Abstract
Sequestosome 1 (SQSTM1)/p62 is an adapter protein mainly involved in the transportation, degradation and destruction of various proteins that cooperates with components of autophagy and the ubiquitin-proteasome degradation pathway. Numerous studies have shown that SQSTM1/p62 functions at multiple levels, including involvement in genetic stability or modification, post-transcriptional regulation and protein function. As a result, SQSTM1/p62 is a versatile protein that is a critical core regulator of tumor cell genetic stability, autophagy, apoptosis and other forms of cell death, malignant growth, proliferation, migration, invasion, metastasis and chemoradiotherapeutic response, and an indicator of patient prognosis. SQSTM1/p62 regulates these processes via its distinct molecular structure, through which it participates in a variety of activating or inactivating tumor-related and tumor microenvironment-related signaling pathways, particularly positive feedback loops and epithelial-mesenchymal transition-related pathways. Therefore, functioning as a proto-oncogene or tumor suppressor gene in various types of cancer and tumor-associated microenvironments, SQSTM1/p62 is capable of promoting or retarding malignant tumor aggression, giving rise to immeasurable effects on tumor occurrence and development, and on patient treatment and prognosis.
Keywords: SQSTM1/p62, autophagy, ubiquitin-proteasome degradation, EMT, tumor
1. Introduction
SQSTM1/P62, an adapter and scaffolding protein described cursorily by Park (1) in 1995, is a 62-kD protein that binds to the Src homology 2 domain of p56lck and is involved in the degradation and destruction of ubiquitinated proteins. Over time, an increasing number of studies have extensively shown that SQSTM1/p62 is a multifunctional protein that impinges on a variety of tumor biological behaviors, such as cell growth, proliferation, migration, invasion, metastasis, autophagy, apoptosis and ubiquitination (2-5). SQSTM1/p62 has important and versatile roles in cancer owing to its distinct and important structures, including a ubiquitin-associated (UBA) region and a Phox and Bem1p (PB1) domain, which mediate the ubiquitin-proteasome degradation pathway, a LC3-interacting region (LIR), which mediates the autophagy pathway, and a Kelch-like ECH-associated protein 1 (KEAP1)-interacting region, which is involved in the NF-κB pathway (6). The coexistence of a UBA region and LIR indicates that SQSTM1/p62 serves as a bridge between autophagy and the ubiquitin-proteasome system, which is of the utmost importance for SQSTM1/p62 regulation of various biological behaviors (7,8). In addition, after incorporation into a completed autophagosome, SQSTM1/p62 is quickly degraded in autolysosomes; therefore, it is an important index useful for monitoring autophagic degradation. Generally, increased SQSTM1/p62 expression reflects inhibition of autophagy. Analogously, decreased SQSTM1/p62 expression indicates activation of autophagy, and thus, to a certain degree, it is used to monitor autophagic flux in cells; however, these differences in expression may be context specific (9). For instance, the SQSTM1/p62 level may not change when autophagy is induced, as SQSTM1/p62 may be involved in other signaling pathways, interacting with numerous molecules through other domains as described in the aforementioned literature (9). Additionally, the phosphorylation of SQSTM1/p62 at Ser403 may post-transcriptionally participate in the regulation of the autophagic clearance of ubiquitinated proteins (10). Notably, recent evidence has confirmed that in gastric cancer, SQSTM1/p62 can directly bind and transport the long non-coding RNA ARHGAP5-AS1 as cargo to autophagosomes, ultimately leading to RNA recycling by autolysosomes (11). Furthermore, SQSTM1/p62 may directly recruit and interact with autophagy-linked FYVE protein, a protein similar to SQSTM1/p62 that is essential to the formation and autophagic degradation of ubiquitinated proteins, to degrade ubiquitinated proteins via autophagy (12). In particular, SQSTM1/p62 plays a crucial role in autophagy and the regulation of other transcriptional regulators, including nuclear factor erythroid 2 (NRF2) and NF-κB. For example, NRF2 degradation is primarily powered by the KEAP1, whereas, phosphorylation of SQSTM1/p62 at S349 may tremendously enhance the adhesion to KEAP1 and subsequently disrupts association between KEAP1 and NRF2, and promotes NRF2 stabilization and activation to facilitate growth of tumor cells, implying crosstalk between SQSTM1/p62-mediated autophagy and the KEAP1-NRF2 system (13,14). Moreover, SQSTM1/p62 acts as a direct transcriptional target of NF-κB and can in turn activate the NF-κB pathway by stimulating inhibitor of NF-κB kinase subunit β/IκB by TNF receptor-associated factor 6 (TRAF6) polyubiquitination (15-17). In addition, SQSTM1/p62 p.R321C mutation leads to autophagy inhibition by activating the NF-κB pathway in Paget's disease of the bone (18), and as autophagy is inhibited, positive feedback between SQSTM1/p62 and NF-κB interaction can excessively and sustainably trigger the NF-κB pathway, leading to epithelial-mesenchymal transition (EMT) in various RAS-mutated cells (19).
Recently, an increasing number of studies have confirmed that under conditions of various intracellular or extracellular pressures, including nutrient deficiency, and endoplasmic reticulum, oxidative and metabolic stresses, such as hypoxia and high concentrations of insulin, SQSTM1/p62 may act as either a proto-oncogene or tumor suppressor gene, promoting or inhibiting malignant tumor progression, respectively (20-23). By directly binding to numerous cancer-associated genes, such as Tribbles 3, EGFR, COX-2, MMP1/2, membrane type-MMP, c-Myc, Snail, Twist (20), RAD51 recombinase, filamin A (21), ring finger protein 168 (an E3 ubiquitin-protein ligase) (22) and checkpoint kinase 1 (23), SQSTM1/p62 can mitigate genetic instability and DNA damage foci, and induce DNA repair, thereby playing a role in cancer oncogenesis, malignant progression, senescence and chemoradiotherapeutic sensitivity (20-23). Since SQSTM1/p62 is at the center of a hub of various complicated signaling pathways in different cancer types and as its functions have wide implications in different cellular systems, the underlying regulatory mechanisms of its role in steering tumor progression are not entirely known and remain to be revealed in the future. The present review focuses on the mechanisms and functions of SQSTM1/p62 in regulating diverse biological behaviors in different types of cancer.
2. Gastrointestinal tumors
It has been established that SQSTM1/p62, as a tumor oncogene, is frequently abnormally upregulated and involved in the aggression of gastrointestinal tumors, including gastric, colorectal and pancreatic cancer (24). Several lines of evidence also suggest that specific expression patterns of SQSTM1/p62 in tumor cells are closely related to invasion and metastasis, and can indicate prognosis. Notably, the punctiform expression of SQSTM1/p62 in the cytoplasm and/or cell nucleus may be an independent prognostic factor of esophageal adenocarcinoma, particularly early esophageal adenocarcinoma, in which the expression of SQSTM1/p62 is usually aberrantly upregulated (24,25). However, patients with higher SQSTM1/p62 expression in the tumor cell cytoplasm and nucleus have a better prognosis, while patients with lower SQSTM1/p62 or lower combined LC3 and SQSTM1/p62 expression in tumor cells have more aggressive esophageal adenocarcinoma (25). The overexpression of SQSTM1/p62 in gastric cancer is related to hematogenous and hepatic metastasis, particularly in early gastric cancer, with an unfavorable prognosis. However, SQSTM1/p62 expression in colorectal cancer treated with 5-fluorouracil has not been shown to be significantly associated with prognosis (26,27). All of these data indicate that SQSTM1/p62 dysregulation may be an early and important event in tumorigenesis. SQSTM1/p62 expression in different gastrointestinal tumors has different prognostic significance, possibly due to the location of the cancer lesion and the immune response to cancer cells that express SQSTM1/p62 and to the tumor microenvironment, which exhibits a high density of regulatory forkhead box P3+ T cells (28).
Furthermore, inhibition of SQSTM1/p62 may repress autophagy, impeding tumor aggression by involving the MEK/ERK signaling pathway in KRAS- and BRAFV600E-mutated colorectal cancer cells. The evidence for this comes from the finding of a previous study where positive cytoplasmic SQSTM1/p62 staining for conspicuously present in the majority of colorectal cancer tissues. Although there was no significant association between SQSTM1/p62-positive staining and KRAS mutations, patients with SQSTM1/p62-positive staining in the cytoplasm, particularly those with KRAS mutations, may have better overall survival rates (29). However, KRAS and BRAFV600E mutations have been shown to activate the MEK/ERK signaling pathway to promote autophagy and downregulate SQSTM1/p62 expression in colorectal cancer cells when the phosphoinositol-3 kinase (PI3K)/mTOR signaling pathway is inhibited (30). Therefore, the activation of autophagy and the downregulation of SQSTM1/p62 may indirectly mirror the effects of KRAS and BRAF mutations in cells. However, autophagy can be repressed by SQSTM1/p62 inhibition, leading to the cessation of cancer cell growth and tumor formation (31).
Nevertheless, SQSTM1/p62 expression is not always decreased and has sometimes been increased when autophagy was activated. For instance, a study showed that SQSTM1/p62 expression in colorectal cancer cells was inhibited by the β-catenin/transcription factor (TCF)4 complex, which inhibits autophagosome formation (32). However, under starvation-induced autophagy conditions, SQSTM1/p62 expression was markedly increased, as β-catenin was spontaneously degraded by LC3 binding to the LIR of β-catenin (31). Thus, it seems that detecting SQSTM1/p62 expression alone may not always be a reliable approach for evaluating the autophagy process; it may be necessary to also assess the expression of other autophagy-related molecules, such as Beclin1 or ATG8.
In addition, recent studies have shown that SQSTM1/p62 plays an indispensable role in regulating drug sensitivity in colorectal cancer by inducing autophagy, apoptosis and DNA damage. For example, high expression of SQSTM1/p62 can be induced directly by activated heat shock factor 1 in colorectal cancer cells treated with a heat shock protein 90 (HSP90) inhibitor, which may accelerate the process of autophagy and suppress cell death, ultimately resulting in the weak anticancer effects of HSP90. By contrast, SQSTM1/p62 inhibition may markedly enhance colorectal cancer chemosensitivity to HSP90 (33). Alternatively, it was previously reported that SQSTM1/p62 and LC3 were overexpressed in stage III-IV colon cancer exposed to 5-FU therapy, which indicated that high levels of SQSTM1/p62 and activation of autophagy may represent the self-defense of a tumor against internal and external stress to compromise therapeutic efficacy (27). This outcome may be partially reflected at the genetic level; for example, in escin-treated CRC cells, activated autophagy and SQSTM1/p62 accumulation have been shown to play a protective role against escin-induced apoptosis and DNA damage, as SQSTM1/p62 abrogates the ataxia-telangiectasia mutated/phosphorylated histone family member X pathway (34). However, a high level of SQSTM1/p62 may enhance the efficacy of photodynamic therapy by promoting tumor cell death (35). Therefore, on the one hand, SQSTM1/p62 may act as a protective factor in tumor cell survival by promoting autophagy and inhibiting apoptosis under drug-induced stress, while on the other hand, it may act as a driving force to accelerate the death of tumor cells subjected to photodynamic therapy via mechanisms that remain unknown.
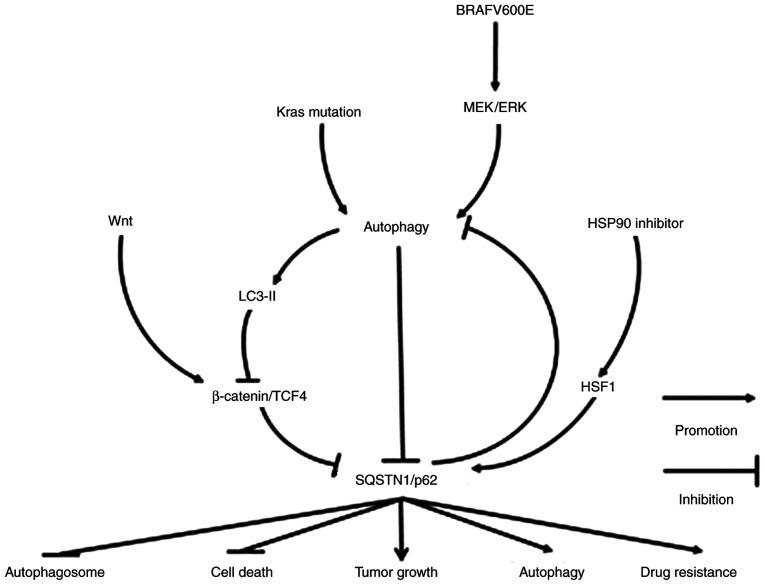
In conclusion, SQSTM1/p62 is involved in the occurrence and development of gastrointestinal tumors through various signaling pathways (Fig. 1). SQSTM1/p62 plays a key role in the process of autophagy. However, increased or decreased SQSTM1/p62 expression does not necessarily represent the inactivation or activation of autophagy. In fact, SQSTM1/p62 expression may be elevated or decreased in association with activated or suppressed autophagy. Additionally, patients with an elevated SQSTM1/p62 level do not necessarily tend to have a poor prognosis, nor does a decreased SQSTM1/p62 expression level indicate a favorable prognosis. Nonetheless, it seems certain that the inhibition of SQSTM1/p62 expression can significantly enhance the therapeutic strategies used in the treatment of gastrointestinal tumors. Therefore, targeting SQSTM1/p62-mediated signaling pathways may be an ancillary strategy used with therapies against these cancer types.
Figure 1.
Molecular mechanisms of SQSTM1/p62 in regulating gastrointestinal tract tumors. SQSTM1/p62 expression may be decreased following the activation of Wnt/β-catenin/TCF4, a pathway inhibiting autophagy, including autophagy caused by KRAS mutation and BRAFV600E activation. However, under conditions of starvation-induced autophagy or in the presence of an HSP90 inhibitor, SQSTM1/p62 expression may also increase due to the degradation of LC-3II-bound β-catenin in autophagosomes or the activation of heat shock factor 1, respectively. The elevated expression of SQSTM1/p62 ultimately not only inhibits the formation of autophagosomes and cell death, but also facilitates autophagy, tumor growth and drug resistance. SQSTM1, sequestosome 1; TCF4, transcription factor 4; HSP90, heat shock protein 90; HSF1, heat shock factor protein 1.
3. Hepatocellular carcinoma (HCC)
The pattern and significance of SQSTM1/p62 expression are different between cancer cells and peripheral stromal cells. SQSTM1/p62 expression has been shown to be higher in HCC cells but lower in peripheral mesenchymal astrocytes, where SQSTM1/p62 directly promotes the interaction between the vitamin D receptor and retinoid X receptor to inhibit the process of liver fibrosis, inflammation and cancer (36). In this respect, SQSTM1/p62 may function as an anti-oncogene in HCC formation by regulating steroid hormone reactions.
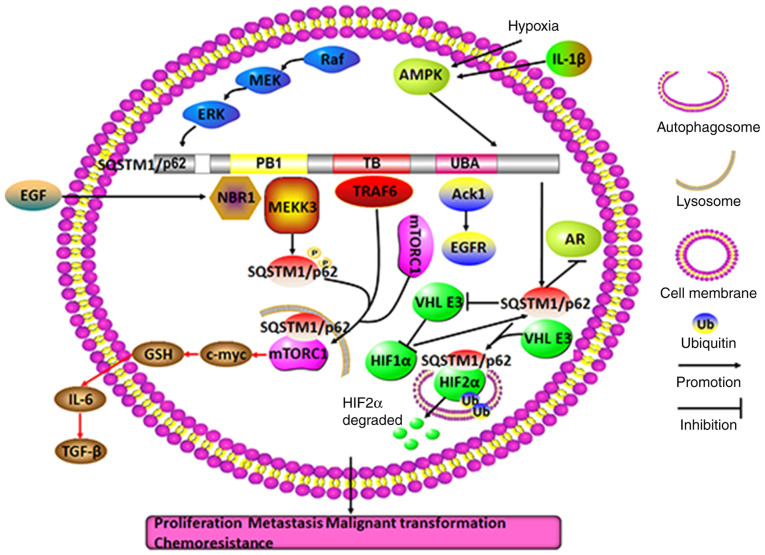
In HCC, SQSTM1/p62 gene amplification, mutation and hepatitis C virus infection can cause excessive SQSTM1/p62 aggregation and phosphorylation. Under these conditions, glucose is carried into the glucuronic acid metabolic pathway, and glutamate is converted into glutathione by activating nuclear factor erythroid 2 (NRF2), which further promotes SQSTM1/p62 expression. The positive feedback induces aggressive cancer cell proliferation and chemoresistance (37-39). In addition, SQSTM1/p62 can bind to the N-terminal region of TRAF6 [a ubiquitin ligase activating the NF-κB cascade by binding to the N-terminal region of ζ-protein kinase C (PKC)] to activate the NF-κB signaling pathway, thus facilitating invasion, migration and distant metastasis of HCC. Enhanced expression of SQSTM1/p62 participates in the formation of benign adenoma by liver cells with autophagy deficiency; furthermore, in addition to activating NRF2, enhanced SQSTM1/p62 expression in conditions of cellular reactive oxygen stress or inflammation in HCC precancerous lesions can induce c-myc expression by initiating the mechanistic target of rapamycin complex 1 (m-TORC1) signaling pathway, rather than the ubiquitination pathway, to promote the occurrence of liver cancer. Even after HCC resection, high expression of SQSTM1/p62 in the remainder of the liver is an indicator of HCC recurrence (40). In summary, accumulation of SQSTM1/p62 as an oncogene in HCC may eventually promote malignant tumor cell progression by either interacting with NRF2 or TRAF6 to activate the NF-κB signaling pathway or to upregulate c-myc expression to initiate the m-TORC1 signaling pathway.
The promoter region of SQSTM1/p62 contains antioxidant response elements (AREs), making SQSTM1/p62 an NRF2 (and NRF1) response gene. Thus, the SQSTM1/p62 gene is induced in response to NRF2 activation. Substantial analyses of a high number of cancer biopsy samples of different forms of cancer have indicated that the NRF2 pathway is frequently activated by somatic mutations (41). Thus, in cancer cells where NRF2 is constitutively activated, the transcription of SQSTM1/p62 and a number of other protective oxidative stress response genes is expected to be increased. Thus, elevated SQSTM1/p62 gene expression is considered to be a consequence of mutations that frequently occur in cancer. A study showed that autophagy-deficient mouse livers exhibited aberrant SQSTM1/p62 accumulation and developed severe liver damage as SQSTM1/p62 accumulation disrupted the KEAP1-NRF2 association and promoted NRF2 stabilization and accumulation. However, in SQSTM1/p62-deficient mouse livers, KEAP1 was primarily degraded via the autophagy pathway in a SQSTM1/p62-dependent manner, and NRF2 accumulation caused severe liver dysfunction through the autophagy pathway, not the proteasome pathway, independent of SQSTM1/p62 expression (42).
Additionally, SQSTM1/p62 regulates the chemosensitivity and chemoresistance of HCC in different ways. When dehydroepiandrosterone induces autophagic cell death, unexpectedly, SQSTM1/p62 is not degraded through the autophagic process; it aggregates upon upregulation via the initiation of the Jun N-terminal kinase (JNK)-NRF2-SQSTM1/p62 signaling pathway, in which JNK methylation triggers NRF2 to induce SQSTM1/p62 expression (43). On the other hand, a ferroptosis agonist promotes the expression of SQSTM1/p62 in HCC cells, which can further inactivate the cobinding factor KEAP1 to release and thus activate NRF2. Once released, NRF2 accumulates in the cell nucleus and induces the expression of various downstream oncogenes, such as ferritin heavy chain 1, NAD(P)H dehydrogenase quinone 1 (NQO1) and heme oxygenase-1 (HO-1), which suppresses cellular ferroptosis and causes drug resistance (38). Notably, SQSTM1/p62 also regulates both apoptosis and pyroptosis via acetylation. Researchers have confirmed that SQSTM1/p62 colocalizes with histone deacetylase 6 (HDAC6) during ubiquitylation in mitochondria and abolishes HDAC6 deacetylation (44). The inhibition of SQSTM1/p62 may facilitate HDAC6-mediated deacetylation of α-tubulin and cortactin to disrupt the stability of microtubes and the formation of autolysosomes. Unexpectedly, the inhibition of autolysosomes can cause the downregulation not the upregulation of SQSTM1/p62 (45), suggesting that the underlying mechanisms for regulating SQSTM1/p62-related networks under autophagy conditions should be studied further. Therefore, it seems that SQSTM1/p62 may weaken the curative effects of some anticancer drugs primarily through the SQSTM1/p62-NRF2 axis.
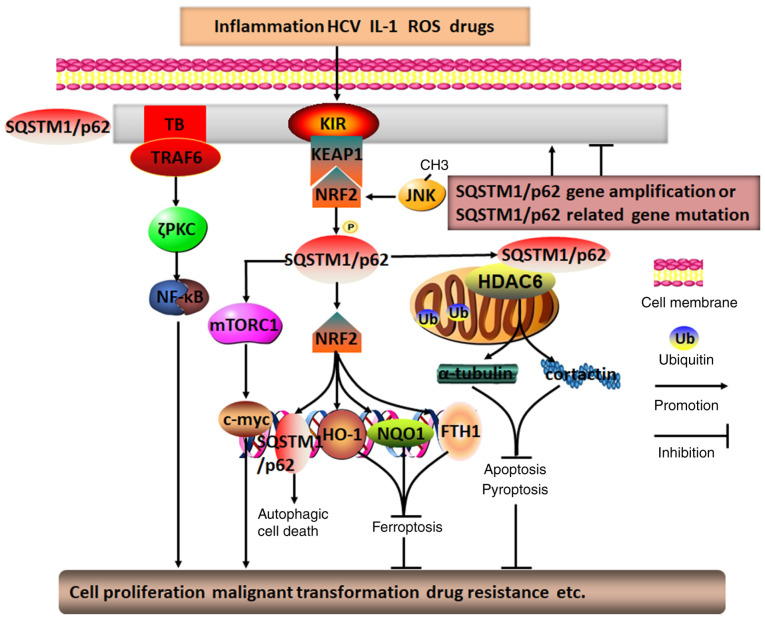
In summary, it appears that SQSTM1/p62 in HCC cells may act primarily as an oncogene, while it may also function as a tumor repressor gene in peripheral stromal cells; in these cells, SQSTM1/p62 regulates chemosensitivity and chemoresistance through a variety of processes, including autophagic cell death, apoptosis, ferroptosis and pyroptosis, via different signaling pathways, particularly protein deacetylation and the SQSTM1/p62-NRF2 positive feedback loop (Fig. 2). Therefore, targeting SQSTM1/p62 in intricate networks may be a good adjuvant therapeutic approach to HCC treatment.
Figure 2.
Molecular mechanisms of SQSTM1/p62 in regulating HCC. Diverse intracellular and extracellular risk factors, including SQSTM1/p62 gene amplification, SQSTM1/p62-related gene mutation, inflammation, HCV infection, ROS-induced stress, drugs and IL-1, may promote SQSTM1/p62 aggregation and phosphorylation. Subsequently, SQSTM1/p62 is capable of activating the mTOR/c-myc, TRAF6/ζPKC/NF-κB, and SQSTM1/p62/KEAP1/NRF2 pathways through its distinctive TB and KIR domains. The positive feedback loop of SQSTM1/p62/KEAP1/NRF2, involving JNK methylation-induced NRF2 activation, may generate various secondary messengers, such as SQSTM1/p62, HO-1, NQO1 and FTH1. In addition, by colocalizing with HDAC6 during ubiquitylation in mitochondria, SQSTM1/p62 may inhibit HDAC6 deacetylation of α-tubulin and cortactin, which may ultimately result in p62 directly or indirectly regulating HCC autophagic cell death, apoptosis, pyroptosis, ferroptosis, cell proliferation, malignant transformation and drug resistance, among others. TRAF6, TNF receptor-associated factor 6; TB, TRAF6-binding domain; KIR, KEAP1-interacting region; PKC, protein kinase C; SQSTM1, sequestosome 1; ROS, reactive oxygen species; FTH1, ferritin heavy chain 1; KEAP1, Kelch-like ECH-associated protein 1; NRF2, nuclear factor erythroid 2; HO-1, heme oxygenase 1; NQO1, NAD(P)H dehydrogenase quinone 1; HDAC6, histone deacetylase 6; HCC, hepatocellular carcinoma; HCV, hepatitis C virus.
4. Breast cancer
A mechanism of SQSTM1/p62 function as an oncogene similar to that involved in HCC has been observed in breast cancer; that is, expression of SQSTM1/p62 in the presence of NRF2 can be decreased in the cytoplasm and increased in the cell nucleus through the SQSTM1/p62-KEAP1-NRF2 axis, which causes NRF2 to activate the expression of numerous downstream oncogenes. Furthermore, the aggregation of NRF2 in the cell nucleus may also significantly promote SQSTM1/p62 expression. In particular, SQSTM1/p62 mediates CD44-NRF2 activation during autophagy in cancer stem cells (CSCs), leading to aggressive cancer growth and drug resistance (46). Thus, the formation of the SQSTM1/p62 and the NRF2 positive feedback loop ultimately endows CSCs with self-renewal capacity and chemoradiotherapy resistance (47,48). Notably, SQSTM1/p62 has been shown to interact with microRNAs to regulate breast cancer malignancy. That is, in a previous study, SQSTM1/p62 prevented myc degradation by directly abrogating the expression of the microRNAs let7a and let7b, resulting in persistent overexpression of myc in the cancer cells (49).
To some degree, it seems that SQSTM1/p62 plays a central role in in complex networks to regulate the occurrence and development of breast cancer. Recent data have demonstrated that SQSTM1/p62, as a bridge between Notch1 intracellular domain (Notch1-IC) and LC-3-II in the autophagosome, has the ability to abolish the Notch1 signaling pathway through its role in the autophagosome, which engulfs and degrades Notch1-IC (50,51). Moreover, HER-2-mediated activation of SQSTM1/p62 can maintain cancer cell vitality and markedly accelerate cell proliferation and malignant growth through various signaling pathways, including the PI3K/AKT signaling pathway (which is initiated through SQSTM1/p62-induced PTEN degradation), the AKT/glycogen synthase kinase 3β/β-catenin signaling pathway, the Wnt/β-catenin signaling pathway, the NF-κB signaling pathway and the KEAP1-NRF2 signaling pathway (51-53). Among these pathways, the SQSTM1/p62-activated NF-κB signaling pathway under conditions of autophagy deficiency may lead to high rates of p65 phosphorylation and transport into the cell nucleus, inducing the upregulation of NF-κB target genes, such as IFN-γ, TNF-α and IL-6, which promote cancer cell proliferation (54). In addition, SQSTM1/p62 can recruit and phosphorylate JNK to facilitate the development of cancer cells by directly binding to Van Gogh-like 2 in the Wnt/PCP signaling pathway (55). In addition, SQSTM1/p62 dysfunction may cause autophagy deficiency and initiate the mitochondrial apoptosis pathway by inducing the accumulation of the apoptosis-related BH3-only protein NBK/Bik on endoplasmic reticulum membranes, thus converting autophagic cells to apoptotic cells and suppressing cancer cell proliferation (56). Furthermore, in a previous study, SQSTM1/p62 degradation was diminished due to the inhibition of cathepsin in the autolysosome in cancer cells treated with chemicals that induce autophagy, and apoptosis occurred due to the increased reactive oxygen species (ROS) level that was induced upon SQSTM1/p62 accumulation (57).
SQSTM1/p62 aggregation in breast cancer cells usually acts as an indicator of chemoresistance to multiple drugs, such as Adriamycin, PI3K/AKT inhibitors and Pseudomonas aeruginosa mannose-sensitive hemagglutinin (46). Breast cancer with high expression of SQSTM1/p62 has been linked to distant lymphatic and vessel metastasis, and is correlated with an unfavorable prognosis in these patients, particularly those with triple-negative breast cancer (58-61). As a result, the inhibition of SQSTM1/p62 expression in these cancer cells may lead to the abrogation of cell growth and proliferation, enhancing the efficacy of therapeutic treatments. Nevertheless, an exception has also been reported in gemcitabine-treated estrogen receptor (ER)-positive breast cancer cells, where ERK phosphorylation may promote SQSTM1/p62 expression to accelerate the process of autophagic cell death. By contrast, SQSTM1/p62 silencing may abrogate the ER/ERK/SQSTM1/p62 signaling pathway to interrupt excessive autophagy, thereby protecting cancer cells from cytotoxic effects (62). Taken together, these findings imply that SQSTM1/p62 may enhance either chemosensitivity or chemoresistance depending on the type of breast cancer, information of importance for implementing precision therapies to treat breast cancer.
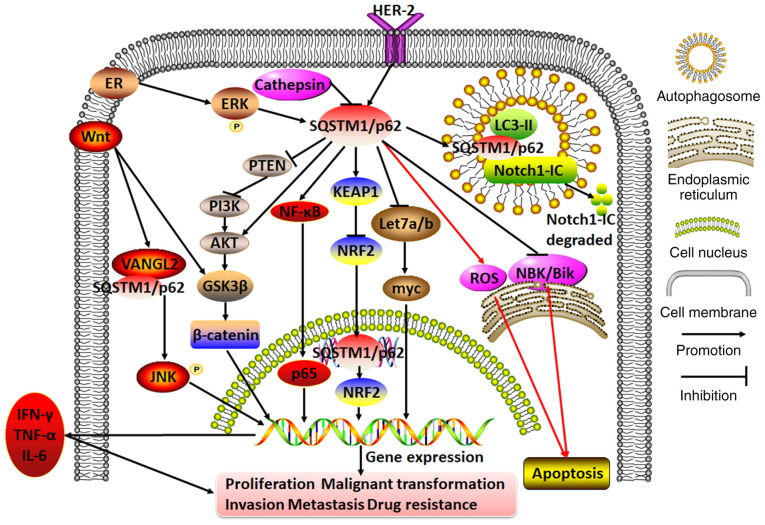
In general, SQSTM1/p62 can regulate breast cancer cell proliferation, malignant transformation, invasion, migration and distant metastasis through multiple apoptosis- and autophagy-associated signaling pathways; thus, it plays a dual role in breast cancer, acting both as an oncogene to confer cancer cell chemoresistance and as a tumor repressor to confer chemosensitivity (Fig. 3).
Figure 3.
Molecular mechanisms of SQSTM1/p62 in regulating breast cancer. In breast cancer cells, cathepsin on the autolysosome may repress SQSTM1/p62 expression. By contrast, ER or HER-2 can initiate SQSTM1/p62 expression. The hyperexpression of SQSTM1/p62 exacerbates cell proliferation, malignant transformation, invasion, metastasis and drug resistance by participating in diverse cascades, including the PTEN/PI3K/AKT, Wnt/β-catenin, AKT/glycogen synthase kinase 3β/β-catenin, KEAP1/NRF2, NF-κB (which releases IFN-γ, TNF-α and IL-6), Wnt/VANGL2/SQSTM1/p62/JNK and Let7a/b/myc pathways. Moreover, servicing as a bridge between Notch1-IC and LC3-II, SQSTM1/p62 mediates Notch1-IC degradation in autophagosomes. In addition, SQSTM1/p62 aggregation may either promote or inhibit apoptosis by repressing NBK/Bik expression at the endoplasmic reticulum or inducing ROS expression. VANGL2, Van Gogh-like 2; ROS, reactive oxygen species; SQSTM1, sequestosome 1; ER, estrogen receptor; PI3K, phosphoinositol-3 kinase; KEAP1, Kelch-like ECH-associated protein 1; NRF2, nuclear factor erythroid 2; Notch1-IC, Notch1 intracellular domain.
5. Hematological malignancy
In multiple myeloma cells treated with TNF-α, high-risk myelodysplastic syndromes/acute myeloid leukemia (AML) del(5q) cells and hematopoietic stem cells/progenitor cells with miR-146a deletion, the sustained expression of SQSTM1/p62 has an oncogenic effect that leads to the marked activation of NF-κB or p38MAPK cascades through the following mechanisms: i) Direct binding to and ubiquitination of TRAF-6 through its TRAF-binding domain to activate the NF-κB signaling pathway (63-65); ii) direct binding with receptor-interacting protein 1 through its C-terminal region ZZ domain, as well as with atypical PKCs through its N-terminal PB1 domain to activate the NF-κB signaling pathway (66,67); and iii) direct binding to p38MAPK through its p38 domain to activate the p38MAPK signaling pathway (68). Activated NF-κB can further enhance SQSTM1/p62 gene expression, ultimately generating predominately positive feedback loops to continuously amplify its deleterious effect in exacerbating unrestrained growth, proliferation and malignant transformation, and inhibiting osteoblast activation to hamper bone formation (66,69). As a result, the deprivation of SQSTM1/p62, leading to the inhibition of SQSTM1/p62 ZZ domain activity, may attenuate the NF-κB signaling pathway by downregulating phospho-focal adhesion kinase, p-IκBα and NF-κB expression, promoting osteoblast differentiation and generating myeloid progenitor cells and osteoclasts, thereby inhibiting the growth of multiple myeloma cells (67,69).
Given that SQSTM1/p62 is closely linked to autophagy and ubiquitylation-mediated proteasomal processes, defective autophagy in multiple myeloma cells may lead to an increase in undigested and toxic proteins with SQSTM1/p62 located in the endoplasmic reticulum; thus, SQSTM1/p62 may be regarded as a reliable biomarker for the chemosensitivity of multiple myeloma to proteasome inhibitors (70). Furthermore, both SQSTM1/p62 and Kruppel-like factor 4 (KLF4), an intranuclear transcription factor, are upregulated in multiple myeloma cells that are resistant to the proteasome inhibitor carfilzomib. Notably, KLF4 has been found to bind to the promoter regions encoding ubiquitin-binding domains to upregulate SQSTM1/p62 and trigger the ubiquitin-proteasome and autophagy pathways, thus inducing cancer cell resistance to carfilzomib (71). In addition, all-trans retinoic acid (ATRA)-induced NF-κB activation may upregulate SQSTM1/p62 expression. By contrast, SQSTM1/p62 may decrease the response to ATRA in AML patients with a poor prognosis (72,73). However, SQSTM1/p62 may function as a tumor suppressor in chronic myeloid leukemia, as the proto-oncogene BCR-ABL can be translocated to autophagosomes by SQSTM1/p62 for degradation by the cysteine protease cathepsin B in autolysosomes (74). Notably, a recent article reported that in mice lacking SQSTM1/p62 expression in all hematopoietic cells, including macrophages, no changes in NF-κB signaling were found (75). These data query the importance of the signaling role of SQSTM1/p62 in non-transformed cells. On the other hand, another paper emphasized the possible autophagy-independent roles of SQSTM1/p62 in cancer (76). In summary, SQSTM1/p62 may either transmit deleterious messages to induce resistance to chemotherapeutic drugs through the ubiquitin-proteasome, autophagy or ATRA/NF-κB pathways in patients with multiple myeloma or AML, or provide attenuating signals through the autophagy pathway in patients with chronic myeloid leukemia.
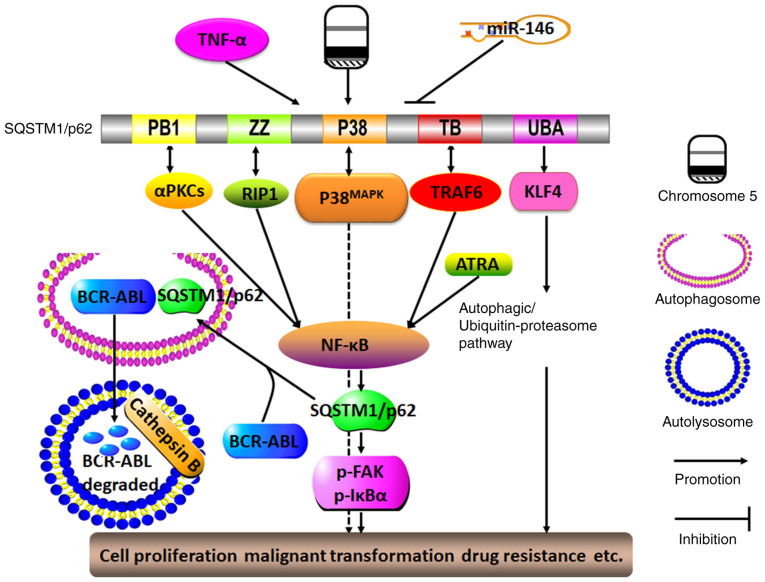
In summary, by participating in multiple signaling pathways, particularly in the positive feedback loop cascade with NF-κB, SQSTM1/p62 may serve as a proto-oncogene to induce hematological malignancy with resistance to various anti-tumor drugs. Unexpectedly, SQSTM1/p62 may, under certain circumstances, sensitize cancer cells to some chemotherapeutics (Fig. 4). Therefore, silencing SQSTM1/p62 expression may enhance chemosensitivity, but notably, elevated SQSTM1/p62 expression does not necessarily indicate that cancer cells will become resistance to antitumor reagents; therefore, these observations should be used only to guide the treatment of specific hematological malignancies.
Figure 4.
Molecular mechanisms of SQSTM1/p62 in regulating hematological malignancy. The presence of TNF-α, del(5q) and miR-146a deletions may lead to the sustained expression of SQSTM1/p62. By interacting with TRAF-6, RIP1 and αPKCs via its TB, ZZ and PB1 domains, respectively, SQSTM1/p62 may activate the NF-κB signaling pathway, which in turn promotes SQSTM1/p62 expression. ATRA may also trigger the NF-κB signaling pathway, thereby forming a positive feedback loop with SQSTM1/p62 and NF-κB. By interacting with p38MAPK via the P38 domain, SQSTM1/p62 can function in conjunction with the activated autophagy/ubiquitin-proteasome pathway as triggered by the interaction between SQSTM1/p62 and KLF4 through the SQSTM1/p62 UBA domain, and SQSTM1/p62 can ultimately promote cell proliferation, malignant transformation and drug resistance. By contrast, SQSTM1/p62 may function as a tumor suppressor by driving the oncogene BCR-ABL into autophagosomes, and it is degraded by cathepsin B in autolysosomes. aPKC, atypical protein kinase C; PB1, Phox and Bem1p; ZZ, ZZ-type zinc finger domain; P38, P38MAPK domain; RIP1, receptor-interacting protein 1; TB, TRAF6-binding domain; UBA, ubiquitin-associated domain; KLF4, Kruppel-like factor 4; SQSTM1, sequestosome 1; TRAF6, TNF receptor-associated factor 6.
6. Lung cancer
Recent investigations have confirmed that SQSTM1/p62 is usually upregulated in the cytoplasm of non-small cell lung cancer (NSCLC) cells, which indicates a more progressive phenotype for these cancer cells and a shorter survival period for patients (77). However, when autophagy is defective, the expression of SQSTM1/p62 is negatively associated with Tumor-Node-Metastasis stage and lymph node metastases, which indicates that it may be considered an independent prognostic factor for NSCLC (78).
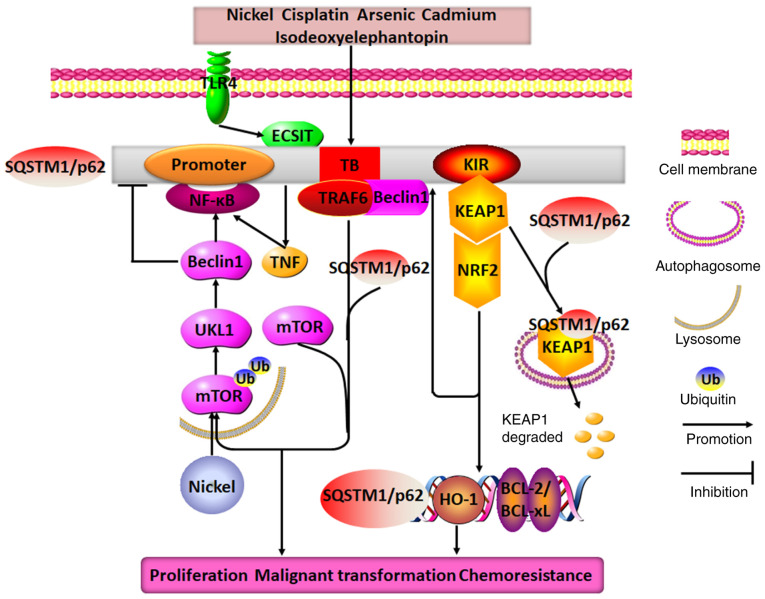
It has been confirmed that SQSTM1/p62 plays an oncogenic role through its involvement with different pathways, such as the NF-κB signaling pathway and SQSTM1/p62-KEAP1-NRF2 cascade, to regulate autophagy and apoptosis. For instance, emerging evidence has indicated that nickel exposure may cause the degradation of SQSTM1/p62 through the autophagy-related mTOR-Unc-51 like autophagy activating kinase1-Beclin1 cascade. However, nickel exposure increases SQSTM1/p62 expression by stabilizing TNF mRNA, which in turn promotes SQSTM1/p62 mRNA transcription due to the activation of the NF-κB/REL-associated protein signaling pathway; thus, TNF may function as a key transcription regulator binding to the promoter regions of the SQSTM1/p62 gene (79). Similarly, in cisplatin-treated lung adenocarcinoma cells or lung adenocarcinoma cells in nutrient-rich environments, the binding of SQSTM1/p62 and TRAF6 through the TRAF6-binding motif may also activate the NF-κB signaling pathway through TRAF6-mediated ubiquitination of mTORC1, which is then transported to the lysosome by SQSTM1/p62 and thereby inactivated; this positive feedback loop promotes the malignant transformation of bronchial epithelial cells, the abolishment of autophagy, the proliferation of cancer cells and the secondary chemoresistance of cells to cisplatin (80,81). These results indicate that, in some cases, increased SQSTM1/p62 expression may indirectly result in the inhibition of autophagy. In addition, a number of other factors contribute to the occurrence of lung cancer. For instance, arsenic, cadmium and isodeoxyelephantopin may activate the SQSTM1/p62-KEAP1-NRF2 cascade, in which SQSTM1/p62 transports KEAP1 to autophagosomes, where KEAP1 is spontaneously digested in the cytoplasm; alternatively, exposure to these compounds might activate the NRF2-KEAP1-SQSTM1/p62 cascade, in which NRF2 in the nucleus directly triggers SQSTM1/p62 binding to KEAP1 to release NRF2. NRF2 is subsequently imported into nucleus where it actively stimulates the expression of secondary downstream molecules such as HO-1, SQSTM1/p62 and Bcl-2/Bcl-xL by binding to ARE promoter regions. Thus, the formation of another positive feedback loop between SQSTM1/p62 and NRF2 leads to the increased expression of antiapoptotic proteins and antioxidases, ultimately causing cancer cell proliferation, growth and malignant transformation (82-84). Unexpectedly, SQSTM1/p62 is capable of weakening the oncolytic effects of measles virus in lung cancer, as seen through its ability to evoke mitochondrial autophagy to degrade the Edmonton strain (85).
Since SQSTM1/p62 acts as an oncogene to facilitate malignant cancer progression, the inhibition of excessive SQSTM1/p62 aggregation may represent an effective anti-cancer strategy. For instance, silencing SQSTM1/p62 may lead to the initiation of the autophagic cell death process and the abrogation of cell proliferation not only in lung cancer, but also in pancreatic cancer and squamous cell carcinoma of the esophagus, oral cavity and skin (86). The silencing or degradation of SQSTM1/p62 apparently disrupts the SQSTM1/p62/TRAF6/NF-κB cascade and activates the apoptosis pathway. Moreover, as SQSTM1/p62 may directly block the formation of the Fas/Cav-1 complex, non-functional SQSTM1/p62 may result in the escape of the Fas/Cav-1 complex to activate caspase-8 and cleave Beclin-1, and produce diverse mediators, including ROS, that eventually enhance the killing effects of resveratrol or the combined effects of the Chinese herb Yu Ping Feng San and cisplatin (81,87). Furthermore, silencing SQSTM1/p62 expression to downregulate neuronal precursor cell expression of developmentally downregulated gene 9 may increase the response to cisplatin treatment in small cell lung cancer cells (88). Alternatively, SQSTM1/p62 can bind with both the TRAF-C domain of TRAF6 and the coiled-coil domain of Beclin1 to disrupt the interaction of the TRAF6-Beclin1 and TRAF6-evolutionarily conserved signaling intermediate in Toll pathways (ECSIT); thus, SQSTM1/p62 may simultaneously abolish TRAF6-ECSIT signaling and activate the NF-κB pathway in response to toll-like receptor 4 stimulation, thereby abrogating Beclin1 and ECSIT ubiquitination, along with the activation of autophagy and cancer invasion of both lung and breast cancer cells (89,90).
In conclusion, SQSTM1/p62 functions as an oncogene to disrupt the malignant transformation of lung cancer by forming several positive feedback loops, particularly loops involving SQSTM1/p62 and NF-κB or NRF2 (Fig. 5). Therefore, interrupting deleterious cycles of these amplifying feedback loops may be considered a feasible way to obtain marked clinical benefits for patients suffering from lung cancer.
Figure 5.
Molecular mechanisms of SQSTM1/p62 in regulating lung cancer. Nickel exposure may cause SQSTM1/p62 degradation through the mTOR/ULK1/Beclin1 pathway, and it upregulates p62 expression by stimulating the TNF/NF-κB/SQSTM1/p62 positive feedback loop. Additionally, when exposed to different drugs, such as cisplatin, arsenic, cadmium, isodeoxyelephantopin and TLR4, SQSTM1/p62 expression is observed to be increased in lung cancer cells, where it may directly interact with TRAF6, KEAP1, Beclin1 and ECSIT through its TB and KIR domains to form two distinct positive feedback loops: SQSTM1/p62/TRAF6/mTORC1/NF-κB and SQSTM1/p62/KEAP1/NRF2. These feedback loops promote the expression of SQSTM1/p62, HO-1 and BCL2/BCL-xL. Therefore, the end result of the combined effects of these positive feedback loops with SQSTM1/p62 at their core is the promotion of lung cancer cell proliferation, malignant transformation and chemoresistance. TRAF6, TNF receptor-associated factor 6; TB, TRAF6-binding domain; KIR, KEAP1-interacting region; ECSIT, evolutionarily conserved signaling intermediate in Toll pathways; SQSTM1, sequestosome 1; ULK1, Unc-51 like autophagy activating kinase; TLR4, toll-like receptor 4; KEAP1, Kelch-like ECH-associated protein 1; HO-1, heme oxygenase 1.
7. Genital system cancer
Recent evidence has confirmed that SQSTM1/p62 expression is markedly higher in epithelial ovarian cancer, particularly in cisplatin-resistant ovarian cancer, than in benign ovarian tumors (91). The positive rates of SQSTM1/p62 are also significantly higher in the advanced stage than in the early stage of ovarian cancer (92). However, a study has reported that compared with patient-matched primary tumor tissues, metastatic and recurrent ovarian cancer samples had lower SQSTM1/p62 expression levels, and a low level of SQSTM1/p62 was closely related to cancer recurrence, metastasis and paclitaxel resistance; furthermore, the positive rates of SQSTM1/p62 in cisplatin-resistant ovarian cancer cells were also lower than those in cisplatin-sensitive cells (93,94). In some specific cancer types, such as seminomas and non-seminomas, SQSTM1/p62 expression is negligibly changed (95). Therefore, the function and mechanism of SQSTM1/p62 in ovarian cancer should be further explored.
Moreover, high SQSTM1/p62 expression in the cytoplasm and low expression in the nucleus is associated with an advanced stage, a residual tumor and unfavorable survival in patients with endometrial cancer, epithelial ovarian cancer, serous carcinoma or cervical cancer (92,96,97). The possible underlying mechanisms might be that by binding NRF2 and the cotranscription factor TCF20 through its binding domain containing the ARE element, activated SQSTM1/p62 can initiate the activation of NRF2 and TCF20, which subsequently enter the nucleus to stimulate SQSTM1/p62 expression; this outcome leads to resistance to cisplatin or sulforaphane by initiating cascades of antioxidant gene expression and facilitating differentiation, proliferation and anti-apoptotic behavior in drug-resistant cancer cells (97). Additionally, SQSTM1/p62-mediated upregulation of the cell cycle protein Skp2 through autophagy, not proteasomal processes, can cause the degradation of p21 and p27, which results in abrogation of quinacrine-induced apoptosis (98).
Upon treatment with vitamin K3, ovarian cancer cells may escape ROS-mediated oxidative damage by activating SQSTM1/p62/KEAP1/NRF2 signaling (99). By contrast, after treatment with cisplatin to block autophagy, SQSTM1/p62 may function as an anti-oncogene to accelerate the apoptosis of ovarian cancer cells by interacting with the apoptosis-related protein caspase-8 (100).
In summary, SQSTM1/p62 expression varies in different genital system cancer types and different cellular locations. However, depending largely on the type of cancer, SQSTM1/p62 can display various functions by engaging in diverse signaling events, and it may be a new reliable prognostic biomarker for endometrial cancer and ovarian cancer, and a drug-sensitive biomarker for use in neoadjuvant chemotherapy.
8. Urological cancer
Similar to that in genital system cancer, high expression of SQSTM1/p62 may be an indicator of poor prognosis for patients with prostate cancer. SQSTM1/p62 expression is substantially higher in prostate cancer, particularly prostate cancer at an advanced stage, with resistance to androgen, and with low androgen expression, compared with that in benign prostatic hyperplasia (101,102); moreover, its expression in the cytoplasm is highly associated with advanced Gleason grade, a positive resection margin and the recurrence of prostate cancer. The higher the SQSTM1/p62 expression level in the cytoplasm, the poorer the prognosis in these patients. Thus, by targeting SQSTM1/p62-induced constitutive NRF2 activation and Bcl-xL expression, verteporfin, a drug used to treat macular degeneration, can effectively block autophagy and enhance drug sensitivity in prostate cancer (103). More importantly, at the genetic level, elevated SQSTM1/p62 levels may also be relevant to the gene fusion of TMPRSS2 and ERG, and gene deletions in PTEN, 3p13, 5q21 and 6q15 (104), which suggests that crosstalk between SQSTM1/p62 and prostate cancer-related genes may play a pivotal role in the occurrence and development of prostate cancer.
SQSTM1/p62 can drive the autophagy process in response to the activation of the Raf/MEK/ERK signaling pathway in both malignant melanoma and prostate cancer (105). In addition, SQSTM1/p62 can bind to MEKK3 and neighbor of BRCA1 (NBR1) through its PB1 domain, activate Cdc42-associated kinase 1 (Ack1) through its UBA domain and then stimulate the following downstream events: i) Under nutrient-deprived conditions, MEKK3 induces SQSTM1/p62 phosphorylation, and phosphorylated SQSTM1/p62 recruits TRAF6, which directs mTORC1 to lysosomes (106); and ii) in the presence of EGF, Ack1 dissociation from SQSTM1/p62 and NBR1 in autophagosome precursors results in prolonged survival of EGFR through slow endocytosis rather than lysosomal degradation pathways (107). These cascades markedly promote autophagy, proliferation and malignant processes in PTEN-deficient prostate organoids, prostate cancer and cervical cancer cells. Furthermore, by directly simulating the KEAP1/NRF2/ARE signaling pathway, SQSTM1/p62 may contribute to the aggressive behavior of prostate cancer (108).
In addition, it seems that SQSTM1/p62 expression in prostate cancer cells may have some underlying connections with the surrounding microenvironment. SQSTM1/p62 expression is frequently upregulated in cancer cells, but downregulated in nearby microenvironments. For example, emerging studies have validated that, in contrast to prostate cancer cells, in cancer-associated fibroblasts (CAFs), SQSTM1/p62 expression is usually decreased. The lower the expression of SQSTM1/p62, the higher the Gleason grade in prostate cancer. Notably, SQSTM1/p62 deficiency in adipocytes near prostate cancer cells can be beneficial to tumor nutrient availability, the EMT and invasiveness, by increasing osteopontin secretion and inactivating mTORC1-related energy-consuming pathways (109). Furthermore, SQSTM1/p62 deletion stimulates the downstream SQSTM1/p62/mTORC1/c-Myc/glutathione (GSH)/IL-6/TGFβ pathway, in which mTORC1 and c-Myc inactivation resulting from SQSTM1/p62 deletion decreases GSH expression and stimulates the IL-6/TGFβ pathway to generate a CAF phenotype that favors malignant transformation, proliferation and invasion (110). Furthermore, it has been revealed that both IL-1β and hematopoietic HS-5 derived from cancer-related bone marrow stromal cells upregulate and phosphorylate SQSTM1/p62 by activating the AMPK pathway in androgen receptor (AR)-independent prostate cancer, and that phosphorylation of SQSTM1/p62 recruits and degrades AR, eventually causing repression of apoptosis, promotion of proliferation and resistance to multiple drugs (101,111). In summary, downregulation of SQSTM1/p62 in cancer-related surrounding cells may assist tumor cells in promoting malignant transformation in prostate cancer.
In renal cell carcinoma, in response to hypoxia, SQSTM1/p62 can directly integrate and inactivate the ubiquitin-protein ligase VHL E3 to improve the activity and stability of hypoxia-inducible factor 1α (HIF1α), which in turn forces SQSTM1/p62 to trigger the glycolytic pathway. Therefore, the collaboration of SQSTM1/p62 and HIF1α may provide more energy to facilitate renal cancer cell survival in harsh environments (112). By contrast, when the proteasome degradation pathway is mitigated, SQSTM1/p62 can directly bind to and direct ubiquitylated HIF2α to autophagosomes, where HIF2α is degraded through the SQSTM1/p62-dependent autophagic degradation pathway. Conversely, when the autophagic degradation pathway is inhibited, ubiquitylated HIF2α can be degraded via the proteasome degradation pathway (113). These data indicate that SQSTM1/p62 may function as an oncogene in manipulating the occurrence and development of renal cell carcinoma by inducing the action of HIF as a switch between the autophagic degradation pathway and proteasome degradation pathway.
Moreover, in bladder cancer, overexpressed SQSTM1/p62 may protect bladder cancer cells from oxidative stress by stimulating the KEAP1/NRF2 signaling pathway, which upregulates the expression of several antioxidant genes, such as glutamate-cysteine ligase catalytic subunit, glutathione S-transferase Mu 5 and glutathione peroxidase 2, promoting cell proliferation and inhibiting apoptosis (114).
In summary, SQSTM1/p62 is closely linked not only to oncogenic characteristics or tumor repressor gene deletions in tumor cells, but also to tumor microenvironments. For example, under nutrient-rich or nutrient-deprivation conditions, inflammation, hypoxia and growth factors can upregulate SQSTM1/p62 expression to stimulate downstream signal transduction, which activates a variety of oncogenes and accelerates malignant transformation, growth, proliferation, distant metastasis and drug resistance in urinary system cancer (Fig. 6).
Figure 6.
Molecular mechanisms of SQSTM1/p62 in regulating urological cancer. In prostate cancer, SQSTM1/p62 aggregation and phosphorylation may facilitate autophagy and AR degradation owing to the activation of the Raf/MEK/ERK pathway and the IL-1β and hypoxia-activated AMPK pathway, respectively. Moreover, in a nutrient-rich environment or in the presence of EGF, SQSTM1/p62 can promote autophagy, proliferation and malignant transformation by tightly binding either to MEKK3 or NBR1 to recruit TRAF6 and direct mTORC1 to lysosomes through the SQSTM1/p62 PB1 domain or to Ack1 to prolong EGFR survival through the SQSTM1/p62 UBA domain. In CAFs, SQSTM1/p62 deletion may cause stimulation of the mTORC1/c-myc/GSH/IL-6/TGF-β pathway, which facilitates the action of nearby prostate cancer cells in combating malignant transformation, proliferation and invasion (red arrow). In renal cell carcinoma, hypoxia-induced SQSTM1/p62 may not only directly bind to and drive ubiquitylated HIF2α to autophagosomes for degradation, but may also form a positive feedback loop with HIF1α by inactivating VHL E3, facilitating renal cell survival in unfavorable environments. TRAF6, TNF receptor-associated factor 6; PB1, Phox and Bem1p; NBR1, neighbor of BRCA1 gene 1; TB, TRAF6-binding domain; UBA, ubiquitin-associated domain; SQSTM1, sequestosome 1; NBR1, neighbor of BRCA1; Ack1, activate Cdc42-associated kinase 1; CAFs, cancer-associated fibroblasts; mTORC1, mechanistic target of rapamycin complex 1; GSH, glutathione; HIF1α, hypoxia-inducible factor 1α.
9. Head and neck neoplasms and skin cancer
Accumulating evidence has indicated that SQSTM1/p62 may play roles via positive feedback between SQSTM1/p62 and NRF2 in squamous cell carcinoma (115-119). Highly aggressive oral squamous carcinoma cells have high SQSTM1/p62 expression in the cytoplasm and low SQSTM1/p62 expression in the nucleus, but not the opposite expression trend (115). In addition, SQSTM1/p62 aggregation in the cytoplasm has been found to be associated with poor prognosis and drug resistance to PI3K/AKT inhibitors in autophagy-deficient head and neck squamous cell carcinoma (116,117). Usually, the change in SQSTM1/p62 expression, to some degree, reflects the change in LC3-II expression. As autophagy is initiated, LC3-II is formed on autophagosome; meanwhile SQSTM1/p62 incorporates into the autophagosome by interacting with LC3-II and then will be degraded in autolysosomes (9). Autophagy is triggered in oral squamous cell carcinoma cells, but SQSTM1/p62 expression may not fluctuate with LC3-II. By contrast, in skin squamous cell carcinoma, strong activation of autophagy and low expression of SQSTM1/p62 have been correlated with a poor prognosis (118). Similarly, arsenic-mediated overexpression of SQSTM1/p62 triggers positive feedback between SQSTM1/p62 and NRF2, which causes the expression of numerous secondary messengers, including the glutamate-cysteine ligase catalytic subunit, HO-1 and NQO1, and promotes keratinocyte proliferation and malignant transformation (119). Therefore, inducing the oncolytic virus ΔPK to degrade SQSTM1/p62 by activating the calcium protease calpain pathway, not the autophagy pathway, may be a novel therapeutic approach for treating tumor stem cell-rich malignant melanomas (120). Nevertheless, SQSTM1/p62, as a direct target of miR-372, can suppress cancer cell migration by inducing NQO1 expression to repress ROS activation in head and neck squamous cell carcinoma (121).
Collectively, these observations imply that SQSTM1/p62 expression is typically higher in the cytoplasm than in the nucleus, usually indicating that SQSTM1/p62 participates in a series of downstream cascades involved in oncogene activation. Thus, inhibiting SQSTM1/p62 activation in different ways or decreasing its expression may be an effective treatment against head and neck carcinoma and skin cancer.
10. Nerve and brain tumors
The SQSTM1/p62/NRF2 axis may play a central role in nerve and brain tumors by participating in autophagy. Increasing evidence shows that SQSTM1/p62 can cause neuronal degeneration and neural stem cell differentiation through its abnormal aggregation and the inhibition of superoxide dismutase (122-124). Additionally, SQSTM1/p62 overexpression, along with NRF2 activation, is capable of stimulating classical macroautophagy and mitochondrial autophagy, which allow neuroblastoma cell and glioblastoma multiforme cell survival by antagonizing apoptosis, resulting in drug resistance to proteasome inhibitors (125,126). As a result, deleting the SQSTM1/p62 UBA domain to abolish the defensive function of autophagy and promote apoptosis may be a potential and powerful way to inhibit the proteasome in these tumors (127).
In summary, SQSTM1/p62 plays an oncogenic role in these tumors by engaging in macroautophagy and mitochondrial autophagy to prevent apoptosis, thereby contributing to uncontrolled cell growth and unresponsiveness to chemotherapeutic drugs.
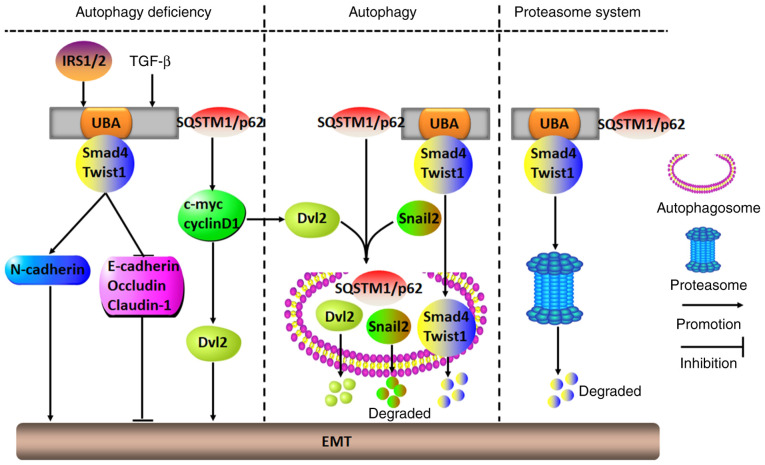
11. EMT
Recently, numerous studies have shown that SQSTM1/p62 participates in EMT-modulated malignant transformation (128-132). Tumor cells undergoing EMT shift from being in an epithelial state to being in a mesenchymal state, and in the mesenchymal state, a variety of signaling pathways are activated, including TGF-β/Smad and Wnt/β-catenin pathways, which is followed by decreased expression of epithelial markers, including E-cadherin and occludin, and by increased expression of mesenchymal markers, including vimentin, Twist1, snail1/2 and N-cadherin. The EMT renders most cancer cells aggressive (128). For instance, in squamous cell carcinoma of the skin, malignant melanoma, hepatic carcinoma, breast cancer and bladder cancer, TGF-β-mediated SQSTM1/p62 aggregation in the cytoplasm of autophagy-defective cells can initiate the EMT process to exacerbate cancer cell growth, proliferation, migration, invasion and distant metastasis. As SQSTM1/p62 binds to Smad4 and Twist1 through its UBA domain to prevent degradation by autophagy and the proteasome pathway, the activation of Smad4 and Twist1 causes increased N-cadherin and decreased E-cadherin, occludin and claudin-1 expression, ultimately stimulating the EMT (129-131). In addition, SQSTM1/p62 can inhibit autophagy and promote the EMT to accelerate tumor invasion and metastasis by directly binding with HDAC6 to impair the acetylation of α-tubulin and microtubules, which leads to disrupted fusion of autophagosomes and lysosomes, and subsequently to the impairment of autophagosomal degradation (132). Similarly, in breast cancer MCF-7 cells, in the presence of insulin receptor substrate 1/2, SQSTM1/p62 releases disheveled 2 (Dvl2) to trigger the EMT by activating the EMT-related molecules c-Myc and cyclinD1, which are involved in the Wnt/β-catenin signaling pathway. However, when autophagy is activated, SQSTM1/p62 transports both Dvl2 and snail2 into autophagosomes, where they are degraded, thereby inhibiting EMT initiation (133,134).
In summary, functioning as a proto-oncogene or tumor suppressor gene, SQSTM1/p62 can play a dual role in activating or silencing EMT. The prerequisite condition for exerting its biological functions depends largely on whether autophagy is activated. When autophagy is ongoing, SQSTM1/p62 can block the EMT process and malignant cancer progression by promoting the degradation of EMT-related factors through the autophagy pathway. By contrast, when autophagy is inhibited or damaged, SQSTM1/p62 activates the EMT signaling pathways and promotes cancer cell malignant transformation by binding to and stabilizing EMT-related factors (Fig. 7).
Figure 7.
Molecular mechanisms of SQSTM1/p62 in regulating the tumor EMT. In numerous tumor cells with deficient autophagy, IRS1/2- or TGF-β-induced SQSTM1/p62 aggregation may initiate the EMT to accelerate malignant aggression by either binding to and stabilizing Smad4 and Twist1 through its UBA domain or stimulating c-myc and cyclinD1 to release Dvl2. However, when cells show autophagic ability, SQSTM1/p62 may act as a carrier to transport EMT-related molecules, including Dvl2, snail2, Smad4 and Twist1, to autophagosomes for their degradation, which results in inhibition of the EMT. Alternatively, SQSTM1/p62 may also repress the EMT by directing Smad4 and Twist1 into proteasomes for degradation. UBA, ubiquitin-associated domain; Dvl2, Disheveled 2; EMT, epithelial-mesenchymal transition; SQSTM1, sequestosome 1; IRS1/2, insulin receptor substrate 1/2.
12. Conclusions and prospects
To date, the molecular mechanisms and functions of SQSTM1/p62 in regulating malignant progression are unclear, but SQSTM1/p62 has been shown to participate in the occurrence and development of various tumors in multiple ways, including by affecting genetic stability, transcription and post-transcriptional regulation, to modify and interact with different molecules.
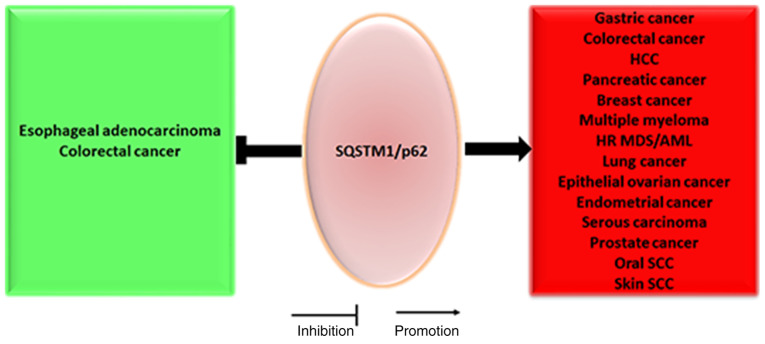
For instance, at the transcriptional level, either NRF2 or TNF may upregulate SQSTM1/p62 mRNA expression to facilitate hepatocellular malignant transformation of hepatocytes or bronchial epithelial cells by directly binding the promoter region of SQSTM1/p62 (40,79). At the post-transcriptional level, SQSTM1/p62 may be directly targeted by miR-372 to inhibit cancer cell migration in head and neck squamous cell carcinoma (121), and it may also be targeted by miR-487a to promote malignant progression of esophageal cancer cells (135). Notably, high levels of the lncRNA associated with small nucleolar RNA host gene 16 may increase SQSTM1/p62 expression by directly inhibiting its target molecule miR-17-5p, a miRNA that directly targets SQSTM1/p62, to facilitate HCC cell proliferation, migration and invasion (136). Since SQSTM1/p62 can function as an oncogene or tumor suppressor gene in different tumors under different circumstances (Fig. 8), SQSTM1/p62 can be considered to be like a multifunctional ship, ferry or car trafficking different bad or good cargos to different factories for further processing.
Figure 8.
SQSTM1/p62 plays an oncogenic and tumor-suppressive role in malignancy. SQSTM1/p62 plays an oncogenic role in esophageal adenocarcinoma and colorectal cancer, whereas SQSTM1/p62 primarily plays a tumor-suppressive role in a number of other tumors, such as gastric cancer, lung cancer, breast cancer and serous carcinoma. HCC, hepatocellular carcinoma; SQSTM1, sequestosome 1; HR MDS, high-risk myelodysplastic syndrome; AML, acute myeloid leukemia; SCC, squamous cell carcinoma.
Under normal conditions, SQSTM1/p62 expression is inversely associated with LC3-II expression when autophagy is activated; however, to a certain extent, elevated SQSTM1/p62 expression not only induces autophagy and autophagic cell death, but also inhibits autophagy, and vice versa. These results demonstrate that, in addition to the classical autophagy pathway, SQSTM1/p62 participates in a variety of signaling pathways to regulate tumor progression. However, until recently, the relationships between autophagy and SQSTM1/p62 had not been well known. Therefore, although guidelines for monitoring autophagy have been described, these data indicate that monitoring autophagic flux merely by assessing the fluctuation in SQSTM1/p62 and LC3-II levels is insufficient, and it is advisable to assess the functional status of the ubiquitin proteasome system (137). As SQSTM1/p62 acts as a bridge or an axis between the autophagy pathway and the ubiquitin-proteasome degradation pathway, when either pathway is disrupted, SQSTM1/p62 activity may be diverted to the other pathway.
In an overwhelming majority of cases, cancer cells with SQSTM1/p62 aggregation in the cytoplasm are more aggressive than those in which SQSTM1/p62 resides only in the nucleus, implying that excessively activated SQSTM1/p62 shuttling from the nucleus to the cytoplasm is involved in diverse signaling pathways, particularly the EMT-related TGF-β/Smad and Wnt/β-catenin, SQSTM1/p62/KEAP1/NRF2 and SQSTM1/p62/TRAF6/NF-κB pathways. The formation of positive feedback loops clearly amplifies the effects of carcinogenesis, which results in pleiotropic effects on the clinicopathological parameters of human tumors (Table I). As drug resistance in malignant cancer is the most critical challenge to effective treatment encountered thus far, it is of great significance to search for specific molecule-targeted drugs. It is promising that recent studies have demonstrated that XRK3F2, a novel small-molecule inhibitor specifically targeting the SQSTM1/p62-ZZ domain, can selectively impair leukemia-initiating cells in AML and multiple myeloma cells to inhibit tumor aggression by inhibiting the SQSTM1/p62-ZZ domain-dependent autophagic or TNFα plus IL17 signaling pathways, respectively; therefore, XRK3F2 may be, to some extent, a potentially useful tool for targeting therapies specifically to these tumors in the future (138,139). In addition, the crosstalk between components in tumor microenvironments (SQSTM1/p62 and various autocrine or paracrine factors originating from tumor stromal cells, etc.) and tumor cells may markedly increase the expression of SQSTM1/p62 in tumor cells by augmenting or inhibiting specific signaling pathways, as well as by boosting tumor stem cell self-renewal, malignant tumor growth and aggression, and chemoradiotherapeutic resistance. To date, the communication between tumor microenvironments and tumor cells has been unclear, and further understanding may elucidate the underlying mechanisms of SQSTM1/p62 serving as a bridge between tumor microenvironments and tumor cells. A SQSTM1/p62 DNA vaccine or gene fusion vaccine based on the SQSTM1/p62 gene and tumor antigens has already exhibited complementary antitumor effects and few adverse effects in experimental subjects, such as dogs, mice, and rats, as well as in breast cancer, lung cancer, melanoma, ovarian cancer, renal cancer and sarcoma; these effects were achieved by augmenting immunoreactions among CD3-positive cells surrounding the tumor cells and stimulating a fibrotic response around the tumor cells (140-143). Therefore, unmasking the underlying mechanisms of SQSTM1/p62 in regulating the tumor microenvironment and tumor cells may be very worthwhile and lead to an increase in the effective precision therapy treatments available for various tumors in the future.
Table I.
Clinicopathological significance and biological pathways of SQSTM1/p62 in human tumors.
| Tumors | Location and expression | Pathways | Vessel invasion | Lymph node and distant metastasis | TNM stage | Chemoresistance | Prognosis | (Refs.) |
|---|---|---|---|---|---|---|---|---|
| Esophageal adenocarcinoma | Cytoplasm ↑, Nucleus↑ | N.A. | N.A. | Positive | N.A. | N.A. | Good | (25) |
| Gastric cancer | Cytoplasm ↑, Nucleus↑ | N.A. | Positive | Positive or Negative | Positive or No | N.A. | Poor | (24,26) |
| Colorectal cancer | Cytoplasm↑, Nucleus↑or↓ | Autophagy, MEK/ERK PI3K/mTOR, Wnt/β-catenin, ATM/γH2AX | N.A. | No | No | Yes | Good or No | (24,27, 29-32,34) |
| Pancreatic adenocarcinoma | Cytoplasm ↑, Nucleus↑ | N.A. | N.A. | No | No | N.A. | No | (24) |
| HCC | Cytoplasm↑ | Vitamin D, SQSTM1/p62/KEAP1/NRF2, SQSTM1/p62/TRAF6/NF-κB, SQSTM1/p62/HDAC6 | N.A. | N.A. | N.A. | Yes | Poor | (36-40, 42-44) |
| Breast cancer | Cytoplasm ↑, Nucleus↑ | CD44/SQSTM1/p62/NRF2, SQSTM1/p62/KEAP1/NRF2, SQSTM1/p62/TRAF6/NF-κB, PI3K/AKT Wnt/β-catenin, AKT/GSK3β/β-catenin, ER/ERK, Wnt/PCP, IRS1/2/SQSTM1/P62/Dvl2, TRAF6-Beclin1 | Positive | Positive | Positive or No | Yes | Poor | (46-55, 57-61, 90,133) |
| Multiple myeloma | Cytoplasm↑ | SQSTM1/P62/TRAF6/NF-κB, SQSTM1/P62/RIP1/NF-κB, SQSTM1/P62/aPKCs/NF-κB, SQSTM1/p62/P38MAPK | N.A. | N.A. | N.A. | Yes | Poor | (62-66, 69,70) |
| HR MDS/AML | Cytoplasm↑, Nucleus↑ | SQSTM1/p62/TRAF6/NF-κB | N.A. | N.A. | N.A. | N.A. | Poor | (62,63) |
| Chronic myeloid leukemia | Cytoplasm↑ | Autophagy | N.A. | N.A. | N.A. | No | N.A. | (73) |
| Lung cancer | Cytoplasm↑ | mTOR-ULK1-Beclin1, SQSTM1/p62/TRAF6/NF-κB, SQSTM1/p62/KEAP1/NRF2, TRAF6-Beclin1 | N.A. | Negative | Negative | Yes | Poor | (77-83,90) |
| Epithelial ovarian cancer | Cytoplasm↑ or↓ | SQSTM1/p62/KEAP1/NRF2, SQSTM1/p62/caspase 8 | N.A. | Positive | Positivea | Yes | Poor | (91,92, 94,99, 100) |
| Seminoma | Cytoplasm/Nucleus, NC | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. | (95) |
| Non-seminoma | Cytoplasm/Nucleus, NC | N.A. | N.A. | N.A. | N.A. | N.A. | N.A. | (95) |
| Endometrial cancer | Cytoplasm↑ | N.A. | N.A. | Positive | No | N.A. | Poor | (89) |
| Serous carcinoma | Cytoplasm↑ | N.A. | N.A. | Positive | No | N.A. | Poor | (96) |
| Cervical cancer | Cytoplasm ↑, Nucleus↑ | SQSTM1/p62/NRF2 | N.A. | N.A. | N.A. | N.A. | N.A. | (97) |
| Prostate cancer | Cytoplasm↑ | SQSTM1/p62/NRF2, Raf/MEK/ERK, SQSTM1/p62/MEKK3, SQSTM1/P62/TRAF6/mTORC1, SQSTM1/p62/mTORC1/c-myc/GSH/IL-6/TGFβ, AMPK/SQSTM1/p62/AR | N.A. | Positive | Positive | Yes | Poor | (101,103, 105-111) |
| Malignant melanoma | Cytoplasm↑ | Raf/MEK/ERK, EGF/TGFβ/Twist1 | N.A. | N.A. | N.A. | N.A. | N.A. | (105,128) |
| Renal cell carcinoma | Cytoplasm↑ | Glycolytic pathway, Autophagic pathway, Proteasome degradation pathway | N.A. | N.A. | N.A. | N.A. | N.A. | (112,113) |
| Bladder cancer | Cytoplasm↑ | TGFβ/Smad4, SQSTM1/p62/KEAP1/NRF2 | N.A. | N.A. | N.A. | N.A. | N.A. | (115,129) |
| Oral SCC | Cytoplasm↑, Nucleus↓ | N.A. | Positive | Positive | Positive | N.A. | Poor | (115) |
| Head and neck SCC | Cytoplasm↑ | PI3K/AKT, SQSTM1/p62/NRF2 | N.A. | N.A. | N.A. | Yes | N.A. | (116) |
| Skin SCC | Cytoplasm↓ or↑ | SQSTM1/P62/NRF2, EGF/TGFβ/Twist1 | N.A. | Negative | Negative | N.A. | Poor | (117-119, 129) |
| Neuroblastoma mitophagy | Cytoplasm↑ | SQSTM1/P62/NRF2, | N.A. | N.A. | N.A. | Yes | N.A. | (125) |
| Glioblastoma | Cytoplasm↑ | SQSTM1/P62/NF-κB, autophagy | N.A. | N.A. | N.A. | Yes | N.A. | (126) |
FIGO stage. SQSTM1, sequestosome 1; NC, no change; SCC, squamous cell carcinoma; HCC, hepatocellular carcinoma; HR MDS, higher-risk myelodysplastic syndromes; AML, acute myeloid leukemia; ↑, upregulation; ↓, downregulation; N.A, not available; No, no relationship; MEK, mitogen-activated protein kinase kinase; ERK, extracellular-signal-regulated kinase; MEKK3, mitogen-activated protein kinase kinase kinase 3, PI3K, phosphoinositol-3 kinase; AKT, protein kinase B; AMPK, AMP-activated protein kinase; mTOR, mammalian target of rapamycin; mTORC1, mTOR complex 1; RIP1, receptor interacting protein 1; ULK1, Unc-51 like autophagy activating kinase; NRF2, nuclear factor erythroid 2; TRAF6, TNF receptor-associated factor 6; KEAP1, Kelch-like ECH-associated protein 1; HDAC6, histone deacetylase 6; ATM, ataxia-telangiectasia mutated; γH2AX, phosphorylated histone family member X; GSH, glutathione; PCP, planar cell polarity; Dvl2, disheveled 2; KLF4, Kruppel-like factor 4; GSK3β, glycogen synthase kinase-3β; TGFβ, transforming growth factor-beta; EGF, epidermal growth factor; NF-κB, nuclear factor-κB; IRS1/2, insulin receptor substrate 1/2; aPKCs, atypical protein kinase Cs; Raf, resistance allele frequencies; AR, androgen receptor.
Acknowledgments
The authors would like to thank Professor Iain Charles Bruce (College of Medicine, Zhejiang University, Hangzhou, China) for providing English language assistance.
Abbreviations
- SQSTM1
sequestosome 1
- UBA
ubiquitin-associated domain
- PB1
Phox and Bem1p
- LIR
LC3-interacting region
- HSP90
heat shock protein 90
- HCC
hepatocellular carcinoma
- NRF2
nuclear factor erythroid 2
- TRAF6
TNF receptor-associated factor 6
- ARE
antioxidant response element
- JNK
Jun N-terminal kinase
- KEAP1
Kelch-like ECH-associated protein 1
- NQO1
NAD(P)H dehydrogenase quinone 1
- HDAC6
histone deacetylase 6
- CSC
cancer stem cell
- ROS
reactive oxygen species
- PI3K
phosphoinositol-3 kinase
- KLF4
Kruppel-like factor 4
- ATRA
all-trans retinoic acid
- AML
acute myeloid leukemia
- NSCLC
non-small cell lung cancer
- NBR1
neighbor of BRCA1
- Ack1
activate Cdc42-associated kinase 1
- CAFs
cancer-associated fibroblasts
- EMT
epithelial-mesenchymal transition
- AR
androgen
- HIF1α
hypoxia-inducible factor 1α
- Dvl2
disheveled 2
- ECSIT
evolutionarily conserved signaling intermediate in toll pathways
Funding Statement
This study was supported by the Key Technology Research and Development Program of Zhejiang Province (grant no. 2017C03017), the National Natural Science Foundation of China (grant no. 81772545), the Project of the Regional Diagnosis and Treatment Center of the Health Planning Committee (grant no. JBZX-201903), the Zhejiang Provincial Natural Science Foundation of China (grant no. LZ21H160001) and the CAMS Innovation Fund for Medical Sciences (grant no. 2019-I2M-5-044).
Availability of data and materials
Not applicable.
Authors' contributions
JT drafted the manuscript and YL created the figures; all the images have been drawn using with Pathway Builder Tool 2.0 (www.proteinlounge.com) and ScienceSlides 2016 version (www.visiscience.com). SX and YL participated in the literature collection, JL and QY designed and created the tables, and HZ and KD designed the study and supervised the creation of the manuscript. All authors have read and approved the final manuscript. Data authentication is not applicable.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Park I, Chung J, Walsh CT, Yun Y, Strominger JL, Shin J. Phosphotyrosine-independent binding of a 62-kDa protein to the src homology 2 (SH2) domain of p56lck and its regulation by phosphorylation of Ser-59 in the lck unique N-terminal region. Proc Natl Acad Sci USA. 1995;92:12338–12342. doi: 10.1073/pnas.92.26.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishaq M, Khan MA, Sharma K, Sharma G, Dutta RK, Majumdar S. Gambogic acid induced oxidative stress dependent caspase activation regulates both apoptosis and autophagy by targeting various key molecules (NF-κB, Beclin-1, p62 and NBR1) in human bladder cancer cells. Biochim Biophys Acta. 2014;1840:3374–3384. doi: 10.1016/j.bbagen.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Li S, Yang G, Zhu X, Cheng L, Sun Y, Zhao Z. Combination of rapamycin and garlic-derived S-allylmercaptocysteine induces colon cancer cell apoptosis and suppresses tumor growth in xenograft nude mice through autophagy/p62/Nrf2 pathway. Oncol Rep. 2017;38:1637–1644. doi: 10.3892/or.2017.5849. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Zhang N, Zhang L, Li R, Fu W, Ma K, Li X, Wang L, Wang J, Zhang H, et al. Autophagy regulates chromatin ubiquitination in DNA damage response through elimination of SQSTM1/p62. Mol Cell. 2016;63:34–48. doi: 10.1016/j.molcel.2016.05.027. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y, Weihl CC. Regulation of SQSTM1/p62 via UBA domain ubiquitination and its role in disease. Autophagy. 2017;13:1615–1616. doi: 10.1080/15548627.2017.1339845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohen-Kaplan V, Livneh I, Avni N, Fabre B, Ziv T, Kwon YT, Ciechanover A. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc Natl Acad Sci USA. 2016;113:E7490–E7499. doi: 10.1073/pnas.1615455113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, Abeliovich H, Abildgaard MH, Abudu YP, Acevedo-Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition) 1. Autophagy. 2021;17:1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumoto G, Wada K, Okuno M, Kurosawa M, Nukina N. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011;44:279–289. doi: 10.1016/j.molcel.2011.07.039. [DOI] [PubMed] [Google Scholar]
- 11.Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D, Xu W, Jiang T, Feng L, Shin VY, et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019;10:383. doi: 10.1038/s41419-019-1585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clausen TH, Lamark T, Isakson P, Finley K, Larsen KB, Brech A, Øvervatn A, Stenmark H, Bjørkøy G, Simonsen A, Johansen T. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010;6:330–344. doi: 10.4161/auto.6.3.11226. [DOI] [PubMed] [Google Scholar]
- 13.Ichimura Y, Waguri S, Sou YS, Kageyama S, Hasegawa J, Ishimura R, Saito T, Yang Y, Kouno T, Fukutomi T, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Park JY, Sohn HY, Koh YH, Jo C. Curcumin activates Nrf2 through PKCδ-mediated p62 phosphorylation at Ser351. Sci Rep. 2021;11:8430. doi: 10.1038/s41598-021-87225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ling J, Kang Y, Zhao R, Xia Q, Lee DF, Chang Z, Li J, Peng B, Fleming JB, Wang H, et al. KrasG12D-induced IKK2/β/NF-κB activation by IL-1α and p62 feedforward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:105–120. doi: 10.1016/j.ccr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura K, Kimple AJ, Siderovski DP, Johnson GL. PB1 domain interaction of p62/sequestosome 1 and MEKK3 regulates NF-kappaB activation. J Biol Chem. 2010;285:2077–2089. doi: 10.1074/jbc.M109.065102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Usategui-Martin R, Gestoso-Uzal N, Calero-Paniagua I, De Pereda JM, Del Pino-Montes J, González-Sarmiento R. A mutation in p62 protein (R321C), associated to Paget's disease of bone, causes a blockade of autophagy and an activation of NF-kB pathway. Bone. 2020;133:115265. doi: 10.1016/j.bone.2020.115265. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Xiong H, Liu D, Hill C, Ertay A, Li J, Zou Y, Miller P, White E, Downward J, et al. Autophagy inhibition specifically promotes epithelial-mesenchymal transition and invasion in RAS-mutated cancer cells. Autophagy. 2019;15:886–899. doi: 10.1080/15548627.2019.1569912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua F, Hu ZW. TRIB3-P62 interaction, diabetes and autophagy. Oncotarget. 2015;6:34061–34062. doi: 10.18632/oncotarget.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hewitt G, Carroll B, Sarallah R, Correia-Melo C, Ogrodnik M, Nelson G, Otten EG, Manni D, Antrobus R, Morgan BA, et al. SQSTM1/p62 mediates crosstalk between autophagy and the UPS in DNA repair. Autophagy. 2016;12:1917–1930. doi: 10.1080/15548627.2016.1210368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Zhu WG, Zhao Y. Autophagy substrate SQSTM1/p62 regulates chromatin ubiquitination during the DNA damage response. Autophagy. 2017;13:212–213. doi: 10.1080/15548627.2016.1245262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Howell MEA, Sparks-Wallace A, Hawkins C, Nicksic CA, Kohne C, Hall KH, Moorman JP, Yao ZQ, Ning S. p62-mediated selective autophagy endows virus-transformed cells with insusceptibility to DNA damage under oxidative stress. PLoS Pathog. 2019;15:e1007541. doi: 10.1371/journal.ppat.1007541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohamed A, Ayman A, Deniece J, Wang T, Kovach C, Siddiqui MT, Cohen C. P62/Ubiquitin IHC expression correlated with clinicopathologic parameters and outcome in gastrointestinal carcinomas. Front Oncol. 2015;5:70. doi: 10.3389/fonc.2015.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams O, Dislich B, Berezowska S, Schläfli AM, Seiler CA, Kröll D, Tschan MP, Langer R. Prognostic relevance of autophagy markers LC3B and p62 in esophageal adenocarcinomas. Oncotarget. 2016;7:39241–39255. doi: 10.18632/oncotarget.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuda GO, Yashiro M, Kitayama K, Miki Y, Kasashima H, Kinoshita H, Morisaki T, Fukuoka T, Hasegawa T, Sakurai K, et al. Clinicopathological correlations of autophagy-related proteins LC3, beclin 1 and p62 in gastric cancer. Anticancer Res. 2016;36:129–136. [PubMed] [Google Scholar]
- 27.Park JM, Huang S, Wu TT, Foster NR, Sinicrope FA. Prognostic impact of beclin 1p62/sequestosome 1 and LC3 protein expression in colon carcinomas from patients receiving 5-fluorouracil as adjuvant chemotherapy. Cancer Biol Ther. 2013;14:100–107. doi: 10.4161/cbt.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosumi K, Masugi Y, Yang J, Qian ZR, Kim SA, Li W, Shi Y, da Silva A, Hamada T, Liu L, et al. Tumor SQSTM1 (p62) expression and T cells in colorectal cancer. Oncoimmunology. 2017;6:e1284720. doi: 10.1080/2162402X.2017.1284720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitz KJ, Ademi C, Bertram S, Schmid KW, Baba HA. Prognostic relevance of autophagy-related markers LC3, p62/sequestosome 1, beclin-1 and ULK1 in colorectal cancer patients with respect to KRAS mutational status. World J Surg Oncol. 2016;14:189. doi: 10.1186/s12957-016-0946-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goulielmaki M, Koustas E, Moysidou E, Vlassi M, Sasazuki T, Shirasawa S, Zografos G, Oikonomou E, Pintzas A. BRAF associated autophagy exploitation: BRAF and autophagy inhibitors synergise to efficiently overcome resistance of BRAF mutant colorectal cancer cells. Oncotarget. 2016;7:9188–9221. doi: 10.18632/oncotarget.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren F, Shu G, Liu G, Liu D, Zhou J, Yuan L, Zhou J. Knockdown of p62/sequestosome 1 attenuates autophagy and inhibits colorectal cancer cell growth. Mol Cell Biochem. 2014;385:95–102. doi: 10.1007/s11010-013-1818-0. [DOI] [PubMed] [Google Scholar]
- 32.Petherick KJ, Williams AC, Lane JD, Ordóñez-Morán P, Huelsken J, Collard TJ, Smartt HJ, Batson J, Malik K, Paraskeva C, Greenhough A. Autolysosomal β-catenin degradation regulates Wnt-autophagy-p62 crosstalk. EMBO J. 2013;32:1903–1916. doi: 10.1038/emboj.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samarasinghe B, Wales CT, Taylor FR, Jacobs AT. Heat shock factor 1 confers resistance to Hsp90 inhibitors through p62/SQSTM1 expression and promotion of autophagic flux. Biochem Pharmacol. 2014;87:445–455. doi: 10.1016/j.bcp.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Z, Chen Q, Li B, Xie JM, Yang XD, Zhao K, Wu Y, Ye ZY, Chen ZR, Qin ZH, et al. Escin-induced DNA damage promotes escin-induced apoptosis in human colorectal cancer cells via p62 regulation of the ATM/γH2AX pathway. Acta Pharmacol Sin. 2018;39:1645–1660. doi: 10.1038/aps.2017.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JH, Kim IW. p62 manipulation affects chlorin e6-mediated photodynamic therapy efficacy in colorectal cancer cell lines. Oncol Lett. 2020;19:3907–3916. doi: 10.3892/ol.2020.11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duran A, Hernandez ED, Reina-Campos M, Castilla EA, Subramaniam S, Raghunandan S, Roberts LR, Kisseleva T, Karin M, Diaz-Meco MT, Moscat J. p62/SQSTM1 by binding to vitamin D receptor inhibits hepatic stellate cell activity, fibrosis, and liver cancer. Cancer Cell. 2016;30:595–609. doi: 10.1016/j.ccell.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimizu T, Inoue K, Hachiya H, Shibuya N, Aoki T, Kubota K. Accumulation of phosphorylated p62 is associated with NF-E2-related factor 2 activation in hepatocellular carcinoma. J Hepatobiliary Pancreat Sci. 2016;23:467–471. doi: 10.1002/jhbp.364. [DOI] [PubMed] [Google Scholar]
- 38.Saito T, Ichimura Y, Taguchi K, Suzuki T, Mizushima T, Takagi K, Hirose Y, Nagahashi M, Iso T, Fukutomi T, et al. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun. 2016;7:12030. doi: 10.1038/ncomms12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Øvervatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Umemura A, He F, Taniguchi K, Nakagawa H, Yamachika S, Font-Burgada J, Zhong Z, Subramaniam S, Raghunandan S, Duran A, et al. p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell. 2016;29:935–948. doi: 10.1016/j.ccell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoadley KA, Yau C, Wolf DM, Cherniack AD, Tamborero D, Ng S, Leiserson MDM, Niu B, McLellan MD, Uzunangelov V, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taguchi K, Fujikawa N, Komatsu M, Ishii T, Unno M, Akaike T, Motohashi H, Yamamoto M. Keap1 degradation by autophagy for the maintenance of redox homeostasis. Proc Natl Acad Sci USA. 2012;109:13561–13566. doi: 10.1073/pnas.1121572109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vegliante R, Desideri E, Di Leo L, Ciriolo MR. Dehydroepiandrosterone triggers autophagic cell death in human hepatoma cell line HepG2 via JNK-mediated p62/SQSTM1 expression. Carcinogenesis. 2016;37:233–244. doi: 10.1093/carcin/bgw003. [DOI] [PubMed] [Google Scholar]
- 44.Yan J, Seibenhener ML, Calderilla-Barbosa L, Diaz-Meco MT, Moscat J, Jiang J, Wooten MW, Wooten MC. SQSTM1/p62 interacts with HDAC6 and regulates deacetylase activity. PLoS One. 2013;8:e76016. doi: 10.1371/journal.pone.0076016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Q, Yue F, Li W, Zou J, Xu T, Huang C, Zhang Y, Song K, Huang G, Xu G, et al. Potassium bisperoxo(1,10-phenanthroline) oxovanadate (bpV(phen)) induces apoptosis and pyroptosis and disrupts the P62-HDAC6 protein interaction to suppress the acetylated microtubule-dependent degradation of autophagosomes. J Biol Chem. 2015;290:26051–26058. doi: 10.1074/jbc.M115.653568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryoo IG, Choi BH, Ku SK, Kwak MK. High CD44 expression mediates p62-associated NFE2L2/NRF2 activation in breast cancer stem cell-like cells: Implications for cancer stem cell resistance. Redox Biol. 2018;17:246–258. doi: 10.1016/j.redox.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryoo IG, Choi BH, Kwak MK. Activation of NRF2 by p62 and proteasome reduction in sphere-forming breast carcinoma cells. Oncotarget. 2015;6:8167–8184. doi: 10.18632/oncotarget.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jain A, Rusten TE, Katheder N, Elvenes J, Bruun JA, Sjøttem E, Lamark T, Johansen T. p62/sequestosome-1, autophagy-related gene 8, and autophagy in drosophila are regulated by nuclear factor erythroid 2-related factor 2 (NRF2), independent of transcription factor TFEB. J Biol Chem. 2015;290:14945–14962. doi: 10.1074/jbc.M115.656116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu LZ, Li SS, Zhou W, Kang ZJ, Zhang QX, Kamran M, Xu J, Liang DP, Wang CL, Hou ZJ, et al. p62/SQSTM1 enhances breast cancer stem-like properties by stabilizing MYC mRNA. Oncogene. 2017;36:304–317. doi: 10.1038/onc.2016.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao C, Cao W, Bao L, Zuo W, Xie G, Cai T, Fu W, Zhang J, Wu W, Zhang X, Chen YG. Autophagy negatively regulates Wnt signalling by promoting dishevelled degradation. Nat Cell Biol. 2010;12:781–790. doi: 10.1038/ncb2082. [DOI] [PubMed] [Google Scholar]
- 51.Ahn JS, Ann EJ, Kim MY, Yoon JH, Lee HJ, Jo EH, Lee K, Lee JS, Park HS. Autophagy negatively regulates tumor cell proliferation through phosphorylation dependent degradation of the Notch1 intracellular domain. Oncotarget. 2016;7:79047–79063. doi: 10.18632/oncotarget.12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cai-McRae X, Karantza V. p62: A hub of multiple signaling pathways in HER2-induced mammary tumorigenesis. Mol Cell Oncol. 2015;2:e975035. doi: 10.4161/23723556.2014.975035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cai-McRae X, Zhong H, Karantza V. Sequestosome 1/p62 facilitates HER2-induced mammary tumorigenesis through multiple signaling pathways. Oncogene. 2015;34:2968–2977. doi: 10.1038/onc.2014.244. [DOI] [PubMed] [Google Scholar]
- 54.Wei H, Wang C, Croce CM, Guan JL. p62/SQSTM1 synergizes with autophagy for tumor growth in vivo. Genes Dev. 2014;28:1204–1216. doi: 10.1101/gad.237354.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Puvirajesinghe TM, Bertucci F, Jain A, Scerbo P, Belotti E, Audebert S, Sebbagh M, Lopez M, Brech A, Finetti P, et al. Identification of p62/SQSTM1 as a component of non-canonical Wnt VANGL2-JNK signalling in breast cancer. Nat Commun. 2016;7:10318. doi: 10.1038/ncomms10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen S, Zhou L, Zhang Y, Leng Y, Pei XY, Lin H, Jones R, Orlowski RZ, Dai Y, Grant S. Targeting SQSTM1/p62 induces cargo loading failure and converts autophagy to apoptosis via NBK/Bik. Mol Cell Biol. 2014;34:3435–3449. doi: 10.1128/MCB.01383-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi YK, Cho SG, Woo SM, Yun YJ, Park S, Shin YC, Ko SG. Herbal extract SH003 suppresses tumor growth and metastasis of MDA-MB-231 breast cancer cells by inhibiting STAT3-IL-6 signaling. Mediators Inflamm. 2014;2014:492173. doi: 10.1155/2014/492173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo RZ, Yuan ZY, Li M, Xi SY, Fu J, He J. Accumulation of p62 is associated with poor prognosis in patients with triple-negative breast cancer. Onco Targets Ther. 2013;6:883–888. doi: 10.2147/OTT.S46222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei Y, Liu D, Jin X, Gao P, Wang Q, Zhang J, Zhang N. PA-MSHA inhibits the growth of doxorubicin-resistant MCF-7/ADR human breast cancer cells by downregulating Nrf2/p62. Cancer Med. 2016;5:3520–3531. doi: 10.1002/cam4.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fuchinoue F, Hirotani Y, Nakanishi Y, Yamaguchi H, Nishimaki H, Noda H, Tang XY, Iizuka M, Amano S, Sugitani M, et al. Overexpression of PGC1α and accumulation of p62 in apocrine carcinoma of the breast. Pathol Int. 2015;65:19–26. doi: 10.1111/pin.12235. [DOI] [PubMed] [Google Scholar]
- 61.Nozaki F, Hirotani Y, Nakanishi Y, Yamaguchi H, Nishimaki H, Noda H, Tang X, Yamamoto H, Suzuki A, Seki T, Masuda S. p62 regulates the proliferation of molecular apocrine breast cancer cells. Acta Histochem Cytochem. 2016;49:125–130. doi: 10.1267/ahc.16013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen P, Chen M, He M, Chen L, Song Y, Xiao P, Wan X, Dai F, Pan T, Wang Q. Inhibition of ERα/ERK/P62 cascades induces 'autophagic switch' in the estrogen receptor-positive breast cancer cells exposed to gemcitabine. Oncotarget. 2016;7:48501–48516. doi: 10.18632/oncotarget.10363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang J, Starczynowski DT. Genomic instability establishes dependencies on acquired gene regulatory networks: A novel role of p62 in myeloid malignancies with del(5q) Mol Cell Oncol. 2015;2:e1014219. doi: 10.1080/23723556.2015.1014219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fang J, Barker B, Bolanos L, Liu X, Jerez A, Makishima H, Christie S, Chen X, Rao DS, Grimes HL, et al. Myeloid malignancies with chromosome 5q deletions acquire a dependency on an intrachromosomal NF-κB gene network. Cell Rep. 2014;8:1328–1338. doi: 10.1016/j.celrep.2014.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1TRAF6 pathway. EMBO J. 2000;19:1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Teramachi J, Silbermann R, Yang P, Zhao W, Mohammad KS, Guo J, Anderson JL, Zhou D, Feng R, Myint KZ, et al. Blocking the ZZ domain of sequestosome1/p62 suppresses myeloma growth and osteoclast formation in vitro and induces dramatic bone formation in myeloma-bearing bones in vivo. Leukemia. 2016;30:390–398. doi: 10.1038/leu.2015.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sanz L, Sanchez P, Lallena MJ, Diaz-Meco MT, Moscat J. The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J. 1999;18:3044–3053. doi: 10.1093/emboj/18.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rubio N, Verrax J, Dewaele M, Verfaillie T, Johansen T, Piette J, Agostinis P. p38(MAPK)-regulated induction of p62 and NBR1 after photodynamic therapy promotes autophagic clearance of ubiquitin aggregates and reduces reactive oxygen species levels by supporting Nrf2-antioxidant signaling. Free Radic Biol Med. 2014;67:292–303. doi: 10.1016/j.freeradbiomed.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 69.Chang KH, Sengupta A, Nayak RC, Duran A, Lee SJ, Pratt RG, Wellendorf AM, Hill SE, Watkins M, Gonzalez-Nieto D, et al. p62 is required for stem cell/progenitor retention through inhibition of IKK/NF-κB/Ccl4 signaling at the bone marrow macrophage-osteoblast niche. Cell Rep. 2014;9:2084–2097. doi: 10.1016/j.celrep.2014.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Milan E, Perini T, Resnati M, Orfanelli U, Oliva L, Raimondi A, Cascio P, Bachi A, Marcatti M, Ciceri F, Cenci S. A plastic SQSTM1/p62-dependent autophagic reserve maintains proteostasis and determines proteasome inhibitor susceptibility in multiple myeloma cells. Autophagy. 2015;11:1161–1178. doi: 10.1080/15548627.2015.1052928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Riz I, Hawley TS, Hawley RG. KLF4-SQSTM1/p62-associated prosurvival autophagy contributes to carfilzomib resistance in multiple myeloma models. Oncotarget. 2015;6:14814–14831. doi: 10.18632/oncotarget.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trocoli A, Bensadoun P, Richard E, Labrunie G, Merhi F, Schläfli AM, Brigger D, Souquere S, Pierron G, Pasquet JM, et al. p62/SQSTM1 upregulation constitutes a survival mechanism that occurs during granulocytic differentiation of acute myeloid leukemia cells. Cell Death Differ. 2014;21:1852–1861. doi: 10.1038/cdd.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ségal-Bendirdjian E, Tschan MP, Reiffers J, Djavaheri-Mergny M. Pro-survival role of p62 during granulocytic differentiation of acute myeloid leukemia cells. Mol Cell Oncol. 2014;1:e970066. doi: 10.4161/23723548.2014.970066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goussetis DJ, Gounaris E, Wu EJ, Vakana E, Sharma B, Bogyo M, Altman JK, Platanias LC. Autophagic degradation of the BCR-ABL oncoprotein and generation of antileukemic responses by arsenic trioxide. Blood. 2012;120:3555–3562. doi: 10.1182/blood-2012-01-402578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhong Z, Sanchez-Lopez E, Karin M. Autophagy, inflammation, and immunity: A Troika governing cancer and its treatment. Cell. 2016;166:288–298. doi: 10.1016/j.cell.2016.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moscat J, Karin M, Diaz-Meco MT. p62 in cancer: Signaling adaptor beyond autophagy. Cell. 2016;167:606–609. doi: 10.1016/j.cell.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang X, Du Z, Li L, Shi M, Yu Y. Beclin 1 and p62 expression in non-small cell lung cancer: Relation with malignant behaviors and clinical outcome. Int J Clin Exp Pathol. 2015;8:10644–10652. [PMC free article] [PubMed] [Google Scholar]
- 78.Schläfli AM, Adams O, Galván JA, Gugger M, Savic S, Bubendorf L, Schmid RA, Becker KF, Tschan MP, Langer R, Berezowska S. Prognostic value of the autophagy markers LC3 and p62/SQSTM1 in early-stage non-small cell lung cancer. Oncotarget. 2016;7:39544–39555. doi: 10.18632/oncotarget.9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang H, Zhu J, Li Y, Zhang L, Gu J, Xie Q, Jin H, Che X, Li J, Huang C, et al. Upregulation of SQSTM1/p62 contributes to nickel-induced malignant transformation of human bronchial epithelial cells. Autophagy. 2016;12:1687–1703. doi: 10.1080/15548627.2016.1196313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Linares JF, Duran A, Yajima T, Pasparakis M, Moscat J, Diaz-Meco MT. K63 polyubiquitination and activation of mTOR by the p62-TRAF6 complex in nutrient-activated cells. Mol Cell. 2013;51:283–296. doi: 10.1016/j.molcel.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lou JS, Yan L, Bi CW, Chan GK, Wu QY, Liu YL, Huang Y, Yao P, Du CY, Dong TT, Tsim KW. Yu Ping Feng San reverses cisplatin-induced multi-drug resistance in lung cancer cells via regulating drug transporters and p62/TRAF6 signalling. Sci Rep. 2016;6:31926. doi: 10.1038/srep31926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lau A, Zheng Y, Tao S, Wang H, Whitman SA, White E, Zhang DD. Arsenic inhibits autophagic flux, activating the Nrf2-Keap1 pathway in a p62-dependent manner. Mol Cell Biol. 2013;33:2436–2446. doi: 10.1128/MCB.01748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Son YO, Pratheeshkumar P, Roy RV, Hitron JA, Wang L, Zhang Z, Shi X. Nrf2/p62 signaling in apoptosis resistance and its role in cadmium-induced carcinogenesis. J Biol Chem. 2014;289:28660–28675. doi: 10.1074/jbc.M114.595496. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 84.Wang Y, Zhang J, Huang ZH, Huang XH, Zheng WB, Yin XF, Li YL, Li B, He QY. Isodeoxyelephantopin induces protective autophagy in lung cancer cells via Nrf2-p62-keap1 feedback loop. Cell Death Dis. 2017;8:e2876. doi: 10.1038/cddis.2017.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xia M, Gonzalez P, Li C, Meng G, Jiang A, Wang H, Gao Q, Debatin KM, Beltinger C, Wei J. Mitophagy enhances oncolytic measles virus replication by mitigating DDX58/RIG-I-like receptor signaling. J Virol. 2014;88:5152–5164. doi: 10.1128/JVI.03851-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nihira K, Miki Y, Ono K, Suzuki T, Sasano H. An inhibition of p62/SQSTM1 caused autophagic cell death of several human carcinoma cells. Cancer Sci. 2014;105:568–575. doi: 10.1111/cas.12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang J, Ma K, Qi T, Wei X, Zhang Q, Li G, Chiu JF. P62 regulates resveratrol-mediated Fas/Cav-1 complex formation and transition from autophagy to apoptosis. Oncotarget. 2015;6:789–801. doi: 10.18632/oncotarget.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu L, Xu F, Kong Q, Yang T, Tan D, Zhang X, Li N, Zhao S, Zhao J, Li M. Inhibition of p62/SQSTM1 sensitizes small-cell lung cancer cells to cisplatin-induced cytotoxicity by targeting NEDD9 expression. Mol Carcinog. 2020;59:967–979. doi: 10.1002/mc.23215. [DOI] [PubMed] [Google Scholar]
- 89.Kim MJ, Min Y, Im JS, Son J, Lee JS, Lee KY. p62 is negatively implicated in the TRAF6-BECN1 signaling axis for autophagy activation and cancer progression by toll-like receptor 4 (TLR4) Cells. 2020;9:1142. doi: 10.3390/cells9051142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim MJ, Min Y, Kwon J, Son J, Im JS, Shin J, Lee KY. p62 negatively regulates TLR4 signaling via functional regulation of the TRAF6-ECSIT complex. Immune Netw. 2019;19:e16. doi: 10.4110/in.2019.19.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li S, Wei Y. Association of HMGB1, BRCA1 and P62 expression in ovarian cancer and chemotherapy sensitivity. Oncol Lett. 2018;15:9572–9576. doi: 10.3892/ol.2018.8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Iwadate R, Inoue J, Tsuda H, Takano M, Furuya K, Hirasawa A, Aoki D, Inazawa J. High expression of SQSTM1/p62 protein is associated with poor prognosis in epithelial ovarian cancer. Acta Histochem Cytochem. 2014;47:295–301. doi: 10.1267/ahc.14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ju LL, Zhao CY, Ye KF, Yang H, Zhang J. Expression and clinical implication of beclin1, HMGB1, p62, survivin, BRCA1 and ERCC1 in epithelial ovarian tumor tissues. Eur Rev Med Pharmacol Sci. 2016;20:1993–2003. [PubMed] [Google Scholar]
- 94.Wang J, Garbutt C, Ma H, Gao P, Hornicek FJ, Kan Q, Shi H, Duan Z. Expression and role of autophagy-associated p62 (SQSTM1) in multidrug resistant ovarian cancer. Gynecol Oncol. 2018;150:143–150. doi: 10.1016/j.ygyno.2018.04.557. [DOI] [PubMed] [Google Scholar]
- 95.Bartsch G, Jennewein L, Harter PN, Antonietti P, Blaheta RA, Kvasnicka HM, Kögel D, Haferkamp A, Mittelbronn M, Mani J. Autophagy-associated proteins BAG3 and p62 in testicular cancer. Oncol Rep. 2016;35:1629–1635. doi: 10.3892/or.2015.4505. [DOI] [PubMed] [Google Scholar]
- 96.Iwadate R, Inoue J, Tsuda H, Takano M, Furuya K, Hirasawa A, Aoki D, Inazawa J. High expression of p62 protein is associated with poor prognosis and aggressive phenotypes in endometrial cancer. Am J Pathol. 2015;185:2523–2533. doi: 10.1016/j.ajpath.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 97.Darvekar SR, Elvenes J, Brenne HB, Johansen T, Sjøttem E. SPBP is a sulforaphane induced transcriptional coactivator of NRF2 regulating expression of the autophagy receptor p62/SQSTM1. PLoS One. 2014;9:e85262. doi: 10.1371/journal.pone.0085262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jung D, Khurana A, Roy D, Kalogera E, Bakkum-Gamez J, Chien J, Shridhar V. Quinacrine upregulates p21/p27 independent of p53 through autophagy-mediated downregulation of p62-Skp2 axis in ovarian cancer. Sci Rep. 2018;8:2487. doi: 10.1038/s41598-018-20531-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xia MH, Yan XY, Zhou L, Xu L, Zhang LC, Yi HW, Su J. p62 suppressed VK3-induced oxidative damage through Keap1/Nrf2 pathway in human ovarian cancer cells. J Cancer. 2020;11:1299–1307. doi: 10.7150/jca.34423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yan XY, Zhong XR, Yu SH, Zhang LC, Liu YN, Zhang Y, Sun LK, Su J. p62 aggregates mediated caspase 8 activation is responsible for progression of ovarian cancer. J Cell Mol Med. 2019;23:4030–4042. doi: 10.1111/jcmm.14288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chang MA, Morgado M, Warren CR, Hinton CV, Farach-Carson MC, Delk NA. p62/SQSTM1 is required for cell survival of apoptosis-resistant bone metastatic prostate cancer cell lines. Prostate. 2014;74:149–163. doi: 10.1002/pros.22737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Falasca L, Torino F, Marconi M, Costantini M, Pompeo V, Sentinelli S, De Salvo L, Patrizio M, Padula C, Gallucci M, et al. AMBRA1 and SQSTM1 expression pattern in prostate cancer. Apoptosis. 2015;20:1577–1586. doi: 10.1007/s10495-015-1176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang L, Kim D, Wise JT, Shi X, Zhang Z, DiPaola RS. p62 as a therapeutic target for inhibition of autophagy in prostate cancer. Prostate. 2018;78:390–400. doi: 10.1002/pros.23483. [DOI] [PubMed] [Google Scholar]
- 104.Burdelski C, Reiswich V, Hube-Magg C, Kluth M, Minner S, Koop C, Graefen M, Heinzer H, Tsourlakis MC, Wittmer C, et al. Cytoplasmic accumulation of sequestosome 1 (p62) is a predictor of biochemical recurrence, rapid tumor cell proliferation, and genomic instability in prostate cancer. Clin Cancer Res. 2015;21:3471–3479. doi: 10.1158/1078-0432.CCR-14-0620. [DOI] [PubMed] [Google Scholar]
- 105.Kim JH, Hong SK, Wu PK, Richards AL, Jackson WT, Park JI. Raf/MEK/ERK can regulate cellular levels of LC3B and SQSTM1/p62 at expression levels. Exp Cell Res. 2014;327:340–352. doi: 10.1016/j.yexcr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Linares JF, Duran A, Reina-Campos M, Aza-Blanc P, Campos A, Moscat J, Diaz-Meco MT. Amino acid activation of mTORC1 by a PB1-domain-driven kinase complex cascade. Cell Rep. 2015;12:1339–1352. doi: 10.1016/j.celrep.2015.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jones S, Cunningham DL, Rappoport JZ, Heath JK. The non-receptor tyrosine kinase Ack1 regulates the fate of activated EGFR by inducing trafficking to the p62/NBR1 pre-autophagosome. J Cell Sci. 2014;127:994–1006. doi: 10.1242/jcs.136895. [DOI] [PubMed] [Google Scholar]
- 108.Jiang G, Liang X, Huang Y, Lan Z, Zhang Z, Su Z, Fang Z, Lai Y, Yao W, Liu T, et al. p62 promotes proliferation, apoptosis-resistance and invasion of prostate cancer cells through the Keap1/Nrf2/ARE axis. Oncol Rep. 2020;43:1547–1557. doi: 10.3892/or.2020.7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang J, Duran A, Reina-Campos M, Valencia T, Castilla EA, Müller TD, Tschöp MH, Moscat J, Diaz-Meco MT. Adipocyte p62/SQSTM1 suppresses tumorigenesis through opposite regulations of metabolism in adipose tissue and tumor. Cancer Cell. 2018;33:770–784.e6. doi: 10.1016/j.ccell.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Valencia T, Kim JY, Abu-Baker S, Moscat-Pardos J, Ahn CS, Reina-Campos M, Duran A, Castilla EA, Metallo CM, Diaz-Meco MT, Moscat J. Metabolic reprogramming of stromal fibroblasts through p62-mTORC1 signaling promotes inflammation and tumorigenesis. Cancer Cell. 2014;26:121–135. doi: 10.1016/j.ccr.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang MA, Patel V, Gwede M, Morgado M, Tomasevich K, Fong EL, Farach-Carson MC, Delk NA. IL-1β induces p62/SQSTM1 and represses androgen receptor expression in prostate cancer cells. J Cell Biochem. 2014;115:2188–2197. doi: 10.1002/jcb.24897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen K, Zeng J, Xiao H, Huang C, Hu J, Yao W, Yu G, Xiao W, Xu H, Ye Z. Regulation of glucose metabolism by p62/SQSTM1 through HIF1α. J Cell Sci. 2016;129:817–830. doi: 10.1242/jcs.178756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu XD, Yao J, Tripathi DN, Ding Z, Xu Y, Sun M, Zhang J, Bai S, German P, Hoang A, et al. Autophagy mediates HIF2α degradation and suppresses renal tumorigenesis. Oncogene. 2015;34:2450–2460. doi: 10.1038/onc.2014.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li T, Jiang D, Wu K. p62 promotes bladder cancer cell growth by activating KEAP1/NRF2-dependent antioxidative response. Cancer Sci. 2020;111:1156–1164. doi: 10.1111/cas.14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu JL, Chen FF, Lung J, Lo CH, Lee FH, Lu YC, Hung CH. Prognostic significance of p62/SQSTM1 subcellular localization and LC3B in oral squamous cell carcinoma. Br J Cancer. 2014;111:944–954. doi: 10.1038/bjc.2014.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kuo WL, Sharifi MN, Lingen MW, Ahmed O, Liu J, Nagilla M, Macleod KF, Cohen EE. p62/SQSTM1 accumulation in squamous cell carcinoma of head and neck predicts sensitivity to phosphatidylinositol 3-kinase pathway inhibitors. PLoS One. 2014;9:e90171. doi: 10.1371/journal.pone.0090171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liang L, Luo H, He Q, You Y, Fan Y, Liang J. Investigation of cancer-associated fibroblasts and p62 expression in oral cancer before and after chemotherapy. J Craniomaxillofac Surg. 2018;46:605–610. doi: 10.1016/j.jcms.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 118.Yoshihara N, Takagi A, Ueno T, Ikeda S. Inverse correlation between microtubule-associated protein 1A/1B-light chain 3 and p62/sequestosome-1 expression in the progression of cutaneous squamous cell carcinoma. J Dermatol. 2014;41:311–315. doi: 10.1111/1346-8138.12439. [DOI] [PubMed] [Google Scholar]
- 119.Shah P, Trinh E, Qiang L, Xie L, Hu WY, Prins GS, Pi J, He YY. Arsenic induces p62 expression to form a positive feedback loop with Nrf2 in human epidermal keratinocytes: Implications for preventing arsenic-induced skin cancer. Molecules. 2017;22:194. doi: 10.3390/molecules22020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Colunga A, Bollino D, Schech A, Aurelian L. Calpain-dependent clearance of the autophagy protein p62/SQSTM1 is a contributor to ΔPK oncolytic activity in melanoma. Gene Ther. 2014;21:371–378. doi: 10.1038/gt.2014.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yeh LY, Liu CJ, Wong YK, Chang C, Lin SC, Chang KW. miR-372 inhibits p62 in head and neck squamous cell carcinoma in vitro and in vivo. Oncotarget. 2015;6:6062–6075. doi: 10.18632/oncotarget.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yamanaka T, Tosaki A, Kurosawa M, Matsumoto G, Koike M, Uchiyama Y, Maity SN, Shimogori T, Hattori N, Nukina N. NF-Y inactivation causes atypical neurodegeneration characterized by ubiquitin and p62 accumulation and endoplasmic reticulum disorganization. Nat Commun. 2014;5:3354. doi: 10.1038/ncomms4354. [DOI] [PubMed] [Google Scholar]
- 123.Wang C, Chen S, Yeo S, Karsli-Uzunbas G, White E, Mizushima N, Virgin HW, Guan JL. Correction: Elevated p62/SQSTM1 determines the fate of autophagy-deficient neural stem cells by increasing superoxide. J Cell Biol. 2016;212:879. doi: 10.1083/jcb.20150702303082016c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang C, Chen S, Yeo S, Karsli-Uzunbas G, White E, Mizushima N, Virgin HW, Guan JL. Elevated p62/SQSTM1 determines the fate of autophagy-deficient neural stem cells by increasing superoxide. J Cell Biol. 2016;212:545–560. doi: 10.1083/jcb.201507023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ivankovic D, Chau KY, Schapira AH, Gegg ME. Mitochondrial and lysosomal biogenesis are activated following PINK1/parkin-mediated mitophagy. J Neurochem. 2016;136:388–402. doi: 10.1111/jnc.13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Su J, Liu F, Xia M, Xu Y, Li X, Kang J, Li Y, Sun L. p62 participates in the inhibition of NF-κB signaling and apoptosis induced by sulfasalazine in human glioma U251 cells. Oncol Rep. 2015;34:235–243. doi: 10.3892/or.2015.3944. [DOI] [PubMed] [Google Scholar]
- 127.Zeng RX, Zhang YB, Fan Y, Wu GL. p62/SQSTM1 is involved in caspase-8 associated cell death induced by proteasome inhibitor MG132 in U87MG cells. Cell Biol Int. 2014;38:1221–1226. doi: 10.1002/cbin.10311. [DOI] [PubMed] [Google Scholar]
- 128.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 129.Qiang L, Zhao B, Ming M, Wang N, He TC, Hwang S, Thorburn A, He YY. Regulation of cell proliferation and migration by p62 through stabilization of Twist1. Proc Natl Acad Sci USA. 2014;111:9241–9246. doi: 10.1073/pnas.1322913111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 130.Qiang L, He YY. Autophagy deficiency stabilizes TWIST1 to promote epithelial-mesenchymal transition. Autophagy. 2014;10:1864–1865. doi: 10.4161/auto.32171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bertrand M, Petit V, Jain A, Amsellem R, Johansen T, Larue L, Codogno P, Beau I. SQSTM1/p62 regulates the expression of junctional proteins through epithelial-mesenchymal transition factors. Cell Cycle. 2015;14:364–374. doi: 10.4161/15384101.2014.987619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jiang X, Huang Y, Liang X, Jiang F, He Y, Li T, Xu G, Zhao H, Yang W, Jiang G, et al. Metastatic prostate cancer-associated P62 inhibits autophagy flux and promotes epithelial to mesenchymal transition by sustaining the level of HDAC6. Prostate. 2018;78:426–434. doi: 10.1002/pros.23487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Grassi G, Di Caprio G, Santangelo L, Fimia GM, Cozzolino AM, Komatsu M, Ippolito G, Tripodi M, Alonzi T. Autophagy regulates hepatocyte identity and epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions promoting snail degradation. Cell Death Dis. 2015;6:e1880. doi: 10.1038/cddis.2015.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Geng Y, Ju Y, Ren F, Qiu Y, Tomita Y, Tomoeda M, Kishida M, Wang Y, Jin L, Su F, et al. Insulin receptor substrate 1/2 (IRS1/2) regulates Wnt/β-catenin signaling through blocking autophagic degradation of dishevelled2. J Biol Chem. 2014;289:11230–11241. doi: 10.1074/jbc.M113.544999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ma JB, Hu SL, Zang RK, Su Y, Liang YC, Wang Y. MicroRNA-487a promotes proliferation of esophageal cancer cells by inhibiting p62 expression. Eur Rev Med Pharmacol Sci. 2019;23:1502–1512. doi: 10.26355/eurrev_201902_17108. [DOI] [PubMed] [Google Scholar]
- 136.Zhong JH, Xiang X, Wang YY, Liu X, Qi LN, Luo CP, Wei WE, You XM, Ma L, Xiang BD, Li LQ. The lncRNA SNHG16 affects prognosis in hepatocellular carcinoma by regulating p62 expression. J Cell Physiol. 2020;235:1090–1102. doi: 10.1002/jcp.29023. [DOI] [PubMed] [Google Scholar]
- 137.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Adamik J, Silbermann R, Marino S, Sun Q, Anderson J L, Zhou D, Xie XQ, Roodman GD, Galson DL. XRK3F2 inhibition of p62-ZZ domain signaling rescues myeloma-induced GFI1-driven epigenetic repression of the Runx2 gene in pre-osteoblasts to overcome differentiation suppression. Front Endocrinol (Lausanne) 2018;9:344. doi: 10.3389/fendo.2018.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li Y, Li Y, Yin J, Wang C, Yang M, Gu J, He M, Xu H, Fu W, Zhang W, et al. A mitophagy inhibitor targeting p62 attenuates the leukemia-initiation potential of acute myeloid leukemia cells. Cancer Lett. 2021;510:24–36. doi: 10.1016/j.canlet.2021.04.003. [DOI] [PubMed] [Google Scholar]
- 140.Andersen AN, Landsverk OJ, Simonsen A, Bogen B, Corthay A, Øynebråten I. Coupling of HIV-1 antigen to the selective autophagy receptor SQSTM1/p62 promotes T-cell-mediated immunity. Front Iunol. 2016;7:167. doi: 10.3389/fimmu.2016.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Venanzi F, Shifrin V, Sherman M, Gabai V, Kiselev O, Komissarov A, Grudinin M, Shartukova M, Romanovskaya-Romanko EA, Kudryavets Y, et al. Broad-spectrum anti-tumor and anti-metastatic DNA vaccine based on p62-encoding vector. Oncotarget. 2013;4:1829–1835. doi: 10.18632/oncotarget.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gabai V, Venanzi FM, Bagashova E, Rud O, Mariotti F, Vullo C, Catone G, Sherman MY, Concetti A, Chursov A, et al. Pilot study of p62 DNA vaccine in dogs with mammary tumors. Oncotarget. 2014;5:12803–12810. doi: 10.18632/oncotarget.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ponomarenko DM, Klimova ID, Chapygina YA, Dvornichenko VV, Zhukova NV, Orlova RV, Manikhas GM, Zyryanov AV, Burkhanova LA, Badrtdinova II, et al. Safety and efficacy of p62 DNA vaccine ELENAGEN in a first-in-human trial in patients with advanced solid tumors. Oncotarget. 2017;8:53730–53739. doi: 10.18632/oncotarget.16574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.