Abstract
The Std1 protein modulates the expression of glucose-regulated genes, but its exact molecular role in this process is unclear. A two-hybrid screen for Std1-interacting proteins identified the hydrophilic C-terminal domains of the glucose sensors, Snf3 and Rgt2. The homologue of Std1, Mth1, behaves differently from Std1 in this assay by interacting with Snf3 but not Rgt2. Genetic interactions between STD1, MTH1, SNF3, and RGT2 suggest that the glucose signaling is mediated, at least in part, through interactions of the products of these four genes. Mutations in MTH1 can suppress the raffinose growth defect of a snf3 mutant as well as the glucose fermentation defect present in cells lacking both glucose sensors (snf3 rgt2). Genetic suppression by mutations in MTH1 is likely to be due to the increased and unregulated expression of hexose transporter genes. In media lacking glucose or with low levels of glucose, the hexose transporter genes are subject to repression by a mechanism that requires the Std1 and Mth1 proteins. An additional mechanism for glucose sensing must exist since a strain lacking all four genes (snf3 rgt2 std1 mth1) is still able to regulate SUC2 gene expression in response to changes in glucose concentration. Finally, studies with green fluorescent protein fusions indicate that Std1 is localized to the cell periphery and the cell nucleus, supporting the idea that it may transduce signals from the plasma membrane to the nucleus.
The STD1 gene was identified in two very different genetic screens. In one screen, increased gene dosage of STD1 was found to partially suppress the growth defects associated with overexpression of TBPΔ57, a dominant negative mutant of the TATA binding protein (TBP) (11). In the second screen, increased gene dosage of STD1 was shown to partially suppress the Snf− phenotype (sucrose nonfermenting) of a snf4 mutation (17). Hubbard et al. (17) used low-stringency hybridization to identify a homologue of Std1, designated Mth1, that shares 61% amino acid identity. In silico analysis of the yeast genome and other available sequence databases indicates that there are no other known proteins closely related to Std1 and Mth1. Deletion of either STD1 or MTH1 had no apparent deleterious effects on cell growth or gene expression. However, deletion of both genes resulted in a strain with a mild Snf− phenotype and a three- to fourfold reduction in the derepression of invertase (17). This finding suggests that these homologous genes are functionally redundant. In wild-type cells, overexpression of Std1 partially relieves glucose repression (17). Mutagenesis and deletion analysis of the STD1 gene demonstrated that mutations that abrogated its ability to suppress TBPΔ57 were also unable to relieve glucose repression of invertase (37), suggesting that these two assays may measure the same biological activity.
Genetic analysis has identified a number of genes required for the fermentation of sucrose (5, 25). The Snf− phenotype is characterized by the inability to grow by fermentation on media containing raffinose (a trisaccharide related to sucrose) and antimycin A. The drug antimycin A, a Streptomyces antibiotic, blocks mitochondrial function by preventing electron transport from cytochrome b to cytochrome c1. When present in yeast media, antimycin A prevents cell growth by respiration of sugars and amino acids. Only cells that can generate energy by fermentation are able to grow. In order to ferment raffinose, yeast cells must be able to carry out two distinct processes. First, they must be able to express and secrete the enzyme invertase which hydrolyzes raffinose, thereby allowing the products to be imported. Many of the mutations that confer a Snf− phenotype inactivate either the Snf1 kinase complex or the Swi-Snf chromatin remodeling complex and block expression of invertase. Second, the cells must also express sufficient high-affinity hexose transporter proteins for the import of hydrolyzed raffinose. Null mutations in the SNF3 gene have little effect on invertase expression (26), yet they generate a Snf− phenotype due to impaired expression of the high-affinity hexose transporters (28, 29).
Genetic studies of STD1 have not been able to determine whether Std1 regulates gene expression through interactions with TBP, with Snf1 kinase complex, or with both. Indeed, biochemical studies of Std1 found that it was able to interact directly with both TBP (33) and Snf1 kinase (17). In an effort to understand the role in gene regulation played by the Std1 protein, we undertook a two-hybrid screen to identify proteins that interact with Std1. The results of that screen are reported here. Two strong Std1-interacting proteins were found to be the glucose sensors, Snf3 and Rgt2 (28).
The yeast glucose sensors are members of a family of hexose transporter proteins (HXT) that in Saccharomyces cerevisiae consists of HXT1 to HXT17, SNF3, RGT2, and GAL2 (19). The hexose transporter proteins are integral membrane proteins that promote the facilitated diffusion of hexoses, the metabolic step that may in fact be the rate-limiting step of fermentation (4). Hexose transporters found in bacterial (2), plant (31), and mammalian (24) species all have an approximately 500-residue domain that spans the plasma membrane 12 times (16). Snf3 and Rgt2, however, are structurally distinct from the other 18 members of this family in yeast by the presence of a large, hydrophilic C-terminal domain (28). Several lines of evidence suggest that C-terminal tails of Snf3 and Rgt2 are essential for glucose sensing and signal transduction. Deletion of the tail domain reduces Snf3 function (22, 27); fusion of the tail domain to Hxt1 or Hxt2 proteins confers glucose-sensing ability to those proteins (27); and expression of the Snf3 tail domain by itself can suppress the defects in glucose transport observed in a snf3 strain (7).
The process of glucose sensing and signal transduction in yeast are likely to be similar to receptor-ligand binding and signal transduction characterized in mammalian cells. This hypothesis is suggested by several observations. First, the Snf3 and Rgt2 proteins do not actually transport hexoses themselves (21). Rather, Snf3 and Rgt2 control hexose transport by regulating the expression of high- and low-affinity transporters (28). Second, the fact that a dominant mutation in RGT2 could signal changes in gene expression in the absence of glucose argues strongly that glucose transport and metabolism are not required for glucose signaling (28). Thus it seems possible that the yeast glucose sensors have adapted the glucose transporter domain into a glucose binding domain that can transmit extracellular information about glucose concentration to an intracellular signal transduction apparatus.
MATERIALS AND METHODS
Yeast strains, media, and genetic techniques.
S. cerevisiae strains used in this study are described in Table 1. Except where indicated, yeast strains were grown on standard media (30) at 30°C. For carbon sources, glucose or raffinose was present at 2% (grams per 100 ml) unless otherwise indicated, and a glycerol-ethanol mixture was present at 3% (vol/vol) and 2% (vol/vol), respectively. Antimycin A was included at 1 μg/ml where indicated. Standard procedures were used for genetic crosses, sporulation, and tetrad analysis (30). Transformations of yeast strains were done by the lithium acetate procedure (13).
TABLE 1.
S. cerevisiae strains used
| Strain | Genotype | Reference or source |
|---|---|---|
| Y153 | MATa ura3-52 leu2-3,112 trp1-901 his3-Δ200 ade2-101 gal4Δ gal80Δ URA3::GAL-lacZ LYS2::GAL-HIS3 | 10 |
| FY14 | MATa ura3-52 trp1-Δ63 | 36 |
| FY86 | MATα ura3-52 leu2-Δ1 his3-Δ200 | 36 |
| FY1193 | MATα ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 snf1-Δ10 | Fred Winston |
| MSY192 | MATa ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 std1::HIS3 mth1-Δ2 | 12 |
| MSY401 | MATα ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 | This study |
| MSY403 | MATa ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 rgt2::HIS3 | This study |
| MSY441 | MATα ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 snf3::hisG rgt2::HIS3 | This study |
| MSY443 | MATα ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 snf3::hisG rgt2::HIS3 std1::LEU2 | This study |
| MSY445 | MATα ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 snf3::hisG rgt2::HIS3 mth1-Δ2 | This study |
| MSY447 | MATα ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 snf3::hisG rgt2::HIS3 std1::LEU2 mth1-Δ2 | This study |
| MSY449 | MATα ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 snf3::hisG | This study |
| MSY451 | MATa ura3-52 leu2Δ1 his3-Δ200 trp1-Δ63 snf3::hisG std1::LEU2 | This study |
| MSY453 | MATα ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 snf3::hisG mth1-Δ2 | This study |
| MSY455 | MATα ura3-52 leu2-Δ1 his3-Δ200 snf3::hisG std1::HIS3 mth1-Δ2 | This study |
| MSY465 | MATα ura3-52 leu2-Δ1 his3-Δ200 | This study |
| MSY467 | MATα ura3-52 leu2-Δ1 his3-Δ200 trp1Δ63 std1::LEU2 | This study |
| MSY469 | MATα ura3-52 leu2-Δ1 his3-Δ200 trp1Δ63 mth1-Δ2 | This study |
| MSY471 | MATα ura3-52 leu2-Δ1 his3-Δ200 trp1-Δ63 std1::LEU2 mth1-Δ | This study |
| MSY478 | MATα ura3-52 his3-Δ200 trp1-Δ63 snf1-Δ10 mth1-Δ2 | This study |
Two-hybrid screen.
S. cerevisiae Y153 (10) was transformed with the bait plasmid, pGBT9-STD1, containing the entire STD1 reading frame fused to the Gal4 DNA binding domain. Positive interactors were then selected from a complex library of yeast genomic DNA (18) by histidine prototrophy. Transformation efficiency was monitored by selection on synthetic complete (SC) medium lacking tryptophan and leucine. Clones that were able to grow on medium lacking histidine and supplemented with 25 mM 3-aminotriazole were screened for lacZ expression. DNA from clones that were positive for expression of both the lacZ and the HIS3 reporter genes was prepared, and the library plasmid was amplified in Escherichia coli, using selection for leucine prototrophy. Approximately one half of these isolates (out of a total of approximately 200) remained positive when retransformed into yeast bearing the Gal4-Std1 bait plasmid. Representatives from this set of positive isolates were then subjected to DNA sequence analysis. Plasmids which contained out-of-frame fusions or fusions outside of a reading frame were discarded. Multiple clones were found to contain fusions to the hydrophilic C-terminal domains of the yeast glucose sensors Snf3 and Rgt2 (28). Colony hybridization confirmed that the majority (63 of 98) of the Std1-interacting clones contained sequences encoding the C-terminal domain of Snf3 and Rgt2. All remaining isolates were discarded based on positive hybridization to sequenced clones containing out-of-frame fusions. The fusion junctions for two independent clones of both Snf3 (amino acids 576 to 825 and 663 to 806) and Rgt2 (amino acids 617 to 763 and 647 to 763) were determined by DNA sequencing. Interactions were assayed by growth on SC medium (30) containing 2% glucose and 25 mM 3-aminotriazole and lacking histidine, leucine, and tryptophan. Plasmid pGAD-Snf4 (17) was used as a negative control.
Plasmid constructions and gene knockouts.
pGBT9-Std1 contains the entire STD1 reading frame cloned as a PCR product with EcoRI termini cloned in frame into pGBT9 (3). Strains with null alleles were created in a diploid strain (FY86 × FY14) that is isogenic to S288c (36). The STD1, MTH1, and SNF3 genes were disrupted by one-step gene replacement using plasmids pMU1 (11), pJH124 (17), and pBM3103 (28). The RGT2 gene was disrupted by using a PCR fragment containing the HIS3 gene flanked by RGT2 sequences (28). All disruptions were confirmed by Southern blot analysis. The mth1Δ2 allele was constructed in a previous study (12). All strains containing null alleles in the glucose sensors were constructed and maintained on SC medium with glycerol-ethanol as the carbon source in order to prevent selection of suppressing mutations. The Std1-green fluorescent protein (GFP) fusion was constructed by PCR amplification of the GFP gene on a BamHI fragment and insertion in frame at the 3′ end of an engineered STD1 gene. The resulting plasmid, p6A5-GFP, expresses the Std1-GFP fusion protein from the endogenous STD1 promoter on the 2μm LEU2 vector, YEp351 (15). The SNF3-GFP fusion was constructed by PCR amplification of the GFP gene on a BamHI fragment and insertion in frame at the 3′ end of an engineered SNF3 gene. The resulting plasmid, pSnf3-GFP, expresses the Snf3-GFP fusion protein from the ADH1 promoter on the 2μm URA3 vector, pRS426 (6).
Enzyme assays.
For invertase assays, repressed and derepressed cells (25) were harvested in mid-log phase and normalized for equal optical density at 600 nm (OD600). Cells were harvested, washed in cold 10 mM sodium azide, and assayed for invertase activity (14). Specific activity was defined in terms of milliunits of invertase activity (1 U being equal to the activity required to release 1 μmol of glucose per min) per OD600 of cells assayed. For β-galactosidase assays, cells transformed with HXT-LacZ fusion plasmids (28) were grown in SC medium lacking uracil supplemented with 3% glycerol and 2% ethanol as the carbon source. Logarithmically growing cells (OD600 of <0.4) were harvested, resuspended in the same medium with either no glucose or 0.1 or 6% glucose, and grown an additional 4 h. Protein extracts were then prepared and assayed for β-galactosidase activity (1). Results are expressed in Miller units (23).
Western analysis.
Cultures of yeast cells (40 ml) were harvested in log-phase (OD600 of 0.1 to 0.4), and proteins extracts were prepared by vortexing with glass beads in a solution containing 40 mM HEPES (pH 7.3), 350 mM NaCl, 0.1% Tween 20, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, and 1 μg each of benzamidine, pepstatin A, leupeptin, and aprotinin per ml. The concentration of soluble protein was determined by the Bradford method, using bovine serum albumin as a standard. An equal aliquot (25 μg) from each extract was resolved on a sodium dodecyl sulfate–10% polyacrylamide gel and transferred to Hybond ECL nitrocellulose. The nitrocellulose membrane was blocked with 10% milk and 0.1% Tween 20 in 1× Tris-buffered saline (TBS; 20 mM Tris-HCl [pH 7.6], 135 mM NaCl) for 1 h at 65°C and washed in 1× TBS with 0.1% Tween 20. The membrane was incubated with monoclonal mouse anti-hemagglutinin (HA) antibody (12CA5; Boehringer Mannheim) at 0.2 μg/ml in 1× TBS for 2 h at room temperature. The membrane was washed and then incubated with sheep anti-mouse immunoglobulin antibody linked with horseradish peroxidase (Amersham) at a 1:5,000 dilution in 1× TBS with 0.1% Tween 20 for 1 h at room temperature and developed according to Amersham’s protocol.
Northern analysis.
Liquid cultures (5 to 10 ml) were harvested in log phase, and total RNA was prepared by using a Purescript RNA isolation kit (Gentra Systems, Minneapolis, Minn). RNA samples (15 μg) were subjected to electrophoresis in formaldehyde–1% agarose gels. The RNA was transferred to nylon membrane by capillary action and hybridized to 32P-labeled DNA sequences. The DNA probe for MTH1 was the 562-bp EcoRI-to-NcoI genomic fragment encompassing the 3′ half of the open reading frame. The probe for actin mRNA was the 563-bp ClaI genomic DNA fragment that includes the 5′ region of the actin open reading frame. All probes were radiolabeled by the random priming method in the presence of [α-32P]dATP.
Microscopy.
Cells expressing GFP fusion proteins were analyzed by fluorescence microscopy using a Zeiss Axiovert135 microscope equipped with epifluorescence optics and computer-controlled shutters, stage, and cameras. GFP was visualized with a green filter (Chroma). Images were collected using a cooled CCD (charge-coupled device) camera (Photometrics) at a pixel resolution of 1,300 by 1,000 with a 100× 1.3NA plan apochromat lens. Microscope control and image collection were managed by ONCORimage (ONCOR, Gaithersburg Md.). Following collection, images were assembled with Photoshop 4.0 (Adobe). The images shown in this report were collected at the microscope with no electronic filtration or enhancement. The camera used is a highly sensitive, monochrome device. Thus, to collect multicolor images, images were collected for appropriate times (set to achieve optimal saturation of the CCD camera) for each color (green and blue cubesets). This was performed automatically by the control system for the microscope. Generation of the through-focus series was managed by using the microscope control software. The top and bottom of cells were defined, and all intermediate image planes were collected automatically at 0.2-μm intervals. Illumination was shuttered between frames to minimize photobleaching of the fluorochromes. To remove out-of-focus blur, the image stack was postprocessed via an exhaustive photon reassignment algorithm using the measured point spread function of the 100× objective used for image collection. To counterstain nuclear DNA, cells were stained by Hoechst dye by first being fixed in 2% paraformaldehyde in 1× phosphate-buffered saline (PBS) for 1 h at room temperature. Fixed cells were permeabilized and stained by incubation in 1× PBS containing 5% Triton X-100 and 2 μg of Hoechst dye per ml for 1 h at room temperature. Cells were then washed for an additional hour in 1× PBS prior to image collection.
RESULTS
Two-hybrid screen for Std1-interacting proteins.
To identify proteins that interact with the Std1, approximately 107 clones from a complex library of yeast genomic DNA (18) were screened in the two-hybrid system using full-length Std1 fused downstream of the DNA binding domain of Gal4 as bait. Multiple independent clones that showed strong two-hybrid interaction with Std1 contained in-frame fusions to the hydrophilic C-terminal domains of the yeast glucose sensors Snf3 and Rgt2 (28). Snf3 and Rgt2 are structurally distinct from all other members of the HXT family in yeast by the presence of a large, hydrophilic C-terminal domain. Independent library isolates captured by the Std1 bait encoded most or all of the hydrophilic C-terminal domains of Snf3 and Rgt2. None of the library isolates contained any of the predicted transmembrane regions, perhaps a reflection of the structural limitations of the two-hybrid assay. Studies of glucose signal transduction indicate that the hydrophilic C-terminal domains of Snf3 and Rgt2 are required for glucose signal transduction (7, 27, 28).
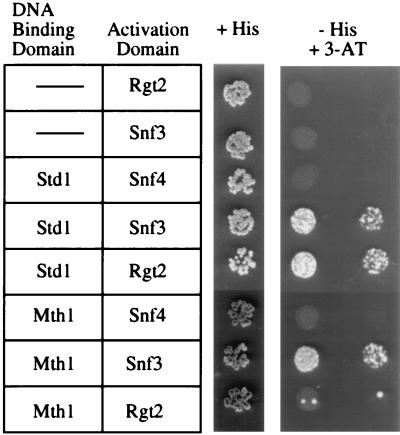
The Gal4-Std1 fusion interacted equally well with the C-terminal domain of either Snf3 or Rgt2 as judged by the ability to grow on SC-His medium supplemented with 25 mM 3-aminotriazole (Fig. 1) as well as quantitative β-galactosidase assays (data not shown). Since the STD1 and MTH1 genes are homologues, we tested the ability of the Mth1 to interact with the C-terminal tails of Snf3 and Rgt2. Mth1 interacted strongly with the C-terminal tail of Snf3 but failed to interact significantly with Rgt2. This result was the first indication that the Std1 and the Mth1 may not be functionally redundant. Instead, the possibility arose that these proteins, while homologous, may have evolved to play distinct roles with respect to glucose signaling.
FIG. 1.
Two-hybrid interactions between Std1, Mth1, and the C-terminal domains of the glucose sensors. S. cerevisiae Y153 was transformed with the indicated two-hybrid fusion constructs. Positive two-hybrid interactions are indicated by the growth of cells on medium lacking histidine and containing 25 mM 3-aminotriazole (3-AT).
We have thus far been unable to detect a direct physical interaction between these proteins. Experiments with GST-Snf3 and GST-Rgt2 fusions have not shown any specific interaction with Std1 or Mth1 tagged with three copies of the HA epitope (Std1-3HA or Mth1-3HA) (data not shown).
Analysis of snf3 and rgt2 null mutants.
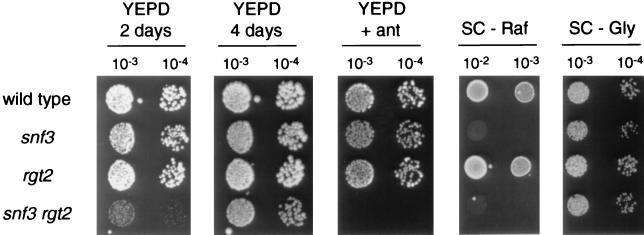
Our first step toward understanding the role played by the SNF3 and RGT2 genes in glucose signaling was to analyze the effects of null alleles in these genes. A diploid strain isogenic to S288c (36) was used to create a double heterozygote bearing one copy of a snf3::hisG allele and one copy of a rgt2::HIS3 allele. Multiple tetrads dissected from this strain resulted in four viable haploid progeny, and marker analysis demonstrated that the double mutant lacking both glucose sensors was indeed viable. Similar results have been reported previously (27). The single mutants behaved as expected from earlier studies (22, 28), with the snf3 mutant showing a growth defect on raffinose-antimycin medium (Fig. 2) and a small defect in glucose derepression of invertase (SUC2) expression (see Fig. 4A). The rgt2 mutant showed no detectable growth defect on any media tested (Fig. 2) and displayed wild-type levels of derepression of the SUC2 gene. However, we were able to reproducibly detect a minor defect in glucose repression of the SUC2 gene (see Fig. 4A) in rgt2 mutants. A snf3 defect in derepression by low glucose concentrations and an rgt2 defect in repression by high glucose concentrations is consistent with the model proposed by Ozcan and Johnston (29) in which these two glucose sensors have become specialized in function, with Rgt2 acting as the high-glucose sensor and Snf3 acting as the low-glucose sensor.
FIG. 2.
Effects of mutations in SNF3 and RGT2 on cell growth. Serial dilutions of wild-type cells and cells with various null alleles as indicated were spotted onto agar plates containing YEPD, YEPD containing antimycin (Ent), SC-glycerol, or SC-raffinose containing antimycin as indicated. Cells were grown for 4 days (except as indicated) and photographed. The strains used were MSY465 (wild type) MSY449 (snf3), MSY403 (rgt2), and MSY441 (snf3 rgt2).
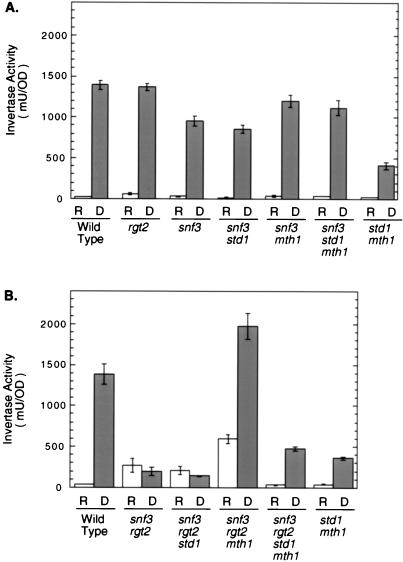
FIG. 4.
Invertase expression in cells with mutations in SNF3, RGT2, STD1, and MTH1. Quantitative invertase assays were performed on cells grown under repressing (R) and derepressing (D) conditions (25) as indicated. At least three independent colonies of each strain were assayed, and the error bars represent 1 standard error. The strains used for panels A and B were the same as those used for Fig. 3A and B, respectively.
Analysis of the double snf3 rgt2 mutant revealed a new synthetic phenotype not present in either single mutant. The snf3 rgt2 mutant grew very slowly with glucose as the carbon source. This growth defect was easily seen on YEPD medium after 2 days of incubation (Fig. 2). Furthermore, the snf3 rgt2 cells could not grow on glucose in the presence of antimycin, a drug which blocks mitochondrial function. This finding suggests that the snf3 rgt2 cells are unable to grow by fermentation. Consistent with a defect in fermentation, the snf3 rgt2 cells grew with wild-type rates on nonfermentable carbon sources such as glycerol and ethanol. The double mutant was also unable to grow by fermentation of raffinose, indicating that the Snf− phenotype of the snf3 mutant was not suppressed by loss of RGT2 function.
Genetic interactions of STD1 and MTH1 with the glucose sensors.
Since Std1 was found to interact with the glucose sensors in the two-hybrid system, we tested for genetic interactions between the genes encoding the glucose sensors and the STD1 and MTH1 genes. A diploid strain that was heterozygous for a wild-type and a null allele at all four loci was constructed and induced to sporulate. Twenty-five tetrads that produced four viable haploid progeny were analyzed. Since all possible combinations of the four null alleles were represented in these 100 segregants, we conclude that there was no synthetic lethality generated in this cross. However, two phenotypes observed in parental strains, the Snf− phenotype of the snf3 strains and the glucose-antimycin growth defect of the snf3 rgt2 strains did not segregate as expected. For instance, one-half of the segregants should inherit the snf3 null allele and therefore be Snf− on raffinose-antimycin medium. This was not observed. Instead, some of the snf3 strains grew as well as wild-type strains on raffinose-antimycin medium. Analysis of the genotypes of the snf3 strains revealed that loss of the MTH1 gene function was able to suppress the raffinose growth defect associated with the snf3 mutation (Fig. 3A). The suppression of snf3 was independent of the RGT2 gene but was somewhat dependent on STD1 since a snf3 mth1 std1 strain grew more slowly on raffinose-antimycin medium than did a snf3 mth1 STD1 strain. Thus, the STD1 gene and the MTH1 gene play distinct and seemingly antagonistic roles with respect to suppression of the snf3 mutation.
FIG. 3.
Suppression of snf3 and snf3 rgt2 phenotypes by mutation of STD1 and MTH1. Serial dilutions of wild-type cells and cells with various null alleles as indicated were spotted onto agar plates containing YEPD, YEPD plus antimycin (ant), SC-glycerol, or SC-raffinose plus antimycin as indicated. The strains used: (A) MSY465 (wild type), MSY449 (snf3), MSY451 (snf3 std1) MSY453 (snf3 mth1), MSY455 (snf3 std1 mth1), and MSY471 (std1 mth1); (B) MSY465 (wild type), MSY441 (snf3 rgt2), MSY443 (snf3 rgt2 std1), MSY445 (snf3 rgt2 mth1), and MSY447 (snf3 rgt2 std1 mth1).
A second phenotype in the std1 mth1 × snf3 rgt2 cross that did not segregate as expected was the glucose-antimycin growth defect of the snf3 rgt2 mutant. Approximately one-half of the snf3 rgt2 strains derived from this cross regained the ability to grow at the wild-type rate on glucose-antimycin medium. Analysis of the genotypes of the snf3 rgt2 strains revealed that loss of MTH1 gene function was responsible for the suppression of this phenotype (Fig. 3B). Suppression of this snf3 rgt2 phenotype was not dependent on the STD1 gene. Since these results depend on comparison of growth rates between related but not isogenic strains, we sought to confirm that the MTH1 gene was solely responsible for the changes in growth rates. To do this, a snf3 rgt2 std1 mth1 strain was transformed with centromeric plasmids bearing either no insert or complete copies of the STD1 or MTH1 gene (data not shown). The growth rates of these three strains which are isogenic except at the STD1 and MTH1 loci confirmed that the MTH1 gene is solely responsible for the suppression of the snf3 rgt2 growth defect.
Invertase expression in glucose signaling mutants.
The SUC2 gene encodes secreted invertase and is a paradigm for the study of glucose repression. Many of the mutations which produce a Snf− phenotype show large defects in the derepression of SUC2 (25). In contrast, disruption of the SNF3 gene causes a Snf− phenotype with relatively little effect on SUC2 expression (26). Since mutations in MTH1 suppress the Snf− phenotype in a snf3 disruption, we analyzed invertase expression in cells lacking different combinations of SNF3, STD1, and MTH1. Deletion of SNF3 has only a modest effect on invertase depression (Fig. 4A). Mutations in either STD1 or MTH1 in a snf3 background also show relatively normal regulation of invertase expression. Interestingly, the std1 mth1 strain displays a Snf− growth phenotype that correlates with low invertase depression and which is suppressed by mutation of SNF3 (Fig. 3A and 4A). These data support the conclusion by Neigeborn et al. (26) that the Snf− phenotype in snf3 cells is likely to due to problems in hexose transport rather than invertase expression. We conclude that the genetic suppression of snf3 by mutations in MTH1 is not to be due to any changes in invertase expression.
Invertase expression was also analyzed in strains lacking both glucose sensors and either STD1, MTH1, or both (Fig. 4B). Loss of both glucose sensors resulted a severe defect in invertase regulation. The repressed level of invertase expression is much higher than in wild-type strains or either of the single glucose sensor mutants. In addition, invertase depression is much more defective in the snf3 rgt2 strains than in either single mutant. We conclude that the glucose sensors have overlapping roles with respect to invertase expression, with either Snf3 or Rgt2 being sufficient for both repression and derepression. Loss of the STD1 gene had little effect on invertase expression in the snf3 rgt2 background. However, loss of MTH1 caused a large increase in invertase expression, under both repressing and derepressing conditions. This large increase in invertase expression required the Std1 protein (compare the derepressed level in the snf3 rgt2 mth1 strains with the level in the snf3 rgt2 mth1 std1 strain). We conclude that Std1 and Mth1 play distinct and antagonistic roles in a snf3 rgt2 background. Mth1 inhibits expression of invertase, while Std1 is required for high-level induction. Lastly, it is worth noting that the strain lacking all four genes (snf3 rgt2 std1 mth1) is still able to regulate invertase expression 15-fold in response to changes in glucose concentration (32 versus 478 mU/OD). Therefore, some additional glucose-sensing mechanism that is independent of the glucose sensors must exist.
Regulation of HXT gene expression.
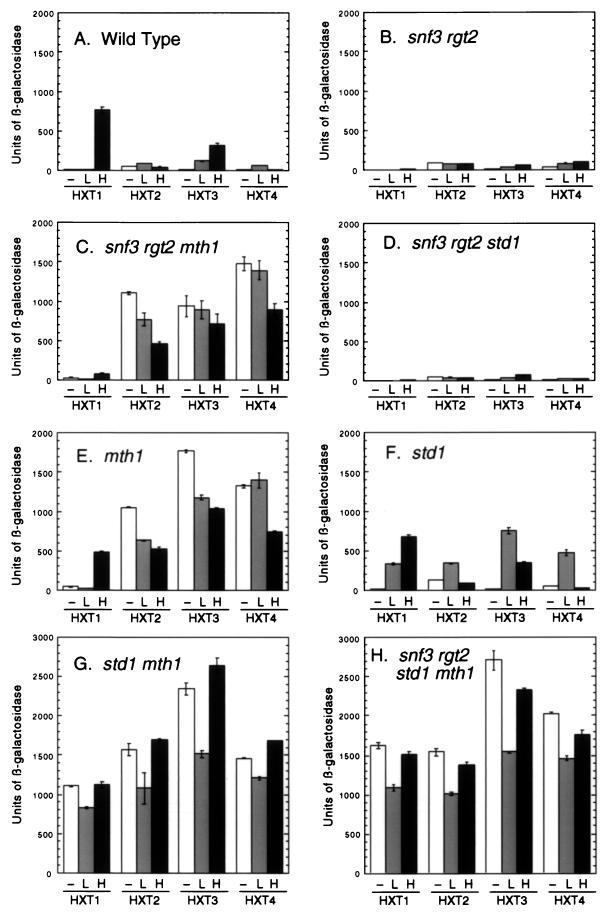
The finding that mutations in MTH1 could suppress the Snf− phenotype of the snf3 strain without affecting invertase expression suggested that these phenotypes were mediated by changes in the expression of the hexose transporter genes. To analyze this, we used a set of reporter plasmids with the lacZ gene cloned downstream of HXT promoters (29). Cells were grown in the absence of glucose and then shifted for 4 h to medium either lacking glucose or containing 0.1 or 6% glucose (Fig. 5). In wild-type cells, the predominant HXT expressed in the presence of high glucose concentrations is HXT1 (Fig. 5A). Cells lacking both glucose sensors lose HXT1 expression (Fig. 5B and reference 27) and lose the ability to ferment glucose. Mutation of MTH1 in a snf3 rgt2 background (Fig. 5C) causes a large increase in the expression of HXT2, HXT3, and HXT4. The high-level HXT gene expression in the snf3 rgt2 mth1 strain correlates with the restoration of the ability to grow on glucose-antimycin medium (Fig. 3B). Mutation of STD1 in the snf3 rgt2 background did not restore HXT expression (Fig. 5D) or growth on glucose-antimycin medium (Fig. 3B). Mutations in MTH1, in STD1, or in both also produced deregulated expression of the HXT genes (Fig. 5E to G) even in the presence of the glucose sensors. Mutations in STD1 affected HXT expression primarily in the presence of low glucose, while mutations in MTH1 affected HXT2, HXT3, and HXT4 expression both in the absence of glucose and in the presence of low and high glucose concentrations. These data suggest that the Std1 and Mth1 proteins act as a repressors of HXT gene expression. While the expression of the HXT2, HXT3, and HXT4 genes is affected by loss of either STD1 or MTH1, expression of the HXT1 gene is observed only in the absence of glucose when both STD1 and MTH1 are deleted.
FIG. 5.
HXT gene expression in cells with mutations in SNF3, RGT2, STD1, and MTH1. Cultures were grown in SC medium containing 3% glycerol and 2% ethanol and lacking uracil. Cells were collected and resuspended in the same medium containing either no glucose (−), 0.1% glucose (L), or 6% glucose (H). After 4 h in this medium, cells were harvested and protein extracts were assayed for β-galactosidase activity. Extracts from three independent transformants of each culture were assayed, and the mean value is plotted; the error bars represent 1 standard error. All bar graphs are drawn to the same scale, allowing direct comparisons between the different panels. The strains used were MSY465 (wild type), MSY441 (snf3 rgt2), MSY445 (snf3 rgt2 mth1), MSY443 (snf3 mth1 std1), MSY460 (mth1), MSY467 (std1), MSY471 (std1 mth1), and MSY447 (snf3 rgt2 std1 mth1).
Perturbation of Snf3 and Std1 stoichiometry affects SUC2 expression.
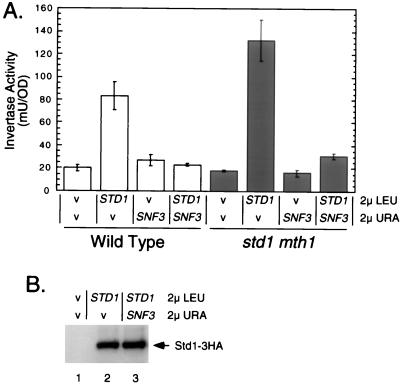
We sought additional evidence that the interaction of Std1 with the glucose sensors affects glucose-regulated gene expression. Previous studies have shown that increased expression of Std1 protein causes the induction of invertase expression (17, 37). We tested whether altering the relative stoichiometry of Std1 and Snf3 proteins affected invertase expression. Increased gene dosage of STD1 causes an induction of invertase even under repressing conditions (Fig. 6A). When the Snf3 protein is also overexpressed, increased gene dosage of STD1 is no longer able to induce invertase expression. Overexpression of Snf3 did not have any effect on the accumulation of Std1 protein, as judged by a Western blotting of epitope-tagged Std1 (Fig. 6B). Thus, Snf3 acts antagonistically to Std1 with respect to invertase induction, and the relative stoichiometry of these proteins can determine the level of invertase expression.
FIG. 6.
Snf3 inhibits Std1-mediated gene induction. (A) Invertase activity was measured in repressed cells (25) transformed with 2μm plasmids containing either no insert (v) or complete genomic copies of STD1 or SNF3 as indicated. Invertase activity from three independent transformants was measured, and the mean value is plotted; the error bars represent one standard error. The strains used were MSY401 (wild type) and MSY192 (std1 mth1). (B) Western blot analysis of Std1-3HA. Wild-type cells (MSY401) were transformed with the 2μm plasmids containing either no insert, the SNF3 gene, or an epitope-tagged STD1 gene. Protein extracts were prepared, and the level of the Std1-3HA protein (15 μg per lane) was detected with monoclonal antibody directed against the HA epitope.
Gene regulation by Std1 and Mth1 differ in the requirement for Snf1.
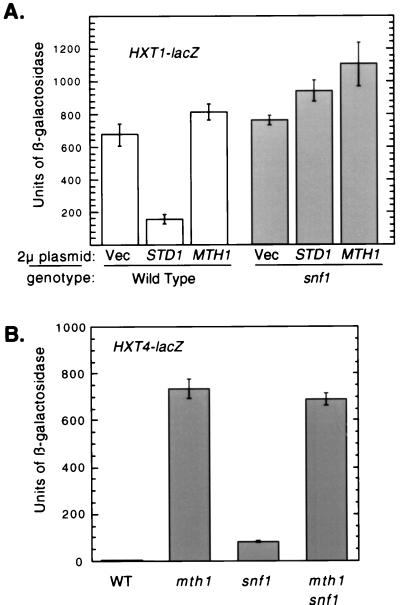
Since HXT1 expression is repressed under low glucose conditions whereas SUC2 expression is induced, we tested whether increased gene dosage of STD1 could repress HXT1 expression. Indeed, overexpression of Std1 but not Mth1 caused repression of HXT1 expression under high glucose conditions (Fig. 7A). Similar to the induction of SUC2, this activity required the Snf1 kinase. We conclude that Std1 acts in the same pathway and upstream of the Snf1 kinase.
FIG. 7.
Std1 and Mth1 act through distinct pathways that are Snf1 dependent and Snf1 independent, respectively. (A) Cultures containing the HXT1-lacZ reporter and the indicated 2μm plasmid were grown in SC medium containing 6% glucose lacking uracil and leucine. Cells from mid-logarithmic-phase cultures were collected, and protein extracts were assayed for β-galactosidase activity. Extracts from three independent transformants of each culture were assayed, and the mean value is plotted; the error bars represent 1 standard error. The strains used in this experiment, MSY465 (wild type) and FY1193 (snf1Δ10), were transformed with plasmid YEP351 (Vec), p6A5 (STD1), or pMT51 (MTH1). (B) Cultures containing the HXT4-lacZ reporter were grown in SC medium containing 6% glucose lacking uracil. Cells from mid-logarithmic-phase cultures were collected, and protein extracts were assayed for β-galactosidase activity. Extracts from three independent transformants of each culture were assayed, and the mean value is plotted; the error bars represent 1 standard error. The strains used were MSY465 (wild type), MSY469 (mth1), FY1193 (snf1), and MSY479 (mth1 snf1).
Mth1 acts as a potent repressor of HXT gene expression. We tested whether the ability of Mth1 to repress gene expression required the Snf1 kinase. HXT4 expression is relatively low in the presence of high concentrations of glucose but is greatly increased in cells lacking MTH1 (Fig. 5). Mth1-mediated repression of HXT4 did not require the Snf1 kinase since snf1 cells express low levels of HXT4 (Fig. 7B). Thus, Mth1-mediated repression of HXT4 is not dependent on a functional Snf1 kinase.
Expression patterns of STD1 and MTH1.
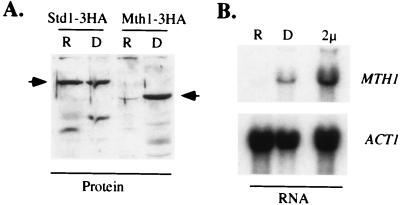
Since the STD1 and MTH1 genes play a large role in modulating the signals coming from the glucose sensors, we tested whether these genes are themselves regulated by glucose. Wild-type cells were transformed with centromeric plasmids encoding epitope-tagged STD1 or MTH1 genes. Protein extracts were prepared from cells grown under repressing and derepressing conditions and analyzed by Western blotting. Std1 accumulation was at a low but constitutive level independent of glucose concentration (Fig. 8A). In contrast, the Mth1 was glucose repressed. Neither protein displayed an altered electrophoretic mobility in response to changes in glucose concentration, suggesting that these proteins may not be subject to posttranslational modification in response to the glucose signal. To determine if the accumulation of Mth1 is regulated at the level of transcription, a Northern blot of total yeast RNA was probed with MTH1 sequences (Fig. 8B). MTH1 mRNA was not detectable in repressed cells but was clearly present in derepressed cells and was overexpressed in cells that contained a 2μm plasmid copy of MTH1. Therefore, the glucose-mediated regulation of Mth1 occurs at the level of mRNA accumulation.
FIG. 8.
MTH1 expression is subject to glucose repression. (A) Western blot of protein extracts (25 μg per lane) from cells bearing centromere plasmids encoding either Std1-3HA or Mth1-3HA as indicated. Wild-type cells (MSY401) were grown under repressing (R) and derepressing (D) conditions (25). Arrows indicate the mobility of the full-length proteins. (B) Northern blot of total cellular RNA (15 μg per lane) extracted from wild-type cells (MSY401) under repressing (R) and derepressing (D) conditions or from wild-type cells bearing a 2μm MTH1 plasmid (2μ), as indicated. The blot was first probed with 32P-labeled DNA complementary to MTH1 and then stripped and reprobed with [32P]DNA complementary to yeast actin mRNA (ACT1).
Subcellular localization of Std1-GFP.
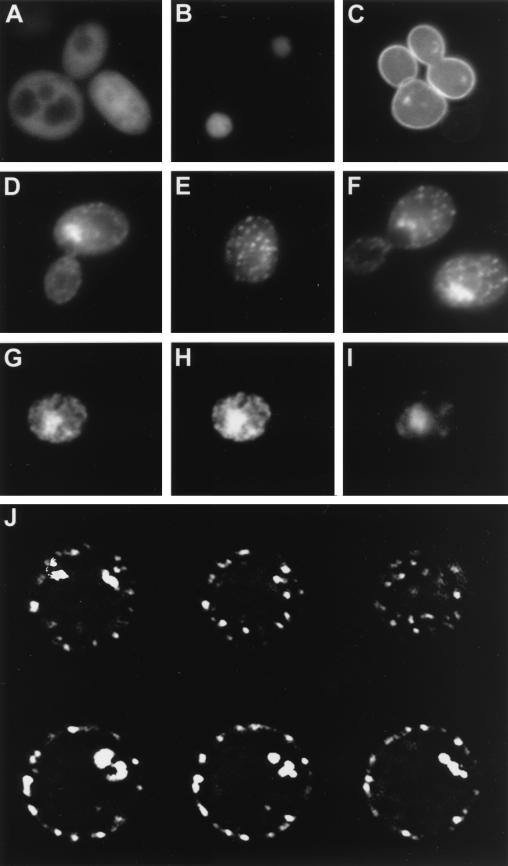
To determine the subcellular localization of the Std1, a fusion of GFP to the C terminus of the Std1 was engineered and expressed in yeast cells from a high-copy-number plasmid. The Std1-GFP protein used in these experiments was functional in two assays: its ability to induce SUC2 expression under repressing conditions and its ability to suppress the growth defect of a std1 mth1 strain on raffinose-antimycin medium (data not shown). The Std1-GFP protein was observed in both the cytoplasm and the nucleus (Fig. 9). Nuclear localization of the Std1-GFP fusion was confirmed by fixing cells and detecting the precise colocalization of the GFP fluorescence with Hoechst dye fluorescence (Fig. 9G to I). The Std1-GFP observed in the cytoplasm was punctate in nature (Fig. 9D to F). The punctate staining was due to the Std1 moiety since it was not observed when GFP was expressed by itself (Fig. 9A), nor was it observed when GFP was fused to histone H4 protein (Fig. 9B). Std1-GFP protein did not colocalize with Snf3 since a functional Snf3-GFP fusion (Fig. 9C) showed a distinct ring pattern of fluorescence, consistent with localization to the cytoplasmic membrane. The localization pattern of Std1-GFP was not affected by mutations in the glucose sensors (Fig. 9F), nor was it affected by the glucose concentration in the media (not shown). The subcellular localization of the punctate cytoplasmic staining was examined in more detail by focal plane composite imaging. In this experiment, a single cell showing both nuclear and punctate cytoplasmic staining was analyzed in a series of images at various focal planes. The six images shown (Fig. 9J) show that the punctate staining was localized at the cell periphery and not randomly scattered throughout the cytoplasm. However, the subcellular localization of Std1-GFP was not affected by deletion of the SNF3 and RGT2 genes. Therefore, the peripheral localization of the cytoplasmic Std1-GFP protein does not require the glucose sensors. Nonetheless, the peripheral localization of the cytoplasmic Std1 indicates that direct interactions between these proteins are possible.
FIG. 9.
Std1 is localized in cell nucleus and at the plasma membrane. Wild-type (A to E) or snf3 rgt2 (F) cells were analyzed by fluorescence microscopy. Cells expressed either unfused GFP from the GAL1 promoter (20) (A), histone-GFP fusion (34) (B), Snf3-GFP fusion (C), or Std1-GFP fusion (D to F). Fluorescence images were collected of a single Std1-GFP-expressing cell (G to I) that had been fixed with formaldehyde and stained with Hoechst dye. (G) Std1-GFP fluorescence; (I) Hoechst dye fluorescence; (H) composite image of both showing colocalization of the Hoechst and GFP fluorescence. (J) A single Std1-GFP-expressing cell that showed both nuclear and punctate cytoplasmic fluorescence was analyzed sequentially at six different focal planes.
DISCUSSION
We report here the interaction of the Std1 and Mth1 proteins with the glucose sensors Snf3 and Rgt2. The evidence for interaction is primarily genetic. First, these proteins interact in the two-hybrid system. The region of the glucose sensors identified in the two-hybrid system, the hydrophilic C-terminal tail domains have been implicated in glucose signal transduction in other studies (7, 27). The results of the two hybrid screen suggest that the glucose sensor tail domains may signal glucose availability through interactions with the Std1 and Mth1 proteins.
An additional line of genetic evidence for the interaction of the Std1 and Mth1 proteins with the glucose sensors came from phenotypic suppression studies. We constructed a set of strains with null alleles in STD1, MTH1, SNF3, and RGT2 in all 16 possible combinations. The Snf− phenotype (poor growth on raffinose-antimycin medium) of cells lacking snf3 function was suppressed by mutations in MTH1 but not by mutations in STD1. Second, cells lacking both glucose sensors (snf3 rgt2) are unable to grow by fermentation of glucose. This phenotype was not observed in either single mutant and was suppressed by mutation of the MTH1 gene. The suppression of this fermentation defect is independent of the STD1 gene. These data further support the existence of genetic interactions between these loci and the idea that the STD1 and MTH1 genes are functionally distinct.
Analysis of gene regulation in this set of mutant strains further supported the idea that interactions between the Std1, Mth1, Snf3, and Rgt2 proteins determines glucose signal transduction. Regulation of invertase expression is relatively normal in cells lacking either one of the glucose sensors; however, derepression is severely defective in cells lacking both Snf3 and Rgt2 proteins (Fig. 4). This derepression defect in snf3 rgt2 cells is suppressed by mutations in MTH1 but requires the STD1 gene. These data suggest that in a snf3 rgt2 background, Mth1 plays a role in invertase repression whereas Std1 is required for its activation. The idea that Std1 and Mth1 play distinct roles in the gene regulation is further supported by analysis of HXT gene expression. Mutations in STD1 specifically affect low-glucose signaling (Fig. 5F), while mutations in MTH1 affect HXT expression even in the absence of glucose (Fig. 5E). Lastly, Ozcan and Johnston have proposed that the HXT genes are regulated by distinct pathways (29). With respect to the STD1 and MTH1 genes, it is clear that the HXT1 gene is regulated by a mechanism that is distinct from that used for the HXT2, HXT3, and HXT4 genes. Deletion of MTH1 has no effect on HXT1 expression under any of the glucose conditions that we tested, while HXT2, HXT3, and HXT4 were induced as much as 400-fold by this mutation. Expression of HXT1 in the absence of glucose was observed only when both MTH1 and STD1 were deleted. Thus, either Std1 or Mth1 protein was sufficient to repress HXT1 expression in the absence of glucose. The HXT expression data suggest that the glucose sensors and the Std1 and Mth1 proteins act antagonistically, with the sensors being required for HXT induction and the Std1 and Mth1 proteins being required for their repression. A physical antagonism between these proteins is supported by the data presented in Fig. 6. Overexpression of Snf3 protein has no effect on Std1 protein levels yet it blocks the ability of Std1 to induce SUC2 expression.
A key component in glucose signaling is the Snf1 kinase. We found that the Std1 protein acts upstream of the Snf1 kinase. This is true both for the induction of invertase expression (17) as well as for the repression of HXT1 expression (Fig. 7A) caused by increased STD1 gene dosage. In contrast, we show that the Mth1 protein can act through a Snf1-independent pathway. HXT4 expression is repressed in the presence of high glucose concentrations, growth conditions under which the Snf1 kinase is inactive (34). Repression of HXT4 requires MTH1 but not SNF1 (Fig. 7B), demonstrating that Mth1 can mediate repression via a Snf1-independent pathway.
Analysis of the expression of these two sets of homologous genes, STD1/MTH1 and SNF3/RGT2, shows some interesting parallels. Earlier studies have shown that SNF3 is glucose repressed (26), while RGT2 is constitutively expressed (28). An identical pattern was found for STD1 and MTH1. In this case, STD1 was expressed constitutively, independent of glucose concentration (Fig. 8), while the MTH1 gene was subject to glucose repression. Indeed, analysis of global patterns of gene expression indicated that both SNF3 and MTH1 mRNAs accumulated when glucose was depleted from the medium and both were subject to repression by Tup1 (8). The expression data for these proteins correlate well with the two-hybrid interaction data. Std1 is expressed constitutively and interacts with both Rgt2 and Snf3, while Mth1 is subject to glucose repression interacts only with the glucose-repressed Snf3.
While we have demonstrated considerable genetic interactions between the STD1, MTH1, SNF3, and RGT2 loci, our studies are not consistent with a stable complex between the glucose sensor tails and the Std1 or Mth1 proteins. First, experiments with GST-Snf3-tail fusions have not been able to detect a complex with Std1 or Mth1 protein (32). Second, these proteins have distinct patterns of subcellular localization. A GFP fusion to the C terminus of Snf3 protein produced a functional Snf3-GFP protein that localized to the cytoplasmic membrane. In contrast, a functional Std1-GFP protein showed nuclear localization and punctate staining at the cytoplasmic periphery that is not affected by glucose concentration or the absence of the glucose sensors. Similar localization patterns were observed with a Mth1-GFP fusion (data not shown). Thus, our data do not support a model in which the Std1 and Mth1 proteins form a stable complex with the tail domains of the glucose sensors. Instead, we hypothesize a dynamic interaction between these proteins. Alternatively, it is possible that Std1 associates with additional membrane signaling proteins. Recently a 12-transmembrane-domain protein with a hydrophilic N-terminal extension was shown to be involved in signaling amino acid availability (9). Furthermore, there is evidence that Std1 also plays a role in cation stress response (12), and the signaling molecule(s) in that pathway has not been identified.
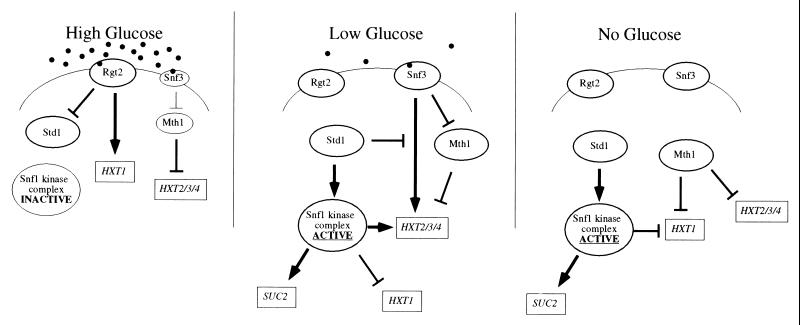
Taken together, our data are consistent with the model of glucose signaling presented in Fig. 10. We hypothesize that the sensors signal only when they are bound by glucose and that Rgt2 has a higher Km for glucose than Snf3. In the presence of high glucose concentrations, the Rgt2 protein is bound by glucose and signals activation of HXT1 (28, 29). Rgt2 also inhibits the activity of the Std1 protein; however, this can be overcome in cells with increased gene dosage of STD1. Snf3 and Mth1 are represented by smaller symbols in the presence of high glucose since they are both subject to glucose repression. However, the Mth1 protein is still functional in the presence of high glucose since deletion of the MTH1 gene results in significantly increased expression of HXT2, HXT3, and HXT4 under these conditions. In the presence of low glucose concentrations (0.1%), Snf3 but not Rgt2 is bound by glucose and capable of signaling. Snf3 activates expression of the high-affinity hexose transporters (27, 28) and inhibits the activity of Mth1. Std1 acts upstream of the Snf1 kinase, which relieves gene repressive forces at both SUC2 and the high-affinity HXT’s (29). Activated Snf1 also plays a role in signaling repression of the low-affinity transporter, HXT1. Lastly, Std1 acts as a damper on Snf3-mediated activation, perhaps by direct competition with Mth1 for binding to the Snf3 tail domain. In the absence of any glucose, neither Snf3 not Rgt2 is capable of signaling. Either Std1 or Mth1 is sufficient to signal repression to HXT1, while the Mth1 protein by itself plays an essential role in the repression of the high-affinity transporters. The appropriate regulation of gene expression in response to changes in glucose concentrations is thereby accomplished through the complex interactions of these two homologous pairs of proteins.
FIG. 10.
Model for the glucose signal transduction in yeast. Arrows indicate activation, and lines with perpendicular bars indicate repression. Proteins are represented by ovals, and genes are represented by rectangles. Filled circles represent glucose.
ACKNOWLEDGMENTS
We are grateful to Eckhard Boles, Arle Kruckeberg, Mark Johnston, and Sabire Ozcan for gifts of plasmids and strains and for discussion of results prior to publication.
This work was supported by grant GM46443 from the National Institutes of Health.
REFERENCES
- 1.Ausubel A M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 2.Baldwin S A, Henderson P J F. Homologies between sugar transporters from eukaryotes and prokaryotes. Annu Rev Physiol. 1989;51:459–471. doi: 10.1146/annurev.ph.51.030189.002331. [DOI] [PubMed] [Google Scholar]
- 3.Bartel P L, Chien C, Sternglanz R, Fields S, editors. Using the two-hybrid system to detect protein-protein interactions. Oxford, England: Oxford University Press; 1993. [Google Scholar]
- 4.Becker J U, Betz A. Membrane transport as controlling pacemaker of glycolysis in Saccharomyces carlsbergensis. Biochim Biophys Acta. 1972;274:584–597. doi: 10.1016/0005-2736(72)90205-2. [DOI] [PubMed] [Google Scholar]
- 5.Carlson M, Osmond B C, Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 7.Coons D M, Vagnoli P, Bisson L F. The C-terminal domain of Snf3p is sufficient to complement the growth defect of snf3 null mutations in Saccharomyces cerevisiae: SNF3 functions in glucose recognition. Yeast. 1997;13:9–20. doi: 10.1002/(SICI)1097-0061(199701)13:1<9::AID-YEA51>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 8.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 9.Didion T, Regenberg B, Jurgensen M U, Kielland-Brandt M C, Andersen H A. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol Microbiol. 1998;27:643–650. doi: 10.1046/j.1365-2958.1998.00714.x. [DOI] [PubMed] [Google Scholar]
- 10.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 11.Ganster R, Shen W, Schmidt M C. Isolation of STD1, a high-copy-number suppressor of a dominant negative mutation in the yeast TATA-binding protein. Mol Cell Biol. 1993;13:3650–3659. doi: 10.1128/mcb.13.6.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganster R W, McCartney R R, Schmidt M C. Identification of a calcineurin-independent pathway required for sodium ion stress response in Saccharomyces cerevisiae. Genetics. 1998;150:31–42. doi: 10.1093/genetics/150.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 14.Goldstein A, Lampen J O. β-D-Fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42C:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- 15.Hill J E, Meyers A M, Koerner T J, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 16.Hresko R C, Kruse M, Strube M, Mueckler M. Topology of the Glut 1 glucose transporter deduced from glycosylation scanning mutagenesis. J Biol Chem. 1994;269:20482–20488. [PubMed] [Google Scholar]
- 17.Hubbard E J A, Jiang R, Carlson M. Dosage-dependent modulation of glucose repression by MSN3 (STD1) in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:1972–1978. doi: 10.1128/mcb.14.3.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James P, Halladay J, Craig E A. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruckeberg A L. The hexose transporter family of Saccharomyces cerevisiae. Arch Microbiol. 1996;166:283–292. doi: 10.1007/s002030050385. [DOI] [PubMed] [Google Scholar]
- 20.Lee M, Henry M, Silver P A. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 21.Liang H, Gaber R F. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol Biol Cell. 1996;7:1953–1966. doi: 10.1091/mbc.7.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall-Carlson L, Celenza J L, Laurent B C, Carlson M. Mutational analysis of the SNF3 glucose transporter of Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1105–1115. doi: 10.1128/mcb.10.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 24.Mueckler M, Caruso C, Baldwin S A, Panico M, Blench I, Morris H R, Allard W J, Lienhard G E, Lodish H F. Sequence and structure of a human glucose transporter. Science. 1985;229:941–945. doi: 10.1126/science.3839598. [DOI] [PubMed] [Google Scholar]
- 25.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neigeborn L, Schwartzberg P, Reid R, Carlson M. Null mutations in the SNF3 gene of Saccharomyces cerevisiae cause a different phenotype than do previously isolated missense mutations. Mol Cell Biol. 1986;6:3569–3574. doi: 10.1128/mcb.6.11.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ozcan S, Dover J, Johnston J. Glucose sensing and signaling by two glucose receptors in the yeast S. cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ozcan S, Dover J, Rosenwald A G, Woelfl S, Johnston M. Two glucose transporters in S. cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozcan S, Johnston S. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose M D, Winston F, Hieter P, editors. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 31.Sauer N, Friedlander K, Graml-Wicke U. Primary structure, genomic organization and heterologous expression of a glucose transporter from Arabidopsis thaliana. EMBO J. 1990;9:3045–3050. doi: 10.1002/j.1460-2075.1990.tb07500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solimeo, H., and M. C. Schmidt. Unpublished data.
- 33.Tillman T S, Ganster R W, Jiang R, Carlson M, Schmidt M C. STD1 (MSN3) interacts directly with the TATA-binding protein and modulates transcription of the SUC2 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:3174–3180. doi: 10.1093/nar/23.16.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 35.Wilson W A, Hawley S A, Hardie D G. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 36.Winston F, Dollard C, Ricupero-Hovasse S L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1996;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Shen W, Schmidt M C. Amino acid residues in Std1 protein required for induction of SUC2 transcription are also required for suppression of TBPΔ58 growth defect in Saccharomyces cerevisiae. Gene. 1998;215:131–141. doi: 10.1016/s0378-1119(98)00276-5. [DOI] [PubMed] [Google Scholar]