ABSTRACT
This study describes the characterization of conjugation sites for a random, lysine conjugated 2-iminothiolane (2-IT) based antibody-drug-conjugate synthesized from an IgG1 antibody and a duocarmycin analog-based payload-linker. Of the 80 putative lysine sites, 78 were found to be conjugated via tryptic peptide mapping and LC-HRMS. Surprisingly, seven cysteine-linked conjugated peptides were also detected resulting from the conjugation of cysteine residues derived from the four inter-chain disulfide bonds during the reaction. This unexpected finding could be attributed to the free thiols of the 2-IT thiolated antibody intermediates and/or the 4-mercaptobutanamide by-product resulting from the hydrolysis of 2-IT. These free thiols could cause the four inter-chain disulfide bonds of the antibody to scramble via intra- or inter-molecular attack. The presence of only pair of non-reactive (unconjugated) lysine residues, along with the four intact intra-chain disulfide bonds, is attributed to their poor accessibility, which is consistent with solvent accessibility modeling analysis. We also discovered a major by-product derived from the hydrolysis of the amidine moiety of the N-terminus conjugate. In contrast, the amidine moiety in lysine-linked conjugates appeared stable. Based on our results, we propose plausible formation mechanisms of cysteine-linked conjugates and the hydrolysis of the N-terminus conjugate, which provide scientific insights that are beneficial to process development and drug quality control.
KEYWORDS: Antibody drug conjugate, lysine-linked conjugation, cysteine-linked conjugation, mass spectrometry
Introduction
Therapeutic monoclonal antibody-drug conjugates (ADCs) use highly target-selective antibodies to carry potent cytotoxic compounds (payloads) to tumor cells, which enhances the therapeutic index for the treatment of a wide range of cancer types.1,2 Payloads are covalently bound to the antibody through stable linkers to reduce cytotoxicity to healthy cells during the circulation of ADCs in the bloodstream. Two common methods to conjugate payload-linkers to antibodies are through cysteine (Cys) or lysine (Lys) residues on the antibody, although other site-specific technologies have also been developed. In the case of cysteine-based conjugates, it is well known that the 4 inter-chain disulfide bonds of antibodies can be partially reduced to generate a maximum of 8 free cysteine thiols for drug conjugation to yield “random cysteine-linked” ADCs.3,4 Hamblett et al. reported that incorporating an average of 2–4 drugs relative to the antibody (drug-to-antibody ratio (DAR)) would achieve the best balance between the slow clearance from circulation and maximal potency.5 In lysine-based conjugates, the primary amines of the N-termini and lysine residues of antibodies are used as conjugation sites. A typical antibody usually contains 80–100 lysine residues and most of them are accessible for conjugation.6 However, each antibody can usually achieve 1–8 lysine-linked conjugations, with an average DAR of 3–4,7 resulting in heterogeneous lysine conjugates. Given the complexity of ADCs, the two most critical quality attributes (CQAs), i.e., the average DAR and the conjugations location, must be addressed during their development.
Several synthetic protocols for lysine-linked ADCs have been described by Brun et al. The “one-step conjugation” approach is often used for the conjugation of antibodies with non-cleavable linkers that are already connected to payloads. These conjugations also can be carried out using a two-step protocol. In the first step, the lysine residues on the antibody are modified to enable a subsequent preferential reaction with the payload-linker. This initial modification typically converts the lysine residue into an N-hydroxysuccinimide ester in preparation for reaction with the payload-linker. After the initial modification of lysine residues, the payload-linker is introduced to complete the conjugation. Variations of this “two-step conjugation” approach have been widely used in the synthesis of lysine-linked ADCs. The in-depth characterization of such conjugates has been reported.8–11 Wang et al. detected 20 lysine conjugated sites in their maytansinoid ADC via a cleavable linker, N-succinimidyl 4-(20-pyridyldithio) pentanoate (SPP). Luo et al. reported 76 of 92 putative conjugation sites in a monoclonal antibody−maytansinoid immunoconjugate, and emphasized that the cysteine residues in the native antibody were fully disulfide-linked and no cysteine/tyrosine conjugation was found. In contrast, Chih et al.12 detected side-reactions between the residual cysteine/tyrosine of the antibody and the N-hydroxysuccinimide ester analog from an antibody-coumarin-linker conjugate. This observation was confirmed using surrogate peptides to mimic the conjugation. These studies indicated that native antibody scaffold structures, conjugation chemistries, and variations in the conjugation processes could greatly affect the ADC product heterogeneity.
Iminothiolane reagents, such as 2-iminothiolane (2-IT, also named Traut’s reagent13,14), can be used in “two-step conjugation”. The primary amines of lysine residues and N-termini of the antibodies are modified by 2-IT to form amidine moieties with sulfhydryl groups, which can subsequently react with specific reactive functional groups, such as a maleimido moiety, as illustrated in Figure 1a.15 It is known that the sulfhydryl group of the thiolated antibody intermediate can undergo intra-molecular attack of the amidine group, resulting in an unwanted cyclic 2-IT derived by-product. However, to our knowledge, there are no known reports on the scrambling of the inter-chain disulfide bonds by the sulfhydryl group of the thiolated antibody intermediate or the 4-mercaptobutanamide, a hydrolysis product of 2-IT during the reaction. During the synthesis of a duocarmycin analog-based ADC16 with 2-IT via the “two-step conjugation”, we found that the scrambling of the inter-chain disulfide bonds may occur by the sulfhydryl group of the thiolated antibody intermediate and/or the 4-mercaptobutanamide, as postulated in Figure 1b. We also found that the amidine groups derived from the lysine residues appeared stable, while those from the N-terminal amines of the antibody were prone to hydrolysis.
Figure 1.
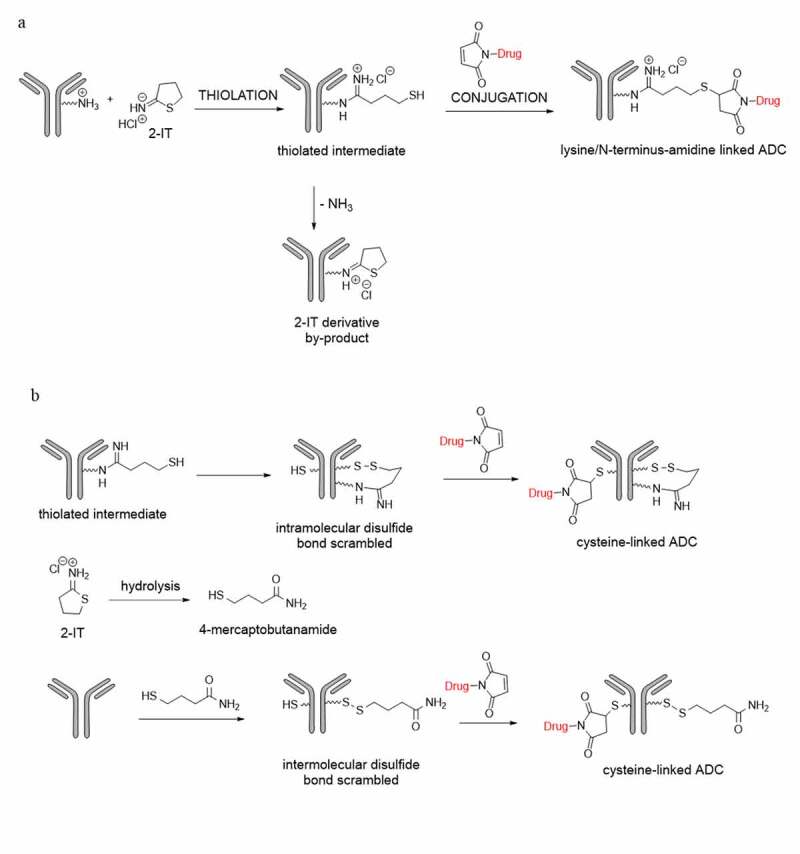
Illustration of an antibody (a) 2-IT thiolation and payload-linker conjugation on lysine or N-terminus residues; (b) direct payload-linker conjugation on cysteine
The aim of this study was to thoroughly characterize the duocarmycin analog-based ADC in support of drug development and ensure the drug product quality. The structural characterization of the lysine-, cysteine-, and N-terminus-linked conjugation sites of the ADC were conducted via tryptic peptide mapping. Based on the results, we have postulated plausible formation mechanisms of the unexpected cysteine-linked ADC and the hydrolysis of the amidine moiety in N-terminus-linked ADC mediated by the proximal glutamic acid group. We also evaluated the solvent accessibility of the native antibody that is commonly used in assessing the exposure/reactivity of the lysine and cysteine residues via modeling analysis.17
Results
Identification of conjugation sites on lysine residues
In this study, the naked antibody contains 40 pairs of lysine residues. As long as a lysine residue is thiolated with 2-IT or subsequently conjugated with payload-linker, its peptide backbone becomes “non-cleavable” during trypsin digestion. This outcome of the digestion was used for the identification of the conjugation sites using the procedure described in the Data Processing section and exemplified by the identification of the anticipated alkylated tryptic peptide, named as (H227)K for SC(alk)D(H227)K(2-IT-drug)THTC(alk)PPC(alk)PAPELLGGPSVFLFPP(H251)KP(H253)K with the conjugation site at (H227)K. In this peptide, only (H227)K was thiolated with 2-IT and subsequently conjugated with payload-linker, while (H251)K, and (H253)K were neither thiolated with 2-IT nor conjugated with the payload-linker. Note, (H251)K is non-cleavable by trypsin digestion due to its adjacency to a proline moiety. When searching for this tryptic peptide based on its accurate protonated mass, five peaks were observed in the extracted ion chromatogram (XIC) using the most abundant ion at m/z 777.2214 ([M + 6 H]6+), as shown in Figure 2a. While one of these peaks was anticipated to be the (H227)K conjugated peptide, each individual peak was thoroughly examined by the following steps to ascertain its identity.
Figure 2.
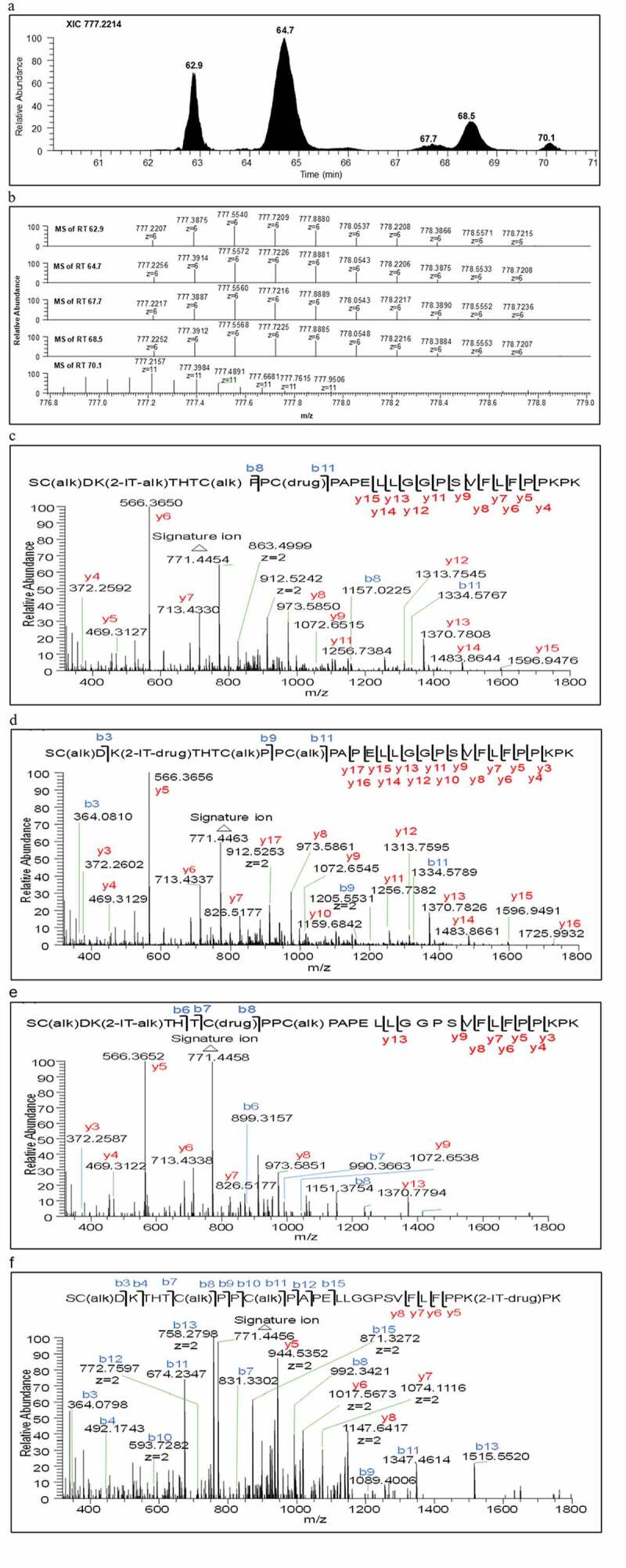
(a) Extracted ion chromatogram at m/z 777.2214, (b) Zoomed accurate mass spectra of the ion at m/z 777.2214 of RT 62.9, RT 64.7, RT 67.7, RT 68.6 and RT 70.1 minutes respectively, (c) Tandem mass spectrum of peak at RT 62.9, (d) Tandem mass spectrum of peak at RT 64.7, (e) Tandem mass spectrum of peak at RT 67.7, and (f) Tandem mass spectrum of peak at RT 68.5 minutes
Next, the high-resolution mass spectrum of each peak in the XIC was inspected to observe the accurate protonated mass, isotopic distribution and charge state (e.g., m/z 777.2214 in Figure 2b). As shown in Figure 2b, the retention time (RT) 70.1 min peak was clearly a false-positive conjugated peptide based on its incorrect charges state (z = 11 instead of 6) and isotopic distribution, while the remaining four peaks at RTs 62.9, 64.7, 67.7 and 68.5 min exhibited the expected charge state and nearly identical isotopic distribution. Careful examination of the tandem mass spectra of these four peaks shown in Figure 2c–f was required to further elucidate their structures. The key fragment ion detected was the signature ion at m/z 771 (Figure 3) along with other fragments corresponding to the payload-linker (see Data Processing section for detail), and the peptide backbone fragments (y and b ions). The signature ion at m/z 771 was clearly detected in each of the four spectra (Figure 2c–f), indicating they were isomeric drug conjugated peptides. In this case, key fragmentation patterns of the peptide backbone were essential for the identification of the anticipated (H227)K conjugated peptide. From the tandem mass spectrum of RT 64.7 min peak shown in Figure 2d, we found that the fragment ions of C-terminal from y3 to y17 and N-terminal b3, b9, and b11 ions were in agreement with the (H227)K conjugated peptide. Thus, the major peak at RT 64.7 min in Figure 2a was assigned as the expected lysine-linked conjugated peptide (H227)K.
Figure 3.
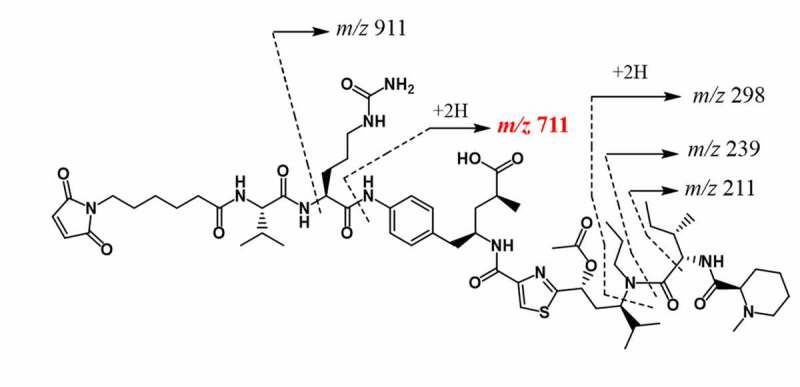
The chemical structure of the payload-linker and assigned mass fragments
For the identification of the remaining three isomeric conjugated peptides of (H227)K, the following logic was applied, based on the conjugation chemistry. The (H227)K conjugated peptide also contains another lysine residue, (H251)K. If (H251)K, instead of (H227)K, was thiolated with 2-IT and conjugated with the payload-linker, while the (H227)K was miss-cleaved during trypsin digestion, it would be the isomer of the (H227)K conjugated peptide, SC(alk)D(H227)KTHTC(alk)PPC(alk)PAPELLGGPSVFLFPP(H251)K(2-IT-drug)P(H253)K. This peptide was designated as (H251)K’ to differentiate it with the typical (H251)K conjugated peptide, THTC(alk)PPC(alk)PAPELLGGPSVFLFPP(H251)K(2-IT-drug)P(H253)K, where the unthiolated (H227)K in the (H251)K’ peptide was cleaved during trypsin digestion. By following this lead, we examined the tandem mass spectrum of the RT 68.5 min peak (Figure 2f) and found that it was consistent with the (H251)K’ conjugated peptide. Therefore, the (H251)K lysine conjugation site was likely distributed in two tryptic peptides (H251)K’ and (H251)K. The (H251)K was indeed unambiguously identified at RT 69.77 min by using this manual multi-step procedure (data not shown). Similarly, a total of 78 of 80 putative lysine-linked conjugated sites were identified. The relative area percent of the identified lysine conjugated peptides derived from peptide mapping analysis and the solvent accessibility area (%SAA) of the lysine side chains from modeling analysis (vide infra) are shown in Table 1.
Table 1.
Comparison of the solvent accessible area from modeling analysis and the relative area percent of conjugated peptides from mass spectrometry
| No. | Conjugation site | Observed mass (da) | Theoretic mass (da) | Accuracy (ppm) | Saa (%) | Conjugated peptide (%) | No. | Conjugation site | Observed mass (da) | Theoretic mass (da) | Accuracy (ppm) | Saa (%) | Conjugated peptide (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | L1E | 3217.7188 | 3217.7130 | 1.8 | 89.0 | <0.1 | 43 | L214C | 1526.7556 | 1526.7506 | 3.3 | 34.0 | 4.8 |
| 2 | L1E+1 | 3218.7080 | 3218.6971 | 3.4 | 89.0 | 12.3 | 44* | H225C | 1670.8446 | 1670.8406 | 2.4 | 40.1 | 6.5 |
| 3 | L39K | 3671.8764 | 3671.8632 | 3.6 | 29.7 | 0.7 | H225C | 2283.1252 | 2283.1353 | 4.4 | 0.3 | ||
| 4 | L103K | 3053.6220 | 3053.6122 | 3.2 | 31.5 | 1.0 | 45* | H231C | 4007.0904 | 4007.0796 | 2.7 | 43.4 | 1.0 |
| 5 | L107K | 1964.1027 | 1964.0991 | 1.8 | 49.0 | 2.7 | H231C | 4657.2834 | 4657.2843 | −0.2 | <0.1 | ||
| 6 | L126K | 5045.5825 | 5045.5785 | 0.8 | 85.1 | 3.0 | 46* | H234C | 4007.0904 | 4007.0796 | 2.7 | 83.7 | 1.2 |
| 7 | L145K | 2208.1944 | 2208.1839 | 4.8 | 74.4 | 2.3 | H234C | 4657.2642 | 4657.2843 | 4.3 | 0.4 | ||
| 8 | L149K | 3996.9720 | 3996.9601 | 3.0 | 25.9 | 1.0 | 47*** | L23C | nd | 1868.9893 | nd | 0.0 | nd |
| 9 | 169 K | 4939.4150 | 4939.3994 | 3.2 | 94.1 | 2.6 | 48*** | L88C | nd | 4524.1546 | nd | 0.0 | nd |
| 10 | L183K | 3428.7305 | 3428.7135 | 5.0 | 47.5 | 1.8 | 49*** | L134C | nd | 2959.5340 | nd | 0.0 | nd |
| 11 | L188K | 2210.1345 | 2210.1268 | 3.5 | 73.1 | 1.5 | 50*** | L194C | nd | 3037.5657 | nd | 0.0 | nd |
| 12 | L190K | 3461.7580 | 3461.7550 | 0.9 | 88.9 | 3.5 | 51*** | H22C | nd | 2372.2636 | nd | 0.0 | nd |
| 13 | L207K | 3700.8575 | 3700.8460 | 3.1 | 44.6 | 5.2 | 52*** | H96C | nd | 2494.2276 | nd | 0.0 | nd |
| 14 | H43K | 4388.1688 | 4388.1692 | 0.1 | 56.5 | 0.4 | 53*** | H149C | nd | 2483.3168 | nd | 0.0 | nd |
| 15 | H65K | 4120.0420 | 4120.0267 | 3.7 | 59.1 | 1.0 | 54*** | H205C | nd | 7874.9532 | nd | 0.0 | nd |
| 16 | H76K | 3116.5916 | 3116.5861 | 1.8 | 59.3 | 3.0 | 55*** | H266C | nd | 3300.6662 | nd | 0.0 | nd |
| 17 | H126K | 5456.6533 | 5456.6512 | 0.4 | 52.0 | 1.8 | 56*** | H326C | nd | 1468.7822 | nd | 0.0 | nd |
| 18 | H138K | 3809.9880 | 3809.9810 | 1.8 | 15.7 | 1.0 | 57*** | H372C | nd | 2323.2684 | nd | 0.0 | nd |
| 19 | H152K | 9337.6390 | 9337.6328 | 0.7 | 1.8 | 0.1 | 58*** | H430C | nd | 3962.9059 | na | 0.0 | nd |
| 20 | H210K | 8034.0075 | 8033.9886 | 2.4 | 76.2 | 2.5 | *detected in multiple peptides | ||||||
| 21 | H215K | 8376.1902 | 8376.1789 | 1.3 | 79.5 | 1.1 | ** nd: not detected | ||||||
| 22 | H218K | 1836.9996 | 1836.9994 | 0.1 | 33.3 | 1.3 | *** intra-chain cystein | ||||||
| 23 | H223K | 2283.1356 | 2283.1353 | 0.1 | 27.9 | 0.2 | |||||||
| 24 | H227K | 4657.2732 | 4657.2843 | 2.4 | 82.2 | 1.6 | |||||||
| 25* | H251K | 4657.2720 | 4657.2843 | 2.6 | 67.7 | 0.6 | |||||||
| H251K | 4166.1248 | 4166.1157 | 2.2 | 1.8 | |||||||||
| 26 | H253K | 4982.5325 | 4982.5321 | 0.1 | 23.4 | 0.8 | |||||||
| 27 | H279K | 5118.4998 | 5118.4857 | 2.8 | 79.3 | 1.5 | |||||||
| 28 | H293K | 3479.8030 | 3479.7886 | 4.1 | 60.0 | 1.4 | |||||||
| 29 | H295K | 1821.0069 | 1821.0045 | 1.3 | 45.1 | 1.5 | |||||||
| 30 | H322K | 3547.9115 | 3547.8975 | 3.9 | 29.4 | 2.0 | |||||||
| 31 | H325K | 2048.0229 | 2048.0185 | 2.1 | 36.0 | 0.6 | |||||||
| 32 | H327K | 2056.0617 | 2056.0559 | 2.8 | 26.7 | 5.8 | |||||||
| 33 | H331K | 2586.4338 | 2586.4317 | 0.8 | 66.2 | 2.4 | |||||||
| 34 | H339 K | 2587.4586 | 2587.4521 | 2.5 | 24.8 | 2.4 | |||||||
| 35 | H343 K | 1967.0940 | 1967.0988 | 2.4 | 4.4 | 0.4 | |||||||
| 36 | H345K | 1976.0751 | 1976.0739 | 0.6 | 68.8 | 1.8 | |||||||
| 37 | H365 K | 3100.5750 | 3100.5721 | 0.9 | 32.0 | 2.6 | |||||||
| 38 | H375K | 5007.4385 | 5007.4173 | 4.2 | 6.9 | 2.3 | |||||||
| 39 | H397K | 5718.7625 | 5718.7255 | 6.5 | 44.6 | 1.6 | |||||||
| 40 | H414K | nd** | 3749.9340 | nd | 0.3 | nd | |||||||
| 41 | H419K | 2138.1668 | 2138.1631 | 1.7 | 32.0 | 3.4 | |||||||
| 42 | H444K | 4763.2944 | 4763.2797 | 3.1 | 30.8 | 0.9 | |||||||
However, the remaining two isomeric conjugated peptides peaks at RTs 62.9 and 67.7 min in Figure 2a could not be assigned as lysine conjugates because no more lysine residues other than (H227)K and (H251)K were present in this peptide. Therefore, the investigation was extended to potential cysteine conjugation sites, as discussed below.
Identification of conjugation sites on cysteine residues
The anticipated (H227)K conjugated peptide includes three cysteine residues that would be expected to be in an inter-chain disulfide configuration and would not react with the maleimido moiety for drug conjugation. However, the disulfide bonds could be scrambled in the presence of free thiols.18 In other words, the thiolated lysine intermediates (Figure 1b) produced during the conjugation reaction could cause the proximal disulfide bonds to scramble, resulting in cysteine-linked conjugated peptides. Therefore, it could be reasonably assumed that the two un-assigned peaks mentioned above could be produced by direct cysteine conjugation from two of the three cysteine residues, (H225)C, (H231)C, and (H234)C, in conjunction with the (H227)K residue thiolated by 2-IT, which formed a new disulfide bonds with one of the three cysteine residues through disulfide bond scrambling, as postulated in Figure 4a.
Figure 4.
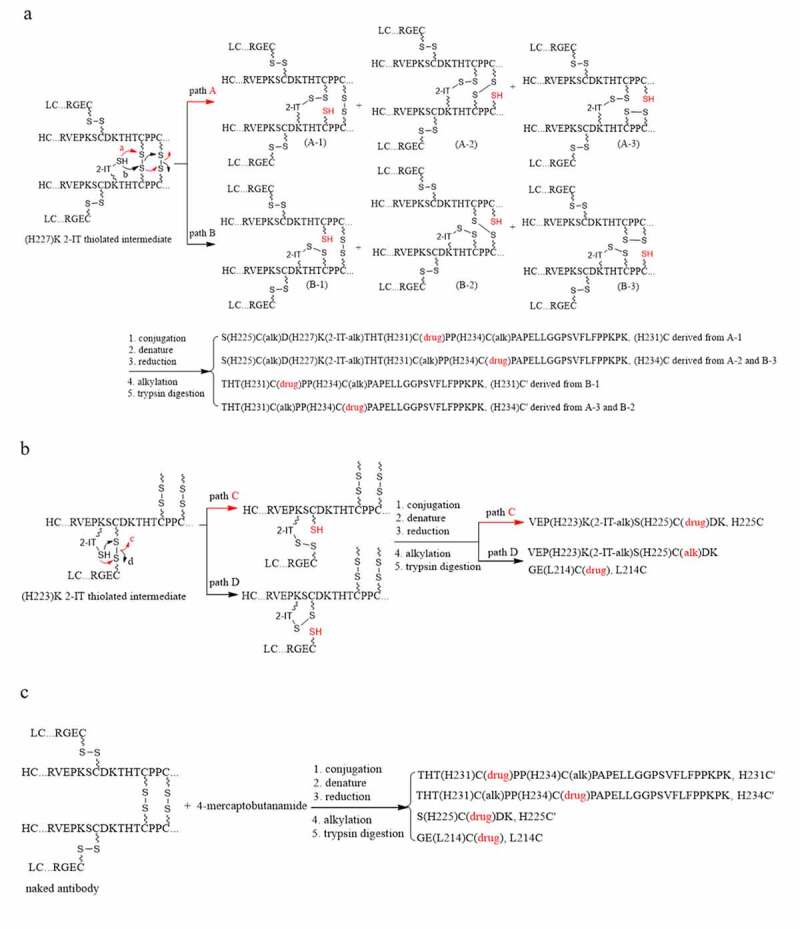
Plausible formation mechanisms of the cysteine-linked conjugates. (a) (H227)K-2-IT pathways, (b) (H223)K-2-IT pathways, (c) Free thiol pathway
As shown in Figure 4a, the scrambling of the (H231)C-(H231)C and (H234)C-(H234)C inter-chain disulfide bonds initiated by the neighboring (H227)K-2-IT thiolated intermediate could produce six cysteine free thiols through pathway A (A-1 – A-3) and pathway B (B-1 – B-3) for conjugation with the payload-linker. The targeted isomeric cysteine-linked conjugated peptide, (H231)C for S(H225)C(alk)D(H227)K(2-IT-alk)THT(H231)C(drug)PP(H234)C(alk)PAPELLGGPSVFLFPP(H251)KP(H253)K, could be derived from A-1 intermediate, while the other one, (H234)C for S(H225)C(alk)D(H227)K(2-IT-alk)THT(H231)C(alk)PP(H234)C(drug)PAPELLGGPSVFLFPP(H251)KP(H253)K, could be derived from A-2 and B-3. In fact, the tandem mass spectra with key fragment assignments of RT 62.9 min peak in Figure 2c and RT 67.7 min peak in Figure 2e are in agreement with the postulated cysteine-linked conjugated peptides (H234)C and (H231)C, respectively. Therefore, the four isomeric conjugated peptides observed at RTs 64.7, 68.5, 62.9 and 67.7 min in Figure 2a were assigned to the two lysine-linked conjugated peptides, (H227)K and (H251)K’, and two cysteine-linked conjugated peptides, (H234)C and (H231)C, respectively.
Additionally, the scrambling of the heavy-heavy inter-chain disulfide bonds (labeled as C’) by the (H227)K-2-IT thiolated intermediate could also generate another two cysteine-linked conjugated peptides designated as (H231)C’ for THT(H231)C(drug)PPC(alk)PAPELLGGPSVFLFPPKPK and (H234)C’ for THTC(alk)PP(H234)C(drug)PAPELLGGPSVFLFPPKPK, respectively, as shown in Figure 4a. The scrambling of the light-heavy inter-chain disulfide bond by the (H223)K 2-IT thiolated intermediate could yield two cysteine-linked peptides designated as (H225)C for VEP(H223)K(2-IT-alk)S(H225)C(drug)DK and (L214)C for GE(L214)C(drug), as postulated in Figure 4b. These tryptic cysteine-linked conjugated peptides, (H231)C’, (H234)C’, (H225)C and (L214)C, were indeed identified and are listed in Table 1.
The inter-chain disulfide bonds could also be subject to scrambling via intermolecular attack by other free thiols, such as 4-mercaptobutanamide derived from the hydrolysis of 2-IT during the conjugation reaction, to yield the (H231)C’, (H234)C’ and (L214)C cysteine-linked conjugated peptides, as postulated in Figure 4c. Furthermore, a characteristic cysteine-linked conjugated peptide designated as (H225)C’ for S(H225)C(drug)DK, was identified, which could only be derived from the light-heavy inter-chain disulfide bond scrambled by free thiols, such as 4-mercaptobutanamide, via intermolecular, but not intramolecular, attack by the neighboring lysine (H223)K thiolated intermediate based on the chemistry. This was strong evidence in support of the hypothesis that the inter-chain disulfide bonds were indeed scrambled by 4-mercaptobutanamide free thiol. On the other hand, the 4-mercaptobutanamide free thiol was unlikely to be involved in the formation of the (H231)C and (H234)C (pathways A and B in Figure 4a) and the (H225)C (pathway C in Figure 4b) because the (H227)K-2-IT or (H223)K-2-IT thiolated intermediates needed to form disulfide bonds with free thiols through oxidation during drug conjugation reaction so as to form the (H227)K-2-IT-alk or (H223)K-2-IT-alk moieties in these tryptic peptides.
Identification of conjugation site on N-terminus of light chain
N-terminus residues with a free amino group can be directly conjugated with payload-linker unless it has been converted to pyroglutamate.19,20 The strategy for the identification of lysine linked conjugated peptides was also applied to the light chain N-terminus linked conjugated peptide (L1)E, (L1)E(2-IT-drug)IVLTQSPATLSLSPGER. The m/z 805.4355 ([M + 4 H]4+) was the most abundant ion corresponding to (L1)E conjugated peptide in a 4+ charge state. Its XIC is shown in Figure 5a. Two major peaks at RTs 38.4 and 64.3 min were observed, but only the RT 64.3 min peak showed the correct 4+ charge state (Figure 5b). The tandem mass spectrum of RT 64.3 min peak shown in Figure 5e was also consistent with this N-terminal conjugated peptide.
Figure 5.
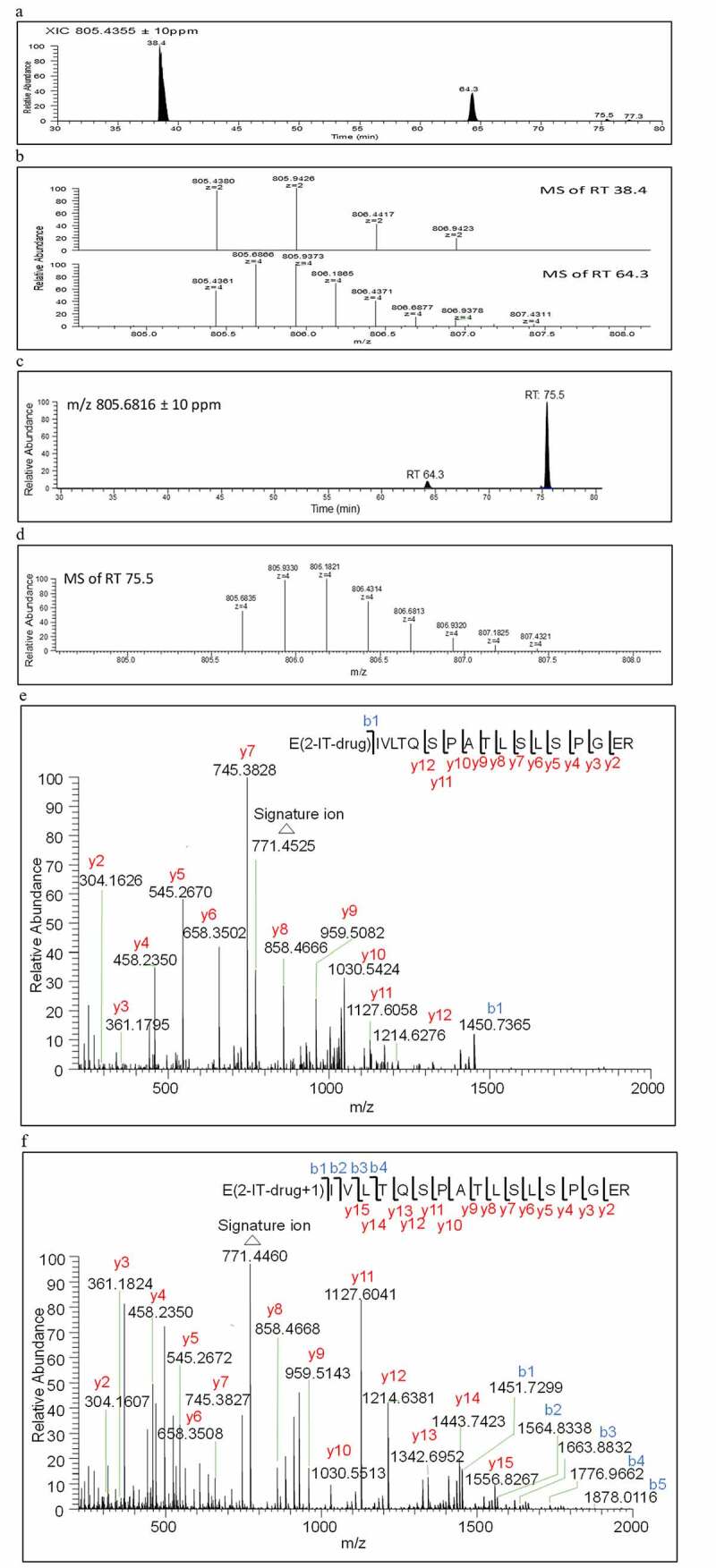
(a) Extracted ion chromatogram of conjugated peptide of the light chain N-terminus (L1E, XIC m/z 805.4355 ± 10 ppm), (b) Zoomed-in accurate mass spectra of the ion at m/z 805 from RT 38.4, and 64.3 minutes, respectively, (c) Extracted ion chromatogram of the hydrolyzed conjugated peptide of the light chain N-terminus (L1E + 1, XIC m/z 805.6816 ± 10 ppm), (d)Zoomed-in accurate mass spectra of the ion at m/z 805 from RT 75.6, (e) Tandem mass spectrum of N-terminal conjugated peptide (L1E) of RT 64.3 minutes, and (f) the hydrolyzed peptide (L1E + 1) of RT 75.5 minutes
However, the level of detection of (L1)E conjugated peptide was considered unusually low (less than 0.1%) considering the high solvent accessibility area of 89% SAA shown in Table 1. It was recognized that the amidine moiety of (L1)E conjugated peptide might be susceptible to hydrolysis to yield an amide analog (L1E+1 or M’), resulting in 1 Da higher mass in the tryptic peptide as compared to that of the (L1)E conjugated peptide. Extracting m/z 805.6816 ± 10 ppm, corresponding to [M’+ 4 H]4+, was conducted. As a result, a new peak at RT 75.5 min with high intensity (12.3% normalized area percentage shown in Table 1) appeared in the XIC as shown in Figure 5c. The RT 75.5 min peak also showed the correct 4+ charge state, as depicted in Figure 5d. The tandem mass spectra of (L1)E at RT 64.3 min and (L1)E + 1 at RT 75.5 min shown in Figure 5e and Figure 5f exhibited the same signature ion at m/z 771 and nearly identical C-terminal fragment ions from y2 to y12. The one Da higher in N-terminal b ions (b1 to b3) of the RT 75.5 min peak (Figure 5f) in comparison with those of (L1)E at RT 64.3 min (Figure 5e) could be attributed to the hydrolysis of the amidine moiety of the N-terminus linked conjugated peptide to its amide analog.
Interestingly, the amidine moiety appeared stable in all of the lysine residue conjugates because the anticipated M + 1 counterparts of each individual identified lysine conjugates were not detected. The only amidine moiety prone to hydrolysis was located in the N-terminal conjugate (L1)E, under the same conjugation conditions. Despite the (L1)E having a high solvent accessibility, as illustrated in the 3D modeling structure in Figure 6a, some of the light chain lysine residues also exhibited high solvent accessibility, such as (L169)K with 94.1% SAA in Table 1. Thus, the hydrolysis of (L1)E to (L1)E + 1 was unlikely due to the solvent accessibility alone. The glutamic acid residue in (L1)E may also play an important role in mediating the hydrolysis of the amidine to amide, as illustrated in Figure 6b. The dissociated glutamate anion could intra-molecularly attack the protonated amidine moiety to form a seven-membered ring intermediate, which would undergo deamination followed by hydrolysis and ring opening to form the amide analog.
Figure 6.
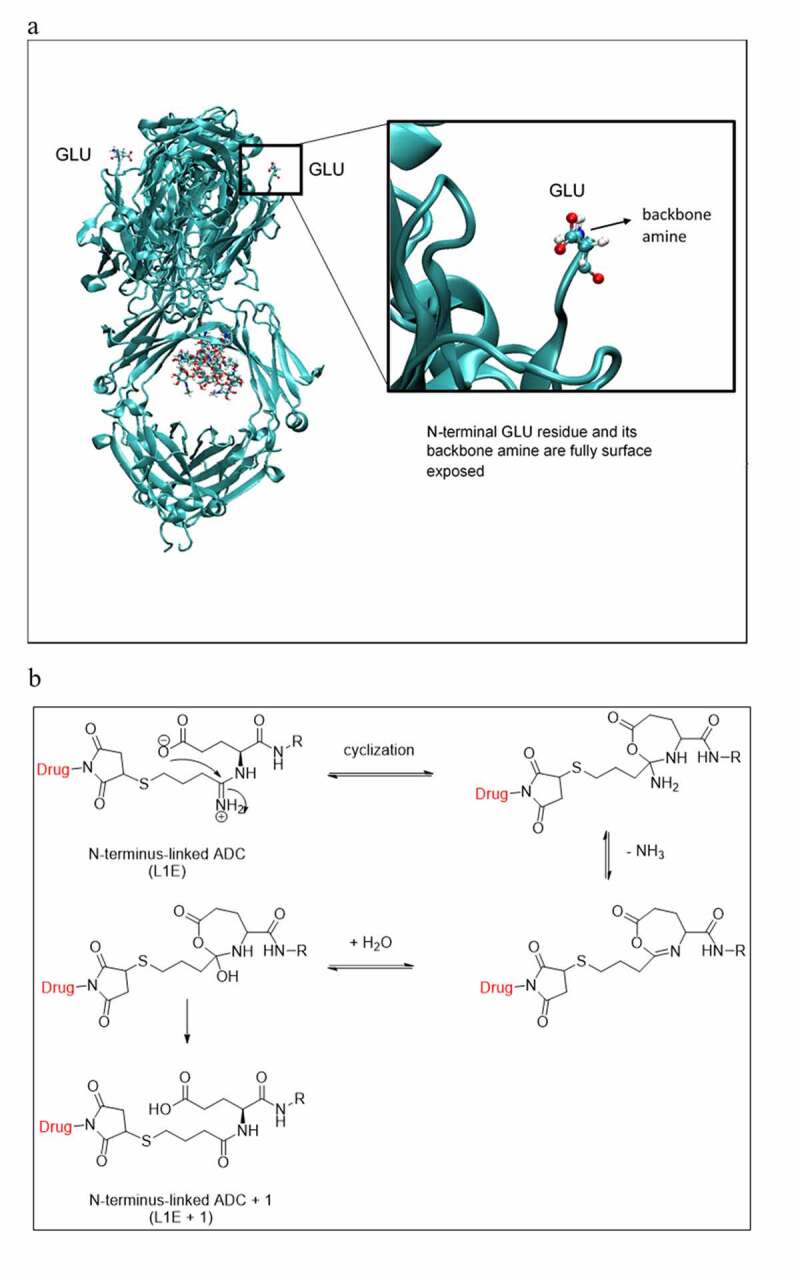
(a) Modeling 3D structure showing the light chain N-terminal glutamic acid is fully surface exposed, (b) Plausible degradation mechanism of the amidine moiety in L1E to the amide analogue in L1E + 1
Identification of unconjugated lysine and cysteine residues
The lysine residue (H414)K located in a constant region (Fc) or CH3 domain was identified as the only unconjugated site. Similarly, a thorough search for all possible cysteine conjugations corresponding to the intra-chain disulfide bonds was conducted, and none were detected. These observations were consistent with their low solvent accessibility, as predicted by 3D-modeling (0.3% SAA for (H414)K and zero solvent accessibility for those intra-chain disulfide bonds shown in Table 1).
Discussion
In this study, we adapted the “two-step” conjugation chemistry described in Brun et al.6 in order to characterize and understand the formation of cysteine conjugates and other by-products in a random, lysine-linked antibody drug conjugate. Thus, the lysine residues in an IgG1 antibody were thiolated with 2-IT to create lysine-linked 2-IT free thiols, which were subsequently conjugated with a maleimido moiety in the payload linker of a duocarmycin analog. The conjugated sites of the resultant ADC product were characterized via tryptic peptide mapping using a manual multi-step procedure. A total of 78 of the 80 anticipated lysine conjugated sites were identified. The only two unconjugated lysine sites remained, likely due to their poor solvent accessibility, as predicted by modeling analysis. Solvent accessibility (%SAA), however, is only one of many factors that could affect conjugation processes. As shown in Table 1, the correlation between some of the conjugated peptides in percentile to their %SAA was poor. Caution should also be taken regarding the comparison of the tryptic conjugated peptide area percent listed in Table 1, as their quantitation relies on the MS response, which may vary among the different conjugated peptides. On the other hand, although 78 of the 80 anticipated lysine sites were conjugated with payload link, the DAR was controlled at around 3.0 for this product. The DAR numbers and high conjugated site distribution are key CQAs of this ADC.
During the identification of the (H227)K conjugated peptide located in the hinge region, the detection of three additional isomers was unexpected. One of them was identified as (H251)K’ conjugated peptide with the unthiolated (H227)K site miss-cleaved in this tryptic peptide. The finding of (H251)K’ was not anticipated, but still possible due to the nature of the experimental conditions. The remaining two isomers were identified as cysteine linked conjugated peptides, (H231)C and (H234)C, respectively. The identification of the latter two isomers was important because it revealed that a small number of the heavy-heavy inter-chain disulfide bonds of the antibody were scrambled by the (H227)K-2-IT free thiol. This could yield six cysteine-linked ADC species, i.e., A-1 to A-3 and B-1 to B-3 (Figure 4a) after payload linker conjugation, further increasing heterogeneity of the ADC.
The heavy-heavy inter-chain disulfide bonds scrambled by the (H227)K-2-IT free thiol could also yield another two cysteine-linked conjugated peptides, (H231)C’ and (H234)C’ (Figure 4a). Similarly, the heavy-light inter-chain disulfide bonds could be scrambled by the (H223)K-2-IT free thiol (Figure 4b). As a result, two cysteine-linked conjugated peptides, (H225)C and L(214)C, along with the no-drug conjugated peptide, VEP(H223)K(2-IT-alk)S(H225)C(alk)DK, were detected. The detection of the no-drug conjugated, but thiolated (H223)K peptide was particularly interesting because, besides pathway D (Figure 4b), it could also be derived from the alkylation of the (H223)K-2-IT free thiol during sample preparation for peptide mapping. In comparison with VEP(H223)K(2-IT-drug)SC(alk)DK (i.e., the (H223)K-2-IT free thiol conjugated with payload linker) and VEP(H223)K(2-IT-alk)SC(drug)DK (i.e., the (H223)K-2-IT free thiol scrambled with the inter-chain disulfide), we found that the no-drug conjugated, but thiolated, (H223)K peptide was only at 1.4%, while the VEP(H223)K(2-IT-drug)SC(alk)DK and VEP(H223)K(2-IT-alk)SC(drug)DK were at 31.0% and 67.6%, respectively. This finding indicated that the 2-IT thiolated (H223)K free thiol was at less than 1.4%, if present, among the three relevant tryptic peptides. Most of the (H223)K-2-IT free thiols were either conjugated with payload linker or scrambled with the heavy-light inter-chain disulfide bond.
The other free thiol presented in the conjugation reaction mixture was likely the 4-mercaptobutanamide derived from the hydrolysis of 2-IT, which could scramble the four inter-chain disulfide bonds of the antibody to generate four cysteine-linked conjugated peptides, (H231)C’, (H234)C’, (H225)C’ and (L214)C (Figure 4c). It is worth mentioning that (H231)C’, (H234)C’ and (L214)C could also be derived from the scrambling of the inter-chain disulfide bonds by the (H233)K-2-IT free thiol and (H227)K-2-IT free thiol (vide supra). Conversely, the (H225)C’ conjugated peptide could only be derived from the scrambling of the heavy-light inter-chain disulfide bond by the 4-mercaptobutanamide free thiol via inter-molecular attack.
It was reported that a small number of free thiols (ca. 0.05%) could exist at cysteines in the naked antibody measured under nondenaturing conditions.21 Such free thiols could also directly conjugate with the payload linker to form (H231)C’, (H234)C’, (H225)C’ and (L214)C cysteine-linked conjugated peptides. Additionally, the (H231)C and (H234)C cysteine-linked conjugated peptides could be derived from these cysteine free thiols provided that the cysteine free thiols were conjugated with the payload linker in conjunction with the (H227)K-2IT free thiol neither conjugated with the payload linker nor scrambled with the inter-chain disulfide bond. However, we believed the contribution of these cysteine free thiols to the formation of (H231)C and (H234)C cysteine-linked conjugated peptides would be minimal because they need to compete with (H227)K-2-IT free thiols upon conjugation with the payload linker. Thus, the quantification of such free thiols at cysteines in the hinge region of the naked antibody was not pursued. The intra-chain disulfide bonds were found to be intact because their anticipated cysteine-linked conjugated peptides were not detected, presumably due to their poor solvent accessibility as predicted by modeling analysis.The identification of the conjugation site on N-terminus of light chain, (L1)E, was straightforward. However, the low-level detection of (L1)E-2-IT-linked conjugated peptide at less than 0.1% caught our attention. Further investigation revealed that most of the amidine moiety in the (L1)E-2-IT-linked conjugated peptide was hydrolyzed to its amide counterpart, (L1)E + 1 (12.3%). This finding led to a thorough search for the amide counterparts of the lysine linked conjugated peptides, but none were detected. Therefore, the amidines in the lysine-linked conjugates were deemed stable. The hydrolysis of (L1)E-2-IT linked conjugated peptide to its amide analog could be attributed to the mediation of the glutamic acid residue in (L1)E.
In conclusion, we conducted a comprehensive structural study of an ADC modified by 2-IT and subsequent conjugation with a duocarmycin analog-based payload-linker. Plausible formation mechanisms of cysteine-linked conjugates and the hydrolysis of the N-terminus conjugate were proposed to guide the identification of the unexpected conjugated peptides, as well as to provide scientific insight for process development and drug quality control.
Materials and methods
Reagents and materials
Guanidine hydrochloride (8 M) and CaCl2 · 2H2O were purchased from Thermo Fisher Scientific (Waltham, MA); Trizma (pH 7.6), sodium iodoacetate, formic acid and acetonitrile were purchased from Sigma-Aldrich (Burlington, MA); NAP-5 columns were purchased from GE Healthcare (Waltham, MA); sequencing grade modified trypsin was purchased from Promega (Madison, WI); dithiothreitol (DTT) was supplied by Bio-Rad (Hercules, CA); trifluoroacetic acid (TFA) was purchased from Pierce Chemical (Rockford, IL).
Preparation of ADC
The ADC used in this study is composed of a potent cytotoxic prodrug covalently linked with a human anti-CD70 antibody (IgG1). CD70 is expressed in renal cell carcinoma, leukemias, lymphomas, and other cancers. Both antibody and cytotoxic prodrug (duocarmycin analog) were produced by Bristol Myers Squibb Co. The frozen naked antibody was thawed and pooled in a reactor, then exchanged with thiolation buffer to adjust pH from 7.0 to 7.4, following by treatment with 2-IT (20.5 equivalent). The resulting solution was agitated for 90 min at 23°C. Upon completion, the thiolated antibody was reacted with cytotoxic prodrug at pH 5.5 in agitation for 60 min at 23°C. Finally, the crude conjugate was exchanged with formulation buffer (pH 6.0) and diluted to 10 mg/mL with DAR 2.5 ~ 3.0 and 98% monomeric purity as determined by SEC.
Trypsin digestion
The reduction of antibody disulfide bonds was performed by mixing 1 mg of ADC with 200 μL of 8 M guanidine hydrochloride, 18 μL of 0.2 M DTT and 60 μL of 0.8 M Trizma, then incubated at 37°C for 30 min. Alkylation of cysteine residues was conducted by the addition of 18 µL of 0.4 M sodium iodoacetate to the reduced ADC and incubation in the dark for 15 min at room temperature. The desalting was accomplished by transferring the reduced and alkylated solution into a NAP-5 column, then exchanging with digestion buffer (10 mM CaCl2 in 50 mM Trizma). Finally, the digestion was carried out by mixing the desalted solution with trypsin (protein:trypsin ratio = 25:1) and incubating at 37°C for 3 hours.
LC-MS analysis
LC-MS for peptide mapping was conducted using a Thermo Fisher Scientific Q Exactive™ Hybrid Quadrupole-Orbitrap Mass Spectrometer (Waltham, MA) coupled to a Waters ACQUITY UPLC H-Class (Milford, MA) with a C18 column (Waters UPLC BEH130, C18, 1.7 μm, 2.1x100mm). In positive ion scanning mode, both probe heater temperature and capillary temperature of 275°C was used. The spray voltage and S-lens RF level were set to 3500 eV and 50 eV, respectively. Sheath gas and auxiliary gas were tuned to 50 and 13 (arbitrary units), respectively. A mass range of 400–2000 Daltons, collision gas of 27 (arbitrary unit) and resolution of 35k were optimized to identify conjugated peptides. Mobile phase A (aqueous) contained 0.1% formic acid in water, while mobile phase B (organic) consisted of 0.085% formic acid in acetonitrile. The flow rate and column temperature were maintained at 0.4 mL/min and 45°C, respectively. The gradient began with a 3 min hold at 2% B followed by an increase to 20%, 30%, 40%, 95% B at 30, 70, 90, 92 min respectively.
Data processing
The data processing for peptide confirmation and quantitation was performed using commercially available software. In order to obtain a unique fingerprint for conjugated peptides and related by-products, such as the amide analog of the light chain N-terminus conjugate and 2-IT modified peptides, a manual sequencing method in Thermo Protein Discovery was implemented to search not only for the peptide backbone fragments within a ± 10 ppm window, but also the possible ions from the cleavage of payload-linker. The signature ion at m/z 771, generated from the cleavage of Valine-Citrulline linker as shown in Figure 3, appeared in all tandem mass spectra of conjugated peptides.
Additionally, a manual, multi-step data processing procedure was developed and demonstrated with examples in this study to systematically search, assess and distinguish conjugated peptides with the help of commercially available software, such as GPMAW (General Protein/Mass Analysis for Windows) and Thermo Biopharma (Pinpoint and Protein discovery). Although the procedure is not simple or quick, these multiple steps, in combination with pertinent knowledge of chemistry, provided sufficient information to identify the conjugated peptides and related degradants by a single trypsin digestion in a highly heterogeneous mixture.
Individual peak areas derived from XIC were integrated for relative quantitation of conjugated peptides. The conjugated peptide percent was determined by taking a ratio of the individual conjugated peptide area against the total peak area of all conjugated peptides. The peak areas of identified conjugated peptides were derived from one or multiple charge states of accurate mass XICs with a ± 10 ppm mass accuracy window and automatically integrated through Thermo Pinpoint software.
Modeling methodology
The homology model for the protein structure was performed by using the antibody modeling protocol within BIOVIA Discovery Studio software. SAA for the side chains of Lysine and Cysteine residues, which is commonly used to assess their reactivity, were calculated using the same Discovery studio software with 1.4 Å probe radius for water. The %SAA were obtained by normalizing the SAA values with the fully exposed SAA values for Lys and Cys based on Ala-Lys-Ala and Ala-Cys-Ala peptides. The final %SAA values were obtained by averaging the two identical heavy and light chains of antibody.
Acknowledgments
The authors would like to acknowledge Annie Tam for her collaboration, Wei Ding, Michael Peddicord and Jonathan Shackman for manuscript review and discussions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Peters C, Brown S.. Antibody–drug conjugates as novel anti-cancer chemotherapeutics. Biosci Rep. 2015;35(4):1–12. doi: 10.1042/BSR20150089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chari RV, Miller ML, Widdison WC. Antibody-drug conjugates: an emerging concept in cancer therapy. Angew Chem Int Ed Engl. 2014;53(15):3796–827. doi: 10.1002/anie.201307628. [DOI] [PubMed] [Google Scholar]
- 3.Janin-Bussat MC, Dillenbourg M, Corvaia N, Beck A, Klinguer-Hamour C. Characterization of antibody drug conjugate positional isomers at cysteine residues by peptide mapping LC–MS analysis. J Chromatogr B. 2015;981–982:9–13. doi: 10.1016/j.jchromb.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 4.Said N, Gahoual R, Kuhn L, Beck A, François YN, Leize-Wagner E. Structural characterization of antibody drug conjugate by a combination of intact, middle-up and bottom-up techniques using sheathless capillary electrophoresis-Tandem mass spectrometry as nanoESI infusion platform and separation method. Anal Chim Acta. 2016;918:50–59. doi: 10.1016/j.aca.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Hamblett KJ, Senter PD, Chace DF, Sun MM, Lenox J, Cerveny CG, Kissler KM, Bernhardt SX, Kopcha AK, Zabinski RF. Effects of drug loading on the antitumor activity of a monoclonal antibody drug conjugate. Clin Cancer Res. 2004;10:7063–70. doi: 10.1158/1078-0432.CCR-04-0789. [DOI] [PubMed] [Google Scholar]
- 6.Brun MP, Gauzy-Lazo L. Antibody-drug conjugates. Editor by Ducry L, Chapter 10 Protocols for lysine conjugation, Humana Press, Totowa, NJ. Vol. 10. 2013. 173–87. doi: 10.1007/978-1-62703-541-5_10. [DOI] [PubMed] [Google Scholar]
- 7.Jain N, Smith SW, Ghone S, Tomczuk B. Current ADC Linker Chemistry. Pharm Res. 2015;32:3526–40. doi: 10.1007/sl1095-015-1657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo Q, Chung HH, Borths C, Janson M, Wen J, Joubert MK, Wypych J. Structural characterization of a monoclonal antibody−maytansinoid Immunoconjugate. Anal Chem. 2016;88:695–702. doi: 10.1080/19420862.2020.1763138. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, Amphlett G, Blättler WA, Lambert JM, Zhang W. Structural characterization of the maytansinoid–monoclonal antibody immunoconjugate, huN901–DM1, by mass spectrometry. Protein Sci. 2005;14:2436–46. doi: 10.1110/ps.051478705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu B, Guo H, Zhang J, Xue J, Yang Y, Qin T, Xu J, Guo Q, Zhang D, Qian W, et al. In-depth characterization of a pro-antibody−drug conjugate by LC−MS. Mol Pharmaceutics. 2016;13:2702–10. doi: 10.1021/acs.molpharmaceut.6b00280. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Wang L, Shion H, Yu C, Yu YQ, Zhu L, Li M, Chen W, Gao K. In-depth structural characterization of Kadcyla (ado-trastuzumab emtansine) and its biosimilar Candidate. mAbs. 2016;8(7):1210–23. doi: 10.1080/19420862.2016.1204502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chih HW, Gikanga B, Yang Y, Zhang B. Identification of amino acid residues responsible for the release of free drug from an antibody-drug conjugate utilizing lysine-succinimidyl ester chemistry. J Pharm Sci. 2011;100(7):2518–25. doi: 10.1002/jps. [DOI] [PubMed] [Google Scholar]
- 13.Traut RR, Bollen A, Sun -T-T, Hershey WB, Sundberg J, Pierce LR. Methyl 4-mercaptobutyrimidate as a cleavable crosslinking reagent and its application to Escherichia coli 30S ribosome. Biochemistry. 1973;12:3266–73. doi: 10.1021/bi00741a019. [DOI] [PubMed] [Google Scholar]
- 14.Singh R, Kats L, Blattler WA, Lambert JM. Formation of N-substituted 2-iminothiolanes when amino groups in proteins and peptides are modified by 2-iminothiolane. Anal Biochem. 1996;236:114–25. doi: 10.1006/abio.1996.0139. [DOI] [PubMed] [Google Scholar]
- 15.Gordon MR, Canakci M, Li L, Zhuang J, Osborne B, Thayumanavan S. Field guide to challenges and opportunities in antibody−drug conjugates for chemists. Bioconjugate Chem. 2015;26:2198–215. doi: 10.1021/acs.bioconjchem.5b00399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Owonikoko T, Hussain A, Stadler W, Smith D, Kluger H, Molina A, Gulati P, Shah A, Ahlers C, Cardarelli P, et al. First-in-human multicenter phase I study of BMS-936561 (MDX-1203), an antibody-drug conjugate targeting CD70. Cancer Chemother Pharmacol. 2016;77:155–62. doi: 10.1007/s00280-015-2909-2. [DOI] [PubMed] [Google Scholar]
- 17.Ahmad S, Gromiha M, Fawareh H, Sarai A. Database and tool for solvent accessibility representation in proteins. BMC Bioinform. 2004;5:1–5. doi: 10.1186/1471-2105-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cuevasanta E, Lange M, Bonanata J, Coitiño EL, Ferrer-Sueta G, Filipovic MR, Alvarez B. Reaction of hydrogen sulfide with disulfides and sulfenic acid to form the strongly nucleophilic persulfide. J Biol Chem. 2015;290(45):26866–80. doi: 10.1074/jbc.M115.672816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dick LW, Kim C, Qiu D, Cheng KC. Determination of the origin of the N-terminal pyro-glutamate variation in monoclonal antibodies using model peptides. Biotechnol Bioeng. 2007;97(3):544–53. doi: 10.1002/bit.21260. [DOI] [PubMed] [Google Scholar]
- 20.Liu Z, Valente J, Lin S, Chennamsetty N, Qiu D, Bolgar M. Cyclization of N-terminal glutamic acid to pyro-glutamic acid impacts monoclonal antibody charge heterogeneity despite its appearance as a neutral transformation. J Pharm Sci. 2018;108(10):3194–200. doi: 10.1016/j.xphs.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 21.Zhang W, Czupryn M. Free sulfhydryl in recombinant monoclonal antibodies. Biotechnol Prog. 2002;18:509–13. doi: 10.1021/bp025511z. [DOI] [PubMed] [Google Scholar]


