Abstract
Globin mRNAs accumulate to 95% of total cellular mRNA during terminal erythroid differentiation, reflecting their extraordinary stability. The stability of human α-globin mRNA is paralleled by formation of a sequence-specific RNA-protein (RNP) complex at a pyrimidine-rich site within its 3′ untranslated region (3′UTR), the α-complex. The proteins of the α-complex are widely expressed. The α-complex or a closely related complex also assembles at pyrimidine-rich 3′UTR segments of other stable mRNAs. These data suggest that the α-complex may constitute a general determinant of mRNA stability. One or more αCPs, members of a family of hnRNP K-homology domain poly(C) binding proteins, are essential constituents of the α-complex. The ability of αCPs to homodimerize and their reported association with additional RNA binding proteins such as AU-rich binding factor 1 (AUF1) and hnRNP K have suggested that the α-complex is a multisubunit structure. In the present study, we have addressed the composition of the α-complex. An RNA titration recruitment assay revealed that αCPs were quantitatively incorporated into the α-complex in the absence of associated AUF1 and hnRNP K. A high-affinity direct interaction between each of the three major αCP isoforms and the α-globin 3′UTR was detected, suggesting that each of these proteins might be sufficient for α-complex assembly. This sufficiency was further supported by the sequence-specific binding of recombinant αCPs to a spectrum of RNA targets. Finally, density sedimentation analysis demonstrated that the α-complex could accommodate only a single αCP. These data established that a single αCP molecule binds directly to the α-globin 3′UTR, resulting in a simple binary structure for the α-complex.
mRNA stability plays an important role in the expression of a wide range of eukaryotic genes (53). The steady-state concentration of a particular mRNA reflects a balance between its rates of synthesis and degradation. Each mRNA species decays at a characteristic rate (half-life [t1/2]). Although the average t1/2 of mRNAs in mammalian cells is 3 to 5 h (26), these values can range from several minutes, as for proto-oncogene and cytokine mRNAs (6, 53), to half a day or more, as for globin mRNAs (3, 54). In general, the t1/2 of a specific mRNA correlates with the function of its protein product. Proteins that must be expressed in a narrow window of time are encoded by short-lived mRNAs, while proteins expressed in large quantities in terminally differentiated cells tend to be encoded by highly stable mRNAs (6, 52, 56, 64). The stability of an mRNA can also change in response to alterations in cellular growth conditions, environmental stress, cell cycle, or developmental cues (reviewed in references 2, 7, 45, and 46).
Determinants of mRNA stability are both general and specific and appear to act through multiple and frequently overlapping pathways. The vast majority of eukaryotic mRNAs share anm7G(5′)ppp(5′)N cap and a 3′ polyadenylate [poly(A)] tail. These structures act, at least in part, to protect the mRNA from rate-limiting exonuclease attack. In yeast, 5′ decapping is believed to be a rate-limiting step for the turnover of most mRNAs, followed by 5′→3′ exonucleolytic degradation (42, 43). This decapping is usually triggered by preceding shortening of the poly(A) tail (14, 15, 50, 58). Decapping can also be triggered directly, as appears to occur in the case of nonsense-mediated mRNA decay (43). Less commonly, mRNA degradation is initiated by site-specific endonucleolytic cleavage (8, 9). In a number of systems, these turnover pathways appear to be controlled by RNA-protein (RNP) complexes that assemble on the target mRNAs (31, 33, 49, 53). Although these complexes could theoretically assemble anywhere on the mRNA strand, the majority of relevant cis elements localize within the 3′ untranslated region (3′UTR) (25).
Globin mRNAs serve as prototypes for long-lived mRNAs. Globin genes are expressed exclusively in cells of the erythroid lineage. Accumulation of globin mRNAs to well over 95% of total cellular mRNA during terminal erythroid differentiation reflects their unusual stability (4, 5, 32). Previous studies from our laboratory suggest that high-level stability of human α-globin (hα-globin) mRNA is conferred by sequences within its 3′ UTR. Mutations that allow ribosomal read-through into this region destabilize hα-globin mRNA, with consequent loss of gene function (11, 24, 35, 69). Our previous studies have mapped a discontinuous, pyrimidine-rich cis-acting stability element in the 3′UTR of hα-globin (α3′UTR) mRNA. Mutations in this element result in direct, translation-independent destabilization of hα-globin mRNA in transfected mouse erythroleukemia (MEL) cells (70). More recent studies have demonstrated in vitro assembly of a sequence-specific RNP complex (α-complex) at this site. This α-complex is operationally defined by its characteristic migration on native gels and by its exquisite sensitivity to poly(C) competition. Mutations that block the in vitro assembly of the α-complex also destabilize the α-globin mRNA in transfected erythroid cells (68, 70). These data suggest that the α-complex controls a rate-limiting step in mRNA degradation.
The α-complex contains one or more proteins with poly(C) binding activity [α-globin mRNA poly(C) binding protein (αCP)] (30, 68). Biochemical studies demonstrate that this activity is encoded by at least three closely related αCP isoforms: αCP-1 and two forms of αCP-2, the full-length protein (αCP-2) and an alternatively spliced form lacking an internal 31-amino-acid segment (αCP2-KL) (19). Each αCP protein contains three repeats of a 50-amino-acid hnRNP K-homology domain (38, 60) that is present in a wide variety of RNA binding proteins (16, 18, 27, 36, 37, 44, 51, 57, 59, 61, 65). αCPs can homodimerize as well as heterodimerize with other RNA binding proteins (references 19, 20, 26, 29, and 68a and our unpublished data). Both αCP-1 and αCP-2 can assemble into the α-complex and in this respect appear to be equivalent in RNA binding (30). Whether αCPs exists in the α-complex as monomers or dimers and/or coassemble in the complex with additional protein partners is not known.
The ability to form the α-complex does not appear to be erythroid cell specific, as this RNP complex can be assembled by using extracts from a wide range of cell types (22, 41, 68). Consistent with this observation is the demonstration that αCPs have a wide tissue distribution (1, 22). These data suggest that αCPs and/or the proteins that constitute the α-complex have broad functions that extend beyond stabilization of hα-globin mRNA. A subset of highly stable mRNAs that share with hα-globin mRNA a pyrimidine-rich motif in their 3′UTRs have been identified (22). These mRNAs, which include 15-lipoxygenase (Lox) (47), α1(I)-collagen (Coll) (63), and tyrosine hydroxylase (TH) (13), each assemble a complex at the pyrimidine-rich regions of their 3′UTRs, and each of the in vitro-assembled complexes contains αCPs (22). In the case of TH and Coll, the high-level stability of the mRNA has been linked to this cis element (13, 63). These data have led to the model that αCP-containing complexes constitute general determinants of mRNA stability (22).
Whether the αCPs alone constitute the α-complex is unclear. While biochemically enriched αCP proteins can reconstitute α-complex formation in a cytosolic extract depleted of poly(C) binding activity, they do not appear to bind directly to the α-globin mRNA 3′UTR (30, 68). This finding has suggested that assembly of αCPs into the α-complex may depend on their interaction with other proteins during complex formation. The presence of coassembling proteins is suggested by the identification of a number of proteins that interact with αCPs by yeast two-hybrid screens (references 19, 29, and 68a and our unpublished results). One of these proteins, AU-rich binding factor 1 (AUF1; also referred to as hnRNP D [28]), has previously been implicated in accelerated decay of immediate-early mRNAs with AU-rich cis elements in their 3′UTRs (17, 66, 72). This same AUF1 was recently identified as one of multiple proteins that comigrate on a native gel with the in vitro-assembled α-complex (29). On the basis of that study, it was proposed that AUF1 constitutes a component of the α-complex. Separate studies suggest that αCP may interact with a second poly(C) binding protein, hnRNP K, at the pyrimidine-rich element within the 3′UTR of the long-lived erythroid cell-specific Lox mRNA (48). This complex formation has been implicated in translational control of the Lox mRNA (47). There is no direct evidence to confirm the incorporation of AUF1, hnRNP K, or any other proteins into the α-complex. The composition and stoichiometry of the proteins in these complexes and their mode of action as determinants of mRNA function remain to be determined. The present study focuses on these questions. The data suggest a simple binary model of α-complex structure.
MATERIALS AND METHODS
Cell extracts.
Human erythroleukemia (K562) and MEL cells were cultured under standard conditions (68). Cell fractionation and preparation of cytosolic extracts (S100) were as previously described (23, 68).
Expression of recombinant αCPs.
Vectors, buffers, and protocols used for expression of αCPs were purchased from Qiagen and Novagen (vector DNA pET-28a). In brief, the coding regions for hαCP-1, hαCP-2, and mouse αCP2-KL (mαCP2-KL) (19, 30, 34) were PCR amplified from plasmids pGBD-αCP1, pGBD-αCP2 (gifts from M. Kiledjian, Rutgers University, New Brunswick, N.J.) (29), and pB1005 (a gift from S. Smale, University of California, Los Angeles) (21), respectively. The gel-purified PCR products were cloned into pET-28a (hαCP-1), pQE-31 (hαCP-2), and pQE-8 (mαCP2-KL), and the fusion proteins were expressed, purified on Ni2+ columns (Novagen), and further purified over a Superdex 200 gel filtration column (Pharmacia). The major column peak, containing the monomeric form of recombinant αCP, was used for RNA binding studies. The concentrations of purified recombinant His6-αCPs were calculated by the Bradford method and verified by comparison with dilutions of ovalbumin, using silver-stained sodium dodecyl sulfate (SDS)-polyacrylamide gels. The recombinant hαCP-1 expressed from the pET-28a vector contained an additional vector-derived 34 amino acids, while hαCP-2 and mαCP2-KL expressed from the pQE series vectors contained an additional 10 vector-derived amino acids.
RNA probes.
3′UTR transcription templates were generated by PCR from the wild type hα-globin gene or from hα-globin genes containing specific 3′UTR mutations (40). Transcription of each full-length 3′UTR was carried out with the amplified fragments as previously reported (68). RNA oligonucleotide probes corresponding to pyrimidine-rich segments within the 3′UTRs of rabbit Lox (47), human Coll (63), rat TH (13), αPR (the 42-nucleotide [nt] RNase-protected region of the α3′UTR protected by the assembled α-complex [22]), and the mutated form of αPR are all as previously reported (22). RNA oligonucleotides were synthesized by the University of Pennsylvania Nucleic Acid Core Facility. Probes were 5′-end labeled by using T4 polynucleotide kinase (New England Biolabs Beverly, Mass.) and [γ-32P]ATP (Amersham).
EMSA.
The RNA electrophoretic mobility shift assay (EMSA) was carried out as described previously (23, 68). RNA probe (∼20,000 cpm) was incubated with 50 μg of S100 proteins (or ∼50 ng of His6-αCP-1, His6-αCP-2, or His6-αCP2-KL purified protein) in a 20-μl total volume at room temperature for 30 min. Binding buffer was 10 mM Tris-HCl (pH 7.4), 150 mM KCl, 1.5 mM MgCl2, and 0.5 mM dithiothreitol. RNase T1 (1 U/μl) was then added, and the mixture was incubated first at room temperature for 10 min and then for additional 10-min incubation in the presence of added heparin (final concentration, 5 mg/ml; Sigma). Samples were subsequently electrophoresed through a 5% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA buffer. Bands were visualized by autoradiography of the dried gel. For EMSA with 32P-labeled synthetic oligonucleotide, 20,000 cpm of probe was mixed with 50 μg of MEL S100 extract (or ∼50 ng of His6-αCP-1, His6-αCP-2, or His6-mαCP2-KL purified protein), incubated, and then gel analyzed as detailed above except that RNase T1 digestion was omitted.
Protein recruitment assay.
S100 extract (80 to 100 μg) was preincubated with 0.5 μl of β-mercaptoethanol (β-ME) and 1.0 U of RNase inhibitor (purchased from 5′ Prime→3′ Prime) for 20 min at room temperature. The extract was then added to a mixture of 30 μg of tRNA, 17 μl of 4× binding buffer (600 mM KCl, 6 mM MgCl2, 40 mM Tris-HCl [pH 7.4], 2 mM dithiothreitol), and 20 ng of [32P]3′UTR. The incubation mixtures also contained 0, 20, or 40 μg of unlabeled 3′UTR in a final volume of 60 μl. After 20 min of incubation at room temperature, RNase T1 (1 U/ml) was added and the mixture was incubated at room temperature for an additional 10 min. Glycerol loading dye containing 200 μg of heparin was added, and equal aliquots of each sample were immediately loaded onto four separate 5% acrylamide–0.5× Tris-borate-EDTA gels and run at 110 V for 3 h. One gel was dried and autoradiographed, while the other gels were transferred to nitrocellulose filters and separately probed or stripped and reprobed with the one of the following five antibodies. The chicken anti-αCP (used at 1:5,000 dilution) lacks isoform specificity (22); mouse monoclonal antibody 3C2 (1:20,000 dilution; gift from G. Dreyfuss) is specific for hnRNP K and J (38); rabbit anti-AUF (used at 1:15,000 dilution) was a gift from G. Brewer, anti-αCP-1 recognizes αCP-1 but not αCP-2; anti-αCP-2 recognizes full-length αCP-2 but not αCP-1 or αCP2-KL. The latter two rabbit antisera (used at 1:6,000 dilutions) were kind gifts from A. Gamarnik, University of California, San Diego (20). To generate sufficient amounts of the anti-αCP-1 and anti-αCP-2 antibodies for all experiments, additional rabbit antisera (lab designations FF1 and FF2) were raised by our laboratory against the same two epitopes as originally described (20) (residues 229 to 243 in the αCP-1 sequence and residues 200 to 214 in the αCP-2 sequence, respectively). An antibody that recognized αCP2-KL was also generated. This antiserum (lab designation FF3) was raised against an epitope (residues 237 to 251 in the αCP-2 sequence) that was common to αCP-2 and αCP2-KL and distinct from αCP-1. After electrotransfer of proteins and incubation of the membrane with primary antibodies, the signals were developed by incubation with appropriate secondary antibodies: horseradish peroxidase-conjugated goat anti-chicken immunoglobulin G (IgG; used at 1:7,000 dilution; Accurate Chemical, Westbury, N.Y.), sheep anti-mouse IgG (used at 1:5,000; Amersham), or donkey anti-rabbit IgG (used at 1:5,000; Amersham). Immune complexes were detected by using the Amersham ECL system.
Reverse transcription (RT)-PCR analysis.
Total cytoplasmic RNA was isolated from MEL and K562 cells by the phenol-detergent method (39). First-strand cDNA was synthesized with 10 U of avian myeloblastosis virus reverse transcriptase (Promega), using 1 μg of the total cytoplasmic RNA and 10 pmol of random hexanucleotide primers (Boehringer Mannheim). Double-stranded DNA was then generated by PCR amplification with Taq DNA polymerase (Perkin-Elmer), using a 32P-end-labeled antisense primer (5′-CAA TAG CCT TTC ACC TCT GGA GA-3′) and an unlabeled sense primer (5′-CRT GAC CAT YCC GTA CC-3′). Conditions for PCR were as follows: preheating at 95°C for 3 min, denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min. Samples were taken after 24 to 32 PCR cycles and applied to 2.5% MetaPhor agarose (FMC) gel. The dried gel was analyzed with a PhosphorImager (Molecular Dynamics), and the radioactivity in each band was determined by using ImageQuant software. The ratio of the different bands was calculated as described from the slope of the line comparing PCR products to the number of PCR cycles (62).
RNA-binding affinities.
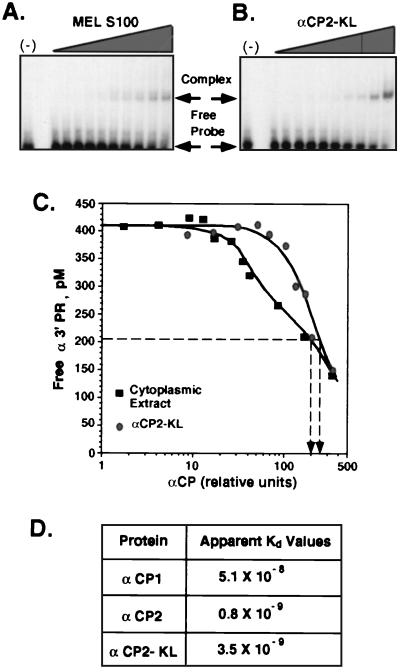
Concentrations of αCP in MEL S100 cytoplasmic extract were normalized to the concentration of recombinant αCP by Western blot analysis with the chicken anti-αCP. RNA-protein binding reactions were carried out by addition of increasing concentrations of protein to a constant RNA concentration. Bound and free RNAs were separated by EMSA (see above), gels were dried, and band activities were quantified by PhosphorImager analysis. Free probe concentrations were plotted versus the relative concentration of His6-αCP2-KL, His6-αCP-1, or His6-αCP-2 or the concentration of αCP in extract. Relative affinities (Fig. 7A to C) were determined as the protein concentration at which 50% of the RNA was bound (10). Absolute values of apparent Kd for the 3′UTR (Fig. 7D) were measured as described elsewhere (55), with minor modifications, by titration of the 3′UTR probe.
FIG. 7.
Binding affinities of recombinant αCPs and native cell extract for the α3′UTR. (A and B) RNA-binding affinities of proteins in the MEL S100 extract (A) and of recombinant αCP2-KL (B). Increasing amounts of each were incubated with a fixed amount of [32P]αPR probe. The amounts of αCP2-KL in the recombinant preparation and in the S100 were normalized by Western blot analysis (see Materials and Methods). The free probe and complexed probe were separated by native gel electrophoresis and quantified. (C) Plot of binding. Results of both sets of experiments are shown; the concentrations of αCP2-KL at which half of the probe is incorporated in the complex are indicated by the vertical arrows. (D) Apparent Kd (molar) for α3′UTR of each of the recombinant αCP isoforms.
Sucrose gradient centrifugation.
Ninety-microliter aliquots of MEL cell S100 extract (1 mg of total protein) or purified recombinant His6-αCPs (30 μg) were incubated for 20 min at room temperature in the presence of 2% β-ME and 2 U of RNase inhibitor (5′ Prime→3′ Prime). Samples were mixed with 50 μl of binding buffer supplemented with 70 μg of yeast tRNA and 4 × 107 cpm of 32P-labeled α3′UTR probe and incubated for additional 20 min at room temperature. An identical mixture in which no probe had been added was prepared in parallel. The mixtures were treated with 150 U of RNase T1 for 5 min, and heparin was added to the final concentration of 2 mg/ml. Samples were loaded onto prechilled 5 to 20% sucrose gradients made in binding buffer and supplemented with 1.5% (vol/vol) β-ME, 2 mg of heparin per ml, 10 μg of leupeptin per ml, 1 μg of pepstatin A per ml, and 2 μg of aprotinin per ml. Gradients were centrifuged in an SW41 rotor at 37,000 rpm for 40 h at 4°C and fractionated from the top. Gradients of S100 extract lacking RNA probe were analyzed for the presence of αCP by Western blot analysis with chicken anti-αCP antibodies (1:5,000 dilution) and for the ability to reconstitute the α-complex by addition of 2 × 105 cpm of 32P-labeled α3′UTR to 20-μl aliquots of the gradient fractions followed by EMSA. Mobility standards were myoglobin (Mr = 17,000, s20,w = 2.04), ovalbumin (Mr = 43,000, s20,w = 3.66), bovine serum albumin (BSA) (Mr = 67,000, s20,w = 4.58), and aldolase (Mr = 158,000, s20,w = 7.35). Peaks were revealed and quantified by exposure of the gels to a PhosphorImager screen (Molecular Dynamics). All data were within the linear range and were analyzed with ImageQuant software (Molecular Dynamics). Sedimentation coefficients (S values) for αCPs and α-complex were determined from the linear curves for the isokinetic gradient.
RESULTS
αCPs are quantitatively recruited into the α-complex in the absence of associated AUF-1 or hnRNP K.
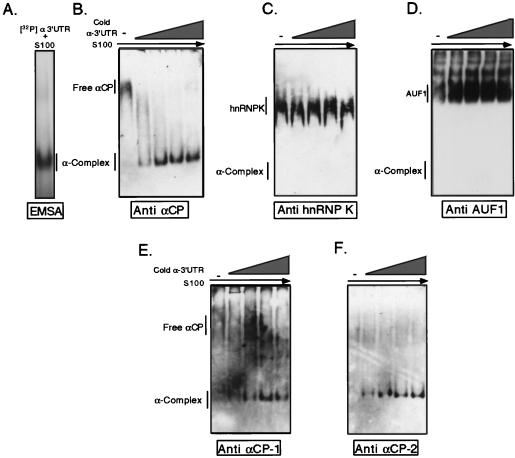
Previous studies have suggested that the 37-kDa AU binding factor AUF1 and the 69-kDa hnRNP K may associate with αCP in RNP complexes (29, 48). To directly test for these two proteins in the α-complex, we carried out a series of RNP recruitment assays (Fig. 1). In these assays, increasing concentrations of a specific 3′UTR RNA target were added to S100 extract followed by RNase digestion and electrophoresis on a native gel. If a protein bound to the added RNA, it would be shifted (recruited) from its uncomplexed position on a native gel to a position corresponding to the resultant RNP complex. Tracer levels of 32P-labeled α-complex, generated in a manner identical to that used for the parallel recruitment reactions, were used to mark the position of this complex (Fig. 1A). Recruitment of each protein in question (αCPs, AUF1, or hnRNP K) was selectively monitored by Western analysis of the recruitment gel by using the corresponding antibody. Addition of increasing amounts of unlabeled α3′UTR probe resulted in a quantitative recruitment of αCPs to the α-complex (Fig. 1B). At levels of mRNA sufficient for full recruitment of all immunoreactive αCPs, there was no corresponding shift in the migration of either hnRNP K or AUF1 from its native positions (Fig. 1C and D, respectively). The presence of both αCP-1 and αCP-2 in the α-complex was specifically demonstrated by reprobing the Western blots with epitope-specific antisera (Fig. 1E and F, respectively). These data demonstrated that a spectrum of αCPs was quantitatively incorporated into the α-complex in the absence of AUF1 and hnRNP K.
FIG. 1.
Selective recruitment of αCP into the α-complex. (A) Position of the α-complex. The α-complex (as indicated) was identified by incubating MEL cytosolic S100 extract with [32P]α3′UTR followed by RNase treatment. The sample was electrophoresed on a native polyacrylamide gel and autoradiographed. (B) Recruitment of α-CP into the α-complex. Increasing concentrations of unlabeled α3′UTR (indicated by the wedge; see Materials and Methods for concentrations) were incubated with MEL S100 extract to form the α-complex. Products of the incubations were analyzed on native gels. The position of the uncomplexed αCP in the S100 and its recruitment to the position of the α-complex were each visualized by Western analysis. (C) AUF1 was not recruited into the α-complex. AUF1 was detected with a monospecific antibody (gift from G. Brewer). The study was carried out as for panel B. (D) hnRNP K was not recruited into the α-complex. hnRNP K was detected with a corresponding monospecific antibody (38). The study was carried out as described above. (E) Recruitment of αCP-1 into the α-complex (determined as detailed for panel B). The antibody used was specific to the αCP-1 isoform (see Materials and Methods and Fig. 6A). (F) Recruitment of αCP-2 into the α-complex (determined as detailed for panel B). The antibody used was specific to the αCP-2 isoform (see Materials and Methods and Fig. 6A).
Quantitative recruitment of αCPs into the Lox mRNA 3′UTR complex in the absence of associated hnRNP K.
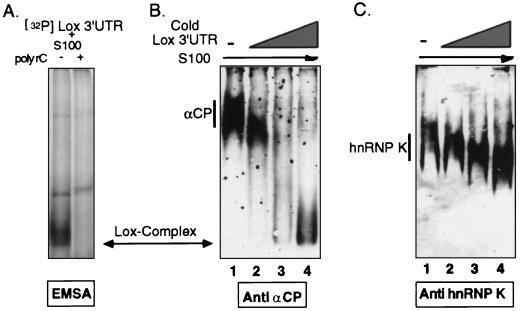
hnRNP K has been specifically reported to associate with αCPs in the translational-silencing RNP complex that forms at the pyrimidine-rich motif of the Lox mRNA 3′UTR (48). Because hnRNP K was not observed to coassemble with αCP in formation of the α-complex (Fig. 1C), the recruitment assay was extended to analyze the complex forming on the Lox 3′UTR (Fig. 2). A position marker for the Lox complex was established by incubating tracer amounts of 32P-labeled Lox 3′UTR with S100 extracts. As expected on the basis of previous studies (22), this complex was sensitive to poly(C) competition (Fig. 2A). Addition of unlabeled Lox 3′UTR to the extract resulted in quantitative recruitment of αCP to the position of the Lox complex (Fig. 2B). Although there was a gradual downward shift in the position of hnRNP K with the addition of increasing amounts of Lox mRNA, this change appeared to be nonspecific, as the band remained well above the position of the Lox complex, as marked by the radioactive RNA probe and the coincident position of the recruited αCP (Fig. 2C). Thus, αCP was identified in the Lox complex, consistent with our prior studies (22), but hnRNP K could not be detected in the same complex.
FIG. 2.
Recruitment of αCP but not hnRNP K into the Lox 3′UTR complex. (A) Identification of the Lox complex by EMSA using a 32P-labeled Lox 3′UTR probe. The position of the Lox complex (a doublet band) is shown in the first lane, and its sensitivity to poly(C) competition is demonstrated in the following lane. (B) Recruitment of αCP into the Lox complex. Increasing concentrations of unlabeled Lox 3′UTR (wedge) were incubated with MEL S100 extract to form the Lox complex. The incubation mixtures were analyzed on native gels. The position of the uncomplexed αCP in the S100 extract and its recruitment to the position of the Lox complex were visualized by Western analysis. (C) hnRNP K was not recruited into the Lox complex. hnRNP K was detected with a monospecific antibody. The study was carried out as described above.
αCPs are sufficient for α-complex formation.
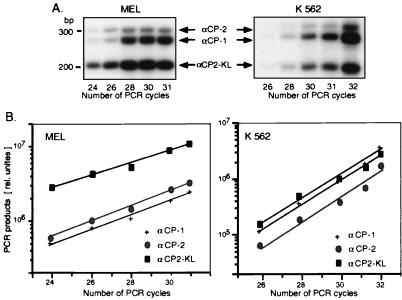
The above data suggested that αCPs might be sufficient for α-complex assembly without heterologous interacting proteins. To test this model, we examined whether recombinant αCPs could directly interact with the α3′UTR. There are three predominant forms of αCP in human and mouse cells: αCP-1, αCP-2, and the alternative-splicing product αCP2-KL. The majority of reported αCP cDNAs cloned to date lack an internal 93-bp exonic segment (αCP2-KL splice variant [19]), and the αCP2-KL isoform is the highest-frequency αCP cDNA in the expressed sequence tag database (data not shown). The relative abundance of the mRNAs corresponding to the various αCP isoforms was directly assessed in mouse (MEL) and human (K562) erythroid cell lines by an RT-PCR analysis. The RT-PCR primers corresponded to regions conserved in all known αCP-1 and αCP-2 sequences and bracketed the alternatively spliced central exons of the transcripts (see Materials and Methods). This combination of primers generated three cDNA products corresponding to the mRNAs encoding full-length αCP-2 and αCP-1 (top two bands) and the αCP2-KL isoform (lower band) (Fig. 3A). The identity of each of these bands was confirmed by excision and sequencing (data not shown). The quantitative data for the mouse and human erythroid cells (Fig. 3B) revealed substantial levels of mRNA for each isoform, with predominance of αCP2-KL mRNA in the MEL cells and a more equal distribution of the mRNAs encoding the three isoforms in the K562 cells. These mRNA data were consistent with the presence of all three protein isoforms in both human and mouse cells.
FIG. 3.
mRNA representation of mαCP isoforms. (A) Autoradiograph of RT-PCR products. MEL RNA (left) or K562 RNA (right) was reverse transcribed and PCR-amplified by using primers that were perfect matches to all known αCP mRNAs. The reverse primer was 32P end labeled. The reaction products were electrophoresed on a 2.5% MetaPhor agarose gel and quantified by PhosphoImager analysis. The identities of the cDNA fragments encoding αCP-2, αCP-1, and αCP2-KL are as indicated and were confirmed by direct sequencing (not shown). (B) RT-PCR amplification kinetics. Relative quantities of the RT-PCR products representing each of the three αCP mRNAs in MEL and K562 cells (left and right, respectively) were determined by PhosphorImager analysis. Logarithms of band intensities were plotted against the number of PCR cycles; these plots formed straight lines for the exponential phase of amplification, and the slopes reflect amplification efficiencies (67). The similarity in slopes for different αCP isoforms shows that the efficiencies of their amplification were similar.
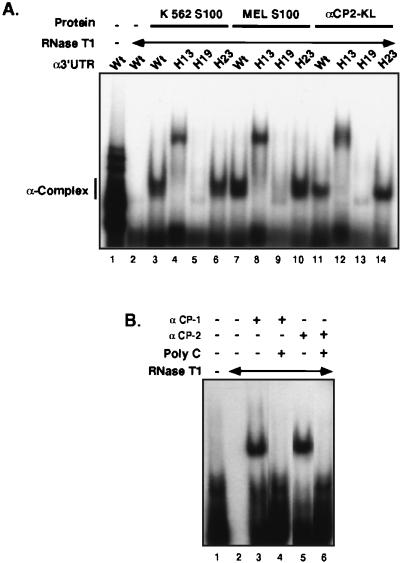
The ability of the recombinant αCP2-KL to form the α-complex was next determined (Fig. 4). An epitope (His6)-tagged αCP2-KL protein was expressed in Escherichia coli and affinity enriched, and the major Superdex 200 gel filtration column peak, containing the monomeric form of recombinant protein, was used for RNA binding studies. The apparent molecular mass of this recombinant αCP-KL as determined by SDS-polyacrylamide gel electrophoresis (PAGE) and sucrose density gradient was in full agreement with that of the native αCP in the S100 extract (data not shown; see below). Incubation of the recombinant αCP2-KL with the [32P]α3′UTR probe resulted in formation of an RNase-resistant complex (Fig. 4A, lane 11). This complex comigrated on the native gel with the α-complex generated with S100 extracts from the human (K562) and mouse (MEL) erythroid cell lines (Fig. 4A, lanes 3 and 7, respectively). Thus, the αCP2-KL binds directly to the α3′UTR.
FIG. 4.
Direct, sequence-specific association of each of the recombinant αCP isoforms with the α3′UTR. (A) Direct sequence-specific binding of recombinant αCP2-KL to the α-3′UTR. 32P-labeled α3′UTR (wild type [Wt]) or homologous RNAs containing specific sets of linker scanning base substitutions (H13, H19, and H23) that either disrupt (H13 and H19) or do not interfere (H23) with α-complex formation (68, 70) were incubated with S100 extracts from K562 cells or MEL cells or with recombinant αCP (αCP2-KL). The complexes were digested with RNase T1 and applied to a native acrylamide gel. Lanes 1 and 2 represent the 3′UTR probe incubated without and with RNase T1, respectively. The subsequent lanes contain labeled RNA incubated with the indicated extracts. The position of the α-complex is noted at the left. (B) Direct binding of recombinant αCP-1 and αCP-2 to the α3′UTR (determined as detailed for panel A). In each case, the complex was fully competed by added poly(C).
The binding specificity of recombinant αCP2-KL was tested against a series of mutant α3′UTR probes and was directly compared to that of the S100 extract (Fig. 4A). The full-length α3′UTR (109 nt) contains a discontinuous pyrimidine-rich and C-rich cis-acting stability element located in the region 27 to 63 nt downstream of the termination codon (69). Clusters of base substitutions (HindIII linker scanning mutations [68, 70]) result in defined effects on α-complex formation in vitro and result in parallel effects on the stability of hα-globin mRNA in transfected erythroid cells (68, 69). Recombinant αCP2-KL formed the normally migrating α-complex when incubated with an RNA containing base substitutions located outside the region critical for α-complex assembly (mutant probe H23 in Fig. 4A; lane 14 compared with lanes 6 and 10). As previously demonstrated, the H13 and H19 mutants, both situated within the cis-acting stability element, are unable to assemble the α-complex with S100 extract; the H13 mutation assembles an aberrantly migrating complex that does not comigrate with the α-complex and does not parallel mRNA stability, and the H19 mutation forms no complex (68, 69, 70). S100 and recombinant αCP-KL both failed to form any complex with H19, whereas the H13 mutation resulted in loss of the normal complex and generation of an aberrantly migrating RNP complex (Fig. 4A). Therefore, recombinant αCP2-KL and the cytosolic S100 extract demonstrated the same sequence specificity for interaction with the α3′UTR.
The two remaining αCP isoforms were tested for direct RNA interaction. Incubations of the α-3′UTR probe with recombinant αCP-1 and αCP-2 resulted in formation of poly(C)-sensitive RNase-resistant complexes (Fig. 4B). Each of these proteins was also incubated with a defined panel of mutant α-3′UTR probes (as in Fig. 4A), and in both cases they showed sequence specificity identical to that of the native K562 extract, MEL extract, and recombinant αCP2-KL (data not shown). Thus, all three of the αCP isoforms appear to be able to directly bind to the α-3′UTR in a manner identical to that of cellular S100 extracts.
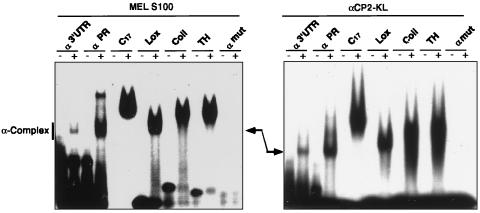
A 42-nt segment of the α3′UTR is protected from RNase T1 digestion by the α-complex. This segment (αPR) is capable of assembling the α-complex (22) and is closely related to pyrimidine-rich motifs in the 3′UTRs of three other highly stable mRNAs, Coll, Lox, and TH. Each of the three corresponding 3′UTR segments can form an RNP complex when incubated in S100 extract from a variety of cell types, and each of these complexes contains αCP (22, 68). The ability of recombinant αCP2-KL to interact with each of these 3′UTR segments was determined by EMSA (Fig. 5; see Materials and Methods). Remarkably, recombinant αCP2-KL bound to each of these mRNA segments. Moreover, as judged by migration of the complexes on a native gel, the organization and composition of each complex were the same whether assembled from recombinant αCP2-KL or from S100 extract. Taken together, these data suggested that recombinant αCP2-KL was sufficient for the assembly of the same α-complex on at least four different long-lived mRNAs.
FIG. 5.
Direct association of recombinant αCP2-KL with the 3′UTR-derived sequences from four highly stable mRNAs. [32P]RNA representing each of the following mRNAs was incubated with either MEL S100 extract (left) or recombinant αCP2-KL (right): α3′UTR, αPR, a poly(C) homoribopolymer (C17), the pyrimidine-rich 3′UTR segments of Lox, Coll, or TH, or a mutant α3′UTR (αmut). An aliquot of each incubation mixture was subjected to native gel electrophoresis. The gel was then dried and autoradiographed. The wild-type α-complex is shown in the second lane of each gel and is indicated by the arrow.
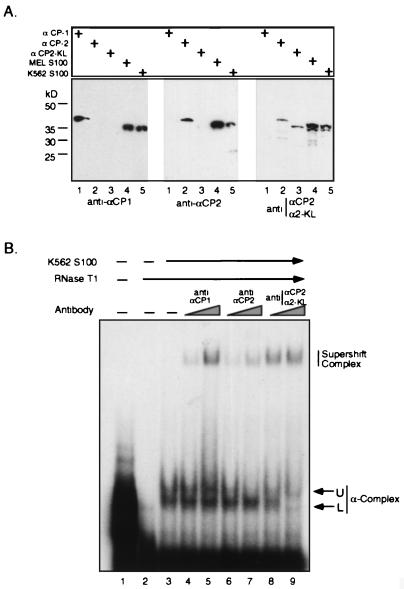
The recombinant protein binding studies demonstrate that all three αCP isoforms can form the α-complex. As these proteins differ in sequence and the αCP2-KL is smaller than the other two, one might expect some heterogeneity in the migration pattern of the α-complex. Indeed, close examination of the complex formed with the cell S100 extract often revealed a broad or split band. To directly document the contribution of each of the isoforms to α-complex formation, we carried out supershift analysis on the α-complex with a set of three epitope-specific rabbit antisera. These three antisera were raised against epitopes specific for human and mouse αCP-1, αCP-2, or an epitope present in αCP-2 and αCP2-KL but missing from αCP-1 (see Materials and Methods). The specificities of these antisera were confirmed by Western blotting (Fig. 6A). Of note, when used against mouse and human cell extracts, the first two antisera recognized a single band (αCP-1 and αCP-2, respectively) whereas the third recognized a band comigrating with αCP-2 and an additional, smaller band. The size of this additional band was consistent with that of αCP2-KL. Thus, these Western analyses confirmed the predicted specificities of the three antisera and were consistent with the presence of the three αCP isoforms in both human and mouse cells.
FIG. 6.
The α-complexes formed with S100 extracts represent a heterogeneous population containing the three αCP isoforms. (A) Selective detection of the three αCP isoforms with epitope-specific antisera. Shown are results of Western analyses of recombinant αCP-1, αCP-2, and αCP2-KL and of S100 extracts from mouse (MEL) or human (K562) erythroid cells probed with antisera specific for αCP-1, αCP-2, or an epitope shared by αCP-2 and αCP2-KL. (B) Supershift analysis of α-complexes. [32P]α3′UTR (lane 1) was incubated with K562 S100 extract and run on a native gel either alone (lane 3) or in the presence of increasing amounts of anti-αCP-1 (lanes 4 and 5), anti-αCP-2 (lanes 6 and 7), or antisera specific to αCP-2 and αCP2-KL (lanes 8 and 9). The native complex is composed of two subbands (upper [U] and lower [L]); the position of the antibody supershifted complex is indicated. Lane 2 contains probe digested with RNase prior to protein addition.
Each of the epitope-specific antibodies was used in a supershift assay to detect a contribution of the three αCP isoforms to α-complex formation (Fig. 6B). The α-complex in this gel can be clearly visualized as split bands (upper and lower). Addition of each of the three αCP antisera supershifted a subpopulation of the α-complex. In the case of the αCP-1 antisera, it was not possible to be sure whether the supershifted complex derived from the top or bottom band. In contrast the αCP-2 antibody selectively shifted the upper band of the complex, while the antisera recognizing both αCP-2 and αCP2-KL quantitatively shifted the lower band as well as decreased the intensity of the upper band. The combined Western and EMSA/supershift data were fully consistent with the contribution of all three αCP isoforms to α-complex assembly and with assignment of the αCP-2 subcomplex to the upper band and αCP2-KL subcomplex to the lower band within the α-complex.
Recombinant αCPs and cytosolic S100 extracts displayed similar RNA-binding affinities.
The αCP proteins are known to be targets of posttranslational modifications (34). Such modifications, which would not be present on the recombinant mαCP produced in E. coli, might affect binding affinity (34). To detect such an effect, we determined the relative binding affinities of each of the purified recombinant αCPs (Fig. 7). The amount of MEL S100 extract used in the study was normalized to its content of total αCP by Western blotting and compared to an equivalent amount of recombinant protein (see Materials and Methods). Results of representative studies comparing αCP2-KL with MEL S100 extract are shown in Fig. 7A and B. A constant amount of RNA (the 42-nt αPR [see Materials and Methods]) of the 3′UTR as the RNA probe (see above) was incubated with increasing amounts of S100 extract or recombinant αCP2-KL. The bound and free RNAs were then separated by EMSA and quantified, and free RNA probe concentrations were plotted versus total αCP concentrations (Fig. 7C). The relative binding affinities of αCPs in S100 extract or of recombinant αCP2-KL were determined as the concentrations at which 50% of RNA was bound (10). The 50% binding concentration was in the same range for recombinant αCP2-KL and S100 extract. Similar results were obtained when poly(C17) homoribopolymer and full-length α3′UTR were used as RNA probes (data not shown); the binding affinities of recombinant αCP2-KL and S100 for the RNA target were equivalent in each case.
The difference in the shapes of titration curves between recombinant αCP2-KL and the S100 extract (Fig. 7C) most likely reflected the fact that the MEL S100 extract contained a mixture of αCP isoforms in addition to αCP2-KL (see above). It was very possible that each of these isoforms could bind αPR with different affinities, resulting in the observed distortion of the composite (extract) binding curve (34). To test this possibility, we determined absolute values of apparent Kd for all three recombinant αCP proteins (see Materials and Methods). As summarized in Fig. 7D, the Kds for α-3′UTR-binding affinities of the three αCP isoforms, while all substantial, differed within a 64-fold range, with a rank order of αCP-2 > αCP2-KL > > αCP-1.
Sedimentation analysis of the α-complex.
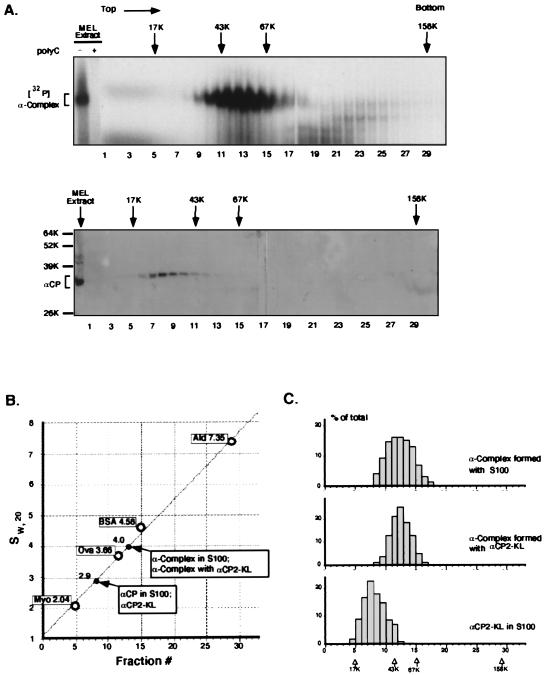
The data detailed above demonstrated the sufficiency of αCP2-KL for α-complex assembly. As the αCPs are capable of forming homodimers (references 19, 29, and 68a and our unpublished results), the stoichiometry of αCPs in the α-complex remained in question. The presence of multiple αCP molecules and/or the presence of additional proteins in the complex would be directly reflected in its sedimentation characteristics during sucrose gradient centrifugation (Fig. 8). A 32P-labeled α-complex was assembled in vitro by incubating ([32P]α3′UTR) with MEL S100 cytosolic extract (68) (Fig. 8A, top). As expected, this complex was sensitive to poly(C) competition (Fig. 8A, top, second lane). The reaction mix was applied to a 5 to 20% sucrose gradient, and the sedimentation profile of the [32P]α-complex was determined by analysis of each gradient fraction on a native gel. The S100 extract assembled a single complex on the RNA target that sedimented just below the ovalbumin marker (ovalbumin Mr = 43,000, S = 3.66). A second gradient in which the [32P]α3′UTR was incubated with recombinant αCP2-KL revealed a single peak complex in the same position (data not shown; see Fig. 8C). A third gradient containing S100 extract without added RNA probe (Fig. 8A, bottom) was run in parallel with these gradients. Western analysis of this gradient revealed that the uncomplexed αCP sedimented at a position several fractions above the ovalbumin marker. From the standard curve (Fig. 8B), the sedimentation coefficients for αCP and α-complex were determined as 2.9 and 4.0, respectively. This difference in sedimentation between the α-complex and the αCP was consistent with the presence of the previously defined 42-nt (13-kDa) RNase T1-resistant RNA fragment (22) bound to a single αCP molecule. These data ruled out the presence of an αCP dimer in the complex because such a complex would have sedimented well above the BSA (68-kDa) marker. Therefore, these studies were most consistent with an α-complex containing a single molecule of αCP.
FIG. 8.
Analysis of αCP2-KL and the α-complex sedimentation on 5 to 20% sucrose gradients. (A) Top, sedimentation analysis of the α-complex and αCP2-KL. 32P-labeled α-complex was assembled by incubating [32P]α3′UTR probe with MEL S100 extract. The gradients were centrifuged and fractionated as detailed in Materials and Methods. Aliquots of each fraction were analyzed on a native gel, and the 32P-labeled complex was detected by autoradiography. The first gel lane contained an aliquot of the loaded material. The position of the [32P]α-complex is indicated by the bracket at the left, and its poly(C) sensitivity was confirmed in the second lane. Fraction numbers are noted below the lanes. Positions of the molecular weight markers run in a parallel gradient are indicated above the gradients. Bottom, MEL S100 with no added RNA probe. The gradient containing MEL S100 extract and no added RNA probe was run as described above. Gradient fractions were analyzed by SDS-PAGE, and αCP was detected by Western blotting with chicken anti-αCP antibody. Positions of molecular weight standards are indicated at the left. (B) Standard curve for sucrose gradient sedimentation. Positions of standards (see Materials and Methods) are indicated by the open circles. Positions of respective peak centers for αCP2-KL and α-complex are indicated by arrows. Sedimentation coefficients of standards, sw,20 (10−13 s−1), are also indicated. Ald, aldolase; Ova, ovalbumin; myo, myoglobin. (C) Sedimentation profiles of the α-complexes assembled with S100 extract (top) or with recombinant αCP2-KL (middle) compared with uncomplexed αCP (bottom). Note identical sedimentation profiles of the native and recombinant α-complexes and their positioning to the right of the uncomplexed αCP.
DISCUSSION
This present study was designed to establish the components and stoichiometry of the α-complex. Previous studies of this complex have identified αCPs as critical components of this complex (30). Multiple lines of investigation demonstrate that αCPs can form protein-protein contacts with themselves and with a number of additional proteins (see the introduction). Contrary to expectations based on these previous reports, the data in the present study are most consistent with a simple binary composition of the α-complex in which a single molecule of αCP interacts with the α3′UTR. This conclusion is supported by several observations: (i) The absence of AUF1 and/or hnRNP K was directly demonstrated by an RNP recruitment assay. This approach revealed a quantitative and selective recruitment of αCPs to the α-complex independent of, and to the exclusion of, either of these other two proteins (Fig. 1 and 2). (ii) All three αCP isoforms bound directly to the α3′UTR. This interaction demonstrated the same sequence specificity as that of the native S100 extract with respect to a set of informative α3′UTR mutants as well as for the 3′UTRs from three additional long-lived mRNAs (Fig. 4 and 5). (iii) The affinities (relative Kds) of recombinant αCPs for the α3′UTR were comparable (over a defined span) to that of the S100 extract (Fig. 7). (iv) The sedimentation characteristics of the α-complex indicated that only a single molecule of an αCP could be accommodated in the α-complex (Fig. 8). These data effectively excluded the possibility that multiple molecules of αCP cooperate in α-complex formation. Taken together, these data support the conclusion of a 1:1 stoichiometry of an αCP with the RNA target in the α-complex.
The simple binary model for the α-complex is at odds with the failure of our previous attempts to demonstrate direct binding of biochemically enriched cytosolic αCP to the α3′UTR (30). These negative results might have reflected technical difficulties such as denaturation or partial degradation of αCP during purification from the extract. Alternatively, the αCPs may have undergone some sort of modification during isolation. It is well established, for example, that hyperphosphorylation of αCPs can ablate binding activity (reference 34 and our unpublished data). However, the ability of the αCP-enriched fractions to complement extracts depleted in poly(C) binding activity (30) renders these explanations unlikely and instead suggests additional variables in the assembly process. Future studies aimed at delineating the functional difference(s) between native and recombinant αCP may therefore be informative.
We were unable to confirm the presence of AUF1 in the α-complex as had been reported by others (29). Direct interaction of αCP-1 and to a lesser extent αCP-2 with AUF1 was initially documented by a yeast two-hybrid analysis and subsequently confirmed by glutathione S-transferase pull-down studies (29). However, evidence for actual coassembly of AUF1 with αCPs in the context of the α-complex was limited to AUF1 enrichment in the α-complex region of a native gel after addition of target RNA to an S130 extract. While there are a number of technical differences between our study and this previous report in how the α-complex was assembled and assayed, these differences are relatively minor. In this study, there was no evidence for incorporation of AUF1 into the α-complex in the recruitment assay despite quantitative recruitment of αCPs (Fig. 1D).
Previous studies by others have identified a multiple-repeat polypyrimidine tract in the Lox mRNA 3′UTR responsible for translational arrest of Lox mRNA during erythroid development (48). We subsequently reported that αCP interacts with the single repeat unit of this pyrimidine-rich Lox mRNA determinant to form a complex that appears to be very similar to the α-complex (22). More recently, studies suggested that in addition to αCPs, the nuclear RNA binding protein hnRNP K also interacts with this region. Functional assays suggest that αCP and hnRNP K mediate a cooperative suppression of Lox mRNA translation in vitro and in vivo. While our results (reference 22 and Fig. 2) agree that αCP has the capacity to bind to the Lox 3′UTR, there was no evidence for significant incorporation of hnRNP K into the α-complex. However these in vitro assays of RNA-protein interactions do not necessarily conflict with the prior report of a translation control effect by hnRNP K (48), as the underlying mechanism of such control may be unrelated to coassembly of hnRNP K with αCP in a functional RNP complex.
Several observations indicate that α-complex, and/or closely related αCP-containing complexes, may represent a generally active mRNA stability determinant(s). αCP proteins have a wide tissue distribution, and the α-complex is formed from S100 extracts isolated from nonerythroid as well as erythroid cell lines (1, 19, 34). Furthermore, the αCP-containing complex forms on the 3′UTR pyrimidine-rich elements of a subset of stable erythroid and nonerythroid mRNAs (13, 22, 47, 63). While these data suggest a general role of αCP in mRNA metabolism, it is not clear how the α-complex contributes to the longevity of target mRNAs. The binding of αCP may be the initial event, serving to identify the target mRNAs. Subsequent involvement of additional proteins may then mediate interactions between the 3′ and 5′ ends of the mRNA to stabilize the mRNA, prevent endo- or exonuclease digestion, and/or direct the mRNA to a specific subcellular site. It has been recently reported that αCP proteins can interact with poly(A) binding protein and that this interaction influences the rate of deadenylation in an in vitro system (68a). In vivo data from our own laboratory further supports a role of the poly(A) tail in the stabilization of hα-globin mRNA (41). Subsequent interaction of poly(A) binding protein, possibly bound to an αCP, with the translation initiation factor eIF4G (71) or a closely related homologue (PAIP [12]) may provide a physical link between the mRNA 5′ and 3′ termini. The delineation of the α-complex as a binary complex should facilitate subsequent formulation of such testable models for the establishment of mRNA stabilization.
ACKNOWLEDGMENTS
We thank Faith Cash for valuable contributions to the RNP recruitment assays and Chenglu Liu for skillful generation of recombinant αCP used in several of the assays. The manuscript was assembled with the expert secretarial assistance of Jessie Harper. The critical reading and constructive comments by Nancy E. Cooke are gratefully acknowledged.
This work was supported in part by grants HL38632-6 and CA72765-01 (S.A.L.). S.A.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Aasheim H C, Loukianova T, Deggerdal A, Smeland E B. Tissue specific expression and cDNA structure of a human transcript encoding a nucleic acid binding [oligo(dC)] protein related to the pre-mRNA binding protein K. Nucleic Acids Res. 1994;22:959–964. doi: 10.1093/nar/22.6.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atwater J A, Wisdom R, Verma I M. Regulated mRNA stability. Annu Rev Genet. 1990;24:519–641. doi: 10.1146/annurev.ge.24.120190.002511. [DOI] [PubMed] [Google Scholar]
- 3.Aviv H, Voloch Z, Bastos R N, Levy S. Biosynthesis and stability of globin mRNA in cultured erythroleukemic Friend cells. Cell. 1976;8:495–503. doi: 10.1016/0092-8674(76)90217-8. [DOI] [PubMed] [Google Scholar]
- 4.Bastos R N, Aviv H. Theoretical analysis of a model for globin messenger RNA accumulation during erythropoiesis. J Mol Biol. 1977;110:205–218. doi: 10.1016/s0022-2836(77)80069-7. [DOI] [PubMed] [Google Scholar]
- 5.Bastos R N, Volloch Z, Aviv H. Messenger RNA population analysis during erythroid differentiation: a kinetical approach. J Mol Biol. 1977;110:191–203. doi: 10.1016/s0022-2836(77)80068-5. [DOI] [PubMed] [Google Scholar]
- 6.Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. doi: 10.1016/0092-8674(95)90326-7. [DOI] [PubMed] [Google Scholar]
- 7.Belasco J, Brawerman G. Control of mRNA stability. San Diego, Calif: Academic Press; 1993. [Google Scholar]
- 8.Binder R, Horowitz J A, Basilion J P, Koeller D M, Klausner R D, Harford J B. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J. 1994;13:1969–1980. doi: 10.1002/j.1460-2075.1994.tb06466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown B D, Zipkin I D, Harland R M. Sequence-specific endonucleolytic cleavage and protection of mRNA in Xenopus and Drosophila. Genes Dev. 1993;7:1620–1631. doi: 10.1101/gad.7.8.1620. [DOI] [PubMed] [Google Scholar]
- 10.Carey J. Gel retardation. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 11.Clegg J B, Weatherall D J, Milner P F. Haemoglobin Constant Spring—a chain termination mutant? Nature (London) 1971;234:337–340. doi: 10.1038/234337a0. [DOI] [PubMed] [Google Scholar]
- 12.Craig A W B, Haghighat A, Yu A T K, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature (London) 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- 13.Czyzyk-Krzeska M F, Beresh J E. Characterization of the hypoxia-inducible protein binding site within the pyrimidine-rich tract in the 3′-untranslated region of the tyrosine hydroxylase mRNA. J Biol Chem. 1996;271:3293–3299. doi: 10.1074/jbc.271.6.3293. [DOI] [PubMed] [Google Scholar]
- 14.Decker C J, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 15.Decker C J, Parker R. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem Sci. 1994;19:336–340. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 16.Ebersole T A, Chen Q, Justice M J, Artzt K. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- 17.Ehrenman K, Long L, Wagner B J, Brewer G. Characterization of cDNAs encoding the murine A+U-rich RNA-binding protein AUF1. Gene. 1994;149:315–319. doi: 10.1016/0378-1119(94)90168-6. [DOI] [PubMed] [Google Scholar]
- 18.Fumagalli S, Totty N F, Hsuan J J, Courtneidge S A. A target for Src in mitosis. Nature (London) 1994;368:871–874. doi: 10.1038/368871a0. [DOI] [PubMed] [Google Scholar]
- 19.Funke B, Zuleger B, Benavente R, Schuster T, Goller M, Stevenin J, Horak I. The mouse poly(C)-binding protein exists in multiple isoforms and interacts with several RNA-binding proteins. Nucleic Acids Res. 1996;24:3821–3828. doi: 10.1093/nar/24.19.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gamarnik A, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- 21.Hahm K, Kim G, Turck C W, Smale S T. Isolation of a murine gene encoding a nucleic acid-binding protein with homology to hnRNP K. Nucleic Acids Res. 1993;21:3894. doi: 10.1093/nar/21.16.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holcik M, Liebhaber S A. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis and trans components. Proc Natl Acad Sci USA. 1997;94:2410–2414. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holcik M, Liebhaber S A. Analysis of mRNP complexes assembled in vitro. In: Richter J, editor. mRNA formation and function. San Diego, Calif: Academic Press; 1997. pp. 196–208. [Google Scholar]
- 24.Hunt D M, Higgs D R, Winichagoon P, Clegg J B, Weatherall D J. Haemoglobin Constant Spring has an unstable alpha chain messenger RNA. Br J Haematol. 1982;51:405–413. doi: 10.1111/j.1365-2141.1982.tb02796.x. [DOI] [PubMed] [Google Scholar]
- 25.Jackson R J. Cytoplasmic regulation of mRNA function: the importance of the 3′ untranslated region. Cell. 1993;74:9–14. doi: 10.1016/0092-8674(93)90290-7. [DOI] [PubMed] [Google Scholar]
- 26.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 27.Jones A R, Schedl T. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 1995;9:1491–1504. doi: 10.1101/gad.9.12.1491. [DOI] [PubMed] [Google Scholar]
- 28.Kajita Y, Nakayama J, Aizawa M, Ishikawa F. The UUAG-specific RNA binding protein, heterogeneous nuclear ribonucleoprotein D0. Common modular structure and binding properties of the 2×RBD-Gly family. J Biol Chem. 1995;270:22167–22175. doi: 10.1074/jbc.270.38.22167. [DOI] [PubMed] [Google Scholar]
- 29.Kiledjian M, DeMaria C T, Brewer G, Novick K. Identification of AUF1 (heterogeneous nuclear ribonucleoprotein D) as a component of the alpha-globin mRNA stability complex. Mol Cell Biol. 1997;17:4870–4876. doi: 10.1128/mcb.17.8.4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiledjian M, Wang X, Liebhaber S A. Identification of two KH domain proteins in the α-globin mRNA stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klausner R D, Rouault T A, Harford J B. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- 32.Krowczynska A, Yenofsky R, Brawerman G. Regulation of messenger RNA stability in mouse erythroleukemia cells. J Mol Biol. 1985;181:231–239. doi: 10.1016/0022-2836(85)90087-7. [DOI] [PubMed] [Google Scholar]
- 33.Kuhn L C. mRNA-protein interactions regulate critical pathways in cellular iron metabolism. Br J Haematol. 1991;79:1–5. doi: 10.1111/j.1365-2141.1991.tb07998.x. [DOI] [PubMed] [Google Scholar]
- 34.Leffers H, Dejgaard K, Celis J E. Characterization of two major cellular poly(rC)-binding human proteins, each containing three K-homologous (KH) domains. Eur J Biochem. 1995;230:447–453. [PubMed] [Google Scholar]
- 35.Liebhaber S A, Kan Y W. Differentiation of the mRNA transcripts originating from the alpha 1- and alpha 2-globin loci in normals and alpha-thalassemics. J Clin Investig. 1981;68:439–446. doi: 10.1172/JCI110273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu K, Hanna M M. NusA contacts nascent RNA in Escherichia coli transcription complexes. J Mol Biol. 1995;247:547–558. doi: 10.1006/jmbi.1994.0161. [DOI] [PubMed] [Google Scholar]
- 37.Mahone M, Saffman E E, Lasko P F. Localized Bicaudal-C RNA encodes a protein containing a KH domain, the RNA binding motif of FMR1. EMBO J. 1995;14:2043–2055. doi: 10.1002/j.1460-2075.1995.tb07196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matunis M J, Michael W M, Dreyfuss G. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol Cell Biol. 1992;12:164–171. doi: 10.1128/mcb.12.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menhra M. RNA isolation from cell and tissues. In: Krieg P A, editor. A laboratory guide to RNA: isolation, analysis, and synthesis. New York, N.Y: Wiley-Liss, Inc.; 1996. pp. 1–20. [Google Scholar]
- 40.Milligan J F, Uhlenbeck O C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 41.Morales J, Russell J E, Liebhaber S A. Destabilization of human α-globin mRNA by translation anti-termination is controlled during erythroid differentiation and is paralleled by phased shortening of the poly(A) tail. J Biol Chem. 1997;272:6607–6613. doi: 10.1074/jbc.272.10.6607. [DOI] [PubMed] [Google Scholar]
- 42.Muhlrad D, Decker C J, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 43.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature (London) 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 44.Nandabalan K, Price L, Roeder G S. Mutations in U1 snRNA bypass the requirement for a cell type-specific RNA splicing factor. Cell. 1993;73:407–415. doi: 10.1016/0092-8674(93)90239-m. [DOI] [PubMed] [Google Scholar]
- 45.Nielson D A, Shapiro D J. Insights into hormonal control of messenger RNA stability. Mol Endocrinol. 1990;4:953–957. doi: 10.1210/mend-4-7-953. [DOI] [PubMed] [Google Scholar]
- 46.Osley M A. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 47.Ostareck-Lederer A, Ostareck D H, Standart N, Thiele B J. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J. 1994;13:1476–1481. doi: 10.1002/j.1460-2075.1994.tb06402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostareck D H, Ostareck-Lederer A, Wilm M, Thiele B J, Mann M, Hentze M W. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 49.Peltz S W, Brewer G, Bernstein P, Hart P A, Ross J. Regulation of mRNA turnover in eukaryotic cells. Crit Rev Eukaryotic Gene Expr. 1991;1:99–126. [PubMed] [Google Scholar]
- 50.Peltz S W, Jacobson A. mRNA stability: in trans-it. Curr Opin Cell Biol. 1992;4:979–983. doi: 10.1016/0955-0674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 51.Regnier P, Grunberg-Manago M, Portier C. Nucleotide sequence of the pnp gene of Escherichia coli encoding polynucleotide phosphorylase. Homology of the primary structure of the protein with the RNA-binding domain of ribosomal protein S1. J Biol Chem. 1987;262:63–68. [PubMed] [Google Scholar]
- 52.Ross J. Messenger RNA turnover in eukaryotic cells. J Mol Biol Med. 1988;5:1–14. [PubMed] [Google Scholar]
- 53.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ross J, Sullivan T D. Half-lives of beta and gamma globin messenger RNAs and of protein synthetic capacity in cultured human reticulocytes. Blood. 1985;66:1149–1154. [PubMed] [Google Scholar]
- 55.Russell J E, Morales J, Makeyev A, Liebhaber S A. Sequence divergence in the 3′ untranslated regions of human ζ- and α-globin mRNAs mediates a difference in their stabilities and contributes to efficient α-to-ζ gene developmental switching. Mol Cell Biol. 1998;18:2173–2183. doi: 10.1128/mcb.18.4.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sachs A B. Messenger RNA degradation in eukaryotes. Cell. 1993;74:413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt C, Henkel B, Poschl E, Zorbas H, Purschke W G, Gloe T R, Muller P K. Complete cDNA sequence of chicken vigilin, a novel protein with amplified and evolutionary conserved domains. Eur J Biochem. 1992;206:625–634. doi: 10.1111/j.1432-1033.1992.tb16967.x. [DOI] [PubMed] [Google Scholar]
- 58.Shyu A B, Belasco J G, Greenberg M E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 59.Siebel C W, Admon A, Rio D C. Soma-specific expression and cloning of PSI, a negative regulator of P element pre-mRNA splicing. Genes Dev. 1995;9:269–283. doi: 10.1101/gad.9.3.269. [DOI] [PubMed] [Google Scholar]
- 60.Siomi H, Matunis M J, Michael W M, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res. 1993;21:1193–1198. doi: 10.1093/nar/21.5.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siomi H, Siomi M C, Nussbaum R L, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 62.Smith C W J, Gooding C, Roberts G C, Scadden A D J. Analyzing patterns of alternative splicing. In: Krieg P A, editor. A laboratory guide to RNA: isolation, analysis, and synthesis. New York, N.Y: Wiley-Liss, Inc.; 1996. pp. 411–440. [Google Scholar]
- 63.Stefanovic B, Hellerbrand C, Holcik M, Briendl M A, Liebhaber S, Brenner D A. Posttranscriptional regulation of collagen alpha1(I) mRNA in hepatic stellate cells. Mol Cell Biol. 1997;17:5201–5209. doi: 10.1128/mcb.17.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Surdej P, Riedl A, Jacobs-Lorena M. Regulation of mRNA stability in development. Annu Rev Genet. 1994;28:263–282. doi: 10.1146/annurev.ge.28.120194.001403. [DOI] [PubMed] [Google Scholar]
- 65.Urlaub H, Kruft V, Bischof O, Muller E C, Wittmann-Liebold B. Protein-rRNA binding features and their structural and functional implications in ribosomes as determined by cross-linking studies. EMBO J. 1995;14:4578–4588. doi: 10.1002/j.1460-2075.1995.tb00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wagner B J, Long L, Rao P N, Pettenati M J, Brewer G. Localization and physical mapping of genes encoding the A+U-rich element RNA-binding protein AUF1 to human chromosomes 4 and X. Genomics. 1996;34:219–222. doi: 10.1006/geno.1996.0269. [DOI] [PubMed] [Google Scholar]
- 67.Wang A M, Doyle M V, Mark D F. Quantitation of mRNA by the polymerase chain reaction. Proc Natl Acad Sci USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X, Kiledjian M, Weiss I M, Liebhaber S A. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human α-globin mRNA stability. Mol Cell Biol. 1995;15:1769–1777. doi: 10.1128/mcb.15.3.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68a.Wang Z, Day N, Trifillis P, Kiledjian M. An mRNA stability complex functions with the poly(A)–binding protein to stabilize mRNA in vitro. Mol Cell Biol. 1999;19:4552–4560. doi: 10.1128/mcb.19.7.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiss I M, Liebhaber S A. Erythroid cell-specific determinants of α-globin mRNA stability. Mol Cell Biol. 1994;14:8123–8132. doi: 10.1128/mcb.14.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weiss I M, Liebhaber S A. Erythroid cell-specific mRNA stability elements in the α2-globin 3′ nontranslated region. Mol Cell Biol. 1995;15:2457–2465. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wells S E, Hillner P E, Vale R D, Sachs A B. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 72.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]