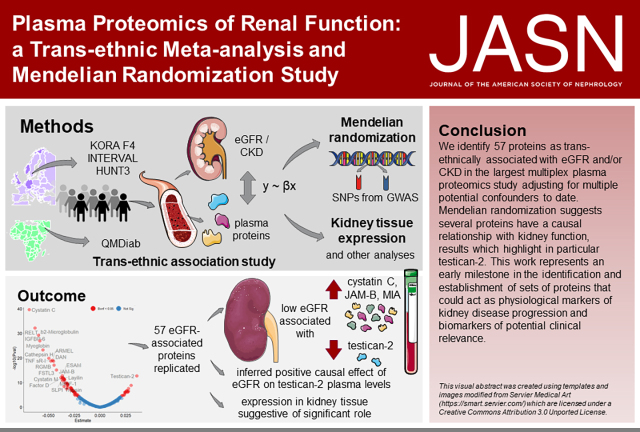
Significance Statement
Studies on the plasma proteome of renal function have identified several biomarkers, but have lacked replication, were limited to European populations, and/or did not investigate causality with eGFR. Among four cohorts in a transethnic cross-sectional study, 57 plasma proteins were associated with eGFR, 23 of them also with CKD. Furthermore, Mendelian randomization and gene expression analyses in kidney tissue highlighted testican-2 as a physiological marker of kidney disease progression with potential clinical relevance, and identified a few additional proteins warranting further investigation.
Keywords: Mendelian randomization, chronic kidney disease, glomerular filtration rate, epidemiology and outcomes, plasma proteomics, biomarker discovery, testican-2, gene expression, genetic epidemiology
Visual Abstract
Abstract
Background
Studies on the relationship between renal function and the human plasma proteome have identified several potential biomarkers. However, investigations have been conducted largely in European populations, and causality of the associations between plasma proteins and kidney function has never been addressed.
Methods
A cross-sectional study of 993 plasma proteins among 2882 participants in four studies of European and admixed ancestries (KORA, INTERVAL, HUNT, QMDiab) identified transethnic associations between eGFR/CKD and proteomic biomarkers. For the replicated associations, two-sample bidirectional Mendelian randomization (MR) was used to investigate potential causal relationships. Publicly available datasets and transcriptomic data from independent studies were used to examine the association between gene expression in kidney tissue and eGFR.
Results
In total, 57 plasma proteins were associated with eGFR, including one novel protein. Of these, 23 were additionally associated with CKD. The strongest inferred causal effect was the positive effect of eGFR on testican-2, in line with the known biological role of this protein and the expression of its protein-coding gene (SPOCK2) in renal tissue. We also observed suggestive evidence of an effect of melanoma inhibitory activity (MIA), carbonic anhydrase III, and cystatin-M on eGFR.
Conclusions
In a discovery-replication setting, we identified 57 proteins transethnically associated with eGFR. The revealed causal relationships are an important stepping stone in establishing testican-2 as a clinically relevant physiological marker of kidney disease progression, and point to additional proteins warranting further investigation.
The kidneys’ ability to filter blood and maintain homeostasis is reflected in the GFR.2 Blood levels of serum creatinine, a filtration marker, can be used for eGFR.3,4 CKD, characterized by reduced eGFR (<60 ml/min per m2) and proteinuria, has a global prevalence of 10% to 16%5,6 and is expected to be increasingly common in aging populations.2 Increased serum creatinine is not evident until approximately 50% of the renal filtration function is lost,7 making CKD a silent disease and creating a blind spot for early kidney disease detection.8 Its rising prevalence, in addition to the lack of therapeutic options,8 imposes a significant burden on health systems and individuals worldwide.2,6
A number of biomarker research studies have been conducted in regard to early detection, diagnosis, and/or progression prediction of kidney diseases.7,8 Early efforts in proteome research focused on urine biomarkers, where combining multiple urinary biomarkers was successful (e.g., CKD273 classifier).7,9 More recent studies have focused on blood, an easily accessible tissue mirroring the metabolic status of multiple organs, with a complex profile requiring sensitive techniques for its study. SOMAscan, a platform using DNA aptamers to measure hundreds of plasma proteomic biomarkers,10 has been successfully used in different epidemiological settings.11-15 However, kidney disease has not been sufficiently investigated: prior studies have tested a limited number of proteins,16 relied on small samples without replication,10,17 or have not investigated causality.18 Mendelian randomization (MR), a causal inference method relying on the random allocation of alleles at conception to estimate causal effects on outcomes, is an increasingly popular method used in genetic epidemiology studies to address causality.19,20
We present a cross-sectional study using a multiplexed aptamer-based proteomics platform to investigate associations between 1095 plasma proteins and eGFR/CKD, and other renal parameters in a discovery cohort (Cooperative Health Research in the Region of Augsburg S4 prospective cohort follow-up; KORA F4), with replication in three independent studies of European and admixed ancestry (INTERVAL, Nord-Trøndelag Health Study [HUNT], and Qatar Metabolomics Study on Diabetes [QMDiab]). To better understand the biological significance of the identified proteins, we conducted enrichment, protein, and transcriptome analyses across tissues, and investigated their interconnection using protein-protein interaction (PPI) network analysis. We also investigated causal effects between eGFR and the replicated proteins using two-sample bidirectional MR. We further examined the correlation between their gene expression in kidney tissue, eGFR, and histological parameters using both publicly available datasets and transcriptomic data from a kidney resource.
Methods
Study Populations
The KORA study is a population-based sample from the general population living in the region of Augsburg, Southern Germany. The KORA F4 survey, a follow-up of the KORA S4 prospective cohort (1999–2001), was conducted from 2006 to 2008, and included a total of 3080 participants. Clinical and demographic information, and peripheral blood for “omics” analyses, were collected; details on the standardized examinations, interviews, and tests conducted in the KORA study have been previously described.21,22 This study acted as discovery cohort in the cross-sectional association study of plasma proteins and renal function (Figure 1A).
Figure 1.
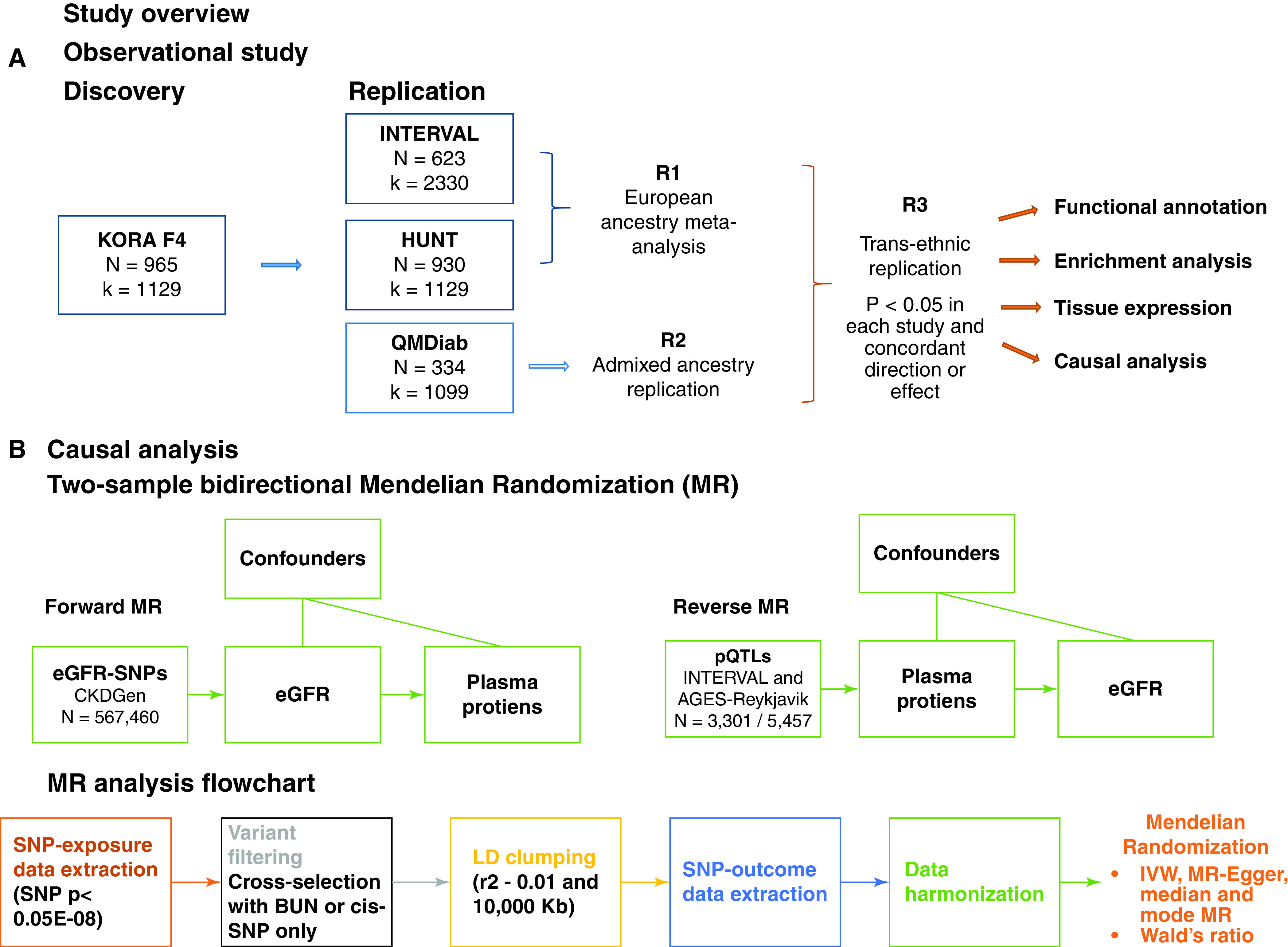
(A) Cross-sectional association study. Data from 995 participants and 1129 proteins from KORA F4 was used in the discovery phase of a proteome-wide association study of renal function using confounder-adjusted regression models. The replication studies were INTERVAL, HUNT, and QMDiab. Three rounds of replication are shown: R1, replication based on the meta-analysis of P values from the linear regression results of the studies with European ancestry; R2, replication based on the results of linear regression models performed in the Arab, South Asian, and Filipino descent sample QMDiab; R3, identification of proteins consistently associated with eGFR across samples and ethnicities. The set of proteins identified in R3 was then functionally annotated and brought forward to the causal analysis phase. (B) Causal analysis. Two-sample bidirectional MR using data on participants from European ancestry studies in the Chronic Kidney Disease (CKDGen) Consortium to instrument the forward analysis (eGFR causal to protein level) and data from INTERVAL and AGES-Reykjavik to instrument the reverse analysis (protein level causal to eGFR). Details on the data processing workflow for MR are shown.
Included in the replication phase were HUNT, namely the third survey (HUNT3) from this population-based study from Norway with data on participants of European descent;23 the INTERVAL study, a randomized trial assessing blood donation practices across the United Kingdom with extensive phenotyping available for 50,000 participants of European descent;24 and the QMDiab, a cross-sectional case-control study on type 2 diabetes from participants of Arab, South Asian, and Filipino descent in Qatar.25 Population characteristics from the four studies are shown in Table 1. Information on data availability is given in Supplemental Note 1.
Table 1.
Population characteristics of association studies
| Trait | KORA | HUNT3 | INTERVAL | QMDiab |
|---|---|---|---|---|
| N | 995 | 930 | 623 | 334 |
| Age, yr | 59.31 (7.81) | 68.94 (10.29) | 47.36 (13.35) | 47.10 (12.57) |
| Male | 480 (48.2) | 688 (74.0) | 343 (55.1) | 169 (50.6) |
| BMI, kg/m2 | 27.78 (4.58) | 28.38 (3.97) | 27.16 (10.05) | 29.66 (5.95) |
| Smokers | 572 (57.5) | 699 (75.16) | 99 (15.89) | 60 (18.0) |
| Serum creatinine (mg/dl) | 0.85 (0.18) | 0.92 (0.32) | 0.70 (0.14) | 0.85 (0.22) |
| eGFR, ml/min per 1.73m2 | 85.98 (14.06) | 80.25 (18.75) | 108.27 (16.21) | 96.00 (18.36) |
| CKD | 38 (3.8) | 138 (14.8) | 1 (0.16) | 19 (5.7) |
| UACR, mg/dl | 5.64 (3.61, 9.94) | NA | NA | NA |
| MA | 58 (5.9) | NA | NA | NA |
| HDL cholesterol, mg/dl | 57.32 (15.20) | 45.12 (11.24) | 74.33 (24.42) | 47.58 (13.75) |
| Triglycerides, mg/dl | 107 (75, 155.5) | 141.27 (106.28, 194.85) | 132.75 (97.35,194.70) | 169 (99.20, 215.23) |
| Lipid lowering medication use | 142 (14.3) | NA | 33 (5.30) | NA |
| Hypertension | 397 (39.9) | 355 (38.2) | 48 (7.70) | 103 (30.8) |
| Diabetes | 68 (6.8) | 128 (13.8) | 2 (0.32) | 172 (51.5) |
Measurement units are shown in parentheses in the trait column, where the absence of units means it is a categorical trait. The mean and (SD) are presented for non-skewed continuous variables, whereas for skewed continuous variables median (first and third quartile) are presented. Count and % are shown for categorical variables.
Sample Collection and Proteomic Profiling
EDTA plasma samples collected by the studies following standardized procedures were centrifuged, aliquoted, and stored at -80°C.26-28 Samples for proteomic profiling and GFR estimation were taken at the same time.
Proteomic profiling in all participating studies was done using SOMAscan (SomaLogic, Inc.), an aptamer-based, affinity proteomics platform.10,29-31 Plasma samples from KORA F4, HUNT3, and INTERVAL were shipped on dry ice to SomaLogic (Boulder, CO), and proteomic profiling was performed using a SOMAscan panel with 1129 protein-specific SOMAmer probes for KORA,26 3622 for INTERVAL,27 and 5000 for HUNT3.28 In the QMDiab cohort, the kit-based SOMAscan platform was run by the Weill Cornell Medicine Qatar proteomics core following protocols and instrumentation provided by SomaLogic Inc., under supervision of SomaLogic personnel, to measure 1129 proteins in plasma samples.26 The samples were all measured by individuals blinded to the identities corresponding to the samples. In summary, fluorescently labeled single-stranded synthetic nucleotides (Slow Off-rate Modified Aptamers, SOMAmers) immobilized on streptavidin-coated beads are incubated with plasma samples to capture proteins and generate SOMAmer-protein complexes. Washing steps eliminate unbound SOMAmers and unbound/nonspecifically bound proteins. The next steps are biotin labeling and photocleavage to liberate SOMAmer-protein complexes from the beads. This is followed by incubation in a buffer disrupting nonspecific interactions, recapturing the biotin-labeled protein/aptamer complexes in streptavidin-coated beads, and additional washing steps to remove nonspecific SOMAmers. These are then eluted from the target proteins and quantified on custom DNA microarrays using deposited SOMAmer-complementary oligonucleotides, which produces measurements in relative fluorescence units as proxies to protein concentrations. Quality control (QC) at the sample and SOMAmer levels using control aptamers and calibrator samples was conducted by the manufacturer. In brief, based on standard samples included on each plate, the resulting raw intensities were processed using a workflow including hybridization normalization, median signal normalization, and signal calibration to control for interplate differences. In the discovery cohort (KORA F4), QC resulted in the exclusion of 29 proteins and one sample, and five proteins were further excluded due to crossreactivity (Assay Change Log SSM-064_Rev_0_DCN_16-263 issued by SomaLogic, available at the integrated web-server at http://proteomics.gwas.eu),26 producing data on k=1095 proteins in 999 participants (Supplemental Table 1). The same QC conducted in each study resulted in the inclusion of 3301 participants in INTERVAL,27 2432 individuals in HUNT3,12 and 352 participants in QMDiab,26 and a set of 993 proteins passing QC in all studies (column “R” in Supplemental Table 1); further details on the proteomics profiling from the samples included in this study are described elsewhere.12,26,27 Protein mapping to several identifiers was provided by the manufacturer (Supplemental Table 1).
Outcome Definitions
Our first analysis is a proteome wide association study: we investigated associations between proteins and renal traits as outcomes, using linear regression models with adjustment for potential confounders. The primary outcomes studied in this analysis were eGFR from serum creatinine and CKD, given their availability in all included studies.
eGFR was calculated using the CKD Epidemiology Collaboration equation with serum creatinine4 with the R package nephro v1.2.32 Serum creatinine was measured using the modified kinetic Jaffé reaction in KORA, HUNT, and QMDiab (and calibrated by multiplying by 0.95),33 and a nuclear magnetic resonance platform (Nightingale Health) in the INTERVAL study. Pearson’s correlation between serum creatinine–based eGFR and this nuclear magnetic resonance–based eGFR variable was estimated in KORA (Supplemental Figure 1). CKD was defined as eGFR <60 ml/min per 1.73 m2.34
We performed some analyses for outcomes available only in the discovery study. Urinary albumin and urinary creatinine were used to calculate urinary albumin-creatinine ratio (uACR) and its derived parameter microalbuminuria (MA, defined as uACR >30 mg/g). eGFR decline was defined as log(eGFR)follow-up – log(eGFR)baseline divided by the follow-up time, where KORA F4 (2006–2008) was used as baseline and KORA FF4 (2013–2014) as its follow-up survey. Sensitivity analyses were also run using eGFRcys (derived from the CKD Epidemiology Collaboration equation using cystatin C).13
Definition of Covariates
Covariates used in the regression analyses were age at the time of examination, sex, body mass index (BMI), smoking status, diabetes (yes/no), hypertension (yes/no), log-transformed triglycerides, HDL, and intake of lipid-lowering drugs (yes/no). See Supplemental Note 2 for precise cohort-specific definitions of covariates used.
Statistical Analysis
Data preprocessing and statistical analyses were conducted using the R language for statistical computing v.3.6.0.35 Before statistical analysis, proteomic data were log transformed and standardized. Linear regression was used to examine the association between protein levels and continuous kidney traits (log-transformed eGFR, uACR, eGFR change), whereas logistic regression was used for binary kidney traits (CKD, MA). Multiple testing was accounted for using a Bonferroni correction considering the total number of investigated proteins at each stage (k=1095 in discovery).
Sensitivity analyses in the discovery sample included regression models with serum creatinine–based eGFR as an outcome and no adjustment for BMI or diabetes, and models including cystatin C–based eGFR as outcome and the same set of covariates from the main model. Pearson’s correlations between the regression coefficients resulting from the sensitivity and the main analyses were calculated. Interaction analyses were also conducted for the proteins identified at discovery by adding an interaction term (each of age, sex, and smoking status individually) to the fully adjusted model (Supplemental Note 3).
For those protein-outcome pairs significantly associated in the discovery, two replications were conducted: a European replication (R1) and a replication in an admixed population (R2) (Figure 1A). Replication was defined as P<0.05 and consistent direction of effect as in the discovery study. The European replication for eGFR consisted of the meta-analysis of results from HUNT and INTERVAL using Stouffer’s method, a P value combination method especially useful when raw data cannot be pooled across studies, which is the case with aptamer-based measurements, where data in relative fluorescence units is not directly comparable across studies. Also known as “inverse normal” or weighted Z-test, this method takes the P values for the i-th study (pi), transforms them by using the inverse normal transformation, and weights them according to the square root of the sample sizes (wi). The sum is then computed, and the combined P value is obtained using the distribution of the resulting statistic, T=ΣwiH(pi).36
For CKD, only the HUNT study was used in the European replication (INTERVAL had only one patient), and the admixed population replication was based on the results of QMDiab. Our final set of transethnic associations (R3) were those pairs of proteins-outcomes that were replicated in both R1 and R2. Replicated eGFR-associated proteins were taken to the next stages of the analysis: proteomic target validation, enrichment analyses, and MR.
Validation of Proteomic Targets
We examined the plasma levels of proteins measured using Proximity Extension Assay (PEA) technology (Olink) in a subgroup of randomly selected participants from the KORA F4 study (n=173).37,38 In brief, protein abundance was quantified using real-time PCR in the PEA proteomic technology (Olink), producing relative quantification data in NPX units (normalized protein expression levels, on log2 scale); NPX values were intensity normalized with the plate median for each assay as the normalization factor, and samples and proteins that did not pass QC were excluded.38 Eight of the most relevant proteins (cystatin C, RELT, IGFBP-6, myoglobin, TNF sR-I, RGMB, FSTL3, contactin-4), and three of the proteins identified in the causal inference analysis (carbonic anhydrase 3, melanoma inhibitory activity [MIA], cystatin M) were available in this subset of proteomic measurements. Of note, testican-2 was not available for measurement using this technology. Scatterplots of the aptamer-based and PEA measurements, annotated with their Pearson’s correlations and P values, are shown in Supplemental Figure 2. The lack of immunoassays fully validated for specificity, linearity, and possible interference for most of the measured analytes in SOMAScan (including testican-2) limits the investigation into the concordance between these two methods.39
Information on specificity and crossreactivity of the aptamers was available from three independent studies27,40,41 for 54 of the 57 proteins identified to be transethnically associated with eGFR. Target specificity issues (i.e., comparable binding observed to a target that is not the product of the same gene) were observed in four proteins (ephrin-A5, IGFBP-5, hemojuvelin, and cystatin SA)27,40 (Supplemental Tables 2–4). Moreover, in previous studies, 23 of the 57 proteins were directly validated via mass spectrometry in blood plasma/serum, and other biological matrices41 (Supplemental Table 2), and 49 using solution affinity measurements27, 40 (Supplemental Tables 3–4).
Functional Annotation, Enrichment, and Expression Analyses
Annotation was conducted using the R package InterMineR v1.6.1,42 a tool facilitating access to data from the HumanMine release 6.0 (May 2019). DAVID v.6.843 was used to look for annotations for Gene Ontology Terms (molecular function, biological process) and pathway information, and to identify publications relevant to the set of 57 replicated proteins. Gene information was retrieved from the human assembly GRCh37 (hg19) using BioMart v.4,44 and shown in Supplemental Table 5.
To investigate the expression patterns of the 57 eGFR-associated proteins and their corresponding protein coding genes across tissues, we used proteomics and RNA-seq expression data from the ProteomicsDB45,46 and the Genotype-Tissue Expression (GTEx) database.47 The data presented and described in this manuscript were generated on October 2, 2020 through a multigene query on the ProteomicsDB Analytics Toolbox portal from: https://www.proteomicsdb.org/proteomicsdb/#analytics/expressionHeatmap and GTEx portal https://www.gtexportal.org/home/multiGeneQueryPage.
PPI Network Analysis
We queried STRING,48 the PPI server, to examine the relationship between the proteins that were identified as robustly associated with eGFR across studies and ethnicities (k=57 transethnically eGFR-associated proteins). We used the set of SOMAscan proteins available across studies as background (k=993), adding no additional interactors (proteins) to the network during the analyses, and considered a minimum required interaction score for a medium confidence (0.400) (Supplemental Note 4).
MR
MR, an instrumental variable method used to infer causality, leverages the natural randomization inherent in the (random) assortment of genes during gamete formation to assess the effect of lifelong exposures on health outcomes.49 Single-nucleotide polymorphisms (SNPs) are used as instrumental variables (IV; or instruments), given their alleles are randomly assigned to individuals before any exposures/outcome and they are nonmodifiable, thus minimizing the risk of reverse causation and confounding.49 The idea behind MR is that if genetic variation produces differences mirroring the biological effects of environmental exposures that alter disease risk, then genetic variation itself should be related to disease risk by having an influence on the exposure.49,50 MR uses SNPs as surrogates for an exposure of interest, allowing the estimation of the effects of life-long, genetically determined “exposures” on health outcomes.49 MR produces robust causal inference estimates if the SNPs used are valid instruments—that is, if they meet the three assumptions on which MR relies: SNPs must be strongly associated with the exposure, and not associated with either (measured or unmeasured) confounders or with the outcome except potentially through the exposure.51 Causality in MR is thus defined as the modification of an exposure leading to a change in the outcome, where the inferred causal effects by MR do not necessarily imply the existence of a straightforward interpretation with respect to direct causal factors,50 nor do they offer information on the time interval (e.g., during development) or target tissue in which such modification of the exposure or intervention would need to be delivered.52
To investigate whether a genetic liability to lower or higher eGFR causally alters plasma protein levels or vice versa, MR was conducted in the set of 57 proteins whose associations with eGFR showed transethnic replication. Two-sample bidirectional MR19 was used to infer the causal effect of renal function (eGFR as proxy thereof) on plasma protein levels (forward MR) and vice versa (reverse MR, Figure 1B). Results from publicly available genome-wide association studies (GWAS) for (1) eGFR from the CKDGen consortium (meta-analysis of European-ancestry populations),53 and (2) plasma proteins from INTERVAL27 and Age, Gene/Environment Susceptibility-Reykjavik Study (AGES-Reykjavik)41 were used to perform MR using MRBase.54 A detailed account on the MR methods, data sources, and analyses conducted is available in Supplemental Note 5.
Instrument Selection
In the forward MR (i.e., assessing the effect of renal filtration on protein levels), 256 SNPs associated with eGFR at genome-wide significance in the CKDGen results were selected as candidate IV. These SNPs were then filtered based on their relevance to renal function (associated with BUN, a complementary renal trait, with an opposite direction of effect, NIV=47) and clumped based on linkage disequilibrium (r2=0.01 and Kb=10,000) to identify independent variants (NIV=41). Summary statistics on 41 SNP-eGFR associations were extracted from the CKDGen results, and corresponding SNP-protein associations were extracted from the INTERVAL results for 47 proteins. For investigating the causal effects of eGFR on proteins, 47 eGFR-protein relationships were instrumented by 41 SNPs.
For the reverse MR (i.e., interrogating the causal effect of proteins on renal filtration), gene positions (GChr37, Supplemental Table 5) were used to identify genome-wide significant cis-SNPs for 28 proteins in the INTERVAL results as candidate IV and LD clumped (same criteria as forward MR). Summary statistics on SNP-protein associations for 28 proteins were extracted from the INTERVAL results, and its corresponding SNP-eGFR associations were extracted from the CKDGen results. The same strategy was followed to identify instruments in the AGES-Reykjavik results; SNP-protein results were extracted from this dataset for 29 proteins, and SNP-eGFR results were extracted for 26 proteins from the CKDGen data. Further details of the genetic instrument selection and data harmonization process are shown in Supplemental Figure 3 and Supplemental Table 6. Thus, for investigating the causal effect of proteins on eGFR, 35 protein-eGFR relationships were instrumented by 1–5 SNPs, of which 17 proteins were examined using data from both INTERVAL and AGES-Reykjavik (Supplemental Figure 4).
Data Harmonization, Phenotypic Variance Explained, and Instrument Specificity
Details on data harmonization, the handling of palindromic SNPs, and calculating the phenotypic variance explained by the SNPs are given in Supplemental Note 5. Harmonized datasets used in the MR analyses are available in Supplemental Table 7.
To look for further evidence of horizontal pleiotropy, association between our SNPs and other traits were searched for in the GWAS Catalog55 (Supplemental Table 8).
MR and Sensitivity Analyses
The primary MR analysis used inverse variance weighted (IVW) regression. In this method, the coefficient of the gene-outcome association is regressed on the coefficient of the gene-exposure association with the intercept constrained to zero, assuming no directional pleiotropy.56,57 Because IVW requires two or more SNPs, in patients where only one SNP instrumented the analysis, Wald’s ratio (coefficient of the gene-outcome association divided by the gene-exposure association) was calculated whenever only one SNP instrumented the analyses, as IVW MR requires two or more SNPs.57
For MR analyses instrumented by more than two SNPs, three further MR methods were used as sensitivity analyses.58 MR-Egger regression was used to assess pleiotropy, because this method allows for horizontal pleiotropy and provides an estimate of the unbalanced horizontal pleiotropic effects in its intercept.59 Weighted median60 and weighted mode MR,61 methods less sensitive to the presence of invalid instruments and to pleiotropic SNPs behaving as outliers, were also used. A number of additional analyses were run to check for outliers, directional pleiotropy and heterogeneity, as recommended.58,62 Details are given in Supplemental Note 5.
Causal estimates were assessed at a Bonferroni-corrected significance level, namely, 0.05 divided by the number of proteins assessed in each MR direction (47 in forward and 51 in reverse MR). Causal effects were considered robust if they were significant at Bonferroni P<0.05 in the IVW or Wald estimator, and results from the pleiotropy-robust sensitivity MR analyses examined to test for violations to MR assumptions.
Expression Analyses in Human Kidney Tissue
The correlation between expression of SPOCK2, one of the genes coding for proteins showing evidence for a causal relationship with eGFR, was calculated with data from microdissected tubulointerstitial components of human renal biopsies from 26 individuals with CKD at different disease stages (I–IV)63 (GEO accession: GSE69438). Gene expression of the protein-coding genes identified in MR (SPOCK2, CA3, CST6, MIA) and renal traits was further assessed in (1) data from Nephroseq v5 (n=458), a platform of comprehensive kidney disease gene expression datasets,64 and (2) a human kidney tissue resource based on RNA sequencing (n=427, see Supplemental Note 6).1
Within Nephroseq, univariate correlation analyses between eGFR and gene expression were conducted separately in study-defined histological compartments of the human kidney (i.e., glomerular and tubulointerstitial) in 458 available kidney samples from three datasets of patients with kidney disease (Ju et al., Sampson et al., and Reich et al.),63,65,66 and one dataset of “apparently” healthy renal tissue (Rodwell et al.).67 The correlations were meta-analyzed using inverse variance weighted random effects models,68 and heterogeneity was assessed using Cochran’s Q test.
Multivariable regression analyses were conducted in the human kidney resource (n=427).1 In brief, we constructed linear regression models with renal expression of each candidate as the response variable; whereas eGFR and histologically confirmed measures of structural kidney damage were used as independent variables together with age, sex, BMI, three genetic principal components, diabetes, and a variable number of surrogate variables (29 for eGFR and 26 for all histology phenotypes).1,69 eGFR was based on circulating levels of creatinine, as reported before.1 Histologic measures of structural integrity (glomerular sclerosis, glomerular Bowman’s capsule thickening, tubular atrophy, interstitial fibrosis, interstitial inflammation, and vascular lesions) were assessed microscopically and scored on a semiquantitative scale (whereby 0 indicates no or minimal damage and 3 is consistent with the highest degree of structural injury), as reported previously.70
Results
Figure 1 illustrates the design of this study. First, a cross-sectional association study was performed to identify proteins associated with renal function parameters in a discovery-replication setting: KORA F4 acted as the discovery, and INTERVAL, HUNT3, and QMDiab as replication studies (Figure 1A). Replicated transethnic protein associations were then assessed for causality using two-sample MR, using data from the largest GWAS available for the traits of interest (CKDGen, INTERVAL, and AGES-Reykjavik) (Figure 1B).
Cross-sectional Association of Plasma Proteins and Renal Function
Population characteristics of the four cohorts included in the cross-sectional association study are shown in Table 1.
Results from the Discovery Study
The association between 1095 plasma proteins and eGFR/CKD was assessed in the KORA F4 study (n=995). A total of 80 proteins were significantly associated with eGFR (P<0.05/1095) (Supplemental Figure 5A). The top three negative associations (i.e., higher eGFR associated with lower plasma protein levels) were observed with cystatin C (β=-0.068; 95% confidence interval [95% CI], -0.078 to -0.059] change in log-transformed eGR per standard deviation increase in protein level, P=2.63E-40), TNF receptor superfamily member 19L (RELT; β=-0.063; 95% CI, -0.073 to -0.053; P=7.82E-33) and β2-microglobulin (β=-0.059; 95% CI, -0.070 to -0.050, P= 6.16E-30), and the strongest positive association (i.e., higher eGFR associated with higher plasma protein levels) was that of testican-2 (β=0.036; 95% CI, 0.026 to 0.045, P=2.066E-13) (Supplemental Table 1). Of note, 34 of these 80 proteins were also associated with CKD (Supplemental Table 1).
Sensitivity analyses showed that 71 of the 80 eGFR-associated-proteins identified in the main analysis were consistently associated with cystatin C–based eGFR, with a high correlation between regression coefficients (r=0.841, P<0.001). Models with no adjustment for BMI or diabetes produced highly similar estimates to those obtained in the main analysis (r=0.99, P<0.001 for both; Supplemental Table 9). Likewise, the exclusion of individuals with CKD (n=38) did not significantly affect the correlation between the plasma levels of proteins and eGFR (Supplemental Figures 6 and 7). Interaction analyses suggested associations with five plasma proteins were accentuated with age (Supplemental Table 10).
Additional renal outcomes were assessed in the discovery cohort: eGFR change was associated with five proteins, three proteins were associated with uACR, and no proteins were associated with MA (Supplemental Table 11).
Results from Replication Studies
Serum creatinine was the only available trait across all replication cohorts (Figure 1A), thus only associations with eGFR/CKD were further explored.
The European replication (R1) was conducted using HUNT3 and INTERVAL; results from this analysis confirmed the association of 62 of the 76 proteins available across studies (Supplemental Figure 5). The second replication round (R2) was performed in QMDiab, a population of admixed ancestry; this confirmed 65 eGFR-protein associations (Supplemental Figure 5). High correlations between z values from the discovery study and replication studies were observed (correlations ranging from 0.66 with INTERVAL to 0.93 with HUNT3, Supplemental Figure 8). The overlap of the proteins replicated in R1 and R2 produced the set of 57 robustly replicated transethnic eGFR-protein associations (Supplemental Table 12). Figure 2 shows the cross-sectional effect estimates for the top 10 protein-eGFR associations across the four cohorts; INTERVAL, a largely healthy and younger population, showed the smallest effect sizes, whereas the strongest effects were observed in HUNT3, a cohort of older individuals with lower mean eGFR and higher CKD prevalence. One novel protein, contactin-4, was identified. All 57 proteins were replicated in the cystatin C–based eGFR sensitivity analysis (Supplemental Table 9).
Figure 2.
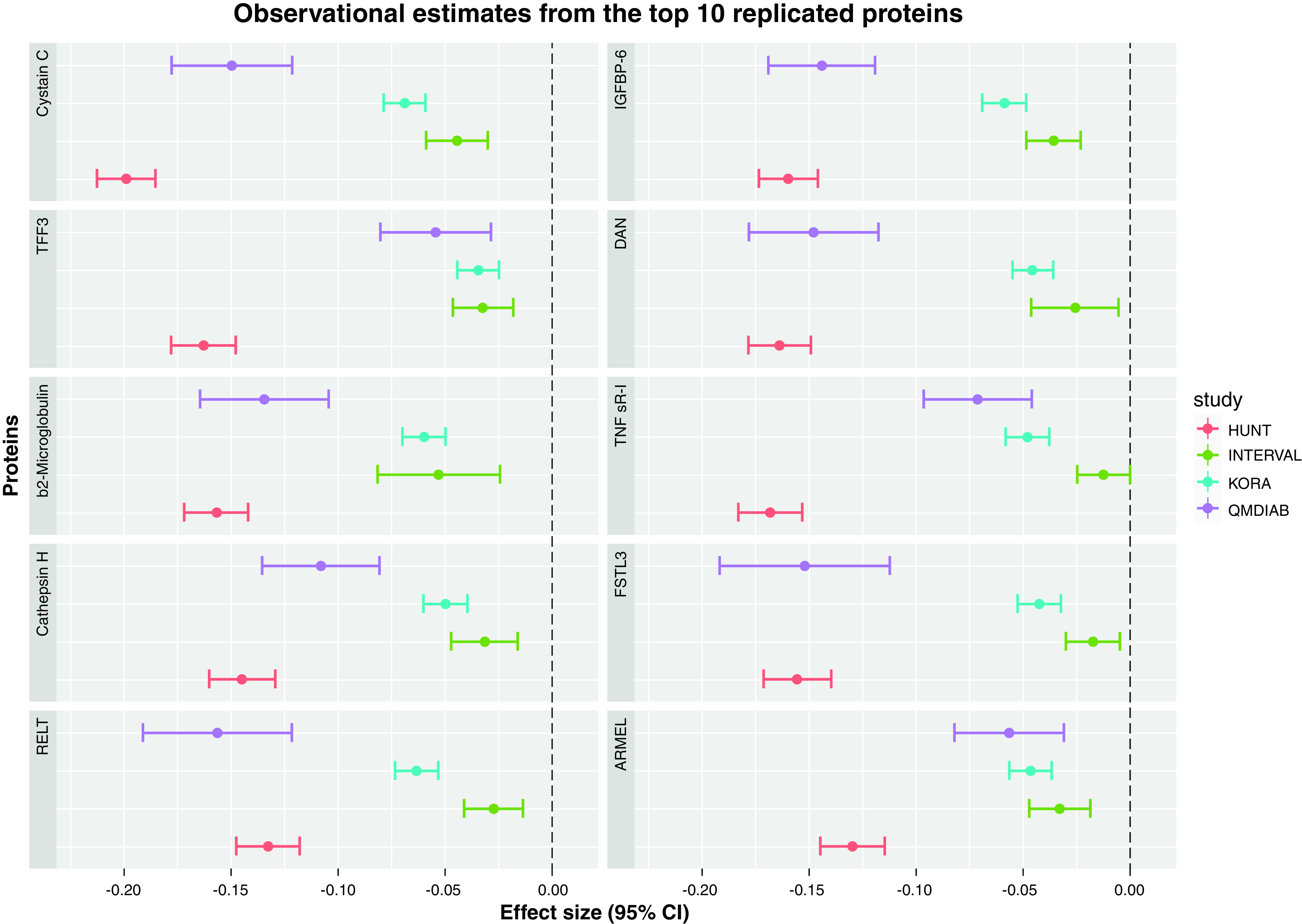
Regression coefficient estimates from the top 10 proteins identified in the cross-sectional association transethnic study on eGFR. The x axis shows the estimates and 95% CI for the regression coefficients (i.e., change in log-transformed eGFR per standard deviation increase in protein level), and each panel corresponds to one protein. Estimates are color coded according to the specific study: HUNT3 in red, INTERVAL in green, KORA in blue, and QMDiab in purple. TFF3, Trefoil factor 3; RELT, TNF receptor superfamily member 19L; IGFBP-6, Insulin-like growth factor-binding protein 6; DAN, Neuroblastoma suppressor of tumorigenicity 1; TNF sR-I: TNF receptor superfamily member 1A; FSTL3, Follistatin-related protein 3; ARMEL, Cerebral dopamine neurotrophic factor.
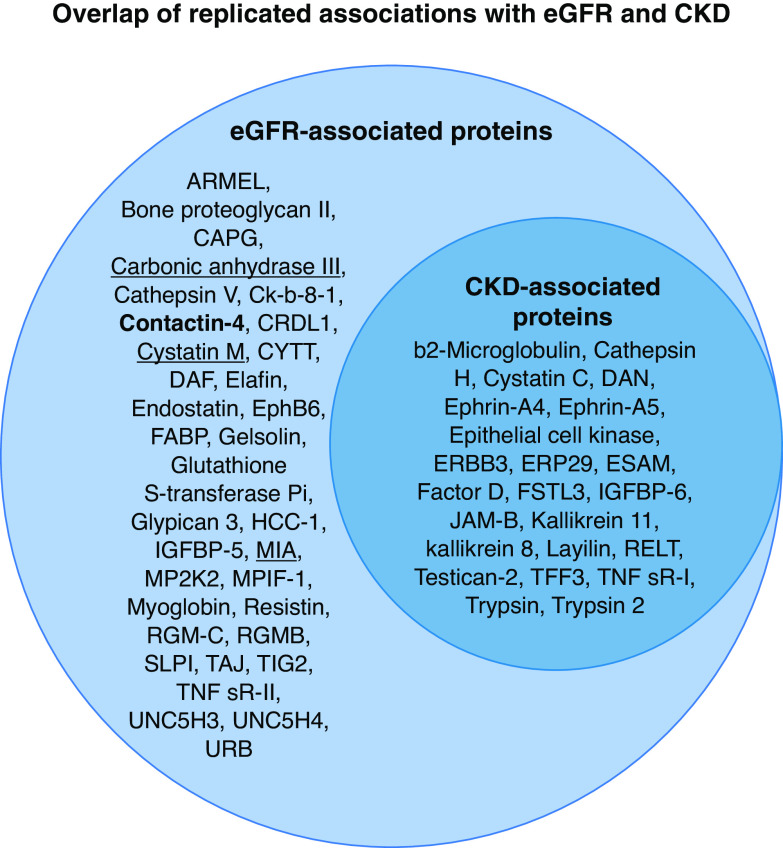
All 34 CKD-protein associations from the discovery phase were replicated in HUNT3 and 23 in QMDiab; these 23 were thus considered transethnically robust (Supplemental Table 13). Figure 3 shows the overlap between proteins associated with eGFR/CKD.
Figure 3.
Results from the transethnic discovery-replication observational study. Depicted in the left circle are the 57 proteins associated with eGFR, the continuous measurement of renal function; the 34 eGFR-specific proteins reflect associations along the full range of renal function, whereas the 23 proteins also associated with CKD reflect a direct association with a clinically relevant low eGFR (<60 ml/min per 1.73m2). Shown in bold is contactin-4, a novel protein identified by this study, and underlined are the four proteins for which evidence on causal effects was identified by MR.
Functional Annotation, Enrichment and Expression Analyses
Several pathways, biological processes and molecular functions were represented in the set of replicated proteins (Supplemental Table 14). No enrichment was observed, perhaps due to the coverage of the analytical platform.10,30,71 Peptides for most of the replicated proteins were detected in multiple tissues and body fluids, including kidney tissue (Supplemental Figure 9). Most genes showed ubiquitous expression across the human tissues represented in the ProteomeDB (Supplemental Figure 10) and GTEx datasets (Supplemental Figure 11).
PPI Network Analysis
We queried STRING to examine PPIs in the set of 57 replicated proteins. More interactions than expected were observed in the resulting network (P=1.12E-04, Supplemental Figure 12). Because the inclusion of proteins in the main PPI analysis was conditional on the platform’s coverage and study design, less stringent sensitivity analyses allowing for the inclusion of additional proteins connected additional nodes (e.g., SPOCK2 or MIA) not connected in the main analysis to the network (Supplemental Note 4).
MR
To assess whether genetically determined higher or lower plasma levels of the 57 proteins identified as transethnically associated with eGFR may affect this renal trait, and whether genetically determined eGFR causally alters circulating levels of plasma proteins, two-sample bidirectional MR was conducted (Figure 1B).
Forward MR: eGFR Has an Effect on Testican-2
In the forward direction of the MR (i.e., inferring the effect of eGFR on levels of 47 proteins), 40 SNPs explaining 1.59% of the variance of eGFR were used as instruments (Supplemental Table 15).
Plasma levels of seven proteins were identified as causally affected by eGFR according to the IVW MR model (Supplemental Table 16 and Supplemental Figures 13–19). Although no evidence of directional pleiotropy, influential SNPs, or instrument heterogeneity was observed (with the exception of IGFB6, Supplemental Table 17–19), pleiotropy-robust sensitivity MR analyses did not provide further evidence of causality for six of them, suggesting IVW findings may be driven by undetected horizontal pleiotropy.72 In contrast, a positive causal effect of eGFR on testican-2 was identified by multiple MR methods (weighted median P=2.84E-04, Figure 4). Assuming this robust evidence reflects a true relationship, the results suggest that if eGFR is altered by an intervention mimicking the effect of the SNP on eGFR, plasma levels of testican-2 will also increase.
Figure 4.
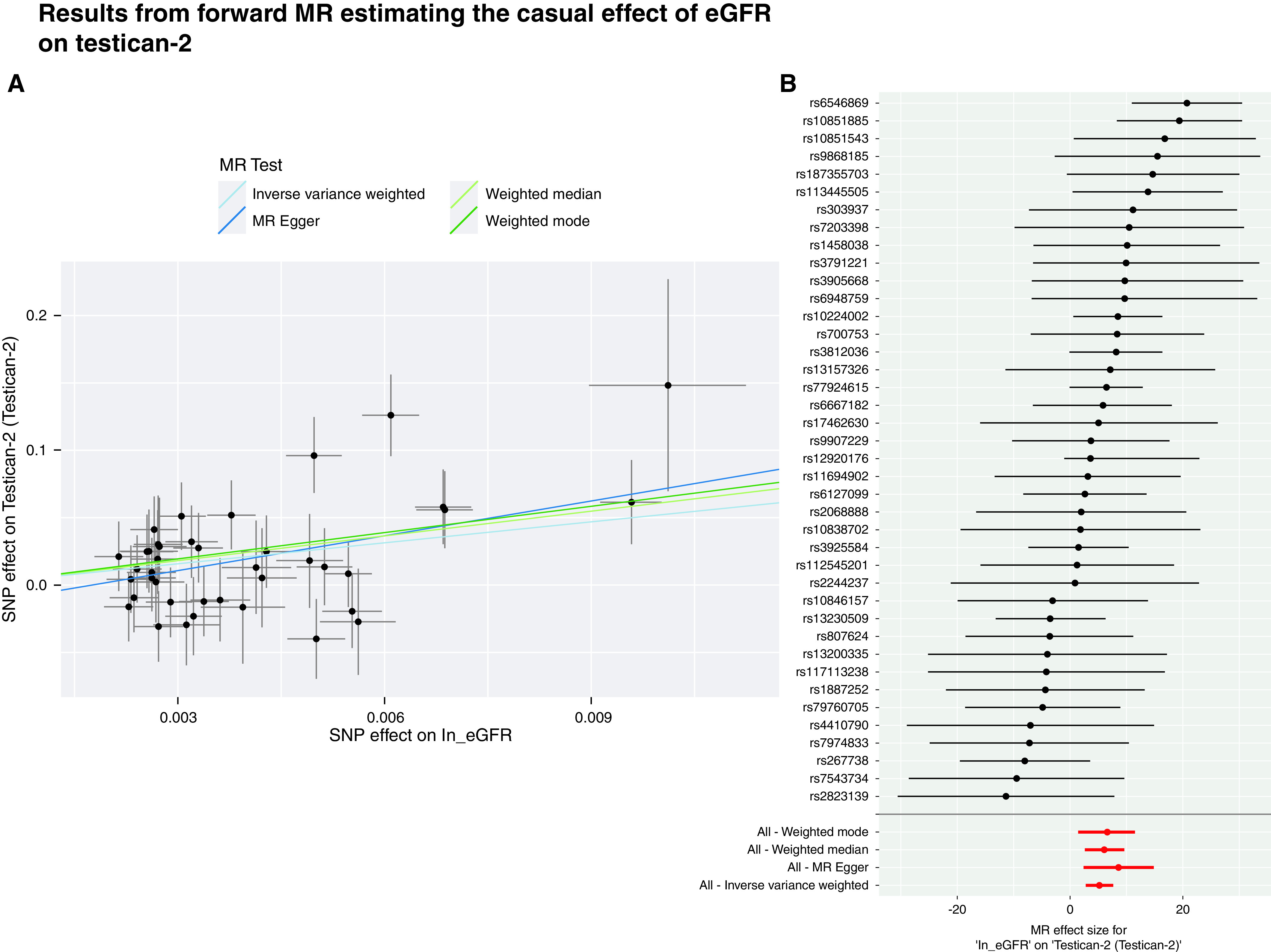
(A) Scatter plot showing the individual genetic effects of the selected IVs on log transformed eGFR (coefficient of the SNP-exposure association) on the x axis and on testican-2 plasma levels (coefficient of the SNP outcome) on the y axis, along with their 95% CI. Each data point corresponds to an individual SNP. The lines correspond to the slopes of the different MR methods, which can be interpreted as the change in testican-2 levels per unit increase in log-transformed eGFR, and are color coded as follows: IVW-MR in light blue, MR-Egger in dark blue, weighted median in light green, weighted mode in dark green. (B) Forest plot showing the individual causal estimates of each of the 40 genetic instruments. The red points show the pooled estimates using all SNPs in the four methods. 95% CI are shown.
In total, 11 of the SNPs instrumenting the forward MR analysis were identified as potentially pleiotropic (associations at P<5E-08 with other traits, Supplemental Table 8). Restrictive MR was conducted excluding these SNPS; although not significant, perhaps due to reduced statistical power derived from using fewer SNPs as instruments and/or the exclusion of SNPs that might be on the actual causal pathway of interest, these results were in agreement with those from the main analysis (same direction and size of effect, Supplemental Table 20).
Reverse MR: MIA, Cystatin M, and Carbonic Anhydrase III Affect eGFR
In the reverse direction of the MR (i.e., assessing the effect of 35 proteins on eGFR), one to five cis-SNPs explaining 0.91%–29.33% of phenotypic variance were used as genetic instruments (Supplemental Table 15).
A negative effect of MIA on eGFR was identified by multiple MR models (Table 2), results that suggest if plasma protein levels are lowered by means of an intervention mimicking the effect of the SNP on MIA, eGFR will increase. No evidence of influential SNPs was observed (Supplemental Tables 17–19), yet the funnel plot suggested directional pleiotropy (Supplemental Figure 20). One SNP instrumenting this analysis was identified as potentially pleiotropic (Supplemental Table 8).
Table 2.
Causal estimates across MR methods
| Association | IVW | MR-Egger | Weighted Median | Weighted Mode | Wald Ratio |
|---|---|---|---|---|---|
| eGFR → SPOCK2 | |||||
| β | 5.21 | 8.61 | 6.10 | 6.51 | |
| CI | 2.78 to 7.66 | 2.41 to 14.8 | 2.81 to 9.40 | 1.19 to 11.8 | -- |
| P | 2.95E-05 | 0.01 | 2.84E-04 | 0.021 | |
| NIV | 40 | 40 | 40 | 40 | |
| CA3 → eGFR | |||||
| Β | 0.007 | ||||
| CI | -- | -- | -- | -- | 0.003 to 0.01 |
| P | 5.04E-04 | ||||
| NIV | 1 | ||||
| CST6 → eGFR | |||||
| Β | 0.007 | ||||
| CI | -- | -- | -- | -- | 0.004 to 0.01 |
| P | 8.41E-05 | ||||
| NIV | 1 | ||||
| MIA → eGFR | |||||
| β | -0.002 | -0.001 | -0.002 | -0.001 | |
| CI | -0.003 to -0.001 | -0.002 to 0.000 | -0.003 to -0.001 | -0.002 to -0.001 | -- |
| P | 8.79E-04 | 0.299 | 2.00E-04 | 0.081 | |
| NIV | 3 | 3 | 3 | 3 |
Results from the forward MR (effect of eGFR on protein levels, i.e. eGFR → protein) are based on the 40 instruments retrieved from Wuttke et al., 2019,53 whereas the reverse MR (protein → eGFR) are based on the one to three instruments retrieved from the INTERVAL pGWAS reported in Sun et al., 2018.27 In bold are significant P values at a Bonferroni-corrected level (0.05/47 for the forward analysis, 0.05/28 for the reverse analysis). SPOCK2, testican-2; CA3, carbonic anhydrase III; CST6, cystatin-M; MIA, melanoma-derived growth regulatory protein; β, causal estimate.
Positive effects of carbonic anhydrase III and cystatin M on eGFR were also identified (Wald’s ratio P=5.04E-04 and 8.41E-05, respectively) (Table 2). Although further sensitivity analyses could not be conducted given the availability of one cis-SNP for each protein, no gene-trait associations were found for these SNPs in the GWAS Catalog,55 indicating a lack of evidence for pleiotropic effects. MR estimates obtained with different pGWAS data (see Methods, Supplemental Figure 4) had the same direction of effect (Supplemental Table 16). Of note, no statistically significant effect of testican-2 on eGFR was identified (Supplemental Table 16).
Gene Expression in Kidney Tissue
No correlation between eGFR and expression of SPOCK2 (protein-coding gene for testican-2) in tubulointerstitial components of human renal biopsies from 26 individuals with CKD63 was found (Supplemental Figure 21). Further univariate analyses conducted with Nephroseq data showed a statistically significant correlation between eGFR and SPOCK2 gene expression in glomerular compartment/kidney cortex (r=0.242, P=0.033) (Supplemental Table 21). We then conducted multivariable analyses using RNA-sequencing–characterized human kidney transcriptomes from up to 427 individuals1,69,70 (Supplemental Note 6, Supplemental Table 22). Whereas no association between gene expression and eGFR was observed (Supplemental Table 23), SPOCK2 expression was negatively associated with tubular atrophy and interstitial fibrosis (P=0.03 for both), and CST6 expression was negatively associated with glomerular sclerosis (P=0.02, Supplemental Table 23).
Discussion
We conducted a cross-sectional association study of plasma proteomics and eGFR/CKD in four independent cohorts, identifying known and potential novel biomarkers. Two-sample bidirectional MR suggested the existence of causal effects in four eGFR-protein associations.
A total of 80 proteins were associated with eGFR in our discovery analysis, with transethnic replication confirming 57 of these; 23 were also found to be associated with CKD. Although our analyses use serum creatinine–based eGFR due to its availability across studies and its utility in clinical practice, models using cystatin C–based eGFR show the results are robust to the GFR estimation method. Likewise, further sensitivity analyses indicate the associations are largely independent of adjustment for BMI or diabetes.
Additional analyses in the discovery cohort produced associations between eGFR decline and DAN, TNF sR-1 and FSTL3, in line with those previously reported.18 No overlap between the set of proteins associated with uACR and the proteins previously reported73 was observed, which may be explained by the different time points of the eGFR and albuminuria measurements.73 No proteins were significantly associated with MA, possibly due to its low prevalence (5.9%) in KORA.
We identify several well-known biomarkers of renal function,10,16,17,74,75 supporting the validity of our eGFR analyses. Our results are also in line with previous proteomic studies on eGFR,10,16-18 inflammation in ESKD,75 and a “standalone” renal health test40 (Supplemental Table 12). Contactin-4, involved in neuronal network development, was identified as a novel eGFR-associated protein. Its plasma (but not urine) detection suggests it is either not filtered at the glomerular capillaries, or filtered but later reabsorbed into blood from the tubules, so that variations in its plasma levels may reflect changes in glomerular and tubule function.76,77 The age-interaction effects identified for four proteins may be a consequence of their age-varying trajectories,78 and require future investigation. The ubiquitous expression of our proteins across tissues, and our PPI network results, suggests the proteins are involved in cellular functions relevant to multiple tissues (Supplemental Table 14),79 potentially mirroring the systemic nature of kidney disease.80 Interestingly, podocyte-exosome enrichment in urine identified 23 of our proteins, pointing to their participation in processes underlying glomerular filter permeability.81
To investigate whether genetically determined renal function (using eGFR as a proxy thereof) or plasma protein levels may have a causal effect on the other, bidirectional MR with publicly available GWAS data was conducted. Our findings in the forward MR direction, supported by pleiotropy-robust sensitivity MR methods, identified a causal effect of eGFR on plasma levels of testican-2. Considering the MR definition of causality, these results suggest lifestyle or pharmacological interventions designed to improve eGFR (as a proxy of renal function) have the potential to increase plasma testican-2 levels.
Testican-2 is a secreted protein of the SPARC family,82 a group of matricellular proteins regulating extracellular matrix–cell interactions and extracellular matrix processing,83 and is involved in a number of biological processes (Supplemental Table 24). In line with our results, higher testican-2 plasma levels have also been associated with less eGFR loss over time and reduced odds of incident CKD.18 Its protein-coding gene, SPOCK2, is associated with both normal maintenance of organ and tissue integrity, and with wound healing and other responses to injury.85 Despite the ubiquitous expression of this gene, its enriched expression in human glomeruli in comparison to other nonrenal tissues18,86-89 and its renal downregulation in diabetic kidney disease86 suggest it has a particularly relevant role in renal cellular mechanisms. Furthermore, recent evidence from arteriovenous sampling demonstrated renal release of this protein into the bloodstream,18 which in addition to its urine detection,84 suggests changes in its plasma levels may be indicative of kidney function.18,76 However, the contribution other organs might have in its plasma levels cannot be ruled out.
Although the association between eGFR and SPOCK2 renal expression did not reach statistical significance in some of our analyses (which may be partly explained by low statistical power in the dataset with 26 patients with CKD), the directionality of the coefficient (i.e., positive association) was consistent across datasets. Moreover, multivariable analyses showed higher scores of histologic measures of renal structural damage to be negatively associated with SPOCK2 renal expression, consistent with prior evidence.86
All in all, the agreement between the cross-sectional results reported by us and by others,17,18,40 and our MR findings and the associations with histologic measures, indicate testican-2 and its protein-coding gene SPOCK2 may have an important role in kidney function. Low plasma levels of testican-2 may be indicative of poor renal function, meaning this protein may be a physiological biomarker of kidney health and disease progression.17,18 Although the reverse direction of this causal association did not reach statistical significance in our MR analyses, a reverse causal effect (i.e., testican-2 on renal function) is biologically plausible given the role matricellular proteins play in extracellular matrix repair83,90 and its in vitro effects on human glomerular endothelial cells.18 The utility of testican-2 as a biomarker with regard to its potential functional effects, tissue of origin, or the mechanisms influencing its blood levels, requires further study.
Three proteins (MIA, cystatin M, and carbonic anhydrase III) were identified as potentially having a causal effect on eGFR, effects biologically plausible given their known roles (Supplemental Table 24). Nevertheless, the precise mechanisms through which these proteins could be exerting effects on eGFR remain to be elucidated. Discordant directions of effect from the observational and the causal estimates (cystatin M, carbonic anhydrase III) could be explained due to differences in sample size/characteristics, reverse causation, or confounding in the case of the observational estimates, or due to limitations inherent to the MR methods.91,92 A further explanation might be that they represent different effects: MR examines lifelong exposures to higher or lower protein levels, whereas results from observational studies could be reflective of acute or short-term effects.92
The strengths of our study include the use of a multiplex proteomics platform and large sample size. This is the first report of eGFR-protein associations adjusted for multiple potential confounders, replicated in independent samples of diverse ancestries, and assessed using causal inference. MR was conducted with the largest available GWAS results from nonoverlapping European ancestry populations, thus avoiding issues derived from population stratification and sample overlap. We reduced the possibility of horizontal pleiotropy by using GWAS summary statistics from a complementary renal trait (blood urea nitrogen) to improve the specificity of the genetic instruments for eGFR, by focusing on cis-SNPs, and by using multiple pleiotropy-robust sensitivity analyses.
Our study also has several limitations. Aptamer-based proteomic methods may be affected by probe cross-reactivity and nonspecific binding,79 although the aptamer-based measurements of most of the reported proteins have been validated in multiple independent studies.27,40,41 This platform does not produce absolute plasma concentrations or cover post-translational modifications, limiting the interpretability of the regression coefficients and the scope of the studied plasma proteome.79 Our findings are based on cross-sectional data, so studies examining their longitudinal changes are warranted. Despite the multiethnic nature of our study, our results may not extend to ethnic groups not represented in our analyses. We avoided weak instrument bias in MR, but cannot discount the possibility of having incurred selection bias in the case of the SNP-protein data. The sample size in which genetic associations with protein levels were calculated was significantly smaller than the sample used to identify genetic associations with eGFR, which likely resulted in differences in power. Finally, knowledge of the biological role of the proteins identified is insufficient for our findings to suggest mechanistic insights. A follow-up of our findings in appropriate experimental models would provide additional evidence on the inferred causal associations reported here and help to unravel the molecular mechanisms underlying our findings. Likewise, future validation studies using validated absolute quantitative assays with increased sensitivity for the detection of testican-2, and other proteins identified here, are warranted to establish reference ranges and to explore their suitability as prognostic and diagnostic biomarkers in clinical settings.
In summary, our transethnic population-based study of plasma proteomics and renal function identified multiple markers of kidney function. Our MR findings are a stepping stone in establishing testican-2 as a physiological marker of kidney disease progression, and further identify proteins warranting additional investigation. Our results may serve as the starting point for future translational work on the utility of these proteins as diagnostic or prognostic biomarkers of disease, and for research on mechanistic insights at the tissue and single-cell levels.
Disclosures
A. Butterworth reports receiving research funding from AstraZeneca, Bayer, Biogen, BioMarin, Bioverativ, Merck, Novartis, and Sanofi; and reports receiving honoraria from Novartis. A. Fornoni reports having consultancy agreements with Dimerix, Gilead, Janssen, Novartis, ONO Pharmaceutical, and Zyversa Therapeutics; reports having an ownership interest as CSO and Vice-President of L&F Health LLC., being a shareholder in River 3 Renal Corp and Zyversa Therapeutics; reports receiving research funding from Boheringer Ingelheim and Roche; reports having patents and inventions for the use of cyclodextrin for the treatment of kidney diseases, a patent for use of small molecule inducers of cholesterol efflux; reports being a scientific advisor or member of the Journal of Clinical Investigation and Kidney International; and reports having other interests/relationships as an inventor on five pending US patents and one published patent. A. Kottgen reports receiving honoraria from Sanofi Genzyme; repost being a scientific advisor or member of the American Journal of Kidney Diseases, American Kidney Fund, Journal of the American Society of Nephrology, Kidney International, and Nature Reviews Nephrology. A. Teumer reports being a scientific advisor or member of the Editorial Board of the Journal Endocrine Connections; reports having other interests/relationships as a member of the American Society of Human Genetics. C. Herder reports receiving research funding from Sanofi-Aventis; reports receiving honoraria from Lilly, Sanofi-Aventis; and reports being a scientific advisor or member of the Editorial Boards for Diabetologia, Diabetic Medicine, and Diabetes/Metabolism Research and Reviews (DMRR). C. Jonasson reports receiving personal fees for research consultancy work from Bayer and Pfizer outside of the submitted work. E. Di Angelantonio reports receiving research funding from a British Heart Foundation research grant, a National Health Service Blood and Transplant research grant, National Institute for Health Research grant, and a United Kingdom Medical Research Council research grant; reports being a scientific advisor or member as Chair of the working group for the European Society of Cardiology Cardiovascular Risk Collaboration, Member of the World Obesity Federation and World Heart Federation Expert Group on Obesity and CVD, Member of the National Health Service Blood and Transplant Clinical Trial Unit Steering Committee, European Society of Cardiology/European Atherosclerosis Society Task Force for Guidelines on Primary Prevention of Cardiovascular Disease and Management of Dyslipidaemia, and Member of the World Health Organization Risk Chart Working Group. J. Dormer reports consultancy agreements with Royal College of Pathologists; reports being on the AstraZeneca Genomics Advisory Board (2018), International Cardiovascular and Metabolic Advisory Board for Novartis (since 2010), the International Cardiovascular and Metabolism Research and Development Portfolio Committee for Novartis, the Medical Research Council International Advisory Group (ING) member, London (since 2013), the MRC High Throughput Science ‘Omics Panel Member, London (since 2013); the Scientific Advisory Committee for Sanofi (since 2013), and the Steering Committee of UK Biobank (since 2011). M. Prunotto is employed by Galapagos Ltd. M. Waldenberger reports being a scientific advisor or member of the Editorial board of Genes and International Journal of Molecular Science. Q. Guo is employed by BenevolentAI. S. Zaghlool reports receiving research funding from the Qatar Foundation. W. Koenig reports having consultancy agreements with Amgen, AstraZeneca, Corvidia, DalCor, Daiichi-Sankyo, Genentech, Kowa, Novartis, Pfizer, and The Medicines Company; reports receiving research funding from Abbott, Beckmann, Roche Diagnostics, Singulex; reports receiving honoraria from Amgen, AstraZeneca, Berlin-Chemie, Bristol-Myers Squibb, Novartis, and Sanofi; reports being a scientific advisor or member of the Advisory Boards for Amgen, AstraZeneca, Corvidia, Daiichi-Sankyo, DalCor, Esperion, Genentech, Kowa, Novartis, Pfizer, The Medicines Company; and reports being an Editorial Board member for Cardiovascular Drugs and Therapy and Clinical Chemistry. J. Danesh reports Scientific Advisor or Membership with the Scientific Advisory Board of Oxford BHF Centre for Research Excellence (2020), Scientific Advisory Board for Nightingale Health (2020), Genomics Advisory Board for AstraZeneca (2018), 2015 International Cardiovascular and Metabolism Research and Development Portfolio Committee for Novartis, MRC Global Health Group member in London (2014-16), Scientific Advisory Committee for Sanofi (2013), MRC High Throughput Science Omics panel member in London (2013), and MRC International Advisory Gro (2013). D. Roberts reports Honoraria from Wiley. All remaining authors have nothing to disclose. The HUNT part of the project re-used protein data that were originally analyzed and paid for by Somalogic Inc., CO, USA. Somalogic had no role in the design and conduct of the study; collection of phenotypic data, statistical analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or the decision to submit the manuscript for publication.
Funding
The KORA study was initiated and financed by the Helmholtz Zentrum München German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research and the State of Bavaria. This work was also supported by Weill Cornell Medicine in Qatar Biomedical Research Program, the Qatar Foundation, and Qatar National Research Fund grant NPRPC11-0115-180010 (to K. Suhre). The HUNT Study is a collaboration between HUNT Research Centre (Norwegian University of Science and Technology), Nord-Trøndelag County Council, Central Norway Health Authority, and the Norwegian Institute of Public Health. J. Danesh is supported by the National Institute for Health Research Senior Investigator Award. RNA-sequencing experiments and kidney gene expression studies were supported by British Heart Foundation project grants PG/17/35/33001 and PG/19/16/34270, and Kidney Research UK grants RP_017_20180302 and RP_013_20190305 (to M. Tomaszewski). The German Diabetes Center is funded by the German Federal Ministry of Health (Berlin, Germany), the Ministry of Culture and Science of the state North Rhine-Westphalia (Düsseldorf, Germany), and grants from the German Federal Ministry of Education and Research (Berlin, Germany) to the German Center for Diabetes Research. The work of A. Köttgen is supported by he Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – Project-ID 431984000 – SFB 1453.
Supplementary Material
Acknowledgments
C. Geiger, P. Matías-García, M. Waldenberger, R. Wilson, and J. Winkelmann conceptualized and designed the study; J. Graumann, D. Mook-Kanamori, H. Maalmi, and A. Petrera performed experiments; F. Charchar, J. Eales, J. Graumann, Q. Guo, P. Matías-García, M. Prunotto, M. Tomaszewski, M. Waldenberger, R. Wilson, X. Xu, and S. Zaghlool analyzed data or interpreted results; A. Butterworth, F. Charchar, J. Danesh, E. Di Angelantonio, J. Dormer, J. Eales, M. Elhadad, A. Fornoni, C. Geiger, J. Graumann, Q. Guo, S. Hauck, C. Herder, Human Kidney Tissue Resource, K. Hveem, C. Jonasson, W. Koenig, A. Köttgen, H. Maalmi, D. Mook-Kanamori, W. Ouwehand, A. Peters, A. Petrera, M. Prunotto, D. Roberts, P. Schlosser, S. Sharma, K. Suhre, A. Teumer, M. Tomaszewski, M. Waldenberger, N. Watkins, J. Winkelmann, X. Xu, and S. Zaghlool contributed reagents/materials/analysis tools; P. Matías-García wrote the paper; F. Charchar, J. Danesh, J. Eales, J. Graumann, Q. Guo, S. Hauck, C. Herder, W. Koenig, A. Köttgen, J. Nano, A. Petrera, P. Schlosser, A. Teumer, M. Tomaszewski, M. Waldenberger, R. Wilson, X. Xu, and S. Zaghlool edited and improved the clarity of the manuscript; all authors discussed the results and reviewed and approved the final manuscript. We are grateful to all study participants of KORA, HUNT, INTERVAL, and QMDiab for their invaluable contributions to these studies, and all members of field staff conducting the studies. An early version of the abstract was sent to the 53rd European Society of Human Genetics Conference, and is listed online as P03.24.C at the European Society of Human Genetics Conference 2020.2 Abstract Library (https://2020.eshg.org/index.php/abstract-library/). The visual abstract was created using templates and images modified from Servier Medical Art (https://smart.servier.com/), which are licensed under a Creative Commons Attribution 3.0 Unported License. The views expressed are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research, or the Department of Health and Social Care.
ETHICS APPROVAL AND INFORMED CONSENT
All participants provided written consent. Details on institutions providing ethics approval from each study are: the KORA cohort ethical approval was granted by the ethics committee of the Bavarian Medical Association (REC reference number F4: 06068); HUNT: Regiona Committee for Medical Research Ethics (REK) and Data Inspectorate; INTERVAL: National Research Ethics Service Committee East of England - Cambridge East (Research Ethics Committee (REC) reference 11/EE/0538); and QMDiab: Institutional Review Boards of HMC and Weill Cornell Medicine Qatar under research protocol number 11131/11).
Footnotes
The Human Kidney Tissue Studies investigators who contributed to recruitment and/or phenotyping of human kidney gene expression studies reported in reference 1 are Wojciech Wystrychowski, Monika Szulinska, Andrzej Antczak, Maciej Glyda, Robert Król, Joanna Zywiec, Ewa Zukowska-Szczechowska, and Pawel Bogdanski.
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.13039/100007273/-/DCSupplemental.
Supplemental Note 1. Data availability.
Supplemental Note 2. Covariate definition.
Supplemental Note 3. Interaction analyses.
Supplemental Note 4. PPI network analysis.
Supplemental Note 5. MR analysis.
Supplemental Note 6. Expression analyses in human kidney samples.
Supplemental Table 1. Results from cross-sectional analysis of eGFR and CKD in KORA F4.
Supplemental Table 2. Validation of proteomic targets from Emilsson et al., 2018.
Supplemental Table 3. Validation of proteomic targets from Sun et al., 2018.
Supplemental Table 4. Validation of proteomic targets from Williams et al., 2019.
Supplemental Table 5. Gene information from proteins included in MR.
Supplemental Table 6. Genetic instrument selection and data harmonization (INTERVAL and AGES- Reykjavik).
Supplemental Table 7. Harmonized summary statistics used in MR.
Supplemental Table 8. Gene-trait information retrieved from GWAS Catalog for pleiotropic SNPs.
Supplemental Table 9. Sensitivity analyses with CysC-based eGFR, no adjustment for BMI and no adjustment for T2D.
Supplemental Table 10. Sensitivity analyses (interaction with age, sex, and smoking) in KORA F4.
Supplemental Table 11. Results from observational analysis of supplementary renal phenotypes (eGFR decline, log(uACR) and MA) in KORA F4.
Supplemental Table 12. Replication of cross-sectional eGFR-protein associations.
Supplemental Table 13. Replication of cross-sectional CKD-protein associations.
Supplemental Table 14. Extended annotation file (DAVID).
Supplemental Table 15. Phenotypic variance explained by MR instruments.
Supplemental Table 16. Results from MR (IVW, MBE, weighted median, and MR-Egger).
Supplemental Table 17. Sensitivity analyses: Heterogeneity (Cochran's Q test).
Supplemental Table 18. Sensitivity analyses: Pleiotropy in MR-Egger.
Supplemental Table 19. Sensitivity analyses: Leave-one-out analyses.
Supplemental Table 20. Sensitivity analyses: Results from restrictive MR.
Supplemental Table 21. Results from correlation analyses between gene expression and eGFR from Nephroseq datasets.
Supplemental Table 22. Clinical characteristics of studies included in gene expression analyses.
Supplemental Table 23. Results from multivariate regression analyses on gene expression, eGFR and histological characteristic scoring from human kidney resource.
Supplemental Table 24. Description and biological roles of selected proteins.
Supplemental Figure 1. Correlation of serum creatinine variables in KORA F4.
Supplemental Figure 2. Correlation between aptamer-based and other measurements for proteins in KORA F4.
Supplemental Figure 3. Genetic instrument selection and data harmonization.
Supplemental Figure 4. Protein overlap in pGWAS datasets used in reverse direction of MR.
Supplemental Figure 5. Proteins and log(eGFR) distribution in discovery dataset.
Supplemental Figure 6. Cross sectional results for eGFR-protein associations across studies.
Supplemental Figure 7. Proteins and log(eGFR) distribution in discovery dataset after CKD exclusion.
Supplemental Figure 8. Correlation between Z-values for eGFR-protein associations across studies.
Supplemental Figure 9. Tissue expression of 57 eGFR-associated proteins (ProteomeDB).
Supplemental Figure 10. Tissue expression of 56 eGFR-associated protein coding genes (ProteomeDB).
Supplemental Figure 11. Expression of 56 eGFR-associated protein coding genes across tissues (GTEx).
Supplemental Figure 12. PPI network of 57 replicated eGFR-associated proteins.
Supplemental Figures 13–19. Forward MR results for effects of eGFR on proteins.
Supplemental Figure 20. Reverse MR analysis for MIA-eGFR.
Supplemental Figure 21.SPOCK2 gene expression in renal tissue from 26 patients with CKD.
References
- 1.Jiang X, Eales JM, Scannali D, Nazgiewicz A, Prestes P, Maier M, et al. : Hypertension and renin-angiotensin system blockers are not associated with expression of angiotensin-converting enzyme 2 (ACE2) in the kidney. Eur Heart J 41: 4580–4588, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckardt KU, Coresh J, Devuyst O, Johnson RJ, Kottgen A, Levey AS, et al. : Evolving importance of kidney disease: from subspecialty to global health burden. Lancet 382: 158–169, 2013 [DOI] [PubMed] [Google Scholar]
- 3.Levey AS, Inker LA, Coresh J: GFR estimation: From physiology to public health. Am J Kidney Dis 63: 820–834, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF 3rd, Feldman HI, et al. : A new equation to estimate glomerular filtration rate. Ann Intern Med 150: 604–612, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill NR, Fatoba ST, Oke JL, Hirst JA, O’Callaghan CA, Lasserson DS, et al. : Global prevalence of chronic kidney disease: A systematic review and meta-analysis. PLoS One 11: e0158765, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levey AS, Atkins R, Coresh J, Cohen EP, Collins AJ, Eckardt KU, et al. : Chronic kidney disease as a global public health problem: Approaches and initiatives - a position statement from Kidney Disease Improving Global Outcomes. Kidney Int 72: 247–259, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Mischak H, Delles C, Vlahou A, Vanholder R: Proteomic biomarkers in kidney disease: Issues in development and implementation. Nat Rev Nephrol 11: 221–232, 2015 [DOI] [PubMed] [Google Scholar]
- 8.Sanchez-Nino MD, Sanz AB, Ramos AM, Fernandez-Fernandez B, Ortiz A: Clinical proteomics in kidney disease as an exponential technology: Heading towards the disruptive phase. Clin Kidney J 10: 188–191, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Good DM, Zurbig P, Argiles A, Bauer HW, Behrens G, Coon JJ, et al. : Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Molecular Cellular Proteomics 9: 2424–2437, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, et al. : Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 5: e15004, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ngo D, Sinha S, Shen D, Kuhn EW, Keyes MJ, Shi X, et al. : Aptamer-based proteomic profiling reveals novel candidate biomarkers and pathways in cardiovascular disease. Circulation 134: 270–285, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganz P, Heidecker B, Hveem K, Jonasson C, Kato S, Segal MR, et al. : Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA 315: 2532–2541, 2016 [DOI] [PubMed] [Google Scholar]
- 13.Zaghlool SB, Sharma S, Molnar M, Matías-García PR, Elhadad MA, Waldenberger M, et al.: Revealing the role of the human blood plasma proteome in obesity using genetic drivers. Nature communications 12: 1279, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pena MJ, Mischak H, Heerspink HJ: Proteomics for prediction of disease progression and response to therapy in diabetic kidney disease. Diabetologia 59: 1819–1831, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elhadad, MA, Jonasson, C, Huth, C, Wilson, R, Gieger, C, Matias, P, et al.: Deciphering the Plasma Proteome of Type 2 Diabetes. Diabetes 69: 2766–2778, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlsson AC, Ingelsson E, Sundstrom J, Carrero JJ, Gustafsson S, Feldreich T, et al. : Use of proteomics to investigate kidney function decline over 5 years. Clin J Am Soc Nephrol 12: 1226–1235, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensson A, Ash JA, DeLisle RK, Gaspar FW, Ostroff R, Grubb A, et al. : The impact of the glomerular filtration rate on the human plasma proteome. Proteomics Clin Appl 12: e1700067, 2018 [DOI] [PubMed] [Google Scholar]
- 18.Ngo D, Wen D, Gao Y, Keyes MJ, Drury ER, Katz DH, et al. : Circulating testican-2 is a podocyte-derived marker of kidney health. Proc Natl Acad Sci USA 117: 25026–25035, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierce BL, Burgess S: Efficient design for Mendelian randomization studies: Subsample and 2-sample instrumental variable estimators. Am J Epidemiol 178: 1177–1184, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekula P, Del Greco M: F, Pattaro, C, Köttgen, A: Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol 27: 3253–3265, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holle, R, Happich, M, Lowel, H, Wichmann, HE: KORA: A research platform for population based health research. Gesundheitswesen 67: S19–S25, 2005 [DOI] [PubMed] [Google Scholar]
- 22.Wichmann, HE, Gieger, C, Illig, T: KORA-gen–resource for population genetics, controls and a broad spectrum of disease phenotypes. Das Gesundheitswesen 67: S26–S30, 2005 [DOI] [PubMed] [Google Scholar]
- 23.Krokstad S, Langhammer A, Hveem K, Holmen T, Midthjell K, Stene T, et al. : Cohort profile: The HUNT Study, Norway. Int J Epidemiol 42: 968–977, 2013 [DOI] [PubMed] [Google Scholar]
- 24.Moore C, Bolton T, Walker M, Kaptoge S, Allen D, Daynes M, et al. : Recruitment and representativeness of blood donors in the INTERVAL randomised trial assessing varying inter-donation intervals. Trials 17: 458, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mook-Kanamori DO, Selim MM, Takiddin AH, Al-Homsi H, Al-Mahmoud KA, Al-Obaidli A, et al. : 1,5-Anhydroglucitol in saliva is a noninvasive marker of short-term glycemic control. J Clin Endocrinol Metab 99: E479–E483, 2014 [DOI] [PubMed] [Google Scholar]
- 26.Suhre K, Arnold M: Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun 8: 14357, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun BB, Maranville JC, Peters JE, Stacey D, Staley JR, Blackshaw J, et al. : Genomic atlas of the human plasma proteome. Nature 558: 73–79, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nayor M, Short MI, Rasheed H, Lin H, Jonasson C, Yang Q, et al. : Aptamer-based proteomic platform identifies novel protein predictors of incident heart failure and echocardiographic traits. Circ Heart Fail 13: e006749, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gold L, Walker JJ, Wilcox SK, Williams S: Advances in human proteomics at high scale with the SOMAscan proteomics platform. N Biotechnol 29: 543–549, 2012 [DOI] [PubMed] [Google Scholar]
- 30.Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, et al. : Nucleic acid ligands with protein-like side chains: Modified aptamers and their use as diagnostic and therapeutic agents. Mol Ther Nucleic Acids 3: e201, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraemer S, Vaught JD, Bock C, Gold L, Katilius E, Keeney TR, et al. : From SOMAmer-based biomarker discovery to diagnostic and clinical applications: A SOMAmer-based, streamlined multiplex proteomic assay. PLoS One 6: e26332–e26332, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pattaro C, Riegler P, Stifter G, Modenese M, Minelli C, Pramstaller PP: Estimating the glomerular filtration rate in the general population using different equations: Effects on classification and association. Nephron Clin Pract 123: 102–111, 2013 [DOI] [PubMed] [Google Scholar]
- 33.Coresh J, Turin TC, Matsushita K, Sang Y, Ballew SH, Appel LJ, et al. : Decline in estimated glomerular filtration rate and subsequent risk of end-stage renal disease and mortality. JAMA 311: 2518–2531, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.National Kidney Foundation: K/DOQI clinical practice guidelines for chronic kidney disease: Evaluation, classification, and stratification. Am J Kidney Dis 39: S1–S266, 2002. [PubMed] [Google Scholar]
- 35.R Core Team: R: A language and environment for statistical computing, Vienna, Austria, R Foundation for Statistical Computing, 2019 [Google Scholar]
- 36.Zaykin DV: Optimally weighted Z-test is a powerful method for combining probabilities in meta-analysis. J Evol Biol 24: 1836–1841, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huth C, von Toerne C, Schederecker F, de Las Heras Gala T, Herder C, Kronenberg F, et al. : Protein markers and risk of type 2 diabetes and prediabetes: A targeted proteomics approach in the KORA F4/FF4 study. Eur J Epidemiol 34: 409–422, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrera A, von Toerne C, Behler J, Huth C, Thorand B, Hilgendorff A, et al. : Multiplatform approach for plasma proteomics: Complementarity of olink proximity extension assay technology to mass spectrometry-based protein profiling. J Proteome Res 20: 751–762, 2021 [DOI] [PubMed] [Google Scholar]
- 39.Raffield LM, Dang H, Pratte KA, Jacobson S, Gillenwater LA, Ampleford E, et al. : Comparison of proteomic assessment methods in multiple cohort studies. Proteomics 20: e1900278, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams SA, Kivimaki M, Langenberg C, Hingorani AD, Casas JP, Bouchard C, et al. : Plasma protein patterns as comprehensive indicators of health. Nat Med 25: 1851–1857, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emilsson V, Ilkov M, Lamb JR, Finkel N, Gudmundsson EF, Pitts R, et al. : Co-regulatory networks of human serum proteins link genetics to disease. Science 361: 769–773, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kyritsis KA, Wang B, Sullivan J, Lyne R, Micklem G: InterMineR: An R package for InterMine databases. Bioinformatics 35: 3206–3207, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang W, Sherman BT, Lempicki RA: Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009 [DOI] [PubMed] [Google Scholar]
- 44.Durinck S, Spellman PT, Birney E, Huber W: Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 4: 1184–1191, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt T, Samaras P, Frejno M, Gessulat S, Barnert M, Kienegger H, et al. : ProteomicsDB. Nucleic Acids Res 46: D1271–D1281, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samaras P, Schmidt T, Frejno M, Gessulat S, Reinecke M, Jarzab A, et al. : ProteomicsDB: A multi-omics and multi-organism resource for life science research. Nucleic Acids Res 48: D1153–D1163, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.GTEx Consortium: The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. : STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47: D607–D613, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davey Smith G, Hemani G: Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 23: R89–R98, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith GD, Ebrahim S: ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 32: 1–22, 2003 [DOI] [PubMed] [Google Scholar]
- 51.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G: Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat Med 27: 1133–1163, 2008 [DOI] [PubMed] [Google Scholar]
- 52.Bretherick AD, Canela-Xandri O, Joshi PK, Clark DW, Rawlik K, Boutin TS, et al. : Linking protein to phenotype with Mendelian randomization detects 38 proteins with causal roles in human diseases and traits. PLoS Genet 16: e1008785, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wuttke M, Li Y, Li M, Sieber KB, Feitosa MF, Gorski M, et al. : A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet 51: 957–972, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. : The MR-Base platform supports systematic causal inference across the human phenome. eLife 7: e34408, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. : The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47: D1005–D1012, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burgess S, Butterworth A, Thompson SG: Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37: 658–665, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teumer A: Common methods for performing Mendelian randomization. Front Cardiovasc Med 5: 51, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG: Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology 28: 30–42, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bowden J, Davey Smith G, Burgess S: Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int J Epidemiol 44: 512–525, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bowden J, Davey Smith G, Haycock PC, Burgess S: Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 40: 304–314, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hartwig FP, Davey Smith G, Bowden J: Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol 46: 1985–1998, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verbanck M, Chen C-Y, Neale B, Do R: Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 50: 693–698, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ju W, Nair V, Smith S, Zhu L, Shedden K, Song PXK, et al. : Tissue transcriptome-driven identification of epidermal growth factor as a chronic kidney disease biomarker. Sci Transl Med 7: 316ra193, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martini S, Eichinger F, Nair V, Kretzler M: Defining human diabetic nephropathy on the molecular level: Integration of transcriptomic profiles with biological knowledge. Rev Endocr Metab Disord 9: 267–274, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sampson MG, Robertson CC, Martini S, Mariani LH, Lemley KV, Gillies CE, et al. : Integrative genomics identifies novel associations with APOL1 risk genotypes in Black NEPTUNE subjects. J Am Soc Nephrol 27: 814–823, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reich HN, Tritchler D, Cattran DC, Herzenberg AM, Eichinger F, Boucherot A, et al. : A molecular signature of proteinuria in glomerulonephritis. PLoS One 5: e13451, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, et al. : A transcriptional profile of aging in the human kidney. PLoS Biol 2: e427, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balduzzi S, Rücker G, Schwarzer G: How to perform a meta-analysis with R: A practical tutorial. Evid Based Ment Health 22: 153–160, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu X, Eales JM, Akbarov A, Guo H, Becker L, Talavera D, et al. : Molecular insights into genome-wide association studies of chronic kidney disease-defining traits. Nat Commun 9: 4800, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rowland J, Akbarov A, Eales J, Xu X, Dormer JP, Guo H, et al. : Uncovering genetic mechanisms of kidney aging through transcriptomics, genomics, and epigenomics. Kidney Int 95: 624–635, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Billing AM, Ben Hamidane H, Bhagwat AM, Cotton RJ, Dib SS, Kumar P, et al. : Complementarity of SOMAscan to LC-MS/MS and RNA-seq for quantitative profiling of human embryonic and mesenchymal stem cells. J Proteomics 150: 86–97, 2017 [DOI] [PubMed] [Google Scholar]
- 72.Richmond RC, Davey Smith G: Commentary: Orienting causal relationships between two phenotypes using bidirectional Mendelian randomization. Int J Epidemiol 48: 907–911, 2019 [DOI] [PubMed] [Google Scholar]
- 73.Carlsson AC, Sundström J, Carrero JJ, Gustafsson S, Stenemo M, Larsson A, et al. : Use of a proximity extension assay proteomics chip to discover new biomarkers associated with albuminuria. Eur J Prev Cardiol 24: 340–348, 2017 [DOI] [PubMed] [Google Scholar]
- 74.Jovanovic D, Krstivojevic P, Obradovic I, Durdevic V, Dukanovic L: Serum cystatin C and beta2-microglobulin as markers of glomerular filtration rate. Ren Fail 25: 123–133, 2003 [DOI] [PubMed] [Google Scholar]
- 75.Niewczas MA, Pavkov ME, Skupien J, Smiles A, Md Dom ZI, Wilson JM, et al. : A signature of circulating inflammatory proteins and development of end-stage renal disease in diabetes. Nat Med 25: 805–813, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schenk S, Schoenhals GJ, de Souza G, Mann M: A high confidence, manually validated human blood plasma protein reference set. BMC Med Genomics 1: 41, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jia L, Zhang L, Shao C, Song E, Sun W, Li M, et al. : An attempt to understand kidney’s protein handling function by comparing plasma and urine proteomes. PLoS One 4: e5146, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lehallier B, Gate D, Schaum N, Nanasi T, Lee SE, Yousef H, et al. : Undulating changes in human plasma proteome profiles across the lifespan. Nat Med 25: 1843–1850, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suhre K, McCarthy MI, Schwenk JM: Genetics meets proteomics: Perspectives for large population-based studies. Nat Rev Genet 22: 19–37, 2021 [DOI] [PubMed] [Google Scholar]
- 80.Zoccali C, Vanholder R, Massy ZA, Ortiz A, Sarafidis P, Dekker FW, et al. : The systemic nature of CKD. Nat Rev Nephrol 13: 344–358, 2017 [DOI] [PubMed] [Google Scholar]
- 81.Prunotto M, Farina A, Lane L, Pernin A, Schifferli J, Hochstrasser DF, et al. : Proteomic analysis of podocyte exosome-enriched fraction from normal human urine. J Proteomics 82: 193–229, 2013 [DOI] [PubMed] [Google Scholar]
- 82.Clark CJ, Sage EH: A prototypic matricellular protein in the tumor microenvironment: Where there’s SPARC, there’s fire. J Cell Biochem 104: 721–732, 2008 [DOI] [PubMed] [Google Scholar]
- 83.Feng D, Ngov C, Henley N, Boufaied N, Gerarduzzi C: Characterization of matricellular protein expression signatures in mechanistically diverse mouse models of kidney injury. Sci Rep 9: 16736, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Marimuthu A, O’Meally RN, Chaerkady R, Subbannayya Y, Nanjappa V, Kumar P, et al. : A comprehensive map of the human urinary proteome. J Proteome Res 10: 2734–2743, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Francki A, Sage EH: SPARC and the kidney glomerulus: matricellular proteins exhibit diverse functions under normal and pathological conditions. Trends Cardiovasc Med 11: 32–37, 2001 [DOI] [PubMed] [Google Scholar]
- 86.Woroniecka KI, Park AS, Mohtat D, Thomas DB, Pullman JM, Susztak K: Transcriptome analysis of human diabetic kidney disease. Diabetes 60: 2354–2369, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lindenmeyer MT, Eichinger F, Sen K, Anders HJ, Edenhofer I, Mattinzoli D, et al. : Systematic analysis of a novel human renal glomerulus-enriched gene expression dataset. PLoS One 5: e11545, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nystrom J, Fierlbeck W, Granqvist A, Kulak SC, Ballermann BJ: A human glomerular SAGE transcriptome database. BMC Nephrol 10: 13, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ju W, Greene CS, Eichinger F, Nair V, Hodgin JB, Bitzer M, et al. : Defining cell-type specificity at the transcriptional level in human disease. Genome Res 23: 1862–1873, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wynn TA, Ramalingam TR: Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat Med 18: 1028–1040, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Haycock PC, Burgess S, Wade KH, Bowden J, Relton C, Davey Smith G: Best (but oft-forgotten) practices: The design, analysis, and interpretation of Mendelian randomization studies. Am J Clin Nutr 103: 965–978, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Davies NM, Holmes MV, Davey Smith G: Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 362: k601, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.