Abstract
Novel techniques were used to determine when in the cell cycle of proliferating NIH 3T3 cells cellular Ras and cyclin D1 are required. For comparison, in quiescent cells, all four of the inhibitors of cell cycle progression tested (anti-Ras, anti-cyclin D1, serum removal, and cycloheximide) became ineffective at essentially the same point in G1 phase, approximately 4 h prior to the beginning of DNA synthesis. To extend these studies to cycling cells, a time-lapse approach was used to determine the approximate cell cycle position of individual cells in an asynchronous culture at the time of inhibitor treatment and then to determine the effects of the inhibitor upon recipient cells. With this approach, anti-Ras antibody efficiently inhibited entry into S phase only when introduced into cells prior to the preceding mitosis, several hours before the beginning of S phase. Anti-cyclin D1, on the other hand, was an efficient inhibitor when introduced up until just before the initiation of DNA synthesis. Cycloheximide treatment, like anti-cyclin D1 microinjection, was inhibitory throughout G1 phase (which lasts a total of 4 to 5 h in these cells). Finally, serum removal blocked entry into S phase only during the first hour following mitosis. Kinetic analysis and a novel dual-labeling technique were used to confirm the differences in cell cycle requirements for Ras, cyclin D1, and cycloheximide. These studies demonstrate a fundamental difference in mitogenic signal transduction between quiescent and cycling NIH 3T3 cells and reveal a sequence of signaling events required for cell cycle progression in proliferating NIH 3T3 cells.
The cell cycle requirements for peptide growth factors have been demonstrated in cells rendered quiescent by growth factor deprivation (24). At a certain point following stimulation to reenter the cell cycle, the continued presence of a variety of growth factors became unnecessary for continuation through G1 phase and the initiation of DNA synthesis (25). This point of commitment to cell cycle progression, termed the restriction point, is generally observed several hours before the initiation of S phase. The restriction point is thus defined as the last point at which positive proliferative signals are required for completion of the cell cycle. Interestingly, in quiescent cells the restriction point is identical to the G1 arrest point induced by other conditions, including protein synthesis inhibitors and amino acid deprivation (29).
While results obtained with quiescent cells are important, there is reason to believe the situation might be different for cells continuously progressing through the cell cycle. For example, passage through G1 phase in quiescent and proliferating cells exhibits several important molecular differences, including expression of the cyclin-inhibitory protein p27kip1 (3), Cdc6 (42), and Fos and related proteins (14), and formation of a complex between E2F and the retinoblastoma protein (Rb)-related p130 (19, 34). In addition, the time required between the restriction point and initiation of S phase (near 4 h in the case of the cells used in this study) is nearly as long as the entire G1 phase when these cells are actively cycling. In cells with a short G1 phase, a prolonged period between completion of all required signaling events and the initiation of S phase would not appear warranted or perhaps even allowed. Indeed, since the overall length of the cell cycle appears to depend in large part on the length of the G1 phase, it is likely that this cell cycle period must be compressed as much as possible for rapid proliferation, potentially requiring the transfer of activities present in the G1 phase of quiescent cells to other cell cycle periods when they continuously cycle (26).
Foremost among the positive proliferative signaling pathways in fibroblast-like cells is the one involving peptide growth factors and cellular Ras (30). Upon binding to their receptors, peptide growth factors initiate a cascade of signaling events which leads to the activation of cellular Ras, which in turn functions as a molecular switch to transmit mitogenic signals to a cascade of cytoplasmic kinases (35), ultimately resulting in the activation of mitogen-activated protein kinases (MAPKs) (20). MAPKs stimulate transcription factors which in many cells turn on the expression of cyclin D1 (1). Cyclin D1 binds to the cyclin-dependent kinases 4 and 6 to form an Rb kinase (31). Upon phosphorylation, Rb loses its repressive activity for the E2F-DP transcription factor complex, which then activates transcription of many genes required for transition from G1 to S phase and for DNA replication (22). Therefore, cyclin D1 is essential for G1/S transition in Rb-positive cells (17, 23). It is believed that this molecular sequence forms a cascade vital to the positive stimulation of cell cycle progression.
Because Ras and its downstream targets play a central role in regulating mitogenic signaling within the cell, it is conceivable that Ras plays a role in progression past the restriction point. This has been verified by using microinjection of an efficiently neutralizing anti-Ras antibody. While Ras activity is required multiple times during the stimulation of quiescent cells to proliferate, the final of these requirements appears to be at or near the restriction point (5, 21, 38). In addition, cyclin D1, the cyclin-inhibitory protein p27kip1 (32), and phosphorylation of Rb (41) have each been proposed to be critical for passage through the restriction point.
Although it is apparent that growth factors and Ras activity are required for proliferation in actively cycling cells (21, 27), the timing of these requirements remains to be determined. Such analyses are complicated on one hand by the fact that cell cycle synchronization induces alterations in cell cycle characteristics and on the other hand by the difficulty in performing cell cycle analyses without synchronization. Time-lapse analyses have been used to determine in asynchronous cultures a point in G1 phase at which growth factors become unnecessary for continued proliferation (15, 44). These studies avoided the necessity of synchronization and perturbation of the cell cycle resulting therefrom. We have extended these studies by using time-lapse analyses combined with quantitative fluorescence cytochemistry and microinjection to analyze the requirements for signaling molecules in continuously proliferating cells. The results reveal a requirement for positive signaling molecules fundamentally different from that previously reported for quiescent cells.
MATERIALS AND METHODS
Materials.
Cy3-conjugated anti-rat immunoglobulin G (IgG), Cy2-conjugated anti-rat IgG with minimal cross-reactivity against mouse IgG, Cy3-conjugated anti-mouse IgG, and Cy3-conjugated anti-mouse IgG with minimal cross-reactivity against rat IgG were purchased from Jackson ImmunoResearch (West Grove, Pa.). Anti-ACTIVE MAPK polyclonal antibody was obtained from Promega (Madison, Wis.). Mouse monoclonal anti-mouse cyclin D1 antibody 72-13G (Santa Cruz Biotechnology, Santa Cruz, Calif.) was used for all staining applications. Nonimmune rat IgG was obtained from Sigma (St. Louis, Mo.). [methyl-3H]thymidine (80 Ci/mmol) was purchased from Amersham (Arlington Heights, Ill.). Autoradiography emulsion-type NTB2 was obtained from Eastman Kodak Company (New York, N.Y.). DAPI (4′,6-diamidino-2-phenylindole, dilactate) was a product of Molecular Probes (Eugene, Oreg.).
Antibody preparation.
Hybridomas expressing neutralizing antibodies against either Ras (Y13-259, rat IgG) or cyclin D1 (DCS-6, mouse IgG; a generous gift from Michele Pagano) were grown in serum-free medium (Hybridoma-SFM; Gibco BRL, Gaithersburg, Md.). Each secreted antibody was purified accordingly, using ammonium sulfate precipitation followed by DEAE chromatography (13). The purified antibodies were dialyzed against 25 mM Tris-HCl (pH 7.5) and concentrated to 10 mg/ml in a Centricon 30 (Amicon, Bedford, Mass.). These antibodies were used exclusively for microinjection.
Cell culture and microinjection.
NIH 3T3 cells were maintained and made quiescent as described elsewhere (11). To determine the requirement for protein synthesis, cells were treated with 2 μM cycloheximide, which suppressed overall protein synthesis by 95 to 98% (33). The antibodies indicated were injected into cells grown on a coverslip as described previously (37).
Time-lapse video photography.
Digital images were obtained with a charge-coupled device (CCD) camera (Sony; Tokyo, Japan) attached to a frame capture board controlled by the NIH Image program. Individual frames of 640 by 480 pixels were captured every 5 to 15 min. Up to 310 individual frames were captured in a single stack, which was then replayed by the NIH Image program at various speeds in the forward or backward direction to create a movie for analysis. Images of cells were obtained with phase-contrast optics and a 4× objective to allow analysis of the largest possible area of the coverslip. The area of the coverslip to be analyzed was marked with two contiguous circles of varying size by using a diamond object marker (Leitz). This allowed realignment of the area of analysis and identification of individual cells following immunostaining, DAPI staining, and/or autoradiography. When a second time-lapse analysis followed microinjection, care was taken to ensure that the same area of the coverslip and same alignment were viewed in each movie. Antibodies to be studied were microinjected into all the cells, and only the cells, within the designated circular area. In this way it was possible to follow the analysis from the beginning and determine which cells had arisen from originally injected cells. The injection was further confirmed by immunofluorescence staining for injected antibody.
To avoid possible complications with this constant illumination, several experiments were duplicated with a similar approach except that the light was shuttered. In this case, the cells were illuminated only during the time of exposure, less than 1 s every 10 min. These images were collected with a highly sensitive, cooled CCD camera (Princeton Instrument) and controlled by the Metamorph software package (Universal Imaging). Because of the sensitivity of the cooled CCD camera, the cells were illuminated with extremely faint light. In these cases, there is little possibility that the illumination of cells would have any effects on the results obtained. In no case were the results obtained with the two methods of illumination different.
Immunofluorescence staining and detection of DNA synthesis.
For immunostaining, the cells were fixed with methanol for 5 min at −20°C. The cells were rehydrated with phosphate-buffered saline (PBS) and blocked with PBS containing 3% (vol/vol) normal goat serum (Gibco BRL) for 1 h at room temperature. The antibody solution was made in blocking solution. After incubation with primary antibody for 1 h at room temperature or overnight at 4°C, the monolayer was rinsed with PBS for 5 min thrice. The cells were incubated with fluorochrome-conjugated specific secondary antibody (1:1,000 dilution). After three washes with PBS, cells were stained with DAPI (10 mg/ml) and mounted with 50% glycerol in PBS. To detect DNA synthesis, 5-bromo-2′-deoxyuridine (BrdU) labeling was performed with a 5-Bromo-2′-deoxy-uridine Labeling and Detection Kit II (Boehringer Mannheim, Indianapolis, Ind.) according to the manufacturer’s instructions except that positive cells were visualized by Cy3-conjugated anti-mouse IgG antibody. S phase was also detected by labeling with [3H]thymidine followed by autoradiography as described before (11). Measurement of immunofluorescence or DAPI signal was performed before autoradiography.
Measurement of fluorescent signal of each stained cell.
A Leica model DM 900 fluorescent microscope was used to quantitate fluorescence intensity. The filter cubes A, L4, and N2.1 were used to detect signals from DAPI, Cy2, and Cy3, respectively. With those cubes, no significant crossover signal was detected. Digital images were captured with a cooled (−25°C) CCD camera (Princeton Instrument) controlled with Metamorph software. The exposure time was adjusted so that the brightest signal in the specimen gave less than 90% of the maximum linear range for the camera (a gray scale of 0 to 4,096; 12-bit gray scale). Integration of the fluorescence signal from each nucleus was obtained by processing the image by using Metamorph analysis features.
RESULTS
Determination of the restriction point in quiescent cells.
For the purposes of comparison with cycling cells, it was first necessary to carefully determine the point at which various inhibitory treatments lose their ability to block cell cycle progression following serum addition to quiescent cells. Quiescence was induced by culture for 48 h in 0.5% serum. One set of these plates was stimulated with serum and injected with anti-Ras antibody Y13-259 at various times thereafter. Cells were treated with tritiated thymidine immediately following injection until 24 h after the original addition of serum. Thus, labeling of these cells with thymidine would indicate that the injected antibody had failed to block cell cycle progression to S phase. Thymidine labeling would indicate that the injection had taken place after the critical point in G1 phase at which cellular Ras is last required prior to commitment to enter DNA synthesis (Fig. 1E). In the second set of plates, tritiated thymidine was added together with serum, and the cells were fixed various times thereafter. In these plates, thymidine labeling would indicate that the cells had progressed into S phase by the time of fixation (Fig. 1E). From the injected set of plates, the point of Ras requirement following serum addition was determined; from the second set of plates, the timing of entry into S phase was determined.
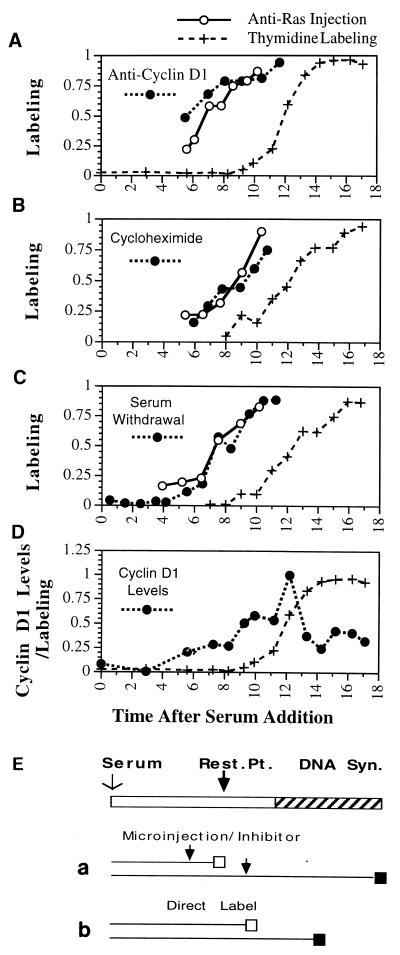
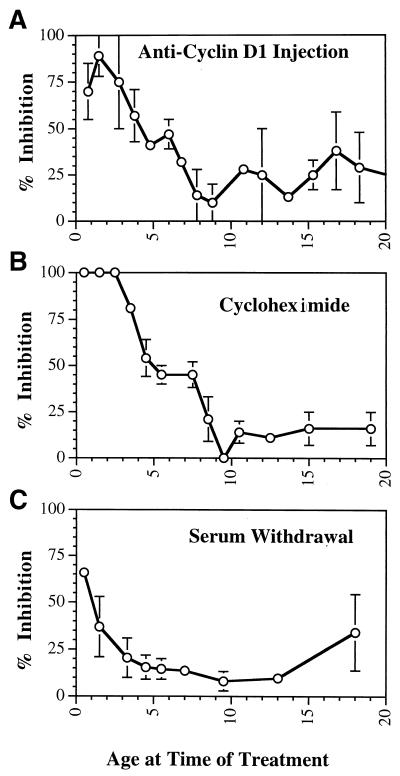
FIG. 1.
Restriction point determination in quiescent cells. Inhibitory treatments were applied at the indicated times following serum addition to quiescent NIH 3T3 cells. Thymidine was added following antibody microinjection (or addition of the inhibitor) until 24 h following the initial addition of serum to the culture, at which time the cells were fixed and autoradiographed. The proportion of cells which were labeled with thymidine following treatment is plotted versus the time (in hours) following serum addition at which the injection (inhibitory treatment) took place. To identify the time these cells first enter S phase, labeled thymidine was added together with serum to a parallel set of cultures, which were fixed at the indicated times. (A to C) In each experiment, an inhibitor as indicated (solid circles in each panel) was analyzed together with anti-Ras injection (○), while thymidine labeling (+) was performed in each experiment to determine the timing of entry into S phase. (D) Accumulation of nuclear cyclin D1 protein determined by antibody staining at the indicated times following serum addition to quiescent cells, reported as the proportion of maximal antibody staining at each time point. For each point, the average of approximately 100 individual cells is given. (E) Procedures used. The top bar represents cell cycle progression following serum addition from G0 phase into S phase; the restriction point (Rest. Pt.) is indicated together with DNA synthesis. The lower lines indicate progression of cells through the cell cycle. An open box indicates the cell cycle point to which the cell had progressed, and a solid box indicates thymidine labeling. (a) Treatment with inhibitors or injection of antibody. Cells treated prior to the restriction point (arrow, top line) would be unable to progress past the restriction point and thus unable to incorporate thymidine. Treatment after the cells had passed the restriction point (arrow, lower line) would be unable to inhibit entry into S phase and would result in thymidine labeling of the cells. Direct labeling of cells at the time of serum addition (b) would result in thymidine labeling of any cell in S phase at the time the cells were fixed.
Anti-Ras efficiently blocked thymidine labeling when injected within 5 h of serum addition and inhibited by 50% entry into S phase when injected near 8 h following serum addition. In these cultures, half of the cells entered S phase near 12 h following addition of serum (Fig. 1A). Thus, the last point at which cellular Ras was required was approximately 4 h prior to the initiation of S phase. For comparison, in the same experiment, a third set of plates was treated as described above except that anti-cyclin D1 antibody (from M. Pagano) was injected instead of anti-Ras antibody. In these cultures, the pattern of inhibition of entry into S phase was indistinguishable from that observed with anti-Ras injection, indicating that within the limits of this determination, the last requirement for cyclin D1 prior to entry into S phase was identical to the last point at which cellular Ras was required (Fig. 1A). Similar analyses were performed to determine the last point at which protein synthesis or serum was required by treatment with 2 μM cycloheximide (Fig. 1B) or by serum removal (Fig. 1C). The timing of inhibition with these two treatments was indistinguishable from the timing of inhibition by injected anti-Ras or anti-cyclin D1 antibody.
Finally, the inhibition points identified above were compared to the timing of appearance of cyclin D1 in the nuclei of quiescent cells following treatment with serum. The level of cyclin D1 in individual cells was determined by indirect immunofluorescence using anti-cyclin D1 at various times following serum addition. The amounts of cyclin D1 were then determined by quantitating the fluorescence levels with a sensitive CCD camera and image analysis using the Metamorph software program. Nuclei were identified by DAPI staining, and only fluorescence over the nuclei was measured. Numbers reported represent the averages of approximately 300 cells. The levels of cyclin D1 in these cultures began to increase at 5.5 h following serum addition and reached a peak at 11 h, after which the cyclin D1 returned to modest levels (Fig. 1D). Half-maximal cyclin D1 levels were observed slightly after the time of half-maximal inhibition by anti-Ras and anti-D1 (Fig. 1A and D). It is not possible to determine the minimal level of cyclin D1 required to promote cell cycle progression, and therefore it is not possible to strictly relate the data on D1 levels to inhibition by anti-D1 antibody, but the timing of cyclin D1 expression and the requirement for cyclin D1 as determined by antibody injections were similar. To confirm the analyses described above, the total amount of cellular cyclin D1 protein was determined at various times following serum addition to quiescent NIH 3T3 cultures by Western analysis. The total amounts of cellular cyclin D1 as determined by Western analysis were similar to the results obtained as described above with antibody staining (data not shown). The close correlation between total cellular cyclin D1 protein and nuclear staining confirms the quantitative nature of the fluorescence staining measurements.
Cell cycle characteristics of cycling NIH 3T3 cells.
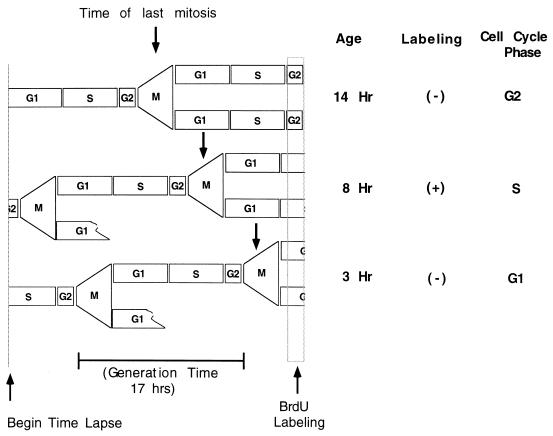
Any previously identified means of synchronizing continuously proliferating cells would result in alterations of their cell cycle characteristics. For example, mitotic shake-off of NIH 3T3 cells resulted in a G1 phase of near 10 h (unpublished data), while the G1 phase determined by time-lapse analysis was about 5 h (see below). We therefore performed all studies without synchronization. Instead, time-lapse analyses of an asynchronous culture was used to identify the approximate cell cycle position of individual cells. The time-lapse analysis made it possible to determine how long prior to the termination of the experiment an individual cell had passed through mitosis. This value, termed the age of the cell, was used as an indication of the approximate cell cycle position (Fig. 2) (44). To relate age with cell cycle position, a culture was followed in time lapse for more than 20 h, with the final 30 min in the presence of BrdU. The cells were then fixed and stained with a fluorescent antibody against BrdU. The age of each cell was determined individually by following that cell in the time-lapse movie beginning at the time of fixation and running the movie backwards until the cell could be observed to pass through mitosis (Fig. 2). Labeling with BrdU would indicate that the cell had been in S phase during the last 30 min. This approach allowed an analysis of cells in all cell cycle phases at the same time.
FIG. 2.
Time-lapse determination of approximate cell cycle position. The diagram illustrates the cycling characteristics of three typical cells in an asynchronous culture. At the beginning of the time-lapse analysis, these cells were in G1, G2, and S phases, respectively. The passage of these cells and their daughters through cell cycle periods is illustrated, together with the position of the final mitosis prior to the BrdU labeling period at the termination of the experiment. The age is the time between this final mitosis and the end of the BrdU labeling period, which is followed immediately by fixation of the cells. Labeling with BrdU identified the cell in S phase at the end of the experiment, while unlabeled cells, which could be in G1 or G2 phase, were distinguished by cell age.
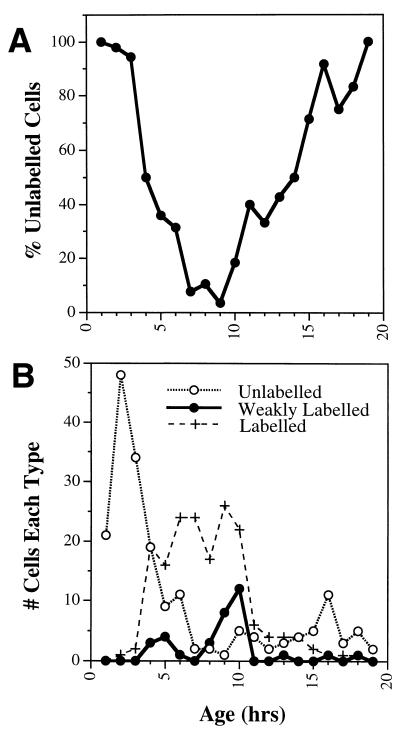
To determine the age at the time of entry into S phase, we grouped together all cells within a given age period and determined the proportion of these which remained unlabeled with BrdU (Fig. 3A). Cells began to enter S phase within 4 h, approximately half were labeled at 5 h, and most cells had entered S phase within 7 h following mitosis. At 17 h following mitosis (the average generation time of the culture), the proportion of BrdU-labeled cells had declined to near zero (Fig. 3A). This indicates that cells from 4 to 7 h of age were in G1 phase, while G2 phase was first observed in cells 11 to 12 h of age. Cells weakly labeled with BrdU would have been in transitions to or from S phase at the time of labeling. Their appearance was taken as an indication of early and late S phase, and they were observed with peaks at 5 and 10 h of age, which indicates the general boundaries of S phase (Fig. 3B). While the cycling characteristics of individual cells vary (with cell cycle times of 13 to 23 h), the overall characteristics of the entire culture were quite consistent between analyses. Therefore, from 100 to 250 cells were analyzed in each experiment to compensate for random variations between individual cells.
FIG. 3.
Cell age and cell cycle position. Asynchronous NIH 3T3 cells were followed for 20 h in time lapse, with the last 30 min in the presence of BrdU. The age of each cell was determined from the time-lapse movie by measuring how long before fixation each cell had passed through mitosis (Fig. 2). Cells in S phase at the termination of the analysis were identified by staining with a fluorescent antibody against BrdU. (A) Proportion of BrdU-unlabeled cells determined at each hour of age; (B) total numbers of unlabeled (○), labeled (+), or weakly labeled (●) cells in each hour shown separately. The peaks in weakly labeled cells indicate entry into and from S phase.
Ras and cyclin D1 requirements in cycling cells.
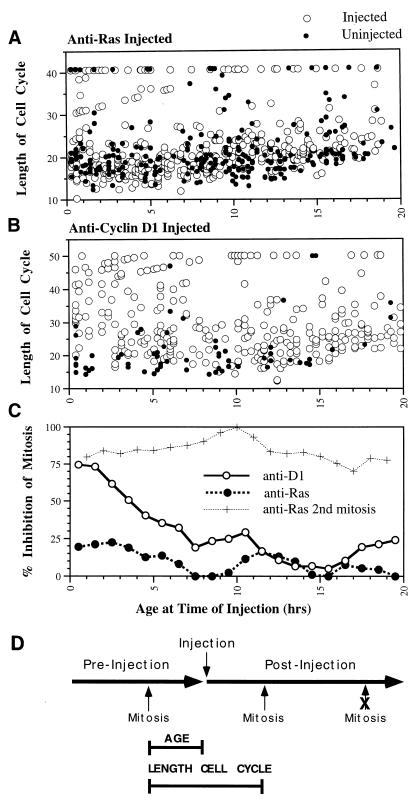
To determine the cell cycle point at which Ras is required in proliferating NIH 3T3 cells, a culture was followed in two sequential time-lapse movies of the same area of the coverslip. Anti-Ras antibody was microinjected between the two movies into cells in the area of analysis (localized within a circle scribed on the back of the coverslip). The age of cells at the time of injection was determined from the first movie, while the fate of injected cells was determined from the second (Fig. 4D). For comparison, the behavior of neighboring uninjected cells was also determined. Surprisingly, almost all of the injected cells passed through mitosis following anti-Ras injection, regardless of the cell cycle position at the time of injection. To more carefully analyze the data, the length of the cell cycle in which the injection took place was calculated by determining the time of the mitoses before and after injection for each cell (Fig. 4D). The combined data from five separate, highly similar experiments clearly indicate that while some injected as well as uninjected cells were delayed in passage through mitosis following treatment, most injected cells passed through the cell cycle without delay compared to uninjected cells, regardless of the cell cycle position at the time of injection (Fig. 4A).
FIG. 4.
Requirement for Ras and cyclin D1 in cycling cells. Cycling NIH 3T3 cells received injections of anti-Ras antibody (A) or anti-cyclin D1 antibody (B). The age of each injected cell is determined from a time-lapse analysis before injection; the occurrence and time of mitosis after injection are determined from a time-lapse analyses after injection. The length of the cell cycle is the time between the mitoses before and after injection (D). The data for uninjected cells (closed circles) are presented for comparison to the behavior of antibody-injected cells (open circles). These data are the compiled data from multiple highly consistent individual experiments. (C) Proportion of cells in each age period which exhibited a delay in entry into mitosis (cell cycle time greater than 30 h) following antibody injection presented in graph form. Note that in most cases anti-Ras-injected cells divided without delay, while those injected with anti-cyclin D1 during the G1 period experienced a substantial delay in passage through mitosis compared to uninjected cells. In panel D, the large horizontal arrows indicate time-lapse analyses before and after microinjection of antibody. The timing of typical mitoses is indicated, together with the determination of cell age and the length of the cell cycle in which injection took place. Note that the second mitosis following injection is efficiently inhibited by the injected antibody.
These data were summarized by calculating the proportion of cells with lengthened cell cycle times (greater than 30 h) versus the age at the time of injection. The level of inhibition was generally less than 20% even for cells soon after mitosis (Fig. 4C). Surprisingly, these data indicate that there was no requirement for cellular Ras during the first G1 phase of the cell cycle following injection in asynchronous NIH 3T3 cells. This conclusion was apparent from the combined data and from each of the five individual experiments. The effectiveness of the injected antibody was confirmed by the fact that while most all injected cells passed through one mitosis following injection, very few cells passed through a second mitosis (Fig. 4C). Thus, anti-Ras was inhibitory for the progression of cycling cells, but only if it was introduced into the cell prior to mitosis, in the preceding cell cycle.
The cell cycle requirement for cyclin D1 was next determined by microinjecting a neutralizing, highly specific monoclonal antibody. The results from three highly similar (Fig. 4A) experiments were combined to indicate that the inhibitory characteristics of anti-cyclin D1 were quite different from those of anti-Ras. Cells injected with anti-D1 within the first few hours following mitosis were essentially all delayed in passage through the current cell cycle compared to uninjected cells. By 8 h of age, however, the proportion of cells which were inhibited in cell cycle passage by anti-D1 injection was low (Fig. 4B). Summary of these data (Fig. 4C) shows that the inhibitory behavior of injected anti-D1 was strikingly similar to the profile of entry into S phase (Fig. 3A). Inhibition by anti-cyclin D1 antibody was high in the first 3 h following mitosis, when the cells were all in G1 phase. From 4 to 7 h after mitosis, the proportion of cells in S phase increased, while inhibition by anti-cyclin D1 decreased. Therefore, cyclin D1 was required during G1 phase of cycling NIH 3T3 cells and was apparently required up until near the beginning of S phase. Clearly, cell cycle progression in cycling cells is much different with respect to molecular requirements from that in quiescent cells.
Based upon previous studies (7, 36), it would appear extremely unlikely that the delay in cell cycle inhibition by anti-Ras compared to anti-D1 resulted from a delayed action of anti-Ras antibody following introduction into the cell. To directly test this possibility, however, we determined how rapidly anti-Ras antibody was able to block ERK phosphorylation (one of the most rapid effects of Ras activity). Quiescent NIH 3T3 cells were injected with anti-Ras antibody and stimulated with serum at various time thereafter. At 15 min following serum addition, the cells were fixed and stained with an antibody which recognized only phosphorylated ERK proteins. The anti-Ras antibody blocked serum-induced phosphorylation in all plates analyzed, including those cells which were stimulated with serum immediately following antibody injection (Fig. 5). As a control, nonspecific immunoglobulin injected at the same time had no inhibitory effects on ERK phosphorylation (Fig. 5). Thus, the above results must not have been due to delayed activity of injected anti-Ras antibody. Moreover, the effects of anti-Ras differed morphologically from the effects of anti-cyclin D1. Even within the first cell cycle period, the anti-Ras antibody caused the cells to flatten and stop migrating. This fact is curious when it is considered that these morphological changes did not lead to altered cell cycle progression, while anti-cyclin D1, which did not cause altered morphology or interfere with cellular migration, dramatically inhibited cell cycle progression when introduced early in the cell cycle (data not shown).
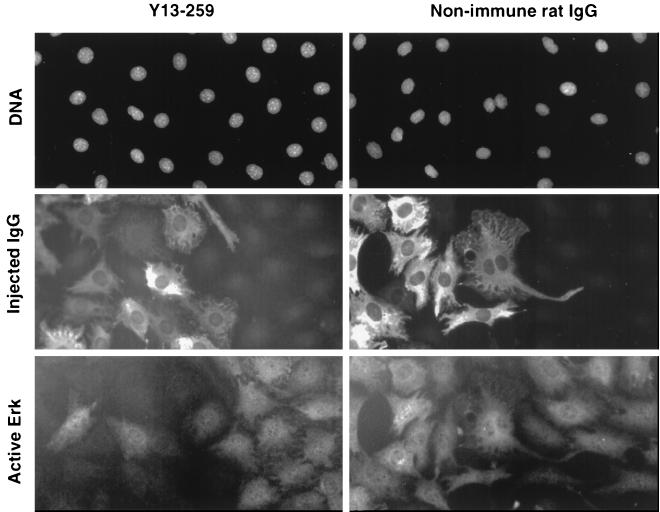
FIG. 5.
Rapid inhibition of ERK activation by injected anti-Ras. To demonstrate the speed at which injected anti-Ras antibody is able to neutralize cellular Ras, quiescent NIH 3T3 cells were injected with anti-Ras antibody (left) or a nonspecific control antibody (right). Cells were then treated with serum immediately following injection; after 15 min, the cells were fixed and stained with an antibody able to selectively recognize phosphorylated ERK, a downstream target of Ras activity. The location of all cells within the field is revealed by a photograph of DAPI-stained nuclei (top row). Antibody-injected cells are revealed by staining with a fluorescent antibody against the injected immunoglobulin (middle row). Note that most, but not all, cells at the left in each row were injected, while cells at the right were not injected. The ability of the added serum to stimulate ERK activation is revealed by staining with an antibody labeled with a separate fluorochrome against the phosphorylated ERK (bottom row). It can be seen that while the nonspecific antibody had no effect on ERK phosphorylation, the injected anti-Ras blocked this phosphorylation even when injected immediately prior to a 15-min stimulation with serum.
Inhibition of cycling cells with cycloheximide and serum removal.
To determine the restriction point in cycling cells for cycloheximide and serum removal, experiments similar to those described above were performed. The results of two separate experiments with each reagent were summarized and compared to the results obtained as reported above with anti-cyclin D1 (Fig. 4B). The inhibition characteristics of anti-D1 (Fig. 6A), cycloheximide (Fig. 6B), and entry into S phase (Fig. 3A) were indistinguishable within the level of accuracy possible in these types of experiment. Cells within 3 h of mitosis were essentially all inhibited in cell cycle progression by anti-cyclin D1 or cycloheximide, while both inhibitors lost activity by the time cells in the culture had entered S phase (Fig. 6B). On the other hand, the inhibitory effect of serum removal was intermediate between the effects of anti-Ras and anti-cyclin D1 inhibitions. In this case, cells were consistently and efficiently inhibited only within the first hour following mitosis, after which cell cycle inhibitions became minimal (Fig. 6C). In all cases, few cells passed through a second mitosis following any of these treatments, indicating that each was an efficient inhibitor of cell cycle progression in cycling cells. These results not only emphasize the differences in cell cycle points of inhibition in cycling compared to quiescent cells but also reveal differences even between separate inhibitors. It is apparent that a complex signaling sequence operates to regulate cell cycle progression in cycling cells.
FIG. 6.
The restriction point in cycling cells. Asynchronous NIH 3T3 cultures were treated with inhibitors as described in the legend to Fig. 3. The age (in hours) at the time of treatment (as determined from a time-lapse analysis prior to treatment) is plotted versus the ability of that treatment to delay passage through the cell cycle and mitosis (as determined from a posttreatment time-lapse analysis [Fig. 4D]). Results from multiple analyses for each inhibitor are presented as the proportion of cells in each age period which experienced a delay in entry into mitosis (±standard error). Both injection with anti-cyclin D1 (A) and treatment with 2 μM cycloheximide (B) resulted in inhibition profiles which were highly similar to each other and highly similar to the profile of entry of cells into S phase (Fig. 3A), indicating that cells in G1 phase are inhibited by both treatments. In contrast, the profile of inhibition following serum removal (C) is quite different from the other profiles and more similar to the profile of inhibition following anti-Ras antibody injection (Fig. 4).
Direct comparison of the timing of Ras, cyclin D1, and cycloheximide requirements.
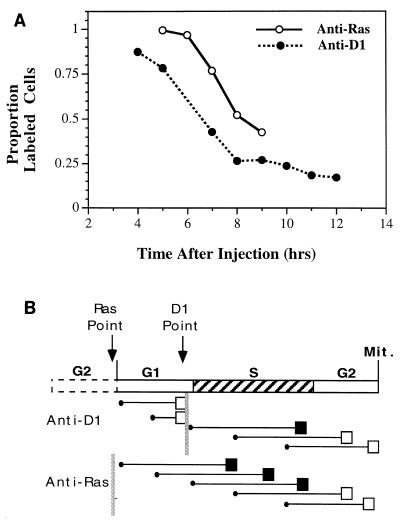
The above results reveal for the first time differences in the cell cycle time at which Ras and cyclin D1 are required in continuously proliferating cells. To confirm this result, and to more accurately determine the differences between these times, anti-Ras and anti-cyclin D1 were each injected into several plates of asynchronous NIH 3T3 cells. After incubation for various lengths of time, individual plates were then pulsed for 1 h with tritiated thymidine. The cells were then fixed and stained for injected immunoglobulin (to identify injected cells), followed by autoradiography to detect cells in S phase. Little inhibition of thymidine labeling was observed soon after either of these injections, since neither antibody would be expected to interfere with an ongoing S phase, and most labeling soon after injection would be by cells already in S phase when injection took place. On the other hand, with increasing incubation time between injection and thymidine labeling, cells synthesizing DNA at the time of injection would complete S phase, such that new cells would be required to initiate DNA synthesis if the proportion of labeled cells were to be maintained. Inhibition of thymidine labeling in this experiment would be seen most rapidly following injection of an antibody able to immediately inhibit entry into S phase (Fig. 7B). In individual experiments, anti-cyclin D1 reduced the labeling of injected cells between 2 and 4 h sooner than did injected anti-Ras. These results indicate that anti-Ras had to be in the cells from 2 to 4 h longer than anti-cyclin D1 in order to block entry into S phase (Fig. 7). In other words, anti-Ras inhibited cell cycle progression 2 to 4 h prior to the point at which anti-cyclin D1 was inhibitory.
FIG. 7.
Kinetics of anti-Ras and anti-cyclin D1 inhibition. (A) To determine the cell cycle time at which Ras is required in cycling cells compared to cyclin D1, both antibodies were injected into several cultures of proliferating NIH 3T3 cells. These cultures were then pulsed with thymidine for 1 h at various times thereafter, fixed, stained with fluorescent antibodies against the injected antibody, and finally autoradiographed. The proportion of injected cells labeled with thymidine is plotted versus the time following injection at which the cells were fixed. (B) Results expected in a specific circumstance: when thymidine labeling followed injection by 5 h. The cell cycle is represented by the top bar, which indicates each cell cycle period and the points at which Ras and cyclin D1 are last required for cell cycle progression. Below this bar, each circle represents the position of individual cells in the cell cycle at the time of injection, horizontal lines represent progress through the cell cycle, boxes indicate whether the cells were labeled (filled box) or not (open box), and vertical lines indicate the points in the cell cycle at which Ras or cyclin D1 is last required prior to entry into S phase. Because the Ras requirement precedes mitosis, cells in G1 and early S phase at the time of injection become labeled following anti-Ras injection. Because anti-cyclin D1 blocks cell cycle progression just prior to S phase, on the other hand, only cells in early S phase at the time of injection are labeled following anti-cyclin D1 injection. This results in a lower labeling efficiency 5 h following injection of anti-cyclin D1 compared to anti-Ras.
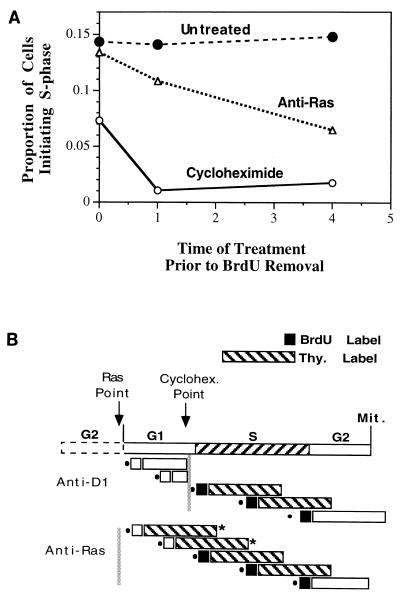
In the above experiment, the time of anti-Ras and anti-cyclin D1 inhibition were compared to each other, without direct reference to the cell cycle. For example, the same result would be obtained if the restriction point for Ras and cyclin D1 were separated by 2 to 4 h in G2 phase. Therefore, we designed a separate experimental strategy to directly determine the time of inhibitor action relative to the beginning of S phase. A double-labeling procedure was used to identify cells which initiated a new cycle of DNA synthesis as follows. Cells were pulsed for 1 h with BrdU, washed, and immediately pulsed with tritiated thymidine for the next 5 h. A cell labeled with thymidine but not with BrdU must have initiated a new cycle of DNA synthesis during the 5 h of thymidine labeling following removal of BrdU. To relate the point of Ras and protein synthesis requirements to the beginning of S phase, cells were injected with anti-Ras or treated with cycloheximide (2 μM) and at various times thereafter were subjected to the double-labeling procedure (Fig. 8B). With this protocol, approximately 15% of untreated cells scored positive for entry into S phase at all times tested. (The slight underestimate from the expected number was apparently due to the manipulations required.) Cycloheximide treatment, as predicted based on the above data, blocked entry into S phase almost immediately. Even cells treated with cycloheximide immediately after removal of BrdU were reduced to approximately half of the level of untreated cells; cells were efficiently inhibited from entering S phase if treated with cycloheximide just prior to the 1-h BrdU treatment. On the other hand, anti-Ras required longer to inhibit entry into S phase. Cells injected immediately following BrdU treatment exhibited little inhibition, while the number of cells able to initiate S phase 4 h after anti-Ras injection was reduced to approximately half of the level of untreated cells (Fig. 8).
FIG. 8.
Inhibition of initiation of S-phase by anti-Ras and cycloheximide. (A) Cells which initiate a new S phase were identified by being pulsed with BrdU for 1 h, washed, and then pulsed with tritiated thymidine for 5 h. Cells labeled with thymidine but not BrdU would have initiated S phase after removal of BrdU. This procedure was used at various times after injection of anti-Ras or treatment with cycloheximide (2 μM). The proportion of cells which initiated S phase is plotted versus the time (in hours) of thymidine addition following injection or treatment. Thus, the 0-h time point indicates that injection or treatment took place immediately after BrdU removal and just before addition of thymidine. (B) Results for one particular circumstance: when the 1-h BrdU labeling period began immediately following injection or treatment (indicated as 1 h in panel A). All representations are as in Fig. 7B except that the long hatched box represents the thymidine labeling period and the short solid box represents the BrdU labeling period (with empty boxes indicating unlabeled cells). A cell which initiates S phase in this analysis must be negative for BrdU and positive for thymidine labeling (indicated by an asterisk). Under these experimental conditions, where treatment immediately precedes BrdU labeling, because cycloheximide is inhibitory just prior to entry into S phase, essentially no cells scored positive for entry into S phase. Cells in G1 phase at the time of cycloheximide treatment would be labeled with neither BrdU nor thymidine, while cells treated in S phase would be labeled with both. Since the inhibition point for anti-Ras precedes the beginning of S phase, on the other hand, cells in early to mid-G1 phase at the time of injection would fail to be labeled with BrdU but would pass into S phase without inhibition and become labeled with thymidine. Such a cell would score positive for entry into S phase following thymidine addition.
These data confirm previous results and indicate that while cycloheximide blocks entry into S phase up until just prior to the beginning of DNA synthesis, cellular Ras activity becomes unnecessary to the cell several hours prior to the beginning of S phase. Taken together these results confirm the difference between cycling and quiescent cells and between the timing at which different inhibitory treatments block the progression of cycling cells.
DISCUSSION
When serum is added to quiescent cells, it is continuously required until the cells progress to a point in G1 phase after which the cells become committed to entry into S phase even in the absence of serum (25). A number of other inhibitory treatments also lose their ability to block entry into S phase at this same point, several hours prior to the beginning of DNA synthesis (5, 11, 12, 23, 26). Consistent with this pattern, we found that injection of anti-Ras and anti-cyclin D1 antibodies together with serum removal and cycloheximide treatment all blocked entry into S phase following release from quiescence at essentially the same point, approximately 4 h prior to the beginning of S phase in these NIH 3T3 cells. It was the purpose of this study to extend this type of analysis to cells continuously passing through the cell cycle without introducing the perturbations required for inducing cell synchrony. For this purpose, the approximate cell cycle position of individual cells in an asynchronous culture was determined by following them in time-lapse prior to treatment or injection. This allowed determination of the age of each cell and therefore its approximate cell cycle position without synchronization. In addition, time-lapse analysis after injection allowed a determination of the fate of injected cells.
Surprisingly, we found that the injected anti-Ras antibody did not delay passage of injected cells through the next scheduled mitosis, regardless of the cell cycle position at the time of injection. Efficient inhibition was eventually observed, but only after each injected cell had passed through one mitosis. Thus, in order to block entry into S phase, the anti-Ras antibody had to be present within the cell prior to the preceding mitosis. By contrast, anti-cyclin D1 antibody efficiently inhibited the number and timing of the next mitoses when introduced during G1 phase of asynchronous cells. Our concern that these differences might result from a delay in the ability of the injected anti-Ras to neutralize cellular Ras activity following microinjection was unfounded, based on work performed here as well as previous studies (7, 36). Another possibility is that Y13-259 could not rapidly neutralize Ras because a Ras effector molecule, Raf-1, had already bound to Ras preventing antibody binding. This, however, is not likely because of the following lines of evidences. (i) The Raf-Ras complex has an off rate of about 1 min in an equilibrium system (8). If this were the case in vivo, injected anti-Ras antibody would quickly replace Raf already bound to Ras. (ii) Binding of Raf to Ras does not change the intrinsic GTPase activity of Ras (50% conversion of GTP to GDP in about 30 min) (10) or slightly enhances it (40). Because Raf would be released upon hydrolysis of the GTP bound to Ras, the half-life of the Ras-Raf complex in Y13-259 injected cells could not be longer than 30 min. Once the Ras-Raf complex dissociates, anti-Ras would be able to bind and inactivate the Ras protein (9). (iii) The anti-Ras antibody Y13-259 used here can inhibit the proliferation of oncogenic Ras-transformed cells with kinetics similar to those of nontransformed cells (35). Moreover, injection of anti-Ras antibody into cells transformed by oncogenic Ras weas found to stop migration within 15 min (unpublished data), despite the fact that oncogenic Ras has very slow intrinsic GTPase activity, and would be expected to be continuously bound to Raf protein prior to injection of anti-Ras. Migration has previously been shown to be dependent on Ras activity and to be stimulated by oncogenic Ras (7). Therefore, even though Raf was in a complex with Ras at the time of anti-Ras injection, the antibody was able to replace Raf and neutralize Ras activity in a time frame much shorter than the delay in antibody neutralization reported here. Binding of Ras to Raf could not, therefore, account for the delay in cell cycle inhibition following anti-Ras injection compared to anti-cyclin D1 injection. Thus, cellular Ras and cyclin D1 were apparently required at different times in cycling cells, while cycloheximide was similar to anti-cyclin D1 and serum removal was intermediate between anti-Ras and anti-cyclin D1.
This conclusion was confirmed with two separate experimental strategies. In the first, a kinetic labeling procedure was used to determine the time required for each antibody to block thymidine labeling in an asynchronous culture. In the second, a novel BrdU-tritiated thymidine labeling sequence was utilized to identify cells which initiate S phase within a given time period. In both cases, the requirement for cellular Ras activity was found to precede that for cyclin D1 or cycloheximide. For example, it required almost 4 h for anti-Ras to block entry into S phase by 50%, while cycloheximide (which functions at a time similar to that of anti-cyclin D1) inhibited entry into S phase almost immediately. Therefore, these data from three separate techniques together indicate that in cycling NIH 3T3 cells, cellular Ras activity and cyclin D1 are required at distinct cell cycle points.
The difference in timing of inhibition between cycling and quiescent cells most probably results from the fact that stimulation of quiescent cells with serum not only involves a stimulation of cell cycle progression but also requires a change in growth phase. This fact might also account for the long period between (i) the point when all protein synthesis required for the initiation of S phase is complete and (ii) the actual initiation of DNA synthesis. Such a delay would apparently not be tolerated in cycling cells with a short G1 phase. Indeed, a short delay between the cyclin D1 or the protein synthesis requirement and the beginning of S phase might be essential to reduce the overall length of the cell cycle in rapidly proliferating cells.
The time at which serum is required in cycling NIH 3T3 cells is slightly different from than for Ras. Serum was required during early G1 phase. Similar results have been obtained with a time-lapse approach similar to the one used in this study (43, 44). In that work, the G1 phase was divided into two parts by the requirement for serum. The fact that anti-Ras and serum removal have profiles of inhibition in cycling cells which show subtle but important differences indicates that Ras must not be the only important target of serum stimulation. Thus, another signaling pathway induced by serum, which is distinct from the Ras pathway, is apparently required after Ras activity becomes unnecessary for continued cell cycle progression. This alternative pathway might control the stability of cyclin D1. Serum withdrawal is reported to activate the calpain-dependent degradation of cyclin D1 protein (2). In addition, the phosphatidylinositol 3-kinase-Akt-glycogen synthase kinase-3β pathway has been shown to involve posttranslational control of cyclin D1 protein stability (4). Phosphatidylinositol 3-kinase is activated by Ras through direct interaction with its catalytic subunit, p110 (28). However, it can also be activated independently of Ras through interaction of its regulatory subunit with the growth factor receptor (39). Thus, it is possible that the continued presence of activated growth factor receptors can stabilize cyclin D1 proteins, or promote their continued synthesis, even in the absence of Ras activity.
Cyclin D1, however, is not the only target of growth factor action. Cell cycle progression requires a number of other signaling activities, including the suppression of cyclin-inhibitory proteins, the activation and coordination of multiple MAPK pathways, the activation of transcription factors, the regulation of other small G proteins such as those involved in controlling the cytoskeleton, and controls over translation and apoptosis. It will be essential to consider a variety of other signaling molecules before we are able to fully understand these results. Moreover, the cyclin D family contains three members, at least two of which are likely to be expressed in fibroblastic cells. The anti-cyclin D1 antibody used here has been shown in previous studies to be highly specific for the D1 isoform in neutralizing activity (18). The potential role of other cyclin D molecules will also have to be determined by future experimentation.
These data, therefore, provide novel insight into the control of cell cycle progression in actively cycling cells. Cellular Ras activity is required prior to mitosis to allow a cell to make a commitment to progress through the entire subsequent cell cycle, while cyclin D1 activity is required for the G1/S-phase transition. These data suggest the following model for control of cell cycle progression in continuously cycling cells. Continued cell cycle progression requires the presence of growth factors and other appropriate proliferative conditions. These conditions are communicated to the cell, at least in part, through the activation of cellular Ras. In a cell with a relatively short G1 phase, such as the NIH 3T3 cells studied here, Ras must become activated during G2 phase to allow the cell to proceed into the next cell cycle after passage through mitosis. Since Ras activity becomes largely unnecessary by the time a cell has passed through mitosis, it must stimulate the production of another molecule(s) whose continued production is independent of continued Ras activity and which is required until the cell begins DNA synthesis, after which completion of the cell cycle is normally ensured. The data suggest the possibility that cyclin D1 is this downstream target of Ras. It is reported to be stimulated by Ras activity (1, 6, 16, 27), and these studies demonstrate that it is required through G1 phase. This possibility is supported by data which indicate that cyclin D1 is induced during G2 phase of the NIH 3T3 cell cycle in a Ras-dependent manner. This expression of cyclin D1 then continues independently of Ras for over 6 h and is required for initiation of S phase (unpublished data).
ACKNOWLEDGMENTS
We thank Michele Pagano for the generous gift of a neutralizing anti-cyclin D1 monoclonal antibody. We also thank Guan Chen for helpful suggestions throughout this work and Alan Wolfman and Seiji Kondo for helpful suggestions in preparation of the manuscript.
This work was supported by NIH grant GM52271.
REFERENCES
- 1.Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell R G. Transforming p21ras mutants and c-ETS-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- 2.Choi Y H, Lee S J, Nguyen P, Jang J S, Lee J, Wu M-L, Takano E, Maki M, Henkart P A, Trepel J B. Regulation of cyclin D1 by calpain protease. J Biol Chem. 1997;272:28479–28484. doi: 10.1074/jbc.272.45.28479. [DOI] [PubMed] [Google Scholar]
- 3.Coats S, Flanagan M, Nourse J, Roberts J M. Requirement of p27kip1 for restriction point control of the fibroblast cell cycle. Science. 1996;272:877–880. doi: 10.1126/science.272.5263.877. [DOI] [PubMed] [Google Scholar]
- 4.Diehl J A, Cheng M, Roussel F M, Sherr C J. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 1998;12:3499–3511. doi: 10.1101/gad.12.22.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobrowolski S, Harter M, Stacey D W. Cellular ras activity is required for passage through multiple points of the G0/G1 phase in BALB/c 3T3 cells. Mol Cell Biol. 1994;14:5441–5449. doi: 10.1128/mcb.14.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filmus J, Robles A I, Shi W, Wong M J, Colombo L L, Conti C J. Induction of cyclin D1 overexpression by activated ras. Oncogene. 1994;9:3627–3633. [PubMed] [Google Scholar]
- 7.Fox P L, Sa G, Dobrowolski S F, Stacey D W. The regulation of endothelial cell motility by p21 ras. Oncogene. 1994;9:3519–3526. [PubMed] [Google Scholar]
- 8.Gorman C, Skinner R H, Skelly J V, Neidle S, Lowe P N. Equilibrium and kinetic measurements reveal rapidly reversible binding of Ras to Raf. J Biol Chem. 1996;271:6713–6719. doi: 10.1074/jbc.271.12.6713. [DOI] [PubMed] [Google Scholar]
- 9.Hattori S, Clanton D J, Satoh T, Nakamura S, Kaziro Y, Kawakita M, Shih T Y. Neutralizing monoclonal antibody against ras oncogene product p21 which impairs guanine nucleotide exchange. Mol Cell Biol. 1987;7:1999–2002. doi: 10.1128/mcb.7.5.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrmann C, Martin G A, Wittinghofer A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. J Biol Chem. 1995;270:2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- 11.Hitomi M, Shu J, Strom D, Hiebert S W, Harter L M, Stacey D W. Prostaglandin A2 blocks the activation of G1 phase cyclin-dependent kinase without altering mitogen-activated protein kinase stimulation. J Biol Chem. 1996;271:9376–9383. doi: 10.1074/jbc.271.16.9376. [DOI] [PubMed] [Google Scholar]
- 12.Howe P H, Draetta G, Leof E B. Transforming growth factor β1 inhibition of p34cdc2 phosphorylation and histone H1 kinase activity is associated with G1/S-phase growth arrest. Mol Cell Biol. 1991;11:1185–1194. doi: 10.1128/mcb.11.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurn B A L, Chantler S M. Production of reagent antibodies. Methods Enzymol. 1980;70:104–142. doi: 10.1016/s0076-6879(80)70044-7. [DOI] [PubMed] [Google Scholar]
- 14.Kovary K, Bravo R. Existence of different Fos/Jun complexes during the G0-to-G1 transition and during exponential growth in mouse fibroblasts: differential role of Fos proteins. Mol Cell Biol. 1992;12:5015–5023. doi: 10.1128/mcb.12.11.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsson O, Zetterberg A, Engstrom W. Consequences of parental exposure to serum-free medium for progeny cell division. J Cell Sci. 1985;75:259–268. doi: 10.1242/jcs.75.1.259. [DOI] [PubMed] [Google Scholar]
- 16.Liu J-J, Chao J-R, Jiang M-C, Ng S-Y, Yen J-J, Yang-Yen H-F. Ras transformation results in an elevated level of cyclin D1 and acceleration of G1 progression in NIH 3T3 cells. Mol Cell Biol. 1995;15:3654–3663. doi: 10.1128/mcb.15.7.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukas J, Bartkova J, Rohde M, Strauss M, Bartek J. Cyclin D1 is dispensable for G1 control in retinoblastoma gene-deficient cells independently of cdk4 activity. Mol Cell Biol. 1995;15:2600–2611. doi: 10.1128/mcb.15.5.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukas J, Pagano M, Staskova Z, Draetta G, Bartek J. Cyclin D1 protein oscillates and is essential for cell cycle progression in human tumor cell lines. Oncogene. 1994;9:707–718. [PubMed] [Google Scholar]
- 19.Mayol X, Garriga J, Grana X. G1 cyclin/CDK-independent phosphorylation and accumulation of p130 during the transition from G1 to G0 lead to its association with E2F-4. Oncogene. 1996;13:237–246. [PubMed] [Google Scholar]
- 20.Moodie S A, Wolfman A. The 3Rs of life: ras raf and growth regulation. Trends Genet. 1994;10:44–48. doi: 10.1016/0168-9525(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 21.Mulcahy L S, Smith M R, Stacey D W. Requirement for ras proto-oncogene function during serum-stimulated growth of NIH 3T3 cells. Nature. 1985;313:241–243. doi: 10.1038/313241a0. [DOI] [PubMed] [Google Scholar]
- 22.Nevins J R. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 23.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Keefe E J, Pledger W J. A model of cell cycle control: sequential events regulated by growth factors. Mol Cell Endocrinol. 1983;31:167–186. doi: 10.1016/0303-7207(83)90147-8. [DOI] [PubMed] [Google Scholar]
- 25.Pardee A B. A restriction point for control of normal animal cell proliferation. Proc Natl Acad Sci USA. 1974;71:1286–1290. doi: 10.1073/pnas.71.4.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardee A B. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 27.Peeper D S, Upton T M, Ladha M H, Neuman E, Zalvide J, Bernard R, DeCaprio J A, Ewen M E. Ras signaling linked to the cell-cycle machinery by the retinoblastoma protein. Nature. 1997;386:177–181. doi: 10.1038/386177a0. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez-Viciana P, Warne P H, Dhand R, Vanhaesebroeck B, Gout I, Fry M J, Waterfield M D, Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–534. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 29.Rossow P W, Riddle V G H, Pardee A B. Synthesis of labile, serum-dependent protein in early G1 controls animal cell growth. Proc Natl Acad Sci USA. 1979;76:4446–4450. doi: 10.1073/pnas.76.9.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlessinger J. How receptor tyrosine kinases activate ras. Trends Biochem Sci. 1993;18:273–275. doi: 10.1016/0968-0004(93)90031-h. [DOI] [PubMed] [Google Scholar]
- 31.Sherr C J. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 32.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 33.Shu J, Hitomi M, Stacey D W. Activation of JNK/SAPK pathway is not directly inhibitory for cell cycle progression in NIH3T3 cells. Oncogene. 1996;13:2421–2430. [PubMed] [Google Scholar]
- 34.Smith E J, Leone G, DeGregori J, Jakoi L, Nevins J R. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 cell state from a G1 cell state. Mol Cell Biol. 1996;16:6965–6976. doi: 10.1128/mcb.16.12.6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith M R, DeGudicibus S J, Stacey D W. Requirement for c-ras proteins during viral oncogene transformation. Nature. 1986;320:540–543. doi: 10.1038/320540a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stacey D, Watson T, Kung H, Curran T. Microinjection of transforming Ras protein induces c-fos expression. Mol Cell Biol. 1987;7:523–527. doi: 10.1128/mcb.7.1.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stacey D W. Microinjection of mRNA and other macromolecules into living cells. Methods Enzymol. 1981;79:76–88. doi: 10.1016/s0076-6879(81)79016-5. [DOI] [PubMed] [Google Scholar]
- 38.Taylor S J, Shalloway D. Cell cycle-dependent activation of Ras. Curr Biol. 1996;6:1621–1627. doi: 10.1016/s0960-9822(02)70785-9. [DOI] [PubMed] [Google Scholar]
- 39.van Weering D, de Rooij J, Marte B, Downward J, Bos J L, Burgering B M. Protein kinase B activation and lamellipodium formation are independent phosphoinositide 3-kinase-mediated events differentially regulated by endogenous Ras. Mol Cell Biol. 1998;18:1802–1811. doi: 10.1128/mcb.18.4.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warne P H, Viciana P R, Downward J. Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro. Nature. 1993;364:352–355. doi: 10.1038/364352a0. [DOI] [PubMed] [Google Scholar]
- 41.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 42.Yan Z, DeGregori J, Shohet R, Leone G, Stiliman B, Nevins J R, Williams R S. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci USA. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yen A, Pardee A B. Exponential 3T3 cells escape in mid-G1 from their high serum requirement. Exp Cell Res. 1978;116:103–113. doi: 10.1016/0014-4827(78)90068-x. [DOI] [PubMed] [Google Scholar]
- 44.Zetterberg A, Larsson O. Kinetic analysis of regulatory events in G1 leading to proliferation or quiescence of Swiss 3T3 cells. Proc Natl Acad Sci USA. 1985;82:5365–5369. doi: 10.1073/pnas.82.16.5365. [DOI] [PMC free article] [PubMed] [Google Scholar]