Abstract
The human immunodeficiency virus type 1 (HIV-1) Tat protein (hTat) activates transcription initiated at the viral long terminal repeat (LTR) promoter by a unique mechanism requiring recruitment of the human cyclin T1 (hCycT1) cofactor to the viral TAR RNA target element. While activation of equine infectious anemia virus (EIAV) gene expression by the EIAV Tat (eTat) protein appears similar in that the target element is a promoter proximal RNA, eTat shows little sequence homology to hTat, does not activate the HIV-1 LTR, and is not active in human cells that effectively support hTat function. To address whether eTat and hTat utilize similar or distinct mechanisms of action, we have cloned the equine homolog of hCycT1 (eCycT1) and examined whether it is required to mediate eTat function. Here, we report that expression of eCycT1 in human cells fully rescues eTat function and that eCycT1 and eTat form a protein complex that specifically binds to the EIAV, but not the HIV-1, TAR element. While hCycT1 is also shown to interact with eTat, the lack of eTat function in human cells is explained by the failure of the resultant protein complex to bind to EIAV TAR. Critical sequences in eCycT1 required to support eTat function are located very close to the amino terminus, i.e., distal to the HIV-1 Tat-TAR interaction motif previously identified in the hCycT1 protein. Together, these data provide a molecular explanation for the species tropism displayed by eTat and demonstrate that highly divergent lentiviral Tat proteins activate transcription from their cognate LTR promoters by essentially identical mechanisms.
All primate lentiviruses, as well as a subset of ungulate lentiviruses, encode transcriptional transactivator (Tat) proteins that potently upregulate the level of gene expression from their cognate long terminal repeat (LTR) promoters (8, 18). Uniquely, these Tat proteins act via a promoter-proximal, structured RNA element, termed TAR, rather than via a DNA target, as is the case for all other known eukaryotic transcriptional activators. Also unusual is the fact that Tat proteins predominantly act by increasing the elongation competence of initiated RNA polymerase II (RNAPII) molecules, rather than by increasing transcription initiation (10, 22, 23). Recently, it has become clear that an essential step in transactivation by the human immunodeficiency virus type 1 (HIV-1) Tat protein (hTat) is the recruitment of a protein complex, termed positive transcription elongation factor b (P-TEFb), to the HIV-1 TAR (hTAR) element (17, 26, 35, 37, 39). The human cyclin T1 (hCycT1) component of P-TEFb binds to the cysteine-rich and core domains of hTat and then participates with hTat in a highly cooperative interaction with the nascent hTAR element (35). The subsequent phosphorylation of the C-terminal domain (CTD) of RNAPII by cdk9, a second P-TEFb component that is also bound by hCycT1 (29, 35), is then believed to be critical for activation of efficient transcription elongation (7, 20, 28, 36). Importantly, it is the ability of hTat to recruit P-TEFb to hTAR that primarily determines the host range for hTat function. Thus, while the murine homolog of hCycT1 (mCycT1) can interact with the hTat activation domain, the resulting hTat-mCycT1 complex is not efficiently recruited to hTAR, and hTat is therefore poorly active in murine cells (1, 2, 14, 15, 24). Recently, it has been demonstrated that a single amino acid difference between hCycT1 and mCycT1 determines the distinct RNA binding properties of the two hTat-CycT1 complexes and thereby governs the differential ability of these species to support hTat-mediated transactivation of the HIV-1 LTR promoter (2, 14, 15).
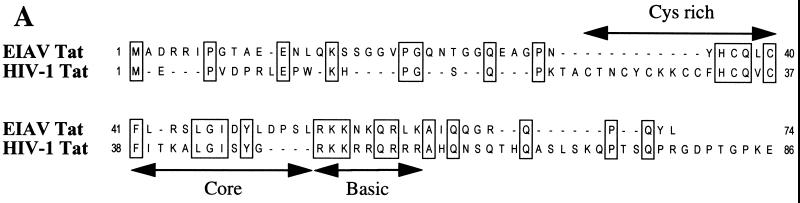
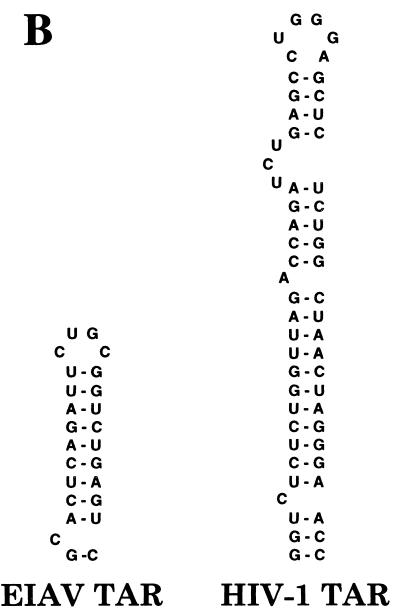
The equine infectious anemia virus (EIAV) Tat protein (eTat) shares little sequence homology with hTat (Fig. 1A) (32). Specifically, while both eTat and hTat contain a readily recognizable basic domain, i.e., the Tat domain involved in binding to TAR, plus the hTat core and cysteine-rich domains, both of which are critical for binding to hCycT1 (2, 24, 30), are only poorly conserved (Fig. 1A). Particularly noteworthy is the absence in eTat of five of the seven critical cysteine residues present in hTat (30). Similarly, the EIAV TAR element (eTAR) is a simple 25-nucleotide RNA stem-loop structure that lacks both the conserved U-rich bulge and terminal hexanucleotide loop that are critical for the biological activity of the 59-nucleotide hTAR element (Fig. 1B) (6, 9, 11, 31). In addition, eTat function displays a species tropism very different from that of hTat. Specifically, while hTat is a potent activator of the HIV-1 LTR in human cells, the ability of eTat to activate the EIAV LTR promoter is highly attenuated in human cells compared to permissive equine and canine cells (4, 24, 27). Thus, while both eTat and hTat have the ability to activate transcription via an RNA target, it has remained unclear whether the activities of these two proteins involve similar or different mechanisms, given the sequence and tropism differences outlined above. In this context, it is noteworthy that in addition to the hTat cofactor P-TEFb, at least two other proteins, cdk8 and the herpes simplex virus transactivator VP16, have also been shown to activate transcription when artificially recruited to a promoter-proximal RNA element (13, 16, 17, 33). Furthermore, both the VP16 and the adenovirus E1A transcriptional activators have been shown to interact with protein kinases capable of hyperphosphorylating the RNAPII CTD (21). Thus, RNA sequence-dependent activation of transcription can apparently be achieved by more than one mechanism.
FIG. 1.
Comparison of the Tat proteins and TAR elements of HIV-1 and EIAV. (A) This alignment of the hTat and eTat proteins shows reasonable conservation of the basic domain required for TAR binding. In contrast, the cysteine-rich and core motifs, essential for CycT1 binding by hTat, are poorly conserved. Alignment of the amino- and carboxy-terminal regions of these Tat proteins is essentially arbitrary. (B) The eTAR element lacks both the U-rich bulge, required for Tat binding, and the terminal hexanucleotide loop, required for CycT1 recruitment, that are critical for hTAR function.
To address whether these highly divergent Tat proteins indeed act via a common mechanism, and to determine the molecular basis for the species restriction to eTat function, we have cloned the equine homolog of the essential hTat cofactor hCycT1 (eCycT1). We demonstrate that defective eTat function in human cells can be overcome by expression of eCycT1 and that the species specificity of eTat function is entirely determined by the ability eTat-CycT1 complexes to bind to eTAR. Thus, recruitment of CycT1–P-TEFb to a promoter-proximal RNA element represents a general mechanism by which highly divergent lentiviral Tat proteins activate transcription directed by either primate or ungulate lentivirus LTR promoter elements.
MATERIALS AND METHODS
Molecular cloning of eCycT1.
Total RNA was extracted from primary fetal equine fibroblasts, and mRNA was purified by using oligo(dT)-Sepharose columns (Stratagene). Negative-strand cDNA was synthesized by using random hexanucleotide primers and amplified by using PCR and oligonucleotide primers derived from the 5′ and 3′ ends of the hCycT1 sequence (the corresponding amino acid sequences are identical in hCycT1 and mCycT1). These primers included HindIII and SalI restriction sites that were then used to insert the eCycT1 cDNA into the mammalian expression plasmid pBC12/CMV. The complete sequences of three independent clones of eCycT1 were determined and aligned with the known hCycT1 and mCycT1 sequences by using Clustal W.
Plasmid construction.
Mammalian expression plasmids pcTat, pTat-Rev, peTat, and pRev-eTat, encoding hTat and eTat expressed either as unfused proteins or as fusions to the HIV-1 Rev protein, have been described elsewhere (24, 33). The reporter plasmids pHIV/CAT, pEIAV/CAT, and pSLIIB/CAT have been described elsewhere (24, 33) and are referred to here as pHIV/hTAR/CAT pEIAV/eTAR/CAT and pHIV/SLIIB/CAT, respectively. pHIV/eTAR/CAT was constructed by replacement of hTAR sequences in pHIV/hTAR/CAT, located between BglII and SacI sites, with synthetic oligonucleotides encoding the eTAR RNA stem-loop. Plasmids expressing chimeric hCycT1-eCycT1 proteins in mammalian cells were constructed by exchange of restriction fragments between vectors pBC12/CMV/hCycT1 and pBC12/CMV/eCycT1. The complete coding sequence of eCycT1 was inserted into the yeast expression vector pVP16 (3), resulting in a plasmid expressing a VP16-eCycT1 fusion protein in yeast that is similar to the previously described pVP16/hCycT1 (2). The yeast expression plasmid pPGK/eTat, encoding the wild-type eTat protein, was constructed as previously described for pPGK/hTat (2). Yeast expression plasmid pIII/MS2/eTAR, encoding a hybrid MS2-eTAR RNA, was constructed as described for pIII/MS2/TAR (2), which encodes an MS2-hTAR hybrid RNA. For expression of recombinant CycT1 proteins in bacteria, sequences encoding the N-terminal 300 amino acids of hCycT1 or eCycT1 were inserted into pGEX 4T-1 and expressed as glutathione S-transferase (GST) fusion proteins. Similar GST-hTat and GST-eTat expression plasmids have been previously described (12). For in vitro synthesis of TAR RNA molecules, oligonucleotides encoding hTAR and eTAR were inserted into pGEM 3Zf(+), thereby generating pGEM/hTAR and pGEM/eTAR, respectively.
Mammalian cell transfection assays.
Human 293T and canine D17 cells were transfected with a total of 350 ng of DNA comprising 200 ng of reporter plasmid, 100 ng of effector plasmid, and 50 ng of pBC12/CMV/lacZ by calcium phosphate coprecipitation. In experiments assaying the ability of CycT1 proteins to rescue Tat activity, 400 ng of a pBC12/CMV/CycT1 expression plasmid was included. Murine LmTK-cells were similarly transfected, except that the DEAE-dextran method was used. Forty-eight hours after transfection, chloramphenicol acetyltransferase (CAT) enzyme levels in cell lysates were determined (2) and normalized for minor fluctuations in transfection efficiency as determined by assay of the β-galactosidase (β-Gal) internal control.
Protein-protein and protein-RNA interaction assays in yeast.
For two-hybrid protein-protein interaction assays, Saccharomyces cerevisiae Y190 cells were transformed with pGBT9, pGBT9/eTat, or pGAL4/hTat (3, 12) together with pVP16, pVP16/hCycT1, or pVP16/eCycT1. For three-hybrid protein-RNA interaction assays, yeast L40-coat cells were transformed with plasmids expressing a hybrid MS2-TAR RNA, a VP16-CycT1 fusion protein, and a Tat expression plasmid. For both assays, β-Gal activity in yeast cell lysates was determined quantitatively after growth on appropriate selective media.
RNA gel shift analyses.
Recombinant GST-Tat and GST-CycT1 proteins were expressed in Escherichia coli BL21 cells and purified by using glutathione-agarose. 32P-labeled TAR RNA probes were generated by in vitro transcription using SP6 or T7 polymerase and linearized pGEM/eTAR or pGEM/hTAR, respectively. For RNA gel shift analyses, 100 ng of a GST-Tat protein was incubated with 250 ng of a GST-CycT protein in 30 μl of binding buffer (30 mM Tris-HCl [pH 7.5], 10% glycerol, 1.3 mM dithiothreitol, 0.01% Nonidet P-40, 5 mM MgCl2), containing 8 U RNAsin and 500 ng of yeast tRNA, for 5 min on ice; 4 × 104 cpm of a 32P-labeled TAR RNA probe (∼20 ng) was then added, and the reaction mixture was incubated for a further 10 min on ice. RNA protein complexes were visualized by autoradiography following electrophoresis on a native 5% polyacrylamide gel containing 3% glycerol and 50 mM Tris-glycine (pH 8.8).
Nucleotide sequence accession number.
The eCycT1 sequence reported here has been assigned GenBank accession no. AF137509.
RESULTS
Poor eTat function in human cells results from defective recruitment to eTAR.
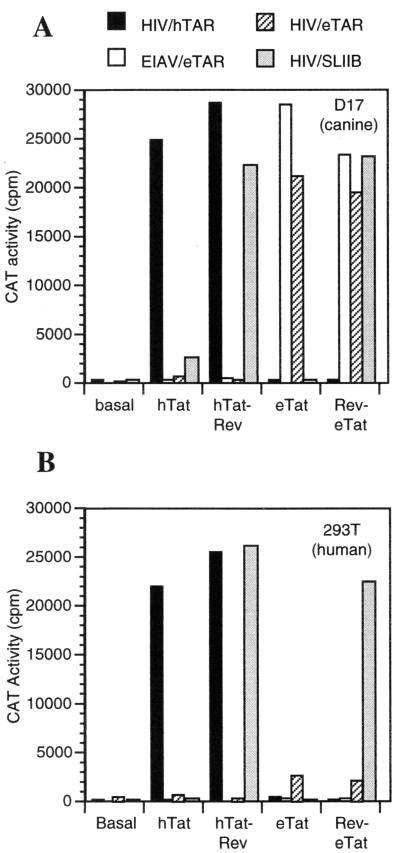
It has previously been demonstrated that eTat is a poor transactivator of the EIAV LTR in human cells but a potent activator in canine cells (4, 24, 27). To investigate the basis for this human cell-specific defect, we used reporter plasmids based on the wild-type EIAV LTR (pEIAV/eTAR/CAT) or on an HIV-1 LTR in which the TAR element remained intact (pHIV/hTAR/CAT), was replaced with eTAR sequences (pHIV/eTAR/CAT), or was replaced by the SLIIB minimal HIV-1 Rev RNA binding site (pHIV/SLIIB/CAT). We then determined whether eTat and hTat could transactivate LTR-driven gene expression via these RNA targets when expressed as wild-type proteins or when fused to the Rev protein. Importantly, it has previously been shown that Rev binds with high affinity to the SLIIB RNA target, in the absence of any cellular cofactors (25, 33, 38), and that Rev can therefore efficiently recruit either the eTat or hTat protein to an SLIIB-containing LTR promoter element (24, 33).
In both canine D17 cells (Fig. 2A) and human 293T cells (Fig. 2B), hTat and hTat-Rev potently transactivated the pHIV/hTAR/CAT reporter plasmid, as expected. Neither hTat nor hTat-Rev displayed significant activity on plasmid pEIAV/eTAR/CAT or the pHIV/eTAR/CAT, while hTat-Rev also efficiently transactivated pHIV/SLIIB/CAT. Thus, both cell types are permissive for hTat-hTAR function, and hTat-Rev is approximately equivalently active when recruited to the HIV-1 LTR via either an hTat-hTAR or a Rev-SLIIB protein-RNA interaction.
FIG. 2.
The species specificity of eTat transactivation is determined by eTAR and can be overcome by heterologous recruitment. Canine D17 (A) and human 293T (B) cells were transfected with the indicated reporter plasmids, pBC12/CMV/lacZ, and hTat or eTat either unfused or fused to the SLIIB RNA binding protein, Rev. Forty-eight hours after transfection, levels of CAT enzyme activity in cell lysates were determined and normalized according to the level of β-Gal internal control measured in parallel.
The eTat protein differed from hTat in terms of both its species and RNA target specificity. The intact eTat protein potently transactivated CAT expression from both the pEIAV/eTAR/CAT and pHIV/eTAR/CAT indicator plasmids in canine cells but not from either pHIV/hTAR/CAT or pHIV/SLIIB/CAT (Fig. 2A). These data demonstrate that replacement of the hTAR element with the eTAR equivalent is, as previously reported (4), entirely sufficient to confer eTat responsiveness in permissive cells and therefore confirm that no DNA sequence specific to the EIAV LTR is required for eTat function. In contrast to the canine cells, both the pEIAV/eTAR/CAT and pHIV/eTAR/CAT plasmids proved only poorly responsive to eTat when tested in human cells (Fig. 2B). This defect in eTat-TAR function in human cells could be overcome by recruitment of eTat to the HIV-1 LTR via a heterologous RNA-protein interaction. Thus, while the Rev-eTat fusion protein, like eTat, was able to activate the pEIAV/eTAR/CAT and pHIV/eTAR/CAT reporter constructs in canine cells (Fig. 2A) but not human cells (Fig. 2B), the pHIV/SLIIB/CAT construct was efficiently transactivated by Rev-eTat in both species contexts. These data therefore indicate that the transactivation potential of eTat is fully functional in human cells, but that recruitment to the eTAR element is defective.
Molecular cloning of eCycT1.
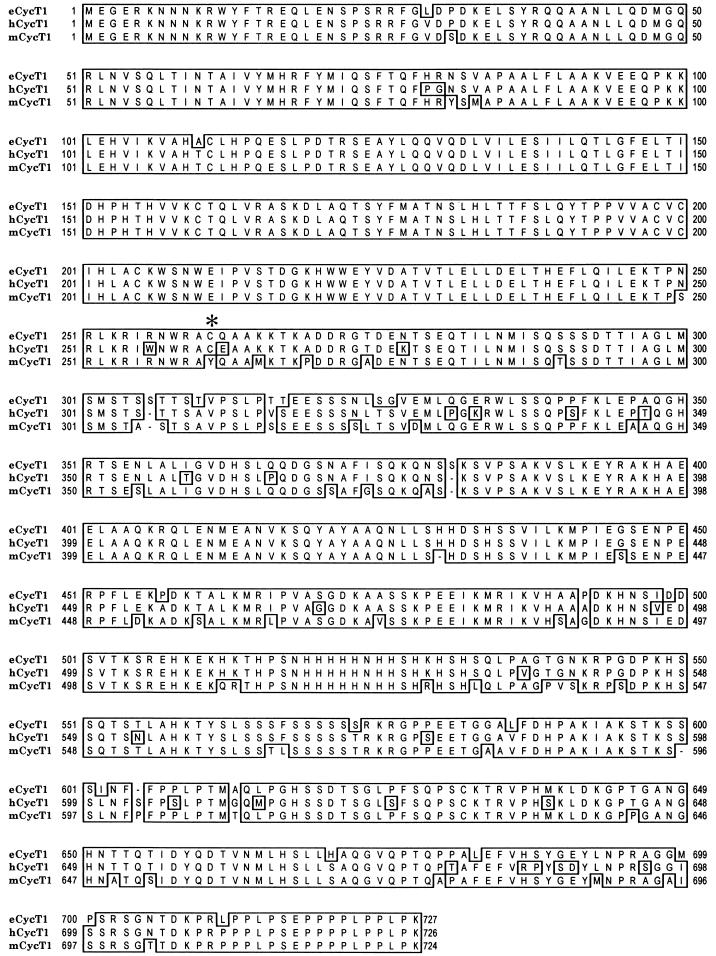
We and others have previously demonstrated that a mechanism similar to that described above, i.e., a defect in recruitment to TAR, accounts for poor hTat function in murine cells (2, 14, 15), and further that a single amino acid difference between hCycT1 and mCycT1 determines the ability of hTat-CycT1 to bind TAR and therefore governs the species specificity of hTat function. To determine whether a similar mechanism might account for the species restriction to eTat function, we cloned a cDNA from fetal equine fibroblasts encoding eCycT1. Sequence analysis revealed an open reading frame of 727 amino acids that was 94 and 91% identical to the hCycT1 and mCycT1 proteins, respectively (Fig. 3). In particular, the amino-terminal cyclin homology domain was well conserved, with scattered regions of sequence identity throughout the remainder of these proteins.
FIG. 3.
Predicted amino acid sequence of eCycT1. The sequences of three independent eCycT1 clones were determined, and the consensus (with correction for a small number of Taq-induced errors) was aligned with sequences of hCycT1 and mCycT1. The critical cysteine residue in hCycT1 is indicated by an asterisk.
eCycT1 expression rescues eTat-eTAR function in human cells.
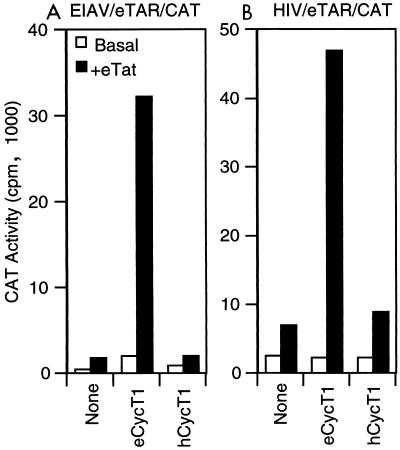
To determine whether CycT1 is the species specific determinant of eTat-TAR function, human 293T cells were transfected with either the pEIAV/eTAR/CAT or pHIV/eTAR/CAT reporter plasmid in the presence or absence of plasmids expressing eTat and either eCycT1 or hCycT1. As shown in Fig. 4, in the absence of any added CycT1 expression plasmid, eTat again proved to be a poor transactivator of reporter plasmids containing an eTAR response element, irrespective of 5′ promoter sequences. However, expression of eCycT1 in human cells dramatically enhanced the responsiveness of both pEIAV/eTAR/CAT and pHIV/eTAR/CAT to eTat. This is a specific property of the equine form of CycT1; cotransfection of an hCycT1 expression plasmid previously shown to fully rescue hTat function in murine cells (2) did not enhance the ability of either eTAR containing construct to be activated by eTat (Fig. 4).
FIG. 4.
Expression of eCycT1 in human cells rescues the defect in eTat transactivation via eTAR. Human 293T cell were transfected with pEIAV/eTAR/CAT (A) or pHIV/eTAR/CAT (B), pBC12/CMV/lacZ, peTat, and either pBC12/CMV/eCycT1 or pBC12/CMV/hCycT1. In control transfections, the parental vector pBC12/CMV replaced either pBC12/CMV/CycT1 or peTat, or both. Normalized CAT enzyme activities were determined as for Fig. 2.
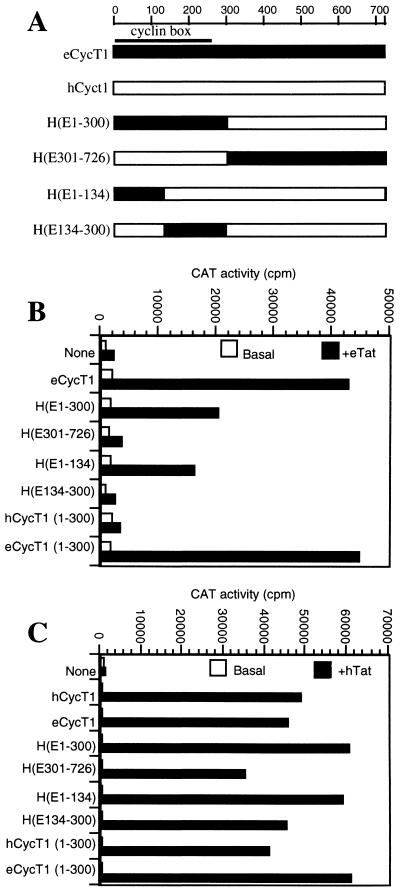
The N-terminal 300 amino acids of eCycT1 are sufficient to support eTat function.
To map the sequence differences between hCycT1 and eCycT1 that account for their different abilities to support eTat/eTAR function, a series of eCycT1-hCycT1 chimeras was constructed (Fig. 5A). These were designated according to which sequences are of eCycT1 origin. Thus, for example, H(E1-134) contains amino acid residues 1 to 134 of eCycT1 in the context of an otherwise full-length hCycT1 protein. It has previously been shown that a critical cysteine residue at position 261 in hCycT1 is essential for the ability of hCycT1 to support hTat function (2, 14, 15). Since eCycT1 is highly homologous to hCycT1, and furthermore contains a cysteine at amino acid residue 261, we anticipated that eCycT1 might be able to support hTat function and, therefore, rescue hTat/hTAR function in murine cells. This indeed proved to be the case; hTat activates the pHIV/hTAR/CAT construct very poorly in murine LmTK-cells, but this defect can be rescued by expression of either hCycT1 or eCycT1, resulting in equivalent, high levels of CAT expression (Fig. 5C). This shared property was used to confirm the functional integrity and expression of chimeric eCycT1-hCycT1 proteins. As shown in Fig. 5C, all of the chimeric and truncated CycT1 proteins were able to rescue hTat-TAR function in murine cells, resulting in levels of transactivation close to those observed in the presence of either intact hCycT1 or eCycT1. In contrast, the eCycT1-hCycT1 chimeras varied dramatically in their ability to rescue eTat function in human cells, as determined using the pEIAV/eTAR/CAT reporter plasmid (Fig. 5B). Essentially identical results were obtained with the pHIV/eTAR/CAT reporter plasmid (data not shown). The chimeras H(E301-726) and H(E134-300), which were fully able to support hTat function in murine cells, were unable to increase the low level of eTat function in human cells. In contrast, chimeras H(E1-300) and H(E1-134) were at least partially active, thus demonstrating that eCycT1 residues 1 to 134, where there are four differences between eCycT1 and hCycT1 (Fig. 3), contain critical determinants of eTat-TAR function. The inability of the H(E134-300) chimera to support eTat function in human cells was surprising, given previous work using hCycT1-mCycT1 chimeras to map residues in hCycT1 that are essential for hTat function (2, 14, 15). These residues were shown to include not only the critical cysteine residue at position 261 but also a proposed hTat-TAR recognition motif located between hCycT1 residues 250 to 262 (15). However, eCycT1 is similar to hCycT1 in that eCycT1 residues 1 to 300 were fully sufficient to support eTat function in human cells and hTat function in murine cells (Fig. 5B). As expected, the equivalent 300-amino-acid hCycT1 protein possessed only the latter activity. Indeed, hCycT1 residues C terminal of position 301 appear to be inhibitory for eTat-eTAR function in the context of eCycT1-hCycT1 chimeras.
FIG. 5.
The N-terminal 300 amino acids of eCycT1 are sufficient to rescue eTat activity in human cells. (A) Schematic representation of chimeric CycT1 proteins that were generated by exchange of restriction fragments. (B) 293T cells were transfected as for Fig. 2, using the pEIAV/eTAR/CAT reporter and chimeric CycT1 expression plasmids. (C) Murine LmTK-cells were transfected with pHIV/hTAR/CAT, pBC12/CMV/lacZ, chimeric CycT1 expression plasmids, and either pcTat or the parental control vector. Normalized CAT enzyme levels were determined as for Fig. 2.
Recruitment of eTat-CycT1 complexes to eTAR determines the species specificity of eTat.
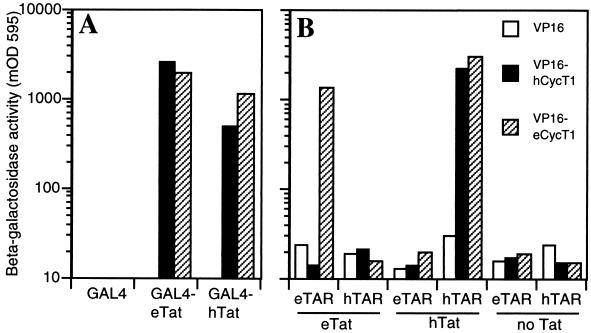
Since we have determined that only the eCycT1 protein is able to rescue eTat activity an human cells, it follows that hCycT1 must be defective for this function in some way. To determine whether there is a difference in the ability of hCycT1 and eCycT1 to bind to eTat, we performed two-hybrid protein-protein interaction assays in yeast. This analysis revealed that both hTat and eTat are able to efficiently interact with both eCycT1 and hCycT1 (Fig. 6A). Therefore, the inability of eTat to efficiently transactivate gene expression in human cells is not due to a block to the interaction of eTat with hCycT1. This result is, in fact, predictable given the data presented in Fig. 2, which showed that eTat is fully functional in human cells provided that it is recruited to the promoter via a heterologous RNA-protein interaction. Therefore, the cellular factors that mediate the transcriptional activation that follows the recruitment of eTat to eTAR must be present in human cells and able to interact with eTat. The defect in eTat function in human cells must therefore be at the level of recruitment to the eTAR RNA element. Thus, because an eTat-CycT1 complex readily forms in both the yeast (Fig. 6A) and the mammalian (Fig. 2) cell nucleus, we predicted that binding of this complex to eTAR would be able to proceed efficiently only when the CycT1 component is of equine origin.
FIG. 6.
Interactions between CycT1, Tat proteins, and TAR RNA elements in yeast cells. (A) Two-hybrid interactions between Tat proteins and CycT1 proteins. Yeast cells were transformed with plasmids expressing the indicated Tat proteins fused to the GAL4 DNA binding domain and CycT1 proteins fused to herpes simplex virus VP16 activation domain. (B) Yeast cells were transformed with plasmids expressing the indicated MS2-TAR fusion RNAs, VP16-CycT1 fusion proteins, and the indicated, unfused Tat proteins. After growth on selective media, β-Gal activities induced in lysed yeast cells were determined as described previously (2). The approximate limit of detection of this assay is 10 milli-optical density units at 595 nm (mOD 595).
To test this prediction, we first performed a modified three-hybrid assay in yeast, as previously described (2). Yeast cells were transformed with plasmids expressing a hybrid MS2-TAR RNA, a VP16 activation domain fused to a CycT1 protein, and an unfused, wild-type Tat protein. Since we have already determined that Tat-CycT1 complexes form in the absence of TAR in the yeast cell nucleus (Fig. 6A), this assay determines the ability of the resultant protein complex to bind to a promoter-tethered TAR element. As shown in Fig. 6B, VP16-eCycT1 bound to the eTAR element in the presence, but not in the absence, of eTat. This interaction was RNA sequence specific, in that no interaction was observed with the hTAR element, and protein sequence specific, in that no interaction could be detected between VP16-hCycT1 and either TAR element in the presence of eTat. Therefore, it is apparent that eTat and hCycT1 form a dead-end complex that cannot be recruited to either TAR element. In contrast, both hCycT1 and eCycT1 bound to hTAR in the presence of hTat. Again this interaction is specific—neither complex could bind eTAR, and these observations fully correlate with the shared ability of hCycT1 and eCycT1 to rescue hTat function in nonpermissive murine cells (Fig. 5C). Finally, neither CycT1 has any detectable affinity for either TAR in the absence of Tat.
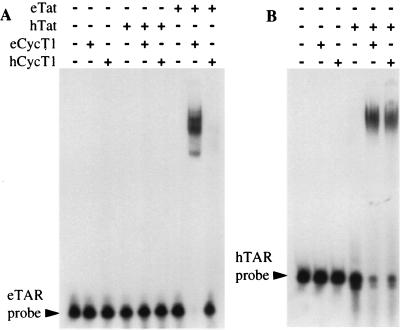
To confirm these observations in vitro, we next performed RNA gel shift analyses using purified recombinant GST-Tat and GST-CycT1 proteins. Since the full-length CycT1 proteins are poorly expressed in E. coli, the amino-terminal 300 amino acid residues, which are fully sufficient to support eTat and hTat function in vivo (Fig. 5) were used. As shown in Fig. 7A, neither Tat protein, and neither CycT1 protein, proved able to form a detectable RNA-protein complex when incubated alone with an eTAR RNA probe. Similarly, hTat also proved unable to interact with eTAR when incubated in the presence of either hCycT1 or eCycT1. In contrast, a ternary complex consisting of eTAR bound to both eTat and eCycT1 was efficiently formed when both equine proteins were incubated with the eTAR probe (Fig. 7A). This represents the first reported demonstration of an in vitro interaction between eTAR and the eTat protein. Finally, we also observed a very low but detectable level of a retarded complex consisting of eTAR bound by eTat and hCycT1. The inefficient formation of this complex, compared to the very efficient formation of the equivalent complex containing eCycT1, correlates with the low but detectable level of eTat function observed in human cells (Fig. 2 and 4).
FIG. 7.
In vitro interaction between different TAR elements and Tat proteins in the presence and absence of CycT1 proteins. (A) A 32P-labeled eTAR RNA probe was incubated in the presence of the indicated GST fusion proteins, and RNA-protein complex formation was visualized by nondenaturing polyacrylamide gel electrophoresis followed by autoradiography. (B) As for panel A except that an hTAR RNA probe was used.
To confirm that the hTat and hCycT1 proteins used in this in vitro assay were, in fact, fully active, we also performed an RNA gel shift analysis using an hTAR probe (Fig. 7B). Others (35) have previously reported weak binding to hTAR by hTat, and no binding by hCycT1, when these proteins were tested alone in vitro. In fact, under the conditions used in our assay, we failed to see any binding to hTAR by either of these proteins (Fig. 7B), although weak binding by hTat could be visualized when markedly higher levels of hTat were added (data not shown). In contrast, and as previously reported (35), a ternary complex consisting of hTAR bound by both hTat and hCycT1 formed readily in vitro. As predicted from the ability of eCycT1 to rescue hTat function in murine cells (Fig. 5C), eCycT1 also proved able to effectively bind to hTAR in the presence, but not in the absence, of hTat (Fig. 7B). Overall, these in vitro data fully confirm the yeast data (Fig. 6B) showing that only eCycT1 can cooperate with eTat to bind eTAR while both eCycT1 and hCycT1 can bind to hTAR when complexed with hTat.
DISCUSSION
Recent evidence has indicated that recruitment of CycT1–P-TEFb to TAR is an essential component of the mechanism by which hTat transactivates viral gene expression (2, 14, 15, 35, 37). In this study, we show that this scheme is fully generalizable, i.e., that the most divergent of the known RNA-targeted lentiviral Tat proteins and TAR elements (Fig. 1) enhance viral gene expression by essentially identical mechanisms. Importantly, and taken in conjunction with previous studies of the species-restricted nature of HIV-1 Tat transactivation, these data demonstrate that the ability of a given CycT1 protein to support high levels of transactivation by a given Tat protein is predominantly determined by the ability of the resultant Tat-CycT1 complex to bind to the cognate TAR element, and not simply by the ability of the Tat protein to bind to CycT1. Indeed, the studies presented herein represent the second demonstration of a Tat-CycT1 complex that is essentially nonfunctional because it cannot bind to the relevant TAR element. In this case, the block to eTat function in human cells results from the inability of an eTat-hCycT1 complex to effectively bind eTAR (Fig. 6 and 7), while it has previously been demonstrated that the block to hTat function in murine cells largely reflects the inability of an hTat-mCycT1 complex to bind to hTAR (2, 14, 15). Thus, while formation of Tat-CycT1 complexes is not species restricted, both components of the resultant heterodimer clearly contribute critical protein contacts that determine both the RNA sequence specificity and the RNA binding affinity of the heterodimer. As a result, binding by uncomplexed Tat or CycT1 proteins to TAR is very inefficient (Fig. 6 and 7), while formation of a complex with an inappropriate partner precludes binding to TAR. Thus, while the eTat-eCycT1 heterodimer binds eTAR efficiently, binding is poor or nonexistent for an eTat-hCycT1 or hTat-eCycT1 heterodimer (Fig. 7A).
While the overall mechanisms of transactivation by hTat and eTat are therefore very similar, critical residues in the CycT1 protein that determine the ability of the Tat-CycT1 complex to bind TAR do not appear to be identically positioned. Specifically, while residues 250 to 261 of hCycT1 have been shown to be critical for the hTat-mediated recruitment to TAR, substitution of these hCycT1 residues with eCycT1 sequences does not result in a CycT1 protein that can support eTat-eTAR function (Fig. 5). Instead, residues closer to the amino terminus of the eCycT1 protein proved to be the major determinant of the ability of eTat to be recruited to eTAR. Therefore, protein-RNA contacts formed by different Tat-CycT1/TAR complexes may well involve different critical CycT1 amino acid residues.
These data explain a number of previous genetic and biochemical observations. (i) Because the Tat-CycT1 interaction is not species restricted, recruitment of a Tat protein to a lentiviral LTR via a heterologous RNA-protein interaction inevitably results in CycT1–P-TEFb recruitment and thereby abrogates the species specificity of Tat transactivation (1, 24). Thus, hTat is fully active in otherwise nonpermissive murine cells, and eTat in otherwise nonpermissive human cells, when recruited to a responsive promoter via a heterologous RNA binding protein such as Rev (Fig. 2) (1, 24). (ii) The ability of the eTat protein to inhibit hTat function in human cells when expressed at high levels (5, 24), even though eTat is not functional on the EIAV LTR promoter in human cells, can now be explained by the ability of eTat to sequester hCycT1–P-TEFb into a nonfunctional protein complex (Fig. 6). (iii) Diverse Tat proteins can coprecipitate Tat-associated kinase activity (subsequently identified as the cdk9 component of P-TEFb) even from nonpermissive cells (19, 20). Clearly, this reflects the formation of nonfunctional Tat–P-TEFb complexes in these cells. Overall, these data suggest a simple and general mechanism by which Tat proteins transactivate lentiviral gene expression and imply that all lentiviral RNA sequence-specific transcriptional activators must have evolved from a single initial precursor that first developed the ability to recruit CycT1–P-TEFb to a promoter-proximal RNA target. An interesting remaining question is whether this biological activity evolved de novo in lentiviruses or whether it is instead a biological activity utilized by host cells to control the expression of host genes that was stolen by these viral parasites.
ACKNOWLEDGMENTS
We thank Fred Fuller for the gift of equine fibroblasts.
This research was sponsored by the Howard Hughes Medical Institute.
REFERENCES
- 1.Alonso A, Derse D, Peterlin B M. Human chromosome 12 is required for optimal interactions between Tat and TAR of human immunodeficiency virus type 1 in rodent cells. J Virol. 1992;66:4617–4621. doi: 10.1128/jvi.66.7.4617-4621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieniasz P D, Grdina T A, Bogerd H P, Cullen B R. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 1998;17:7056–7065. doi: 10.1093/emboj/17.23.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogerd H P, Fridell R A, Blair W S, Cullen B R. Genetic evidence that the Tat proteins of human immunodeficiency virus types 1 and 2 can multimerize in the eukaryotic cell nucleus. J Virol. 1993;67:5030–5034. doi: 10.1128/jvi.67.8.5030-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carroll R, Martarano L, Derse D. Identification of lentivirus tat functional domains through generation of equine infectious anemia virus/human immunodeficiency virus type 1 tat gene chimeras. J Virol. 1991;65:3460–3467. doi: 10.1128/jvi.65.7.3460-3467.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll R, Peterlin B M, Derse D. Inhibition of human immunodeficiency virus type 1 Tat activity by coexpression of heterologous transactivators. J Virol. 1992;66:2000–2007. doi: 10.1128/jvi.66.4.2000-2007.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carvalho M, Derse D. Mutational analysis of the equine infectious anemia virus Tat-responsive element. J Virol. 1991;65:3468–3474. doi: 10.1128/jvi.65.7.3468-3474.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun R F, Jeang K T. Requirements for RNA polymerase II carboxyl-terminal domain for activated transcription of human retroviruses human T-cell lymphotropic virus I and HIV-1. J Biol Chem. 1996;271:27888–27894. doi: 10.1074/jbc.271.44.27888. [DOI] [PubMed] [Google Scholar]
- 8.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 9.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A, Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci USA. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feinberg M B, Baltimore D, Frankel A D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc Natl Acad Sci USA. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng S, Holland E C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988;334:165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 12.Fridell R A, Harding L S, Bogerd H P, Cullen B R. Identification of a novel human zinc finger protein that specifically interacts with the activation domain of lentiviral Tat proteins. Virology. 1995;209:347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- 13.Fujinaga K, Cujec T P, Peng J, Garriga J, Price D H, Grana X, Peterlin B M. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J Virol. 1998;72:7154–7159. doi: 10.1128/jvi.72.9.7154-7159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujinaga K, Taube R, Wimmer J, Cujec T P, Peterlin B M. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc Natl Acad Sci USA. 1999;96:1285–1290. doi: 10.1073/pnas.96.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garber M E, Wei P, KewalRamani V N, Mayall T P, Herrmann C H, Rice A P, Littman D R, Jones K A. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 1998;12:3512–3527. doi: 10.1101/gad.12.22.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gold M O, Rice A P. Targeting of CDK8 to a promoter-proximal RNA element demonstrates catalysis-dependent activation of gene expression. Nucleic Acids Res. 1998;26:3784–3788. doi: 10.1093/nar/26.16.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gold M O, Yang X, Herrmann C H, Rice A P. PITALRE, the catalytic subunit of TAK, is required for human immunodeficiency virus Tat transactivation in vivo. J Virol. 1998;72:4448–4453. doi: 10.1128/jvi.72.5.4448-4453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones K A. Taking a new TAK on tat transactivation. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann C H, Rice A P. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- 20.Herrmann C H, Rice A P. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herrmann C H, Gold M O, Rice A P. Viral transactivators specifically target distinct cellular protein kinases that phosphorylate the RNA polymerase II C-terminal domain. Nucleic Acids Res. 1996;24:501–508. doi: 10.1093/nar/24.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kao S Y, Calman A F, Luciw P A, Peterlin B M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 23.Laspia M F, Rice A P, Mathews M B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell. 1989;59:283–292. doi: 10.1016/0092-8674(89)90290-0. [DOI] [PubMed] [Google Scholar]
- 24.Madore S J, Cullen B R. Genetic analysis of the cofactor requirement for human immunodeficiency virus type 1 Tat function. J Virol. 1993;67:3703–3711. doi: 10.1128/jvi.67.7.3703-3711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malim M H, Tiley L S, McCarn D F, Rusche J R, Hauber J, Cullen B R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 26.Mancebo H S, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maury W J, Carpenter S, Graves K, Chesebro B. Cellular and viral specificity of equine infectious anemia virus Tat transactivation. Virology. 1994;200:632–642. doi: 10.1006/viro.1994.1226. [DOI] [PubMed] [Google Scholar]
- 28.Okamoto H, Sheline C T, Corden J L, Jones K A, Peterlin B M. Trans-activation by human immunodeficiency virus Tat protein requires the C-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA. 1996;93:11575–11579. doi: 10.1073/pnas.93.21.11575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng J, Zhu Y, Milton J T, Price D H. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice A P, Carlotti F. Mutational analysis of the conserved cysteine-rich region of the human immunodeficiency virus type 1 Tat protein. J Virol. 1990;64:1864–1868. doi: 10.1128/jvi.64.4.1864-1868.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selby M J, Bain E S, Luciw P A, Peterlin B M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989;3:547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- 32.Stephens R M, Derse D, Rice N R. Cloning and characterization of cDNAs encoding equine infectious anemia virus Tat and putative Rev proteins. J Virol. 1990;64:3716–3725. doi: 10.1128/jvi.64.8.3716-3725.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tiley L S, Madore S J, Malim M H, Cullen B R. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 1992;6:2077–2087. doi: 10.1101/gad.6.11.2077. [DOI] [PubMed] [Google Scholar]
- 34.Tiley L S, Malim M H, Tewary H K, Stockley P G, Cullen B R. Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc Natl Acad Sci USA. 1992;89:758–762. doi: 10.1073/pnas.89.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei P, Garber M E, Fang S M, Fischer W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Herrmann C H, Rice A P. The human immunodeficiency virus Tat proteins specifically associate with TAK in vivo and require the carboxyl-terminal domain of RNA polymerase II for function. J Virol. 1996;70:4576–4584. doi: 10.1128/jvi.70.7.4576-4584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X, Gold M O, Tang D N, Lewis D E, Aguilar-Cordova E, Rice A P, Herrmann C H. TAK, an HIV Tat-associated kinase, is a member of the cyclin-dependent family of protein kinases and is induced by activation of peripheral blood lymphocytes and differentiation of promonocytic cell lines. Proc Natl Acad Sci USA. 1997;94:12331–12336. doi: 10.1073/pnas.94.23.12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zapp M L, Green M R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature. 1989;342:714–716. doi: 10.1038/342714a0. [DOI] [PubMed] [Google Scholar]
- 39.Zhu Y, Pe’ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews M B, Price D H. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]