Decreasing” patterns if the pre-CICI and early on-CICI ctDNA measurements changed at least 2 fold or were statistically distinguishable (i.e., non-overlapping confidence intervals) when comparing mean ctDNA concentrations and associated 95% confidence intervals from negative binomial estimates. Inferred tumor mutational burden from CAPP-Seq was calculated as described previously12.
Statistics and reproducibility
Our primary aim was to test the hypothesis that patients with ctDNA detected after chemoradiation therapy whose ctDNA concentration decreased during consolidation immunotherapy had improved FFP compared to patients with ctDNA detected after chemoradiation therapy who did not receive consolidation immunotherapy. FFP was defined as the time from the start of chemoradiation to the date of any progression defined radiographically by RECIST 1.1 criteria. Freedom from distant progression was defined as the time from the start of chemoradiation to the date of progression at a new lesion outside the radiation field defined radiographically by RECIST 1.1 criteria. FFP and freedom from distant progression were calculated using the Kaplan-Meier method censoring patients without progression at the time of last imaging follow up. LUP814 developed a biopsy-proven local recurrence and underwent repeat irradiation prior to meeting RECIST 1.1 criteria for progression, so local progression was scored at the time of biopsy. Statistical significance for Kaplan-Meier analyses was determined using the two-sided log-rank test, and hazard ratios were calculated using the Mantel-Haenszel approach. Two-tailed Mann-Whitney tests were used to compare distributions, and two-sided Fisher’s exact tests were used to compare proportions. Statistical significance was assumed at P<0.05. Statistical analyses were performed using Prism 8 (GraphPad Software) or R version 3.5.1 through the RStudio environment. All experiments were performed once, and the experiments were not randomized. Outcome and cohort allocation were not considered during sequencing library preparation and bioinformatic analysis. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Sequencing results supporting the findings are available on request from the lead corresponding author (M.Diehn). Supporting variant level data for all figures are available in Supplementary Tables 2-5. Source data for Fig. 2, 3, 4, and 5, and Extended Data 1, 2, and 3 have been provided as Source Data files. Computer code used for the analysis was described previously26 and is available at http://cappseq.stanford.edu.
Extended Data
Extended Data Fig. 1: Validation of predictive value of ctDNA MRD after chemoradiation therapy (CRT) alone.
a, Event chart showing timing of therapy, progression based on RECIST 1.1 evaluation of imaging, and results of ctDNA testing for each patient. b, Kaplan-Meier analysis of freedom from progression stratified by ctDNA detection within 4 months of completing CRT (n=6 not detected, n=6 detected). P values were calculated using a two-sided log-rank test.
Extended Data Fig. 2: ctDNA dynamics during consolidation immune checkpoint inhibition (CICI) after chemoradiation therapy (CRT) predicts disease progression.
Kaplan-Meier analysis of (a) freedom from progression and (b) freedom from distant progression in patients with ctDNA decreasing early on-CICI (n=5) and ctDNA increasing early on-CICI (n=5). P values were calculated using two-sided log-rank tests.
Extended Data Fig. 3: Pretreatment tumor mutational burden is not significantly correlated with response to chemoradiation or consolidation immune checkpoint inhibition (CICI).
Pretreatment tumor mutational burden in non-synonymous mutations per megabase extrapolated from CAPP-Seq in (a) patients with ctDNA detected (n=26) and not detected (n=25) after chemoradiation therapy and (b) patients with ctDNA increasing early on-CICI (n=5) and decreasing early on-CICI (n=5). P values were calculated using two-sided Mann-Whitney tests. Bars represent medians.
Supplementary Material
Acknowledgements
We thank the patients and families who participated in this study. We thank R. Tibshirani for biostatistics advice. This work was supported by the Stanford Radiation Oncology Kaplan Fellowship (E. Moding) and grants from the American Society for Radiation Oncology (E. Moding), the Radiological Society of North America (E. Moding), the National Cancer Institute (M. Diehn and A. Alizadeh; R01CA188298), the US National Institutes of Health Director’s New Innovator Award Program (M. Diehn; 1-DP2-CA186569), the Virginia and D.K. Ludwig Fund for Cancer Research (M. Diehn and A. Alizadeh), and the CRK Faculty Scholar Fund (M. Diehn). The schematic in Fig. 1a was produced using Servier Medical Art (https://smart.servier.com) licensed under CC BY 3.0.
Competing Interests
E.J.M., Y.L., B.Y.N., J.J.C., A.B.H., R.F.B, R.B.K., C.H.Y., L.G., C.D.J., J.H., Y.Q., T.X., J.V.H., A.T., Z.L., D.R.G., S.K.P., K.J.R., and B.W.L. declare no competing interests. A.A.C. has served as an advisor/consultant for Roche, Tempus Labs, Geneoscopy, and Oscar Health, and has received speaker honoraria from Roche, Varian Medical Systems, and Foundation Medicine. M.Das has received research funding from Verily, Abbvie, United Therapeutics, and Celgene and has served as a consultant and received honoraria from Bristol Myer Squibb and Astra Zeneca. J.W.N. has served as an advisor/consultant for ARIAD/Takeda, AstraZeneca, Genentech/Roche, Lilly, Exelixis, Loxo, and Jounce. J.W.N. has received research funding from Genentech/Roche, Merck, Novartis, Boehringer Ingelheim, Exelixis, ARIAD/Takeda, and Nektar. H.A.W. has served on the advisory board for AstraZeneca, Xcovery, Janssen, Mirati, Merck, Takeda, and Genentech/Roche and has received compensation from AstraZeneca, Xcovery, Janssen, and Mirati. H.A.W. has received research funding from ACEA Biosciences, Arrys Therapeutics, AstraZeneca/Medimmune, BMS, Celgene, Clovis Oncology, Exelixis, Genentech/Roche, Gilead, Lilly, Merck, Novartis, Pfizer, Pharmacyclics, and Xcovery. S.H.L. receives grant funding from Hitachi Chemical Diagnostics, Genentech, Inc., and BeyondSpring Pharmaceuticals, honorarium from Varian Medical Systems, and has served on the advisory board for AstraZeneca and BeyondSpring Pharmaceuticals. A.A.A. and M.Diehn are co-inventors on patent applications related to CAPP-Seq. A.A.A. has equity in FortySeven and CiberMed and has served as a consultant for Roche, Genentech, Chugai, and Pharmacyclics. M.Diehn has equity in CiberMed, has received research funding from Varian Medical Systems, and has served as a paid consultant for Roche, AstraZeneca, BioNTech, Illumina, and RefleXion and an unpaid consultant for Genentech.
References
- 1.Bray F. et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ettinger DS et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 15, 504–535 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Goldstraw P. et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 11, 39–51 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Antonia SJ et al. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N. Engl. J. Med (2018) doi: 10.1056/NEJMoa1809697. [DOI] [PubMed] [Google Scholar]
- 5.Bradley JD et al. Long-Term Results of RTOG 0617: A Randomized Phase 3 Comparison of Standard Dose Versus High Dose Conformal Chemoradiation Therapy +/− Cetuximab for Stage III NSCLC. Int. J. Radiat. Oncol. Biol. Phys 99, S105 (2017). [Google Scholar]
- 6.Moding EJ, Diehn M. & Wakelee HA Circulating tumor DNA testing in advanced non-small cell lung cancer. Lung Cancer 119, 42–47 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Murillas I. et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Science Translational Medicine 7, 302ra133–302ra133 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Sausen M. et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun 6, 7686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tie J. et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 8, 346ra92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tie J. et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut (2018) doi: 10.1136/gutjnl-2017-315852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbosh C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaudhuri AA et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov 7, 1394–1403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen BT et al. Clinical and Histopathologic Features of Immune Checkpoint Inhibitor-related Pneumonitis. Am. J. Surg. Pathol (2019) doi: 10.1097/PAS.0000000000001298. [DOI] [PubMed] [Google Scholar]
- 16.Reinert T. et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol (2019) doi: 10.1001/jamaoncol.2019.0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen E. et al. Early Detection of Metastatic Relapse and Monitoring of Therapeutic Efficacy by Ultra-Deep Sequencing of Plasma Cell-Free DNA in Patients With Urothelial Bladder Carcinoma. J. Clin. Oncol 37, 1547–1557 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Raja R. et al. Early Reduction in ctDNA Predicts Survival in Patients with Lung and Bladder Cancer Treated with Durvalumab. Clinical Cancer Research 24, 6212–6222 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Anagnostou V. et al. Dynamics of Tumor and Immune Responses during Immune Checkpoint Blockade in Non-Small Cell Lung Cancer. Cancer Res. 79, 1214–1225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg SB et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin. Cancer Res 24, 1872–1880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang DY et al. Fatal Toxic Effects Associated With Immune Checkpoint Inhibitors: A Systematic Review and Meta-analysis. JAMA Oncol 4, 1721–1728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coombes RC et al. Personalized Detection of Circulating Tumor DNA Antedates Breast Cancer Metastatic Recurrence. Clin. Cancer Res (2019) doi: 10.1158/1078-0432.CCR-18-3663. [DOI] [PubMed] [Google Scholar]
- 23.Chan TA et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 30, 44–56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 24.Dupont WD & Plummer WD Power and Sample Size Calculations. Control Clin Trials 11, 116–28 (1990). [DOI] [PubMed] [Google Scholar]
- 25.Newman AM et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med 20, 548–554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newman AM et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol 34, 547–555 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mouliere F. et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Science Translational Medicine 10, eaat4921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing results supporting the findings are available on request from the lead corresponding author (M.Diehn). Supporting variant level data for all figures are available in Supplementary Tables 2-5. Source data for Fig. 2, 3, 4, and 5, and Extended Data 1, 2, and 3 have been provided as Source Data files. Computer code used for the analysis was described previously26 and is available at http://cappseq.stanford.edu.