Abstract
Seven Enhancer of split genes in Drosophila melanogaster encode basic-helix-loop-helix transcription factors which are components of the Notch signalling pathway. They are expressed in response to Notch activation and mediate some effects of the pathway by regulating the expression of target genes. Here we have determined that the optimal DNA binding site for the Enhancer of split proteins is a palindromic 12-bp sequence, 5′-TGGCACGTG(C/T)(C/T)A-3′, which contains an E-box core (CACGTG). This site is recognized by all of the individual Enhancer of split basic helix-loop-helix proteins, consistent with their ability to regulate similar target genes in vivo. We demonstrate that the 3 bp flanking the E-box core are intrinsic to DNA recognition by these proteins and that the Enhancer of split and proneural proteins can compete for binding on specific DNA sequences. Furthermore, the regulation conferred on a reporter gene in Drosophila by three closely related sequences demonstrates that even subtle sequence changes within an E box or flanking bases have dramatic consequences on the overall repertoire of proteins that can bind in vivo.
The basic helix-loop-helix (bHLH) family of transcription factors includes many members that mediate cell fate allocation during animal development (30, 39, 45, 71). Their expression and activity can be regulated in response to cell-cell signalling, leading to the transcription of the specific set of genes required for a cell to adopt a particular fate. One pathway whose effect on cell fate decisions involves modulation of bHLH proteins is the Notch signalling pathway (reviewed in references 2 and 25). The most immediate transcriptional target genes of Notch activation in Drosophila melanogaster encode seven bHLH proteins (Mδ, Mβ, Mγ, M3, M5 M7, and M8) which are clustered in the Enhancer of split complex [E(spl)-C] (14, 36, 37). A number of closely related genes, known as Hes, Her, or ESR genes (44, 55, 60, 62), have now been isolated from vertebrates, and like the Drosophila E(spl) genes, many of the vertebrate homologues are expressed in response to Notch activity (3, 13, 32, 34, 38). The products of these genes are essential to implement many of the cell fate decisions mediated by Notch signalling, such as the selection of cells to become neural precursors (2, 25). Thus, a knowledge of the functional characteristics of the E(spl)bHLH proteins should lead to a greater understanding of how the activation of Notch mediates cell fate decisions via changes in gene transcription.
The E(spl) proteins represent a subset of bHLH proteins that also includes the Drosophila proteins Hairy and Deadpan (19). One distinguishing feature of this class of bHLH proteins is the presence of a proline residue in the basic domain. The basic domain confers on bHLH proteins DNA binding specificity (5, 10, 17, 18) for which the canonical target sequence is the E box (5′-CANNTG-3′). Initially it was postulated that the proline residue found in E(spl)-like bHLH proteins would impede DNA binding ability. Subsequently however, the E(spl) M5, M7, and M8 proteins have been shown to bind DNA in vitro, using a fortuitously identified sequence known as the N box (5′-CACNAG-3′) (48, 65) or another noncanonical bHLH target sequence which is a target for Hairy (49, 67) (5′-CACGCG-3′). Another feature shared by the E(spl)bHLH proteins is the C-terminal tetrapeptide WRPW. This conserved motif is required for these proteins to interact with Groucho, a putative corepressor protein (50, 51), and is sufficient to confer repressive functions when fused to heterologous proteins (20).
In several different developmental processes, such as neurogenesis and myogenesis, a primary role of the E(spl)bHLH proteins is to antagonize the activity of another family of bHLH proteins, the proneural proteins. During neurogenesis, proneural genes, which include achaete, scute, and lethal of scute (l’sc), encoded within the achaete-scute complex (AS-C) in Drosophila (70), provide the activity that promotes neural fate. These genes are initially expressed in groups of cells (proneural clusters), and within each cluster, proneural gene expression subsequently becomes refined so that the mRNAs and protein accumulate only in single cells, the neural precursors (9, 42, 54, 59). Mutations in genes encoding components of the Notch signalling pathway, including deletions that remove E(spl)bHLH genes, result in a failure of this refinement so that all the cells within proneural clusters accumulate high levels of proneural proteins and adopt the neural fate, giving rise to hypertrophy of the nervous system in the embryo and massive clusters of sensory organs in the peripheral nervous system (2, 25). The E(spl)bHLH proteins normally accumulate in cells which are inhibited from adopting the neural fate (33, 34), and when these proteins are artificially expressed in presumptive neural precursors, neural development is abolished (12, 22, 47, 63). Thus, ultimately whether or not a cell adopts the neural fate depends on the relative levels of the E(spl) and proneural bHLH proteins. The proneural proteins appear to act as transcriptional activators (8, 48, 69), and their activity is augmented by heterodimerization with the related Daughterless (Da) protein (46, 68). The different effects of proneural and E(spl) proteins can therefore be equated with their actions as repressors [E(spl)] or activators (proneural).
Several mechanisms have been proposed to explain the antagonism between E(spl) and proneural proteins, including direct protein-protein interactions, direct repression of AS-C gene expression by E(spl)bHLH proteins, and competition between the proteins for binding sites in the same genes (19, 35, 47, 49, 67). Experiments in yeast have demonstrated that proneural proteins and E(spl)bHLH proteins can interact with each other (1). Experiments in Drosophila, where E(spl)M7 was converted into an activator by replacing the terminal WRPW with an activation domain (35), support the notion that these proteins can also directly regulate the transcription of the proneural gene achaete.
To investigate the function of the E(spl) proteins, we have determined their DNA binding preference in vitro and have investigated the ability of the consensus site to respond to E(spl) and proneural proteins in vivo. Our results demonstrate that the different E(spl) proteins can recognize the same target site in vitro and in vivo, that this differs from the optimal target of proneural proteins, but that there is an overlap in the sequences recognized by the two classes of protein. In addition, our results demonstrate that subtle differences in nucleotide sequences in and around an E box can have dramatic effects on the profile of transcription factors that act on that site in vivo.
MATERIALS AND METHODS
Expression of fusion proteins in Escherichia coli.
Coding regions of E(spl)bHLH, da, and l’sc genes were amplified by using Taq polymerase (Cetus) (34). The upstream primers corresponded to sequences spanning the initiation codon and BamHI or BglII sites were included to facilitate cloning. Primer sequences are available on request. The amplified products were cloned into appropriate pRSET expression vectors (Invitrogen), transformed into E. coli BL21(DE3)pLysS cells or into appropriate pGex vectors (Pharmacia), and transformed into E. coli DH5α cells. Production of pRSET and pGex fusion proteins was as follows. Overnight cultures or single colonies were diluted into fresh culture medium containing ampicillin (200 μg/ml) and grown at 37°C to an optical density at 600 nm of 0.6 to 0.8. Then 2 mM isopropyl-β-d-thiogalactopyranoside was added, and the cells were grown for approximately 45 min. The harvested cells were resuspended in 1/50 culture volume of MTPBS (150 mM NaCl, 16 mM Na2HPO4, 4 mM NaH2PO4) containing 1% Nonidet P-40 and lysed by mild sonication. Inclusion bodies containing E(spl)bHLH proteins were isolated by centrifugation, washed in 10 volumes of 50 mM Tris (pH 8)–100 mM NaCl–10 mM NaEDTA–0.5% Triton X-100 and then dissolved in 8 M urea–0.1 M NaH2PO4–10 mM Tris (pH 8). After dialysis against MTPBS, the concentration of protein was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Coomassie blue staining. Soluble extracts of fusion proteins were prepared by taking the supernatant after sonication. The soluble pRSET fusion proteins were enriched by using Ni-nitrilotriacetic acid agarose beads (Qiagen). After absorption of 1 volume of 50% Ni-nitrilotriacetic acid agarose beads with 10 volumes of supernatant at 4°C for 60 min, the beads were washed three times in MTPBS containing 10% glycerol. Fusion protein was eluted by an equal volume of 500 mM imidazole in MTPBS–10% glycerol.
Random oligonucleotide binding site selection.
Selection of DNA sequences by E(spl)bHLH proteins was performed according to the protocol of Gogos et al. (23), using the following oligonucleotides: primer 1 (5′-AAGCGGCCGTGCGAGGATCC-3′), primer 2 (5′-TGTAAGCTTCCCGGGAATTC-3′), and degenerate (5′-CCGTGCGAGGATCC[N]16GAATTCCCGGGAAG-3′). The degenerate oligonucleotide was annealed with 10-fold excess of primer 2, and a complementary strand was synthesized by using Klenow fragment. After end labeling with [γ-32P]ATP, protein binding and electrophoretic mobility shift assay were performed as described below. The selected sequences were eluted from the region of dried gel containing protein-DNA complexes and precipitated as described elsewhere (23). Approximately one-fifth of the eluted sample was amplified for 23 cycles in a 100-μl PCR using primers 1 and 2. All experiments included a control PCR without template which did not yield a product. Approximately 1 μg of refolded fusion protein was used for binding in the first to third rounds of selection; 500 ng was used in subsequent rounds. Selected oligonucleotides were cloned into pBluescript (Stratagene) and sequenced with T3 or T7 primers.
Electrophoretic mobility shift assay.
Fifty picomoles of each complementary single-stranded oligonucleotide (Oswel) (Table 1) was annealed in a final volume of 50 μl of 50 mM Tris-HCl (pH 7.9)–10 mM MgCl2–1 mM spermidine–0.1 mM EDTA–1 mM dithiothreitol. Annealed oligonucleotides were end labeled with [γ-32P]ATP by using T4 polynucleotide kinase and separated from free nucleotides by precipitation with ethanol after addition of 5 μg of glycogen carrier and 0.5 M ammonium acetate. Protein-DNA complexes were formed by incubation of protein with 50 fmol of radiolabeled nucleotides in 10 μl of buffer (25 mM HEPES [pH 7.5], 100 mM KCl, 20% [vol/vol] glycerol, 0.1% [vol/vol] Nonidet P-40, 10 μM ZnZO4, 1 mM dithiothreitol). Poly(dI-dC) was included as a nonspecific competitor (0.5 U/ml). After incubation on ice for 30 min, DNA-protein complexes were resolved by electrophoresis on a 5% acrylamide gel. Phosphorimaging was performed with a STORM860 PhosphorImager and ImageQuant software (Molecular Dynamics).
TABLE 1.
Sequences of oligonucleotides used for DNA binding and construction of pGbe-lacZ derivatives
| Name | Sequencea |
|---|---|
| A1 | 5′-tcgagGGTGGCAGGTGCCATTg |
| A2 | 5′-tcgagAGATCTACGCAGGTGGTTCTTGTg |
| B1 | 5′-tcgagGGTGGCACGTGCCATTg |
| B2 | 5′-tcgagTTCTAGCACGTGTCACCAGg |
| B3 | 5′-tcgagGGGTCCACGTGAGCTTg |
| Hairy | 5′-tcgagAGCCGGCACGCGACAGGg |
| N box | 5′-GATCACGCCACGAGCCACAAGGATTG |
Only one strand is shown. The E-box elements are in bold, and E boxes along with three flanking bases are underlined. Additional bases used to create restriction enzyme sites at the termini are in lowercase letters. Sequences of the N-box oligonucleotides are the same as those used previously (65), and the Hairy oligonucleotide sequence is derived from the regulatory region of the achaete gene (49, 67).
Gal4/UAS misexpression experiments.
The Gal4/upstream activating sequence (UAS) misexpression system was first described in reference 7. DNA encoding the protein to be ectopically expressed (e.g., MβACT) is cloned downstream of UASs that are binding sites for the Saccharomyces cerevisiae Gal4 transcriptional activator protein. Stable transformed Drosophila lines are generated and crossed to transgenic flies that express Gal4 protein in a limited domain. In the progeny expression from the UAS construct is induced wherever Gal4 is present.
To create pUAS-mβACT, the coding region of E(spl)-mβ was amplified by PCR. The upstream primer included a BglII site to facilitate cloning. The PCR product was cloned into BglII and PstI sites of pBMTL22 polylinker to create pBMTL22-mβ. The activation domain (amino acids 415 to 490) of VP16 was amplified from pHK3NVP16 (gift from Tony Kouzarides; primer sequences for PCR available on request) and ligated into the BamHI and PstI sites of pBMTL22-mβ such that the coding region of mβ and the VP16 activation domain were fused in frame at the PstI site of mβ. The mβ-VP16 activation domain sequence was excised by using BamHI and BglII and ligated into the BglII site of a modified pUAST (7). pUAS-mβACT DNA constructs were introduced into y,w Drosophila by standard P-element-mediated germ line transformation (53).
Other UAS lines were UAS-mβ and UAS-mδ (12), UAS-L’sc (28), and UAS-LacZ (7). Gal4 lines used were ptc-Gal4 (61), sal-Gal4 (11, 64), 765-Gal4 (24), and 32B-Gal4 (7). UAS-L’sc was recombined with 765-Gal4 and UAS-mδ was recombined with 32B-Gal4 before crossing to flies containing the pGbe-lacZ derivatives.
Construction of B1, A1, and A2 reporter constructs.
Approximately 10 pmol of each of the B1, A1, and A2 double-stranded oligonucleotides was kinase treated and then allowed to ligate for 30 min. Trimers of each oligonucleotide were gel purified and cloned into the KpnI (filled-in) site of pGbe-lacZ (previously described as pGRHbe-2-lacZ [66]), a derivative of pHZ50PL (29). These constructs were sequenced to check the orientation of the inserts and injected into cn,ry embryos to generate ry+ transformants (53). Multiple lines were analyzed for each construct.
Histochemistry.
Wings were prepared by dissection in ethanol and then mounted in a 1:1 mix of ethanol and lactic acid. Expression of the lacZ reporter gene was detected by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining as described previously (66), and immunohistochemical staining with the Achaete monoclonal antibody (gift from S. Carroll) was performed as described previously (33, 59).
RESULTS
Optimal DNA target sites for E(spl)bHLH proteins.
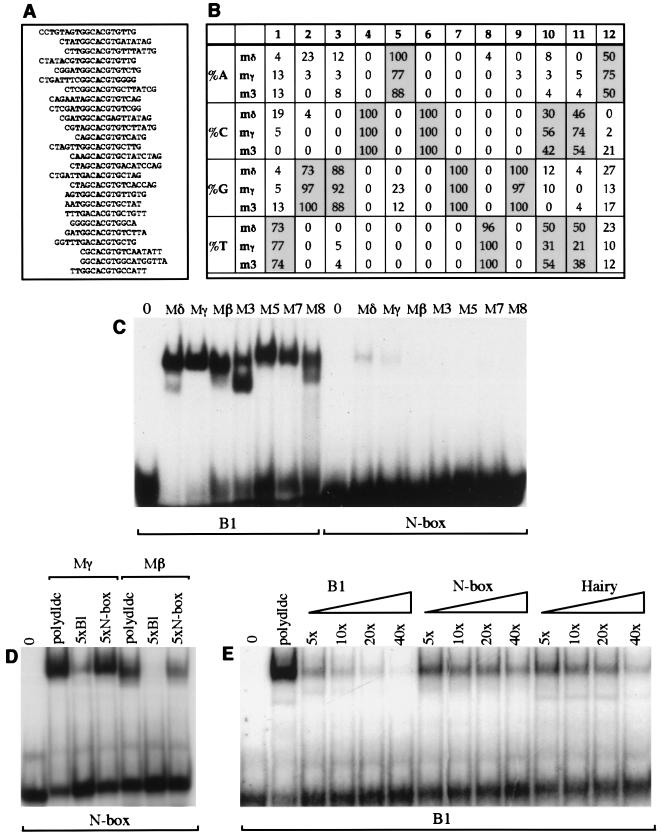
Previous analysis of E(spl)bHLH DNA binding activity used sequences from the promoter regions of the E(spl)bHLH-m8 and achaete genes (48, 49, 65, 67). However, it has not been established whether these are optimal binding sites for E(spl)bHLH proteins or if they are target sites for these proteins in vivo. To investigate whether different E(spl)bHLH proteins prefer the same DNA target sequence and how their binding sites relate to those of other bHLH proteins, we determined their optimal DNA target sequences through random oligonucleotide site selection (23). Using bacterially produced E(spl) M3, Mγ, and Mδ proteins, we carried out five cycles of selection followed by amplification of the interacting oligonucleotides (the protein concentrations were decreased for the fourth and fifth cycles to increase the specificity of selection). Between 20 and 40 oligonucleotides selected by M3, Mδ, and Mγ were sequenced, and a comparison between them established clear consensus binding sites (Fig. 1A and B). All three proteins selected very similar DNA sequences, consistent with the observation that their basic domains differ by only one amino acid residue. The three sets of sequences can be combined to give a palindromic 12-bp consensus sequence (5′-TGGCACGTG[C/T][C/T]A-3′) which we have called the ESE box [E(spl) E box]. The core of the ESE box, 5′-CACGTG-3′, is a canonical E box of the class B type, according to the classification system of Dang et al. (10).
FIG. 1.
Identification of optimal targets for E(spl)bHLH proteins. (A and B) Binding site selection was used to identify optimal E(spl)bHLH DNA targets. (A) Alignment of 26 sequences obtained after five rounds of selection with the Mδ protein. Selection of oligonucleotides with Mδ was performed in two separate experiments; soluble Mδ protein extract was used to select the first 17 oligonucleotides listed. The soluble and refolded Mδ extracts show no obvious differences in DNA binding preferences. (B) The nucleotides present at each position of fifth-round sequences selected by the Mδ (n = 26), Mγ (n = 39), and M3 (n = 24) proteins (n is the number of sequences analyzed). Nucleotides selected with a frequency of 50% or higher are highlighted. (C) Binding of all seven E(spl)bHLH proteins to the optimal E(spl) consensus (B1; contains a palindromic version of the ESE box) or the N-box oligonucleotide. Identical amounts of protein (∼50 ng) and labeled oligonucleotides were used for the equivalent reactions. (D) Addition of a fivefold molar excess of unlabeled B1 oligonucleotide has a greater effect on binding of the Mγ and Mβ proteins (approximately 500 ng of soluble pGex fusion protein) to the N-box probe than addition of a fivefold excess molar excess of unlabeled N-box oligonucleotide. (E) Binding of Mγ and Mβ proteins (approximately 50 ng of pGex fusion protein in a soluble bacterial extract) to labeled B1 probe in the presence of nonspecific competitor DNA [poly(dI-dC)] or 5-, 10-, 20-, or 40-fold molar excess of unlabeled B1, N-box, or Hairy oligonucleotide as indicated. Sequences of all oligonucleotides used are listed in Table 1. The lanes 0 in panels C to E contain labeled oligonucleotides in the absence of added protein.
Since the three proteins initially analyzed all yielded sequences containing the ESE box, we tested whether the remaining four E(spl)bHLH proteins were able to bind this sequence efficiently, using a double-stranded oligonucleotide that contains the 12-bp perfect palindrome (B1 [Table 1; Fig. 1C]). All seven proteins bind to the ESE box with much higher affinity than to the previously described E(spl) target site, the N box (5′-CACNAG-3′ [65]) (Fig. 1C). The variation in the amount of binding between the different proteins tested appears to be primarily due to the amount of active renatured protein in each sample, since it could not be reproduced in competition experiments.
The conclusion that E(spl)bHLH proteins bind the B1 site with greater affinity than the N box was confirmed in competition assays using Mγ and Mβ. The B1 oligonucleotide clearly competes more effectively than the N box (Fig. 1D) when present in fivefold excess in reactions containing labeled N-box oligonucleotide as the probe. Likewise, when B1 is used as the labeled probe, a 5-fold molar excess of unlabeled B1 is sufficient to compete 75% of the labeled oligonucleotide from Mγ-DNA complexes (or Mβ-DNA complexes [data not shown]), whereas a 40-fold molar excess of unlabeled N-box DNA is required to achieve a similar reduction. The same assay was used to test binding to a class C E-box site (5′-CACGCG-3′), the optimal binding site for the related protein Hairy (49, 67). Although the Hairy site competes with the B1 probe in a concentration-dependent manner, a 20-fold molar excess of unlabeled Hairy site DNA is needed to reduce the labeled B1-Mγ complexes by 75%. This effect is comparable to that seen with a fivefold excess of B1, confirming the preference for the B1 site indicated by the oligonucleotide site selections (Fig. 1B).
Our observation that the different E(spl)bHLH proteins recognize the same sites in vitro is mirrored by their activity in vivo. Genetic analysis of E(spl)-C suggested that there is redundancy in the functions of the individual proteins (15, 56). Furthermore, Mβ, M8, M7, and Mδ are all able to suppress both veins and sensory organ development when ectopically expressed in the imaginal discs (12, 47, 63), although some degree of specificity in function has been observed (12, 40). To further test whether different E(spl)bHLH proteins are capable of recognizing similar targets in vivo, we used an assay in which the proteins are converted to activators by replacing the WRPW tetrapeptide with the VP16 activation domain (35). This approach has been used to show that M7 regulates Achaete expression. We compared the activity of a converted Mβ protein since Mβ is expressed in a different pattern to M7 and is not associated with sensory organ development in wild-type imaginal discs (12). However, expression of M7ACT and MβACT resulted in similar adult phenotypes, including ectopic bristles on the wing and notum (Fig. 2A to C). Furthermore, MβACT, like M7ACT, induced ectopic Achaete expression (Fig. 2D to F), supporting the notion that the proteins can recognize the same targets in vivo, even though during imaginal development they are associated with different developmental processes.
FIG. 2.
E(spl)Mβ and E(spl)M7 recognize similar targets in vivo. Expression of both M7ACT (B) and MβACT (C) under the control of ptc-Gal4 give rise to ectopic bristles along the anteroposterior boundary of the wing [cf. wild type (A)], corresponding to an induction of Achaete expression in wing imaginal discs (indicated by arrowheads) found in M7ACT (E) and MβACT (F) (cf. wild type [D]). The pattern of Gal4 driving M7ACT and MβACT expression is visualized in Fig. 7A.
Bases flanking the E-box core influence DNA binding by E(spl)bHLH and proneural proteins.
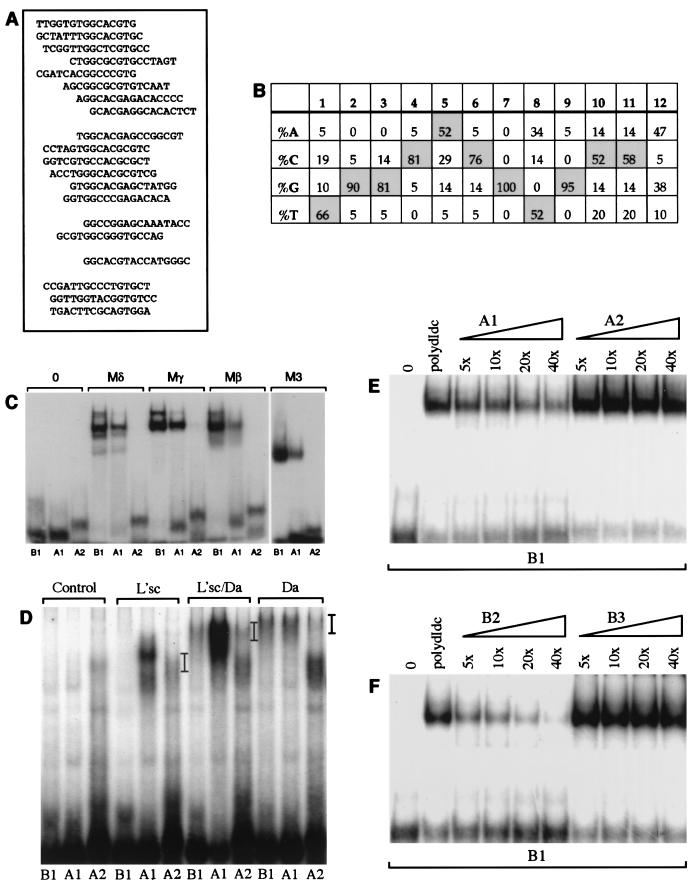
The oligonucleotides present after five rounds of binding site selection represent those bound with the highest affinity. To gain further insight into the parameters influencing binding, we also analyzed oligonucleotides subjected to three rounds of selection by the Mδ protein. This yielded a wider spectrum of sequences, but a strong preference for particular nucleotides in the core E box and in the flanking bases was still observed (Fig. 3A and B). Within the ESE box, the bases at positions 4, 6, 7, and 9 were the most stringently selected, giving a consensus core of CNCGNG (Fig. 1E).
FIG. 3.
Sequences flanking the E-box core influence binding of E(spl)bHLH and proneural proteins. (A and B) Sequences selected after three rounds of binding with the Mδ protein are listed (A) and summarized to show the frequency of nucleotides at each position (B). The top 15 oligonucleotides listed contain a CNCGNG core, followed by 2 with suboptimal bases at position 6 in the E box and 1 with a suboptimal base at position 9. The final three oligonucleotides contain GTG, half of a class B E-box, but may not bind the E(spl)bHLH proteins with high affinity. (B) After three rounds of selection, there is a strong preference for certain nucleotides in the positions flanking the E box (a similar preference is evident if the analysis is restricted to the 15 oligonucleotides containing a CNCGNG core [data not shown]). (C to F) Effect of flanking sequence on DNA binding in vitro. (C and D) Binding of E(spl)bHLH proteins to B1, A1, and A2 (Table 1). The B1 and A1 sites contain optimal flanking bases and differ by a single-base substitution that switches the E box from a class B site (B1) to a class A site (A1) (10). The A2 site has the class A E-box core but suboptimal flanking sequences. (C) Binding of Mδ, Mβ, Mγ, and M3 proteins (75 ng) can be detected with the B1 and A1 sites but not with A2. (D) Binding of proneural proteins to the different E-box oligonucleotides (as indicated) was tested by using L’sc protein extract only, a 1:1 mixture of L’sc and Da extracts, and Da extracts only. Position of the DNA-protein complexes are indicated by bars. Negative control reactions containing soluble bacterial extract (Control) were included for comparison. (E) Effects of 5-, 10-, 20-, or 40-fold molar excess of either unlabeled A1 or A2 oligonucleotide on binding of Mγ (50 ng) to the B1 probe. Addition of unlabeled A1 diminishes binding to B1 in a concentration-dependent manner; 40-fold molar excess of unlabeled A2 did not compete with the B1 probe (confirmed by phosphorimaging analysis). (F) Mγ binding to the B1 probe in the presence of 5-, 10-, 20-, or 40-fold molar excess of either unlabeled B2 and B3 oligonucleotides. B2 differs from B1 by two suboptimal nucleotides in the flanking sequences, and B3 has the least optimal bases at all positions flanking the E-box core (using the information from Fig. 1B). B2 competes with B1 in a concentration-dependent manner, although not as efficiently as B1 (Fig. 1B), while 40-fold molar excess of unlabeled B3 did not compete with the B1 probe (confirmed by phosphorimaging). The amounts of protein and probe used for panels E and F were identical to the amounts used for Fig. 1E. The sequences of the oligonucleotides used in panels C to F are listed in Table 1. Lanes 0 contain labeled oligonucleotides in the absence of added protein.
The strong selection for the bases flanking the E box indicates their importance in target recognition by E(spl)bHLH proteins. This observation prompted us to test whether the presence of optimal nucleotides at the flanking positions can compensate for a suboptimal E-box core. For instance, can the E(spl)bHLH proteins bind a class A E-box site, which is the target of AS-C proteins, if the flanking sequences are optimal? Two double-stranded oligonucleotides containing the class A E-box core (5′-CAGGTG-3′), one with E(spl)bHLH consensus flanking sequences (A1) and the other with nonconsensus flanking sequences (A2), were designed. No interaction between any of the E(spl)bHLH proteins and the A2 oligonucleotide was detected (Fig. 3C and E), in agreement with previous reports (68). However, the A1 site was bound by all seven E(spl)bHLH proteins (Fig. 3C and data not shown), demonstrating that optimal flanking sequences can facilitate binding of these proteins to a suboptimal core. The A1 site was bound less efficiently than B1, confirming that these proteins prefer a class B over a class A E-box core, in line with the results of the site selection.
The identity of the flanking bases also influences binding by E(spl)bHLH proteins in the context of an optimal E-box core. Substitution of two suboptimal bases in the flanking sequences of B1 to create B2 (Table 1) decreases binding by the E(spl)bHLH proteins (compare B1, 5x in Fig. 1E with B2, 5x in Fig. 3F). In addition, we designed an oligonucleotide that contains the optimal CACGTG core flanked by the bases which were selected at the lowest frequency in the site selection experiment (B3 [Table 1]). Even in 40-fold molar excess, this site could not compete with the B1 site for binding to Mγ (Fig. 3F) or Mβ (data not shown). These results clearly demonstrate that the bases flanking the E-box are intrinsic to DNA recognition by E(spl)bHLH proteins.
Similar experiments with proneural proteins demonstrate that their affinity for target sites is also influenced by bases flanking the E-box core (Fig. 3D). Thus, heterodimers of Da and L’sc bind better to the A1 oligonucleotide containing the flanking bases favored by E(spl) proteins than to the A2 oligonucleotide which has the same core. In addition, binding of L’sc-Da heterodimers to the ESE-box oligonucleotide B1 can be detected. Thus, both the E(spl)bHLH and L’sc-Da proteins can interact with either class A or class B E boxes in the context of the optimal flanking sequences.
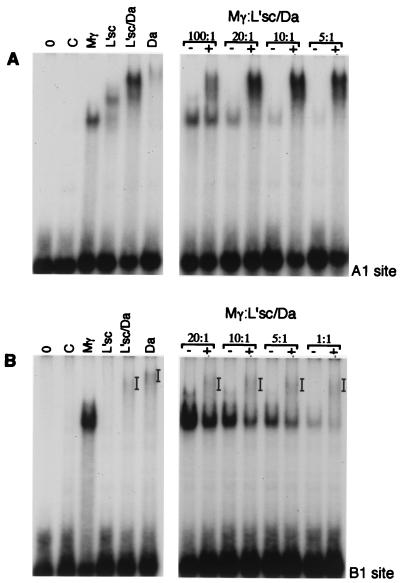
To mimic what occurs in cells where both proneural proteins and E(spl)bHLHs are expressed [e.g., the cells of a proneural cluster which are inhibited from adopting the neural fate by accumulation of E(spl)bHLH proteins and concurrent extinction of proneural protein expression (34)] and to compare more directly the affinities of the two types of proteins for these sites, we mixed the proteins in different ratios and analyzed binding to the A1 and B1 sites (Fig. 4). L’sc and Da (in a 1:1 ratio) were kept constant while the concentration of Mγ was varied, and the mixtures were incubated together for 30 min before the addition of labeled oligonucleotides. Interestingly, we did not detect any obvious formation of heterodimers between Mγ and the proneural proteins and were able to detect complexes containing Mγ and those containing L’sc-Da in the same reaction. Both E(spl)bHLHs and L’sc-Da formed complexes with the A1 and the B1 sites, although with different efficiencies. The ability of these two classes of proteins to bind to the same target sequence raises the possibility that one way that the E(spl)bHLH proteins could antagonize proneural gene activity is by competition for binding to regulatory sequences of downstream genes in vivo.
FIG. 4.
Some sites may be targets for both E(spl) and proneural proteins. Different dilutions of Mγ were mixed with the A1 oligonucleotide (A) or the B1 oligonucleotide (B) in the presence of control or L’sc-Da extracts (20 ng of L’sc and Da combined). The approximate ratios of Mγ protein to L’sc-Da are indicated (assuming 100% of the pRSET-Mγ protein had refolded correctly during preparation). Bars indicate the positions of protein-DNA complexes.
ESE box confers repression in vivo.
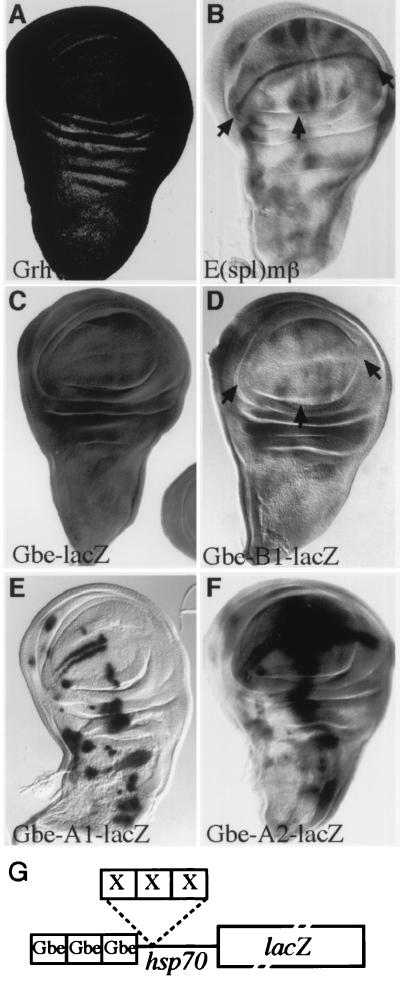
There is substantial evidence to suggest that the E(spl) proteins act to repress transcription. To investigate whether the binding sites that we identified can function as targets for E(spl) proteins in vivo, we designed a reporter gene which would allow us to test for repression. The basic reporter gene (Gbe-lacZ) contained a minimal heat shock promoter upstream of lacZ and three binding sites for the transcriptional activator Grainyhead (66). Grainyhead is expressed ubiquitously in the wing imaginal disc (66) (Fig. 5A), and reflecting this, our basic construct is expressed at high levels throughout the disc, with little modulation (Fig. 5C). Three copies of the optimal ESE-box sequence, the B1 oligonucleotide, were inserted adjacent to the Grainyhead binding sites (Gbe-B1-lacZ [Fig. 5G]). If these sites do represent targets for the E(spl) proteins in vivo, we would expect to see extensive repression of lacZ expression in many regions within the wing disc, particularly at the dorsal-ventral boundary and around proneural clusters where the endogenous E(spl) proteins are expressed (e.g., Fig. 5B [3, 12, 33, 58]). This is indeed what we detect (Fig. 5C and D); the highest levels of repression are detected at the dorsal-ventral boundary, flanking the vein primordia and around presumptive proneural clusters, and the overall extent of repression is >10-fold with respect to expression from Gbe-lacZ (determined by enzymatic assay [data not shown]).
FIG. 5.
Specific regulation conferred by B1, A1, and A2 sites in vivo. Three copies of each of the A1, B1, and A2 sites in tandem were placed adjacent to the Grainyhead binding sites in Gbe-lacZ, and the effects on in vivo expression of lacZ were evaluated. (A and C) Grainyhead (Grh) is expressed ubiquitously in the wing disc (A; detected with a monoclonal antibody) and drives uniform expression of Gbe-lacZ (C). (B and D) Insertion of B1 sites, Gbe-B1-lacZ, leads to lower levels of lacZ expression (D, arrows), with specific repression in regions where E(spl) genes [e.g., E(spl)mβ] are expressed (B). (E) Insertion of A1 sites, Gbe-A1-lacZ, results in a pattern of lacZ expression that resembles proneural clusters in the imaginal wing disc (Fig. 2D), demonstrating that a single base pair change (as in B1) has dramatic effects on the binding of endogenous proteins in vivo. (F) Expression of Gbe-A2-lacZ (E) is strikingly different from that of Gbe-A1-lacZ even though they contain identical E-box cores, illustrating the influence that sequences flanking an E box can have on protein recognition in vivo. In panels C to F, expression was detected by using X-Gal; reactions were terminated after 1 h (C) or ∼16 h (D to F). (G) Diagram illustrating the structure of the transgenes. X represents sites of insertion of B1, A1, and A2 oligonucleotides (not to scale).
To compare the effects of other target sites in the same context, we generated similar reporter gene constructs containing three copies of A1 and A2 oligonucleotides in place of B1. As the A oligonucleotides contain the activator core binding sequences, we expected to detect a composite pattern of activation from the Grainyhead site and the class A sites. This is what was observed with the A2 oligonucleotide (Gbe-A2-lacZ) which confers an additional pattern of activation superimposed on the ubiquitous Grainyhead-driven expression of LacZ (Fig. 5F). Some of the activation conforms to sites where the proneural proteins are expressed. However, in addition there are high levels of activation at other locations (e.g., adjacent to the anterior-posterior boundary) which are not sites of proneural protein expression. The A1 construct Gbe-A1-lacZ, which differs from B1 only by a single base pair within the E box and from A2 by the sequences immediately flanking the E-box gives a strikingly different pattern to either of these constructs. The resulting pattern is most consistent with a combination of activation and repression of the reporter construct (Fig. 5E), since the Grainyhead-dependent activation is no longer detectable and the construct is expressed at high levels in sites that correspond to proneural clusters. The latter finding indicates that this construct is strongly activated by proneural proteins; however, the general repression of lacZ expression elsewhere suggests that the A1 site blocks activation by Grainyhead in the majority of cells either sterically or through the binding of other proteins.
The dramatically different patterns obtained with the constructs illustrates the specificity conferred by subtle changes in and around an E box in vivo. To compare the interactions of the B1, A1, and A2 sequences with the E(spl)bHLH and proneural proteins in vivo with those observed in vitro, we tested whether increasing the dose of E(spl) or proneural proteins, using the UAS Gal4-targeted misexpression system (7), resulted in altered levels of expression. When sal-Gal4 was used to drive ectopic expression of E(spl)Mβ in the wing imaginal disc, Gbe-B1-lacZ was clearly repressed at the sites where Gal4, and thus Mβ, was being expressed at highest levels (Fig. 6A and B), demonstrating that E(spl)bHLH proteins can bind the B1 site and repress transcription in vivo. Similarly, ectopic expression of Mδ driven in a less spatially restricted pattern by 32B-Gal4 caused more widespread repression of Gbe-B1-lacZ (Fig. 6C and D). The same combination, E(spl)Mδ with 32B-Gal4, caused a modest but consistent reduction in Gbe-A1-lacZ expression (Fig. 6E). This could be due to direct binding of Mδ to the A1 site or to an indirect effect caused by Mδ repressing endogenous proneural proteins and thus reducing the activation of Gbe-A1-lacZ. No change in expression of Gbe-A2-lacZ was detected in the presence of the ectopic E(spl)bHLH proteins (Fig. 6F). Conversely, Gal4-driven expression of proneural proteins led to activation of lacZ expression in flies containing Gbe-A1-lacZ and Gbe-A2-lacZ, but not Gbe-B1-lacZ (Fig. 6H to J).
FIG. 6.
Differential responses of target sites to E(spl) and proneural proteins in vivo. The effects of misexpressing E(spl) (B, D, E, and F) and L’sc (H to J) proteins on expression of Gbe-B1-lacZ (B, D, H), Gbe-A1-lacZ (E, I), and Gbe-A2-lacZ (F, J). The patterns of Gal4 expression [and hence ectopic E(spl) and L’sc expression] are visualized by using UAS-LacZ (A, C, and D). Repression of Gbe-B1-lacZ by E(spl)Mβ (B) and E(spl)Mδ (D). LacZ expression from Gbe-A1-lacZ is also repressed by ectopic Mδ (E); however Gbe-A2-lacZ is insensitive to ectopic Mδ (F). Widespread expression of L’sc by using 765-Gal4 has no effect on Gbe-B1-lacZ expression but results in strong activation of Gbe-A1-lacZ and Gbe-A2-lacZ. Expression was detected by X-Gal staining for ∼16 h.
To clarify further the interactions of the E(spl)bHLH proteins with these DNA sequences, we analyzed the effects of expressing M7ACT and MβACT (Fig. 7 and data not shown). Expression of either of these proteins resulted in a dramatic activation of Gbe-B1-lacZ, consistent with this being a direct target of these proteins, a weaker but reproducible activation of Gbe-A1-lacZ was also observed (Fig. 7B and C). The activation of lacZ expression in Gbe-A1-lacZ by M7ACT is comparable to that by L’sc, suggesting that the A1 site is a target for both E(spl) and proneural proteins in vivo. Thus, the behavior of the sites in vivo correlates with their activity in vitro, with B1 being responsive to E(spl), A2 being responsive to proneural proteins and/or other activators, and A1 being responsive to both proneural proteins and E(spl) or other repressors.
FIG. 7.
M7ACT activates reporter gene expression from B1 and A1 sites in vivo. (A) Expression of UAS-LacZ in response to ptc-Gal4 illustrates the domain of UAS-M7ACT expression in panels B and C. (B) M7ACT causes dramatic activation of Gbe-B1-lacZ (X-Gal staining for 6 h) and weaker activation of Gbe-A1-lacZ (X-Gal staining for ∼16 h).
DISCUSSION
Using random oligonucleotide selection, we have established that the consensus binding site for the E(spl)bHLH proteins contains a class B canonical E-box (CACGTG). This is compatible with the presence of the key arginine residue in the basic region that is characteristic of all other bHLH proteins that recognize class B sites (10) and which contacts the central G (18). The selected site differs from the previously identified N box (CANNAG), indicating that the latter may not be generally representative of E(spl) target sites, and we find that in vitro the N box is a much lower affinity target that the class B site. A second key feature which emerged from our analysis is the importance of the three nucleotides immediately flanking the E-box core. The binding site selection identified a 12-bp palindrome (TGGCACGTG[C/T][C/T]A) as the optimal site, which we have called the ESE box. Flanking bases have been implicated in DNA binding by other bHLH proteins including c-Myc and Hairy, based on in vitro assays (6, 21, 26, 67) and X-ray crystallography studies which reveal interactions between bHLH proteins and bases outside the E-box core (17, 18, 57). However, the flanking bases preferred by c-Myc and Hairy differ from those selected by E(spl)bHLH proteins (6, 26, 67), indicating that in vivo, the sequences immediately surrounding an E box are important for determining exactly which bHLH proteins bind there to regulate transcription.
The interactions with flanking bases helps to explain the specificity in vivo of different bHLH proteins, an important factor given the large number of bHLH proteins identified to date. The in vivo expression patterns produced by E boxes with different flanking bases in our experiments emphasizes their significance. For example, a comparison between the A2 and A1 sites demonstrates that the former is a target for many more transcriptional activators. These experiments also illustrate the relevance of different E-box core sequences, since a single-base difference within the E-box core (A1 to B1) is sufficient to prevent binding of proneural proteins and other activators. This is in agreement with earlier studies (49) which argued that proneural proteins and E(spl)bHLH repressors recognize sites with distinct types of E-box core. However, our results show that E(spl)bHLH repressors prefer the class B core, which is recognized by many different bHLH activators and repressors, over the class C core, which was designated the target for repressor bHLH proteins that contain a proline residue in the basic domain (19, 49). The class C site (CACGCG) is the optimal binding site for the Drosophila Hairy protein (49, 67), whose basic domain contains a proline residue but differs from E(spl)bHLH proteins in 7 of the 11 remaining residues, which could account for the different profile of DNA binding specificities. The distinctions in the DNA binding specificities could be significant for studies of the vertebrate homologues of the E(spl)bHLHs and Hairy. Overall, the in vitro binding experiments and the activity of different sites in vivo demonstrate that the bHLH proteins that we tested can recognize a specific range of target sequences and that both core and flanking bases are important for determining the binding specificity.
Similarities in DNA binding of individual E(spl)bHLH proteins.
Although flanking bases may distinguish sites for different types of E-box binding proteins, there are no significant differences in the bases recognized by individual E(spl) proteins; the same consensus binding site was derived for each of three proteins tested. There were subtle differences in the ranges of oligonucleotides, with Mδ selecting a broader range of variants at the flanking sites than Mγ and M3 and the latter two proteins exhibiting more tolerance for variants in the core E box, but experiments comparing the affinity of the proteins for these variant sites revealed no detectable bias.
The binding specificities observed are all for homodimers of individual E(spl) proteins. In places where more than one E(spl)bHLH protein is expressed (e.g., proneural clusters), it is possible that the proteins form heterodimers among themselves to bind DNA and repress transcription. However, given that the amino acid sequences of the DNA binding domains and the DNA binding preferences of the individual E(spl)bHLH proteins are so similar, it seems unlikely that heterodimers between E(spl)bHLH proteins would differ greatly from homodimers in their DNA binding sequence preferences. In addition, during several developmental processes, a single E(spl)bHLH protein predominates (e.g., Mβ in the presumptive intervein region of the wing [12]), indicating that they are likely to function as homodimers. There is also no evidence to suggest that the E(spl)bHLH proteins are required to form heterodimers with other bHLH family members to bind DNA and repress gene transcription in response to Notch signalling. Thus, the homodimers analyzed in our experiments likely represent complexes that are functional in vivo.
The overall similarity in the binding of different E(spl) proteins in vitro suggests that they are capable of recognizing the same targets in vivo and is consistent with the phenotypes observed when the individual proteins are expressed ectopically. Ectopic expression of M8, M5, Mβ, Mδ, and M7 all produce phenotypes of vein and bristle loss (12, 47, 63). Here we demonstrate further that both Mβ and M7 are able to interact with DNA sequences regulating achaete, by assaying the effects of converting both proteins from repressors to activators. The ability to recognize the same DNA target sequences could explain the apparent redundancy between the E(spl) genes (15, 56), as they would all have the potential to act in the same processes. The observation that specific E(spl)bHLH proteins are more or less efficient in regulating different processes, e.g., Mβ more effective at suppressing veins and M8 more effective at suppressing bristles (12), is thus more likely to be consequence of differences in protein:protein interactions than of differences in target recognition.
Relevance of E(spl)bHLH DNA binding to developmental function.
In the absence of E(spl)bHLH proteins, proneural protein expression persists at high levels in all cells of a proneural cluster (41). Thus, one action of E(spl)bHLH proteins is to antagonize the proneural proteins, with the ultimate consequence that proneural gene expression is repressed. It has been proposed that E(spl)bHLH proteins exert their influence by binding to regulatory regions within the AS-C and repressing transcription of the proneural genes (27, 41, 47, 49, 67). This hypothesis is supported by the observations that expression of Achaete is induced by M7ACT and MβACT (35) (Fig. 2) and that induction of ectopic bristles in the Drosophila wing and notum by M7ACT is abolished in the absence of proneural proteins (35). One putative binding site for the E(spl)bHLH proteins upstream of the achaete gene and has the sequence 5′-CGGCACGCGACA-3′ (Hairy site [Table 1]). Mγ will bind this site in vitro (Fig. 1E), and M7 can bind this sequence and repress transcription in a cotransfection assay in Drosophila S2 cultured cells (67). However, mutation of this site in vivo results in a phenotype resembling that caused by mutations in hairy rather than in the E(spl)-C (67). This fits with the observation that this sequence conforms to an optimal Hairy DNA binding site but is a suboptimal site for the E(spl) proteins (Fig. 1E) and indicates that the E(spl) proteins do not recognize this sequence in vivo. Thus, if E(spl) proteins are directly repressing achaete expression, there should be more optimal target sites elsewhere within the AS-C. Indeed, a search of recently available AS-C genomic sequence (14) identifies >10 sequences with good matches to ESE boxes (Table 2), in addition to the sites that have been identified by in vitro binding assays.
TABLE 2.
ESE-box sequences found within the AS-C
| ESE-box sequence | Matrix similarity | Locationb | Position relative to AS-C genes |
|---|---|---|---|
| TGtCtCGTGCa | 0.803 | 11996 (−) | |
| aGGCtCGTGCat | 0.826 | 14201 (−) | |
| aGGCgCGTGTgt | 0.837 | 25034 (+) | 3,219 bp 5′ ac |
| TGtCgCGTGCCg | 0.831 | 27884 (−) | 369 bp 5′ ac |
| TtGCACGTGgCc | 0.871 | 33390 (±) | 4,532 bp 3′ ac |
| TGaCACGTGCCA | 0.921 | 15801 (±) | 1,302 bp 3′ sc |
| aaGCACGTGTCA | 0.877 | 26374 (±) | 708 bp 5′ l’sc |
| aGcCACGTGgat | 0.807 | 28645 (−) | 790 bp 3′ l’sc |
| gGaCACGTGCgt | 0.826 | 28869 (±) | 1,014 bp 3′ l’sc |
| cTGCACGTGTCc | 0.845 | 30297 (±) | 2,422 bp 3′ l’sc |
Sequences within the cosmids 125H10 (contains achaete [ac]; EMBL accession no. AL023873) and 198A6 (contains scute [sc and l’sc; EMBL accession no. AL024453) (16) matching the ESE-box consensus with a matrix similarity of greater than 0.8, using the MatInd and MatInspector programs (52).
−, antisense; +, sense; ±, both sense and antisense.
An alternative hypothesis is that the primary function of the E(spl)bHLH proteins is to antagonize the actions of proneural proteins posttranscriptionally. Evidence in support of this comes from experiments in which L’sc is ectopically expressed using a heterologous promoter that is not subject to direct regulation by E(spl)bHLH proteins (28). Under these conditions L’sc expression results in isolated ectopic bristles, rather than clusters of bristles, demonstrating that lateral inhibition is still able to restrict neural fate to a single cell even though l’sc transcription is insensitive to Notch signalling. This implies that E(spl)bHLH proteins are to antagonize proneural genes in ways other than by repressing their transcription. One possibility is that the E(spl) proteins can interact with the same targets as proneural proteins, but repress rather than activate their transcription. The ability of E(spl) proteins to bind to the B1 and A1 sequences and repress transcription from a heterologous promoter is consistent with this model, as is the observation that M7ACT can induce certain ectopic leg bristles in the absence of the achaete and scute genes (35). In the latter context, M7ACT is likely to be acting on genes with functions downstream of the proneural proteins to cause neural differentiation. In addition, the E(spl)bHLH proteins are involved with developmental processes that do not involve the proneural proteins, e.g. wing vein development; thus, they cannot act solely to repress proneural gene transcription during development.
How might E(spl)bHLH repress transcription of target genes? The closely related protein Hairy has been shown to repress transcription in a dominant manner even when its binding sites are located at some distance from the promoter (4), leading to the hypothesis that Hairy is able to mediate stable, inheritable repression of the target genes. We anticipate that E(spl)bHLH repression will be transitory, so that if Notch signalling were terminated, the E(spl) proteins would decay and the target genes would be susceptible to reactivation. Although proneural and E(spl)bHLH proteins optimally prefer different core E-box binding sites, so that independent binding to target genes appears likely, the importance of the bases flanking the E box in target recognition means that there is potential for overlap in the binding sites of the two groups of proteins. Thus, in cells where expression of E(spl)bHLH proteins is induced by Notch signalling, the proteins accumulate to high levels and could compete for binding to proneural protein target sites of the A1 type described here. Among the E-box sequences recognized by proneural proteins in vitro that have been described, at least a subset have good matches with the ESE consensus and thus could be recognized by both classes of proteins (8, 31, 43, 58, 68). Now that we have identified the sequence preferences of the E(spl)bHLH proteins, when target genes of proneural and E(spl)bHLH proteins have been identified and their regulatory regions analyzed, it will be possible to determine whether the sites present offer the potential for competition (e.g., by resembling our A1 sites) or whether they have the features of completely distinct binding sites for E(spl)bHLH, Hairy, proneural, and other bHLH proteins.
ACKNOWLEDGMENTS
We thank the members of our laboratory for discussions and encouragement and Simon Aspland, Lesley Clayton, Mike Taylor, and Rob White for thoughtful comments on the manuscript. We also benefited from constructive suggestions made by reviewers. We are grateful to Joseph Gogos for advice concerning the site selection procedure; Andrea Brand, Jose de Celis, David Ish-Horowicz, Andreas Prokop for the gift of flies; Sean Carroll for the anti-Achaete antibody; Tony Kouzarides for the VP16 DNA; Emma Harrison for help with DNA sequencing; and John Bashford, Ian Bolton, and Adrian Newman for help preparing the figures.
This work was funded by project grants from the Medical Research Council and the Wellcome Trust. D.M.T. was the recipient of a studentship from the Medical Research Council.
REFERENCES
- 1.Alifragis P, Poortinga G, Parkhurst S M, Delidakis C. A network of interacting transcriptional regulators involved in Drosophila neural fate specification revealed by the yeast two-hybrid system. Proc Natl Acad Sci USA. 1997;94:13099–13104. doi: 10.1073/pnas.94.24.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signalling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 3.Bailey A M, Posakony J W. Suppressor of Hairless directly activates transcription of the Enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 4.Barolo S, Levine M. Hairy mediates dominant repression in the Drosophila embryo. EMBO J. 1997;16:2883–2891. doi: 10.1093/emboj/16.10.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwell T K, Huang J, Ma A, Kretzner L, Alt F W, Eisenman R N, Weintraub H. Binding of Myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA binding by the c-Myc protein. Science. 1990;25:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 7.Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 8.Cabrera C V, Alonso M C. Transcriptional activation by heterodimers of the achaete-scute and daughterless gene products of Drosophila. EMBO J. 1991;10:2965–2973. doi: 10.1002/j.1460-2075.1991.tb07847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cubas P, de Celis J F, Campuzano S, Modolell J. Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal wing discs. Genes Dev. 1991;5:996–1008. doi: 10.1101/gad.5.6.996. [DOI] [PubMed] [Google Scholar]
- 10.Dang C V, Dolde C, Gillison M L, Kato G J. Discrimination between related DNA sites by a single amino acid residue of Myc-related basic-helix-loop-helix proteins. Proc Natl Acad Sci USA. 1992;89:599–602. doi: 10.1073/pnas.89.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Celis J F, Bray S, Garcia-Bellido A. Notch signalling regulates veinlet expression and establishes boundaries between veins and interveins in the Drosophila wing. Development. 1997;124:1919–1928. doi: 10.1242/dev.124.10.1919. [DOI] [PubMed] [Google Scholar]
- 12.de Celis J F, de Celis J, Ligoxygakis P, Preiss A, Delidakis C, Bray S. Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: the E(spl) genes mediate only a subset of Notch activities during imaginal development. Development. 1996;122:2719–2728. doi: 10.1242/dev.122.9.2719. [DOI] [PubMed] [Google Scholar]
- 13.de la Pompa J L, Wakeham A, Correia K M, Samper E, Brown S, Aguilera R J, Nakano T, Honjo T, mak T W, Rossant J, Conlon R A. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 14.Delidakis C, Artavanis-Tsakonas S. The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc Natl Acad Sci USA. 1992;89:8731–8735. doi: 10.1073/pnas.89.18.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delidakis C, Preiss A, Hartley D A, Artavanis-Tsakonas S. Two genetically and molecularly distinct functions involved in early neurogenesis reside within the Enhancer of split locus of Drosophila melanogaster. Genetics. 1991;129:803–823. doi: 10.1093/genetics/129.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Pablos, B., E. Madueno, and J. Modolell. 10 June 1999, posting date. Sequencing the distal X chromosome of Drosophila melanogaster. [On line.] http://www.ebi.ac.uk. [3 May 1999, last date accessed.]
- 17.Ellenberger T, Fass D, Arnaud M, Harrison S C. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8:970–980. doi: 10.1101/gad.8.8.970. [DOI] [PubMed] [Google Scholar]
- 18.Ferre-D’Amare A R, Pendergast G C, Ziff E B, Burley S K. Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature. 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 19.Fisher A, Caudy M. The function of hairy-related bHLH repressor proteins in cell fate decisions. Bioessays. 1998;20:298–306. doi: 10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 20.Fisher A L, Ohshako S, Caudy M. The WRPW motif of the Hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher F, Crouch D H, Jayaraman P, Clark W, Gillespie D A F, Goding C R. Transcription activation by Myc and Max: flanking sequences target activation to a subset of CACGTG motifs in vivo. EMBO J. 1993;12:5075–5082. doi: 10.1002/j.1460-2075.1993.tb06201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giebel B, Campos-Ortega J A. Functional dissection of the Drosophila Enhancer of split protein, a suppressor of neurogenesis. Proc Natl Acad Sci USA. 1997;94:6250–6254. doi: 10.1073/pnas.94.12.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gogos J A, Hsu T, Bolton J, Kafatos F C. Sequence discrimination by alternatively spliced isoforms of a DNA binding zinc finger domain. Science. 1992;257:1951–1955. doi: 10.1126/science.1290524. [DOI] [PubMed] [Google Scholar]
- 24.Gomez-Skarmeta J L, del Corrall R D, de la Calle-Mustienes E, Ferres-Marco D, Modolell J. araucan and caupolican, two members of the novel iroquois complex, encode homeoproteins that control proneural and vein-forming genes. Cell. 1996;85:95–105. doi: 10.1016/s0092-8674(00)81085-5. [DOI] [PubMed] [Google Scholar]
- 25.Greenwald I. LIN-12/Notch signalling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 26.Halazonetis T D, Kandil A N. Determination of the c-MYC DNA-binding site. Proc Natl Acad Sci USA. 1991;88:6162–6166. doi: 10.1073/pnas.88.14.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- 28.Hinz U, Giebel B, Campos-Ortega J A. The basic helix-loop-helix domain of Drosophila Lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell. 1994;76:77–87. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- 29.Hiromi Y, Gehring W J. Regulation and function of the Drosophila segmentation gene fushi turazu. Cell. 1987;50:963–974. doi: 10.1016/0092-8674(87)90523-x. [DOI] [PubMed] [Google Scholar]
- 30.Jan Y N, Jan L Y. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-u. [DOI] [PubMed] [Google Scholar]
- 31.Jarman A P, Brand M, Jan L Y, Jan Y N. The regulation and function of the helix-loop-helix gene, asense, in Drosophila neural precursors. Development. 1993;119:19–29. doi: 10.1242/dev.119.Supplement.19. [DOI] [PubMed] [Google Scholar]
- 32.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 33.Jennings B, de Celis J, Delidakis C, Preiss A, Bray S. Role of Notch and achaete-scute complex in the expression of Enhancer of split bHLH proteins. Development. 1995;121:3745–3752. [Google Scholar]
- 34.Jennings B, Preiss A, Delidakis C, Bray S. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development. 1994;120:3537–3548. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- 35.Jimenez G, Ish-Horowicz D. A chimeric Enhancer-of-split transcriptional activator drives neural development and achaete-scute expression. Mol Cell Biol. 1997;17:4355–4362. doi: 10.1128/mcb.17.8.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klambt C, Knust E, Tietze K, Campos-Ortega J A. Closely related transcripts encoded by the neurogenic gene complex Enhancer of split of Drosophila melanogaster. EMBO J. 1989;8:203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knust E, Schrons H, Grawe F, Campos-Ortega J A. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics. 1992;132:505–518. doi: 10.1093/genetics/132.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lecourtois M, Schweisguth F. The neurogenic Suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the Enhancer of split Complex genes by Notch signalling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 39.Lee J E. Basic helix-loop-helix genes in neural development. Curr Opin Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- 40.Ligoxygakis P, Yu S Y, Delidakis C, Baker N E. A subset of Notch functions during Drosophila eye development require Su(H) and the E(spl) gene Complex. Development. 1998;125:2893–2900. doi: 10.1242/dev.125.15.2893. [DOI] [PubMed] [Google Scholar]
- 41.Martin-Bermudo M D, Carmena A, Jimenez F. Neurogenic genes control gene expression at the transcriptional level in early neurogenesis and in mesectoderm specification. Development. 1995;121:219–224. doi: 10.1242/dev.121.1.219. [DOI] [PubMed] [Google Scholar]
- 42.Martin-Bermudo M D, Martinez C, Rodriguez I, Jimenez F. Distribution and function of the lethal of scute gene product during early neurogenesis in Drosophila. Development. 1991;113:445–454. doi: 10.1242/dev.113.2.445. [DOI] [PubMed] [Google Scholar]
- 43.Martinez C, Modollel J, Garrell J. Regulation of the proneural gene achaete by helix-loop-helix proteins. Mol Cell Biol. 1993;13:3514–3521. doi: 10.1128/mcb.13.6.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muller M V, Weizsacker E, Campos-Ortega J A. Expression domains of a zebrafish homologue of the Drosophila pair-rule gene hairy correspond to primordia of alternating somites. Development. 1996;122:2071–2078. doi: 10.1242/dev.122.7.2071. [DOI] [PubMed] [Google Scholar]
- 45.Murre C, Schonleber McCaw P, Baltimore D. A new DNA binding and dimerization motif in Immunoglobulin Enhancer Binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 46.Murre C, Schonleber McCaw P, Vaessin H, Caudy M, Jan L Y, Jan Y N, Cabrera C V, Buskin J N, Hauschka S D, Lassar A B, Wentraub H, Baltimore D. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 47.Nakao K, Campos-Ortega J A. Persistent expression of genes of the Enhancer of split complex suppresses neural development in Drosophila. Neuron. 1996;16:275–286. doi: 10.1016/s0896-6273(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 48.Oellers N, Dehio M, Knust E. bHLH proteins encoded by the Enhancer of split complex of Drosophila negatively interfere with transcriptional activation mediated by proneural genes. Mol Gen Genet. 1994;244:465–473. doi: 10.1007/BF00583897. [DOI] [PubMed] [Google Scholar]
- 49.Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M. Hairy functions as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 1994;8:2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- 50.Parkhurst S M. Groucho: making its Marx as a transcriptional co-repressor. Trends Genet. 1998;14:130–132. doi: 10.1016/s0168-9525(98)01407-3. [DOI] [PubMed] [Google Scholar]
- 51.Paroush Z, Finley R L, Kidd T, Wainwright M, Ingham P W, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with Hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 52.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rubin G M, Spradling A C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 54.Ruiz-Gomez M, Ghysen A. The expression and role of a proneural gene, achaete, in the development of the larval nervous system of Drosophila. EMBO J. 1993;12:1121–1130. doi: 10.1002/j.1460-2075.1993.tb05753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sasai S, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix loop helix factors structurally related to the Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 56.Schrons H, Knust E, Campos-Ortega J A. The Enhancer of split complex and adjacent genes in the 96F region of Drosophila melanogaster are required for segregation of neural and epidermal progenitor cells. Genetics. 1992;132:481–503. doi: 10.1093/genetics/132.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 1997;16:4689–4697. doi: 10.1093/emboj/16.15.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singson A, Leviten M W, Bang A G, Xuequn H H, Posakony J W. Direct downstream targets of proneural activators in the imaginal disc include genes involved with lateral inhibitory signalling. Genes Dev. 1994;8:2058–2071. doi: 10.1101/gad.8.17.2058. [DOI] [PubMed] [Google Scholar]
- 59.Skeath J, Carroll S. Regulation of proneural gene expression and cell fate during neuroblast segregation in the Drosophila embryo. Development. 1992;114:939–946. doi: 10.1242/dev.114.4.939. [DOI] [PubMed] [Google Scholar]
- 60.Sparrow D B, Jen W, Kotecha S, Towers N, Kintner C, Mohun T J. Thylacine 1 is expressed segmentally within the paraxial mesoderm of the Xenopus embryo and interacts with the Notch pathway. Development. 1998;125:2041–2051. doi: 10.1242/dev.125.11.2041. [DOI] [PubMed] [Google Scholar]
- 61.Speicher S A, Thomas U, Hinz U, Knust E. The Serrate locus of Drosophila and its role in morphogenesis of the wing imaginal disc: control of cell proliferation. Development. 1994;120:535–544. doi: 10.1242/dev.120.3.535. [DOI] [PubMed] [Google Scholar]
- 62.Takebayashi K, Akazawa C, Nakanishi S, Kageyama R. Structure and promoter analysis of the gene encoding the mouse helix-loop-helix factor Hes-5. Identification of the neural precursor cell-specific promoter element. J Biol Chem. 1995;270:1342–1349. doi: 10.1074/jbc.270.3.1342. [DOI] [PubMed] [Google Scholar]
- 63.Tata F, Hartley D A. Inhibition of cell fate in Drosophila by Enhancer of split genes. Mech Dev. 1995;51:305–315. doi: 10.1016/0925-4773(95)00377-0. [DOI] [PubMed] [Google Scholar]
- 64.Thomas U, Jonsson F, Speicher S A, Knust E. Phenotypic and molecular characterisation of SerD, a dominant allele of the Drosophila gene Serrate. Genetics. 1995;139:3431–3440. doi: 10.1093/genetics/139.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tietze K, Oellers N, Knust E. Enhancer of splitD, a dominant mutation of Drosophila, and it’s use in the study of functional domains of a helix-loop-helix protein. Proc Natl Acad Sci USA. 1992;89:6152–6156. doi: 10.1073/pnas.89.13.6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uv A, Harrison E J, Bray S J. Tissue-specific splicing and functions of the Drosophila transcription factor Grainyhead. Mol Cell Biol. 1997;17:6727–6735. doi: 10.1128/mcb.17.11.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Doren M, Bailey A M, Esnayra J, Ede K, Posakony J W. Negative regulation of proneural gene activity: Hairy is a direct repressor of achaete. Genes Dev. 1994;8:2729–2742. doi: 10.1101/gad.8.22.2729. [DOI] [PubMed] [Google Scholar]
- 68.Van Doren M, Ellis H M, Posakony J W. The Drosophila extramacrochaetae protein antagonizes sequence specific DNA binding by Daughterless/Achaete-Scute protein complexes. Development. 1991;113:245–255. doi: 10.1242/dev.113.1.245. [DOI] [PubMed] [Google Scholar]
- 69.Van Doren M, Powell P A, Pasternak D, Singson A, Posakony J W. Spatial regulation of proneural gene activity: auto- and cross-activation of achaete is antagonized by extramacrochaetae. Genes Dev. 1992;6:2592–2605. doi: 10.1101/gad.6.12b.2592. [DOI] [PubMed] [Google Scholar]
- 70.Villares R, Cabrera C V. The achaete-scute gene complex of D. melanogaster conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell. 1987;50:415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- 71.Weintraub H, Davis R, Tapscott S, Thayer M, Krause M, Benezra R, Blackwell T K, Turner D, Rupp R, Hollenberg S, Zhuang Y, Lassar A. The myoD Gene family: nodal point during specification of the muscle lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]