Abstract
Coronavirus disease 2019 (COVID-19) has caused more than 3 million deaths and infected more than 170 million individuals all over the world. Rapid identification of patients with COVID-19 is the key to control transmission and prevent depletion of hospitals. Several networks have been proposed to assist radiologists in diagnosing COVID-19 based on CT scans. However, CTs used in these studies are unavailable for other researchers to do deeper extensions due to privacy concerns. Furthermore, these networks are too heavy-weighted to satisfy the general trend applying on a computationally limited platform. In this paper, we aim to solve these two problems. Firstly, we establish an available dataset COVID-CTx, which contains 828 CT scans positive for COVID-19 across 324 patient cases from three open access data repositories. To our knowledge, it has the largest number of publicly available COVID-19 positive cases compared to other public datasets. Secondly, we propose a light-weighted hybrid neural network: Depthwise Separable Dense Convolutional Network with Convolution Block Attention Module (AM-SdenseNet). AM-SdenseNet synergistically integrates Convolutional Block Attention Module with depthwise separable convolutions to learn powerful feature representations while reducing the parameters to overcome the overfitting problem. Through experiments, we demonstrate the superior performance of our proposed AM-SdenseNet compared with several state-of-the-art baselines. The excellent performance of AM-SdenseNet can improve the speed and accuracy of COVID-19 diagnosis, which is extremely useful to control the spreading of infection.
Keywords: COVID-19 diagnosis, CT scan, Dataset light-weighted, DenseNet, Convolution block attention module, Depthwise separable convolution
1. Introduction
Coronavirus disease 2019 (COVID-19), named SARS-CoV-2 by the International Committee on Taxonomy of Viruses (ICTV), is a highly infectious respiratory disease. More than 170 million confirmed COVID-19 cases and 3 million deaths have been reported in roughly 200 different countries and territories, as of June 7 in 2021. Rapid identification of patients with COVID-19 has been recommended by World Health Organization (WHO) to control transmission and prevent depletion of hospitals. Reverse transcription-polymerase chain reaction (RT-PCR) serves as the gold standard for COVID-19 diagnosis. However, the total positive rate of RT-PCR for throat swab samples was reported to be approximately 30%–60% at the initial presentation [1]. Additionally, it takes 4–6 h to provide results, which is much slower than the speeding of the COVID-19. As a result, some infected patients cannot be identified early and continue to infect others unintentionally.
To mitigate the inefficiency of RT-PCR, chest computed tomography (CT) has been used to supplement RT-PCR testing of patients with suspected COVID-19 [2]. Several studies [3,4] have shown that CT scan manifests clear radiological findings of COVID-19 cases such as ground-glass opacities and consolidations. And CT scans are promised as a more efficient testing tool in serving for COVID-19 diagnosis with higher sensitivity [5]. However, chest CT contains hundreds of slices, which takes a long time for specialists to diagnose. Time pressure, heavy workload, and lack of experienced radiologists result in challenges in the imaging-based analysis of COVID-19 [6]. Fortunately, Artificial Intelligence (AI) has been widely used in the field of medical images, such as lung nodule [7], tuberculosis [8], breast cancer [9], tumor [10], etc. Rapidly developed AI-based automated CT image analysis tools can achieve high accuracy in the detection of Coronavirus positive patients. To alleviate the burden of medical professionals, there have been increasing efforts on developing computer-aided detection (CAD) systems to assist radiologists in diagnosing COVID-19 based on CT scans [[11], [12], [13], [14], [15], [16], [17], [18], [19], [20]].
The CAD systems for the detection of COVID-19 are mainly divided into two modules: segmentation and classification. Segmentation aims to segment lung regions where lesions are located, then the segmented regions could be further fed to a classification module for COVID-19 detection. Segmentation is an essential step in image processing and analysis for the assessment and quantification of COVID-19. It delineates the regions of interest (ROIs), e.g., lung and infected regions, in the CT images. To segment ROIs in CT, U-Net [21], U-Net++ [22], and V-Net [23] are widely used. In the classification module, there are several studies aiming to separate COVID-19 patients from Normal subjects. ResNet [24] is the most widely used network for diagnosing COVID-19 from CT scans [[14], [15], [16], [17], [18]]. Besides being based on a ResNet, some networks further combine with the attention mechanism and feature pyramid network (FPN) [25] to focus on important features [14,16]. Given the problem of limited datasets, studies adopt strategies such as weak label [11], transfer learning [19], human-in-the-loop [13], and data augmentation [20] to improve the evaluation indicators of the CAD systems. Although these studies have achieved good results, there are still two major hurdles: (1) CT scans used in these studies are unavailable for public access and use due to privacy concerns. Consequently, these works are difficult to copy and adapt, which greatly hinders the research and development of deep learning methods; (2) These networks are too heavy-weighted to satisfy the general trend applying on a computationally limited platform.
To address the first problem, we establish a publicly available dataset COVID-CTx. It is composed and modified by three open access data repositories: COVID-SIRM [26], COVID-Seg [27], and COVID19-CT [19]. To the best of our knowledge, COVID-CTx has the largest number of publicly available COVID-19 positive cases compared to other public datasets. All CT scans on these three datasets are confirmed by senior radiologists. For the second, we propose a light-weighted hybrid neural network: Depthwise Separable Dense Convolutional Network with Convolutional Block Attention Module (AM-SdenseNet). AM-SdenseNet uses a Dense Convolutional Network (DenseNet) [28] as the basic network. Compared with ResNet, DenseNet can better satisfy the general trend of application on a limited platform. At the same time, AM-SdenseNet synergistically applies Convolutional Block Attention Module (CBAM) [29] on DenseNet in each block with residual learning. This operation can greatly focus on important features and suppress unnecessary ones. In addition, we replace some regular convolutions in DenseNet with depthwise separable convolutions [30] to reduce training parameters while keeping network performance.
The paper is organized as follows. Section 2 introduces related works regarding existing accessible COVID-19 datasets, COVID-19 diagnosis systems, depthwise separable convolution, and attention module. Section 3 presents the COVID-CTx dataset, data preprocessing, and AM-SdenseNet architecture. In Section 4, we compare the performance of AM-SdenseNet with six state-of-the-art networks: MobileNet [31], InceptionV3 [32], ResNet50, VGG16 [33], DenseNet169, and DenseNet121. Finally, Section 5 concludes the paper and discusses future directions.
2. Related works
2.1. Existing accessible COVID-19 datasets
Due to privacy issues, few data sets with massive CT scans on COVID-19 are available for public access and use until now. Existing larger data sets on COVID-19 are COVID-SIRM, COVID-Seg, and COVID19-CT. The Italian Society of Medical Radiology (SIRM) has publicly provided 100 CT scans across 68 patient cases [26]. Each case in the COVID-SIRM provides detailed information of the lesions in the CT images. In addition, some cases are followed up and treated at intervals of about 4 days. COVID-Seg [27] contains 40 labeled COVID-19 CT scans. Left lung, right lung, and infections are labeled by two radiologists and verified by an experienced radiologist. The University of California constructs a publicly available COVID19-CT data set [19], which contains 349 CT scans that are positive for COVID-19 from 216 COVID-19 cases. All CTs were collected from public websites such as medRxiv, bioRxiv, journals, or papers related to COVID-19. Other existing data sets on COVID-19 are mainly X-ray images. For example, COVID-19 Radiography Database, generated by Chowdhury et al. [20], is comprised of 1200 COVID-19 positive images and 1341 normal images. It is worth noting that the current public data sets still have a very limited number of images for the training and testing of models, and the quality of data sets is insufficient.
2.2. COVID-19 diagnosis systems
During the outbreak of COVID-19, there have been increasing efforts on COVID diagnosis systems to perform screening COVID-19 based on CT scans. Segmentation is an essential step in COVID-19 diagnosis systems to assess and quantify COVID-19. The popular segmentation networks for CT scans in COVID-19 applications include U-Net, U-Net++, and V-Net. The U-Net, a U-shape network, is a popular technique for segmenting lung regions in medical images. For example, Zheng et al. [11] obtain all CT lung masks through a pre-trained U-Net in COVID-19 applications. Various U-Net, meanwhile, has been developed, reaching better segmentation results in COVID-19 image segmentation. Jin et al. [18] propose a two-stage pipeline for diagnosing COVID-19 in CT images, in which the whole lung is first detected by an efficient network based on U-Net++. The U-Net++ can greatly improve the performance of segmentation, as the network inserts a nested convolutional structure between the encoding and decoding path based on U-Net. In addition, CT provides high-quality 3D images for detecting COVID-19. Significantly, V-Net is a 3D image segmentation approach, where volumetric convolutions were applied instead of processing the input volumes slice-wise. To fully utilize the spatial information of CT, Shan et al. [13] propose a 3D segmentation system VB-Net, which combines V-Net with the bottle-neck structure [24]. Obviously, all of these networks can achieve better performance of segmentation. However, their training is difficult without adequate labeled data. In COVID-19 CT segmentation, sufficient labeled data for segmentation tasks is often unavailable since manual delineation for lung regions is labor-intensive and time-consuming.
Segmentation could be used to preprocess the CTs, and classification takes advantage of those segmentation results in the diagnosis. There are many studies aiming to separate COVID-19 patients from non-COVID-19 subjects. For example, Jin et al. [15] establish a deep learning system for COVID-19 detection, which outperforms five radiologists in more challenging tasks at a speed of two orders of magnitude above them. It fully proves that CAD systems can assist radiologists to accelerate the speed of diagnosing COVID-19. ResNet is the most widely used network in CAD systems. For example, Xu et al. [14] propose a diagnostic model, based on a ResNet18, to classify normal, viral, and COVID-19 from CT scans. Furthermore, they add a location-attention mechanism to improve the overall accuracy, which reaches 86.7%. Li et al. [34] preprocess the 2D slices to extract lung regions using U-Net, and a ResNet50 model combined with max-pooling for diagnosis. The model achieves results with a specificity of 96%, a sensitivity of 90%, and an AUC of 0.96 in identifying COVID-19. Similarly, Jin et al. [18] propose a U-Net++ based segmentation model for locating lesions and a ResNet50 based classification model for diagnosis. The specificity and sensitivity using the proposed U-Net++ and ResNet50 combined model are 92.2% and 97.4%.
Given the problem of limited datasets, He et al. [19] propose a self-supervision transfer learning method (Self-Trans). The network integrates contrastive self-supervised learning with transfer learning to learn powerful and unbiased feature representations for reducing over-fitting. The testing dataset shows an AUC of 0.94, even though the number of training CT scans is just a few hundred. Zheng et al. [11] propose a weakly-supervised network: 3D deep CNN (DeCoVNet), which can accurately predict the COVID-19 infectious probability in chest CT volumes without labeled lesions for training. DeepPneumonia, proposed by Song et al. [16], uses a ResNet50 backbone with the FPN and attention module as the feature-extracting part. The network can distinguish the COVID-19 patients from others with an excellent AUC of 99% and sensitivity of 93%. Additionally, those researches [11,19,20] all apply data augmentation to overcome the overfitting problem.
In summary, the above approaches have been proposed for CT-based COVID-19 diagnosis with generally promising results. But most of the mentioned COVID-19 classification techniques were training on large datasets. CT scans used in these studies are unavailable for public access and use, which greatly hinders the research and development of deep learning methods. In addition, these networks are too heavy-weighted to satisfy the general trend applying on a computationally limited platform.
2.3. Depthwise separable convolution
To achieve higher accuracy, the network has become much deeper and more complicated [33]. However, the general trend has been to achieve the recognition tasks in a timely fashion on a computationally limited platform [31]. Depthwise separable convolutions, firstly proposed by Sifre et al. [30], aim to reduce training parameters. The operation is to split the corresponding area and channel of the processed image. Later, Inception V1 and Inception V2 use a depthwise separable convolution as the first layer to lighten the network [35,36]. Howard et al. [31] introduce MobileNets based on depthwise separable convolutions. This network is mostly used in mobile and embedded vision applications, largely due to its lightness. Jin et al. [37] and Wang et al. [38] also do related work aiming at reducing the size and computational cost of convolutional neural networks. Chollet et al. [39] present a novel architecture based on depthwise separable convolutions, named Xception. It shows large gains on the JFT dataset [40].
2.4. Attention module
It's well known that attention plays a crucial role in human perception [41]. Presently, there have been several studies about attention mechanisms to improve the performance of CNNs. Wang et al. [40] propose an encoder-decoder style attention module: Residual Attention Network. The network not only performs well but is also robust to noisy inputs. But directly computing the 3d attention, this network has more computational and parameter overhead. Hu et al. [42] propose a Squeeze-and-Excitation module to exploit the inter-channel relationship. Although using a global average-pooled to compute channel-wise attention, they miss spatial attention. Woo et al. [43] introduce CBAM, which learns channel attention and spatial attention separately. For the qualitative analysis, the authors prove that CBAM outperforms other attention modules, comparing the accuracy improvement from others. Notably, the attention mechanism is reported as an efficient localization method in screening, which can be adopted in COVID-19 applications [14,44].
3. Material and methods
Dataset plays a critical and essential role in deep learning models. However, few datasets with massive CT scans for COVID-19 are available for public access and use until now. To address this problem, we establish a publicly available dataset COVID-CTx. Though the largest of its kind, COVID-CTx still can't meet the training of large networks. For this problem, we propose a light-weighted hybrid neural network AM-SdenseNet for combating overfitting. In the next subsections, we will introduce the COVID-CTx, data preprocessing, and AM-SdenseNet architecture in turn.
3.1. COVID-CTx dataset
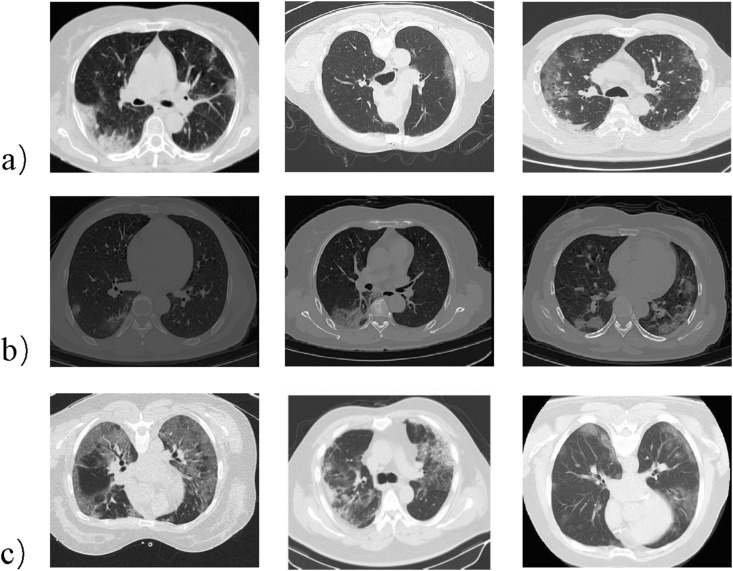
COVID-CTx contains 828 CT scans that are positive for COVID-19 across 324 patient cases. The CT scans from the same patient are visually similar. Hence, our dataset has the largest number of publicly available COVID-19 positive cases and richer features about COVID-19 compared to other public datasets. To build the COVID-CTx, we collected positive CTs for COVID-19 from three open access data repositories: COVID-SIRM, COVID-Seg, and COVID19-CT. Our dataset contains 828 CT scans positive about COVID-19, of which 100 CTs are from COVID-SIRM, 379 CTs are from COVID-Seg, and 349 CTs are from COVID19-CT. In addition, COVID-CTx also contains 1000 negative CT scans negative for COVID-19. Those CT scans, obtained from LUNA16 [45], are normal or containing lung nodules. Fig. 1 shows example CT images from COVID-CTx, of which a) is from COVID-SIRM, b) is from COVID-Seg, and c) is from COVID19-CT.
Fig. 1.
Example CT images from the COVID-CTx dataset, which comprises 828 images across 324 patient cases from three open access data repositories: a). COVID-SIRM, b). COVID-Seg, c). COVID19-CT.
3.2. Data preprocessing
3.2.1. Lung segmentation
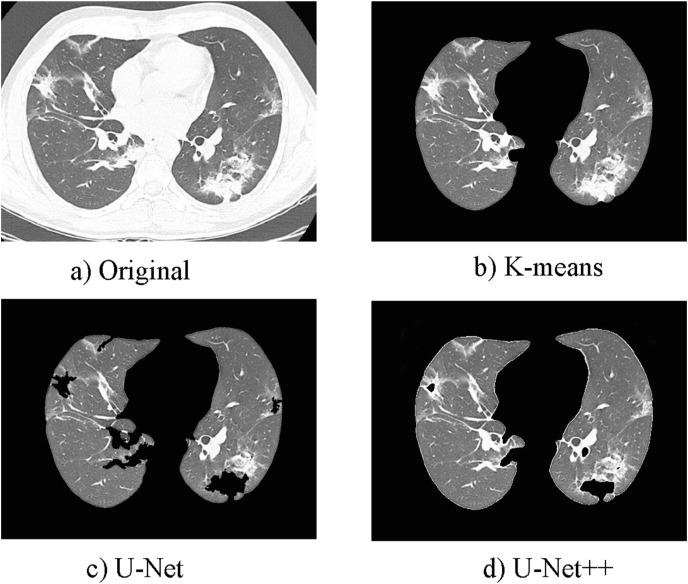
Lung segmentation is an essential step in COVID-19 diagnosis systems to assess and quantify COVID-19. But the training of segmentation networks for CT scans in COVID-19 applications is difficult. Sufficient labeled data for segmentation tasks is often unavailable since manual delineation for lung regions is labor-intensive and time-consuming. Here, U-Net and U-Net++ are first trained from scratch on the LUNA16 and fine-tuned on the COVID-CTx, of which the lung masks are produced by us. In addition, we chose K-means clustering [46] to obtain lung regions due to its simple principle and easy implementation [47,48]. Fig. 2 shows results achieved by the above three lung segmentation methods. Obviously, K-means reaches a better segmentation result on COVID-19 image segmentation. This demonstrates the effectiveness of K-means, which is more suitable for the segmentation task with insufficient labeled data. Consequently, we segment lung regions by K-means clustering in this study.
Fig. 2.
Results achieved by three lung segmentation methods. a). Original CT; b). Lung achieved by K-means; c). Lung achieved by U-Net; d). Lung achieved by U-Net++.
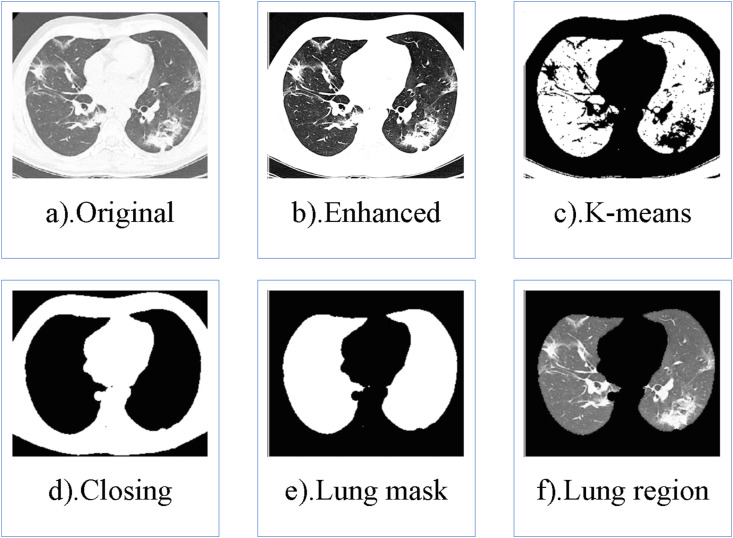
The process of lung segmentation is shown in Fig. 3 , and the specific method is as follows.
-
(1)
The original CT scan is preprocessed to enhance the contrast.
-
(2)
We separate the foreground (opaque tissue) and background (transparent tissue, the lungs) in the image based on the K-means.
-
(3)
We use the morphological closing operation to eliminate the residual trachea.
-
(4)
The hole filling operation algorithm is used to fill the maximum connectivity area in the reverse closing image. Subsequently, the filling image subtracts the reverse closing image to get the lung mask.
-
(5)
The lung region is extracted by its multiplication with the original CT scan.
Fig. 3.
The process of lung segmentation. a). Original CT; b). Contrast-enhanced CT; c). CT after k-means segmentation; d). CT after closing operation; e). Lung mask; f). Lung region.
3.2.2. Data augmentation
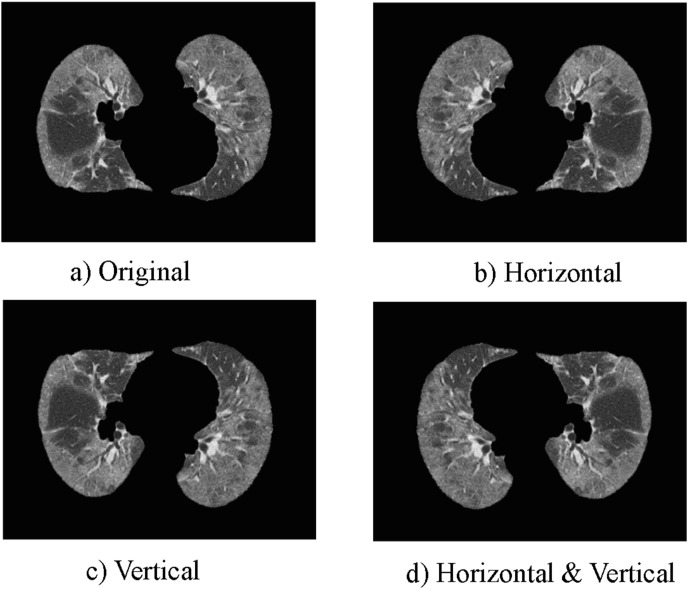
Though the largest of its kind, COVID-CTx may still have the over-fitting problem for data-hungry deep learning models. Therefore, two different image augmentation techniques (horizontal mirror and vertical mirror) are utilized to generate COVID-19 training images, as shown in Fig. 4 . The.
Fig. 4.
The methods of augmentation. a). Original CT; b). CT after horizontal mirror; c). CT after vertical mirror; d). CT after horizontal and vertical mirrors.
horizontal and vertical mirrors used for image augmentation are done by mirroring the training images in the horizontal and vertical directions, respectively. Table 1 summarizes the number of images per class used for training, validation, and testing for each fold. The number of training samples increases from 1098 to 4392.
Table 1.
Number of CTs per class before and after data augmentation.
| Dataset | Total number | Training without augmentation |
Training with augmentation |
||||
|---|---|---|---|---|---|---|---|
| Train | Validation | Test | Train | Validation | Test | ||
| COVID-CTx | 1828 | 1098 | 365 | 365 | 4392 | 365 | 365 |
3.3. AM-SdenseNet architecture
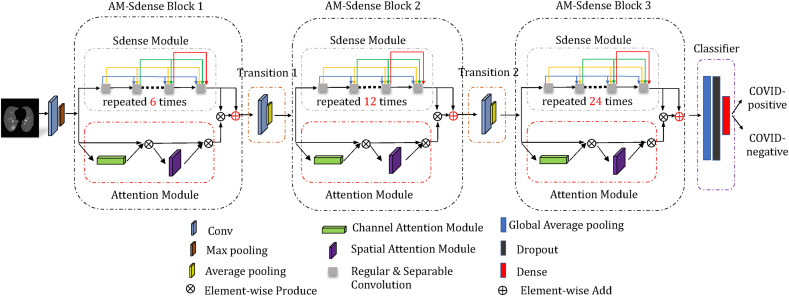
As shown in Fig. 5 , AM-SdenseNet is mainly composed of three AM-Sdense blocks, two transition layers, and one classifier. The AM-Sdense blocks use a DenseNet as the basic network structure to extract image features. Simultaneously, we replace some regular convolutions in the DenseNet with depthwise separable convolutions to reduce training parameters while keeping network performance. In addition, we also apply CBAM on the DenseNet in each AM-Sdense block with residual learning. This operation aims to improve the representation of objective lesion features and suppress the less relevant ones. The layers between two contiguous dense blocks are transition layers, which do convolution and pooling. It consists of batch normalization (BN) [36] layer and 1 × 1 convolution layer followed by 2 × 2 average pooling layer. The output of the last block is fed to the classifier for the final prediction of lung segmentation. The classifier contains a global average pooling layer, a dropout layer with probability p = 0.5, and a dense layer with the sigmoid activation function. We now detail the key components of AM-Sdense blocks.
Fig. 5.
AM-SdenseNet architecture.
3.3.1. a.m.-sdense block
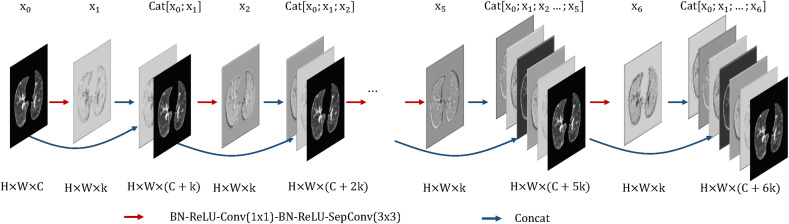
In our network, the number of layers in the three AM-Sdense blocks are 6, 12, 24, respectively. Besides, the growth rate of our network is k = 24 and the compression of our network is θ = 1. A 6-layer dense block with a growth rate of k = 24 is shown in Fig. 6 . Our proposed network takes all preceding feature-maps with direct connections as input. Consequently, the channel count used for the input of the following layer increases with the growth rate k. The layer can be received in Equation (1).
| (1) |
Where (X 0, X 1, …, X i−1) are the feature-maps produced in layers (0, 1, …, i − 1). Cat(⋅) means to concatenate all input in the channel axis. H i(⋅) means four consecutive operations: BN, rectified linear unit (ReLU) [49], convolution (Conv), and depthwise separable convolution (SepConv). The version of H i is BN-ReLU-Conv(1 × 1)-BN-ReLU-SepConv(3 × 3).
Fig. 6.
A 6-layer dense block with a growth rate of k.
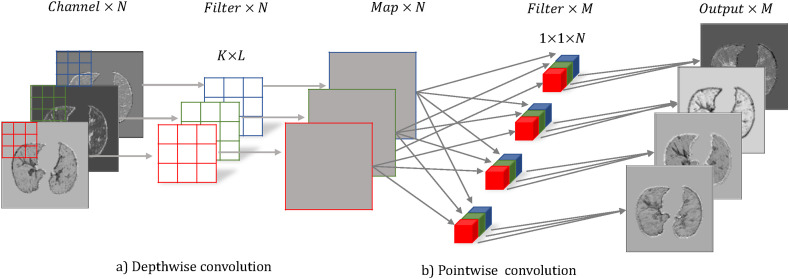
3.3.2. Depthwise separable convolution
Many networks show large gains by using depthwise separable convolutions [35,36,39]. Therefore, we replace some regular convolutions in DenseNet with depthwise separable convolutions to reduce the complexity of the model as much as possible. A depthwise separable convolution consists of two parts: a depthwise convolution and a pointwise convolution. The depth-wise convolution is a spatial convolution performed independently over every channel of input. And the pointwise convolution is a regular convolution with 1 × 1 windows. It projects the channels computed by the depthwise convolution into a new channel space. Fig. 7 shows the schematic diagram.
Fig. 7.
Depthwise separable convolution.
The specific calculation formulas of the two convolutions are as follows.
| (2) |
| (3) |
Where y is an image, and (i, j) represents pixels. The W(k, l, m) is the kernel with size (K, L, M). W p and W d are the parameters of each separable convolution.
The parameters of a regular convolution and a depthwise separable convolution are shown in Table 2 . As can be seen above, the numbers of parameters for a regular convolution and a depthwise separable convolution are M × K × L and M + K × L, respectively. When M is much bigger than 1(as is usually the case), the parameters of separable convolution are much smaller than those of a regular convolution. In this paper, we replace some regular convolutions in DenseNet with depthwise separable convolutions. This operation reduces network parameters by 17.9%.
Table 2.
Parameter count comparison across convolution types.
| Convolution type | Parameters |
|---|---|
| Regular | M × K × L |
| Depthwise Separable | M + K × L |
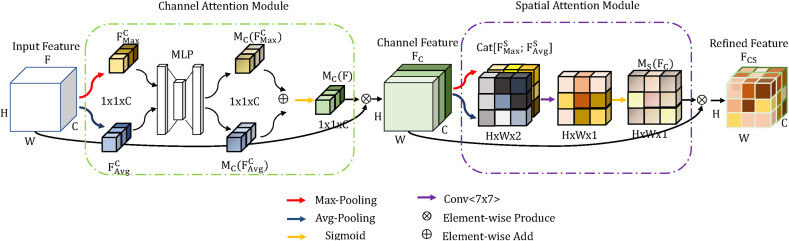
3.3.3. CBAM
Considering the limited data and computationally limited platform, we apply the CBAM on DenseNet to focus on important features and suppress the less relevant ones. The overview of CBAM is shown in Fig. 8 . The channel attention module exploits the inter-channel relationship of features. And the spatial attention module subsequently generates a spatial attention map by utilizing the inter-spatial relationship of features. Given the input feature map F ∈ R H×W×C, the channel attention module uses a global average pooling and a global max pooling to extract the average-pooled features and the max-pooled features respectively. Both features are passed through a multi-layer perceptron (MLP), and then summed element-by-element to get a channel attention map M c(F) ∈ 1 × 1 × C. The channel attention map M c(F) and input feature map F are multiplied element-wise to exploits the inter-channel relationship of the features map F. So that the channel feature map F C ∈ R H×W×C is being generated as Equation (4).
| (4) |
Where σ(⋅) denotes a sigmoid function and ⊗ denotes element-wise multiplication.
Fig. 8.
The overview of CBAM.
In the spatial attention module, the average pooling and max pooling along the channel axis are used to extract two feature maps and . Subsequently, two feature maps concatenated in the channel axis are forward to a convolution layer to produce a spatial attention map M s(F) ∈ H × W × 1. The channel attention map M s(F) and input feature map F C are multiplied element-wise to exploits the inter-spatial relationship of the features map F C. Finally, the refined feature map F CS ∈ H × W × C is computed as Equation (5).
| (5) |
Where Conv(⋅) represents a convolution operation with a filter size of 7 × 7.
In our research, we apply CBAM on DenseNet with residual learning, as shown in Equation (6).
| (6) |
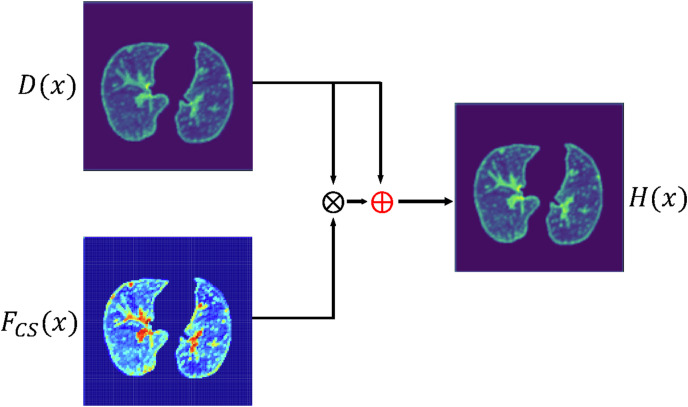
Where H(x) is the output of the AM-Sdense block, x is the input of CBAM and dense block. F CS(x) is the output of CBAM. And D(x) is the output of the dense block.
It is worth noting that residual learning can greatly avoid the problem of interference by the attention mechanism. Even when the CBAM output F CS(x) is 0, H(x) still retains main features in D(x). As clearly seen from Fig. 9 , CBAM can greatly focus on lesion features in F CS(x). Subsequently, lesions in H(x) perform more prominent than those in D(x).
Fig. 9.
The diagram of residual learning method.
3.3.4. The details of AM-SdenseNet architecture
When dataset is limited, conventional shallow CNN models produce better results as compared to deeper models [50]. Therefore, AM-SdenseNet only has three dense blocks. The number of layers in the three AM-Sdense blocks are 6, 12, 24, respectively. In addition, the growth rate for our networks is k = 24 and the compression for our network is θ = 1. AM-SdenseNet takes the lung regions as the input images with size 512 × 512 × 3. Before the data enters the first AM-Sdense block, a convolution layer and a max pooling layer with 48 output channels are performed on the input images. In addition, we use 1 × 1 convolution followed by 2 × 2 average pooling between two contiguous dense blocks to change feature-map sizes. At the end of the thirdly dense block, a global average pooling is performed and then a dropout layer with probability p = 0.5 is attached. Finally, a dense layer with the sigmoid activation function directly outputs the probabilities of being COVID-positive and COVID-negative. Table 3 shows the details of AM-SdenseNet architecture, where each “conv” layer corresponds to the sequence BN-ReLU-Conv and “sconv” corresponds to the sequence BN-ReLU-SepConv.
Table 3.
The parameters of AM-SdenseNet architecture.
| layers | Output size | AM-SdenseNet |
|---|---|---|
| Convolution | 256 × 256 | 7 × 7 conv, s = 2 |
| Pooling | 128 × 128 | 3 × 3 maxpool, s = 2 |
| AM-Sdense Block 1 | 128 × 128 | |
| Transition 1 | 128 × 128 | 1 × 1 conv |
| 64 × 64 | 2 × 2 avgpool, s = 2 | |
| AM-Sdense Block 2 | 64 × 64 | |
| Transition 2 | 64 × 64 | 1 × 1 conv |
| 32 × 32 | 2 × 2 avgpool, s = 2 | |
| AM-Sdense Block 3 | 32 × 32 | |
| Classification | 1 × 1 | global average pool dropout, p = 0.5 Dense, sigmoid |
4. Experiments and discussions
We empirically demonstrate the effectiveness of AM-SdenseNet on the COVID-19 detection and compare it with state-of-the-art architectures, such as DenseNet and its variants. In the following subsections, we will introduce experimental settings, the influence of CBAM and depthwise separable convolution on the AM-SdenseNet, and comparisons between our approach with several state-of-the-art baselines on the COVID-19 classification task.
4.1. Experimental settings
Our COVID-CTx is used in all approaches. Besides COVID-CTx, we also evaluate seven networks trained on COVID-19 Radiology Database. All images are resized to 512 × 512 × 3. The radio of training set, validation set, and test set is 0.6 : 0.2: 0.2. It is worth noting that each patient belongs to a single set. BN is used through all models, and binary cross-entropy serves as the loss function. The optimizer is Adam [51] with an initial learning rate of 5e − 5 and a weight decay of 1e − 7. The models are implemented in Keras and trained with Tesla V100. All models are trained with 40 epochs and a mini-batch size of 16. We evaluate different networks using four metrics: accuracy, precision, recall, and F1-score, which are shown in Table 4 . Where TP and TN mean true positive and negative parameters, respectively. FP and FN are false positive and false negative values respectively. 3-Folds cross-validation is applied by all approaches.
Table 4.
Evaluation metrics used in COVID-19 detection.
| Metrics | Equations | Notes |
|---|---|---|
| Accuracy | ACC = (TP + TN)/(TP + TN + FP + FN) | |
| Precision | PPV = TP/(TP + FP) | Positive Predicted Value (PPV) |
| Recall | SE = TP/(TP + FN) | True Positive Rate (TPR) or Sensitivity |
| F1-score | F1 = 2TP/(2 TP + FP + FN) | Harmonic mean of Precision and Recall |
4.2. Evaluation of two modules
To demonstrate the efficacy of AM-SdenseNet and investigate the effects of CBAM and depthwise separable convolution, we first experiment on the denseNet (initial network), AM-denseNet (combined with CBAM), and AM-SdenseNet. The comparative performance for three CNNs for COVID-19 classification problem with and without augmentation is shown in Table 5 and comparative AUC curves are shown in Fig. 10 . Additionally, we also built the confusion matrices for three models, as shown in Fig. 11 .
Table 5.
Metrics for three networks with and without augmentation.
| Schemes | Models | Accuracy (%) | Precision (%) | Recall (%) | F1 (%) | Parameters(M) |
|---|---|---|---|---|---|---|
| Without augmentation | denseNet | 96.71 | 97.40 | 95.89 | 96.52 | 3.44 |
| AM-denseNet | 97.53 | 97.73 | 97.15 | 97.42 | 4.12 | |
| AM-SdenseNet | 98.36 | 98.19 | 98.40 | 98.29 | 3.38 | |
| With augmentation | denseNet | 97.21 | 97.60 | 97.24 | 97.41 | 3.44 |
| AM-denseNet | 97.55 | 97.29 | 97.60 | 97.44 | 4.12 | |
| AM-SdenseNet | 99.18 | 99.32 | 98.97 | 99.14 | 3.38 |
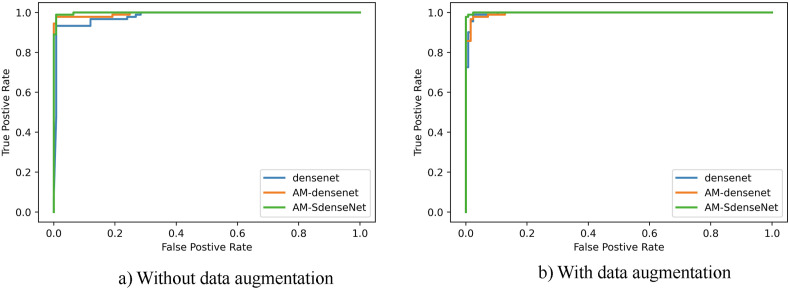
Fig. 10.
Comparison of the ROC curve for COVID-19 classification using CNN based models without a) and with b) image augmentation.
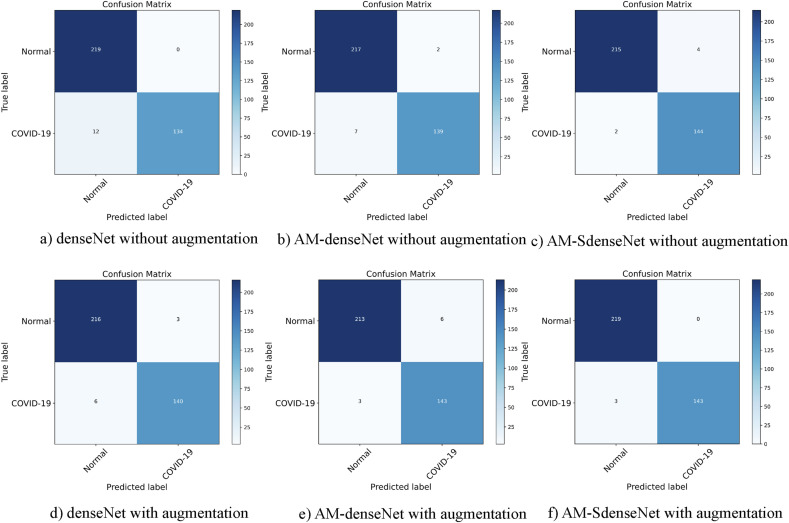
Fig. 11.
Confusion matrices for three models with and without data augmentation.
As shown in Table 5, three models with augmentation all show some increase in four performance metrics. It proves that the benefits of augmentation are highly significant when the data set is limited. Comparing denseNet with AM-denseNet, we can see that the network combined with CBAM yields higher classification performance. Comparing AM-denseNet with AM-SdenseNet, depthwise separable convolution reduces network parameters by 17.9% and improves precision by 2.03% at the same time. We empirically demonstrate depthwise separable convolution can reduce training parameters while keeping network performance. In summary, the performance of AM-SdenseNet benefits a lot from CBAM and depthwise separable convolution. Trained with augmented images, AM-SdenseNet produces the highest accuracy of 99.18%, precision of 99.32%, recall of 98.97%, and F1 score of 99.14% with the least parameters 3.38 M. Fig. 10 shows AM-SdenseNet achieves the best curve through data augmentation. According to the confusion matrices presented in Fig. 11, AM-SdenseNet can classify the Normal class with 0 misclassifications and classify the COVID-19 class with 3 misclassifications.
4.3. Comparison of seven models
Few data sets with massive CT scans on COVID-19 are available for public access and use until now due to privacy concerns. Hence, to illustrate the effectiveness of AM-SdenseNet in COVID-19 diagnosis, we separately investigate the performance of networks trained on COVID-CTx and COVID-19 Radiography Database with different backbones, including MobileNet, InceptionV3, ResNet50, VGG16, DenseNet169, DenseNet121, and AM-SdenseNet.
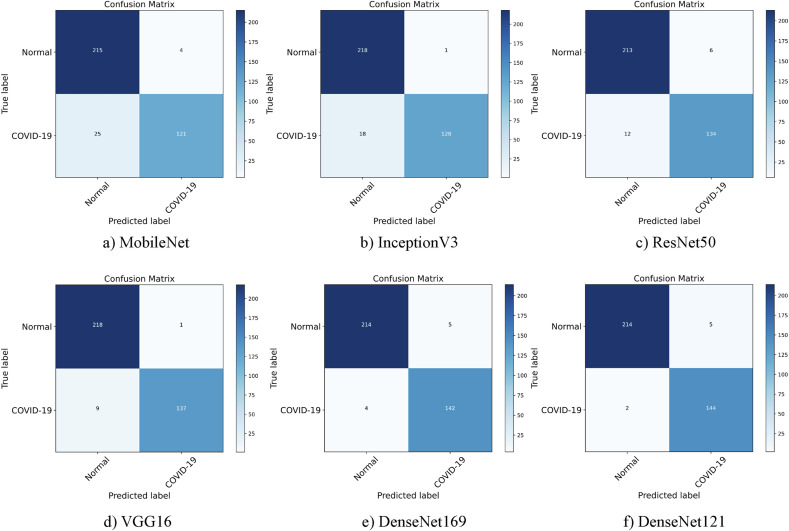
Table 6 shows the metrics for seven networks trained on COVID-CTx and COVID-19 Radiography Database. Trained on COVID-CTx, DenseNet169 and VGG16 present a better performance than InceptionV3 and ResNet50 with fewer parameters. What's more, DenseNet121 outperforms DenseNet169 on the COVID-19 classification task, achieving higher accuracy and precision. This significantly proves that conventional shallow CNN models produce better results as compared to deeper models when the data set is limited. But the network is too simple to extract features well. For instance, MobileNet lacks stronger feature representation learning capabilities, so as to produce low classification performance. Compared with MobileNet, DenseNet can strengthen feature propagation and substantially reduce the number of parameters. The performance benefits a lot from dense connections. For example, DenseNet121 and DenseNet169 achieve good performance on the COVID-19 classification. AM-SdenseNet, however, has a much smaller number of parameters but the performance is better than deeper networks such as DenseNet121 and DenseNet169. The reason is that large-sized networks are more prone to over-fitting, especially considering that our data set is fairly small. Under such circumstances, CBAM and depthwise separable convolution have a better chance to play their value. According to the confusion matrices presented in Figs. 11 and 13, AM-SdenseNet presents the highest radio of classifying the COVID-19 class and the Normal class. Other models have achieved good performance in the classification of the Normal class, but their performance in the classification of COVID-19 is not good enough.
Table 6.
Metrics for seven networks trained on COVID-CTx and COVID-19 Radiography Database.
| Data sets | Models | Accuracy (%) | Precision (%) | Recall (%) | F1 (%) | Parameters(M) |
|---|---|---|---|---|---|---|
| COVID-CTx | MobileNet | 92.05 | 93.19 | 90.53 | 91.49 | 3.23 |
| InceptionV3 | 94.79 | 95.80 | 93.61 | 94.46 | 21.80 | |
| ResNet50 | 95.07 | 95.19 | 94.52 | 94.83 | 24.97 | |
| VGG16 | 97.26 | 97.66 | 96.69 | 97.12 | 14.72 | |
| DenseNet169 | 97.53 | 97.38 | 97.49 | 97.43 | 12.64 | |
| DenseNet121 | 98.08 | 97.85 | 98.17 | 98.00 | 7.04 | |
| AM-SdenseNet | 99.18 | 99.32 | 98.97 | 99.14 | 3.38 | |
| COVID-19 Radiography Database | MobileNet | 94.09 | 93.35 | 92.26 | 92.86 | 3.23 |
| InceptionV3 | 97.24 | 95.25 | 96.86 | 96.23 | 21.80 | |
| ResNet50 | 96.44 | 95.06 | 94.85 | 95.25 | 24.97 | |
| VGG16 | 97.05 | 97.35 | 96.35 | 96.92 | 14.72 | |
| DenseNet169 | 98.03 | 98.45 | 97.59 | 98.03 | 12.64 | |
| DenseNet121 | 97.62 | 97.32 | 96.01 | 96.84 | 7.04 | |
| AM-SdenseNet | 98.62 | 98.66 | 97.56 | 98.33 | 3.38 |
Fig. 13.
Confusion matrices for six models.
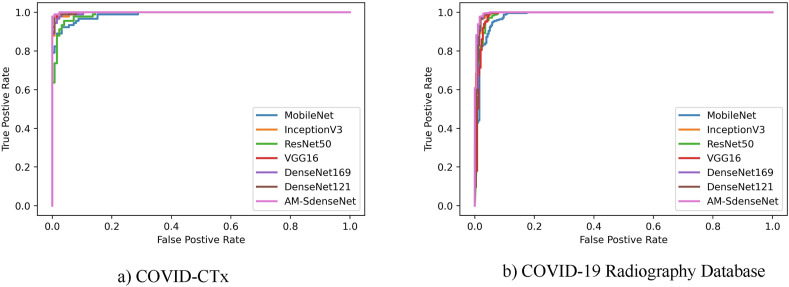
Compared with CT, X-ray is more easily accessible around the world. However, due to the ribs projected onto soft tissues in 2D and thus confounding image contrast, the classification of X-ray images is even more challenging. Trained on the COVID-19 Radiography Database, AM-SdenseNet reaches the best results with an accuracy of 98.62%. It is confirmed that AM-SdenseNet is universal in COVID-19 diagnosis. In Fig. 12 , AM-SdenseNet also achieves the best curve on the COVID-19 classification task. Through the above experiments, AM-SdenseNet achieves the best performance on the COVID-19 classification task compared to other models. Such experimental results not only illustrate the effectiveness of COVID-CTx, but also provide concrete evidence that AM-SdenseNet has stronger capabilities to improve the speed and accuracy of COVID-19 diagnosis.
Fig. 12.
Comparisons of the ROC curve for COVID-19 classification on seven different CNNs.
4.4. Comparison with the latest methods
The comparison of AM-SdenseNet with the latest approaches for COVID-19 classification is presented in this section. Table 7 shows that only limited COVID-19 images are used in most of these approaches. Given the problem of insufficient samples, studies adopt models such as DenseNet, ResNet, and ensemble networks to combat over-fitting. While considering the performance metrics in Table 7, our approach outperforms the considered state-of-the-art approaches, achieving the best classification performance from CT scan and chest X-ray images, respectively. It is further confirmed that AM-SdenseNet is superior to other algorithms in COVID-19 diagnosis and useful to control the spreading of infection.
Table 7.
Related studies with medical images for COVID-19 diagnosis.
| Model | Modality | COVID-19 | Open-source | Parameters (M) | Accuracy (%) |
|---|---|---|---|---|---|
| DeCoVNet [11] | CT | 313 cases | No | – | 90.01 |
| DSAE [52] | CT | 317 cases | No | – | 97.14 |
| AM-SdenseNet | CT | 324 cases | Yes | 3.38 | 99.18 |
| MobileNetV2 + InceptionV3 [50] | X-ray | 1000 images | Yes | 28.50 | 98.45 |
| Covid-ResNet [53] | X-ray | 1200 images | Yes | 25.60 | 96.23 |
| AM-SdenseNet | X-ray | 1200 images | Yes | 3.38 | 98.62 |
5. Conclusions and future works
In our research, we aim to develop a sample and efficient CAD system to diagnose COVID-19 from CT scans. To accelerate the open study in this area, we establish a publicly available data set COVID-CTx. To our knowledge, it has the richest features about COVID-19 compared to other public data sets to date. Although the largest informative, it still has a risk of overfitting for data-hungry deep learning models. For this problem, we propose a light-weighted hybrid neural network: AM-SdenseNet. The network synergistically applies CBAM on DenseNet in each block with residual learning, which can greatly improve the representation of objective lesion features and suppress the less relevant ones. In addition, we replace some regular convolutions in DenseNet with depthwise separable convolutions to reduce training parameters while keeping network performance. Through experiments, it has been proved that AM-SdenseNet can greatly improve the speed and accuracy of COVID-19 diagnosis, which is extremely useful to control the spreading of infection.
Though the largest of its kind, COVID-CTx still can't meet the training of large networks, which need a larger dataset. However, deep learning with small samples is still an important research direction in the future. Unsupervised learning methods such as stack auto-encoder [54] and restricted boltzmann machine [55] can be used for reference. Considering the actual clinical need, the CAD system can be combined with the hospital's imaging system and electronic medical records to achieve the follow-up treatment of patients in the future.
Funding
No funds, grants, or other support was received.
CRediT authorship contribution statement
Qian Li: Conceptualization of this study, methodology, dataset, experiments, and writing paper. Jiangbo Ning: Investigation, validation, writing, review and editing. Jianping Yuan: Data processing. Ling Xiao: Validation and formal analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest ct and rt-pcr testing for coronavirus disease 2019 (covid-19) in China: a report of 1014 cases. Radiology. 2020;296:E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanne J.P., Little B.P., Chung J.H., Elicker B.M., Ketai L.H. 2020. Essentials for Radiologists on Covid-19: an Update—Radiology Scientific Expert Panel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung M., Bernheim A., Mei X., Zhang N., Huang M., Zeng X., Cui J., Xu W., Yang Y., Fayad Z.A., et al. Ct imaging features of 2019 novel coronavirus (2019-ncov) Radiology. 2020;295:202–207. doi: 10.1148/radiol.2020200230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lomoro P., Verde F., Zerboni F., Simonetti I., Borghi C., Fachinetti C., Natalizi A., Martegani A. Covid-19 pneumonia manifestations at the admission on chest ultrasound, radiographs, and ct: single-center study and comprehensive radiologic literature review. European journal of radiology open. 2020;7:100231. doi: 10.1016/j.ejro.2020.100231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest ct for covid-19: comparison to rt-pcr. Radiology. 2020;296:E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang M., Xia C., Huang L., Xu S., Qin C., Liu J., Cao Y., Yu P., Zhu T., Zhu H., et al. Deep learning-based triage and analysis of lesion burden for covid-19: a retrospective study with external validation. The Lancet Digital Health. 2020;2:e506–e515. doi: 10.1016/S2589-7500(20)30199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hua K.-L., Hsu C.-H., Hidayati S.C., Cheng W.-H., Chen Y.-J. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets Ther. 2015;8 doi: 10.2147/OTT.S80733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lakhani P., Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284:574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 9.Bejnordi B.E., Veta M., Van Diest P.J., Van Ginneken B., Karssemeijer N., Litjens G., Van Der Laak J.A., Hermsen M., Manson Q.F., Balkenhol M., et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama. 2017;318:2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohsen H., El-Dahshan E.-S.A., El-Horbaty E.-S.M., Salem A.-B.M. Classification using deep learning neural networks for brain tumors. Future Computing and Informatics Journal. 2018;3:68–71. [Google Scholar]
- 11.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. MedRxiv; 2020. Deep Learning-Based Detection for Covid-19 from Chest Ct Using Weak Label. [Google Scholar]
- 12.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Chen Q., Huang S., Yang M., Yang X., et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shan F., Gao Y., Wang J., Shi W., Shi N., Han M., Xue Z., Shen D., Shi Y. 2020. Lung Infection Quantification of Covid-19 in Ct Images with Deep Learning, arXiv Preprint arXiv:2003. [Google Scholar]
- 14.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yu L., Ni Q., Chen Y., Su J., et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6:1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin C., Chen W., Cao Y., Xu Z., Tan Z., Zhang X., Deng L., Zheng C., Zhou J., Shi H., et al. Development and evaluation of an artificial intelligence system for covid-19 diagnosis. Nat. Commun. 2020;11:1–14. doi: 10.1038/s41467-020-18685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., Chen J., Wang R., Zhao H., Zha Y., et al. Deep learning enables accurate diagnosis of novel coronavirus (covid-19) with ct images. IEEE ACM Trans. Comput. Biol. Bioinf. 2021 doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., et al. Using artificial intelligence to detect covid-19 and community-acquired pneumonia based on pulmonary ct: evaluation of the diagnostic accuracy. Radiology. 2020;296:E65–E71. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin S., Wang B., Xu H., Luo C., Wei L., Zhao W., Hou X., Ma W., Xu Z., Zheng Z., et al. Ai-assisted ct imaging analysis for covid-19 screening: building and deploying a medical ai system in four weeks. MedRxiv. 2020 doi: 10.1016/j.asoc.2020.106897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X., Yang X., Zhang S., Zhao J., Zhang Y., Xing E., Xie P. MedRxiv; 2020. Sample-efficient Deep Learning for Covid-19 Diagnosis Based on Ct Scans. [Google Scholar]
- 20.Chowdhury M.E., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam K.R., Khan M.S., Iqbal A., Al Emadi N., et al. Can ai help in screening viral and covid-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 21.Ronneberger O., Fischer P., Brox T., U-net . International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer; 2015. Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 22.Zhou Z., Siddiquee M.M.R., Tajbakhsh N., Liang J. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer; 2018. Unet++: a nested u-net architecture for medical image segmentation; pp. 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milletari F., Navab N., Ahmadi S.-A., V-net . 2016 Fourth International Conference on 3D Vision (3DV) IEEE; 2016. Fully convolutional neural networks for volumetric medical image segmentation; pp. 565–571. [Google Scholar]
- 24.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 25.Lin T.-Y., Dollár P., Girshick R., He K., Hariharan B., Belongie S. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Feature pyramid networks for object detection; pp. 2117–2125. [Google Scholar]
- 26.The Italian Society of Medical Radiology Covid-19 cases. 2021. https://www.eurorad.org/advanced-search?search=COVID/gt.html accessed.
- 27.Jun M., Ge C., Yixin W., Xingle A., Jiantao G., Minqing Z., Xin L., Xueyuan D., Jian He. Covid-19 ct lung and infection segmentation dataset. 2021. https://zenodo.org/record/3757476/#.YMx8zhP7TDI/gt.html accessed.
- 28.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 29.Woo S., Park J., Lee J.-Y., Kweon I.S. Proceedings of the European Conference on Computer Vision. ECCV); 2018. Cbam: convolutional block attention module; pp. 3–19. [Google Scholar]
- 30.Sifre L., Mallat S. 2014. Rigid-motion Scattering for Texture Classification. arXiv preprint arXiv:1403.1687. [Google Scholar]
- 31.Howard A.G., Zhu M., Chen B., Kalenichenko D., Wang W., Weyand T., Andreetto M., Adam H., Mobilenets . 2017. Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv preprint arXiv:1704. [Google Scholar]
- 32.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [Google Scholar]
- 33.Simonyan K., Zisserman A. 2014. Very Deep Convolutional Networks for Large-Scale Image Recognition; p. 1556. arXiv preprint arXiv:1409. [Google Scholar]
- 34.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., et al. 2020. Artificial Intelligence Distinguishes Covid-19 from Community Acquired Pneumonia on Chest Ct, Radiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szegedy C., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D., Erhan D., Vanhoucke V., Rabinovich A. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2015. Going deeper with convolutions; pp. 1–9. [Google Scholar]
- 36.Ioffe S., Szegedy C. International Conference on Machine Learning. PMLR; 2015. Batch normalization: accelerating deep network training by reducing internal covariate shift; pp. 448–456. [Google Scholar]
- 37.Jin J., Dundar A., Culurciello E. 2014. Flattened Convolutional Neural Networks for Feedforward Acceleration; p. 5474. arXiv preprint arXiv:1412. [Google Scholar]
- 38.Wang M., Liu B., Foroosh H. Proceedings of the IEEE International Conference on Computer Vision Workshops. 2017. Factorized convolutional neural networks; pp. 545–553. [Google Scholar]
- 39.Chollet F. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Xception: deep learning with depthwise separable convolutions; pp. 1251–1258. [Google Scholar]
- 40.Hinton G., Vinyals O., Dean J. 2015. Distilling the Knowledge in a Neural Network. arXiv preprint arXiv:1503.02531. [Google Scholar]
- 41.Corbetta M., Shulman G.L. Control of goal-directed and stimulus-driven attention in the brain. Nat. Rev. Neurosci. 2002;3:201–215. doi: 10.1038/nrn755. [DOI] [PubMed] [Google Scholar]
- 42.Wang F., Jiang M., Qian C., Yang S., Li C., Zhang H., Wang X., Tang X. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2017. Residual attention network for image classification; pp. 3156–3164. [Google Scholar]
- 43.Hu J., Shen L., Sun G. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018. Squeeze-and-excitation networks; pp. 7132–7141. [Google Scholar]
- 44.Gaál G., Maga B., Lukács A. 2020. Attention U-Net Based Adversarial Architectures for Chest X-Ray Lung Segmentation; p. 10304. arXiv preprint arXiv:2003. [Google Scholar]
- 45.Armato S.G., III, McLennan G., Bidaut L., McNitt-Gray M.F., Meyer C.R., Reeves A.P., Zhao B., Aberle D.R., Henschke C.I., Hoffman E.A., et al. The lung image database consortium (lidc) and image database resource initiative (idri): a completed reference database of lung nodules on ct scans. Med. Phys. 2011;38:915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selim S.Z., Ismail M.A. K-means-type algorithms: a generalized convergence theorem and characterization of local optimality. IEEE Trans. Pattern Anal. Mach. Intell. 1984:81–87. doi: 10.1109/tpami.1984.4767478. [DOI] [PubMed] [Google Scholar]
- 47.Zahra S., Ghazanfar M.A., Khalid A., Azam M.A., Naeem U., Prugel-Bennett A. Novel centroid selection approaches for kmeans-clustering based recommender systems. Inf. Sci. 2015;320:156–189. [Google Scholar]
- 48.Peng K., Leung V.C., Huang Q. Clustering approach based on mini batch kmeans for intrusion detection system over big data. IEEE Access. 2018;6:11897–11906. [Google Scholar]
- 49.Glorot X., Bordes A., Bengio Y. Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics. JMLR Workshop and Conference Proceedings; 2011. Deep sparse rectifier neural networks; pp. 315–323. [Google Scholar]
- 50.Ahmad F., Khan M.U.G., Javed K. Deep learning model for distinguishing novel coronavirus from other chest related infections in x-ray images. Comput. Biol. Med. 2021;134:104401. doi: 10.1016/j.compbiomed.2021.104401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kingma D.P., Ba J., Adam . 2014. A Method for Stochastic Optimization. arXiv preprint arXiv:1412.6980. [Google Scholar]
- 52.Cheng J., Zhao W., Liu J., Xie X., Wu S., Liu L., Yue H., Li J., Wang J., Liu J. Automated diagnosis of covid-19 using deep supervised autoencoder with multi-view features from ct images. IEEE ACM Trans. Comput. Biol. Bioinf. 2021 doi: 10.1109/TCBB.2021.3102584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farooq M., Hafeez A. 2020. Covid-resnet: A Deep Learning Framework for Screening of Covid19 from Radiographs; p. 14395. arXiv preprint arXiv:2003. [Google Scholar]
- 54.Yuan X., Qi S., Wang Y. Stacked enhanced auto-encoder for data-driven soft sensing of quality variable. IEEE Transactions on Instrumentation and Measurement. 2020;69:7953–7961. [Google Scholar]
- 55.Bengio Y. Proceedings of ICML Workshop on Unsupervised and Transfer Learning. JMLR Workshop and Conference Proceedings; 2012. Deep learning of representations for unsupervised and transfer learning; pp. 17–36. [Google Scholar]