Abstract
The full-length versions of the Ras-specific exchange factors Ras-GRF1 (GRF1) and Ras-GRF2 (GRF2), which are expressed in brain and a restricted number of other organs, possess an ionomycin-dependent activation of Erk mitogen-activated protein kinase activity in 293T cells (C. L. Farnsworth et al., Nature 376:524–527, 1995; N. P. Fam et al., Mol. Cell. Biol. 17:1396–1406, 1996). Each GRF protein contains a Dbl homology (DH) domain. A yeast two-hybrid screen was used to identify polypeptides that associate with the DH domain of GRF1. In this screen, a positive cDNA clone from a human brain cDNA library was isolated which consisted of the GRF2 DH domain and its adjacent ilimaquinone domain. Deletion analysis verified that the two-hybrid interaction required only the DH domains, and mutation of Leu-263 to Gln (L263Q) in the N terminus of the GRF1 DH domain abolished the two-hybrid interaction, while a cluster of more C-terminally located mutations in the DH domain did not eliminate the interaction. Oligomers between GRF1 and GRF2 were detected in a rat brain extract, and forced expression of GRF1 and GRF2 in cultured mammalian cells formed homo- and hetero-oligomers. Introduction of the L263Q mutation in GRF1 led to a protein that was deficient in oligomer formation, while GRF1 containing the DH cluster mutations formed homo-oligomers with an efficiency similar to that of wild type. Compared to wild-type GRF1, the focus-forming activity on NIH 3T3 cells of the GRF1 DH cluster mutant was reduced, while the L263Q mutant was inactive. Both mutants were impaired in their ability to mediate ionomycin-dependent Erk activity in 293T cells. In the absence of ionomycin, 293T cells expressing wild-type GRF1 contained much higher levels of Ras-GTP than control cells; the increase in Erk activity induced by ionomycin in the GRF1-expressing cells also induced a concomitant increase in Raf kinase activity, but without a further increase in the level Ras-GTP. We conclude that GRF1 and GRF2 can form homo- and hetero-oligomers via their DH domains, that mutational inactivation of oligomer formation by GRF1 is associated with impaired biological and signaling activities, and that in 293T cells GRF1 mediates at least two pathways for Raf activation: one a constitutive signal that is mainly Ras-dependent, and one an ionomycin-induced signal that cooperates with the constitutive signal without further augmenting the level of GTP-Ras.
Ras GTPases, which play a pivotal role as transducers of various mitogenic and differentiation signals, function as molecular switches, cycling between an inactive GDP-bound state and an active GTP-bound state (33). Ras is negatively regulated by GTPase-activating proteins (ras GAPs) that stimulate hydrolysis of GTP-Ras to GDP-Ras. The conversion of the GDP-bound form into the active form is stimulated by Ras-specific guanine nucleotide exchange factors (GNEFs), such as Ras-GRF (GRF, also known previously as CDC25Mm) (10, 44), Sos (45), and Ras-GRP (13, 48). GNEFs function by inducing release of bound GDP from Ras, which results in the rapid binding of GTP, because the concentration of free GTP is much higher than that of free GDP, and Ras has a greater affinity for GTP than for GDP (31). Ras contains several direct downstream targets, including Raf, which in turn activate the Mek and Erk mitogen-activated protein (MAP) kinases (7).
Mammals contain two closely related sos genes, sos1 and sos2 (4), as well as two grf genes which encode homologous proteins, GRF1 and GRF2, respectively (17). While Sos1 and Sos2 are ubiquitously expressed, full-length GRF1 and GRF2 are primarily brain specific, although the full-length protein and various smaller forms have also been observed in other tissues (17, 21, 27, 43, 44, 50). Functionally, GRF1 has been implicated in synaptic transmission and the formation of long-term memory (5), in agreement with its presence in synaptic junctions (47). In mice grf1 has been shown to be imprinted, with only the paternal gene being expressed (40). Animals lacking detectable GRF1 protein are viable but grow more slowly than controls, presumably because of a hypothalamic defect, which is associated with low levels of circulating insulin-like growth factor 1 (30).
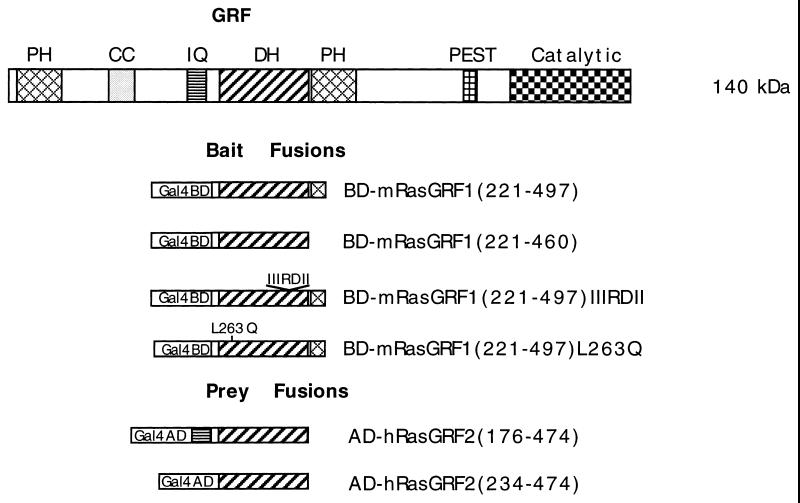
Full-length GRF1 is a 140-kDa protein with many motifs common to other signaling molecules (Fig. 1). In addition to its C-terminally located Ras-catalytic domain, which is responsible for the stimulation of the guanine nucleotide exchange on Ras, GRF contains an N-terminal pleckstrin homology (PH) domain, a coiled-coil (CC) motif, an ilimaquinone (IQ) motif, a Dbl homology (DH) domain adjacent to a second PH domain, and a PEST motif. The N-terminal PH domain of GRF1 has been shown to bind the βγ subunit of heterotrimeric G proteins in vitro (49), and Mattingly and Macara have reported a phosphorylation-dependent activation of GRF1 by muscarinic receptors through the βγ subunit of a heterotrimeric G protein (37). The influx of calcium in human 293T cells, via the calcium ionophore ionomycin, has also been shown to activate GRF1, as measured primarily by an increased Erk1 activity that can be suppressed by a dominant inhibitory Ras mutant (19). The calcium-dependent activation is associated with the binding of calmodulin to the IQ motif, which acts cooperatively with the other motifs in the N terminus of GRF1, including both PH domains as well as the CC and DH domains (6, 24). A PEST-like region renders the protein susceptible in vitro to cleavage by calpain, although it is not clear if this cleavage also occurs in vivo (2). Ionomycin-dependent Erk1 activation has also been reported for GRF2, which is structurally similar to GRF1 (17).
FIG. 1.
Schematic representation of the domain structure of GRF1 and of the baits and preys used in the two-hybrid interactions. The various abbreviations are as defined in the text; PEST is the protein instability motif, and Catalytic refers to the Ras guanine nucleotide exchange domain. In the bait fusions, Gal4BD represents the Gal4-binding domain (BD), while Gal4AD represents the Gal4 activation domain (AD) in the prey fusions. The numbers in parentheses refer to the corresponding amino acid residues of full-length mGRF1 encoded in the bait fusions and of full-length hGRF2 in the prey fusions. BD-mGRF1(221–497) is the starting bait fusion used in the two-hybrid screen. BD-mGRF1(221–460) lacks the PH domain codons in BD-mGRF1(221–497). BD-mGRF(221–497)IIIRDII harbors a cluster of substitutions in codons 394 to 400, while BD-mGRF1(221–497)L263Q contains a point mutation (see the text). AD-hGRF2(176–474) is the insert from clone pGAD10.56 obtained in the two-hybrid screen. In AD-hGRF2(234–474), the IQ motif codons have been deleted from the hGRF2 insert obtained in the two-hybrid screen.
The precise function of the GRF1 DH domain has not been determined. The c-Dbl oncoprotein is the prototype for signaling proteins that contain a DH domain (11). c-Dbl acts as a GNEF for two Rho family GTPases, Cdc42 and RhoA (39). Members of the Rho family of GTPases, which include Rac in addition to CDC42 and Rho, play important roles in mediating various cytoskeletal reorganizations in cells (52). The DH domain of c-Dbl is responsible for its exchange activity in vitro (28). It is believed that other Dbl family members also serve as GNEFs for one or more Rho family GTPases, although this ability has been shown for only a subset of DH domains (39). Mouse GRF2 has been recently shown to display in vitro exchange activity toward Rac (18). However, analogous attempts to show that GRF1 displays exchange activity towards any of the known Rho family GTPases have thus far yielded negative results (11a, 24).
In this study, we included the DH domain of GRF1 as a bait in a yeast two-hybrid screen to identify protein(s) that may interact with this region. We report that the screen unexpectedly identified the DH domain of GRF2 as functionally interacting with the DH domain of GRF1 and that, in mammalian cells, GRF1 and GRF2 formed homo-oligomers as well as hetero-oligomers with each other. We also observed a calcium induced GRF1-dependent Erk activation that was associated with increased Ras activity but, surprisingly, was not correlated with a further increase in GTP-Ras. A point mutation in the DH domain reduced each of these GRF1-dependent activities, as well as the GRF1-dependent cell transformation.
MATERIALS AND METHODS
Plasmid constructions.
The construction of the bait for the two-hybrid screen was as follows: a 0.8-kb PCR product encoding the Dbl homology domain and flanking regions of mGRF1 (amino acids 221 to 497) was synthesized by using Pfu DNA polymerase (Stratagene) with the primers JD79 (GGGGATCCTGTGCCGGCGAAAGTGGAAG) and JD80 (CCGTCGACTCAGAAACACTGGCGCTCACCCTC). The underlined nucleotides indicate a BamHI site and a SalI site, respectively. The fragment was subcloned into pAS1 by using these enzymes to generate pAS1GRFdbl. This plasmid contains the ADH promoter that directs the synthesis of a fusion between the DNA-binding domain of GAL4 (amino acids 1 to 147) and the Dbl homology domain of mGRF1. The mutation changing a leucine residue to a glutamine at position 263 of mGRF1 was created by overlap PCR according to standard techniques. Constructs were verified by DNA sequencing by using Sequenase 2.0 (Amersham). Expression of Gal4-binding domain fusion proteins in yeast strain Y190 was verified by immunoblotting with anti-GAL4 antibodies (Upstate Biotechnology, Inc.). For expression in mammalian cells, full-length wild-type or mutated grf genes were cloned into pEFP (41). For expression as glutathione S-transferase (GST) fusion proteins in mammalian cells, grf genes were cloned into pEBG (obtained from Silvio Gutkind, National Institutes of Health).
Two-hybrid screening.
The yeast strain Y190 and plasmids pAS1 and pSE1112 (12) were provided by S. Elledge (Baylor College, Houston, Tex.). A human brain cDNA library in the pGAD10 vector, used in the two-hybrid screen, was purchased from Clontech. pGAD10 contains the ADH promoter expressing the GAL4 transactivation domain (amino acids 768 to 881). The yeast reporter strain Y190 harbors two reporter gene constructs. Two-hybrid interactions activate the transcription of the HIS3 gene, allowing screening for growth in the absence of histidine, and of the lacZ gene, allowing screening for β-galactosidase activity. Y190 was cotransformed with pAS1GRFdbl and the pGAD10-human brain cDNA library and plated at a density of 5 × 104 colonies per plate on synthetic minimal medium lacking leucine and tryptophan (for plasmid selection) and histidine but containing 25 mM 3-aminotriazole. The plates were incubated at 30°C for 5 to 7 days. Colonies that grew on these plates were transferred to fresh selective plates and assayed for β-galactosidase activity by a filter assay. Liquid β-galactosidase assays were conducted according to standard procedures (26).
GRF2 GenBank accession number and analysis of tissue samples for GRF mRNA and protein.
The GenBank accession number for human GRF2 is AF023130. Two cDNA templates were prepared by PCR for the detection of GRF1 and GRF2, respectively, in various human tissues. For hGRF1 the sequence of the probe corresponded to amino acids Asn-780–Leu-902. For hGRF2 the probe corresponded to Ser-763–Ala-879. The probes, chosen from regions that bear little homology between the two proteins, were labeled with [α-32P]dCTP by random priming by using the Rediprime kit (Amersham) according to the manufacturer’s instructions and hybridized to the membrane in ExpresHyb solution (Clontech) at 68°C for 1 h. Subsequently, the blots were washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.05% sodium dodecyl sulfate (SDS) buffer at room temperature, washed in 0.1× SSC–0.1% SDS at 55°C, and autoradiographed. Northern blots, containing approximately 2 μg of the indicated poly(A)+ RNA in each lane, were purchased from Clontech (blots MTN-1 and MTN-2). Blots were first hybridized with the hGRF2 probe, stripped by incubation in 0.5% SDS at 95°C for 10 min, hybridized with the hGRF1 probe, stripped again, and finally hybridized to an actin probe to verify equal loading of poly(A)+ RNA in each lane.
For hGRF2 antibody preparation, a peptide corresponding to amino acid residues 183 to 200 of human GRF2, which was chosen because it shared only one amino acid residue with GRF1, was coupled to bovine serum albumin (BSA) by using the Imject Activated Immunogen conjugation kit (Pierce). The BSA-conjugated peptide was used to immunize rabbits by subcutaneous injection. Preliminary experiments indicated that it detects human and rodent GRF2 protein but not rodent GRF1 protein. For detection of GRF2, 75 μg of SDS-solubilized proteins from human tissues (brain, kidney, lung, liver, and heart; all from Clontech) were fractionated by SDS-polyacrylamide gel electrophoresis (PAGE) and electroblotted onto polyvinylidine difluoride (PVDF) membranes. Membranes were incubated with anti-GRF2 serum and then with 125I-conjugated goat-anti rabbit immunoglobulin G (ICN). Autoradiography was carried out at −70°C for 2 days.
Analysis of homo- and hetero-oligomerization of GRF1 and GFR2.
NIH 3T3 and 293T cells were transiently transfected with DNA constructs encoding GRF1 or GRF2 with or without a GST tag. At 24 h after transfection, NIH 3T3 cells were metabolically labeled with [35S]methionine (150 μCi/ml; NEN-Dupont) in methionine-free medium for 2 h and chased with Dulbecco modified Eagle medium (DMEM) containing 5% fetal bovine serum (FBS) for another 3 h. The cells were lysed with nondenaturing lysis buffer after two washes with cold phosphate-buffered saline (PBS). Equal amounts of labeled protein, estimated by obtaining trichloroacetic acid-precipitable counts, were immunoprecipitated overnight by anti-GRF antisera or precipitated with glutathione-Sepharose beads (Pharmacia). The pellets were washed twice with nondenaturing lysis buffer, twice with HNTG (20 mM HEPES [pH 7.0], 500 mM NaCl, 0.1% Triton, 10% glycerol), and solubilized in Laemmli sample buffer. After separation by 6 to 13% gradient SDS-PAGE, proteins were detected by autoradiography. For transfected 293T cells, immunoprecipitation followed by immunoblotting was used for the analysis of GRF oligomers. For analysis of more than one protein, blots were stripped according to the manufacturer’s instructions (Amersham) and reprobed with the appropriate antibody.
Whole brains from several 2-day-old Sprague-Dawley rats (Taconic Farms) were homogenized in ice-cold extraction buffer (20 mM Tris-HCl [pH 8.0], 1% Nonidet P-40 [NP-40], 10 nM EGTA, 5 mM MgCl2, 20 mM β-glycerophosphate, 1 mM Na3 VO2, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 10 μg of leupeptin per ml, 10 μg of aprotinin per ml) by 10 strokes with a glass homogenizer. The crude lysates were first cleared by centrifugation at 12,000 × g for 15 min at 4°C and then centrifuged at 100,000 × g for 30 min at 4°C. The supernatants were collected, and the protein concentration was estimated by the BCA kit (Pierce). For the detection of complex formation between GRF1 and GRF2 in the brain extract, antibodies specific for GRF1 (9), rodent GRF2 (Transduction Laboratories), or human GRF2 as described, which in preliminary experiments were found not to recognize the heterologous GRF isoform, were used to obtain high sensitivity and specificity. In the assay, the 4 to 5 mg of extract was first precleared with protein A-Sepharose by rotating for 1 h at 4°C. One-quarter of the extract was then used as a control, i.e., anti-GRF1 immunoprecipitation followed by anti-GRF1 immunoblotting. The remaining three-quarters of the extract was immunoprecipitated with the anti-GRF2 antibodies, followed by immunoblotting with anti-GRF1. The immunocomplexes were washed twice with extraction buffer, twice with HNTG containing 0.5 M NaCl, and twice with low-salt HNTG (containing 150 mM NaCl instead of 500 mM). The beads were then resuspended with sample loading buffer, separated by gradient SDS-PAGE, and immunoblotted as described above.
Cell culture, transfection, and focus formation assays.
NIH 3T3 (clone 7), COS-7, and HEK 293T (293T) cells were maintained in DMEM supplemented with 10% FBS at 37°C in a humidified 5% CO2 atmosphere. NIH 3T3 cells were transfected with 0.4 μg of pEFP vector encoding wild-type or mutant GRF1 or pGV16-MyrSos1 (41) with calcium precipitation as described previously (53). Foci were counted after 14 days.
Calmodulin immunocomplex formation with GRF1 and GRF2.
pEFP vectors encoding GRF1 or GRF2 were transiently transfected into 293T cells by using Lipofectamine Plus (Life Technologies) according to the manufacturer’s instructions and analyzed 48 h after transfection. The day following transfection, cells were serum deprived for 18 to 20 h and stimulated with 5 μM of ionomycin (Sigma) at 37°C for 5 min. Cells were solubilized by nondenaturing lysis buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 10% glycerol, 1% Triton X-100, 1 mM EDTA, 10 mM Na3VO4, 10 mM NaF, 1 mM PMSF, aprotinin [10 μg/ml], leupeptin [10 μg/ml]). Equal amounts of protein from the cleared cell lysates were immunoprecipitated with anti-GRF1- or anti-GRF2-specific antiserum for 2 h on ice. Then, 50 μl of protein A-Sepharose slurry was added and rotated at 4°C for at least 3 h. Immunocomplexes were then washed twice with nondenaturing lysis buffer and twice with HNTG and then denatured in Laemmli sample buffer. After resolution by 6 to 13% gradient SDS-PAGE and transfer to PVDF membranes, immunoblotting was performed to detect the expression of GRF1 and GRF2 by using cognate anti-serum at a 1:1,000 dilution. Calmodulin that had coimmunoprecipitated with the anti-GRF sera was examined by using anti-calmodulin monoclonal antibody (UBI) at a 1:2,000 dilution. For each blot, horseradish peroxidase-conjugated anti-rabbit or anti-mouse immunoglobulin G (Amersham) was used for the second reaction at a 1:10,000 dilution. Immunocomplexes were visualized by enhanced chemiluminescence with an ECL Kit from Amersham.
Erk and Raf-1 kinase assays.
NIH 3T3 cells or 293T cells were transiently cotransfected with pcDNA3-HA-ERK2 (a gift from Silvio Gutkind, National Institutes of Health) and pEFP-GRF1, pEFP-GRF1L263Q, or empty vector, and assayed 2 days after transfection. Transfected cells that had been deprived of serum for 16 to 20 h were either left untreated or were treated with ionomycin (5 μM) for 5 min. For MAP kinase assays, cells were lysed with radioimmunoprecipitation assay buffer (25 mM Tris-HCl [pH 7.5], 300 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 1% Triton X-100, 0.5% NaDOC, 0.1% SDS, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, aprotinin [10 μg/ml], 1 mM PMSF, leupeptin [10 μg/ml]). Protein concentrations were determined by using the BCA kit (Pierce). Equal amounts of protein (100 μg) from cell extracts were immunoprecipitated with anti-HA monoclonal antibody (Babco). After the complexes were washed twice with PBS containing 1% NP-40 and 2 mM Na3VO4, once with buffer 2 (100 mM Tris, [pH 7.5], 0.5 M LiCl2), and once with reaction buffer (12.5 mM MOPS [morpholinepropanesulfonic acid; pH 7.4], 12.5 mM β-glycerophosphate, 7.5 mM MgCl2, 0.5 mM EGTA, 0.5 mM sodium orthovanadate, 0.5 mM NaF), the immunocomplexes were incubated with 50 μl of reaction buffer containing 20 μM ATP, 1 μCi of [γ-32P]ATP (NEN-Dupont), and 2 μg of myelin basic protein (MBP; Upstate Biotechnology, Inc.). After incubation for 20 min at 30°C, kinase reactions were terminated by the addition of 2× Laemmli sample buffer. The samples were then resolved by SDS-PAGE, and the phosphorylated MBP was visualized by autoradiography. For quantitation, dried gels were exposed on Phosphor Screens, and signals were captured on a Molecular Dynamics PhosphorImager and analyzed by using ImageQuant software. Control for equal loading was done by examining exogenous HA-ERK2 proteins by immunoblotting with anti-hemagglutinin (HA) antibody.
For the Raf1 kinase assay, 293T cells were transiently transfected with pEFP-GRF1, pEFP-GRF1L263Q, or empty vector, together with 0.4 μg of c-Raf-1. At 24 h posttransfection, the cells were serum starved overnight and then stimulated with or without 5 μM ionomycin for 5 min at 37°C. The cells were washed twice with ice-cold PBS and lysed by using Gold Lysis Buffer (GLB) containing 20 mM Tris-HCl [pH 7.9], 137 mM NaCl, 5 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM EGTA, 10 mM NaF, 1 mM sodium pyrophosphate, 100 μM β-glycerophosphate, 1 mM leupeptin, aprotinin [10 μg/ml], and 1 mM PMSF. Lysates from 293T cells (180 μg) were immunoprecipitated with 2 μg of Raf-1 antibody (Santa Cruz). The immunocomplexes were washed twice with GLB; once with buffer containing 10 mM HEPES [pH 7.4], 100 mM NaCl, 20 μg of aprotinin per ml, and 0.5% NP-40; and finally with reaction buffer (20 mM Tris [pH 7.4], 20 mM NaCl, 1 mM DTT, 10 mM MgCl2, 1 mM MnCl2). The complexes were then incubated in 40 μl of reaction buffer supplemented with 25 μM PD98059 (RBI), an MEK inhibitor to prevent autophosphorylation, 10 μM ATP, 2 μCi of [γ-32P]ATP, and 100 ng of MEK for 30 min at 30°C. Reactions were stopped and processed as described for the MAP kinase assay.
In vivo analysis of GTP-Ras.
In vivo analysis of GTP-Ras was performed as described previously, with minor modifications (54). Subconfluent NIH 3T3 or 293T cells were transiently transfected with pEFPGRF1, pEFPGRF1L263Q, or empty vector for 24 h, deprived of serum for 16 hours, metabolically labeled with [32P]orthophosphate for 6 h, and stimulated where indicated with 5 μM ionomycin or 20 nM epidermal growth factor (EGF) for 5 min. Cells were rinsed with PBS and lysed in 20 mM Tris-HCl (pH 7.4)–100 mM NaCl–20 mM MgCl2–1% NP-40–0.5% sodium deoxycholate. Ras protein was immunoprecipitated with monoclonal antibody Y13-259 (25), followed with thin-layer chromatography on PEI cellulose plates in 1.3 M LiCl, and analyzed with an AMBIS Radioanalytic Imaging apparatus. Determination of the percentage of GTP (GTP/[GTP + GDP]) bound to Ras was normalized for the moles of phosphate.
RESULTS
Two-hybrid screen identifies a human cDNA insert that encodes the IQ motif and Dbl homology domain of human GRF2.
To identify proteins that interact with the DH domain of GRF1, we employed the yeast two-hybrid system (22) as modified by Durfee et al. (12). As bait for the two-hybrid screen, we used a selectable plasmid, termed pAS1GRFdbl, which encoded a fusion between the Gal4 DNA-binding domain and the DH domain plus flanking regions of mouse (m) GRF1 (amino acids 221 to 497; BD-mGRF1 in Fig. 1). The yeast reporter strain Y190 was cotransformed with pAS1GRFdbl and a selectable human brain cDNA library (Clontech). To select for cells that received a plasmid of each type and also activated the resident His plasmid in the yeast strain, cells were plated on synthetic minimal medium lacking His, Leu, and Trp but containing 25 mM 3-aminotriazole (3-AT) and then incubated at 30°C for 5 to 7 days. The His+ colonies were then screened for activation of β-galactosidase gene activity in the yeast cells, an activity which should also depend on a two-hybrid interaction. Of 2.2 × 106 yeast transformants, 180 grew in the absence of histidine, and 6 of these transformants were positive for β-galactosidase activity as determined by filter assay (not shown). The inserts from these six plasmids were digested by several restriction enzymes, including RsaI and TaqI. This yielded identical patterns for each of the six inserts (not shown). These plasmids were therefore considered identical, and one, designated pGAD10.56, was chosen for further analysis.
To verify the specificity of the interaction between pGAD10.56 and the mGRF1 bait, pAS1GRFdbl, we cotransformed the yeast reporter strain with pGAD10.56 and a number of different plasmid baits expressing various GAL4-binding domain fusion proteins, including a fusion encoding mGRF1 amino acids 464 to 604 (Table 1). Among these baits, only cotransformation of pGAD10.56 with pAS1GRFdbl yielded yeast cells that could grow on minimal medium in the presence of 3-AT and express β-galactosidase activity.
TABLE 1.
Specificity of the interaction of the DH domain of hGRF2 [AD-hGRF2(176–474)] with the DH domain of mGRF1 [BD-mGRF1(221–497)] in the yeast two-hybrid systema
| GAL4 transactivation domain fusion | GAL4 DNA-binding domain fusion | Growth in the presence of 3-AT | β-galactosidase activity |
|---|---|---|---|
| AD-SNF1 | BD-mGRF1(221–497) | − | − |
| AD-hGRF2(176–474) | BD-mGRF1(221–497) | + | + |
| AD-hGRF2(176–474) | BD-mGRF1(464–604) | − | − |
| AD-hGRF2(176–474) | BD-RasC186S | − | − |
| AD-hGRF2(176–474) | BD-SNF4 | − | − |
| AD-hGRF2(176–474) | BD-NF1 | − | − |
Yeast strain Y190 was cotransformed with a GAL4 transactivation domain plasmid and a GAL4 DNA-binding domain plasmid and analyzed for growth in the absence of His and for β-galactosidase activity by a filter assay. AD-hGRF2(176–474) has the IQ and DH domains of hGRF2, expressed from pGAD10.56; BD-mGRF1(221–497) has the DH domain plus flanking amino acids of mGRF1, expressed from pAS1GRFdbl; BD-mGRF1(464–604) has the PH domain 2 plus flanking amino acids of mGRF1 expressed from pAS1GRFPH2; BD-RasC186S has human Ras-p21 with Cys-186 replaced by Ser, expressed from pJD235; BD-SNF1 was expressed from pSE1111 and BD-SNF4 was expressed from pSE1112 (12); BD-NF1 encodes amino acids 1543 to 1875 of neurofibromin (36).
Having determined that the interaction was specific, we obtained the sequence of the human cDNA insert of plasmid pGAD10.56 and found a partial cDNA predicted to encode a polypeptide of 299 amino acids, which was then compared against a protein database by using the basic local alignment sequence tool (1). Unexpectedly, a strong homology was found to the GRF DH domain and IQ motif, with the closest homology being to mGRF2 (17) [AD-hGRF2(176–474) in Fig. 1]. Comparison of the polypeptide sequence encoded by pGAD10.56 with the corresponding region of mGRF2 showed 97% identity, versus 81% identity with mGRF1 (Fig. 2). This result strongly suggested that the human cDNA insert of pGAD10.56 corresponds to a portion of human (h) GRF2.
FIG. 2.
Comparison of the sequences of mouse (m) and human (h) forms of Ras-GRF1 (GRF1) and Ras-GRF2 (GRF2). Alignment is given in comparison with hRas-GRF2 (hGRF2). Going from the N terminus to the C terminus, the boxes indicate the PH, IQ, DH-PH, and the Ras catalytic domains, respectively (see Fig. 1). An asterisk indicates sequence identity with respect to hRas-GRF2; a dash indicates a deletion. The underlined sequence of mRas-GRF1 (mGRF1) corresponds to the bait used in the two-hybrid screen (residues 221 to 497 of mGRF1). The underlined sequence of hGRF2 corresponds to the insert of clone pGAD10.56, the sequence trapped in the two-hybrid screen (residues 176 to 474 of hRas-GRF2). The GenBank accession numbers are as follows: L20899 (mGRF1), U67326 (mGRF2), L26584 (hGRF1), and AF023130 (hGRF2). The following regions are not highly conserved between GRF1 and GRF2: (i) the most N-terminal PH domain (and to a lesser extent the PH domain C terminal to the DH domain); (ii) the region corresponding to residues Ser-180–Glu-200 of hGRF2, in which only 6 of 21 residues are identical between mGRF1 and hGRF2 (this region was used to generate a GRF2-specific peptide antibody); and (iii) the region corresponding to residues Thr761–Pro-878 of hGRF2, in which only 20 of 118 residues are identical between hGRF2 and hGRF1. This region, which is much shorter in mGRF2, was chosen to generate cDNA probes for Northern analysis of human tissues.
Cloning of the full-length cDNA encoding hGRF2.
To confirm that the cDNA was part of the putative hGRF2, the full-length human cDNA was isolated by 5′ rapid amplification of cDNA ends (RACE) and 3′ RACE, performed with a human hippocampal cDNA (Clontech) as a template. The complete open reading frame of the cDNA was found to consist of 3,711 nucleotides encoding a polypeptide of 1237 amino acids, with a predicted molecular mass of 136 kDa (Fig. 2). Comparing the polypeptide sequence with that of mGRF2 confirmed its identity as hGRF2. The overall sequence identity of the two proteins is 90%, versus a 64% identity with mGRF1. Northern analysis of human tissues confirmed a previous report by Fam et al. (17) that GRF2 is more widely expressed than GRF1, and we also found that mRNA for GRF2 was most abundant in the brain, followed by heart tissue and several other tissues such as lung, pancreas, and kidney tissues (data not shown). By Western blot analysis, full-length hGRF2 protein was detected in brain and lung tissue (not shown).
Mapping the interacting regions.
The pGAD10.56 prey, which consists of hGRF2 amino acids 176 to 474, contains the IQ and DH domains, while the pASGRFdbl mGRF1 bait consists of mGRF1 amino acids 221 to 497, which represent the DH domain and the N-terminal portion of the adjacent PH domain (Fig. 1). To determine whether sequences other than the respective DH domains were required for the two-hybrid activity, the IQ domain (amino acids 176 to 233) was deleted from the hGRF2 prey [AD-hGRF2(234–474) in Fig. 1] and the PH domain sequences (amino acids 461 to 497) were deleted from the mGRF1 bait [BD-mGRF1(221–460)], and their functions were examined in the two-hybrid assay. Deletion of the IQ domain from hGRF2 [AD-hGRF2(234–474)] did not prevent the interaction with the mGRF1 bait, while removal of the N-terminal part of the PH domain from the mGRF1 bait [BD-mGRF1(221–460)] resulted in β-galactosidase activity that was more than twofold higher than that of the wild type (Table 2). These results demonstrate that the interaction, as measured in the two-hybrid assay, requires only the DH domain of each peptide.
TABLE 2.
DH-dependent oligomerization of GRF in the yeast two-hybrid systema
| GAL4 transactivation domain fusion | GAL4 DNA-binding domain fusion | Growth in the presence of 3-AT | β-Galactosidase activity |
|---|---|---|---|
| AD-SNF1 | BD-mGRF1(221–497) | − | 0.01 ± 0.002 |
| AD-hGRF2(176–474) | BD-MGRF1(221–497) | + | 0.19 ± 0.02 |
| AD-hGRF2(176–474) | BD-mGRF1(221–497)IIIRDII | + | 0.14 ± 0.04 |
| AD-hGRF2(176–474) | BD-mGRF1(221–497)L263Q | − | ND |
| AD-hGRF2(176–474) | BD-mGRF1(221–460) | + | 0.48 ± 0.04 |
| AD-hGRF2(234–474) | BD-mGRF1(221–497) | + | 0.17 ± 0.02 |
Yeast strain Y190 was cotransformed with a GAL4 transactivation domain plasmid and a GAL4 DNA-binding domain plasmid and analyzed for growth in the absence of His. Cells that grew in the absence of tryptophan, leucine, and histidine but in the presence of 25 mM 3-amino-1, 2, 4-triazole (3-AT) were then tested for β-galactosidase activity (26). AD-hGRF2(234–474) concerns the DH domain without IQ sequences. BD-mGRF1(221–497)IIIRDII and BD-mGRF1(221–497)L263Q concern point mutations in the DH domain as described in the text. BD-mGRF1(221–460) concerns the DH domain without PH sequences. β-Galactosidase activities represent the average for six colonies from two independent transformations. The β-galactosidase activity for the AD-SNF1/BD-mGRF1(221–497) pair was obtained from colonies that grew on an accompanying plate that selected for growth in the absence of tryptophan and leucine only. ND, not determined.
To confirm the essential nature of the mGRF1 DH domain for the interaction, substitution mutations were introduced in either of two regions within the mGRF1 DH domain. Both regions are highly conserved between DH domains of various Dbl-related proteins (3, 46). In one case, a point substitution L263Q was introduced into the most N-terminally located structurally conserved region of the DH domain [BD-mGRF1(221–497)L263Q in Fig. 1]. In the other, a cluster mutation was introduced in the third structurally conserved region (3), replacing residues (394)LTLHELL(400) with residues (394)IIIRDII(400) [BD-mGRF1(221–497)IIIRDII in Fig. 1]. In oncogenic Dbl, an analogous mutation cluster greatly reduces its in vitro exchange activity as well as its transforming activity on NIH 3T3 cells (28), and this cluster mutation in rat GRF1 attenuated its capacity to respond to the ionomycin-dependent activation of Erk (24). However, engineering the DH cluster mutation in the mGRF1 bait did not prevent its interaction with the hGRF2 prey (Table 2). Quantitative analysis of the β-galactosidase assay revealed that, compared with the wild-type mGRF1 bait, the interaction between the cluster mutant and the hGRF2 IQ-DH prey was only marginally lower. By contrast, introduction of the L263Q substitution in the mGRF1 prey abolished the two-hybrid interaction (Table 2).
Oligomerization occurs in mammalian cells.
We wished to determine whether the interaction between the DH domains of GRF1 and GRF2 DH detected by the yeast two-hybrid assay also occurs with the full-length proteins in mammalian cells, whether hetero-oligomerization occurs between GRF1 and other proteins that contain DH regions, and whether GRF1 and GRF2 form oligomers with themselves. Initially, we transferred the hGRF2 coding sequences from the pGAD10.56 prey, tagged with a FLAG epitope, to a mammalian expression vector and cotransfected this construct with full-length GST-mGRF1 or GST alone into COS-7 cells. Immunoprecipitation with antibodies against GST showed a specific coprecipitation of the GRF2 construct, demonstrating that the interaction identified in the yeast two-hybrid assay could be detected as complex formation in mammalian cells (data not shown). These results led us to determine whether the full-length GRF1 and GRF2 proteins form homo-oligomers and/or hetero-oligomers. To carry out these analyses, additional plasmids were constructed for full-length mGRF1 containing either the L263Q mutation or the DH cluster mutation, full-length wild type hGRF2, and full-length hGRF2 fused to GST (GST-GRF2).
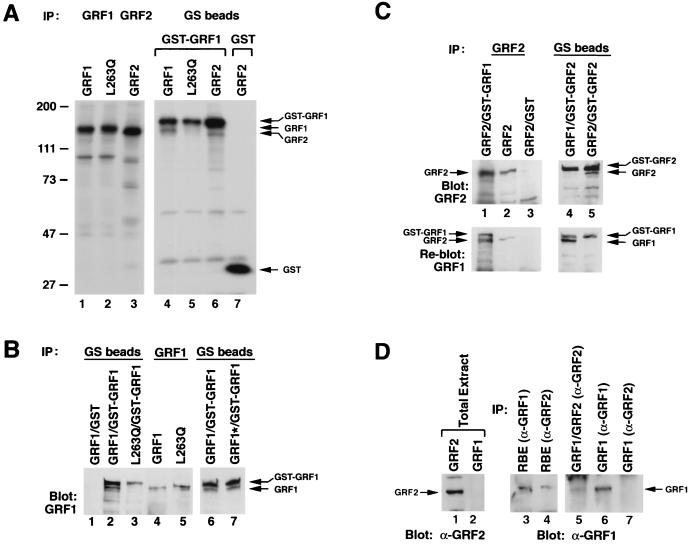
Oligomer formation with full-length proteins was first examined in NIH 3T3 cells, which were transiently cotransfected with (i) GST-GRF1 and (ii) wild-type mGRF1, the L263Q mGRF1 mutant, or wild-type GRF2 (Fig. 3A). Metabolically labeled NIH 3T3 lysates were precipitated with glutathione-Sepharose beads, which recognize GST but not GRF (Fig. 3A, lane 7). As expected, GST-GRF1 signals were present in the precipitates from GST-GRF1 transfectants (Fig. 3A, lanes 4 to 6). In addition to GST-GRF1, the precipitates from the cells cotransfected with GST-GRF1 and wild-type GRF1 (lane 4) or wild-type GRF2 (lane 6) contained GRF1 and GRF2, respectively (note that GRF1 [lane 4] and GRF1 immunoprecipitates [lane 1] migrated slightly more slowly than GRF2 [lane 6] and GRF2 immunoprecipitates [lane 3]). These results indicated that complexes were formed between GST-GRF1 and wild-type GRF1 or wild-type GRF2. By contrast, precipitates from cells cotransfected with GST-GRF1 and the L263Q GRF1 mutant contained almost no mutant protein (lane 5), although control immunoprecipitations with GRF1 and GRF2 antibodies indicated the transfectants contained similar amounts of wild-type or mutant protein (Fig. 3A, lanes 1 to 3). Similar results were also obtained in COS-7 cells (data not shown).
FIG. 3.
Formation of GRF1 and GRF2 oligomers in mammalian cells. (A) GRF oligomerization occurs in NIH 3T3 cells between GST-GRF1 and wild-type GRF1 or wild-type GRF2, but it is deficient between GST-GRF1 and the L263Q GRF1 mutant. Metabolically labeled NIH 3T3 cells transiently expressed mGRF1, mGRF1L263Q, and hGRF2, with or without GST-mGRF1 or GST, as indicated. Cell lysates were immunoprecipitated with antisera to GRF1 (lanes 1 and 2), GRF2 (lane 3) or precipitated with glutathione-Sepharose (GS) beads (lanes 4 to 7). The arrows indicate the location of GRF1 (lanes 4 and 5) or GRF2 (lane 6) coprecipitated with GST-GRF1 (lanes 4 to 6) and the location of GST (lane 7). (B) Wild-type GRF1 and the DH cluster mutant form oligomers in 293T cells, but oligomerization is deficient with the L263Q mutant. Transiently transfected cells expressing wild-type (lanes 1, 2, 4, and 6) or mutant GRF1 (L263Q in lanes 3 and 5; cluster mutant [GRF1*] in lane 7), with or without coexpression of GST-GRF1, were precipitated by glutathione-Sepharose (GS) beads or GRF antibodies as indicated, followed by anti-GRF1 blotting. The designated oligomerized proteins are marked with arrows. (C) Oligomerization in 293T cells. In lanes 1 to 3, lysates from cells expressing GRF2 with or without coexpression of GST-GRF1 or GST were immunoprecipitated and blotted with anti-GRF2. In lanes 4 and 5, lysates from cells coexpressing GST-GRF2 and GRF1 or GRF2 were precipitated with glutathione-Sepharose (GS) beads followed by anti-GRF2 blotting. In the bottom panels, the anti-GRF2 blots were (incompletely) stripped and were reprobed with GRF1 antibodies. The designated oligomerized proteins are marked with arrows. (D) A rat brain extract (RBE) contains GRF1-GRF2 oligomers. Lanes: 1 and 2, extracts from 293T cells expressing GRF2 and GRF1, respectively, Western blotted with anti-GRF2 antibodies; 3 and 4, rat brain extracts immunoprecipitated with anti-GRF1 and anti-GRF2, respectively; 5, 293T cells coexpressing GRF1 and GRF2 immunoprecipitated with anti-GRF2 antibodies; 6 and 7, 293T cells expressing GRF1 immunoprecipitated with anti-GRF1 or anti-GRF2 antibodies, respectively.
GRF oligomerization was also analyzed in unlabeled transiently transfected 293T cells, by precipitation with glutathione-Sepharose beads, followed by Western blotting with antibodies specific for GRF1 or GRF2 (Fig. 3B and C, lanes 3 to 5), or by immunoprecipitation and Western blotting (Fig. 3C, lanes 1 and 2). Lysates from 293T cells cotransfected with GST-GRF1 and wild-type GRF1 formed a readily detectable complex (Fig. 3B, lane 2). This complex required GRF1 in the GST fusion, as the association did not occur in cells cotransfected with GST and wild-type GRF1 (lane 1). As in the NIH 3T3 cells, the L263Q GRF1 mutant was deficient in its association with GST-GRF1 (lane 3), although the cells expressed similar levels of wild-type or L263Q mutant protein (lanes 4 and 5). In contrast to the impaired complex formation seen with the L263Q GRF1 mutant, the efficiency of complex formation between GST-GRF1 and the DH cluster mutant (designated GRF1* in lane 7) was similar to that between the wild type and GST-GRF1 (lane 6). Conversely, specific oligomerization was found between GST-GRF2 and either wild-type GRF1 or wild-type GRF2 (Fig. 3C, lanes 3 to 5). Complex formation between GST-GRF1 and GRF2 was also shown by immunoprecipitation with GRF2 antibodies and Western blotting with GRF1 antibodies (Fig. 3C, lanes 1 and 2; note that the GRF2 blot for these lanes was incompletely stripped). In addition, complex formation between wild-type GRF1 and wild-type GRF2 was also shown for cells expressing both proteins (Fig. 3D, lane 5).
Taken together, the data indicate that hetero-oligomers form between GRF1 and GRF2 in mammalian cells and that homo-oligomers composed of GRF1 or of GRF2 can also form. As in the yeast two-hybrid assay, introduction of the DH cluster mutation did not interfere with oligomerization, while introduction of the L263Q mutation led to a protein that was deficient in oligomer formation. It should also be noted that the oligomerization assays underestimate the overall efficiency of oligomerization because they detect only those oligomers that form between proteins that can be distinguished by virtue of their distinct size (as between GST-GRF1 and GRF1 in Fig. 3A and B) or their distinct antigenic properties (as between GRF1 and GRF2 in Fig. 3C). The assays of the cotransfected cells do not identify the two species of homo-oligomers that must also be forming between molecules of the same protein species, such as GRF1 homo-oligomers and GRF2 homo-oligomers in cells cotransfected with plasmids encoding these two proteins.
To determine whether complex formation might occur between GRF1 and less closely related proteins that also contain DH domains, GRF1 was cotransfected with c-Dbl or with Sos1, each of which contain a Dbl domain. However, under conditions where GRF1 formed oligomers with itself or with GRF2, no complexes were seen between GRF1 and c-Dbl or Sos1 (data not shown). Thus, the ability to form such oligomers appears to be restricted to GRF1 and GRF2, whose DH domains are more closely related to each other than to the DH domains of c-Dbl or Sos.
Rat brain extracts contain GRF1-GRF2 oligomers.
The above-described studies indicated that forced expression of GRF1 and GRF2 led to oligomer formation in cultured cells. To determine whether oligomerization between GRF1 and GRF2 occurs in a physiologic context, a whole-brain extract made from postnatal rats was used to examine this possibility, as Northern blots from various areas of human brains indicated that GRF1 and GRF2 mRNA were expressed in most regions of the brain (data not shown). When the rat brain extract was immunoprecipitated with anti-GRF2 antibodies, GRF1 was also coprecipitated, as demonstrated by Western blotting of the immunoprecipitate with anti-GRF1 antibodies (Fig. 3D, lane 4). Several controls were run with this assay to confirm the specificity of the results. Immunoprecipitation with the anti-GRF2 antibodies of transfectants expressing GRF1 did not precipitate GRF1 detectable by Western blotting with the anti-GRF1 antibodies (lane 7), despite the presence of GRF1 in the transfectants (lane 6). In addition, the anti-GRF2 antibodies did not detect GRF1 in a Western blot of extracts from cells expressing GRF1 (lane 2), although the antibodies could readily detect expressed GRF2 (lane 1). We conclude that GRF oligomerization occurs physiologically in at least some parts of the brain.
Biological activity in NIH 3T3 cells.
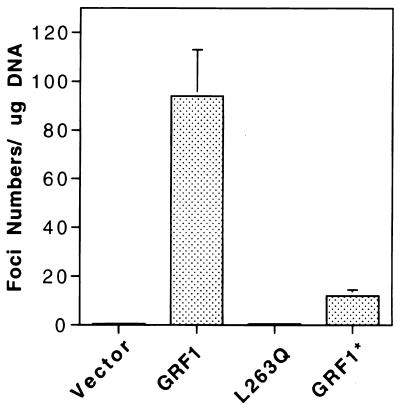
Full-length mGRF1 has been reported previously to induce focus formation in NIH 3T3 cells (9). To determine the effect of the L263Q and DH cluster mutations on this activity, NIH 3T3 cells were transfected with the wild-type gene, the L263Q mutant, or the DH cluster mutant (Fig. 4). The transforming activity of the DH cluster mutant was about 1 order of magnitude lower than that of wild type, while the L263Q mutant was repeatedly transformation defective, although transient transfection indicated that the wild-type and mutant genes expressed similar levels of protein in the cells (Fig. 5A; data shown for wild type and the L263Q mutant). Since the DH cluster mutant formed oligomers with an efficiency similar to that of wild type (Fig. 3B, lanes 6 and 7), whereas the L263Q mutant was deficient in this activity (Fig. 3A, lane 5, and Fig. 3B, lane 3), the cell transformation results support the possibility that GRF oligomerization contributes to the biological activity of the protein. Consistent with their relative transforming activity, the Erk activity in transiently transfected NIH 3T3 cells was highest for the wild-type GRF1 (5.9 arbitrary units versus 1.0 for the vector control), followed by the cluster mutant (4.1 U) and the L263Q mutant (3.4 U) (Fig. 5A; primary data not shown for the cluster mutant). In addition, wild-type GRF1 induced a higher level of GTP-Ras than the L263Q mutant in transiently transfected cells (Fig. 5B).
FIG. 4.
Focal transforming activity of GRF1. NIH 3T3 cells transfected with the indicated GRF1 constructs. The data shown represent the mean of four separate experiments (three readings per experiment). Each bar shows the transforming activity and the standard error.
FIG. 5.
Erk and GTP-Ras in NIH 3T3 cells. Assays were carried out 2 days after transfection of NIH 3T3 cells with the indicated GRF1 plasmid. (A) Erk activity in NIH 3T3 cells. Cells were transiently cotransfected with empty vector, wild-type GRF1 (GRF1), or the L263Q GFR1 mutant (L263Q), along with HA-ERK2. In the upper portion of the panel, the basal Erk activity was determined by precipitating lysates with HA antibody, followed by kinase assay of the immune complex with MBP as the substrate. In the middle portion of the panel, the presence of GRF1 or the mutant was verified by Western blotting of the immunoprecipitates. In the lower portion of the panel, the blots were stripped and probed with the HA antibody to verify the equal expression of HA-Erk2. The arrows show the location of MBP, GRF1, and HA-Erk in the upper, middle, and lower portions, respectively. (B) In vivo measurement of GTP and GDP bound to Ras in transiently transfected NIH 3T3 cells. Cells were metabolically labeled with [32P]orthophosphate, extracts were immunoprecipitated with a Ras-specific monoclonal antibody, and the percentage of GTP-Ras was determined by thin-layer chromatography as described in Materials and Methods.
Ionomycin-dependent stimulation of Erk activity is not associated with an increase in GTP-Ras.
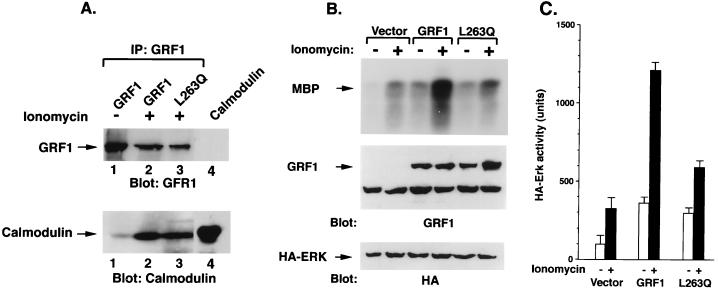
As noted above, treatment of 293T cells expressing rat GRF1 with a calcium ionophore, ionomycin, has been shown to increase the Erk activity in conjunction with calmodulin binding. We confirmed these results for wild-type mouse GRF1 protein in transiently transfected 293T cells (Fig. 6). In the absence of ionomycin treatment, the basal Erk2 activity in 293T cells transfected with wild-type GRF1 was, as expected, higher than in cells transfected with the empty vector, and treatment with ionomycin for 5 min led to a severalfold increase in Erk2 activity (Fig. 6B and C), as well as in calmodulin binding to GRF1 (Fig. 6A, lanes 1 and 2).
FIG. 6.
Calmodulin binding and ERK activity in 293T cells expressing wild-type GRF1 and L263Q mutant. (A) 293T cells were transiently transfected with wild-type GRF1 or the L263Q mutant, serum starved overnight, and either left untreated (−) or treated with ionomycin for 5 min (+). Cell lysates were immunoprecipitated with GRF1 antibodies and blotted with GRF1 antibodies (top portion of the panel) or with a calmodulin antibody (bottom portion of panel). Lane 4 contains only input calmodulin marker. (B) Basal and ionomycin-induced Erk activation in 293T cells transfected with GRF1. Cells were transiently transfected with vector, wild-type GRF1, or the L263Q mutant along with HA-ERK2, serum starved overnight, and either left untreated (−) or treated with ionomycin for 5 min (+). The exogenous ERK activity was determined as described in the text. The cells were processed and analyzed as described for panel A. (C) Quantitation of ERK activity. The amount of radioactivity present in phosphorylated MBP was quantitated with a phosphorimager. The activity of exogenous Erk was based on the mean of two experiments.
Another feature of rat GRF1 is that calcium-dependent Erk activation is markedly reduced in the mutant carrying the DH cluster mutations, as well as for mutants affecting the CC region or either of the PH domains, although these mutants bound calmodulin in an ionomycin-dependent manner (6, 24). We confirmed that this activity was attenuated with the DH cluster mutant (data not shown) and determined whether the L263Q mutant would behave similarly. Compared with cells transfected with wild-type mGRF1, ionomycin treatment of cells transfected with the L263Q mutant induced an attenuated increase in Erk activity (Fig. 6B and C), although calmodulin bound to the mutant protein (Fig. 6A, lane 3). Thus, these results are analogous to those reported for rat GRF1 mutants.
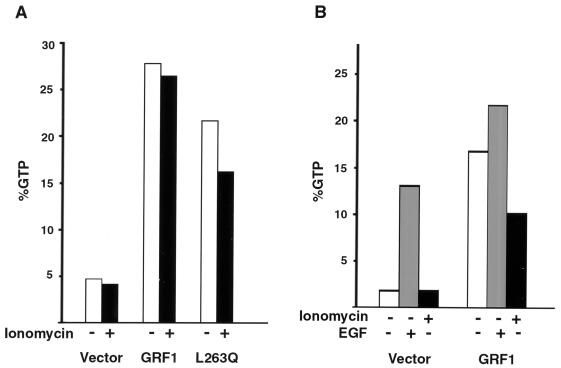
We also examined whether the level of GTP-Ras in the 293T cells would correlate with Erk activity (Fig. 7A). As expected, transient transfection of wild-type GRF1, in the absence of ionomycin, led to a substantial increase in the basal level of GTP-Ras compared to control cells transfected with the empty vector. To our surprise, however, there was no additional increase in GTP-Ras when the GRF1-expressing cells were treated with ionomycin, despite the ability of this treatment to stimulate the substantial increase in Erk activity noted above. Cells transfected with the L263Q mutant also showed an increase in basal GTP-Ras, although it was less than that seen with the wild-type mGRF1, and the GTP-Ras levels did not increase further with ionomycin treatment. To rule out the possibility that the lack of GTP-Ras in response to ionomycin might be secondary to an inability of the transfected cells to increase further their level of GTP-Ras, cells were treated with EGF or with ionomycin. Although EGF treatment of GRF1-expressing cells demonstrated a further increase in GTP-Ras, ionomycin actually led to a small decrease in GTP-Ras in the same experiment (Fig. 7B).
FIG. 7.
GTP-Ras in 293T cells expressing wild-type GRF1 and L263Q mutant. Cells were transiently transfected with the indicated GRF1 gene. Cultures were serum starved and metabolically labeled with [32P]orthophopshate, cells were treated with 5 μM ionomycin or 20 nM EGF for 5 min, extracts were immunoprecipitated with a Ras-specific monoclonal antibody, and the percentage of GTP-Ras was determined by thin-layer chromatography as described in Materials and Methods. Results in panel A represent the average of two experiments, while the results in panel B are from a single experiment.
Ionomycin induces increased Raf activity.
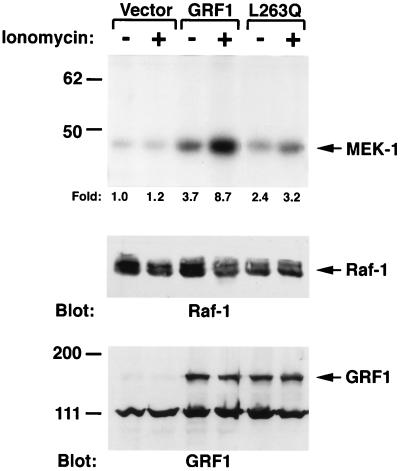
The failure of ionomycin to increase GTP-Ras in the GRF1-expressing cells implied that another mechanism would account for the ionomycin-dependent increase in Erk. Three features of Raf led us to examine this serine-threonine kinase as a possible downstream target for the ionomycin-dependent increase in Erk: Raf is part of the Erk pathway, Raf lies immediately downstream from Ras, and Raf is only partially activated by Ras (reviewed in reference 38). 293T cells were transiently cotransfected with GRF1 and c-Raf, treated with ionomycin, and extracts were immunoprecipitated with anti-Raf antibodies. The kinase activity of the immunoprecipitates was then analyzed by the in vitro phosphorylation of MEK, an authentic Raf substrate located between Raf and Erk (Fig. 8). Consistent with the basal increase in GTP-Ras in cells expressing wild-type GRF1, the basal level of Raf kinase activity in these cells was also increased compared to extracts of cells transfected with the empty vector. Ionomycin treatment of the transfected cells induced a further increase in Raf activity over this already-elevated basal activity. This ionomycin-induced GRF1-dependent increase in Raf kinase probably occurred by a Ras-independent mechanism, given the lack of increase in GTP-Ras seen with ionomycin treatment of GRF1-expressing cells (Fig. 7).
FIG. 8.
GRF1 increased Raf activity in response to ionomycin. Transiently transfected 293T cells were treated with or without ionomycin after serum starvation as in Fig. 6. Cell lysates were immunoprecipitated with Raf antibodies, and the immunocomplexes were assayed for in vitro kinase activity by using MEK-1 as a substrate (upper part of panel). The amount of radioactivity present in phosphorylated MEK-1 was determined with a phosphoimager, and the fold increase, compared with the untreated vector control, is shown underneath the upper part of the panel. The expression of Raf and GRF1 was confirmed by immunoblotting, as shown in the middle and bottom panels, respectively.
To confirm the dependence of this response on wild-type GRF1, the Raf activity was also determined for companion cells cotransfected with the L263Q mutant and c-Raf (Fig. 8). The results obtained in the Raf kinase assay correlated with those observed with this mutant in the Erk assays in that ionomycin treatment induced a smaller increase in Raf kinase than was seen with wild-type GRF1.
DISCUSSION
In this study, we report two unexpected observations about the Ras-specific exchange factor GRF: (i) GRF1 and GRF2 can form homo- and hetero-oligomers, via their respective DH domains, which represents a hitherto-unreported function for a DH domain; and (ii) the ionomycin-induced Erk activity of GRF1 occurs via at least two cooperative GRF1-dependent signaling pathways that converge to activate Raf. In 293T cells under the conditions examined, one pathway is a constitutive GRF1-dependent increase in GTP-Ras, which occurs in the absence of ionomycin. The other pathway is a further GRF1-dependent increase in Raf activity, which is induced by ionomycin. This latter activation occurs via a Ras-independent mechanism in that it is not associated with a further increase in GTP-Ras.
Homo- and hetero-oligomerization of GRF1 and GRF2.
Dimerization via the DH domains was seen initially in a yeast two-hybrid screen developed to identify proteins in a human brain cDNA library that might interact with the mGRF1 DH domain. This screen identified, in the brain library, an encoded polypeptide which turned out to consist of the IQ motif and the DH domain from hGRF2. Deletion analysis of the hGRF2 polypeptide indicated that the IQ motif was dispensable for the interaction, as were the few non-DH domain amino acids present in the mGRF1 bait. Mutation of a highly conserved leucine residue to glutamine (L263Q) in mGRF1 inactivated the interaction with hGRF2. The leucine residue forms part of a structurally conserved N-terminal region of the DH domain (3, 46). By contrast, a cluster of mutations in a C-terminal region of the DH domain did not abolish the interaction. Taken together, the above results implied that the mGRF1 DH domain was capable of interacting with the hGRF2 DH domain, and L263 in mGRF1 was required for this interaction.
When the examination was extended to mammalian cells, the results confirmed that a complex was formed between GRF1 and GRF2 in a rat brain extract or when the full-length proteins were coexpressed in cells and, further, that GRF1 and GRF2 can each form homo-oligomers. The analysis also established that homo-oligomer formation for GRF1 is impaired by the L263Q mutation (the analogous mutation was not introduced into GRF2) but not by the DH cluster mutation. Complex formation between the DH domains of GRF1 and GRF2 appears to be relatively specific, since complexes were not detected between GRF1 and two other proteins that contain DH domains, Sos1 and c-Dbl. It may be relevant that the DH domains in Sos1 and c-Dbl share less than 30% identity with the GRF1 DH domain, while the DH domains of GRF1 and GRF2 are more than 80% identical. It is likely that the oligomerization of GRF1 and GRF2 represents a direct interaction between DH domains that are identical (in the case of homo-oligomerization) or very closely related (in the case of hetero-oligomerization), but it remains possible that the complex formation identified here might depend on an additional unidentified molecule. Such a putative molecule would need to be conserved between yeasts and mammals, given the similarity of the results in both systems. What we are designating as “oligomers” probably represent dimers, but our data do not rule out the possibility of multimers.
Oligomerization has also been reported for the yeast homolog of GRF, Cdc25p, both with itself and with Sdc25p, which is a closely related exchange factor (8). In Cdc25p, the oligomerization domain was localized to its C-terminal portion, which contains the Ras catalytic domain and the signal responsible for membrane localization. Since neither Cdc25p nor SCD25p contain a DH domain, the mechanism of oligomerization involving the yeast factors is distinct from that reported here for mammalian GRF.
Functional relevance of oligomerization.
The possible relevance of oligomerization to GRF function was analyzed by studying the two GRF1 DH mutants for their transforming activity and ability to participate in ionomycin-dependent signaling. In the NIH 3T3 cell transformation assay, the oligomerization-competent DH cluster mutant was less active than wild type, while oligomerization-deficient L263Q mutant lacked any detectable activity. One interpretation of these results is that the DH domain encodes two functions, one being oligomerization and the other possibly involving GDP-GTP exchange involving one or more Rho family GTPases; the oligomerization-competent DH cluster mutant, which retains some biological activity, would be deficient only in the latter function, while the biologically inactive L263Q mutant would be deficient in both functions. Both mutants were deficient for calcium-dependent signaling. The precise relationship between the DH domain of GRFs and GDP-GTP exchange on small GTP-binding proteins remains unclear. As noted in the Introduction, negative GDP-GTP exchange results were reported for the relationship in vitro between GRF1 and Rho family GTPases (11a, 24), but positive results were reported for the relationship between GRF2 and Rac (18). However, while activation of the Rac-dependent JNK/SAPK pathway in 293T cells was reported as being GRF2 dependent, an activated form of Rac (RacV12) was unable to mimic this effect (18).
Oligomerization has been reported for other components involved in signaling. In some instances, oligomerization appears to be primarily inducible, such as with many cell surface receptors (29) and possibly with Raf (20, 34), while in others it appears to be constitutive, as with the 14-3-3 proteins (51). The oligomerization of GRF1 and GRF2 identified here is constitutive. Additional studies will be needed to determine whether it may also have an inducible component.
Raf activation by Ras-dependent and Ras-independent signals.
Although GRF1 induced a constitutive increase in GTP-Ras, we observed that the level of GTP-Ras was not augmented further when 293T cells expressing GRF1 were treated with ionomycin, which induced a substantial increase in Erk activity as previously reported. In seeking an alternate explanation for the Erk activity induced by ionomycin treatment of GRF1-expressing cells, we found that this treatment was associated with a concomitant increase in Raf kinase activity. Given the failure of ionomycin to increase the level of GTP-Ras, it seems most likely that the GRF1 signal induced by ionomycin further activates Raf via a Ras-independent pathway. The Ras-dependent activation of Raf by GRF1 may be a prerequisite for the postulated ionomycin-mediated Ras-independent signal that increases Raf activity, since a dominant inhibitory mutant of Ras has been shown to inhibit the ionomycin-dependent activation of Erk by GRF1 or GRF2 (17, 19). Thus, the ionomycin-dependent signal in the 293T cells acts in cooperation with the constitutive GRF1 signal, which may be largely Ras dependent.
The GTP-Ras results seen here are somewhat at variance with those reported by Farnsworth et al. (19), who found that ionomycin treatment of 293T cells expressing rat GRF1 induced a small additional increase in GTP-Ras from ca. 35% GTP-Ras without ionomycin to ca. 43% GTP-Ras with ionomycin (19). It is likely that minor differences in cells, growth conditions, and/or transfection efficiency might account for this discrepancy. We do not view this discrepancy as being incompatible with the conclusion that ionomycin has induced a Ras-independent effect on Raf, since the small increase in GTP-Ras seen by Farnsworth et al. was associated with a seven- to eightfold increase in Erk activity. Furthermore, our in vivo results are actually consistent with other reported data with GRF1: ionomycin treatment did not lead to a further increase in the in vitro exchange activity on Ras in GRF1 immunoprecipitates (19), and in vitro calcium and calmodulin binding to GRF1 actually inhibited Ras exchange activity GRF up to twofold (2).
Although we do not yet have a detailed molecular explanation for the Ras-independent effect on Raf, it is known that GTP-Ras, by itself, induces only a partial activation of Raf activity. Additional signals, such as the Src protein tyrosine kinase, can cooperate with GTP-Ras to induce higher levels of Raf kinase activity (16, 35). Ca2+ influx can activate the Ras/Raf/MAPK pathway by diverse routes (14, 15, 23). In PC12 cells, which do not express GRF, voltage-dependent Ca2+ influx has been reported to activate Src (42). Activation of a kinase such as Pak3, which can be activated by Cdc42 or Rac, can also increase the activity of Raf via the phosphorylation of serine 338 (32).
It is likely that the ionomycin-dependent activation of Raf depends, at least in part, on a calmodulin-dependent activity, such as calmodulin-dependent kinase, given that calmodulin binding to GRF1 depends on ionomycin, that mutation of the IQ domain in GRF1 or GRF2 abolishes this binding, and that the ionomycin-dependent activation of Erk by GRF1 or GRF2 is abolished by mutation of the IQ domain (17, 19). However, the requirements for calmodulin-dependent activation of GRF1 must be quite complex since, as noted above, mutation of the other motifs in the N terminus of GRF1 attenuate or abolish the ionomycin-dependent activation of Erk, although these mutations do not prevent the ionomycin-dependent binding of calmodulin to GRF1.
In summary, the results reported here imply that full GRF1 activity requires at least two cooperative signals that activate Raf. One is a Ras-dependent activation, which depends primarily on the Ras catalytic domain in the GRF1 C terminus, although the N terminus may also participate. Under the growth and transfection conditions employed here, the Ras-dependent activity is constitutive in NIH 3T3 and 293T cells. However, we speculate that in neuronal cells, where GRF is expressed physiologically, this activity may be repressed in the absence of an extracellular signal and may therefore be inducible. The Ras-independent signal, as detected by ionomycin treatment of 293T cells, depends on calmodulin binding, as well as on other functions that depend on several motifs located in the GRF N terminus. As identified here, oligomerization via its DH domain appears to be one of these functions.
ACKNOWLEDGMENTS
We thank Evelyn Ralston and Christine Winters from The National Institute of Neurological Disorders and Stroke for providing neonatal rat brains and Cheerag D. Upadhyaya for help with the preparation of figures.
P.H.A. and X.Q. contributed equally to this work.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped blast and PSI-Blast: an new generation of protein database search programs. Nucleic Acid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baouz S, Jacquet E, Bernardi A, Parmeggiani A. The N-terminal moiety of CDC25Mm, a GDP/GTP exchange factor of Ras proteins, controls the activity of the catalytic domain: modulation by calmodulin and calpain. J Biol Chem. 1997;272:6671–6676. doi: 10.1074/jbc.272.10.6671. [DOI] [PubMed] [Google Scholar]
- 3.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 4.Bowtell D, Fu P, Simon M, Senior P. Identification of murine homologues of the Drosophila son of sevenless gene—potential activators of ras. Proc Natl Acad Sci USA. 1992;89:6511–6515. doi: 10.1073/pnas.89.14.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brambilla R, Gnesutta N, Minichiello L, White G, Roylance A J, Herron C E, Ramsey M, Wolfer D P, Cestari V, Rossi-Arnaud C, Grant S G N, Chapman P F, Lipp H P, Sturani E, Klein R. A role for the Ras signalling pathway in synaptic transmission and long-term memory. Nature. 1997;390:281–286. doi: 10.1038/36849. [DOI] [PubMed] [Google Scholar]
- 6.Buchsbaum R, Telliez J B, Goonesekera S, Feig L A. The N-terminal pleckstrin, coiled-coil, and IQ domains of the exchange factor Ras-GRF act cooperatively to facilitate activation by calcium. Mol Cell Biol. 1996;16:4888–4896. doi: 10.1128/mcb.16.9.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell S L, Khosravi-Far R, Rossman K L, Clark G J, Der C J. Increasing complexity of Ras signaling. Oncogene. 1998;17:1395–1413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- 8.Camus C, Gemonat M, Garreau H, Baudet-Nessler S, Jacquet M. Dimerization of Cdc25p, the guanine nucleotide exchange factor for Ras from Saccharomyces cerevisiae, and its interaction with Sdc25p. Eur J Biochem. 1997;247:703–708. doi: 10.1111/j.1432-1033.1997.00703.x. [DOI] [PubMed] [Google Scholar]
- 9.Cen H, Papageorge A G, Vass W C, Zhang K, Lowy D R. Regulated and constitutive activity by CDC25(Mm) (GRF), a Ras-specific exchange factor. Mol Cell Biol. 1993;13:7718–7724. doi: 10.1128/mcb.13.12.7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cen H, Papageorge A G, Zippel R, Lowy D R, Zhang K. Isolation of multiple mouse cDNAs with coding homology to Saccharomyces cerevisiae CDC25—identification of a region related to bcr, vav, dbl and CDC24. EMBO J. 1992;11:4007–4015. doi: 10.1002/j.1460-2075.1992.tb05494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cerione R, Zheng Y. The Dbl family of oncogenes. Curr Opin Cell Biol. 1996;6:204–211. doi: 10.1016/s0955-0674(96)80068-8. [DOI] [PubMed] [Google Scholar]
- 11a.DeClue, J. E. Unpublished data.
- 12.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type I catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 13.Ebinu J O, Bottorff D A, Chan E Y W, Stang S L, Dunn R J, Stone J C. RasGRP, a Ras guanyl nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Science. 1998;280:1082–1086. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- 14.Egea J, Espinet C, Comella J X. Calmodulin modulates mitogen-activated protein kinase activation in response to membrane depolarization in PC12 cells. J Neurochem. 1998;70:2554–2564. doi: 10.1046/j.1471-4159.1998.70062554.x. [DOI] [PubMed] [Google Scholar]
- 15.Enslen H, Tokumitsu H, Stork P J S, Davis R J, Soderling T R. Regulation of mitogen-activated protein kinases by a calcium/calmodulin-dependent protein kinase cascade. Proc Natl Acad Sci USA. 1996;93:10803–10808. doi: 10.1073/pnas.93.20.10803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fabian J R, Daar I O, Morrison D K. Critical tyrosine residues regulate the enzymatic and biological activity of raf-1 kinase. Mol Cell Biol. 1993;13:7170–7179. doi: 10.1128/mcb.13.11.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fam N P, Fan W-T, Wang Z, Zhang L-J, Chen H, Moran M F. Cloning and characterization of Ras-GRF2, a novel guanine nucleotide exchange factor for Ras. Mol Cell Biol. 1996;17:1396–1406. doi: 10.1128/mcb.17.3.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan W-T, Koch C A, De Hoog C L, Fam N P, Moran M F. The exchange factor Ras-GRF2 activates Ras-dependent and Rac-dependent mitogen-activated protein kinase pathways. Curr Biol. 1998;8:935–938. doi: 10.1016/s0960-9822(07)00376-4. [DOI] [PubMed] [Google Scholar]
- 19.Farnsworth C L, Freshney N W, Rosen L B, Ghosh A, Greenberg M E, Feig L A. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature. 1995;376:524–527. doi: 10.1038/376524a0. [DOI] [PubMed] [Google Scholar]
- 20.Farrar M A, Alberolaila J, Perlmutter R M. Activation of the Raf-1 kinase cascade by coumermycin-induced dimerization. Nature. 1996;383:178–181. doi: 10.1038/383178a0. [DOI] [PubMed] [Google Scholar]
- 21.Ferrari C, Zippel R, Martegani E, Gnesutta N, Carrera V, Sturani E. Expression of two different products of CDC25Mm, a mammalian Ras activator, during development of mouse brain. Exp Cell Res. 1994;210:353–357. doi: 10.1006/excr.1994.1048. [DOI] [PubMed] [Google Scholar]
- 22.Fields S, Song O-K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 23.Finkbeiner S, Greenberg M E. Ca2+-dependent routes to Ras: mechanisms for neuronal survival, differentiation, and plasticity? Neuron. 1996;16:233–236. doi: 10.1016/s0896-6273(00)80040-9. [DOI] [PubMed] [Google Scholar]
- 24.Freshney N W, Goonesekera S D, Feig L A. Activation of the exchange factor Ras-GRF by calcium requires an intact Dbl homology domain. FEBS Lett. 1997;407:111–115. doi: 10.1016/s0014-5793(97)00309-8. [DOI] [PubMed] [Google Scholar]
- 25.Furth M E, Davis L J, Fleurdelys B, Scholnick E M. Monoclonal antibodies to the p21 products of the transforming gene of Harvey murine sarcoma virus and of the cellular ras gene family. J Virol. 1982;43:294–304. doi: 10.1128/jvi.43.1.294-304.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enyzmol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 27.Guerrero C, Rojas J M, Chedid M, Esteban L M, Zimonjic D B, Popescu N C, Font de Mora J, Santos E. Expression of alternative forms of Ras exchange factors GRF and SOS1 in different human tissues and cell lines. Oncogene. 1996;12:1097–1107. [PubMed] [Google Scholar]
- 28.Hart M J, Eva A, Zangrilli D, Aaronson S A, Evans T, Cerione R, Zheng Y. Cellular transformation and guanine nucleotide exchange activity are catalyzed by a common domain on the dbl oncogene product. J Biol Chem. 1994;269:62–65. [PubMed] [Google Scholar]
- 29.Heldin C. Dimerization of cell surface receptors in signal transduction. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 30.Itier J-M, Tremp G L, Leonard J-F, Multon M C, Ret G, Schweighoffer F, Tocque B, Bluet-Pajot M-T, Cormier V, Dautry F. Imprinted gene in postnatal growth role. Nature. 1998;393:125–126. doi: 10.1038/30120. [DOI] [PubMed] [Google Scholar]
- 31.Jacquet E, Baouz S, Parmeggiani A. Characterization of mammalian C-CDC25(Mm) exchange factor and kinetic properties of the exchange reaction intermediate p21-C-CDC25(Mm) Biochemistry. 1995;34:12347–12354. doi: 10.1021/bi00038a031. [DOI] [PubMed] [Google Scholar]
- 32.King A J, Sun H, Diaz B, Barnard D, Miao W, Bagrodia S, Marshall M S. The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature. 1998;396:180–183. doi: 10.1038/24184. [DOI] [PubMed] [Google Scholar]
- 33.Lowy D R, Willumsen B M. Function and regulation of Ras. Annu Rev Biochem. 1993;62:851–891. doi: 10.1146/annurev.bi.62.070193.004223. [DOI] [PubMed] [Google Scholar]
- 34.Luo Z, Tzivion G, Belshaw P J, Vavvas D, Marshall M, Avruch J. Oligomerization activates c-Raf-1 through a Ras-dependent mechanism. Nature. 1996;383:181–185. doi: 10.1038/383181a0. [DOI] [PubMed] [Google Scholar]
- 35.Marais R, Light Y, Paterson H F, Marshall C J. Ras recruits Raf-1 to the plasma membrane for activation by tyrosine phosphorylation. EMBO J. 1995;14:3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchuk D A, Saulino A M, Tavakkol R, Swaroop M, Wallace M R, Andersen L B, Mitchell A L, Gutmann D H, Boguski M, Collins F S. cDNA cloning of the type 1 neurofibromatosis gene: complete sequence of the NF1 gene product. Genomics. 1991;11:931–940. doi: 10.1016/0888-7543(91)90017-9. [DOI] [PubMed] [Google Scholar]
- 37.Mattingly R R, Macara I G. Phosphorylation-dependent activtaion of the Ras-GRF/CDC25Mm exchange factor by muscarinic receptors and G-protein beta gamma subunits. Nature. 1996;382:268–272. doi: 10.1038/382268a0. [DOI] [PubMed] [Google Scholar]
- 38.Morrison D, Cutler R J. The complexity of Raf-1 regulation. Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 39.Olson M. Guanine nucleotide exchange factors for the Rho GTPases: a role in human disease? J Mol Med. 1996;74:563–571. doi: 10.1007/s001090050060. [DOI] [PubMed] [Google Scholar]
- 40.Plass C, Shibata H, Kalcheva I, Mullins L, Kotelevtseva N, Mullins J, Kato R, Sasaki H, Hirotsune S, Okazaki Y, Held W A, Hayashizaki Y, Chapman V M. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat Genet. 1996;14:106–109. doi: 10.1038/ng0996-106. [DOI] [PubMed] [Google Scholar]
- 41.Qian X, Vass W C, Papageorge A G, Anborgh P H, Lowy D R. N terminus of Sos1 Ras exchange factor: critical roles for the Dbl and pleckstrin homology domains. Mol Cell Biol. 1998;18:771–778. doi: 10.1128/mcb.18.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rusanescu G, Qi H, Thomas S M, Brugge J S H. Calcium influx induces neurite growth through a Src-Ras signaling cassette. Neuron. 1995;15:1415–1425. doi: 10.1016/0896-6273(95)90019-5. [DOI] [PubMed] [Google Scholar]
- 43.Schweighoffer F, Faure M, Fath I, Chevallier-Multon M C, Apiou F, Dutrillaux B, Sturani E, Jacquet M, Tocque B. Identification of a human guanine nucleotide-releasing factor (H-GRF55) specific for Ras proteins. Oncogene. 1993;8:1477–1485. [PubMed] [Google Scholar]
- 44.Shou C, Farnsworth C L, Neel B G, Feig L A. Molecular cloning of cDNAs encoding a guanine-nucleotide-releasing factor for Ras p21. Nature. 1992;358:351–354. doi: 10.1038/358351a0. [DOI] [PubMed] [Google Scholar]
- 45.Simon M A, Bowtell D D L, Dodson G S, Laverty T R, Rubin G M. Ras1 and a putative guanine nucleotide exchange factor perform crucial steps in signaling by the sevenless protein tyrosine kinase. Cell. 1991;67:701–716. doi: 10.1016/0092-8674(91)90065-7. [DOI] [PubMed] [Google Scholar]
- 46.Soisson S M, Nimnual A S, Uy M, Bar-Sagi D, Kuriyan J. Crystal structure of the Dbl and pleckstrin homology domains from the human son of sevenless protein. Cell. 1998;95:259–268. doi: 10.1016/s0092-8674(00)81756-0. [DOI] [PubMed] [Google Scholar]
- 47.Sturani E, Abbondio A, Branduardi P, Ferrari C, Zippel R, Martegani E, Vanoni M, Denis-Donini S. The Ras guanine nucleotide exchange factor CDC25Mm is present at the synaptic junction. Exp Cell Res. 1997;235:117–123. doi: 10.1006/excr.1997.3660. [DOI] [PubMed] [Google Scholar]
- 48.Tognon C E, Kirk H E, Passmore L A, Whitehead I P, Der C J, Kay R J. Regulation of RasGRP via a phorbol ester-responsive C1 domain. Mol Cell Biol. 1998;18:6995–7008. doi: 10.1128/mcb.18.12.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Touhara K, Inglese J, Pitcher J, Shaw G, Lefkowitz R J. Binding of G protein beta gamma-subunits to pleckstrin homology domains. J Biol Chem. 1994;264:10217–10220. [PubMed] [Google Scholar]
- 50.Tung P S, Fam N P, Chen L, Moran M F. A 54-kDa protein related to ras-guanine nucleotide release factor expressed in the rat exocrine pancreas. Cell Tissue Res. 1997;289:505–515. doi: 10.1007/s004410050896. [DOI] [PubMed] [Google Scholar]
- 51.Tzivion G, Luo Z, Avruch J. A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature. 1998;394:88–92. doi: 10.1038/27938. [DOI] [PubMed] [Google Scholar]
- 52.Van Aelst L, D’Souza-Schorey C. The Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 53.Willumsen B M, Vass W C, Velu T J, Papageorge A G, Schiller J, Lowy D R. The BPV E5 oncogne can cooperate with ras: identification of a p21 amino acid segment required for transformation by c-rasH but not v-rasH. Mol Cell Biol. 1991;11:6026–6033. doi: 10.1128/mcb.11.12.6026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang K, Papageorge A G, Lowy D R. Mechanistic aspects of signalling through Ras in NIH 3T3 cells. Science. 1992;257:671–674. doi: 10.1126/science.1496380. [DOI] [PubMed] [Google Scholar]