PURPOSE
Nearly all men with prostate cancer treated with androgen receptor (AR) signaling inhibitors (ARSIs) develop resistance via diverse mechanisms including constitutive activation of the AR pathway, driven by AR genomic structural alterations, expression of AR splice variants (AR-Vs), or loss of AR dependence and lineage plasticity termed neuroendocrine prostate cancer. Understanding these de novo acquired ARSI resistance mechanisms is critical for optimizing therapy.
MATERIALS AND METHODS
A novel liquid biopsy technology was used to collect mRNA from circulating tumor cells (CTCs) to measure expression of AR-Vs, AR targets, and neuroendocrine prostate cancer markers. An institutional review board–approved prospective cohort (N = 99) was used to identify patterns of gene expression. Two prospective multicenter phase II clinical trials of ARSIs for men with castration-resistant prostate cancer (ClinicalTrials.gov: NCT01942837 [enzalutamide, N = 21] and NCT02025010 [abiraterone, N = 27]) were used to further validate these findings.
RESULTS
Hierarchical clustering of CTC transcripts identified two distinct clusters. Cluster 2 (C2) exhibited increased expression of AR-regulated genes and was associated with worse overall survival (median 8.6 v 22.4 months; P < .01; hazard ratio [HR] = 3.45 [1.9 to 6.14]). In multivariable analysis, C2 was prognostic independent of other clinicopathologic variables. AR-V status was not significant when accounting for C2. Upon further validation in pooled multicenter phase II trials, C2 was associated with worse overall survival (15.2 months v not reached; P < .01; HR = 8.43 [2.74 to 25.92]), prostate-specific antigen progression-free survival (3.6 v 12 months; P < .01; HR = 4.64 [1.53 to 14.11]), and radiographic progression-free survival (2.7 v 40.6 months; P < .01; HR = 4.64 [1.82 to 17.41]).
CONCLUSION
We demonstrate that a transcriptional profile detectable in CTCs obtained from liquid biopsies can serve as an independent prognostic marker beyond AR-V7 in patients with metastatic prostate cancer and can be used to identify the emergence of multiple ARSI resistance mechanisms. This is currently being investigated in additional prospective trials.
INTRODUCTION
Androgen Receptor (AR) signaling inhibitors (ARSIs), including abiraterone acetate (AA), enzalutamide, apalutamide, and darolutamide, have improved survival for men with metastatic castration-sensitive prostate cancer (CSPC) and castration-resistant prostate cancer (CRPC).1-5 However, approximately 5%-10% of patients will have primary resistance to ARSI treatment and a majority of initial responders will develop resistance within 1-3 years.6-8 The underlying drivers of treatment resistance can include activating AR genomic alterations (amplifications, mutations, and rearrangements), epigenetic alterations, and expression of truncated constitutively active AR splice variants (AR-V), among others.7,9-11 These alterations culminate in increased AR transcriptional activity and target gene expression despite androgen blockade. Conversely, some patients with CRPC develop AR-independent, neuroendocrine prostate cancer (NEPC) as an alternate escape pathway from androgen blockade.12-15 Comprehensive understanding of the molecular drivers of ARSI resistance and the ability to monitor this evolution over time is critical for early detection and appropriate treatment selection to improve outcomes for these patients.
CONTEXT
Key Objective
Multiple mechanisms of resistance result in disease progression in men with prostate cancer treated with androgen receptor (AR) targeted therapies (AR signaling inhibitor [ARSI]). This study aimed to develop a multiplex liquid biopsy that could detect resistance from AR-driven and AR-independent mechanisms to identify diverse drivers of treatment resistance.
Knowledge Generated
A pattern of gene expression was identified by hierarchical cluster analysis from RNA extracted from circulating tumor cells (CTCs) from patients with prostate cancer. This cluster was shown to be prognostic for decreased overall survival in the training cohort and decreased progression-free survival and overall survival in the validation cohort including patients from two clinical trials with ARSI.
Relevance
Detection of androgen receptor splice variant 7 in CTCs has emerged as a biomarker that is prognostic for treatment resistance to ARSI. Here, we demonstrate that expression of AR-regulated genes in CTCs is also prognostic for survival and radiographic progression that may have broader ability to identify treatment resistance than androgen receptor splice variant 7 alone.
Molecular profiling to detect these resistance mechanisms has historically required serial tumor biopsies, which carry a risk of procedural complication and high likelihood of not obtaining adequate tissue for analysis. The predominance of bone metastases in prostate cancer and the process of decalcification for pathologic assessment also render it challenging to assess RNA expression in metastatic bone biopsy samples. Liquid biopsies, including circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA), have shown prognostic relevance in CRPC and the potential to be used as a surrogate biomarker of survival.16-19 AR-V7 expression or protein in CTCs can be predictive of response to ARSIs such as enzalutamide and abiraterone acetate (AA).20-26 In the PROPHECY trial, which profiled 118 patients with CRPC, 0% and 11% of patients who had detectable CTC AR-V7 by the Epic Sciences nuclear AR-V7 protein assay or the Johns Hopkins AR-V7 mRNA assay, respectively, had confirmed prostate-specific antigen (PSA) responses. The AR-V7–positive patients also had significantly worse progression-free and overall survival (OS) as compared with patients who lacked detectable AR-V7. The prevalence of AR-V7 is low in the first-line setting and many AR-V7–negative patients do not respond to second-line ARSIs, suggesting other mechanisms of resistance.26-28 NEPC is characterized by the loss of AR transcriptional activity in tandem with upregulation of neuroendocrine markers and is observed in 5%-17% of men with CRPC.14,15,29 NEPC has been previously identified using ctDNA methylation profiling and morphologic characterization of CTCs.30,31 To our knowledge, there is no assay capable of assessing AR-Vs, AR activity, and NEPC on a single platform, which is required to understand the timing and emergence of these diverse resistance mechanisms that occur and interact.
Herein, we present a CTC liquid biopsy transcriptional assay capable of simultaneously measuring these three resistance mechanisms. This multiplex panel includes AR splice variants (AR-V7 and AR-V9), a panel of AR-regulated genes (TMPRSS2, KLK2, KLK3, and FOLH1) that may indicate persistent or overactivation of AR transcriptional activity, and NEPC-associated genes (SYP and CHGA). We applied this assay to a large multi-institutional cohort of patients with metastatic prostate cancer where we observed associations with clinical outcomes and described longitudinal changes compared with changes in ctDNA. These findings were subsequently independently validated in two multicenter phase II trials.
MATERIALS AND METHODS
Multi-Institutional Prospective Cohort
Between August 2014 and January 2020, blood samples were collected from 99 patients with prostate cancer treated at the University of Wisconsin Carbone Cancer Center and Dana Farber Cancer Institute (DFCI). Patients were required to have a histologically confirmed metastatic prostate cancer diagnosis. All patients gave informed consent following an institutional review board–approved protocol to participate in prospective blood sample collection and analysis. Study data were managed using approved REDCap electronic database hosted at the University of Wisconsin (UW)–Madison, School of Medicine and Public Health.32 This cohort was used as the training set for the C2 classifier.
Phase II Clinical Trials
The ENZA-CRPC trial (ClinicalTrials.gov: NCT01942837) was a phase II trial of enzalutamide in metastatic CRPC (mCRPC), which accrued 66 patients from 2013 to 2017. The AA-CRPC trial (ClinicalTrials.gov: NCT02025010) was a phase II trial of AA without glucocorticoid therapy for men with mCRPC,33 which accrued 58 patients between 2014 and 2016. Liquid biopsies were prospectively collected before treatment and evaluable on 21 patients from ENZA-CRPC and 27 patients from AA-CRPC, for a total of 48 patients from prospective phase II trials treated with ARSIs. These trials served as independent validation for the C2 classifier.
CTC Isolation, RNA Extraction, and Quantitative Reverse Transcriptase-Polymerase Chain Reaction
Fifteen milliliters of blood was collected from each patient in EDTA vacutainer tubes (BD Biosciences, Franklin Lakes, NJ). Samples were shipped in temperature-controlled packing and processed within 24 hours. The VERSA platform, as described previously,34 was used for CTC capture and extraction of mRNA.34,35 Quantitative reverse transcriptase-polymerase chain reaction was used to assess gene expression. Details are available in the Data Supplement (online only). Expression values of CTCs isolated from 15 mL of blood were calculated as 38 − Ct values and were reported as raw (unnormalized) data.
Circulating Tumor DNA
Plasma was collected from whole blood collected in EDTA tubes within 4 hours of draw and before isolation of CTCs. Plasma (6 mL) was removed after centrifugation at 290 × g for 10 minutes, spun at 2,730 × g for 10 minutes, and stored at –80°C. Cell-free DNA was isolated using the Qiagen Circulating Nucleic Acids kit following manufacturer's instructions. Cell-free DNA sequencing was performed as previously described.36
Statistical Analysis
Hierarchical clustering of the gene expression data from the UW-DFCI samples (training set) was performed using the R hclust function with default parameters to identify distinct clusters. To create a single-sample approach to independently classify samples in the phase II trials, a nearest shrunken centroids classifier37 was trained using the same genes on the baseline CTC samples using the R pamr package with default parameters. This approach computes a centroid for each cluster, applies a shrinkage step to reduce noise, and classifies new samples on the basis of the nearest centroid. This was then locked and used to classify the ENZA-CRPC and AA-CRPC samples (independent validation set). For comparisons between two clusters, Fisher's exact test and Mann-Whitney U test were used for categorical and continuous variables, respectively. OS was the primary clinical end point and was defined as date of death or last contact relative to treatment start. PSA progression-free survival (PSA PFS) was defined as date of PSA progression according to the Prostate Cancer Working Group 2 criteria or death because of any cause or censored at date of last disease assessment relative to treatment start. Radiographic PFS (rPFS) was defined as radiographic progression according to RECIST 1.1 criteria for soft tissue and lymph node disease and Prostate Cancer Working Group 2 for bone disease or death because of any cause or censored at date of last disease assessment relative to treatment start. The Kaplan-Meier method was used to estimate the survival distributions by clusters and AR-V status, and log-rank test was used to compare two groups. Univariate and multivariate Cox proportional hazards models were fitted to quantify the association of molecular and clinical variables with time-to-event end points.
RESULTS
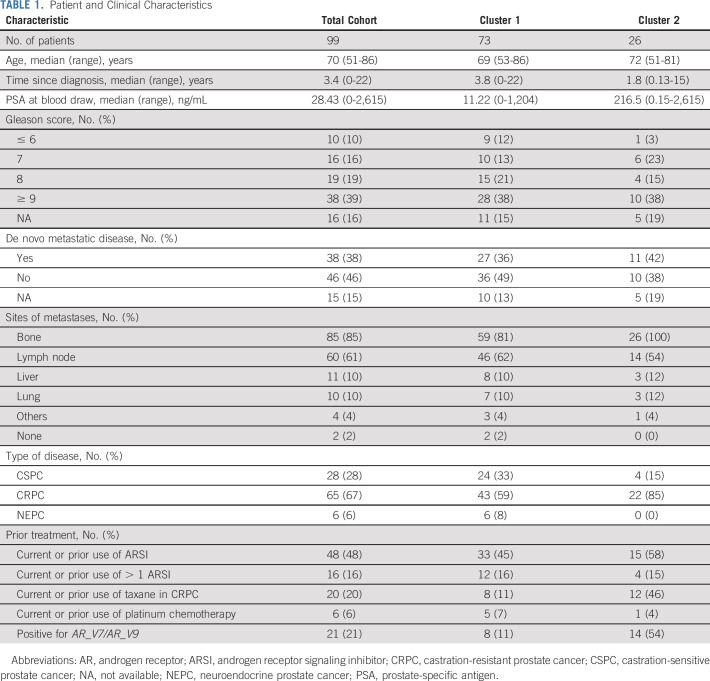
Blood samples from 99 unique patients with metastatic prostate cancer were included in the initial cohort. Patient characteristics and treatment history at the time of blood sample collection are detailed in Table 1. The samples were acquired from a biospecimen protocol that collected blood from any patient with metastatic prostate cancer for correlation with clinical outcomes. As such, the patient cohort consisted of patients with CSPC (28%), CRPC (67%), and NEPC (6%) at the time of blood draw. These patients received various treatments, including androgen deprivation therapy, ARSIs, taxane chemotherapy, platinum doublet therapies, and clinical trials. Forty-eight percent of the patients had current or prior treatment with ARSIs, whereas 20% had current or prior treatment with taxane chemotherapy.
TABLE 1.
Patient and Clinical Characteristics
Transcriptional Analysis of CTCs
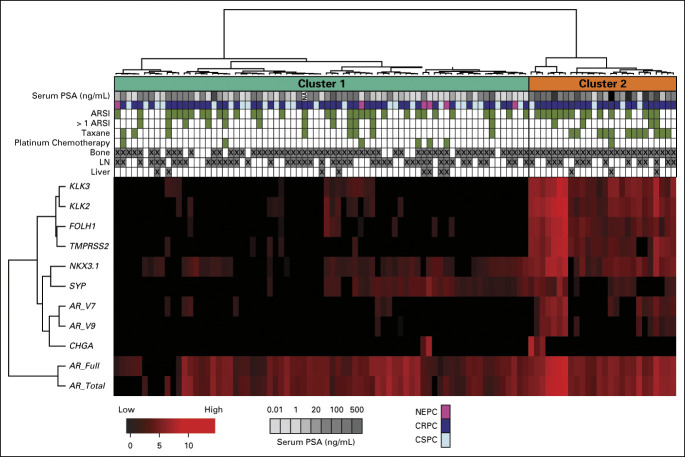
Quantitative multiplex reverse transcriptase-polymerase chain reaction was used to measure the expression of AR, AR-Vs, AR-regulated genes, and genes upregulated in NEPC using RNA isolated from CTCs captured with an anti-EpCAM antibody. Hierarchical clustering of the transcript expression from these samples identified two distinct clusters of patients (Fig 1). Cluster 1 (C1) was characterized by low to absence of detection of AR-regulated genes. C1 included all patients with a histologic diagnosis of NEPC. By contrast, cluster 2 (C2) was enriched for patients who had high expression of transcriptional targets of AR, indicating increased AR transcriptional activity. Consistent with increased AR activity, patients classified as C2 had higher levels of serum PSA (median 216 ng/mL) relative to the patients in cluster 1 (median 11.22 ng/mL, P < .0001) (Data Supplement, Table 1). Patient and disease characteristics for each cluster are detailed in Table 1. Patient age, time since diagnosis, and percent of patients diagnosed with metastatic disease at initial diagnosis were all similar in C1 and C2. Patients in C2 were significantly more likely to have CRPC versus CSPC and bone metastases and had treatment with taxane chemotherapy.
FIG 1.
Two distinct molecular clusters in circulating tumor cells from patients with metastatic prostate cancer. Hierarchical clustering of gene expression data of 11 genes from 99 patients with prostate cancer identified two distinct clusters (cluster 1: C1 and cluster 2: C2). Serum PSA (ng/mL), prostate cancer type, treatment with ARSI or chemotherapy in CRPC setting, sites of metastases at time of blood draw, and serum PSA at time of blood draw are indicated. X indicates presence of metastases at indicted site. AR, androgen receptor; ARSI, androgen receptor signaling inhibitor; AR-V, androgen receptor splice variant; CRPC, castration-resistant prostate cancer; CSPC, castration-sensitive prostate cancer; LN, lymph node; NEPC, neuroendocrine prostate cancer; PSA, prostate-specific antigen.
UW-DFCI Clinical Outcomes
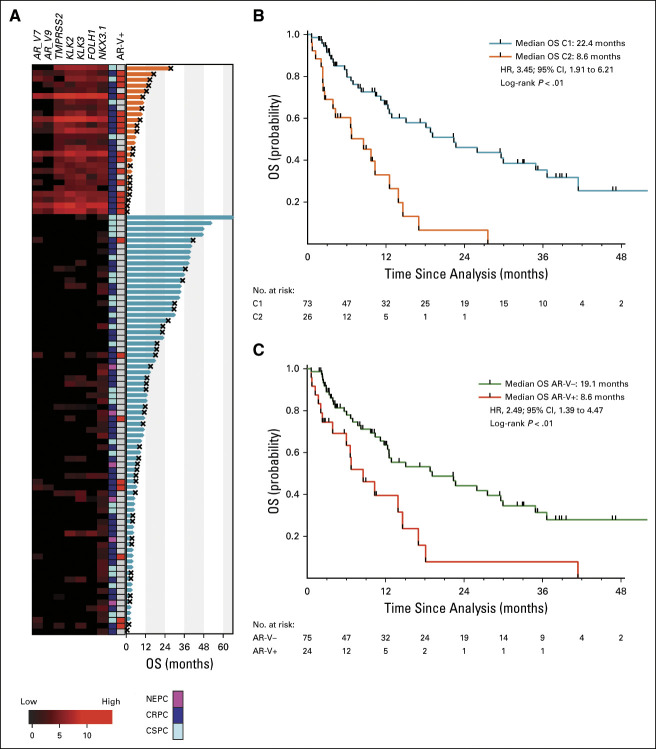
We next examined the clinical end point of OS. At time of analysis, 55 patients (56%) were deceased and the median follow-up time was 9.4 months in this cohort. We observed significantly shorter OS for patients in C2 versus C1 (Figs 2A and 2B). The median OS was 8.6 months in C2 (95% CI, 2.6 to 12.6) (n = 26) versus 22.4 (95% CI, 12.5 to 34.9) months in C1 (n = 73) (P < .01, hazard ratio [HR] = 3.45 [1.91 to 6.21]). As has been reported,20,22,26 AR-V+ patients (AR-V7 and AR-V9) had worse OS (Fig 2C). The median OS for AR-V+ patients was 8.6 (95% CI, 3.9 to 14.6) months (n = 24) versus 19.1 (95% CI, 12.3 to 30) months (n = 75) for AR-V– (P < .01, HR = 2.49 [1.39 to 4.47]). On multivariable analysis (MVA), C2 was independently associated with shorter OS after adjusting for patient age, CRPC versus CSPC, NEPC versus CSPC, PSA, visceral metastasis, and AR-V status. CRPC and NEPC were the only other variables also independently associated with OS on MVA (Fig 3 and Data Supplement). Consistent with the MVA, OS remained shorter for patients in C2 when patients with CRPC (Data Supplement) and CSPC (Data Supplement) were analyzed independently. Unexpectedly, AR-V status was not statistically significant in the MVA. AR-V+ patients were significantly enriched in C2 versus C1 (Fig 2A, 54% v 10%, P < .0001), suggesting that AR-V status may be prognostic because of its association with C2, rather than being independently prognostic. When we examined only C2 patients, no significant differences in OS were detected on the basis of AR-V status (Data Supplement). However, when we examined only AR-V+ patients, significantly shorter OS was observed for C2 patients compared with C1 patients (Data Supplement). This result is consistent with the MVA and suggests that these transcriptional clusters are inclusive of AR-Vs, and the prognostic importance of AR-Vs alone is eliminated when taking into account CTC gene expression clusters.
FIG 2.
C2 is prognostic for OS independent of AR-V status. (A) Swimmer plot ordered by OS for C2 (orange, top) and C1 (cyan, bottom). Gene expression for AR-V and AR pathway genes is indicated. X indicates the patient death. (B) Kaplan-Meier plot of OS by C2 versus C1. (C) Kaplan-Meier plot of OS by AR-V status. AR, androgen receptor; AR-V, androgen receptor splice variant; C1, cluster 1; C2, cluster 2; CRPC, castration-resistant prostate cancer; CSPC, castration-sensitive prostate cancer; HR, hazard ratio; NEPC, neuroendocrine prostate cancer; OS, overall survival.
FIG 3.
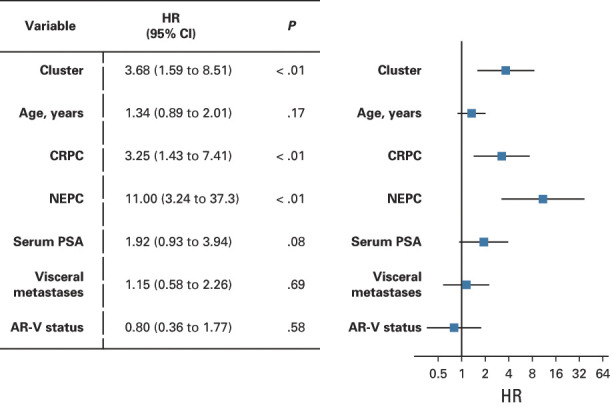
Multivariate survival analysis indicates that the association of cluster and risk of OS remained after adjusting covariates. Association of AR-V status and OS was no longer significant. AR-V, androgen receptor splice variant; CRPC, castration-resistant prostate cancer; HR, hazard ratio; NEPC, neuroendocrine prostate cancer; OS, overall survival; PSA, prostate-specific antigen.
Independent Validation in Prospective Trials With AA or Enzalutamide
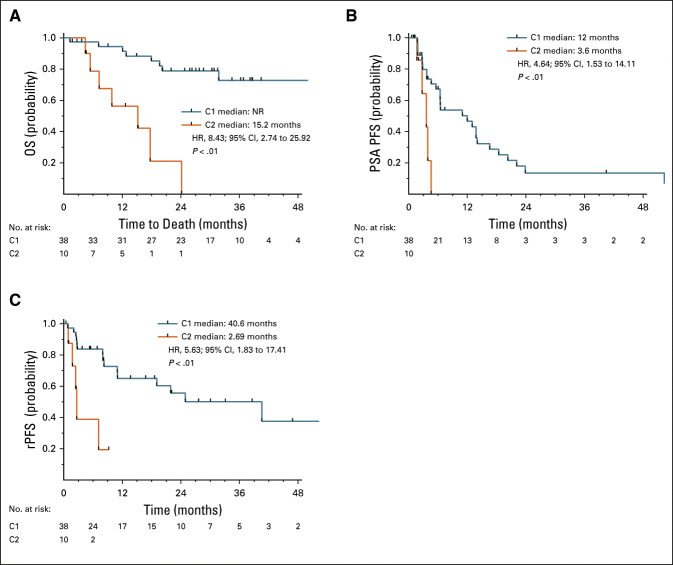
We next sought to validate our findings in two prospective phase II trials of patients with mCRPC (excluding NEPC) treated with ARSIs, enzalutamide (ENZA-CRPC), or AA-CRPC. At time of analysis, 15 patients (31%) were deceased and the median follow-up time was 28 months in this cohort. We applied the classifier trained in UW-DFCI to these cohorts to independently identify and clinically validate C2-like patterns in CTCs. In the combined trials of ENZA-CRPC and AA-CRPC (N = 48), we identified 10 patients (21%) with a C2 expression pattern. This C2 cluster was associated with worse OS (median 15.2 [95% CI, 4.4 to not reached (NR)] months v NR; P < .01; HR = 8.43 [2.74 to 25.92]) (Fig 4A) in the pooled validation cohort. These independent results are consistent with the findings in our multi-institutional UW-DFCI cohort. Similarly, C2 is independently prognostic for OS relative to serum PSA, the presence of visceral metastases, age, and AR-V status using pairwise MVA in the phase II trials (Data Supplement). Importantly, we also observed that patients in C2 had significantly shorter PSA PFS (3.6 [95% CI, 1.7 to NR] v 12 [95% CI, 5.6 to 14] months; P < .01; HR = 4.64 [1.53 to 14.11]) (Fig 4B) and rPFS (2.7 [95% CI, 1 to NR] v 40.6 [95% CI, 11 to NR] months; P < .01; HR = 5.63 [1.82 to 17.41]) (Fig 4C).
FIG 4.
C2 has worse PSA, rPFS, and OS in two multicenter phase II trials of androgen receptor signaling inhibitors. Kaplan-Meier plot of (A) OS, (B) PSA PFS, and (C) rPFS for patients in C2 compared with C1 in patient treated with enzalutamide or abiraterone acetate. C1, cluster 1; C2, cluster 2; HR, hazard ratio; NR, not reached; OS, overall survival; PFS, progression-free survival; PSA, prostate-specific antigen; rPFS, radiographic progression-free survival.
CTC and ctDNA Changes Precede Clinical Progression
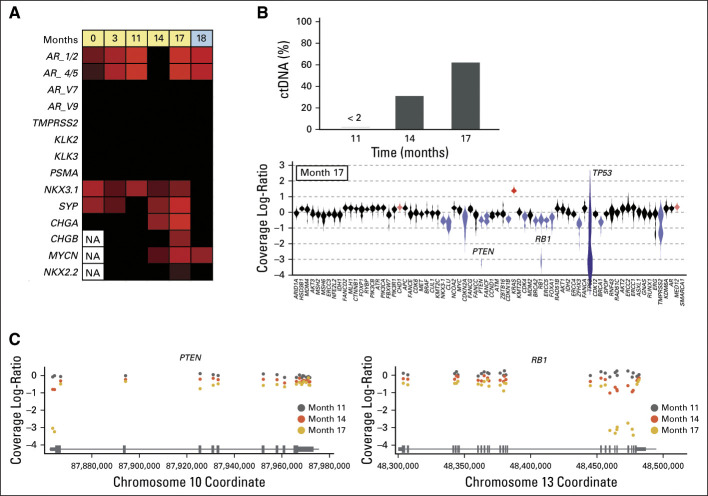
An advantage of liquid biopsy technology is the ability to collect serial samples. We obtained longitudinal CTC and ctDNA data from a patient with CSPC. The patient initially presented with widespread bone metastases and a single liver lesion with a serum PSA of 175 ng/mL. A bone biopsy showed a poorly differentiated adenocarcinoma that was positive for PSA and prostatic acid phosphatase. The patient was treated with combined androgen blockade and docetaxel. Serum PSA decreased to 0.33 ng/mL 1 year after the initial presentation, and complete resolution of the liver lesion was noted. At the 17-month timepoint, the patient developed pancytopenia and elevated liver enzymes, whereas restaging scans identified extensive new bone, liver, and lung metastases. A liver biopsy was performed that showed emergence of NEPC with expression of synaptophysin and chromogranin A, and Ki67 was 80% positive. When we examined gene expression in the patient's longitudinal CTC samples, we observed acquired expression of SYP, MYCN, and CHGA 3 months before clinical symptoms (Fig 5A). These molecular changes in CTCs mirrored changes in ctDNA content, which increased from months 14 to 17. NEPC is enriched for inactivating alterations in TP53, RB1, and PTEN.14,15 We observed focal copy number loss in liquid biopsies from months 11 to 17 for all three and concordant expression of NKX2.2 and CHGB (Figs 5B and 5C). The timing of the CTC and ctDNA changes was concordant, and both preceded clinical progression at month 17. These data demonstrate that integration of ctDNA and CTC molecular profiling is feasible and can promote early identification of NEPC emergence to improve patient stratification for clinical trials and novel therapeutic strategies.
FIG 5.
Treatment-emergent NEPC can be detected before the onset of clinical progression. (A) Heatmap of gene expression data demonstrates the acquisition of genes commonly upregulated in NEPC as a patient is treated with androgen deprivation therapy (yellow) and carboplatin and etoposide (blue). (B) Top panel shows ctDNA fractions (as a proportion of total cell-free DNA) at three sampled timepoints. Bottom panel shows a violin plot of the copy number profile at month 17. Blue indicates genes that pass the thresholds for identifying a copy deletion, and red indicates copy gain. Note that the violin for TP53 is distorted because of very low coverage (because of a deep deletion). For PTEN and RB1, the violins have long tails, suggesting that there may be intragenic differences in copy profiles. (C) Copy number profiles showing RB1 and PTEN exon-level data. All three patient timepoints are shown. Differences between the timepoint are a function of ctDNA fraction. Note that the high ctDNA fraction in the month 17 timepoint enables identification of an exon 1 deep deletion in PTEN and a 3′ deep deletion within RB1. AR, androgen receptor; AR-V, androgen receptor splice variant; ctDNA, circulating tumor DNA; NA, not available; NEPC, neuroendocrine prostate cancer.
DISCUSSION
Liquid biopsies using CTCs or ctDNA represent an attractive alternative to tissue-based molecular profiling. Herein, to our knowledge, we present the first study of a multiplex gene expression biomarker panel in 147 patients with metastatic prostate cancer designed to detect three major mechanisms of resistance to ARSIs in prostate cancer: AR-Vs, AR pathway activation, and NEPC. Although landmark studies of liquid biomarkers for AR-Vs or NEPC individually have been published previously,20,23,25,26,30 to our knowledge, this is the first report of an assay capable of measuring all these components of resistance simultaneously. When we applied this CTC assay to a large cohort of patients with metastatic prostate cancer, we identified a distinct cluster C2 characterized by high expression of AR target genes, which was prognostic for OS independent of other variables including AR-V status, CRPC, and NEPC. We then independently validated the prognostic significance of C2 in a combination of two phase II CRPC ARSI trials.
Many diverse mechanisms of resistance to androgen blockade culminate in increased expression of AR target gene expression. These include mutations, amplifications, and rearrangements of the AR gene;38-44 amplification of an enhancer element upstream of the AR gene;44,45 expression of AR-Vs such as AR-V7 or AR-V9;10,20,46,47 epigenetic modifications (eg, methylation and BET proteins),48-50 or even bypassing AR completely via glucocorticoid receptor.51 C2 appears to be primarily driven by increased expression of AR-regulated genes, whereas total AR levels do not differ dramatically. Interestingly, our work suggests that prior interest in AR-Vs may be incomplete. AR-V positivity was correlated with C2, but after adjusting for C2 membership, AR-V status was no longer prognostic. This may be because multiple mechanisms can result in ARSI resistance and manifest in increased AR signaling activity, including not only AR-Vs but also from a myriad of other etiologies. We also independently validated that C2 was associated with worse prognosis in two phase II ARSI trials. Strikingly, C2 was also associated with worse PSA and rPFS with ARSIs, suggesting that it could be used as a biomarker to identify patients who may benefit from additional or alternative therapies up-front and monitor for the early emergence of molecular resistance. These findings warrant further exploration in larger prospective trials.
Previous studies of CTCs in CRPC identified other prognostic signatures that associate with clinical outcomes. CTC enumeration has been shown to be prognostic for OS in prostate cancer using Cell Search platform.16-18 The PROPHECY trial demonstrated that AR-V7 expression, either by gene expression and protein analysis, is prognostic for PFS and OS with ARSI treatment independent of enumeration.26 It is unknown if the number of CTCs present in each sample may affect the detection or level of gene expression for the genes in this panel. Further analysis is necessary for the panel in this report to evaluate the relative contribution of CTC number to this assay. In addition, broader evaluation of non–AR-driven resistance mechanisms, such as the recently described Double Negative Prostate Cancer classifier, may identify a distinct population of patients with different outcomes following treatment with ARSIs. Expansion of this gene expression panel to incorporate newly identified disease subtypes and mechanisms of resistance may further enhance the utility of liquid biopsies.
Molecular classifiers such as the approach described in this study also represent an attractive way to follow prostate cancer disease status in addition to PSA and imaging. These could be easily integrated into existing clinical workflows, such that, in addition to standard periodic laboratory studies, blood samples could be sent for real-time liquid biomarker studies. We demonstrate the feasibility and concordance of longitudinal CTC and ctDNA profiling simultaneously. Identification of molecular mechanisms of ARSI resistance could trigger a change in therapy or enrollment into clinical trials. Early detection maximizes eligibility for other treatments, and although tumor burden is still low (eg, at month 14, before widespread clinical progression at month 17 in the patient described), the patient's performance status is still optimal, before symptomatic disease progression.
In conclusion, to our knowledge, we demonstrate the first multiplex gene expression liquid biopsy assay that can assess multiple potential ARSI resistance mechanisms simultaneously in metastatic prostate cancer. Early identification of these molecular changes could help guide treatment decisions. The heterogeneous nature of this cohort, which included different disease states and therapies, suggests that these findings may be generalizable to the broader patient population. The validation of our prognostic clustering in two phase II trials with ARSIs suggests clinical utility; however, larger clinical trials are necessary. We are prospectively testing these findings further in multiple ongoing clinical trials for patients with CSPC and CRPC (NCT02445976, NCT01942837, NCT03725761, NCT02025010, and NCT04126070).
ACKNOWLEDGMENT
We would like to thank all patients who participated in this study. We are also grateful for the help of Katie Kovacich for project management; the UWCCC GU clinical research group especially Jamie Wiepz, Emily Nordin, Kelly Bush, and Mary Jane Staab; and the biospecimen team especially Laura Ruelle and Hannah Ranous. The authors thank the University of Wisconsin Translational Research Initiatives in Pathology laboratory, in part supported by the UW Department of Pathology and Laboratory Medicine and UWCCC Grant No. P30 CA014520, for the use of its facilities and services.
Hamid Emamekhoo
Consulting or Advisory Role: Exelixis, Bayer, Bristol Myers Squibb, Cardinal Health, Seattle Genetics
Research Funding: Bristol Myers Squibb, Exelixis, Replimune, Merck, Calithera Biosciences, Roche/Genentech
Travel, Accommodations, Expenses: DAVA Pharmaceuticals
Rana R. McKay
Consulting or Advisory Role: Janssen, Novartis, Tempus, Exelixis, Pfizer, Bristol Myers Squibb, Astellas Medivation, Dendreon, Bayer, Sanofi, Merck, Vividion Therapeutics, Calithera Biosciences, AstraZeneca
Research Funding: Pfizer, Bayer, Tempus
Xiao X. Wei
Honoraria: OncLive
Consulting or Advisory Role: Novartis
Research Funding: Bristol Myers Squibb
Travel, Accommodations, Expenses: Corvus Pharmaceuticals
Rebecca Silver
Research Funding: Bayer
Michael J. Morris
Consulting or Advisory Role: Bayer, Endocyte, Advanced Accelerator Applications, ORIC Pharmaceuticals, Johnson & Johnson, Curium Pharma, Athenex
Research Funding: Bayer, Sanofi, Endocyte, Progenics, Corcept Therapeutics, Roche/Genentech, Janssen
Travel, Accommodations, Expenses: Endocyte, Fujifilm
Felix Y. Feng
Consulting or Advisory Role: Astellas, Bayer, BlueEarth Diagnostics, Celgene, EMD Serono, Genentech, Janssen, Myovant, Ryovant, Bristol Myers Squibb, Exact Sciences, Varian, Bluestar Genomics, Serimmun
Research Funding: Zenith Epigenetics
Howard I. Scher
Consulting or Advisory Role: Janssen, Amgen, Janssen Research & Development, Menarini Silicon Biosystems, WIRB-Copernicus Group, ESSA, Ambry Genetics/Konica Minolta, Pfizer, Bayer, Sun Pharma
Research Funding: Janssen, Illumina, Epic Sciences, Menarini Silicon Biosystems, Thermofisher Scientific Biomarkers
Patents, Royalties, Other Intellectual Property: BioNTech—Intellectual Property Rights, Elucida Oncology—Intellectual Property Rights, MabVAX—Intellectual Property Rights, Y-mAbs Therapeutics Inc—Intellectual Property Rights
Travel, Accommodations, Expenses: Asterias Biotherapeutics, Menarini Silicon Biosystems, Amgen, WIRB-Copernicus Group, Konica Minolta, ESSA, Prostate Cancer Foundation, Sanofi, Bayer, Phosplatin Therapeutics
Dana Rathkopf
Consulting or Advisory Role: Janssen, Genentech, AstraZeneca, Bayer, Myovant Sciences
Research Funding: Janssen Oncology, Medivation, Celgene, Takeda, Millennium, Ferring, Novartis, Taiho Pharmaceutical, AstraZeneca, Genentech/Roche, TRACON Pharma, Bayer, Phosplatin Therapeutics
Scott M. Dehm
Consulting or Advisory Role: Celgene, Oncternal Therapeutics, Janssen Research & Development
Research Funding: Janssen Research & Development, Medivation/Astellas
Patents, Royalties, Other Intellectual Property: Royalties from licensing genome-engineered prostate cancer cell lines
Toni K. Choueiri
Employment: Dana Farber Cancer Hospital
Leadership: Dana Farber Cancer Hospital, NCCN, KidneyCAN, ASCO
Stock and Other Ownership Interests: Pionyr, Tempest Therapeutics
Honoraria: NCCN, UpToDate, Michael J. Hennessy Associates, ASCO, Harborside Press, Analysis Group, AstraZeneca, Alexion Pharmaceuticals, Sanofi/Aventis, Bayer, Bristol Myers Squibb, Genentech/Roche, GlaxoSmithKline, Merck, Novartis, Peloton Therapeutics, Pfizer, Corvus Pharmaceuticals, Ipsen, Foundation Medicine, Eisai, PlatformQ Health, Clinical Care Options, Navinata Health, Kidney Cancer Association, Exelixis, Prometheus, Lpath, The New England Journal of Medicine, Lancet Oncology, Cerulean Pharma, Alligent, EMD Serono, HERON, Lilly, Janssen Oncology, IQvia, Aveo, NCI GU Steering Committee
Consulting or Advisory Role: Pfizer, Bayer, Novartis, GlaxoSmithKline, Merck, Bristol Myers Squibb, Roche/Genentech, Eisai, Foundation Medicine, Cerulean Pharma, AstraZeneca, Exelixis, Prometheus, Alligent, Ipsen, Corvus Pharmaceuticals, Lpath, Alexion Pharmaceuticals, Sanofi/Aventis, Peloton Therapeutics, UpToDate, NCCN, Michael J. Hennessy Associates, Analysis Group, Kidney Cancer Association, Clinical Care Options, PlatformQ Health, Navinata Health, Harborside Press, ASCO, The New England Journal of Medicine, Lancet Oncology, EMD Serono, HERON, Lilly, ESMO
Research Funding: Pfizer, Novartis, Merck, Exelixis, TRACON Pharma, GlaxoSmithKline, Bristol Myers Squibb, AstraZeneca, Peloton Therapeutics, Roche/Genentech, Celldex, Agensys, Eisai, Takeda, Prometheus, Ipsen, Corvus Pharmaceuticals, Cerulean Pharma, Seattle Genetics/Astellas, Bayer, Foundation Medicine, Roche, Calithera Biosciences, Analysis Group, NCI, Gateway for Cancer Research, Congressionally Directed Medical Research Programs (DOD)
Patents, Royalties, Other Intellectual Property: International Patent Application No. PCT/US2018/058430, titled “Biomarkers of Clinical Response and Benefit to Immune Checkpoint Inhibitor Therapy,” and International Patent Application No. PCT/US2018/12209, titled “PBRM1 Biomarkers Predictive of Anti-Immune Checkpoint Response”
Travel, Accommodations, Expenses: Pfizer, Bayer, Novartis, GlaxoSmithKline, Merck, Bristol Myers Squibb, Roche/Genentech, Eisai, Foundation Medicine, Cerulean Pharma, AstraZeneca, Exelixis, Prometheus, Alligent, Ipsen, Corvus Pharmaceuticals, Lpath, Alexion Pharmaceuticals, Sanofi/Aventis, UpToDate, Peloton Therapeutics, NCCN, Michael J. Hennessy Associates, Analysis Group, Kidney Cancer Association, Clinical Care Options, PlatformQ Health, Harborside Press, Navinata Health, The New England Journal of Medicine, Lancet Oncology, EMD Serono, HERON, Lilly, ESMO
Other Relationship: Medical writing and editorial assistance support might have been funded by Communications companies funded by pharmaceutical companies such as ClinicalThinking, Health Interactions, Envision Pharma Group, Fishawack Group of Companies, Parexel
Susan Halabi
Employment: ASCO
Andrew J. Armstrong
Honoraria: Astellas Scientific and Medical Affairs Inc
Consulting or Advisory Role: Bayer, Dendreon, Pfizer, Astellas Scientific and Medical Affairs Inc, Clovis Oncology, AstraZeneca, Merck, Bristol Myers Squibb
Research Funding: Dendreon, Bayer, Pfizer, Novartis, Janssen Oncology, Astellas Pharma, Gilead Sciences, Roche/Genentech, Bristol Myers Squibb, Constellation Pharmaceuticals, Merck, AstraZeneca, BeiGene
Patents, Royalties, Other Intellectual Property: Circulating tumor cell novel capture technology
Travel, Accommodations, Expenses: Astellas Scientific and Medical Affairs Inc
Alexander W. Wyatt
Honoraria: Janssen, Astellas Pharma, AstraZeneca, Merck, AstraZeneca Canada
Consulting or Advisory Role: AstraZeneca
Research Funding: ESSA
Mary-Ellen Taplin
Honoraria: Janssen-Ortho, Clovis Oncology, Astellas Pharma, Incyte, UpToDate, Research to Practice, Pfizer, Bayer, Amgen, AstraZeneca, Progenics, Guidepoint Global, Celgene, Merck, GlaxoSmithKline, Myovant Sciences, Roivant, AbbVie, Arcus Biosciences, Constellation Pharmaceuticals
Consulting or Advisory Role: Janssen-Ortho, Bayer, Guidepoint Global, Best Doctors Inc, UpToDate, Clovis Oncology, Research to Practice, Myovant Sciences, Incyte, Pfizer, AstraZeneca, Arcus Ventures
Research Funding: Janssen-Ortho, Medivation, Bayer, Pfizer
Travel, Accommodations, Expenses: Medivation, Janssen Oncology, Tokai Pharmaceuticals, Astellas Pharma, Incyte, Pfizer, Clovis Oncology, Bayer
Shuang G. Zhao
Employment: Exact Sciences (I)
Patents, Royalties, Other Intellectual Property: Patent applications pending with Decipher Biosciences, Exact Sciences
Joshua M. Lang
Stock and Other Ownership Interests: Salus Discovery
Consulting or Advisory Role: Sanofi, Immunomedics, Janssen, Pfizer/Astellas, 4D Pharma
Research Funding: Medivation, Agensys, GlaxoSmithKline, Immunomedics, Bristol Myers Squibb, Janssen
Patents, Royalties, Other Intellectual Property: I am listed on the patent on a technology for rare cell capture and analysis. This technology has been licensed by Salus Discovery LLC although no commercial products are available
No other potential conflicts of interest were reported.
SUPPORT
Supported by a Movember-Prostate Cancer Foundation Challenge Award (to J.M.L.), NIH Grant No. 1R01CA181648 (to J.M.L.), Department of Defense Synergistic Idea Development Award PC140746 (to J.M.L. and S.M.D.), and a Department of Defense Physician Research Award PC190039 (to S.G.Z.).
CLINICAL TRIAL INFORMATION
J.M.S., H.E., and R.R.M. contributed equally to this work.
DATA SHARING STATEMENT
Individual participant data that are included in the results reported in the manuscript will be available after deidentification beginning 6 months and ending 36 months following publication. Researchers must provide a methodologically sound proposal, and requestors may need to sign a data access agreement. Proposals should be directed to the corresponding author.
AUTHOR CONTRIBUTIONS
Conception and design: Jamie M. Sperger, Hamid Emamekhoo, Lucia Kwak, Xiao X. Wei, Glenn Bubley, Howard I. Scher, Toni K. Choueiri, Mary-Ellen Taplin, Joshua M. Lang
Financial support: Scott M. Dehm, Joshua M. Lang
Administrative support: Toni K. Choueiri, Andrew J. Armstrong, Joshua M. Lang
Provision of study materials or patients: Rana R. McKay, Michael J. Morris, Dana Rathkopf, Toni K. Choueiri, Mary-Ellen Taplin, Joshua M. Lang
Collection and assembly of data: Jamie M. Sperger, Hamid Emamekhoo, Rana R. McKay, Charlotte N. Stahlfeld, Anupama Singh, Cole S. Gilsdorf, Serena K. Wolfe, Rebecca Silver, Zhenwei Zhang, Michael J. Morris, Howard I. Scher, Dana Rathkopf, Alexander W. Wyatt, Mary-Ellen Taplin, Joshua M. Lang
Data analysis and interpretation: Jamie M. Sperger, Hamid Emamekhoo, Rana R. McKay, Anupama Singh, Xinyi E. Chen, Lucia Kwak, Xiao X. Wei, Michael J. Morris, Glenn Bubley, Felix Y. Feng, Scott M. Dehm, Toni K. Choueiri, Susan Halabi, Andrew J. Armstrong, Alexander W. Wyatt, Mary-Ellen Taplin, Shuang G. Zhao, Joshua M. Lang
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Prospective Evaluation of Clinical Outcomes Using a Multiplex Liquid Biopsy Targeting Diverse Resistance Mechanisms in Metastatic Prostate Cancer
The following represents disclosure information provided by the authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Hamid Emamekhoo
Consulting or Advisory Role: Exelixis, Bayer, Bristol Myers Squibb, Cardinal Health, Seattle Genetics
Research Funding: Bristol Myers Squibb, Exelixis, Replimune, Merck, Calithera Biosciences, Roche/Genentech
Travel, Accommodations, Expenses: DAVA Pharmaceuticals
Rana R. McKay
Consulting or Advisory Role: Janssen, Novartis, Tempus, Exelixis, Pfizer, Bristol Myers Squibb, Astellas Medivation, Dendreon, Bayer, Sanofi, Merck, Vividion Therapeutics, Calithera Biosciences, AstraZeneca
Research Funding: Pfizer, Bayer, Tempus
Xiao X. Wei
Honoraria: OncLive
Consulting or Advisory Role: Novartis
Research Funding: Bristol Myers Squibb
Travel, Accommodations, Expenses: Corvus Pharmaceuticals
Rebecca Silver
Research Funding: Bayer
Michael J. Morris
Consulting or Advisory Role: Bayer, Endocyte, Advanced Accelerator Applications, ORIC Pharmaceuticals, Johnson & Johnson, Curium Pharma, Athenex
Research Funding: Bayer, Sanofi, Endocyte, Progenics, Corcept Therapeutics, Roche/Genentech, Janssen
Travel, Accommodations, Expenses: Endocyte, Fujifilm
Felix Y. Feng
Consulting or Advisory Role: Astellas, Bayer, BlueEarth Diagnostics, Celgene, EMD Serono, Genentech, Janssen, Myovant, Ryovant, Bristol Myers Squibb, Exact Sciences, Varian, Bluestar Genomics, Serimmun
Research Funding: Zenith Epigenetics
Howard I. Scher
Consulting or Advisory Role: Janssen, Amgen, Janssen Research & Development, Menarini Silicon Biosystems, WIRB-Copernicus Group, ESSA, Ambry Genetics/Konica Minolta, Pfizer, Bayer, Sun Pharma
Research Funding: Janssen, Illumina, Epic Sciences, Menarini Silicon Biosystems, Thermofisher Scientific Biomarkers
Patents, Royalties, Other Intellectual Property: BioNTech—Intellectual Property Rights, Elucida Oncology—Intellectual Property Rights, MabVAX—Intellectual Property Rights, Y-mAbs Therapeutics Inc—Intellectual Property Rights
Travel, Accommodations, Expenses: Asterias Biotherapeutics, Menarini Silicon Biosystems, Amgen, WIRB-Copernicus Group, Konica Minolta, ESSA, Prostate Cancer Foundation, Sanofi, Bayer, Phosplatin Therapeutics
Dana Rathkopf
Consulting or Advisory Role: Janssen, Genentech, AstraZeneca, Bayer, Myovant Sciences
Research Funding: Janssen Oncology, Medivation, Celgene, Takeda, Millennium, Ferring, Novartis, Taiho Pharmaceutical, AstraZeneca, Genentech/Roche, TRACON Pharma, Bayer, Phosplatin Therapeutics
Scott M. Dehm
Consulting or Advisory Role: Celgene, Oncternal Therapeutics, Janssen Research & Development
Research Funding: Janssen Research & Development, Medivation/Astellas
Patents, Royalties, Other Intellectual Property: Royalties from licensing genome-engineered prostate cancer cell lines
Toni K. Choueiri
Employment: Dana Farber Cancer Hospital
Leadership: Dana Farber Cancer Hospital, NCCN, KidneyCAN, ASCO
Stock and Other Ownership Interests: Pionyr, Tempest Therapeutics
Honoraria: NCCN, UpToDate, Michael J. Hennessy Associates, ASCO, Harborside Press, Analysis Group, AstraZeneca, Alexion Pharmaceuticals, Sanofi/Aventis, Bayer, Bristol Myers Squibb, Genentech/Roche, GlaxoSmithKline, Merck, Novartis, Peloton Therapeutics, Pfizer, Corvus Pharmaceuticals, Ipsen, Foundation Medicine, Eisai, PlatformQ Health, Clinical Care Options, Navinata Health, Kidney Cancer Association, Exelixis, Prometheus, Lpath, The New England Journal of Medicine, Lancet Oncology, Cerulean Pharma, Alligent, EMD Serono, HERON, Lilly, Janssen Oncology, IQvia, Aveo, NCI GU Steering Committee
Consulting or Advisory Role: Pfizer, Bayer, Novartis, GlaxoSmithKline, Merck, Bristol Myers Squibb, Roche/Genentech, Eisai, Foundation Medicine, Cerulean Pharma, AstraZeneca, Exelixis, Prometheus, Alligent, Ipsen, Corvus Pharmaceuticals, Lpath, Alexion Pharmaceuticals, Sanofi/Aventis, Peloton Therapeutics, UpToDate, NCCN, Michael J. Hennessy Associates, Analysis Group, Kidney Cancer Association, Clinical Care Options, PlatformQ Health, Navinata Health, Harborside Press, ASCO, The New England Journal of Medicine, Lancet Oncology, EMD Serono, HERON, Lilly, ESMO
Research Funding: Pfizer, Novartis, Merck, Exelixis, TRACON Pharma, GlaxoSmithKline, Bristol Myers Squibb, AstraZeneca, Peloton Therapeutics, Roche/Genentech, Celldex, Agensys, Eisai, Takeda, Prometheus, Ipsen, Corvus Pharmaceuticals, Cerulean Pharma, Seattle Genetics/Astellas, Bayer, Foundation Medicine, Roche, Calithera Biosciences, Analysis Group, NCI, Gateway for Cancer Research, Congressionally Directed Medical Research Programs (DOD)
Patents, Royalties, Other Intellectual Property: International Patent Application No. PCT/US2018/058430, titled “Biomarkers of Clinical Response and Benefit to Immune Checkpoint Inhibitor Therapy,” and International Patent Application No. PCT/US2018/12209, titled “PBRM1 Biomarkers Predictive of Anti-Immune Checkpoint Response”
Travel, Accommodations, Expenses: Pfizer, Bayer, Novartis, GlaxoSmithKline, Merck, Bristol Myers Squibb, Roche/Genentech, Eisai, Foundation Medicine, Cerulean Pharma, AstraZeneca, Exelixis, Prometheus, Alligent, Ipsen, Corvus Pharmaceuticals, Lpath, Alexion Pharmaceuticals, Sanofi/Aventis, UpToDate, Peloton Therapeutics, NCCN, Michael J. Hennessy Associates, Analysis Group, Kidney Cancer Association, Clinical Care Options, PlatformQ Health, Harborside Press, Navinata Health, The New England Journal of Medicine, Lancet Oncology, EMD Serono, HERON, Lilly, ESMO
Other Relationship: Medical writing and editorial assistance support might have been funded by Communications companies funded by pharmaceutical companies such as ClinicalThinking, Health Interactions, Envision Pharma Group, Fishawack Group of Companies, Parexel
Susan Halabi
Employment: ASCO
Andrew J. Armstrong
Honoraria: Astellas Scientific and Medical Affairs Inc
Consulting or Advisory Role: Bayer, Dendreon, Pfizer, Astellas Scientific and Medical Affairs Inc, Clovis Oncology, AstraZeneca, Merck, Bristol Myers Squibb
Research Funding: Dendreon, Bayer, Pfizer, Novartis, Janssen Oncology, Astellas Pharma, Gilead Sciences, Roche/Genentech, Bristol Myers Squibb, Constellation Pharmaceuticals, Merck, AstraZeneca, BeiGene
Patents, Royalties, Other Intellectual Property: Circulating tumor cell novel capture technology
Travel, Accommodations, Expenses: Astellas Scientific and Medical Affairs Inc
Alexander W. Wyatt
Honoraria: Janssen, Astellas Pharma, AstraZeneca, Merck, AstraZeneca Canada
Consulting or Advisory Role: AstraZeneca
Research Funding: ESSA
Mary-Ellen Taplin
Honoraria: Janssen-Ortho, Clovis Oncology, Astellas Pharma, Incyte, UpToDate, Research to Practice, Pfizer, Bayer, Amgen, AstraZeneca, Progenics, Guidepoint Global, Celgene, Merck, GlaxoSmithKline, Myovant Sciences, Roivant, AbbVie, Arcus Biosciences, Constellation Pharmaceuticals
Consulting or Advisory Role: Janssen-Ortho, Bayer, Guidepoint Global, Best Doctors Inc, UpToDate, Clovis Oncology, Research to Practice, Myovant Sciences, Incyte, Pfizer, AstraZeneca, Arcus Ventures
Research Funding: Janssen-Ortho, Medivation, Bayer, Pfizer
Travel, Accommodations, Expenses: Medivation, Janssen Oncology, Tokai Pharmaceuticals, Astellas Pharma, Incyte, Pfizer, Clovis Oncology, Bayer
Shuang G. Zhao
Employment: Exact Sciences (I)
Patents, Royalties, Other Intellectual Property: Patent applications pending with Decipher Biosciences, Exact Sciences
Joshua M. Lang
Stock and Other Ownership Interests: Salus Discovery
Consulting or Advisory Role: Sanofi, Immunomedics, Janssen, Pfizer/Astellas, 4D Pharma
Research Funding: Medivation, Agensys, GlaxoSmithKline, Immunomedics, Bristol Myers Squibb, Janssen
Patents, Royalties, Other Intellectual Property: I am listed on the patent on a technology for rare cell capture and analysis. This technology has been licensed by Salus Discovery LLC although no commercial products are available
No other potential conflicts of interest were reported.
REFERENCES
- 1.Armstrong AJ: New treatment options in castration-resistant prostate cancer. J Natl Compr Canc Netw 13:690-693, 2015 [DOI] [PubMed] [Google Scholar]
- 2.Mohler JL Armstrong AJ Bahnson RR, et al. : Prostate cancer, version 1.2016. J Natl Compr Canc Netw 14:19-30, 2016 [DOI] [PubMed] [Google Scholar]
- 3.Beer TM Armstrong AJ Rathkopf D, et al. : Enzalutamide in men with chemotherapy-naive metastatic castration-resistant prostate cancer: Extended analysis of the phase 3 PREVAIL study. Eur Urol 71:151-154, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith MR Saad F Chowdhury S, et al. : Apalutamide treatment and metastasis-free survival in prostate cancer. N Engl J Med 378:1408-1418, 2018 [DOI] [PubMed] [Google Scholar]
- 5.Fizazi K Shore N Tammela TL, et al. : Darolutamide in nonmetastatic, castration-resistant prostate cancer. N Engl J Med 380:1235-1246, 2019 [DOI] [PubMed] [Google Scholar]
- 6.Aggarwal R Zhang T Small EJ, et al. : Neuroendocrine prostate cancer: Subtypes, biology, and clinical outcomes. J Natl Compr Canc Netw 12:719-726, 2014 [DOI] [PubMed] [Google Scholar]
- 7.Antonarakis ES Armstrong AJ Dehm SM, et al. : Androgen receptor variant-driven prostate cancer: Clinical implications and therapeutic targeting. Prostate Cancer Prostatic Dis 19:231-241, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armstrong AJ Lin P Higano CS, et al. : Development and validation of a prognostic model for overall survival in chemotherapy-naive men with metastatic castration-resistant prostate cancer. Ann Oncol 29:2200-2207, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dehm SM Schmidt LJ Heemers HV, et al. : Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res 68:5469-5477, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohli M Ho Y Hillman DW, et al. : Androgen receptor variant AR-V9 is coexpressed with AR-V7 in prostate cancer metastases and predicts abiraterone resistance. Clin Cancer Res 23:4704-4715, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo J Attard G Balk SP, et al. : Role of androgen receptor variants in prostate cancer: Report from the 2017 mission androgen receptor variants meeting. Eur Urol 73:715-723, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beltran H Tomlins S Aparicio A, et al. : Aggressive variants of castration-resistant prostate cancer. Clin Cancer Res 20:2846-2850, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beltran H Prandi D Mosquera JM, et al. : Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 22:298-305, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aggarwal RR Quigley DA Huang J, et al. : Whole-genome and transcriptional analysis of treatment-emergent small-cell neuroendocrine prostate cancer demonstrates intraclass heterogeneity. Mol Cancer Res 17:1235-1240, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aggarwal R Huang J Alumkal JJ, et al. : Clinical and genomic characterization of treatment-emergent small-cell neuroendocrine prostate cancer: A multi-institutional prospective study. J Clin Oncol 36:2492-2503, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Bono JS Scher HI Montgomery RB, et al. : Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 14:6302-6309, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Scher HI Jia X de Bono JS, et al. : Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: A reanalysis of IMMC38 trial data. Lancet Oncol 10:233-239, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scher HI Heller G Molina A, et al. : Evaluation of circulating tumor cell (CTC) enumeration as an efficacy response biomarker of overall survival (OS) in metastatic castration-resistant prostate cancer (mCRPC): Planned final analysis (FA) of COU-AA-301, a randomized, double-blind, placebo-controlled, phase III study of abiraterone acetate (AA) plus low-dose prednisone (P) post docetaxel. J Clin Oncol 29, 2011. (abstr LBA4517) [Google Scholar]
- 19.Scher HI Heller G Molina A, et al. : Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol 33:1348-1355, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonarakis ES Lu C Wang H, et al. : AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 371:1028-1038, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markowski MC Silberstein JL Eshleman JR, et al. : Clinical utility of CLIA-grade AR-V7 testing in patients with metastatic castration-resistant prostate cancer. JCO Precis Oncol 2017, 2017. 10.1200/PO.17.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bastos DA, Antonarakis ES: CTC-derived AR-V7 detection as a prognostic and predictive biomarker in advanced prostate cancer. Expert Rev Mol Diagn 18:155-163, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scher HI Lu D Schreiber NA, et al. : Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol 2:1441-1449, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scher HI Graf RP Schreiber NA, et al. : Phenotypic heterogeneity of circulating tumor cells informs clinical decisions between AR signaling inhibitors and taxanes in metastatic prostate cancer. Cancer Res 77:5687-5698, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scher HI Graf RP Schreiber NA, et al. : Nuclear-specific AR-V7 protein localization is necessary to guide treatment selection in metastatic castration-resistant prostate cancer. Eur Urol 71:874-882, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armstrong AJ Halabi S Luo J, et al. : Prospective multicenter validation of AR-V7 and hormone therapy resistance in high risk castration resistant prostate cancer: The PROPHECY study. J Clin Oncol 37:1120-1129, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y Yang R Henzler CM, et al. : Diverse AR gene rearrangements mediate resistance to androgen receptor inhibitors in metastatic prostate cancer. Clin Cancer Res 26:1965-1976, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henzler C Li Y Yang R, et al. : Truncation and constitutive activation of the androgen receptor by diverse genomic rearrangements in prostate cancer. Nat Commun 7:13668, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labrecque MP Coleman IM Brown LG, et al. : Molecular profiling stratifies diverse phenotypes of treatment-refractory metastatic castration-resistant prostate cancer. J Clin Invest 129:4492-4505, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beltran H Romanel A Conteduca V, et al. : Circulating tumor DNA profile recognizes transformation to castration-resistant neuroendocrine prostate cancer. J Clin Invest 130:1653-1668, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beltran H Jendrisak A Landers M, et al. : The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin Cancer Res 22:1510-1519, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris PA Taylor R Thielke R, et al. : Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42:377-381, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKay RR Werner L Jacobus SJ, et al. : A phase 2 trial of abiraterone acetate without glucocorticoids for men with metastatic castration-resistant prostate cancer. Cancer 125:524-532, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sperger JM Strotman LN Welsh A, et al. : Integrated analysis of multiple biomarkers from circulating tumor cells enabled by exclusion-based analyte isolation. Clin Cancer Res 23:746-756, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strotman L O'Connell R Casavant BP, et al. : Selective nucleic acid removal via exclusion (SNARE): Capturing mRNA and DNA from a single sample. Anal Chem 85:9764-9770, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Annala M Vandekerkhove G Khalaf D, et al. : Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov 8:444-457, 2018 [DOI] [PubMed] [Google Scholar]
- 37.Tibshirani R Hastie T Narasimhan B, et al. : Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA 99:6567-6572, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Céraline J Cruchant MD Erdmann E, et al. : Constitutive activation of the androgen receptor by a point mutation in the hinge region: A new mechanism for androgen-independent growth in prostate cancer. Int J Cancer 108:152-157, 2004 [DOI] [PubMed] [Google Scholar]
- 39.Joseph JD Lu N Qian J, et al. : A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov 3:1020-1029, 2013 [DOI] [PubMed] [Google Scholar]
- 40.Korpal M Korn JM Gao X, et al. : An F876L mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov 3:1030-1043, 2013 [DOI] [PubMed] [Google Scholar]
- 41.Li Y Alsagabi M Fan D, et al. : Intragenic rearrangement and altered RNA splicing of the androgen receptor in a cell-based model of prostate cancer progression. Cancer Res 71:2108-2117, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viswanathan SR Ha G Hoff AM, et al. : Structural alterations driving castration-resistant prostate cancer revealed by linked-read genome sequencing. Cell 174:433-447.e19, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson D Van Allen EM Wu YM, et al. : Integrative clinical genomics of advanced prostate cancer. Cell 161:1215-1228, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quigley DA Dang HX Zhao SG, et al. : Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 174:758-769.e9, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dang HX Chauhan PS Ellis H, et al. : Cell-free DNA alterations in the AR enhancer and locus predict resistance to AR-directed therapy in patients with metastatic prostate cancer. JCO Precis Oncol 4:680-713, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu R Lu C Mostaghel EA, et al. : Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res 72:3457-3462, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu Z Chen S Sowalsky AG, et al. : Rapid induction of androgen receptor splice variants by androgen deprivation in prostate cancer. Clin Cancer Res 20:1590-1600, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao SG Chen WS Li H, et al. : The DNA methylation landscape of advanced prostate cancer. Nat Genet 52:778-789, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asangani IA Wilder-Romans K Dommeti VL, et al. : BET bromodomain inhibitors enhance efficacy and disrupt resistance to AR antagonists in the treatment of prostate cancer. Mol Cancer Res 14:324-331, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welti J Sharp A Yuan W, et al. : Targeting bromodomain and extra-terminal (BET) family proteins in castration-resistant prostate cancer (CRPC). Clin Cancer Res 24:3149-3162, 2018 [DOI] [PubMed] [Google Scholar]
- 51.Arora VK Schenkein E Murali R, et al. : Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 155:1309-1322, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Individual participant data that are included in the results reported in the manuscript will be available after deidentification beginning 6 months and ending 36 months following publication. Researchers must provide a methodologically sound proposal, and requestors may need to sign a data access agreement. Proposals should be directed to the corresponding author.